Abstract
The spore-forming bacterium Bacillus subtilis is capable of assembling multicellular communities (biofilms) that display a high degree of spatiotemporal organization. Wild strains that have not undergone domestication in the laboratory produce particularly robust biofilms with complex architectural features, such as fruiting-body-like aerial projections whose tips serve as preferential sites for sporulation. To discover genes involved in this multicellular behavior and to do so on a genome-wide basis, we took advantage of a large collection of mutants which have disruptions of most of the uncharacterized genes in the B. subtilis genome. This collection, which was generated with a laboratory strain, was screened for mutants that were impaired in biofilm formation. This subset of mutated genes was then introduced into the wild strain NCIB 3610 to study their effects on biofilm formation in liquid and solid media. In this way we identified six genes that are involved in the development of multicellular communities. These are yhxB (encoding a putative phosphohexomutase that may mediate exopolysaccharide synthesis), sipW (encoding a signal peptidase), ecsB (encoding an ABC transporter subunit), yqeK (encoding a putative phosphatase), ylbF (encoding a regulatory protein), and ymcA (a gene of unknown function). Further analysis revealed that these six genes play different roles in B. subtilis community development.
Formation of biofilms—surface-associated multicellular assemblages—is an important microbial survival strategy (4, 25). Relative to the rapid, continual, and extreme changes in environmental conditions that can characterize a planktonic existence, biofilms offer the constituent cells some shelter, enabling them to establish long-term relationships with each other and their immediate surroundings. Cells in different regions of a developing biofilm experience diverse environmental conditions, resulting in marked patterns of cellular differentiation (e.g., see references 5 and 16). Such spatiotemporal organization is particularly striking in biofilms formed by wild strains of the spore-forming bacterium Bacillus subtilis. Unlike strains that have been domesticated by decades of propagation in the laboratory (e.g., B. subtilis 168), which form thin and relatively undifferentiated biofilms, wild B. subtilis strains form elaborate multicellular communities that display conspicuous architectural features, such as fruiting-body-like aerial projections that extend from the surfaces of the biofilm. The tips of these fruiting bodies serve as preferential sites for spore formation (2).
We are interested in discovering genes that are involved in biofilm and fruiting body formation. We have focused our efforts on a relatively undomesticated strain of B. subtilis, NCIB 3610 (hereafter referred to as “3610”), which forms robust and highly structured biofilms both in liquid and on solid medium (colonies). When inoculated into a standing culture of minimal medium, 3610 initially grows planktonically as motile, single cells. The cells then migrate to the air-liquid interface, where they proliferate as long chains of nonmotile cells. These chains are highly ordered in parallel patterns and are bound together tightly, presumably by an extracellular matrix, to form a floating biofilm (pellicle). Growth of the bundled chains continues, giving rise to larger structures that include the fruiting bodies. Fruiting bodies are also formed on solid medium, but multicellular development in colonies exhibits distinctive features that are not evident in pellicles, such as concentric rings. Thus, we consider pellicles and colonies as complementary settings for the study of B. subtilis biofilms.
Previously we used a “candidate-gene” approach to identify genes involved in B. subtilis pellicle and colony development (2). Perhaps the most striking result was that genes that regulate entry into the sporulation pathway (spo0A and spo0H) were required for the initial stages of biofilm development. Other genes identified through this directed approach included yveQ and yveR, which appear to be involved in the production of an exopolysaccharide component of the biofilm's extracellular matrix. This matrix acts as the scaffold for biofilm architecture. In addition, genes that mediate production of surfactin (srfAA and sfp) were shown to be required for the erection of fruiting bodies. In a parallel study, Hamon and Lazazzera also used a candidate-gene approach to analyze solid-surface-associated submerged biofilms formed by a domesticated B. subtilis strain. They reported that spo0A and genes involved in the activation of Spo0A (spo0B, spo0F, and kinC to -E) are required for biofilm formation (13). More recently, transcriptional profiling was used to identify transcription factors (Spo0A, Spo0H, LytS, ResE, SigW, YbdK, YcbA, and YfiJ) that affect gene expression during pellicle formation by the domesticated strain (29). All of these studies have provided much information regarding the genetic control of biofilm formation by B. subtilis in diverse settings. We now report a genome-wide mutational approach to identify additional genes involved in these processes.
To facilitate a screen for genes that play a critical role in B. subtilis biofilm formation, we took advantage of the B. subtilis Functional Analysis (BFA) mutant collection, which was constructed by a consortium of laboratories from Europe and Japan. This collection includes mutants in which most B. subtilis genes of unknown function were individually disrupted in the laboratory strain 168 (36, 39). We screened the BFA collection for mutants defective in pellicle formation. The mutant genes were subsequently introduced into 3610 for further analysis. Using this unbiased genetic approach, we identified six genes that play important roles in the development of multicellular communities.
MATERIALS AND METHODS
Bacterial strains and media.
Strain 3610 is B. subtilis NCIB 3610, a prototrophic strain from our laboratory collection (2), originally obtained from A. L. Sonenshein and the Bacillus Genetic Stock Center, Ohio State University, Columbus. Strain 168 (trpC2) was originally obtained from C. Anagnostopoulos. Media used were LB medium (1% tryptone [Difco], 0.5% yeast extract [Difco], 1% NaCl, 1 mM NaOH [14]) and MSgg medium (5 mM potassium phosphate [pH 7], 100 mM morpholinepropane sulfonic acid [pH 7], 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg of tryptophan/ml, 50 μg of phenylalanine/ml [adapted from reference 10]). Media were solidified through addition of agar (Difco) to 1.5%, and the plates were allowed to dry at 25°C for 40 h before use. Antibiotic concentrations (final) were erythromycin (EM) at 0.3 μg/ml, erythromycin plus lincomycin (MLS) at 1 and 25 μg/ml, respectively, spectinomycin at 100 μg/ml, and tetracycline at 5 μg/ml for shaken and standing cultures and 10 μg/ml for transformation and transduction plates, and chloramphenicol at 5 μg/ml.
Screen for pellicle-defective mutants.
The BFA mutant collection was constructed by a consortium of European and Japanese laboratories, and has been described previously (36, 39). In brief, the consortium targeted for disruption the previously uncharacterized genes of B. subtilis strain 168. In each case, an internal fragment of the gene of interest was amplified by PCR and cloned into a pMUTIN integrating plasmid. The construct was introduced into 168 by transformation, selecting for erythromycin resistance (Emr); this resulted in insertional mutation of the gene of interest, through a Campbell-type single-crossover recombination event, as confirmed by Southern analysis or PCR. Because integration within an operon can lead to polar effects on downstream genes, the pMUTIN plasmid carries an inducible promoter that can be used to drive expression of downstream genes; we analyzed only mutants that were viable without induction of this promoter (i.e., mutants in which the integrated plasmid did not have polar effects on essential genes). The genes targeted for disruption in the BFA mutants are listed in the Micado (http://genome.jouy.inra.fr/micado) and BSORF (http://bacillus.genome.ad.jp/bsorf.htm) databases.
