Abstract
In Pseudomonas aeruginosa, type IV pili (TFP)-dependent twitching motility is required for development of surface-attached biofilm (SABF), yet excessive twitching motility is detrimental once SABF is established. In this study, we show that mucin significantly enhanced twitching motility and decreased SABF formation in PAO1 and other P. aeruginosa strains in a concentration-dependent manner. Mucin also disrupted partially-established SABF. Our analyses revealed that mucin increased the amount of surface pilin and enhanced transcription of the pilin structural gene, pilA. Mucin failed to enhance twitching motility in P. aeruginosa mutants defective in genes within the pilin biogenesis operons pilGHI/pilJK-chpA-E. Furthermore, mucin did not enhance twitching motility and reduce biofilm development by chelating iron. We also examined the role of the virulence factor regulator Vfr in the effect of mucin. In the presence or absence of mucin, PAOΔvfr produced a significantly reduced SABF. However, mucin partially complemented the twitching motility defect of PAOΔvfr. These results suggest that mucin interferes with SABF formation at specific concentrations by enhancing TFP synthesis and twitching motility, that this effect, which is iron-independent, requires functional Vfr, and only part of the Vfr-dependent effect of mucin on SABF development occurs through twitching motility.
Keywords: biofilm, mucin, pilin, Pseudomonas aeruginosa, twitching motility
INTRODUCTION
Cystic fibrosis (CF), an inherited disorder that affects approximately 30,000 Americans of primarily Northern European descent, is caused by a mutation in the gene that encodes the cystic fibrosis transmembrane conductance regulator (Gibson et al., 2003; Rommens et al., 1989). Mutations in this protein cause an insufficiency in chloride secretion with resultant accumulation of thick, stagnant mucus within the lung alveoli of patients (Baltch, 1994; Burns et al., 1993; Gibson et al., 2003; Jiang et al., 1993; Rommens et al., 1989). Nutrients in the thick mucus facilitate the colonization of various bacterial pathogens, including Pseudomonas aeruginosa, Haemophilus influenzae, and Staphylococcus aureus (Baltch, 1994; Gibson et al., 2003; Hassett et al., 2002). Colonization by these pathogens leads to decimation of the lung tissue and, eventually, death from respiratory failure (Burns et al., 1993; Gibson et al., 2003; Pier and Ramphal, 2010). P. aeruginosa is one of the significant pathogens involved in the chronic lung infections of CF patients (Gibson et al., 2003; Lyczak et al., 2002).
Among the different factors that contribute to the virulence of P. aeruginosa is its ability to form a biofilm. Biofilms are defined as microbial communities in which bacteria are attached to a substratum or to each other (Donlan and Costerton, 2002). Within a biofilm, bacteria are surrounded by an extracellular polysaccharide matrix which protects them from the effects of the host immune system and a wide variety of antibiotics (Leid et al., 2005; Matsukawa and Greenberg, 2004; Stewart and Costerton, 2001). A number of P. aeruginosa infections are associated with biofilm formation, including chronic wound infections, otitis media, endocarditis, and chronic lung infections in CF patients (Costerton et al., 1999; Donlan and Costerton, 2002). Specific bacterial factors are required at each stage of biofilm development. For instance, during the initial (attachment) stage of biofilm formation, bacteria depend on both the flagellummediated swimming motility and the pilimediated twitching motility (Sauer et al., 2002). However, twitching motility is detrimental to an established biofilm (Hammond et al., 2010).
We previously demonstrated that in the artificial sputum medium ASM+, which resembles the thick mucus that accumulates in the CF lung, P. aeruginosa does not form a biofilm on a surface, but produces biofilm-like structures (BLS) (Haley et al., 2012). The concentration of mucin, a major component of secreted mucus, varies depending on the pathophysiologic condition within the lungs of the CF patient (Henke et al., 2007). In this study, we examined the role of mucin in biofilm development. Our results showed that mucin interferes with biofilm development by significantly enhancing twitching motility. This effect, which is iron-independent, requires functional Vfr.
Materials and methods
Bacterial strains, plasmids, and media
Bacterial strains utilized in this study are described in Table 1. All strains were routinely grown overnight in Luria Bertani (LB) broth under shaking conditions at 37 °C prior to inoculation into different assays. To examine the effect of iron, strains were grown in chelated trypticase soy broth dialysate (TSB-DC) with and without FeCl3 20 μg/mL (Ohman et al., 1980). Type 3 porcine mucin (Sigma-Aldrich, St. Louis, Missouri, USA) was added to LB broth or agar at a concentration of 5 mg/mL (LBM broth or agar or TSB-DC). The ASM+ medium was prepared and utilized as previously described (Haley et al., 2012). To allow visualization of the bacteria, we utilized P. aeruginosa PAO1 strains containing pMRP9-1 from which the gene for green fluorescent protein (GFP) is constitutively expressed (Davies et al., 1998). LB and LBM broth and agar were supplemented with antibiotics where required to maintain plasmids or transposons: carbenicillin 300 μg/mL, tetracycline 60 μg/mL, and gentamicin 60 μg/mL for PAO1 strains and 15 μg/mL for PA14 strains.
Table 1. P. aeruginosa strains and plasmid used in this study.
| Description | Source | |
|---|---|---|
| Strains | ||
| PAO1 | Prototroph; human isolate | (Holloway et al., 1979) |
| PA103 | Human isolate | (Liu, 1966) |
| PAK | Prototroph; human isolate | (Strom and Lory, 1986) |
| PA14 | UCBPP-PA14; burn wound isolate | (Rahme et al., 1995) |
| PAOΔpilA |
pilA inactivated by allelic displacement; tagged with eGFP, in a mini-Tn7 construct; Gmr |
(Klausen et al., 2003) |
| PA14ΔfimV* | PA14 with TnMAR2xT7 insertion in fimV; Gmr | (Liberati et al., 2006) |
| PA14ΔpilG | PA14 with TnMAR2xT7 insertion in pi/G; Gmr | (Liberati et al., 2006) |
| PA14ΔpilH | PA14 with TnMAR2xT7 insertion in pi/H; Gmr | (Liberati et al., 2006) |
| PA14ΔpilI | PA14 with TnMAR2xT7 insertion in pi/I; Gmr | (Liberati et al., 2006) |
| PA14ΔpilJ | PA14 with TnMAR2xT7 insertion in pi/J; Gmr | (Liberati et al., 2006) |
| PA14ΔpilK | PA14 with TnMAR2xT7 insertion in pi/K; Gmr | (Liberati et al., 2006) |
| PA14ΔchpA | PA14 with TnMAR2xT7 insertion in chpA; Gmr | (Liberati et al., 2006) |
| PA14ΔpilR | PA14 with TnMAR2xT7 insertion in pi/R; Gmr | (Liberati et al., 2006) |
| PA14ΔrpoN | PA14 with TnMAR2xT7 insertion in rpoN; Gmr | (Liberati et al., 2006) |
| PW1762† | chpC-F07::IS/acZ/hah at bp 79 or 507 in the ORF; Tcr | (Jacobs et al., 2003) |
| Plasmids | ||
| pMRP9-1 | pUCP18 carrying a gene encoding enhanced green fluorescent protein (GFP); Cbr |
(Davies et al., 1998) |
| pUCP19 | E. coli-P. aeruginosa shuttle vector; Cbr | (Schweizer, 1991) |
| pKF906 | pUCP19 carrying intact vfr; Cbr | (West et al., 1994) |
| pMP220::PpvdS | pMP220 carrying a pvdS promoter transcriptional lacZ fusion, Tcr | (Ambrosi et al., 2002) |
PA14 mutants were purchased from the PA14 Transposon Insertion Mutant Library of non-redundant Pseudomonas aeruginosa transposon insertion mutants (http://ausubellab.mgh.harvard.edu/cgi-bin/pa14/home.cgi [accessed 22 January 2014])
PAO1 transposon mutant purchased from the University of Washington Genome Center (http://www.gs.washington.edu/labs/manoil/libraryindex.htm [accessed 22 January 2014])
resistant; Cb, carbenicillin; Gm, gentamicin; Tc, tetracycline
Swimming and twitching motility
The swimming motility assay was done as previously described (Deziel et al., 2001). Swimming plates (0.3% agar [w/v] with or without mucin 5 mg/mL) were incubated for 16 h at 32 °C. The twitching motility assay was also done as previously described with slight modifications (Deziel et al., 2001). Briefly, bacteria were stab-inoculated to the bottom of twitching plates (1 % agar [w/v] with or without mucin 5 mg/mL). The plates were incubated at 37 °C for 48 h and then at room temperature for an additional 48 h. The agar was removed from the plate and the twitching zone was visualized by staining with 1.0 % crystal violet (w/v) (Deziel et al., 2001). All experiments were performed in triplicate and diameters of the twitching zones were measured and reported in mm.
