Abstract
More than one copy of rRNA operons, which code for both the small-subunit (SSU) and large-subunit (LSU) rRNA, are often found in prokaryotes. It is generally assumed that all rRNA operons within a single cell are almost identical. A notable exception is the extremely halophilic archaeal genus Haloarcula, most species of which are known to harbor highly divergent rRNA operons that differ at ∼5% of the nucleotide positions in the SSU gene and at 1 to 2% of the nucleotide positions in the LSU gene. We report that such intragenomic heterogeneity is not unique to Haloarcula, as high levels of intragenomic sequence variation have been observed for the SSU genes of two other genera of extreme halophiles, Halosimplex and Natrinema. To investigate this in detail, the two rRNA operons of Halosimplex carlsbadense and the four operons of Natrinema sp. strain XA3-1 were cloned and completely sequenced. The SSU and LSU genes of H. carlsbadense show the highest levels of intragenomic heterogeneity observed so far in archaea (6.7 and 2.6%). The operons of Natrinema sp. strain XA3-1 have additional unusual characteristics, such as identical internal transcribed spacers, while one of four SSU genes is 5% divergent and all LSU genes differ from each other by 0.9 to 1.9%. The heterogeneity among the Natrinema sp. strain XA3-1 LSU genes is localized in hot spots, and one of these regions is shown to be the result of a recombination event with a distantly related halophile. This is the first example of interspecies recombination between rRNA genes in archaea, and the recombination occurred over one of the largest phylogenetic distances ever reported for such an event. We suggest that intragenomic heterogeneity of rRNA operons is an ancient and stable trait in several lineages of the Halobacteriales. The impact of this phenomenon on the taxonomy of extremely halophilic archaea is discussed.
The gene coding for the ribosomal small subunit (SSU) has been the “gold standard” for identification and classification of cultivated microorganisms for several decades and is now extensively used to evaluate diversity among uncultivated microbes by direct amplification from DNA samples (17). At the intraspecific level (strains in a species), the internal transcribed spacer (ITS) between SSU and large-subunit (LSU) rRNA genes is convenient for making fine taxonomic distinctions. The SSU rRNA gene is also by far the most common probe for detection of specific strains in the environment by fluorescent in situ hybridization (2).
In placing such reliance on SSU rRNA genes, microbial systematists have assumed (i) that the sequences of multiple SSU rRNA genes often found in any one genome do not differ significantly and (ii) that interspecific (intergenomic) exchange of, or recombination between, SSU rRNA genes does not occur. In the last few years, it has become obvious that these assumptions are not true for all prokaryotes. Low levels of intragenomic variability of the SSU rRNA gene seem to be very common (6), and even more important variability can be present in the ITS between the SSU and LSU genes (5). Even these low levels of divergence can undermine attempts to evaluate prokaryotic diversity at the molecular level, especially when methods as sensitive as denaturing gradient gel electrophoresis are used (7). Also, when the SSU gene sequence is used as a criterion to assign strains to a particular species, even a low level of heterogeneity (∼1.0 to 2.0%) can sometime lead to misidentification, especially when species have been tightly defined (20).
High levels (>5%) of heterogeneity between multiple SSU gene copies found in a single cell have also been determined for some organisms. In eukaryotes, intragenomic SSU gene variability has been reported for the apicomplexan Plasmodium berghei (harboring SSU gene copies that are divergent at 5.0% of their nucleotide positions [13]) and the metazoan Dugesia mediterranea (8.0% divergence [4]). In prokaryotes, divergence levels of more than 5.0% have been found in two different instances: the thermophilic actinomycete Thermomonospora chromogena and the extremely halophilic archaeal genus Haloarcula. However, it seems that these two cases of intragenomic SSU gene variability have different evolutionary origins. T. chromogena harbors six rRNA operons, one of which differs from the others at 6.0% of the nucleotide positions in the SSU gene and at 10.0% of the nucleotide positions in the LSU gene (35). This divergent operon was found to be a mosaic of the other operons of T. chromogena and an rRNA operon from the actinomycete species Thermobispora bispora. This mosaic operon was found in only one species, T. chromogena, and the authors hypothesized that it was originally acquired by lateral gene transfer from T. bispora and was subsequently recombined with the other operons of its new host by gene conversion (35). Presumably, further conversion events ultimately homogenized these operons, and this lateral gene transfer or interspecific recombination event was detected only because it occurred recently.
The situation in Haloarcula is different. Intragenomic rRNA heterogeneity was first detected in Haloarcula marismortui, which had two rRNA operons that were divergent at 5.0% of the positions in the SSU gene and at 1.3% of the positions in the LSU gene (18). The processing of the rRNA product was shown to be different in the two operons; one operon exhibited the canonical archaeal processing, and the other had a unique processing procedure (10). Later, similar levels of divergence were determined for the two to four SSU genes found in numerous species of Haloarcula (12). This showed that the phenomenon is not an isolated event that affects only one species (as was probably the case for T. chromogena) but rather is an evolutionarily stable characteristic. Both of the H. marismortui rRNA operons were shown to be expressed when the organism was grown in rich media in the laboratory (1), suggesting that they are both functional. The recent identification of SSU genes that are ∼7% divergent in a member of the order Halobacteriales, Halosimplex carlsbadense, raises the possibility that this level of heterogeneity might be common in extremely halophilic archaea (32) and that further investigation might reveal why it persists. This prompted us to look in other Halobacteriales genera for high levels of intragenomic rRNA gene heterogeneity. We detected the presence of heterogeneous rRNA operons in strain XA3-1, which most likely belongs to the genus Natrinema, a group that is distantly related to both Halosimplex and Haloarcula according to SSU gene phylogenies.
We isolated the rRNA operons of Natrinema sp. strain XA3-1 by using methods that do not involve PCR amplification, since this can lead to the formation of chimeric molecules if multiple heterogeneous rRNA operons are present in the DNA being amplified. In fact, as we discovered in this work, the three operons reported for H. carlsbadense are actually only two operons plus a PCR chimera (32). We obtained the rRNA operons of H. carlsbadense through a combination of PCR-independent and chimera-limiting PCR methods, and the results clearly showed that only two operons are present and that the third operon that was originally identified is in fact a mosaic of these two operons.
Other than H. marismortui, no haloarchaeon possessing more than one rRNA operon has had all of its copies completely sequenced until now. With the addition of H. carlsbadense and Natrinema sp. strain XA3-1, all heterogeneous rRNA operons of representatives of three divergent groups of the Halobacteriales are available to help us understand the origins and evolutionary importance of intragenomic rRNA variability.