For the purposes of our screen, each BFA mutant was used to inoculate 140 μl of LB plus EM contained within a well of a 96-well microtiter plate (Falcon 35-1172). The microtiter plates were shaken at low speed (160 rpm) at 37°C for 16 h, at which point 5 μl of each culture was used to inoculate 1 ml of MSgg plus EM contained within a well of a 48-well microtiter plate (Falcon 35-1178). These microtiter plates were incubated without shaking at 30°C, and after 60 h of incubation their pellicles were analyzed by visual inspection. It should be noted that the addition of antibiotics to standing cultures (as described here and below) had no detectable effect on pellicle formation by any of the mutants analyzed in detail, as indicated by control experiments.
BFA mutants that reproducibly failed to form wild-type pellicles were tested for their ability to grow in aerated cultures. In these experiments, each mutant was grown in LB plus EM in a 96-well microtiter plate (as described above), and 3 μl of the culture was used to inoculate 3 ml of MSgg plus EM contained within an 18-ml glass tube that was rolled at 30°C for 16 h. Mutants that did not produce wild-type pellicles but did grow robustly (to an optical density at 600 nm of ≥ 2.5) in aerated cultures were considered to exhibit a specific defect in pellicle formation.
To confirm that the observed pellicle defect was conferred by the insertional mutation, genomic DNA isolated from the BFA mutant was reintroduced into parental strain 168 by transformation, using standard methods (14) and selecting for the integrated pMUTIN plasmid (i.e., Emr). At least three transformants were tested for their ability to form pellicles in standing cultures and to grow robustly in aerated cultures, using the methods described above. Specific defects in pellicle formation exhibited by these second-generation BFA mutants were attributed to their insertional mutations.
Each insertional mutation associated with a specific defect in pellicle formation in strain 168 was then introduced into strain 3610, via SPP1 phage transduction (17) and selection for MLSr. In each case, at least three transductants were tested for specific defects in pellicle formation. Transductants were also tested for defects in colony formation: each was grown in 3 ml of LB plus MLS at 37°C for 8 h, and 3 μl of the culture was spotted onto the surface of an MSgg plate; after incubation at 30°C for 96 h, the colonies were analyzed by visual inspection.
Deletion mutant construction, analysis, and complementation.
Deletion of genes of interest from the 168 chromosome and their replacement by an antibiotic resistance marker were achieved using a long-flanking-homology PCR strategy (38). The deletion alleles were then introduced into 3610 by SPP1 phage transduction.
Deletion mutants were tested for their ability to form pellicles, using the following procedure. Each mutant was grown in 3 ml of LB plus antibiotic at 37°C for 8 h, and 3 μl of this culture was used to inoculate 12 ml of MSgg plus antibiotic contained within a well of a six-well microtiter plate; the plate was incubated at 30°C for 60 h, at which point the pellicles were analyzed by visual inspection. Deletion mutants were also tested for their ability to form colonies, using the method described above.
To complement a deletion mutant, we first amplified by PCR a wild-type copy of the deleted gene, using 168 genomic DNA as the template and flanking primers that contained restriction sites at their 5′ ends. In the case of yhxB, the coding region plus 407 bp of upstream sequence and 23 bp of downstream sequence were amplified by using primers amyE::yhxB-1 (TTGGATCCGCGCTGAAAGATGAGGCCGC) and amyE::yhxB-2 (TTGGATCCCAATGACCCAGCTCATACTTG), each of which contains a BamHI site (underlined); in the case of ylbF, the coding region plus 427 bp of upstream sequence and 60 bp of downstream sequence were amplified by using primers amyE::ylbF-1 (TTGGATCCCAGAAGCATTCTCAGCCAGGC) and amyE::ylbF-4 (TTAAGCTTCTCCAATACAATCACCTGCCTG), which contain a BamHI and HindIII site, respectively (underlined). Each PCR product was digested with the appropriate restriction enzyme(s) and ligated into the multiple cloning site of the pDG1662 vector (11), using standard methods (27). The plasmid bearing the wild-type allele was linearized by ClaI digestion and introduced into 168 via transformation (14) and selection for Cmr, which is associated with plasmid integration (11). The transformants were then screened for sensitivity to spectinomycin (Spcs), which is indicative of integration by a double-recombination event (as opposed to a Campbell-type single-crossover recombination event, which is associated with Spcr) (11). To verify that plasmid integration occurred at the amyE locus, this region was amplified by PCR using genomic DNA isolated from Cmr Spcs transformants as the template and flanking primers amyE-1 (GTAAGCGTTAACAAAATTCTCCAG) and amyE-2 (CATTGATGGTTTCTTTCGGTAAG); correct integration was associated with an increase in PCR product size from 2.15 kb (amyE) to 4.27 kb (amyE::yhxB) or 3.09 kb (amyE::ylbF). Finally, the amyE::yhxB and amyE::ylbF constructs were introduced into yhxBΔ and ylbFΔ mutants, respectively, by transformation (168 background) and transduction (3610 background).
Microscopy and photography.
To analyze the initial stages of pellicle formation, the behavior of individual cells was studied. Twelve-milliliter MSgg standing cultures were inoculated as described above, and after incubation at 25°C for 36 h a sample of each pellicle was harvested and examined at high-magnification (×1,000) using an Optiphot-2 phase-contrast microscope (Nikon). Similar cultures were generated for photography of pellicles. Colonies were grown as described above and photographed at low-magnification (×5) using an SMZ1500 stereomicroscope (Nikon) equipped with a charge-coupled device video camera system (Optronics Engineering, Goleta, Calif.) and a computer interface.
Computer analyses.
Gene structures and annotation, chromosome maps, and other information related to the B. subtilis 168 genome (19) were taken from the SubtiList database (http://genolist.pasteur.fr/SubtiList/). Specific B. subtilis proteins were matched with potentially homologous proteins on the basis of sequence similarity using the BLAST program. For the analysis of YlbF and YmcA, however, the more sensitive PSI-BLAST program was used instead. Both programs can be found at http://www.ncbi.nlm.nih.gov/BLAST/. BLAST and PSI-BLAST also cross-checked query sequences against families of known and predicted protein domains, using a number of databases, including COG (http://www.ncbi.nlm.nih.gov/COG/), Pfam (http://pfam.wustl.edu/), and SMART (http://smart.embl-heidelberg.de/); the ProDom database (http://prodes.toulouse.inra.fr/prodom/2002.1/html/form.php) was used as an additional source of information about protein domains. Alignments of multiple protein sequences were generated using the ClustalW program (http://www.ebi.ac.uk/clustalw/). Gene clusters present in multiple species of bacteria were identified and analyzed using the Comprehensive Microbial Resource from The Institute for Genomic Research (http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl).