Pilin preparation
Bacteria from an overnight culture (150 μL) were spread on six freshly prepared 1.8 % LB agar plates and six LBM agar plates and grown overnight at 37 °C. Bacteria were then harvested and prepared as previously described with slight modifications (Giltner et al., 2011). Briefly, bacteria were scraped from the agar surface with a sterile glass rod and resuspended in 4.5 mL of 1X PBS, pH 7.4. Cells from different experimental groups were adjusted with 1X PBS to the same optical density at 600 nm (OD600). Pilin was removed from the cells by vortexing and recovered by centrifugation at 11,688 × g and precipitation in 5M NaCl and 30% polyethylene glycol (MW 8000) (w/v). Protein pellets were resuspended in 100 μL of protein loading buffer (60 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [w/v], 10% glycerol [v/v], 5% - mercaptoethanol [v/v], 0.001% bromophenol blue [w/v]) and the proteins were separated by 15% SDS-PAGE. Pilin proteins were visualized by silver stain.
qRT-PCR analysis of pilin genes
Overnight cultures of PAO1 were spread on four LB agar plates and four LBM agar plates and incubated at 37 °C for 16 h. Cells were harvested from the surface of the plates with a sterile glass rod and resuspended in 2 mL of 1X PBS. The cell suspension was mixed with twice its volume of RNAprotect Bacteria Reagent (QIAGEN, Valencia, California, USA) for 5 min at room temperature. The cells were pelleted and stored at −80 °C. Bacterial pellets were first lysed with lysozyme and proteinase K for 15 min at room temperature. Total RNA was extracted using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s recommendations. The RNA was digested with the RNase-free DNase Set (QIAGEN). RNA was purified from DNase by the RNA cleanup protocol (QIAGEN) with the exception that on-column DNase digestion was applied to eliminate any remaining traces of genomic DNA. Purified RNA was quantified by NanoDrop® spectrophotometer (NanoDrop Products, Wilmington, Delaware, USA) and utilized as a template to synthesize cDNA using the QuantiTect Reverse Transcription Kit (QIAGEN). A 200-ng aliquot of cDNA was mixed with SYBR Green PCR Master Mix (Life Technologies, Carlsbad, California, USA) and 250 nM of specific primers for pilA (forward 5′-AACCTGAACCTGGACTGTGG-3′; reverse 5′-TTGCCTTCGCCATCTTTT-3′). Amplification and detection of the product was conducted using StepOne Plus real-time PCR system (Life Technologies). Three independent biological replicates were used for each experiment. The quantity of cDNA in different samples was normalized using 30S ribosomal RNA (rplS) as an internal standard. Gene expression analysis was performed using StepOne Plus software version 2.2.2 (Life Technologies).
Microtiter plate culture system for development of the biofilms
Biofilms were developed utilizing the microtiter plate culture system as previously described (Hammond et al., 2010). Briefly, plastic cell-culture treated coverslips (Sarstedt, Newton, North Carolina, USA), 13 mm diameter × 0.2 mm thick, were placed into wells of sterile 24-well polystyrene plates (Falcon; Becton Dickinson, Franklin Lakes, New Jersey, USA). Overnight cultures of P. aeruginosa strains were inoculated into wells containing LB or LBM broth to an initial OD600 of 0.02-0.03. Uninoculated wells containing LB or LBM broth were used as sterility controls. The plates were covered and incubated at 37 °C for various time points with slight shaking on a plate shaker (Lab-line Instruments, Melrose Park, Illinois, USA). For analysis of biofilm disruption, PAO1/pMRP9-1 was grown in three sets of microtiter wells containing only LB broth to allow initiation of biofilm formation. One set of wells was examined at 8 h to confirm biofilm initiation. Mucin was added to one set of wells to a final concentration 5 mg/mL from a 0.0625 g/mL stock solution, and the remaining two sets of wells were incubated for an additional 24 h (32 h total).
Crystal violet assay
The assay was conducted as previously described with minor modifications (Hammond et al., 2010). Coverslips were removed from the wells of the microtiter plate, rinsed gently with distilled H2O, and placed into the corresponding wells of a new plate containing 1 mL 1.0 % crystal violet solution. After incubation at room temperature for 30 min, the crystal violet solution was discarded. The coverslips were rinsed, and placed in corresponding wells of a new plate containing 1 mL of 95 % ethanol. The plates were incubated at room temperature for 1 h and the amount of extracted crystal violet, which is proportional to the biofilm biomass, was measured at an absorbance of 595 nm (A595).
Visualization of the biofilms
Plastic coverslips were removed from wells, gently rinsed in 1X PBS, and placed in a Petri plate for visualization by confocal laser scanning microscopy (CLSM) using an IX71 Fluoview 300 confocal laser scanning microscope (Olympus America, Melville, New York, USA). All images were obtained through a 203/0.40 Ph1 NA objective utilizing an argon laser (510-530 nm). Three-dimensional (3D) image reconstructions were performed using NIS-Elements 2.2 (Nikon Instruments, Melville, New York, USA) to visualize the architecture of biofilms. All instrument settings were consistent for each set of experimental conditions tested. Images of the BLS formed by PAO1 and PAOΔvfr strains were acquired on a Nikon Ti-E microscope with A1 confocal and STORM super resolution, and 3D reconstructions were done as above.
Quantitative structural analysis of the biofilms
The number of images obtained in a single stack was based on the greatest depth of the structures formed under the test conditions and was the same for all strains/conditions within an experiment. Each experiment was done in duplicate. Two 10-image stacks were obtained from random positions within each biofilm for a total of four 10-image stacks analyzed for each strain and/or condition. The image stacks were analyzed using the COMSTAT program (Heydorn et al., 2000) for structural features of the biofilm: biovolume, estimates the biomass of the biofilm; mean thickness, a measure of spatial size of the biofilm; roughness coefficient, a measure of how much the thickness of the biofilm varies, or the heterogeneity of the biofilm; total surface area, space occupied in each image stack; and surface to biovolume ratio, estimates the portion of the biofilm exposed to nutrients (biovolume divided by the surface area of the substratum).
Beta-galactosidase assays
Beta-galactosidase assays were performed as previously reported (Gaines et al., 2005; Miller, 1972; Stachel et al., 1985). To determine the level of β-galactosidase activity produced by the chpC::lacZ fusion strain within the BLS, the strain was grown in LBM broth as described above. The BLS were collected, resuspended in LB broth, and gently vortexed to disperse the BLS. The calculation of relative units of β-galactosidase activity accounts for the level of growth.
Statistical analyses
Statistical analyses of the results were done using GraphPad InStat 3.06 (GraphPad Software, San Diego, California, USA). One-way ANOVA with the Tukey-Kramer multiple comparisons post-test was used to determine significant differences among groups of strains or treatments. The two-tailed t-test was used to compare two strains or two treatments.
Results
Mucin prevents surface-attached biofilm formation by P. aeruginosa and enhances its twitching motility
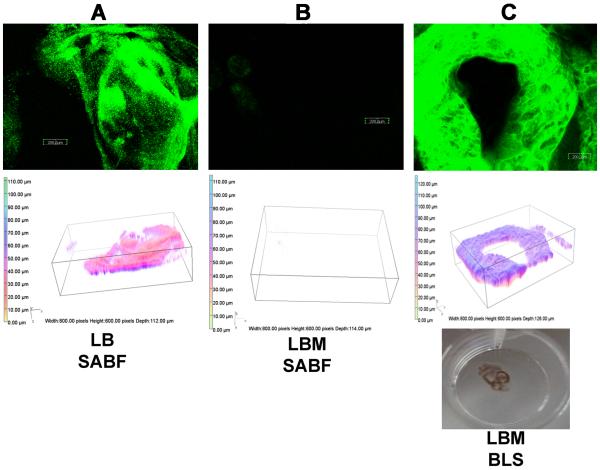
We previously showed that upon their growth in ASM+, different P. aeruginosa strains failed to develop surface-attached biofilms (SABF), instead forming free-floating BLS (Haley et al., 2012). Since a main constituent of ASM+ is mucin, we examined whether mucin itself inhibits biofilm formation and promotes the production of BLS. P. aeruginosa strain PAO1/pMRP9-1, which expresses GFP to allow visualization, was grown in LB or LBM broth in the modified microtiter plate culture system (Haley et al., 2012). The concentration of mucin used in LBM broth (5 mg/mL) is the same as that in the ASM+ we used previously (Haley et al., 2012; Sriramulu et al., 2005). PAO1 formed a mature and well-developed SABF in LB broth (Fig. 1A). However, similar to our observation in ASM+, PAO1 failed to develop SABF when grown in LBM broth (Fig. 1B). Instead, it produced a well-developed BLS (Fig. 1C).