MATERIALS AND METHODS
Archaeal strains and genomic DNA.
Genomic DNA was isolated from pure cultures of Natrinema sp. strain XA3-1 (a gift from P. Zhou) and H. carlsbadense 2-9-1T by using the protocol of Wilson (33).
Genomic DNA digests.
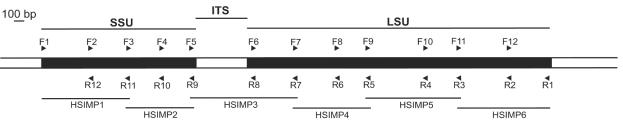
The genomic DNA of Natrinema sp. strain XA3-1 was completely digested with ClaI, while H. carlsbadense genomic DNA was digested with both NotI and ScaI. The digestions were performed overnight at 37°C by following the manufacturer's recommendations (New England Biolabs). The restriction endonucleases used were chosen because the average length of digestion products was between 5 and 10 kb and there was a low probability of cutting within the rRNA operons of the extreme halophiles being investigated. To determine which restriction endonucleases were unlikely to cut within these rRNA operons, we obtained the sequence of one rRNA operon from each species. These sequences were obtained from PCR-amplified fragments (obtained by using primers F1 and R1 [Fig. 1 ]) cloned in plasmid vector Topo-XL (Invitrogen). One clone was sequenced for each strain by using the primers described in Table 1. The resulting sequences were analyzed to identify enzymes that did not cut within them. Also, the enzymes were selected so that they did not cut within other Halobacteriales rRNA operons whose sequences are known, including those of Haloferax volcanii (http://www.ornl.gov/sci/techresources/Human_Genome/publicat/00santa/118.html/), Haloarcula marismortui (http://zdna2.umbi.umd.edu/cgi-bin/blast/blast.pl), Natrialba magadii (16), and Halobacterium sp. strain NRC-1 (19).
FIG. 1.
Schematic diagram of the rRNA operon sequencing strategy. Cloned rRNA operons were sequenced by using primers matching regions conserved in all Halobacteriales SSU and LSU genes. For each operon, both DNA strands were sequenced by using one sequencing primer for every ∼500 bp on each strand. The ITS is the ITS between the SSU and LSU genes. Fragments HSIMP1 to HSIMP6 are the PCR fragments used to obtain the complete sequence of the H. carlsbadense second rRNA operon. The primers used to amplify these fragments are the sequencing primers matching the 5′ and 3′ ends.
TABLE 1.
Primers used for amplification and sequencing of rRNA operons in the Halobacteriales
| Primera | Positionb | Gene | Sequence |
|---|---|---|---|
| F1 | 1-17 | SSU | ATTCCGGTTGATCCTGC |
| F2 | 463-480 | SSU | CCGCGGTAATACCGGCAG |
| F3 | 822-839 | SSU | CCGCCTGGGAAGTACGTC |
| F4X | 1136-1155 | SSU | GCAACGGTAGGTCAGCATGC |
| F4 | 1140-1156 | SSU | CGGTAGGTCAGTATGCC |
| F5 | 1458-1473 | SSU | GGCTGGATCACCTCCT |
| F6 | 2050-2066 | LSU | GGACGTGCCAAGCTGCG |
| F7X | 2435-2454 | LSU | TACTCCTCGAGACCGATAGC |
| F7 | 2460-2479 | LSU | AGTAGTGTGAACGAACGCTG |
| F8 | 2863-2879 | LSU | GGTGAAAGGCCCATCGA |
| F9 | 3189-3207 | LSU | CAGCTTACCGGCCGAGGTT |
| F10 | 3722-3739 | LSU | ACGTTAGGGAATTCGGCA |
| F11 | 4050-4069 | LSU | CCAGTGCGGAGTCTGGAGAC |
| F12 | 4527-4545 | LSU | CGGTTCCCTCCATCCTGCC |
| R1 | 4834-4856 | LSU | CGCGCACACCCCGAGTCTATCGA |
| R2X | 4485-4501 | LSU | CGATATGTACTCTTGCG |
| R2 | 4485-4501 | LSU | CGATATGTGCTCTTGCG |
| R3 | 4078-4094 | LSU | ATAGGGTCTTCGCTTCC |
| R4X | 3722-3741 | LSU | ATTGCCGAATTCCCTAACGT |
| R4 | 3722-3741 | LSU | CTTGCCGAATTCCCTAACGT |
| R5 | 3189-3207 | LSU | AACCTCGGCCGGTAAGCTG |
| R6 | 2863-2879 | LSU | TCGATGGGCCTTTCACC |
| R7X | 2524-2543 | LSU | CAGCGTTCGCTCGCGCTACT |
| R7 | 2519-2537 | LSU | TCGCTCGATCGCCAACTGA |
| R8 | 2050-2066 | LSU | CGCAGCTTGGCACGTCC |
| R9 | 1427-1447 | SSU | CTACGGCTACCTTGTTACGAC |
| R10X | 1241-1261 | SSU | CCTCAATCCGAACTACGACC |
| R10 | 1173-1190 | SSU | CCATTGTAGCCCGCGTGT |
| R11 | 822-839 | SSU | GACGTACTTCCCAGGCGG |
| R12 | 499-516 | SSU | ACGCTTTAGGCCCAATAA |
F, forward; R, reverse; X, specific for Halosimplex.
Nucleotide positions in the Halobacterium sp. strain NRC-1 rRNA operon.
Determination of the rRNA operon copy number by Southern hybridization.
Digested genomic DNA samples were loaded beside a DNA ladder (MBI Fermentas) onto a 0.8% agarose gel, which was electrophoresed for 16 h at 60 V in 1× TAE buffer (4°C) and subsequently transferred onto a positively charged nylon membrane (Roche). SSU rRNA probes were directly amplified from genomic DNA of the species to which they were hybridized. Probes were amplified with universal Halobacteriales primers F1 and R9 (Table 1). Amplified probes were gel purified (MinElute; QIAGEN) and subsequently labeled with digoxigenin-dUTP (Roche). Each probe was individually hybridized to the membrane along with a labeled DNA ladder (GeneRuler 10 Kb; MBI Fermentas). Hybridization of the probes was detected with a CDP-star chemiluminescence system (Roche).
Genomic DNA library construction.