RESULTS AND DISCUSSION
Identification of genes involved in the development of multicellular communities.
Genes in the BFA mutant collection were individually disrupted by single-reciprocal (Campbell-like) recombination using the insertional vector pMUTIN (36, 39). Each mutant was inoculated into standing cultures of minimal medium and incubated at 30°C for 60 h. Under these conditions, the parent strain (B. subtilis 168) formed a wrinkled pellicle (see Fig. 1, top left panel). Ninety-four of the 2,105 mutants tested produced a pellicle of unusual morphology or no pellicle at all. Seventy of the 94 mutants exhibited planktonic growth rates in minimal medium similar to that of 168, indicating that their defects in pellicle formation were not due to a general growth defect. To determine whether their defects in pellicle formation were due to pMUTIN insertion, chromosomal DNA was prepared from each of the 70 mutants and used to transform competent cells of the parental strain; after selection for the antibiotic resistance gene carried by the vector, the resulting transformants were tested for their ability to form pellicles. Twenty-four of these mutants showed defects in pellicle formation similar to those exhibited by their corresponding BFA mutants, while the other 46 showed defects that were less severe and/or reproducible than originally observed. Presumably the latter mutants were derived from BFA mutants that harbored secondary mutations not linked to their pMUTIN insertions; indeed, others reported that 16 of these BFA mutants behaved differently than the reference strain when tested under a variety of conditions and that these mutant phenotypes typically did not segregate with the pMUTIN insertion in genetic backcrosses (http://genome.jouy.inra.fr/micado). The secondary mutations responsible for these phenotypes may have resulted from the use of nonisogenic versions of strain 168 in the construction of the BFA mutant collection; however, the 46 BFA mutants in question were derived from several different isolates of 168, which suggests that their pellicle formation defects may be caused by different secondary mutations.
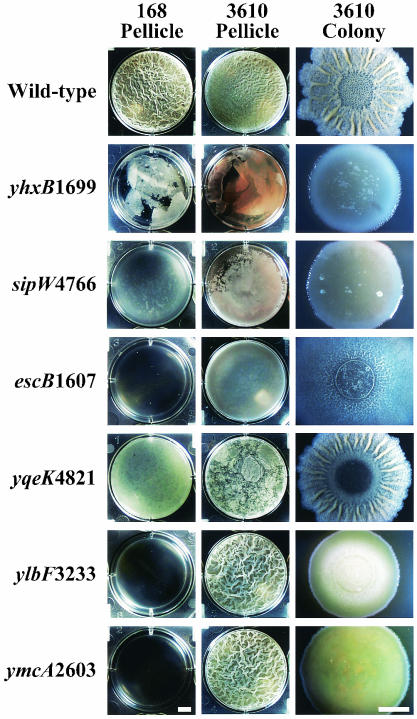
FIG. 1.
BFA mutants defective in B. subtilis community development. Mutants from the BFA collection were isolated on the basis of their failure to form wild-type pellicles. The mutant alleles were then reintroduced into parental strain 168 and also introduced into strain 3610. To assay pellicle formation (first and second columns), each mutant was inoculated at a low density into a standing culture consisting of 12 ml of MSgg plus MLS in a microtiter plate well, and the cultures were incubated at 30°C for 60 h without shaking. To assay colony development (third column), each mutant derived from 3610 was grown overnight in a rolled LB plus MLS culture, and a 3-μl sample of the culture was spotted onto MSgg agar and incubated at 30°C for 96 h. Bars = 5 mm.
With regard to the 24 insertional mutations that did confer upon strain 168 defects in pellicle formation, we wondered whether they might also affect the ability of wild strains to produce robust multicellular communities. To investigate this possibility, each of the mutations was introduced (by phage PBS1-mediated transduction) into strain 3610, and their effects on pellicle formation and colony morphology were assessed. Of these 3610-derived mutants seven showed defects in pellicle formation, two showed defects in colony morphology, and eight showed both types of defects to a limited degree. In contrast, mutations in seven genes—yhxB, sipW, yqxM, ecsB, yqeK, ylbF, and ymcA—caused a conspicuous defect in both pellicle formation and colony morphology when introduced into strain 3610 (Fig. 1 and data not shown). We chose to further characterize these seven genes and their roles in the development of multicellular communities.
Because the insertional mutations were generated by Campbell-like recombination, it was possible that they did not completely inactivate their target genes (14). To address this concern, we deleted the genes of interest from the 168 chromosome, replacing each with an antibiotic resistance gene, to create bona fide null mutations. These null mutations, which are designated by the symbol “Δ”, were introduced into 3610 by phage transduction (see Materials and Methods). In the following sections we describe the effects of these null mutations on pellicle formation and colony morphology in strain 3610; whenever necessary we also describe the effects of the null mutations in strain 168, as well as those of the insertional mutations in strains 168 and 3610. Note that in all cases each insertional mutation and its corresponding null mutation conferred identical phenotypes when introduced into a common parental strain (168 or 3610).
yhxB, a gene that is likely involved in exopolysaccharide synthesis.
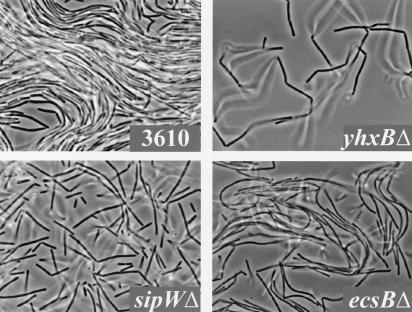
Strain 3610 harboring yhxBΔ formed fragile pellicles when grown in standing liquid cultures and formed flat, undifferentiated colonies when grown on solid media (Fig. 2). Phase-contrast microscopy revealed that in the initial stages of pellicle formation the 3610 yhxBΔ mutant proliferated at the air-medium interface as long chains of cells, but rather than being bound together in parallel the chains instead formed loose aggregates (Fig. 3). This suggested that the yhxB gene product is involved in the production of an extracellular matrix that lends structure to the pellicle.