Fig. 1.
P. aeruginosa PAO1 forms biofilm-like structures (BLS) rather than surface attached biofilm (SABF) in the presence of mucin. PAO1/pMRP9-1, which expresses GFP, was grown aerobically on plastic coverslips in a 24-well microtiter plate in either LB or LBM (5 mg/mL) broth for 24 h at 37 °C under aerobic conditions. CLSM and 3D image analysis of (A) PAO1 SABF in LB broth, (B) PAO1 SABF in LBM broth, (C) PAO1 BLS in LBM broth including photograph of the free-floating BLS within the microtiter plate well (bottom); magnification, 10×; bars, 200.00 nm. Images shown are representative of three independent experiments.
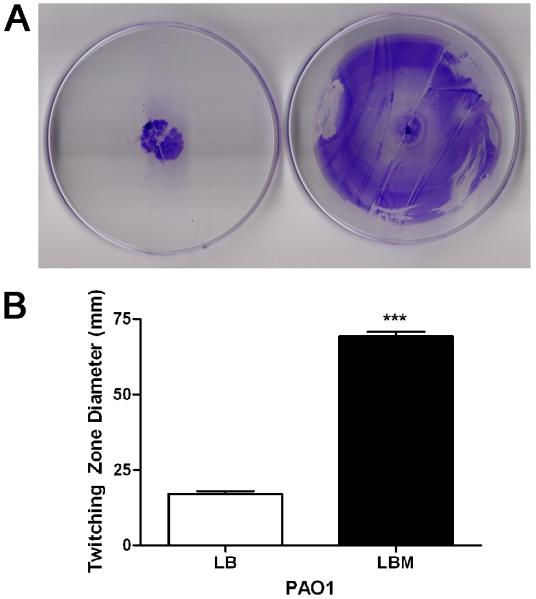
The flagellar-mediated swimming motility and the pilin-mediated twitching motility are essential in the initial establishment of P. aeruginosa biofilms. Mutants defective in either motility form architecturally dissimilar biofilms when compared to wild-type strains (Klausen et al., 2003). However, once the biofilm is established the swimming and/or twitching motility is detrimental for the maintenance of biofilms. The presence of factors that stimulate either motility would prevent the development of a P. aeruginosa biofilm and reduce an already established one. We previously showed that serum, which significantly enhances twitching motility, inhibited biofilm development by PAO1 and significantly reduced established biofilms (Hammond et al., 2010). Therefore, similarly to serum, the presence of mucin in LB agar may enhance PAO1 swimming and/or twitching motility and prevent the maintenance of a stable SABF. We examined the effect of mucin on the swimming and twitching motility of PAO1 using previously described assays (Deziel et al., 2001). Mucin was added to standard swimming or twitching plates and the motilities were examined in the presence and absence of mucin. We detected no significant alteration in the swimming motility (data not shown). However, mucin enhanced PAO1 twitching motility significantly (Fig. 2A-2B).
Fig. 2.
Mucin enhances PAO1 twitching motility. (A) PAO1 was stab-inoculated onto either LB (left) or LBM (right) twitching plates (1 % agar). The plates were incubated at 37 °C for 48 h then at 23°C for an additional 48 h. The agar was removed and the twitching zone revealed by staining with crystal violet. Images shown are representative of three independent experiments. (B) Graphic comparison of twitching zone diameters for PAO1 in LB and LBM broths; values represent the means of 3 independent experiments ± SEM; ***P<0.001.
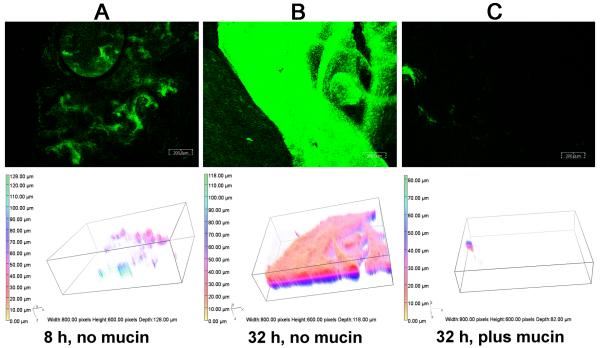
Since mucin enhanced PAO1 twitching motility and prevented the establishment of a mature SABF, the addition of mucin to a partially-developed biofilm should enhance twitching motility leading to disruption of the SABF. PAO1 was grown in three sets of microtiter plate wells without mucin for 8 h to allow initiation of biofilm formation. At 8 h, one set of wells was examined for SABF development. Mucin (5 mg/mL) was added to one of the two remaining sets of wells and incubation was continued for 24 h (32 h total). At 8 h, PAO1 had formed a partially-developed biofilm that covered most of the surface area (Fig. 3A). By 32 h, PAO1 incubated without the addition of mucin produced a dense biofilm with characteristic features of high biomass, thickness and surface area (Fig. 3B; Table 2). In contrast, the addition of mucin resulted in significant reduction of the partially-developed SABF with decreases in biomass, thickness, and surface area covered and increases in roughness and surface to volume ratio (Fig. 3C; Tables 2 and 3). These results strongly support our hypothesis that mucin prevents the establishment of P. aeruginosa SABF by enhancing twitching motility.
Fig. 3.
Mucin disrupts partially established PAO1 biofilms. PAO1/pMRP9-1 was grown aerobically at 37 °C on plastic coverslips in a 24-well microtiter plate in LB broth for 8 h. Mucin was added (5 mg/mL) to one-third of the samples and the growth was continued for an additional 24 h. CLSM micrographs and 3D image analysis of the SABF that were developed for (A) 8 h without mucin, (B) 32 h without mucin, or (C) 32 h with mucin (24 h post addition); magnification, 10×; bars, 200.00 nm. Images shown are representative of three independent experiments.
Table 2. Effect of mucin on P. aeruginosa biofilm development.
| Variable | Images per stack (#)* |
Total biovolume (μm3/μm2)† |
Mean thickness (μm)‡ |
Roughness coefficient§ |
Total surface area × 107 (μm2)|| |
Surface to volume ratio (μm2μm3)¶ |
|---|---|---|---|---|---|---|
| Reduction of SABF | ||||||
| 8 h SABF | 10 | 1.38 ± 0.40 | 2.18 ± 0.49 | 1.34 ± 0.12 | 0.36 ± 0.08 | 1.36 ± 0.13 |
| no mucin# | 10 | 7.90 ± 0.29 | 8.42 ± 0.09 | 0.11 ± 0.01 | 0.80 ± 0.07 | 0.53 ± 0.06 |
| + mucin# | 10 | 0.25 ± 0.06 | 0.35± 0.09 | 1.88 ± 0.03 | 0.10 ± 0.02 | 2.06 ± 0.25 |
| Mucin titration (mg/mL) | ||||||
| 0 | 10 | 5.63 ± 0.30 | 6.12 ± 0.41 | 0.55 ± 0.07 | 0.68 ± 0.03 | 0.63 ± 0.02 |
| 10 | 10 | 0.02 ± 0.00 | 0.00 ± 0.00 | 1.20 ± 0.00 | 0.01 ± 0.00 | 2.89 ± 0.02 |
| 5 | 10 | 0.29 ± 0.06 | 0.40 ± 0.13 | 1.87 ± 0.03 | 0.10 ± 0.18 | 1.88 ± 0.08 |
| 2.5 | 10 | 0.71 ± 0.07 | 0.98 ± 0.09 | 1.58 ± 0.09 | 0.24 ± 0.40 | 1.71 ± 0.21 |
| 1.25 | 10 | 3.84 ± 0.13 | 4.68 ± 0.39 | 1.38 ± 0.05 | 0.45 ± 0.08 | 0.60 ± 0.11 |
| 0.625 | 10 | 4.44 ±0 .57 | 5.43 ± 0.52 | 1.30 ± 0.06 | 0.55 ± 0.10 | 0.69 ± 0.18 |
PAO1/pMRP9-1 was used in all experiments described in the table. Each experiment was done in duplicate. Two 10-image stacks were obtained from random positions within each SABF. Image stacks were analyzed using the COMSTAT program (Heydorn et al., 2000). Values represent the mean ± SEM.