Southern hybridization revealed the locations of the rRNA operons relative to the DNA ladder (between 5 and 10 kb). For both Natrinema sp. strain XA3-1 and H. carlsbadense, genomic DNA was redigested and electrophoresed on an agarose gel under conditions identical to those used for Southern hybridization. Genomic DNA was then extracted from the regions of the gel corresponding to DNA fragments containing an rRNA operon (MinElute; QIAGEN). Before purified DNA fragments could be cloned, protruding 5′ or 3′ overhangs resulting from digestion had to be blunt ended and dephosphorylated in preparation for blunt end cloning according to the manufacturer's instructions (TOPO-Zeroblunt; Invitrogen). The resulting products were ligated into the Topo-Zeroblunt plasmid vector (Invitrogen) and transformed into chemically competent Escherichia coli TOP10 cells (Invitrogen), which were plated on kanamycin-containing media to select for positive transformants.
Library screening.
The libraries were transferred to positively charged nylon membranes (Roche) and screened directly for the presence of rRNA genes by hybridization with a digoxigenin-dUTP-labeled probe (PCR-amplified SSU gene of the species from which the genomic DNA in the library originated). Positive clones were confirmed by end sequencing with the M13 forward and M13 reverse primers and direct sequencing with a universal Halobacteriales SSU gene primer, primer F1 (Table 1).
PCR amplification of rRNA genes not obtained in genomic libraries.
All four rRNA operons of Natrinema sp. strain XA3-1 and one of the two operons of H. carlsbadense were obtained after repeated screening of the libraries. A PCR strategy aimed at minimizing the formation of chimeras was used to obtain the second rRNA operon of H. carlsbadense. Compared to standard PCR conditions, we used five times more of each primer, an extension time that was five times longer, and fewer PCR cycles (25 cycles). This resulted in the following PCR conditions: a 50-μl (final volume) mixture containing 1 to 5 ng of template DNA, 1× PCR buffer, 1 μl of a solution containing each deoxynucleoside triphosphate at a concentration of 10 mM, 5 μl of a solution containing each primer at a concentration of 10 μM, and 1 μl of PFU Turbo DNA polymerase (Stratagene). The initial denaturation took place at 95°C for 2 min, and this was followed by 25 cycles consisting of denaturation at 95°C for 30 s, primer annealing at 55°C for 30 s, and primer extension at 72°C for 5 min. The operon was amplified in six fragments that were 0.8 to 1.0 kb long and overlapped at their extremities, as chimera formation was less likely for shorter products (Fig. 1). After cloning of each fragment (TOPO-ZeroBlunt; Invitrogen), a mixed population of clones (from each of the two operons) was obtained. Twenty-four clones were sequenced for each fragment, and subsequently the sequences were compared to the sequence of the complete H. carlbadense rRNA operon obtained from the library. Sequences that appeared only once (singletons) (∼20%) were flagged as chimeric and discarded. As Southern hybridization confirmed that there were only two operons in H. carlsbadense (see below), a fragment whose sequence was not identical to the sequence in the operon obtained from the library could only belong to a second operon. Such sequences were assembled to obtain the final sequence of the missing H. carlsbadense operon.
DNA sequencing, analysis, and assembly.
Positive clones from the library found to carry an entire rRNA operon were sequenced by using MegaBase technology and BigDye chemistry. Multiple primers were used for sequencing the entire rRNA operon, which gave overlapping data so that a reliable sequence could be obtained (Fig. 1). The sequencing primers targeted conserved regions of rRNA operons, as determined by examination of an alignment of all available Halobacteriales rRNA genes. The nucleotide sequences of these primers are shown in Table 1. Sequencher 4.1.2 (Gene Codes Corporation) was used to analyze sequence chromatograms and assemble sequence fragments.
Multiple-sequence alignment and phylogenetic analysis.
All available nucleotide sequences for SSU and LSU genes of culturable Halobacteriales were retrieved from the GenBank database. Sequences from the database were aligned with novel sequences obtained in this study by using CLUSTALW (29) and were edited manually to remove gaps and ambiguously aligned characters. Phylogenetic analyses were performed with PAUP* 4.04b (28) by using the heuristic search option and the TBR branch-swapping algorithm. Maximum likelihood and maximum-likelihood distances were the tree reconstruction methods used, and the nucleotide substitution model, gamma rate parameter α, proportion of invariable sites, and nucleotide frequencies were determined independently for each gene by using MODELTEST (24). The confidence of each node was determined by constructing a consensus tree for 1,000 bootstrap replicates by using minimum-evolution or maximum-likelihood distances or full maximum likelihood.
Calculation of the among-site rate variation for the SSU gene in Halobacteriales.
TREE-PUZZLE (http://www.tree-puzzle.de) was used to determine the relative evolutionary rates of all nucleotide positions of the SSU gene in Halobacteriales. Rates were calculated from an alignment of the SSU genes of 80 representative taxa belonging to the Halobacteriales under a gamma-distributed model with four rate categories. Such an analysis could not be performed for the LSU gene because sequence data were available for only eight taxa, which was insufficient for calculation of informative relative evolutionary rates.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the EMBL database under accession numbers AJ586107 to AJ586112.
RESULTS
Detection of heterogeneity in the rRNA operons of Halobacteriales.
Genomic DNA was extracted from a pure culture of Natrinema sp. strain XA3-1, and the SSU genes were amplified by PCR, cloned, and sequenced. The multiple clones sequenced displayed high sequence divergence (∼5.0% of nucleotide positions).
H. carlsbadense SSU genes were amplified by PCR as part of the taxonomic identification of this organism by Vreeland et al. (32), who used a DNA polymerase with relatively low fidelity (LA Taq; Takara Shuzo, Kyoto, Japan). Three divergent genes were initially inferred, and one of these genes was different from the other two at ∼7% of the nucleotide positions; this is the highest level of divergence reported so far for multiple copies of the SSU gene within one prokaryotic organism. However, upon close examination, one of the gene copies seemed to be a chimera of the other two. The 5′ end of this chimeric gene was identical to the homologous region in the second copy, and its 3′ end was identical to the third copy.
Determination of the number of rRNA operons in Natrinema sp. strain XA3-1 and H. carlsbadense.