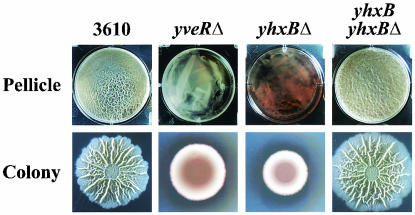
FIG. 2.
Phenotype and complementation of mutant 3610 yhxBΔ. Pellicle and colony development were assayed as described in the legend to Fig. 1.
FIG. 3.
Behavior of individual cells during formation of wild-type and mutant pellicles. Strain 3610 and deletion mutants derived from it were grown in standing cultures, as described in the legend to Fig. 1. After incubation at 25°C for 36 h, samples were withdrawn from the air-medium interface and examined at a magnification of ×1,000X using phase-contrast microscopy.
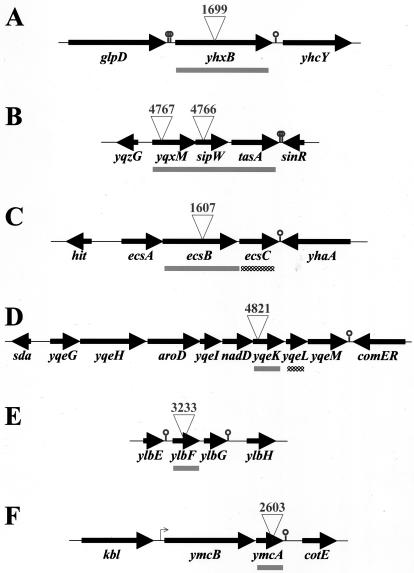
The yhxB coding sequence is located between two predicted Rho-independent transcription terminators (19) (Fig. 4A), consistent with the idea that the yhxB gene lies within a monocistronic operon. Indeed, the 3610 yhxBΔ mutant was fully complemented by a wild-type copy of yhxB that had been introduced at a different site on the chromosome (amyE) (Fig. 2). This indicated that inactivation of yhxB, as opposed to interference with a neighboring gene, was responsible for the observed mutant phenotype.
FIG. 4.
Physical maps of the genes disrupted in BFA mutants of interest. Coding regions are represented as horizontal arrows, a canonical promoter element is shown as a small bent arrow, and canonical Rho-independent transcriptional terminators are shown as stem-loop symbols. Each insertional mutation is represented by an inverted triangle bearing the number of its corresponding BFA mutant. Chromosomal regions that were deleted and replaced by antibiotic resistance markers are represented as horizontal bars—grey if the deletion conferred a defect in community development, checkered if it didn't.
The yhxB gene had not been characterized previously, but it is predicted to encode a protein (YhxB) with sequence similarity to α-phosphoglucomutases (α-PGMs) and phosphomannomutases (PMMs). For example, YhxB is 46% identical to Streptococcus thermophilus PgmA, a bifunctional phosphohexomutase exhibiting both α-PGM and PMM activities (22). α-PGMs (EC 5.4.2.2) interconvert α-glucose-1-phosphate and α-glucose-6-phosphate, and PMMs (EC 5.4.2.8) interconvert mannose-1-phosphate and mannose-6-phosphate. In both cases the forward reaction favors sugar catabolism via glycolysis, while the reverse reaction favors the production of nucleotide sugars: UDP-glucose and GDP-mannose, respectively. The predicted YhxB amino acid sequence contains two motifs highly conserved among α-PGMs and PMMs: “Box 1,” corresponding to the catalytic site; and “Box 2,” corresponding to the binding site for its divalent cation cofactor (see references 8 and 37 and references therein). Whether YhxB acts as an α-PGM or a PMM or as a bifunctional phosphohexomutase cannot be predicted from its sequence because its “Box 3” motif, corresponding to residues that may determine substrate specificity, differs from those of phosphohexomutases that have been characterized at the biochemical level.
Nucleotide sugars are the biosynthetic precursors of exopolysaccharides, which in many cases are important components of extracellular matrices that hold together, and lend structure to, microbial communities (4, 6, 23, 25). In B. subtilis the predicted 16-gene operon yveK-yvfF, which resembles operons whose gene products are involved in exopolysaccharide synthesis (e.g., epsA- to -K of Streptococcus thermophilus Sfi6 [30]), is involved in pellicle formation and colony development (2). In fact, disruption of genes within the yveK-yvfF operon (e.g., yveRΔ) results in a phenotype indistinguishable from that of yhxBΔ mutants (e.g., see Fig. 2). This suggests that YhxB, through its role as a phosphohexomutase, promotes the synthesis of nucleotide sugars which, through processes catalyzed by proteins encoded by the yveK-yvfF operon, are incorporated into exopolysaccharides of the extracellular matrix of B. subtilis communities. Considering that polysaccharide synthesis is apparently of critical importance to B. subtilis biofilm formation, one might have expected other genes involved in this process to have been identified in our screen; however, many such genes are of known function and therefore are not represented in the BFA mutant collection. In any case, our work suggests that a more directed and comprehensive analysis of polysaccharide synthesis and its relationship to biofilm formation should greatly benefit our understanding of B. subtilis communities.
sipW, a gene that encodes a signal peptidase.
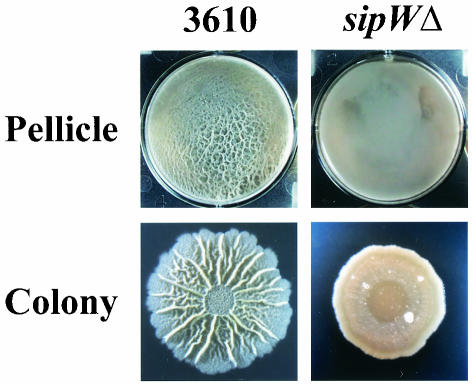
Strain 3610 harboring the sipW4766 or yqxM4767 insertional mutation formed pellicles of inconsistent thickness that failed to colonize all of the available surface area (Fig. 1 and data not shown). These mutant pellicles were composed of floating flocs that only loosely interacted with one another, such that physical manipulations which punctured or ripped other pellicles only parted the floating flocs. Phase-contrast microscopy revealed that the pellicles were composed of aggregates of relatively short chains of cells (data not shown); similar results were obtained using a deletion mutant (Fig. 3; see below). This suggested that the mutants failed to proliferate as long chains of cells and also failed to produce an extracellular matrix capable of holding the cells together in parallel alignment. Similarly, the mutants formed flat, undifferentiated colonies on solid media (Fig. 1 and data not shown), consistent with the idea that they failed to produce a fully functional extracellular matrix.