Estimates the biomass of the SABF
Measures spatial size of the SABF
Assessment of the variation in the thickness of the SABF
Total of the area occupied in each image stack
Estimates the portion of the biofilm exposed to nutrients; biovolume divided by the surface area of the substratum
Three sets of 2 coverslips each were inoculated with PAO1/pMRP9-1 and incubated for 8 h to allow initiation and partial development of SABF. At 8 h, one set of coverslips was harvested for analysis, mucin (5 mg/mL) was added to one set, no mucin to the other and incubation continued for 24 h.
Table 3. Significance of differences in values presented in Table 2.
| Variable | Total biovolume (μm3/μm2)* |
Mean thickness (μm)* |
Roughness coefficient* |
Total surface area × 107 (μm2)* |
Surface to volume ratio (μm2/μm3)* |
|---|---|---|---|---|---|
| Reduction of SABF | |||||
| No mucin vs. 8 h | Increase† | Increase | Decrease | Increase | Decrease |
| 0.0302 | 0.0103 | 0.0045 | 0.0293 | 0.0292 | |
| Mucin vs. 8 h | Decrease | Decrease | Increase | Decrease | Increase |
| <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| Mucin titration (mg/mL) | |||||
| 10 vs. 0 | Decrease | Decrease | Increase | Decrease | Increase |
| <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| 5 vs. 0 | Decrease | Decrease | Increase | Decrease | Increase |
| <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| 2.5 vs. 0 | Decrease | Decrease | Increase | Decrease | Increase |
| <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| 1.25 vs. 0 | Decrease | Decrease | Increase | Decrease | No change |
| <0.0001 | <0.0001 | <0.0001 | <0.0001 | NS‡ | |
| 0.625 vs. 0 | Decrease | Decrease | Increase | Decrease | No change |
| NS | NS | <0.0001 | NS | NS | |
See Table 2 for description of parameters
Significant change with P value indicated
NS, no significant difference
The effect of mucin on SABF formation and twitching motility is concentration-dependent
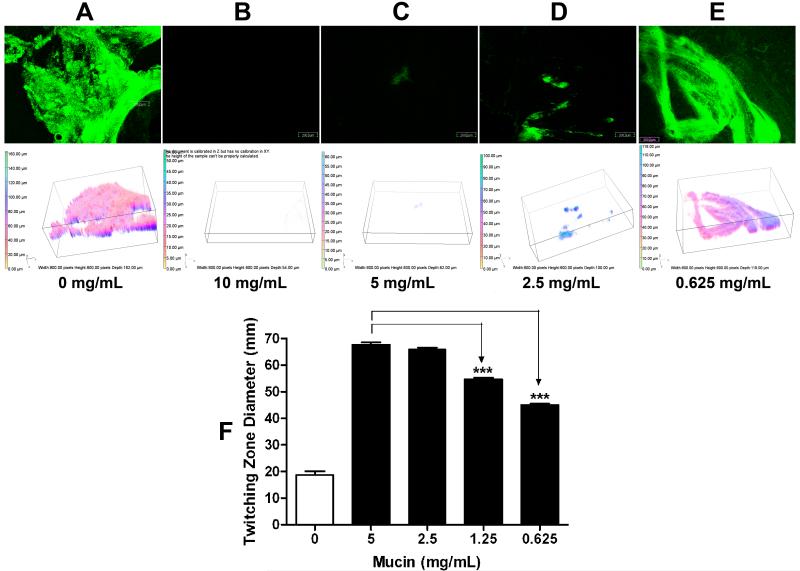
We previously showed that variations in the amount of mucin in ASM+ altered the development of BLS (Haley et al., 2012). When the concentration of mucin was raised or lowered (2.5 or 10 mg/mL), the formation of BLS was significantly reduced (Haley et al., 2012). Therefore, at certain concentrations (2.5 or 10 mg/mL), mucin may favor the formation of SABF at the expense of BLS. To examine this possibility, we analyzed biofilm development by PAO1 in the presence of different concentrations of mucin. Biofilms were developed for 24 h and examined by CLSM. In the mucin-free medium, PAO1 produced a robust SABF (Fig. 4A). At mucin concentrations of 2.5, 5 and 10 mg/mL, PAO1 produced variable BLS (Haley et al., 2012). At these same concentrations, PAO1 still failed to develop a SABF (Fig. 4B-4D). The biomass, mean thickness and total surface area of these biofilms were significantly reduced when compared with those of a PAO1 biofilm developed in mucin-free medium (Tables 2 and 3). Interestingly however, when we decreased the amount of mucin farther to 0.625 mg/mL, PAO1 developed SABF that closely resemble those produced in mucin-free medium (Fig. 4E; Tables 2 & 3). These results suggest that mucin concentration plays a critical role in the development of PAO1 biofilm, with lower mucin concentrations (less than 2.5 mg/mL) favoring development of SABF.
Fig. 4.
Mucin inhibition of biofilm formation by PAO1 is concentration dependent. PAO1/pMRP9-1 was grown aerobically on plastic coverslips in a 24-well microtiter plate in either LB or LBM broth containing variable concentrations of mucin for 24 h at 37 °C under aerobic conditions. CLSM micrographs and 3D image analysis of the PAO1 SABF grown (A) without mucin, and with concentrations of mucin indicated: (B) 10 mg/mL, (C) 5 mg/mL, (D) 2.5 mg mucin/mL, and (E) 0.625 mg/mL; magnification, 10×; bars, 200.00 nm. Images shown are representative of three independent experiments. (F) Twitching motility of PAO1 in different concentrations of mucin. PAO1 was stab-inoculated onto either LB or LBM (5 mg mucin/mL) twitching plates (1 % agar) containing variable concentrations of mucin. The plates were incubated at 37 °C for 48 h then at 23°C for an additional 48 h. The agar was removed and the twitching zones revealed by staining with crystal violet. Values represent the means of 3 independent experiments ± SEM. Twitching motility in decreasing concentrations of mucin was compared to that in 5 mg/ml mucin by one-way ANOVA; ***P<0.001.
If mucin inhibits the development of PAO1 biofilm by enhancing the twitching motility, we expected that a decrease in mucin concentration would also limit twitching motility allowing the development of SABF. To test this possibility, we examined the effect of 2.5, 1.25 and 0.625 mg/mL mucin on PAO1 twitching motility. Compared with the standard 5 mg/mL concentration, 2.5 mg/mL produced a similar enhancement in PAO1 twitching motility (Fig. 4F). However, further decrease in the concentration of mucin to 1.25 or 0.625 mg/mL led to significant reductions in the enhancement of PAO1 twitching motility compared with that observed at 5 mg/mL (Fig. 4F). Thus, the effect of mucin on twitching motility is concentration dependent. At 0.625 mg/mL, mucin still enhanced twitching motility but did not inhibit SABF development (Fig. 4E-4F). This strongly suggests that only part of the mucin effect on SABF development occurs through the twitching motility.
Mucin enhances pilin production and pilA expression
Twitching motility in P. aeruginosa occurs through the coordinated extension, retraction and tethering of the surface filamentous appendages known as type IV pili (TFP). TFP consist of the 15 kDa major subunit protein, pilin, which is encoded by the pilA gene (Paranchych et al., 1985). Several additional P. aeruginosa proteins assemble the synthesized pilin on the outer surface of P. aeruginosa. Therefore, mucin may enhance PAO1 twitching motility by increasing TFP surface pilin. We compared the level of sheared surface pilin produced by PAO1 cells that were grown on LB or LBM agar. As a control, we used the pilin deficient mutant PAOΔpilA (Table 1). As shown in Fig. 5A, PAO1 grown on LBM agar produced a higher amount of sheared surface pilin than PAO1 grown on LB agar, while PAOΔpilA produced no detectable pilin. To determine if the increase is due to the efficient processing, rather than synthesis, of pilin protein, we tried to compare the intracellular pilin in PAO1 grown in LB and LBM by immunoblotting experiments using pilin polyclonal antibody. However, the antibody cross reacted with other cellular proteins (data not shown). We then analyzed the level of pilA transcription in PAO1 grown on either LB or LBM agar using qRT-PCR. In PAO1 grown on LBM agar, the level ofpilA transcription was higher (1.2 fold) than that of PAO1 grown on LB agar (Fig. 5B). For the analysis of pilin protein and pilA transcripts, the cultures were adjusted to a comparable OD600. Thus, the less dramatic increase in surface pilin and pilA expression suggests that multiple mechanisms contribute to the mucin-induced enhancement in PAO1 twitching motility.