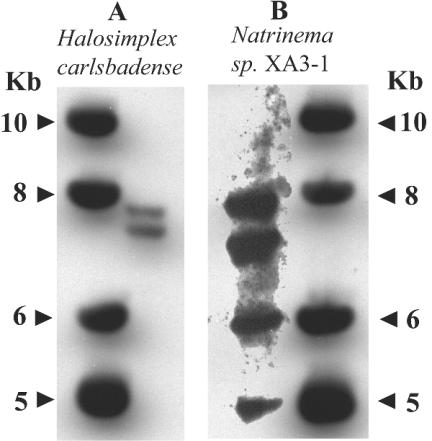
The number of rRNA operons in each of the two organisms in which heterogeneity was detected was determined by Southern hybridization (Fig. 2). Natrinema sp. strain XA3-1 was found to have four rRNA operons. In contrast to previous claims that H. carlsbadense harbors three rRNA operons (based on PCR amplification of three divergent SSU genes from genomic DNA [32]), we found that this haloarchaeon had only two operons. This result was obtained when the genomic DNA was digested to completion with a combination of NotI and ScaI (Fig. 2), as well as with NotI/ClaI and ScaI/ClaI (results not shown).
FIG. 2.
Southern hybridization of SSU gene probes to complete genomic DNA restriction endonuclease digests. The restriction endonucleases used were ClaI for Natrinema sp. strain XA3-1 (A) and NotI and ScaI for H. carlsbadense (B). The SSU gene probes used were directly amplified from genomic DNA of the species to which they were hybridized.
Degree of divergence of the rRNA operons of Halobacteriales containing multiple heterogeneous copies.
Sequence comparisons of the multiple rRNA operons in Natrinema sp. strain XA3-1 and H. carlsbadense confirmed the high degree of divergence detected for the PCR-amplified SSU genes (Table 2). Three of the four SSU genes of Natrinema sp. strain XA3-1 (from operons B, C, and D) are almost identical, and the other (from operon A) is different at 5.0% of the nucleotide positions. The divergence observed for the LSU genes exhibits a different pattern, as the levels of divergence for the genes of all operons, including operon A, are similar to each other, ranging from 0.9 to 1.9%. The sequences of SSU-LSU ITS of all rRNA operons of Natrinema sp. strain XA3-1, including a tRNAAla, are identical.
TABLE 2.
Details of nucleotide substitutions and levels of divergence between the rRNA operons of Halobacteriales species harboring multiple heterogeneous copies
| Comparison | Nucleotide differences
|
% Divergencec | |||
|---|---|---|---|---|---|
| Lengtha | No. of substitutions | No. of indelsb | Total no. | ||
| Haloarcula marismortui | |||||
| SSU-A and -B | 1,472 | 74 | 0 | 74 | 5.0 |
| LSU-A and -B | 2,922 | 39 | 0 | 39 | 1.3 |
| ITS-A and -B | 411 | 65 | 36 | 101 | 24.6 |
| H. carlsbadense | |||||
| SSU-A and -B | 1,476 | 95 | 5 | 100 | 6.8 |
| LSU-A and -B | 2,860 | 76 | 1 | 77 | 2.7 |
| ITS-A and -B | 466 | 110 | 119 | 229 | 49.1 |
| Natrinema sp. strain XA3-1 | |||||
| SSU-A and -B | 1,475 | 68 | 5 | 73 | 4.9 |
| SSU-A and -C | 1,475 | 69 | 5 | 74 | 5.0 |
| SSU-A and -D | 1,475 | 68 | 5 | 73 | 4.9 |
| SSU-B and -C | 1,475 | 1 | 0 | 1 | 0.1 |
| SSU-B and -D | 1,475 | 2 | 0 | 2 | 0.1 |
| SSU-C and -D | 1,475 | 1 | 0 | 1 | 0.1 |
| LSU-A and -B | 2,843 | 53 | 0 | 53 | 1.9 |
| LSU-A and -C | 2,894 | 30 | 0 | 30 | 1.0 |
| LSU-A and -D | 2,867 | 25 | 0 | 25 | 0.9 |
| LSU-B and -C | 2,843 | 35 | 0 | 35 | 1.2 |
| LSU-B and -D | 2,843 | 54 | 0 | 54 | 1.9 |
| LSU-C and -D | 2,867 | 32 | 0 | 32 | 1.1 |
| ITS-A and -B | 434 | 0 | 0 | 0 | 0 |
| ITS-A and -C | 434 | 0 | 0 | 0 | 0 |
| ITS-A and -D | 434 | 0 | 0 | 0 | 0 |
| ITS-B and -C | 434 | 0 | 0 | 0 | 0 |
| ITS-B and -D | 434 | 0 | 0 | 0 | 0 |
| ITS-C and -D | 434 | 0 | 0 | 0 | 0 |
Number of nucleotides at which the sequences were compared.
Number of nucleotide insertions or deletions.
Percentage of divergent nucleotide positions.
The two SSU genes of H. carlsbadense differ at 6.8% of their nucleotide positions. This is the highest level of intragenomic heterogeneity among the SSU genes of a prokaryote reported so far. Similar to the genes of Natrinema sp. strain XA3-1, the LSU genes of H. carlsbadense exhibit lower levels of divergence (2.6%). The two ITS of this organism are extremely divergent (Table 2), exhibiting significant similarity only in the first 33 bp at the 5′ end (which are identical) and in the last 135 bp (39 of the last 135 positions are divergent). The ITS of operon B is 100 bp shorter than its operon A counterpart (357 versus 457 bp) and does not contain a functional tRNAAla (a tRNAAla can clearly be identified in the ITS of operon A).
Locations of the variable positions of heterogeneous rRNA genes on the secondary structure of the rRNA product.
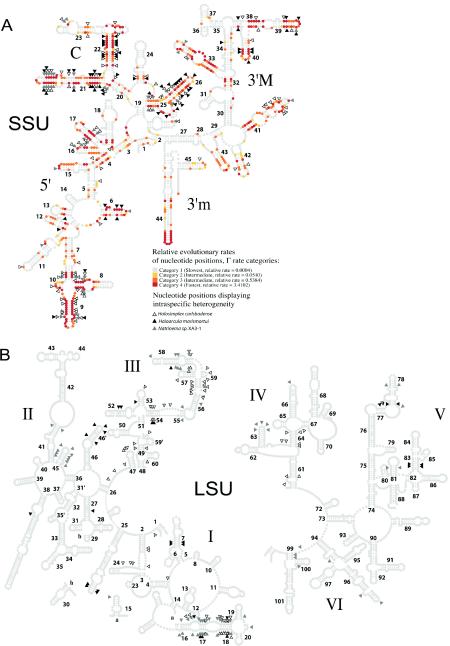
Nucleotide substitutions for the multiple SSU and LSU genes in Halosimplex, Natrinema, and Haloarcula were mapped by using the secondary structures of their rRNA products. Haloarcula was included because it is the only genus of haloarchaea besides those examined in this study known to display intragenomic heterogeneity in its rRNA genes (18). Most substitutions in the heterogeneous rRNA genes found in these haloarchaea are either compensatory mutations that occur in stems or are located in loop regions. This suggests that these substitutions should have little or no effect on the overall secondary structure of the rRNA. Furthermore, all of the ribosomal protein genes of H. marismortui have been cloned and sequenced (26), and all appear to be single-copy genes. This implies that the same protein should have the capacity to bind to rRNA from either of the two heterogeneous genes to form functional ribosomes. When grown in rich media, H. marismortui has indeed been shown (by using fluorescent in situ hybridization probes specific for each of the two SSU genes) to have a mixed ribosome population (1).
