Nearly 100 years ago, Felix d'Herelle, the codiscoverer of bacteriophages, used bacteria to control insect pests and used phages against bacterial disease. His approaches reflected ecological insights before this branch of biology became an established scientific discipline. In fact, one might have predicted that phage research would become the springboard for biotechnology and ecology. However, d'Herelle was ahead of his time, and the zeitgeist in the 1930s pushed physicists into the question “What is life?” Phages as the simplest biological systems were the logical choice for this question, and phage research became the cradle of molecular biology.
Now many researchers speak of a “new age of phage research.” It is now realized that phages play an important role in ecology (e.g., phage impact on the cycling of organic matter in the biosphere at a global level) (27), that phages influence the evolution of bacterial genomes (most obviously in the development of bacterial pathogenicity) (7), and that phages might provide potential tools to face the antibiotic resistance crisis in medicine (59). With this new trend, we now see a clear shift from the reductionist approach, focusing on a handful of phages in carefully controlled laboratory conditions, towards the study of many different phages in the complexity of real-life situations.
In contrast to the molecular biology-oriented phage research where the interaction of molecules took center stage, ecology focuses on the interactions between organisms and their physical environment. Much of ecology is therefore about the evolution of biological diversity in space and time. In contrast to many branches of biology, ecology attributes a great importance to quantitative relationships and numbers and aims at a mathematical formulation of its observations. It is thus appropriate to start this review with an overview of phage titers encountered in the biosphere. Next, we ask how a parasite targets its host if the latter is scarce or not in an appropriate physiological state. Finally, we report on research that tries to bridge phage ecology and genomics and cell biology approaches. It is concluded that the integration of phages into complex networks of interacting biological systems, and analysis by molecular techniques, could give phage research a model character in biology again.
NUMBERS: PHAGE TITERS IN THE BIOSPHERE
Just 14 years ago, a Norwegian group surprised the scientific community with a report on the high concentration of phage-like particles in coastal water and the ocean and even higher concentrations in lakes (5). In eutrophic estuarine water, bacteria are found at a density of 106 cells/ml and viruses with a concentration of 107 particles/ml. These concentrations are estimates that vary with the seasons and the geographical location. In addition, these figures refer to physical and not viable entities. A popular model postulates about 10 to 50 different bacterial species and 100 to 300 different phage strains (70) in this environment. The higher number of phage species was justified by the fact that each bacterial species may be infected by 10 phage species. As one would expect, the production and distribution of marine phages are determined by the productivity and density of the host bacterial populations. This relationship is expressed by the virus-to-bacterium ratio that is frequently at 10 to 1. For a comprehensive review on viruses in water and an extensive reference list see reference 68. Substantial theoretical and experimental research efforts were undertaken to determine the quantitative degree of virus-mediated bacterial mortality and to assess the ratio of phage lysis versus grazing of bacteria by protists and multicellular organisms. The greatest impact of phage lysis was seen in oligotrophic environments, and the ratio of lysis versus grazing changed with water depth both in the ocean and in lakes (66). To summarize the extensive literature, different approaches in various environments yielded a remarkably constant rate of virus-mediated bacterioplankton mortality of about 15% per day (61). The rate seems to be higher for heterotrophic bacteria than for the autotrophic cyanobacteria (61). Ecologically even more important is the profound effect of phages on the relative proportions of different bacterial species or strains in a community.
All these figures have important consequence for our biological view of the world. If phages outnumber bacteria in the ocean, phages are likely to be numerically the most prominent biological systems on earth, with an estimated population size of ≥1030 phage particles. With these numbers, even rare phage-induced events manifest with high frequency. For example, transduction, the accidental packaging of bacterial host DNA into a phage particle, occurs under optimal laboratory conditions about once in every 108 phage infections. When calculated for the global marine phage population, it follows that gene transfer between organisms takes place about 20 million billion times per second in the oceans (16). The actual numbers will probably be lower due to smaller transduction efficiency and more rapid phage decay in the ocean than in the laboratory. If only a small part of this DNA is traveling between different bacterial species, gene transfer via marine phages opens up enormous possibilities for horizontal DNA transfer between bacteria.
High concentrations of phages are not restricted to the ocean water. Counts of up to 109 phage per gram of marine sediment were recorded with bacterial counts higher than in the waters described above (21). Terrestrial ecosystems, e.g., soil associated with plant roots in sugar beet fields, revealed 107 viruses per gram by using transmission electron microscopy (1). Not all viruses will be viable, but hybridization experiments with Serratia and Pseudomonas spp. from the soil showed that about 5% of the bacteria are actually phage infected.