Fig. 5.
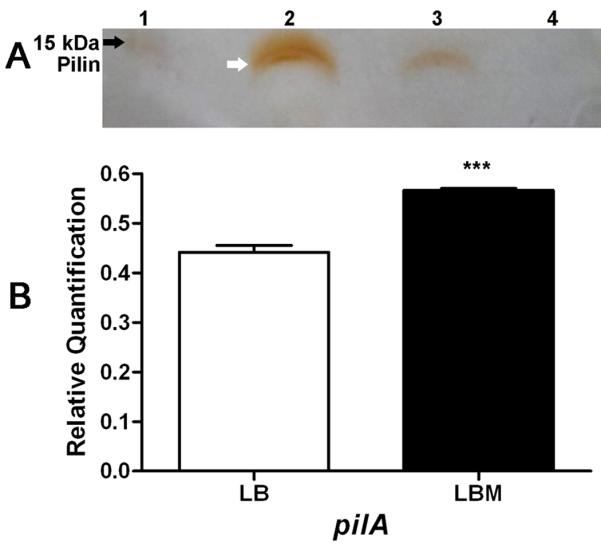
Mucin enhances the production of pilin and the expression of the pilA structural gene. PAO1 was grown on LB and LBM agar for 16 h at 37 °C. (A) Pilin was sheared from the surface of PAO1 and PAOΔpilA and pilin proteins were analyzed by 15% SDS-PAGE and silver staining. Lanes: 1) Molecular weight standard, 2) PAO1 from LBM agar, 3) PAOl from LB agar, 4) PAOΔpilA from LBM agar. (B) Mucin enhances pilA expression. Overnight cultures of PAO1 were spread on four LB agar plates and four LBM agar plates and incubated at 37 °C for 16 h. Cells were harvested from the surface of the plates with a sterile glass rod, RNA was extracted and purified and used in qRT-PCR to analyze the levels of pilA expression. Samples were normalized using 30S ribosomal RNA (rplS) as an internal standard. Values represent the means of 3 independent replicates ± SEM; ***P<0.001.
Mucin requires functional pilin protein to reduce SABF formation
Besides pilA, P. aeruginosa contains more than 40 genes that function in the assembly and biogenesis of the TFP (Mattick, 2002). Together with FimL, the peptidoglycan-binding protein FimV plays a role in pilus assembly (Semmler et al., 2000; Wehbi et al., 2011; Whitchurch et al., 2005). Additionally, the pilin biogenesis gene cluster, encompassed by the pilGHI/pilJK-chpA-E operons, encodes chemotaxis-like chemosensory signal transduction systems that control P. aeruginosa TFP production and twitching motility (Mattick, 2002). To examine the role of these genes in the observed effect of mucin, we utilized P. aeruginosa PA14 transposon mutants defective in each of these genes (Liberati et al., 2006). Therefore, we first determined if mucin affects twitching motility in other P. aeruginosa strains. As with PAO1, mucin significantly enhanced the twitching motility of P. aeruginosa strains PA14, PAK, and PA103 (Fig. 6).
Fig. 6.
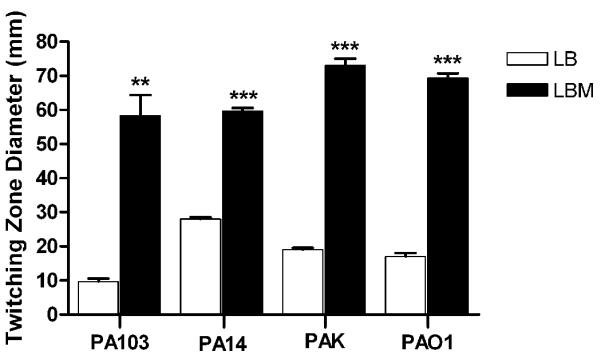
Mucin enhances twitching motility of multiple P. aeruginosa strains. PA103, PA14, PAK and PAO1 were stab-inoculated onto either LB or LBM twitching agar (1 % agar). The plates were incubated at 37 °C for 48 h then at 23°C for an additional 48 h. The agar was removed and the twitching zones revealed by staining with crystal violet. Values represent the means of 3 independent experiments ± SEM. Twitching zones of each strain in LBM were compared to those in LB using the t-test; ***P<0.001, **P<0.01.
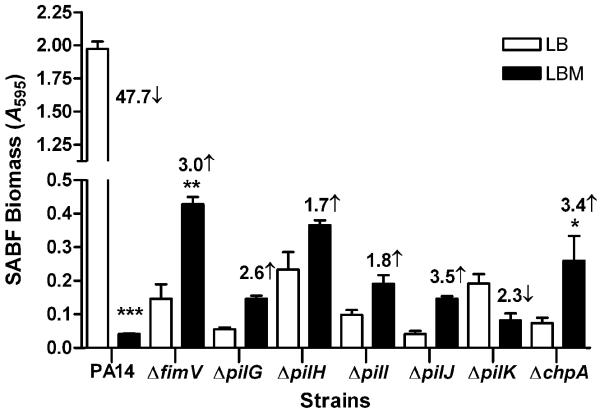
We then selected mutants defective in fimV, pilG, pilH, pilI, pilJ, pilK, and chpA for examination. These mutants produce three specific phenotypes: (1) mutants defective in pilG, pilI, pilJ, chpA or fimV synthesize pilin protein, but fail to assemble pili and are defective in their twitching motilities; (2) the pilH mutant synthesizes pilin protein and assembles it on its outer surface, but is defective in twitching motility; and (3) in contrast, the mutant defective in pilK is piliated and displays twitching motility comparable to its parent strain (Fulcher et al., 2010). Except for PA14ΔpilK, all mutants produced twitching motilities that were significantly lower than that produced by PA14 when grown in the absence of mucin (Fig. S1). The addition of mucin to the twitching medium enhanced the twitching motility of PA14ΔpilK only (data not shown). Additionally, in LB broth and compared with PA14, all mutants including PA14ΔpilK produced significantly reduced SABF (Fig. S2). However, in the presence of mucin, all mutants showed a 1.7-to 3.5-fold increase in SABF formation except PA14ΔpilK, which showed a 2.3-fold decrease (Fig. 7). These results suggest that, depending on the functional status of the pilin protein, mucin affects SABF in P. aeruginosa differently. In the presence of functionally active pilin and twitching motility as in PA14 and PA14ΔpilK, mucin reduces biofilm development. However, in the presence of nonfunctional pilin (twitching motility defective), mucin increases SABF formation. The nonfunctional pilin may be synthesized but not assembled, as in PA14ΔpilG, PA14ΔpilI, PA14ΔpilJ, PA14ΔchpA and PA14Δfim; or synthesized and assembled, as in PA14ΔpilH.
Fig. 7.
In the absence of twitching motility, mucin increases SABF formation by PA14. PA14 and its different PA14 pilin biogenesis mutant strains were grown in LB and LBM broth. SABF formation was determined as described in Fig. 1. In the presence of mucin, the SABF biomass was significantly increased in PA14ΔfimV (3 fold), PA14ΔpilG (2.6 fold), PA14ΔpilH (1.7 fold), PA14ΔpilI (1.8 fold), PA14ΔpilJ (3.5 fold), and PA14ΔchpA (3.4 fold). In contrast, mucin significantly decreased the SABF biomass in PA14ΔpilK (2.3 fold) and PA14 (47.7 fold). Values represent the means of 3 independent experiments ± SEM. SABF of each strain in LBM broth were compared to those in LB broth using the t-test; ***P<0.001, **P<0.01, *P<0.05.
Based on the results of our previous study (Haley et al., 2012) and our current analysis (Figures 5 & 7), mucin may require functional pilin biogenesis genes to reduced SABF formation and enhance BLS development. In this study, we compared the twitching motility of PA14 and different mutants in the pilin biogenesis genes using the twitching motility plate assay (Fig. S1). Therefore, we examined the effect of mucin on the expression of one of the pilin biogenesis genes, chpC (as a representative of the pilGHI/pilJK-chpA-E operon), within the BLS. This was done using PAO1 strain PW1762 that carries an in-frame chpC::lacZ chromosomal fusion. We grew PW1762 in LBM broth for 16 h, and collected pellets of the BLS and planktonic cultures for β-galactosidase assay. The level of β-galactosidase activity produced within the BLS was significantly higher than that within the planktonic cultures (16.5 ± 0.625 [SEM] units vs. 4.3 ± 0.612 units; P <0.001). These results suggest that within the BLS, the expression of pilin biogenesis genes is increased.