Nucleotide substitutions in heterogeneous SSU genes are almost always found in hypervariable regions (Fig. 3A) (that is, regions in which most interspecies divergence occurs). Regions corresponding to helices 21, 22, and 26 (5′ domain) in the SSU gene secondary structure are variable in all three haloarchaeal genera that display high intragenomic rRNA gene heterogeneity. The region corresponding to helices 7, 8, and 9 (5′ domain) is highly variable in both Halosimplex and Natrinema but not in Haloarcula. Multiple regions are heterogeneous in only one genus (e.g., helix 41 is variable in Halosimplex, and helix 6 is variable in Haloarcula).
FIG.3.
Mapping of variable positions in the SSU (A) and LSU (B) genes based on the secondary structure of the rRNA product. Positions displaying intragenomic heterogeneity are indicated. Helices present in the secondary structure are numbered 1 to 45 (SSU gene) and 1 to 101 (LSU gene). Structural domains are indicated as follows: 5′, central (C), 3′M (major), and 3′m (minor) for the SSU gene and I to VI for the LSU gene. The relative evolutionary rate of each nucleotide position was calculated for the SSU gene in the Halobacteriales. The rate categories for the positions are indicated by colors in the secondary structure.
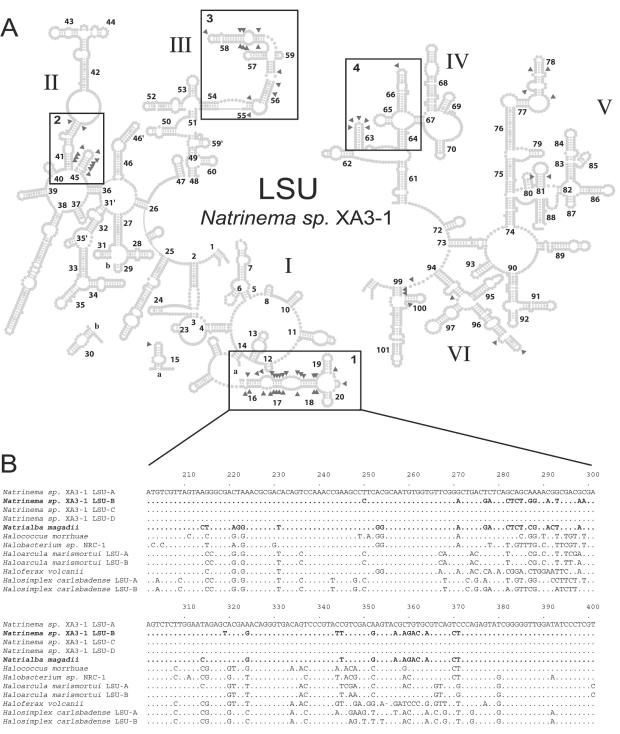
The intragenomic divergence in LSU genes is much lower than the intragenomic divergence in SSU genes and is very patchily distributed across the length of the gene (Fig. 3B). The region containing helices 18, 19, and 20 (domain I) is the only region that displays intragenomic heterogeneity in all three haloarchaea examined; most other variable regions are found in only one of the three organisms. In contrast to the heterogeneity of the SSU gene, the heterogeneity of the LSU genes of Natrinema sp. strain XA3-1 does not occur in one highly divergent copy (SSU-A for the SSU gene) but is spread among the four copies. Each of these copies diverges from the other copies in specific regions (Fig. 4).
FIG. 4.
(A) Locations of the variable positions of Natrinema sp. strain XA3-1 LSU genes in the secondary structure of the rRNA product. Positions displaying intragenomic heterogeneity are indicated by gray triangles. The numbered boxes indicate different regions of the LSU genes in which heterogeneity between the multiple copies is observed, as follows: box 1, divergence of LSU-B at 27 positions between nucleotides 249 and 370; box 2, divergence of LSU-B and -C from LSU-A and -D at 13 positions between nucleotides 1113 and 1284; box 3, divergence of LSU-D at 13 positions between nucleotides 1523 and 1652; and box 4, divergence of LSU-C at 6 positions between nucleotides 1786 and 1856. Variable positions that are not in a box for the most part represent divergence in LSU-A. (B) Alignment of all available Halobacteriales LSU genes from positions 200 to 400, which corresponds to box 1 in the secondary structure diagram. At a given position, a nucleotide identical to the nucleotide in the Natrinema sp. strain XA3-1 LSU-A sequence is indicated by a dot.
Recombination between Natrinema sp. strain XA3-1 and N. magadii LSU genes.
Natrinema sp. strain XA3-1 LSU genes display heterogeneity in several distinct regions (Fig. 4A). Between positions 270 and 370 (corresponding to box 1 [Fig. 4A]), the LSU-B gene of XA3-1 is virtually identical to the N. magadii homolog (differing at only three positions), while it diverges strongly from the three other XA3-1 LSU genes (differing at 26 positions) (Fig. 4B). None of the other regions in which one or two of the LSU genes of XA3-1 differed from the other genes had such strong similarity to the LSU gene(s) of another member of the Halobacteriales.
The surprisingly high level of similarity between positions 270 and 370 of the strain XA3-1 LSU-B gene and the equivalent region in the N. magadii homolog is most easily explained by cross-species homologous recombination. Such a recombination event is made even more likely by the fact that it is limited to a specific structural region of the LSU molecule (helices 18, 19, and 20 in domain I). Most of the nucleotide substitutions caused by the recombination, when they are in stem regions, are complementary to each other, conserving the rRNA secondary structure (Fig. 4A). The maintenance of the secondary structure is important, given the complexity of the ribosome and the numerous interactions of the LSU gene molecule with proteins and other RNA components. To our knowledge, this is the first example of interspecies recombination between rRNA genes in archaea. Also, the recombination took place over one of the largest phylogenetic distances ever reported for recombination of rRNA genes (between two genera that differ at ∼5% of the positions in their SSU genes). Other regions that are heterogeneous in the LSU genes of Natrinema sp. strain XA3-1 might also be the result of recombination. However, this could be verified only if strong similarity was found between these regions and the equivalent LSU genes of other species. The inability to find a donor for these regions, if they indeed originated through recombination, probably is explained by the very limited sampling of the LSU genes of members of the Halobacteriales (such genes are available from only eight species).