The yqxM and sipW genes are the first and second in a three-gene operon whose third member is tasA (31, 33) (Fig. 4B). SipW is a type I signal peptidase that is specifically required for the maturation and secretion of its two known endogenous substrates: the protein products of yqxM and tasA (28, 32-34). Thus, there were a number of possible explanations for the fact that the yqxM4767 and sipW4766 mutant phenotypes were identical. For instance, the mutant phenotypes might result from: (i) lack of YqxM activity, due to disruption of yqxM or loss of processing by SipW; (ii) lack of TasA activity, due to polar effects on tasA or loss of processing by SipW; (iii) accumulation of unprocessed TasA protein in the cytoplasm, due to disruption of, or polar effects on, sipW; (iv) loss of the processing and activity of SipW substrates other than YqxM and TasA, due to disruption of, or polar effects on, sipW; or (v) a combination of these effects. To address the third possibility, we deleted the entire yqxM-sipW-tasA operon in the 3610 genetic background and found that the resulting mutant (yqxM-sipW-tasAΔ; referred to as “sipWΔ”) formed pellicles and colonies that were indistinguishable from those formed by the yqxM4767 and sipW4766 mutants (Fig. 3 and 5 and data not shown). These results effectively rule out the third explanation above. With regard to the remaining explanations, it is interesting that Hamon and colleagues have found that sipW is the only gene in the yqxM-sipW-tasA operon that is required for biofilm formation by JH642 (13a), a B. subtilis strain derived from 168 (3), which itself is thought to be derived from 3610 (D. R. Zeigler, personal communication). This information would seem to support the fourth explanation—that loss of preprotein processing by SipW, but not lack of YqxM or TasA activity, accounts for the mutant phenotypes observed—and we are currently attempting to determine whether SipW plays a similar role in biofilm formation by 3610. In any case, our results suggest that SipW and at least some of its substrates are required for the structural integrity of the extracellular matrix.
FIG. 5.
Phenotype of mutant 3610 yqxM-sipW-tasAΔ (sipWΔ). Pellicle and colony development were assayed as described in the legend to Fig. 1. The parental strain images (first column) are the same as in Fig. 2.
ecsB, a gene encoding the transmembrane subunit of an ABC transporter.
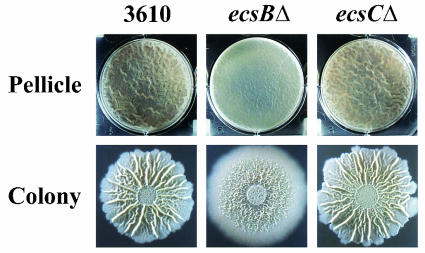
Strain 3610 harboring ecsBΔ formed flat pellicles (Fig. 6). Phase-contrast microscopy revealed that these mutant pellicles were composed of long chains of cells that were bound together rather loosely (Fig. 3). The 3610 ecsBΔ mutant also formed colonies in which certain architectural features were absent or less prominent than those observed in wild-type colonies (Fig. 6).
FIG. 6.
Phenotypes of mutants 3610 ecsBΔ and 3610 ecsCΔ. Pellicle and colony development were assayed as described in the legend to Fig. 1. The parental strain images (first column) are the same as in Fig. 2.
The ecsB gene is the second in a three-gene operon, ecsABC (20, 21) (Fig. 4C). Thus, the ecsBΔ mutant allele could potentially have polar effects on ecsC. However, deletion of ecsC from the 3610 genetic background resulted in a mutant that formed pellicles and colonies that were indistinguishable from those formed by the parental strain (Fig. 6). This suggests that inactivation of ecsB, and not polar effects on ecsC, was responsible for the mutant phenotype observed. However, at this time we cannot rule out the possibility that it was the combination of ecsB inactivation and polar effects on ecsC that gave rise to the mutant phenotype.
The ecsA gene encodes the ATPase subunit and ecsB encodes the transmembrane subunit of an ABC transporter referred to as Ecs (15, 21). The ecsC gene encodes a protein that shows no significant sequence similarity to any previously described ABC transporter component or accessory protein or indeed to any protein of known function. Ecs has been shown to promote the Sec-dependent secretion of several degradative enzymes, primarily through facilitation of their processing by signal peptidases (18, 20, 26). Both EcsA and EcsB are required for this function, whereas EcsC is not (18, 20). EcsA and EcsB are also required for efficient sporulation and development of competence (18 and data not shown), but it is not known whether this is due to the role of Ecs in protein secretion.
Our results indicate that Ecs activity is important for the development of B. subtilis communities. This effect is likely indirect; for example, Ecs could promote the maturation and secretion of protein components of the extracellular matrix and in this way contribute to the structure of pellicles and colonies. An interesting possibility is that some of the Ecs substrates might also be SipW substrates. It is conceivable that Ecs and SipW work together to promote the secretion of proteins that affect B. subtilis community structure. Indeed, Ecs has been shown to facilitate the processing of a heterologously expressed α-amylase by a different type I signal peptidase (26). Identification of the endogenous substrates of Ecs and SipW should lead to a better understanding of their roles in B. subtilis community development.
yqeK, a gene that encodes a predicted phosphatase.
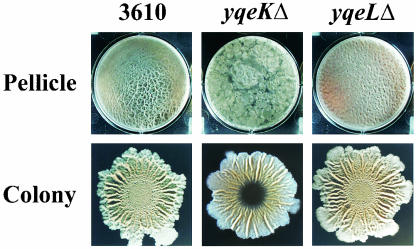
Strain 3610 harboring yqeKΔ formed pellicles that were initially thin and flat; however, these pellicles eventually became much thicker, and their flat surfaces were broken by a number of broad pits, the inner surfaces of which were covered with fruiting bodies (Fig. 7). This heterogeneity in morphology did not appear to be due to accumulation of suppressor mutations, as cells harvested from the invaginations and inoculated into fresh standing cultures produced pellicles that were indistinguishable from their predecessors (data not shown). Phase-contrast microscopy revealed that the mutant pellicles, like those formed by the parental strain, were composed of long chains of cells bound together in parallel (data not shown). The colonies formed by 3610 yqeKΔ also showed an obvious defect in gross morphology, in that their central regions were flat and glossy (Fig. 7).
FIG. 7.
Phenotypes of mutants 3610 yqeKΔ and 3610 yqeLΔ. Pellicle and colony development were assayed as described in the legend to Fig. 1.
The yqeK gene is located within a cluster of eight genes (yqeG to yqeM) that could constitute an operon (19) (Fig. 4D). Thus, the mutant phenotype conferred by yqeKΔ could potentially result from polar effects on downstream genes. However, deletion of the gene located immediately downstream of yqeK (yqeL) from the 3610 genetic background resulted in a mutant that formed wild-type pellicles and colonies (Fig. 7). This suggests that inactivation of yqeK, and not polar effects on downstream genes, accounts for the defects in community development exhibited by the 3610 yqeKΔ mutant.