Mucin does not enhance P. aeruginosa twitching motility by chelating iron
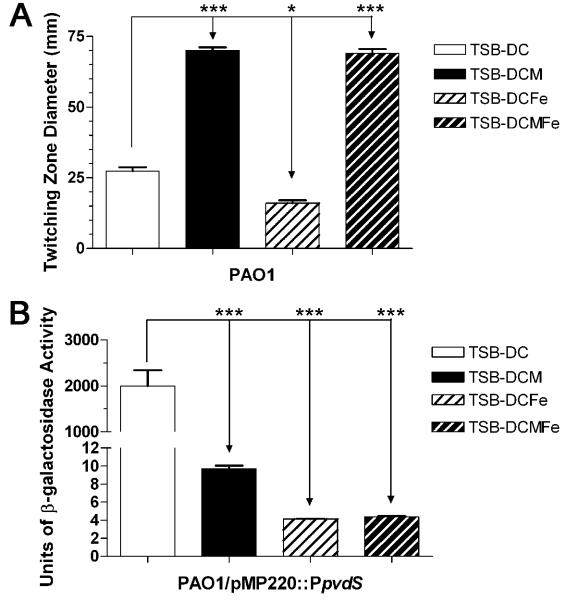
Singh et al. (2002) previously suggested that at concentrations lower than those preventing bacterial growth, lactoferrin chelates iron and stimulates P. aeruginosa twitching motility. As a result of their continuous movement, P. aeruginosa bacteria fail to form stable biofilms (Singh, 2004; Singh et al., 2002). Since lactoferrin exists in sufficient amounts in surface secretions, including airway secretions (Abe et al., 1999; Thompson et al., 1990), mucin may inhibit the development of SABF through the lactoferrin iron-chelating activity. To examine this possibility, we determined the effect of mucin, iron, or both on the twitching motility of PAO1 that was grown in the iron-deficient medium TSB-DC. TSB-DC medium, in which iron has been chelated, is a standard medium for the analysis of different P. aeruginosa iron-regulated genes. As shown in Figure 8A, PAO1 twitching motility was reduced in TSB-DC containing iron but was significantly enhanced in TSB-DC containing mucin or mucin plus iron. Additionally, using a pvdS-lacZ transcriptional fusion plasmid, we determined the effect of mucin, iron, or both on the expression of the PAO1 iron-regulated gene pvdS, which codes for an alternative sigma factor (Vasil and Ochsner, 1999). Iron represses the expression of pvdS (Vasil and Ochsner, 1999). However, pvdS expression was significantly reduced in the presence of mucin, iron, and mucin plus iron (Fig. 8B). This suggests that, although lactoferrin is one of the components of mucin, mucin as a whole does not chelate iron. Rather, it functions like iron and represses the production of P. aeruginosa iron-regulated factors.
Fig. 8.
Mucin does not enhance P. aeruginosa twitching motility by chelating iron. (A) PAO1 was stab-inoculated onto TSB-DC, TSB-DC with mucin (5 mg/mL), TSB-DC with iron (20 μg/mL), or TSB-DC with mucin (5 mg/mL) and iron (20 μg/mL) twitching plates. The plates were incubated at 37 °C for 48 h then at 23 °C for an additional 48 h. The agar was removed and the twitching zone revealed by staining with crystal violet. (B) Either mucin or iron significantly reduced the expression of the iron regulated gene pvdS. PAO1 containing a pvdS-lacZ fusion plasmid (pMP220::PpvdS) was grown in TSB-DC, TSB-DC with mucin, TSB-DC with iron, or TSB-DC with both mucin and iron under shaking conditions for 6 h at 37 °C. Samples were collected and the β-galactosidase activity was determined. Values in A and B represent the means of 3 independent experiments ± SEM. Values obtained in the presence of mucin, iron, or both were compared to values obtained in TSB-DC alone by t- test; ***P<0.001, *P<0.05.
Mucin requires functional Vfr to affect twitching motility and biofilm development in P. aeruginosa
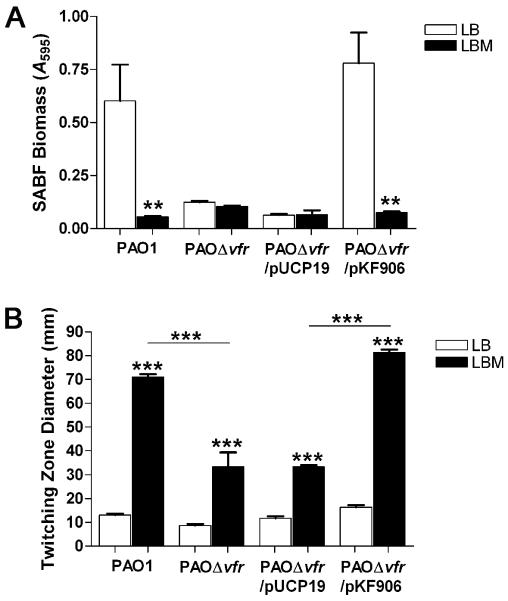
The virulence factor regulator, Vfr, regulates several P. aeruginosa virulence systems including the TFP and twitching motility (Beatson et al., 2002). Compared with its parent strain, a P. aeruginosa vfr mutant was severely defective in twitching motility (Beatson et al., 2002). Vfr also regulates the expression of pilin biogenesis genes (Beatson et al., 2002). Fulcher et al. (2010) suggested that the Pil-Chp system influences twitching motility through the cAMP-dependent activity of Vfr. To determine if Vfr plays a role in the effect of mucin, we compared biofilm development and twitching motility produced by PAO1 and its vfr isogenic mutant PAOΔvfr. Compared with PAO1, PAOΔvfr produced a significantly reduced biofilm (Fig. 9A). This defect was rescued by a plasmid carrying intact vfr (pKF906) but not a cloning vector (Fig. 9A). In the presence of mucin, PAO1 and PAOΔvfr/pKF906 produced a significantly reduced biofilm (Fig. 9A). However, mucin did not eliminate the already reduced biofilm produced by either PAOΔvfr or PAOΔvfr/pUCP19 (Fig.9A).
Fig. 9.
Mucin enhances twitching motility and reduces biofilm development of PAO1 through the virulence factor regulator, Vfr. We assessed biofilm development and twitching motility of PAO1, PAOΔvfr, PAOΔvfr/pUCP19, and PAOΔvfr/pKF906 as described in figures 1 and 2. (A) The effects of mucin and Vfr on SABF development. (B) The effects of mucin and Vfr on twitching motility. Values in A and B represent the means of 3 independent experiments ± SEM. Twitching zones of each strain on LBM agar were compared to those on LB agar using the t-test; and in B, values for the mutant were compared to the parent (upper bar); ***P<0.001, **P<0.01.
We then compared the twitching motility of PAO1 and PAOΔvfr. The defect of PAOΔvfr in twitching motility was not as pronounced as the defect in biofilm formation (13 mm in PAO1 vs. 8.6 mm in PAOΔvfr) (Fig. 9B). Mucin enhanced twitching motility in both strains. However, the enhancement in PAOΔvfr was significantly lower than that in PAO1 (Fig. 9B). The mucin-induced enhancement of the twitching motility of PAOΔvfr/pKF906 paralleled that of PAO1 (Fig. 9B). These results suggest that mucin requires functional Vfr to influence PAO1 twitching motility. We observed similar results with the P. aeruginosa strain PA14 and its vfr mutant. Even at 48 h, mucin significantly reduced biofilm development by PA14 and the complemented mutant (data not shown). Additionally, the mucin-induced enhancement in PA14Δvfr twitching motility was significantly less than that detected in PA14, a defect that was complemented by pKF906 (data not shown).
If Vfr plays a role in the observed mucin-induced reduction in SABF formation, it may also be required for the mucin-induced development of BLS. To examine this possibility, we introduced the GFP plasmid pMRP9-1 plasmid (Table 1) in PAO1 and PAOΔvfr. We grew the transformants (PAO1/pMRP9-1 and PAOΔvfr/pMRP9-1) in the mucin medium ASM+, which we previously utilized to examine different aspects of the BLS (Haley et al., 2012), and compared the BLS formed by the two strains using the COMSTAT. Compared with the BLS formed by PAO1/pMRP9-1, the total biomass, mean thickness, and total surface areas of BLS formed by PAOΔvfr/pMRP9-1 were significantly decreased (Table S1). This suggests that, in addition to its role in the development of SABF, Vfr is critical for the mucin induced development of BLS.