Phylogenetic distribution of haloarchaea with intracellular heterogeneity in the rRNA genes.
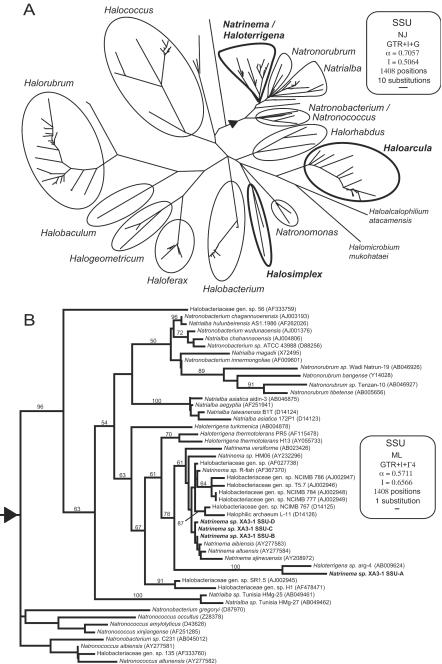
A phylogeny of the order Halobacteriales based on the SSU gene is shown in Fig. 5A. With the addition of data from this study, rRNA heterogeneity is now known to occur in three divergent lineages: Halosimplex, Haloarcula, and Natrinema. These lineages, according to the SSU gene data, do not exhibit specific phylogenetic affinity for each other and are spread across the Halobacteriales diversity (Fig. 5A). Of course, each species with multiple different rRNA operons has many branches on the tree, but assignments at the genus level or higher (perhaps with the exception of Natrinema and Haloterrigena) are not altered.
FIG.5.
(A) Best maximum-likelihood distance tree for the SSU gene for the archaeal order Halobacteriales. (B) Best maximum-likelihood tree for the Halobacteriales subgroup in the triangular area in panel A. The evolutionary models and parameters used for the phylogenetic analyses are indicated in the boxes (NJ, neighbor joining; ML, maximum likelihood; GTR, general time reversible; I, proportion of invariable sites; G, gamma-distributed among-site rate variation; α, gamma rate shape parameter alpha; Γ, number of gamma distribution rate categories). The genera in which single organisms contain highly heterogeneous rRNA operons are indicated by boldface type. Bootstrap values were obtained by using a distance maximum-likelihood tree reconstruction method (only values greater than 50% are indicated at the nodes).
Within the genus Haloarcula, all species that have been investigated show rRNA heterogeneity (12). H. carlsbadense is the only cultured species of its genus. In addition to the heterogeneity of Natrinema sp. strain XA3-1, we detected strong intragenomic heterogeneity in another member of the genus Natrinema, Natrinema versiforme, based on the divergence of the sequences of SSU genes PCR amplified from genomic DNA of this species (data not shown). A phylogeny of the SSU genes from representatives of the haloarchaeal clade to which Natrinema belongs is shown in Fig. 5B, which is a portion of the tree shown in Fig. 5A. The most divergent of the four SSU genes from Natrinema sp. strain XA3-1 (SSU-A) clusters outside the clade formed by the other three genes of this organism and SSU genes from various Natrinema species, including, among others, N. versiforme, along with several uncharacterized Halobacteriales. More specifically, the XA3-1 SSU-A gene clusters very strongly with a homolog from Haloterrigena sp. strain arg-4 (15). In fact, the XA3-1 SSU-A gene differs at only 27 positions from the arg-4 homolog (only 1.8% of the nucleotide positions), compared to the 4.9 to 5.0% divergence of SSU-A from the other SSU genes of XA3-1.
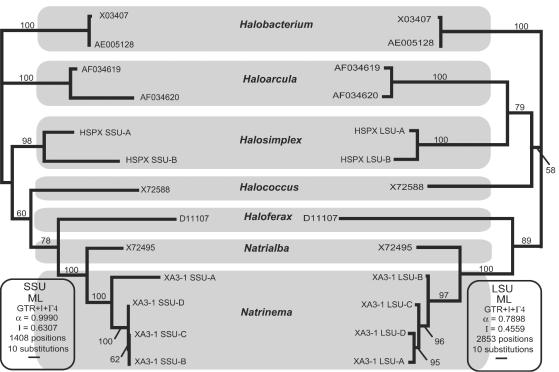
When phylogenetic trees of the SSU and LSU genes of haloarchaea were compared to each other by using the same taxon sampling method, the overall structure was very similar (Fig. 6). The LSU gene tree, which resulted from a data set with more informative positions, seems to be slightly more resolved. The only difference between the two trees is in the branching order of the SSU and LSU genes from Natrinema sp. strain XA3-1. In the SSU gene tree, the SSU-A gene is by far the most divergent and strongly branches in a basal position in relation to the other three copies of the SSU gene. In the LSU gene tree, on the other hand, the most divergent copy is LSU-B, and LSU-A groups strongly with the other two copies.
FIG. 6.
Comparison of best maximum-likelihood trees for the SSU and LSU genes of all genera of the Halobacteriales for which sequences of both genes were available. The evolutionary models and parameters used for the phylogenetic analyses are indicated (ML, maximum likelihood; GTR, general time reversible; I, proportion of invariable sites; α, gamma rate shape parameter alpha; Γ, number of gamma distribution rate categories). The numbers of nucleotide positions present in the edited alignment used in the phylogenetic analysis are also indicated. The bootstrap values at the nodes are the consensus values for maximum-likelihood trees for 1,000 pseudoreplicates.
DISCUSSION
The high intragenomic rRNA heterogeneity in three different genera of Halobacteriales observed here clearly poses a problem to the use of SSU and LSU genes as reliable classification tools for this archaeal order. The degree of intragenomic variability observed (4.9 to 6.8% in the SSU genes) is far greater than the average between-species divergence (∼2.0%) and is more in the range of the divergence usually found between genera in the order Halobacteriales (∼5.0 to 10.0%). One example of taxonomic confusion possibly caused by intragenomic SSU gene heterogeneity is the status of the genera Haloterrigena and Natrinema (30). As discussed above, there is a high level of similarity between one of the Natrinema sp. strain XA3-1 SSU genes and one SSU gene sequence PCR amplified from Haloterrigena sp. strain arg-4. This suggests that this Haloterrigena strain could in fact be a Natrinema strain harboring multiple heterogeneous rRNA operons, only one of which was obtained by PCR. This could also be the case for other species of Haloterrigena, which are close neighbors of the Natrinema in phylogenetic analyses (Fig. 5B).