The yqeG-yqeM gene cluster is well conserved among gram-positive bacterial species. For instance, clusters in which all eight genes are present and in the same order can be found in the genomes of Staphylococcus aureus MW2 (genes MW1549 to MW1542), Listeria innocua CLIP (genes lin1527 to lin1520), and Oceanobacillus iheyensis HTE831 (genes OB1989 to OB1982). Similar yqeK-containing clusters can be found in the genomes of six additional gram-positive bacteria, though in these cases some of the other genes are missing. Aside from yqeK itself, the only other gene present in all of the 11 clusters analyzed is nadD. This conservation of physical proximity suggests a possible functional relationship between yqeK and nadD, an idea consistent with the fact that the coding regions of these genes partially overlap in eight of the clusters, including that of B. subtilis.
The yqeK gene has not been characterized previously, but it is predicted to encode a protein that contains an HD sequence motif characteristic of metal-dependent phosphatases (1). The HD phosphatases that have been analyzed at the biochemical level are involved in a wide variety of biological processes. Thus, membership in this family reveals little about the biological function of YqeK. Clues regarding the function of yqeK may come from its physical, and perhaps functional, link to nadD, which encodes the nicotinic acid mononucleotide adenylyltransferase (NaMN-AT; EC 2.7.7.18) of B. subtilis (24). NaMN-ATs catalyze the penultimate step in the biosynthesis of NAD (NAD+), the essential cofactor that participates in a multitude of redox reactions. It will be of interest to determine whether YqeK plays a role in NAD+ metabolism, and if so, how abrogation of this function leads to the observed effect on B. subtilis community development.
ylbF, a gene that encodes a positive regulator of competence factor ComK.
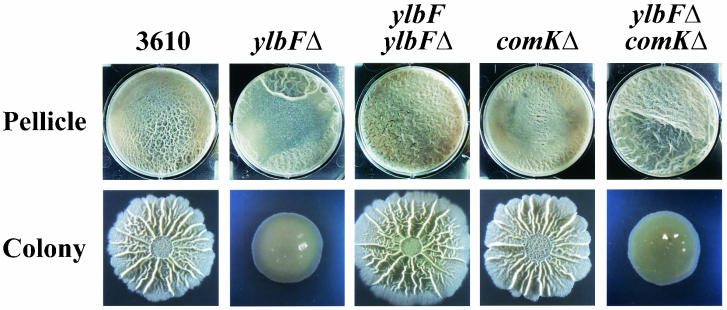
Strain 168 harboring the ylbF3233 insertional mutation failed to form pellicles (Fig. 1). However, when the ylbF3233 allele was introduced into the 3610 strain the resulting mutant formed robust pellicles that featured large, glossy wrinkles as well as some flat regions (Fig. 1). Similarly, strain 168 harboring ylbFΔ did not form pellicles (data not shown), whereas 3610 ylbFΔ formed pellicles that were heterogeneous in morphology (Fig. 8). When samples from 3610 ylbFΔ pellicles were streaked to single cells on selective solid medium and the resulting colonies were used to inoculate standing cultures, each isolate produced pellicles that were homogeneous in morphology, though that morphology differed from isolate to isolate (data not shown). These results strongly suggested that the 3610 ylbF mutants accumulated suppressor mutations that enabled them to form pellicles. Indeed, we have found that such mutations map to a single genetic locus (D. B. Kearns, F. Chu, S. S. Branda, R. Losick, and R. Kolter, unpublished results). It is not yet clear whether similar mutations accumulated in the 168 ylbF mutants and, if so, why these strains failed to form pellicles. Phase-contrast microscopy revealed that the pellicles produced by the 3610 ylbF mutants were composed of long chains of cells aligned in parallel (data not shown) and in this respect were indistinguishable from those produced by the parental strain (Fig. 3). The colonies formed by the 3610 ylbF mutants were essentially flat and undifferentiated (Fig. 1 and 8).
FIG. 8.
Phenotypes of mutants 3610 ylbFΔ, 3610 comKΔ, and 3610 ylbFΔ comKΔ and complementation of 3610 ylbFΔ. Pellicle and colony development were assayed as described in the legend to Fig. 1. The parental strain images (first column) are the same as in Fig. 2.
The ylbF gene appears to lie within a two-gene operon, ylbFG, that is flanked by two predicted Rho-independent transcription terminators (19) (Fig. 4E). Thus, the mutant phenotype conferred by ylbFΔ could potentially result from polar effects on ylbG. However, the 3610 ylbFΔ mutant was fully complemented by a wild-type ylbF allele integrated at the amyE locus (Fig. 8 and data not shown). This indicated that inactivation of ylbF, and not polar effects on ylbG or other downstream genes, accounted for the observed mutant phenotype.
The ylbF gene encodes a protein that promotes the translation and/or stability of ComK (35), a transcription factor that plays a critical role in the induction of natural competence (12). It seemed possible that YlbF might work in conjunction with ComK to coordinate B. subtilis community development. However, deletion of comK from the 3610 genetic background resulted in a mutant that formed wild-type pellicles and colonies (Fig. 8). Moreover, a 3610 ylbFΔ comKΔ double mutant formed pellicles and colonies that were indistinguishable from those formed by 3610 ylbFΔ (Fig. 8), indicating that the mutant phenotype conferred by ylbFΔ was not due to misregulation of ComK. Therefore, in contrast to its role in the development of competence, YlbF controls the development of B. subtilis communities via a pathway that does not include ComK. It has been suggested that in competence development, YlbF may promote ComK stability indirectly, through a positive effect on ComS, a protein that prevents MecA from targeting ComK for degradation by the ClpC/ClpP protease (35). This raises the possibility that YlbF could affect community development via a pathway that includes ComS and/or MecA but not ComK or competence factors acting upstream of ComS (i.e., ComP and ComA), which we and others have shown are not required for biofilm formation (13 and unpublished results). Alternatively, YlbF may control community development through a pathway that is entirely separate from that governing competence. It should be noted that YlbF also plays a role in sporulation, though the mechanism of that action remains unknown (35). We anticipate that the means by which YlbF affects B. subtilis community development will be elucidated through identification of genes whose expression depends upon YlbF and of proteins which interact with YlbF, as well as through further study of the suppression of 3610 ylbF mutants.
ymcA, a gene of unknown function.