Discussion
We previously showed that in the artificial sputum medium ASM+, P. aeruginosa formed free-floating BLS (Haley et al., 2012). Mucin, a major component of this medium, contributed significantly to the formation of BLS (Haley et al., 2012). In this study, we confirmed that regardless of the medium, mucin facilitates the development of BLS at the expense of the development of a surface-attached biofilm. Mucin accomplishes this effect in part by enhancing P. aeruginosa pilin-mediated twitching motility (Figs. 2 and 9). The influence of mucin on twitching motility is not a new finding. Huang et al. (2003) previously showed that mucin as well as bovine serum albumin enhanced P. aeruginosa twitching motility. However, our findings showed that beyond the initiation, the excessive mucin-induced twitching prevented P. aeruginosa from forming SABF. This phenomenon is not limited to the PAO1 strain, but occurs in other P. aeruginosa strains such as PA14, PAK and PA103 (Fig. 6). However, the excessive twitching motility does not seem to interfere with the formation of the BLS, indicating that the mechanism of development of these structures is different from that of SABF (Haley et al., 2012).
Yeung et al. (2012) recently reported that, on swimming agar plates, mucin induced a highly modified P. aeruginosa surface motility termed surfing, in which P. aeruginosa swims relatively more rapidly across the surface of the agar. The phenomenon requires a functional flagellum but not type IV pili as none of the tested chemosensory mutants were surfing defective (Yeung et al., 2012). We did not observe a difference in swimming motility of P. aeruginosa that was inoculated on either regular swimming agar or swimming agar containing mucin (data not shown). The most likely explanation for the variation between the two studies is the difference in the types of media that were used in the swimming agar plates. We utilized a tryptone base for swimming plates (0.3 % agar) and an LB base for twitching motility (1 % agar), media that have been used by other investigators (Deziel et al., 2001), whereas Yeung et al. (2012) used BM2 or MSCFM medium in their swimming plates.
The influence of host factors on biofilm development through the twitching motility was previously proposed by Singh et al. (2002), as they demonstrated that lactoferrin enhances PAO1 twitching motility and compromises biofilm development. What is surprising in our findings, however, is that in the absence of twitching motility, mucin enhances rather than reduces the formation of SABF (Fig. 7). This effect requires the presence of pilin protein. A possible explanation is that mucin interacts with P. aeruginosa outer surface structures including pilin. While this scenario is plausible for PA14ΔpilH, which is piliated but twitching defective, it is not likely for PA14ΔpilI, PA14ΔpilJ, and PA14ΔpilG, which synthesize pilin but are not piliated. At this time, the mechanism for the pilin-dependent increase in SABF by mucin is not known. We have already eliminated a possible role for the flagellum or the flagellum-based swimming motility in this phenomenon. Mucin did not affect the swimming motility of PAO1 (data not shown).
Vfr is required for an efficient twitching motility in P. aeruginosa (Beatson et al., 2002). Compared with its parent strain, P. aeruginosa vfr-deficient mutants displayed a significantly reduced twitching motility (Beatson et al., 2002) (Fig. 9B). Although vfr mutants produced surface pilin, the amount of pilin protein was considerably lower than that produced by the parent strain (Beatson et al., 2002). Based on these results, Beatson et al. (2002) concluded that Vfr does not influence pilA expression or pilin synthesis. Rather, Vfr may affect the pilin assembly or the function of the assembled pilus (Beatson et al., 2002). However, transcriptional analyses demonstrated that, compared with its parent strain, the expression of several genes of the chemosensory system including pilG, pilI, pilJ, pilK and fimV was significantly reduced in the vfr-deletion mutant (Wolfgang et al., 2003). Kanack et al. (2006) demonstrated specific Vfr binding within the upstream region of fimS and pilM. Similarly, we showed that in the absence of functional Vfr both PAO1 and PA14 mutants displayed a deficient twitching motility, a defect that was complemented by a vfr plasmid. Therefore, based on these results and other previous studies, Vfr may not be directly involved in the mucin effect on twitching motility. Rather, Vfr may influence genes through which mucin enhances twitching motility, including some of the chemosensory systems. However, mucin was capable of enhancing twitching motility even in the absence of functional Vfr (Fig. 9B). This partial enhancement in twitching motility suggests that Vfr affects twitching motility through two mechanisms, one Vfr-dependent and one Vfr-independent. The Vfr-dependent mechanism is clearly represented by the increase in the twitching motility of PAO1 and PA14 vs. PAOΔvfr and PA14Δvfr. The Vfr-independent mechanism is represented by the mucin-induced increase in the twitching motility produced in the vfr mutants (Fig. 9B). While evidence suggests that the Vfr-dependent mechanism involves some of the chemosensory genes, the Vfr-independent mechanism is yet to be defined. Our analysis of PA14 and PAO vfr mutants supports the potential two mechanisms.
With respect to twitching motility, mucin’s primary target may be the Pil-Chp system. Fulcher et al. (2010) previously suggested that the Pil-Chp system influences the P. aeruginosa twitching motility by cAMP-independent and cAMP-dependent mechanisms. In the cAMP-dependent one, the Pil-Chp system modulates the activity of the adenylate cyclase that synthesizes cAMP. Vfr requires cAMP to activate the transcription of several factors including those involved in the synthesis and regulation of TFP (Fulcher et al., 2010). Based on our current results, mucin may affect the Pil-Chp system, which in turn enhances twitching motility through the cAMP-independent and cAMP-dependent mechanisms. The cAMP-dependent mechanism occurs through Vfr (Fig. 9). We clearly showed that only part of the mucin effect on twitching motility occurs through Vfr (Fig. 9). Analysis of P. aeruginosa cAMP-deficient mutants is required to confirm this hypothesis.
At this time, we do not know the exact host factors that stimulate P. aeruginosa motility. We excluded the possibility that mucin chelates iron and enhances P. aeruginosa twitching motility. The presence of mucin in TBSD-DC repressed the expression of the P. aeruginosa iron-regulated gene pvdS (Fig. 8B). These results differ from those of previous studies that suggested the presence of an iron-chelating activity within the CF mucus (Fung et al., 2010; Wang et al., 1996). Wang et al (1996) showed that the growth of P. aeruginosa in a minimal medium containing 10% CF sputum induced the expression of the pyochelin receptor gene, fptA. However, no increase in the expression of other iron-regulated genes including toxA, toxR, pvdS, and pvdA was reported (Wang et al., 1996). Fung et al. (2010) also showed that the growth of P. aeruginosa in their modified artificial sputum medium induced the expression of iron acquisition genes. Besides mucin, this synthetic medium contained other components (Fung et al., 2010; Sriramulu et al., 2005). In contrast to these studies, our current analysis was focused on mucin only. Mucin significantly enhanced twitching motility even when added to the iron-deficient medium TSB-DC, an effect that was not influenced by exogenously added iron (Fig. 8A). More importantly, the expression of the iron-regulated gene pvdS was repressed by mucin or iron alone, or both (Fig. 8B). Similar to previous studies (Fung et al., 2010; Sriramulu et al., 2005), we utilized type-3 porcine mucin. Whether other sources of mucin produce a similar effect on the iron-regulated genes is yet to be determined.
Mucin is a complex macromolecule and one or more of its components may induce the enhancement in P. aeruginosa twitching motility. We previously suggested that the binding of P. aeruginosa to certain constituents of mucin contributes to the development of BLS (Haley et al., 2012). Whether such binding affects both the twitching motility and biofilm development is yet to be determined. Five of the 22 known human mucins (MUC) play a key role in respiratory tract health and defense. The membrane-tethered MUC1, which is upregulated by TNFα-mediated inflammatory responses that occur in P. aeruginosa infection in response to TLR5 signaling, is crucial to both protection during infection and in resolution of the anti-inflammatory response (Kim, 2012). Membrane-tethered MUC1 is crucial for the formation of the inner gel formed immediately above the apical cell surfaces by MUC4 and MUC16, as well as formation of the outer, more fluid gel layer by MUC5AC and MUC5B (Kim, 2012). Additionally, the MUC1 ectodomain that is cleaved by neutrophil elastase during acute infection serves as a decoy receptor for P. aeruginosa, keeping the bacteria away from the cell surfaces (Kim, 2012). Certainly, the level of mucin varies within the lungs of CF patients, with decreases of up to 89% (to 0.55 mg or less mucin/mL) during periods of stable disease (no progression) and increases of over 900% (to 45 mg or more mucin/mL) during exacerbations of pulmonary disease (Henke et al., 2007). As a result, the twitching motility would be affected in response to changes in the host, thereby favoring the production of either a surface-attached biofilm at low mucin concentrations or free-floating biofilm-like structures at high mucin concentrations.