The similarity observed in a 100-bp stretch between one of the LSU genes of Natrinema sp. strain XA3-1 and the N. magadii LSU gene, which is likely to be due to an interspecies recombination event, suggests an origin for intragenomic rRNA heterogeneity. As the other heterogeneous regions of Natrinema sp. strain XA3-1 LSU genes are also in 100- to 200-bp stretches and are found in only one or two of the rRNA operons, it is likely that they also originated by recombination (although this cannot be confirmed unless the source of the recombined fragment is identified). The source of recombined fragments is often very difficult to identify, for several reasons. Recombination usually occurs in hypervariable regions, which means that the sequence of the recombined fragment rapidly diverges from the source sequence, making it hard to detect by eye or by using software (which is unable to find a statistically significant match with the source). The limited number of rRNA gene sequences available in databases for Halobacteriales, compared to the diversity found in nature, also makes the search for a recombination donor difficult (especially for the LSU gene, for which less than a dozen sequences are available).
It is clear that the four LSU genes of Natrinema sp. strain XA3-1 did not evolve in coordination with the SSU genes and the ITS partners. First, all LSU gene copies differ from each other by roughly similar numbers of substitutions (0.9 to 1.9% of the nucleotide positions differ). In contrast, only one of the strain XA3-1 SSU genes is really different from the others (it differs at 5.0% of the positions); the other three SSU genes differ from each other at only one or two positions. The ITS between the SSU and LSU genes are identical in all four operons. This is surprising, as the ITS is the fastest evolving part of rRNA operons (23) and usually displays high variability, even at the species level (11). There are two possible explanations (which are not mutually exclusive) for the unequal evolutionary rates of different parts of Natrinema sp. strain XA3-1 rRNA operons: intragenomic homologous recombination (gene conversion) among the four rRNA operons of XA3-1 does not happen uniformly across the length of the operons, and/or interspecies homologous recombination is frequent.
Recombination between rRNA genes of different strains or species could occur at a much higher frequency than originally suspected. Like the intragenomic variability of Natrinema, the intragenomic variability between the LSU genes of Halosimplex and Haloarcula seems likely to have originated through recombination. For both of these taxa, heterogeneous positions are distributed nonrandomly in the LSU gene and usually occur in small patches. Also, although heterogeneity always occurs in hypervariable regions, the regions affected differ from one species to another. This suggests that the origins of regions displaying intragenomic heterogeneity are diverse.
As no widespread survey of the Halobacteriales for intragenomic rRNA variability has been undertaken, the frequency of the phenomenon is still unknown. Such heterogeneity has been detected in only three lineages so far, but it is likely to be present in others. For most haloarchaeal species, only one SSU gene which was amplified by PCR has been sequenced. This approach is likely to miss intragenomic heterogeneity for several reasons: PCR amplification can be biased toward a particular copy of the SSU gene, the amplified gene could be a chimera of the multiple copies present in the organism, and in many studies only a few clones originating from a single PCR are sequenced (and therefore it is likely that heterogeneous PCR products are missed).
Phylogenetic analysis of rRNA genes recovers groups that display intragenomic heterogeneity for these genes as monophyletic entities (e.g., all Haloarcula SSU genes form a clade, and all Natrinema-Haloterrigena LSU genes group together to the exclusion of the LSU genes of other genera). These monophylies suggest that the heterogeneity could have originated in the ancestors of each of the clades, although information about the distribution of this phenomenon in haloarchaea is necessary to verify this claim.
What is the source of the heterogeneity? It could be interspecific recombination, as mentioned above, or simply random divergence of paralogs. It is difficult to distinguish between these possibilities, which again are not necessarily mutually exclusive. Identification of clear traces of a between-genus recombination event in the LSU genes of Natrinema sp. strain XA3-1 established that at least part of the intragenomic rRNA heterogeneity found in some Halobacteriales is due to the latter process.
Could the intragenomic variability of rRNA operons be maintained by evolutionary pressure? Such a link between functionality and intragenomic rRNA divergence has been observed in the apicomplexan Plasmodium berghei. The two types of SSU genes (which differ at 5.0% of their nucleotide positions) are preferentially expressed in different stages of the life cycle of this eukaryotic parasite (13). It has been suggested that in extremely halophilic archaea, a selective advantage could be gained from differential expression of divergent rRNA operons depending on the salt concentration in the environment (10). Indeed, salinity has a significant influence on most biochemical reactions and is very variable in the environments occupied by halophiles, which are subjected to constant fluctuations caused by solubilization-precipitation and dilution-evaporation (9). There is even some experimental evidence of salinity dependence for rRNA expression. Indeed, the promoters used for expression of the unique rRNA operon of Halobacterium cutirubrum vary according to the salt concentration in the medium in which the organism is grown (8). Both rRNA operons of H. marismortui have been shown to be expressed under standard laboratory growth conditions (1), but differential expression under variable growth conditions has yet to be demonstrated.
Intragenomic heterogeneity, as discussed above, causes problems at the level of rRNA data analysis (6). More importantly, it can also cause artifacts at the data acquisition level. Indeed, PCR amplification is known to be susceptible to the formation of chimeric products if the template DNA contains multiple divergent copies of the target gene (25). A certain rate of chimera formation is known to occur when the SSU gene is amplified directly from environmental DNA samples, which usually contain a great diversity of the target gene (31). Intragenomic heterogeneity means that this type of artifact can also occur for PCR amplification of rRNA genes when DNA extracted from a pure culture of an organism is used. Although PCR conditions can be modified to reduce the risk of chimera formation, this problem can never be completely eliminated. The vast majority of the SSU gene sequences of Halobacteriales available in the database have been amplified by PCR without knowledge of the possibility of intragenomic heterogeneity occurring in lineages other than the genus Haloarcula. Several of the sequences found in the database could therefore be chimeric, and thus caution should be used when they are used for phylogenetic analysis (14). Furthermore, rRNA recombination is certainly not a process limited to a specific phylogenetic group. It has been observed in several bacterial lineages at the species or subspecies level (3, 21, 22, 27). The highly evolutionarily conserved stretches of DNA found in rRNA genes, often considered an advantage for tracing the phylogeny of organisms over large evolutionary distances, could also facilitate homologous recombination between divergent organisms (34).