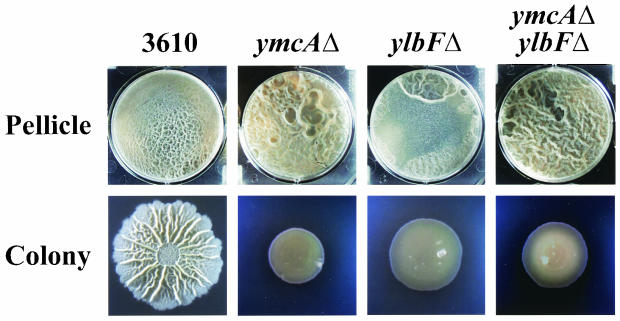
Strain 168 harboring ymcA2603 or ymcAΔ failed to form pellicles (Fig. 1 and data not shown), whereas introduction of either of these mutations into strain 3610 resulted in mutants that formed robust pellicles of heterogeneous morphology (Fig. 1 and 9). Single cells isolated from 3610 ymcAΔ pellicles and grown in standing cultures produced pellicles that were homogeneous in morphology, though that morphology differed from isolate to isolate (data not shown). These results are consistent with the idea that, like 3610 ylbF mutants, the 3610 ymcA mutants accumulated suppressor mutations that enabled them to form pellicles. In fact, we have found that such mutations map to the genetic locus involved in suppression of the 3610 ylbF mutants (Kearns et al., unpublished). Whether comparable mutations accumulated in the 168 ymcA mutants remains to be determined. Phase-contrast microscopy revealed that the 3610 ymcA mutant pellicles were composed of long chains of cells aligned in parallel (data not shown). The 3610 ymcA mutants also produced flat, undifferentiated colonies (Fig. 1 and 9). Thus, the ymcA mutants exhibited a complex phenotype essentially identical to that of the ylbF mutants. Importantly, we found that a 3610 ylbFΔ ymcAΔ double mutant formed pellicles and colonies that were indistinguishable from those formed by the single mutants (Fig. 9). These results suggest that ylbF and ymcA act through a common pathway to control the development of B. subtilis communities.
FIG. 9.
Phenotypes of mutants 3610 ymcAΔ and 3610 ymcAΔ ylbFΔ. Pellicle and colony development were assayed as described in the legend to Fig. 1. The parental strain images (first column) are the same as in Fig. 2, and the 3610 ylbFΔ images (third column) are the same as in Fig. 8.
The ymcA gene appears to lie within a two-gene operon, ymcBA: canonical −35 and −10 promoter elements are located upstream of ymcB, and a predicted Rho-independent transcription terminator lies 7 bp downstream of ymcA (19) (Fig. 4F). Moreover, the gene immediately downstream of ymcA (cotE) is regulated by σE (7), which we and others have shown is not required for biofilm formation (2, 13). Therefore, the ymcAΔ mutant phenotype almost certainly results from inactivation of ymcA rather than polar effects on downstream genes.
The ymcA gene had not been characterized previously, but it is predicted to encode a protein whose sequence resembles that of YlbF (19% identity and 51% similarity). However, programs that are designed to group proteins on the basis of shared domains (COG and ProDom) place YlbF and YmcA in separate ortholog clusters. This implies that sequence similarities between YlbF and YmcA are not concentrated in regions that are predicted to be important for protein function. To examine this issue more closely, we aligned the amino acid sequences of YlbF and YmcA with those of their respective orthologs (48 to 67% identical, with E values of 1 × 10−29 to 8 × 10−40) from Bacillus cereus, Bacillus anthracis, and Bacillus halodurans. We then determined whether residues that are shared among all four orthologs are also shared between B. subtilis YlbF and YmcA. We found that of the 149 residues in B. subtilis YlbF, 58 (39%) are conserved in its three orthologs, and of those 58 residues only 10 (17%) are identical to their corresponding residues in YmcA. Similarly, of the 143 residues in B. subtilis YmcA, 69 (48%) are conserved in its orthologs, but only 12 (17%) of these are identical to their corresponding residues in YlbF. Furthermore, the alignments of YlbF and YmcA orthologs revealed that certain regions of these proteins have been conserved particularly well over the course of evolution (e.g., 17 of 21 residues, starting with L16, in YmcA) and the conserved regions of YlbF do not correspond to those of YmcA. Thus, despite a fair degree of similarity in amino acid sequence, YlbF and YmcA appear to have different domain structures and therefore are unlikely to be paralogs.
Aside from YlbF, no other proteins of known or predicted function show sequence similarity to YmcA. The gene (ymcB) predicted to share an operon with ymcA encodes a protein that exhibits strong sequence similarity (44% identity, with an E value of 1 × 10−103) to MiaB from Escherichia coli and Salmonella enterica serovar Typhimurium. MiaB mediates a posttranscriptional modification of a nucleoside residue within tRNAs that read codons with U in the first position (9). Gram-negative bacteria apparently lack ymcA, but in gram-positive bacteria ymcA is often preceded by a gene encoding an MiaB ortholog. This physical proximity of the genes could reflect a functional relationship. This potential relationship, as well as that between YmcA and YlbF, represents a framework for further study of YmcA and its role in B. subtilis community development.
In summary, using an unbiased, genome-wide approach we identified six genes, some of them previously uncharacterized, that play important roles in the development of B. subtilis communities. Several of the genes appear to mediate production of an extracellular matrix that binds together the cells of these communities. As discussed above, yhxB likely works with the yveK-yvfF operon to synthesize exopolysaccharides that are incorporated into the matrix, while sipW and ecsB may promote the secretion of proteins that assemble, modify, or lend structure to the matrix. Both yhxB and sipW are required for the basic structural integrity of B. subtilis communities, whereas ecsB is required for formation of their distinctive architecture. A fourth gene, yqeK, is predicted to encode a phosphatase, and it too is required for proper formation of community architecture, though not for production of the extracellular matrix. Finally, ylbF and ymcA appear to work together to regulate community development; whether they affect production of the extracellular matrix remains to be determined. Thus, the six genes appear to play different roles in B. subtilis community development.
Considering that the collection screened was composed of mutants representing only 51% of the genome, it is likely that more comprehensive screens will identify additional genes involved in community development. Furthermore, screens utilizing less domesticated strains such as 3610 should identify still more genes, given the observation that mutations can confer different phenotypes when moved from one genetic background to another (Fig. 1). Equally important to our understanding of structured multicellular communities is the observation that phenotypes varied widely depending on whether pellicles or colonies were analyzed. This indicates that under different conditions B. subtilis uses different developmental pathways to form communities. The different pathways do share some genes, however, as mutation of at least six genes had profound effects on both pellicles and colonies, as described above. The relationships between these pathways and genes, and their implications for community development, will be the subject of future study.