Supplementary Material
ACKNOWLEDGEMENTS
We thank James Wang for excellent technical assistance; Matthew R. Parsek for his kind provision of PAO1/pMRP9-1; and Joanna E. Swickard for critical reading of the manuscript. Images were generated in the Imaging Center Core Facility supported by TTUHSC. The construction of PAO1 strain PW1762 wasfunded by grant # NIH P30 DK089507.
References
- Abe T, Nakajima A, Matsunaga M, Sakuragi S, Komatsu M. Decreased tear lactoferrin concentration in patients with chronic hepatitis C. Br. J. Opthalmol. 1999;83(6):684–687. doi: 10.1136/bjo.83.6.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosi C, Leoni L, Visca P. Different responses of pyoverdine genes to autoinduction in Pseudomonas aeruginosa and the group Pseudomonas fluorescens-Pseudomonas putida. Appl. Environ. Microbiol. 2002;68(8):4122–4126. doi: 10.1128/AEM.68.8.4122-4126.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltch AL. Pseudomonas bacteremia. In: Smith RP, Baltch AL, editors. Pseudomonas aeruginosa infection and treatment. Marcel Dekker; New York, NY: 1994. pp. 73–128. [Google Scholar]
- Beatson SA, Whitchurch CB, Sargent JL, Levesque RC, Mattick JS. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 2002;184(13):3605–3613. doi: 10.1128/JB.184.13.3605-3613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JL, Ramsey BW, Smith AL. Clinical manifestations and treatment of pulmonary infections in cystic fibrosis. Adv. Pediatr. Infect. Dis. 1993;8:53–66. PMID:8217005. [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Deziel E, Comeau Y, Villemur R. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 2001;183(4):1195–1204. doi: 10.1128/JB.183.4.1195-1204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol. Microbiol. 2010;76(4):889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Naughton S, Turnbull L, Tingpej P, Rose B, Arthur J, et al. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J. Med. Microbiol. 2010;59(Pt. 9):1089–1100. doi: 10.1099/jmm.0.019984-0. [DOI] [PubMed] [Google Scholar]
- Gaines JM, Carty NL, Colmer-Hamood JA, Hamood AN. Effect of static growth and different levels of environmental oxygen on toxA and ptxR expression in the Pseudomonas aeruginosa strain PAO1. Microbiology. 2005;151(Pt. 7):2263–2275. doi: 10.1099/mic.0.27754-0. [DOI] [PubMed] [Google Scholar]
- Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2003;168(8):918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- Giltner CL, Rana N, Lunardo MN, Hussain AQ, Burrows LL. Evolutionary and functional diversity of the Pseudomonas type IVa pilin island. Environ. Microbiol. 2011;13(1):250–264. doi: 10.1111/j.1462-2920.2010.02327.x. [DOI] [PubMed] [Google Scholar]
- Haley CL, Colmer-Hamood JA, Hamood AN. Characterization of biofilm-like structures formed by Pseudomonas aeruginosa in a synthetic mucus medium. BMC Microbiol. 2012;12:181. doi: 10.1186/1471-2180-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A, Dertien J, Colmer-Hamood JA, Griswold JA, Hamood AN. Serum inhibits P. aeruginosa biofilm formation on plastic surfaces and intravenous catheters. J. Surg. Res. 2010;159(2):735–746. doi: 10.1016/j.jss.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Hassett DJ, Cuppoletti J, Trapnell B, Lymar SV, Rowe JJ, Yoon SS, et al. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 2002;54(11):1425–1443. doi: 10.1016/s0169-409x(02)00152-7. [DOI] [PubMed] [Google Scholar]
- Henke MO, John G, Germann M, Lindemann H, Rubin BK. MUC5AC and MUC5B mucins increase in cystic fibrosis airway secretions during pulmonary exacerbation. Am. J. Respir. Crit. Care Med. 2007;175(8):816–821. doi: 10.1164/rccm.200607-1011OC. [DOI] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146(Pt. 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Holloway BW, Krishnapillai V, Morgan AF. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 1979;43(1):73–102.H. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- uang B, Whitchurch CB, Mattick JS. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 2003;185(24):7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2003;100(24):14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Finkbeiner WE, Widdicombe JH, McCray PB, Jr., Miller SS. Altered fluid transport across airway epithelium in cystic fibrosis. Science. 1993;262(5132):424–427. doi: 10.1126/science.8211164. [DOI] [PubMed] [Google Scholar]
- Kanack KJ, Runyen-Janecky LJ, Ferrell EP, Suh SJ, West SE. Characterization of DNA-binding specificity and analysis of binding sites of the Pseudomonas aeruginosa global regulator, Vfr, a homologue of the Escherichia coli cAMP receptor protein. Microbiology. 2006;152(Pt. 12):3485–3496. doi: 10.1099/mic.0.29008-0. [DOI] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003;48(6):1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Kim KC. Role of epithelial mucins during airway infection. Pulm. Pharmacol. Ther. 2012;25(6):415–419. doi: 10.1016/j.pupt.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 2005;175(11):7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U.S.A. 2006;103(103):2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PV. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. II. Effects of lecithinase and protease. J. Infect. Dis. 1966;116(1):112–116. doi: 10.1093/infdis/116.1.112. [DOI] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002;15(2):194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004;186(14):4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. doi:10.1146/annurev.micro.56.012302.160938. PMID:12142488. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press; New York, NY: 1972. [Google Scholar]
- Ohman DE, Sadoff JC, Iglewski BH. Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect. Immun. 1980;28(3):899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranchych W, Sastry PA, Drake D, Pearlstone JR, Smillie LB. Pseudomonas pili. Studies on antigenic determinants and mammalian cell receptors. Antibiot. Chemother. 1985;36:49–57. PMID:2408561. [PubMed] [Google Scholar]
- Pier GB, Ramphal R. In: Pseudomonas aeruginosa. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Mandell GL, Bennett JE, Dolin R, editors. Livingstone; Philadelphia, PA: 2010. pp. 2835–2860. [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268(5219):1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245(4922):1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002;184(4):1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97(1):109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- Semmler AB, Whitchurch CB, Leech AJ, Mattick JS. Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology. 2000;146(Pt. 6):1321–1332. doi: 10.1099/00221287-146-6-1321. [DOI] [PubMed] [Google Scholar]
- Singh PK. Iron sequestration by human lactoferrin stimulates P. aeruginosa surface motility and blocks biofilm formation. Biometals. 2004;17(3):267–270. doi: 10.1023/b:biom.0000027703.77456.27. [DOI] [PubMed] [Google Scholar]
- Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417(6888):552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- Sriramulu DD, Lunsdorf H, Lam JS, Romling U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 2005;54(Pt. 7):667–676. doi: 10.1099/jmm.0.45969-0. [DOI] [PubMed] [Google Scholar]
- Stachel SE, An G, Flores C, Nester EW. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Strom MS, Lory S. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J. Bacteriol. 1986;165(2):367–372. doi: 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AB, Bohling T, Payvandi F, Rennard SI. Lower respiratory tract lactoferrin and lysozyme arise primarily in the airways and are elevated in association with chronic bronchitis. J. Lab. Clin. Med. 1990;115(2):148–158. [PubMed] [Google Scholar]
- Vasil ML, Ochsner UA. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 1999;34(3):399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Lory S, Ramphal R, Jin S. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol. Microbiol. 1996;22(5):1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- Wehbi H, Portillo E, Harvey H, Shimkoff AE, Scheurwater EM, Howell PL, Burrows LL. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 2011;193(2):540–550. doi: 10.1128/JB.01048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SE, Sample AK, Runyen-Janecky LJ. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 1994;176(24):7532–7542. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Beatson SA, Comolli JC, Jakobsen T, Sargent JL, Bertrand JJ, et al. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 2005;55(5):1357–1378. doi: 10.1111/j.1365-2958.2005.04479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell. 2003;4(2):253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Yeung AT, Parayno A, Hancock RE. Mucin promotes rapid surface motility in Pseudomonas aeruginosa. MBio. 2012;3(3) doi: 10.1128/mBio.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.