The presence of intragenomic heterogeneity, which is at least partially caused by homologous recombination of rRNA genes, can have a significant impact on the acquisition of true (nonchimeric) gene sequences and their use in identification and classification of organisms through phylogenetic analysis. Therefore, efforts to develop non-PCR methods of acquiring rRNA genes and evaluating the frequency of intragenomic heterogeneity and homologous recombination among prokaryotes are needed to ensure that these genes are reliable molecular markers for use in microbiological disciplines.
Acknowledgments
We are grateful to P. Zhou for providing Natrinema sp. strain XA3-1.
REFERENCES
- 1.Amann, G., K. O. Stetter, E. Llobet-Brossa, R. Amann, and J. Anton. 2000. Direct proof for the presence and expression of two 5% different 16S rRNA genes in individual cells of Haloarcula marismortui. Extremophiles 4:373-376. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231-236. [DOI] [PubMed] [Google Scholar]
- 3.Anton, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1999. Intraspecific diversity of the 23S rRNA gene and the spacer region downstream in Escherichia coli. J. Bacteriol. 181:2703-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carranza, S., G. Giribet, C. Ribera, Baguna, and M. Riutort. 1996. Evidence that two types of 18S rDNA coexist in the genome of Dugesia (Schmidtea) mediterranea (Platyhelminthes, Turbellaria, Tricladida). Mol. Biol. Evol. 13:824-832. [DOI] [PubMed] [Google Scholar]
- 5.Cilia, V., B. Lafay, and R. Christen. 1996. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol. Biol. Evol. 13:451-461. [DOI] [PubMed] [Google Scholar]
- 6.Clayton, R. A., G. Sutton, P. S. Hinkle, Jr., C. Bult, and C. Fields. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. [DOI] [PubMed] [Google Scholar]
- 7.Dahllof, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis, P. P. 1999. Expression of ribosomal RNA operons in halophilic archaea, p. 319-329. In A. Oren (ed.), Microbiology and biogeochemistry of hypersaline environments. CRC Press, New York, N.Y.
- 9.Dennis, P. P., and L. C. Shimmin. 1997. Evolutionary divergence and salinity-mediated selection in halophilic archaea. Microbiol. Mol. Biol. Rev. 61:90-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis, P. P., S. Ziesche, and S. Mylvaganam. 1998. Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 180:4804-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Martinez, J., S. G. Acinas, A. I. Anton, and F. Rodriguez-Valera. 1999. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J. Microbiol. Methods 36:55-64. [DOI] [PubMed] [Google Scholar]
- 12.Gemmell, R. T., T. J. McGenity, and W. D. Grant. 1998. Use of molecular techniques to investigate possible long-term dormancy of halobacteria in ancient halite deposits. Ancient Biomol. 2:125-133. [Google Scholar]
- 13.Gunderson, J. H., M. L. Sogin, G. Wollett, M. Hollingdale, V. F. de la Cruz, A. P. Waters, and T. F. McCutchan. 1987. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science 238:933-937. [DOI] [PubMed] [Google Scholar]
- 14.Hugenholtzt, P., and T. Huber. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst Evol. Microbiol. 53:289-293. [DOI] [PubMed] [Google Scholar]
- 15.Ihara, K., T. Umemura, I. Katagiri, T. Kitajima-Ihara, Y. Sugiyama, Y. Kimura, and Y. Mukohata. 1999. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J. Mol. Biol. 285:163-174. [DOI] [PubMed] [Google Scholar]
- 16.Lodwick, D., H. N. M. Ross, J. A. Walker, J. W. Almond, and W. D. Grant. 1991. Nucleotide sequence of the 16S ribosomal RNA gene from the haloalkaliphilic archaeon (archaebacterium) Natronobacterium magadii, and the phylogeny of the halobacteria. Syst. Appl. Microbiol. 14:352-357. [Google Scholar]
- 17.Ludwig, W., and K. H. Schleifer. 1999. Phylogeny of Bacteria beyond the 16S rRNA standard. ASM News 65:752-757. [Google Scholar]
- 18.Mylvaganam, S., and P. P. Dennis. 1992. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ninet, B., M. Monod, S. Emler, J. Pawlowski, C. Metral, P. Rohner, R. Auckenthaler, and B. Hirschel. 1996. Two different 16S rRNA genes in a mycobacterial strain. J. Clin. Microbiol. 34:2531-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker, M. A. 2001. Case of localized recombination in 23S rRNA genes from divergent Bradyrhizobium lineages associated with neotropical legumes. Appl. Environ. Microbiol. 67:2076-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira, M. M., M. Santana, and M. Teixeira. 2001. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta 1505:185-208. [DOI] [PubMed] [Google Scholar]
- 23.Perez Luz, S., F. Rodriguez-Valera, R. Lan, and P. R. Reeves. 1998. Variation of the ribosomal operon 16S-23S gene spacer region in representatives of Salmonella enterica subspecies. J. Bacteriol. 180:2144-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 25.Qiu, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholzen, T., and E. Arndt. 1992. The alpha-operon equivalent genome region in the extreme halophilic archaebacterium Haloarcula (Halobacterium) marismortui. J. Biol. Chem. 267:12123-12130. [PubMed] [Google Scholar]
- 27.Smith, N. H., E. C. Holmes, G. M. Donovan, G. A. Carpenter, and B. G. Spratt. 1999. Networks and groups within the genus Neisseria: analysis of argF, recA, rho, and 16S rRNA sequences from human Neisseria species. Mol. Biol. Evol. 16:773-783. [DOI] [PubMed] [Google Scholar]
- 28.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4.0th ed. Sinauer Associates, Sunderland, Mass.
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tindall, B. J. 2003. Taxonomic problems arising in the genera Haloterrigena and Natrinema. Int. J. Syst. E vol. Microbiol. 53:1697-1698. [DOI] [PubMed] [Google Scholar]
- 31.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 32.Vreeland, R. H., S. Straight, J. Krammes, K. Dougherty, W. D. Rosenzweig, and M. Kamekura.2002. Halosimplex carlsbadense gen. nov., sp. nov., a unique halophilic archaeon, with three 16S rRNA genes, that grows only in defined medium with glycerol and acetate or pyruvate. Extremophiles 6:445-452. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. A. Seidman, K. Struhl, and J. A. Smith (ed.), Current protocols in molecular biology, vol. 1. John Wiley and Sons, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 34.Woese, C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 97:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]