Acknowledgments
We thank the BFA consortium for access to their mutant collection, as well as the Ehrlich lab for generous support during the screening of the collection. We also thank K. Carniol and A. Driks for strains, B. Lazazzera for communication of unpublished results, J. Shuler and the Nikon Imaging Center (Harvard Medical School) for microscopy, D. Kearns for critical reading of the manuscript, and the Kolter and Losick labs for useful discussions.
This work was supported by National Institutes of Health grants GM18568 (to R.L.) and GM58213 (to R.K.), and by National Science Foundation grant MCB-9727234 (to R.L.). S.S.B. was supported by an American Cancer Society Postdoctoral Fellowship. J.E.G.-P. was supported by a Ministerio de Educación y Ciencia Postdoctoral Fellowship (Spain).
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biol. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 2.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brehm, S. P., F. Le Hegarat, and J. A. Hoch. 1974. Developmental modulation of deoxyribonucleic acid-binding proteins of Bacillus subtilis during sporulation stages. J. Bacteriol. 120:1443-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vuyst, L., and B. Degeest. 1999. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol. Rev. 23:153-177. [DOI] [PubMed] [Google Scholar]
- 7.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dwivedi, K., A. F. Post, and S. Bullerjahn. 1996. Cloning and functional analysis of the pmmA gene encoding phosphomannomutase from the photosynthetic prokaryote Prochlorothrix hollandica. Biochim. Biophys. Acta 1290:210-214. [DOI] [PubMed] [Google Scholar]
- 9.Esberg, B., H.-C. E. Leung, H.-C. T. Tsui, G. R. Bjork, and M. E. Winkler. 1999. Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 181:7256-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freese, E., J. E. Heinze, and E. M. Galliers. 1979. Partial purine deprivation causes sporulation of Bacillus subtilis in the presence of excess ammonia, glucose, and phosphate. J. Gen. Microbiol. 115:193-205. [DOI] [PubMed] [Google Scholar]
- 11.Guerout-Fleury, A.-M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 12.Hahn, J., A. Luttinger, and D. Dubnau. 1996. Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol. Microbiol. 21:763-775. [DOI] [PubMed] [Google Scholar]
- 13.Hamon, M. A., and B. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199-1209. [DOI] [PubMed] [Google Scholar]
- 13a.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of Abr-B-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52:847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. Wiley-Interscience, Chichester, United Kingdom.
- 15.Higgins, C. F. 2001. ABC transporters: physiology, structure, and mechanism—an overview. Res. Microbiol. 152:205-210. [DOI] [PubMed] [Google Scholar]
- 16.Huang, C. T., K. D. Xu, G. A. McFeters, and P. S. Stewart. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 64:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 18.Kontinen, V. P., and M. Sarvas. 1988. Mutants of Bacillus subtilis defective in protein export. J. Gen. Microbiol. 134:2333-2344. [DOI] [PubMed] [Google Scholar]
- 19.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S.-K. Choi, J.-J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Düsterhöft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Hénaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauël, C. Médigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 20.Leskela, S., E. Wahlstrom, H.-L. Hyyrylainen, M. Jacobs, A. Palva, M. Sarvas, and V. P. Kontinen. 1999. Ecs, an ABC transporter of Bacillus subtilis: dual signal transduction functions affecting expression of secreted proteins as well as their secretion. Mol. Microbiol. 31:533-543. [DOI] [PubMed] [Google Scholar]
- 21.Leskela, S., V. P. Kontinen, and M. Sarvas. 1996. Molecular analysis of an operon in Bacillus subtilis encoding a novel ABC transporter with a role in exoprotein production, sporulation, and competence. Microbiology 142:71-77. [DOI] [PubMed] [Google Scholar]
- 22.Levander, F., and P. Radstrom. 2001. Requirement for phosphoglucomutase in exopolysaccharide biosynthesis in glucose- and lactose-utilizing Streptococcus thermophilus. Appl. Environ. Microbiol. 67:2734-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack, D. 1999. Molecular mechanisms of Staphylococcus epidermidis biofilm formation. J. Hosp. Infect. 43:S113-S125. [DOI] [PubMed]
- 24.Olland, A. M., K. W. Underwood, R. M. Czerwinski, M.-C. Lo, A. Aulabaugh, J. Bard, M. L. Stahl, W. S. Somers, F. X. Sullivan, and R. Chopra. 2002. Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase. J. Biol. Chem. 277:3698-3707. [DOI] [PubMed] [Google Scholar]
- 25.O'Toole, G. A., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 26.Pummi, T., S. Leskela, E. Wahlstrom, U. Gerth, H. Tjalsma, M. Hecker, M. Sarvas, and V. P. Kontinen. 2002. ClpXP protease regulates the signal peptide cleavage of secretory preproteins in Bacillus subtilis with a mechanism distinct from that of the Ecs ABC transporter. J. Bacteriol. 184:1010-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Serrano, M., R. Zilhao, E. Ricca, A. J. Ozin, C. P. Moran, Jr., and A. O. Henriques. 1999. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J. Bacteriol. 181:3632-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stingele, F., J.-R. Neeser, and B. Mollet. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J. Bacteriol. 178:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stover, A., and A. Driks. 1999. Control of synthesis and secretion of the Bacillus subtilis protein YqxM. J. Bacteriol. 181:7065-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stover, A., and A. Driks. 1999. Regulation of synthesis of the Bacillus subtilis transition-phase, spore-associated antibacterial protein TasA. J. Bacteriol. 181:5476-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stover, A., and A. Driks. 1999. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J. Bacteriol. 181:1664-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tjalsma, H., A. Stover, A. Driks, G. Venema, S. Bron, and J. M. van Dijl. 2000. Conserved serine and histidine residues are critical for activity of the ER-type signal peptidase SipW of Bacillus subtilis. J. Biol. Chem. 275:25102-25108. [DOI] [PubMed] [Google Scholar]
- 35.Tortosa, P., M. Albano, and D. Dubnau. 2000. Characterization of ylbF, a new gene involved in competence development and sporulation in Bacillus subtilis. Mol. Microbiol. 35:1110-1119. [DOI] [PubMed] [Google Scholar]
- 36.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 37.Videira, P. A., L. L. Cortes, A. M. Fialho, and I. Sa-Correia. 2000. Identification of the pgmG gene, encoding a bifunctional protein with phosphoglucomutase and phosphomannomutase activities, in the gellan gum-producing strain Sphingomonas paucimobilis ATCC 31461. Appl. Environ. Microbiol. 66:2252-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573-579. [DOI] [PubMed] [Google Scholar]