There is a dictum that phages are found where bacteria thrive. Thus, we should not be surprised that phages are found on us (skin) and within us (oral cavity, gut). In a survey of stool samples from 600 healthy adults, 34% of the subjects demonstrated coliphages (but only 1% showed high amounts) (28). Most of them were classified as temperate phages related to phage lambda. Ruminants that rely heavily on bacteria for cellulose digestion show intestinal phage concentrations in excess of 107 phages per gram of feces. Stool phages were mainly explored for their potential to trace fecal contamination in the environment and to monitor the intrusion of polluted surface waters into groundwater.
Phages are also present in the food we eat. Many food products from our daily life are the result of fermentation processes by lactic acid bacteria. Cheese factories using Lactococcus lactis can be contaminated with high levels of phages; one study reported up to 109 phage per ml of whey and up to 105 phage per m3 in the air (48). When the phage titers exceeded 103 PFU/ml, the yogurt fermentation process was delayed and came to a stop at higher phage titers, leading to important economical losses (unpublished observations).
CYCLES: DYNAMIC PHAGE-HOST RELATIONSHIPS
Time series experiments revealed the seasonality in marine phage titers, demonstrating a dynamic relationship between phages and their bacterial hosts (8). In fact, at any given moment the marine phage titer is the net result from two opposing processes: synthesis of new phage particles due to ongoing lytic phage infections, which is balanced by phage decay. Sunlight UV was identified as the major destructive factor (69), causing up to 5% phage infectivity loss per hour for surface waters due to thymine dimer formation.
Repeat samples from a Norwegian fjord demonstrated seasonality in the viral titers, with a low in the cold winter season and a high in the summer. As a general rule, bacterioplankton produces greater amounts of phages under environmental conditions favoring fast bacterial growth and productivity. Other phage cycles occur on shorter time scales, as predicted by the “killing the winner populations” hypothesis. This concept states that phages expand on the fastest growing host population in the given ecological setting (54). The phage epidemic ceases when the diminished host population no longer supports phage replication. There is strong ecological evidence that some bloom collapse is in fact mediated by phage lysis: in the prealpine Lake of Constance, a transient increase in bacterial abundance was closely followed by peaks in the frequency of infected bacteria and then free phage (32) (Fig. 1B). Variability in the ocean was shown even over half-hour time intervals, probably reflecting synchronized infection cycles (8).
FIG.1.
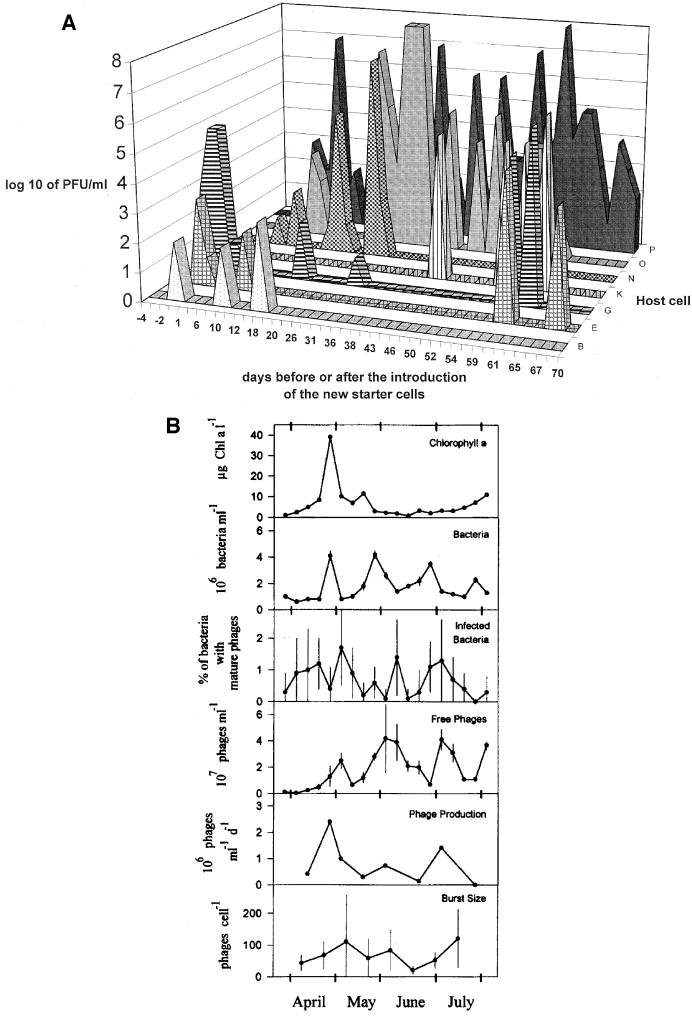
Cycles: dynamic phage-host relationships. (A) Phages in the cheese factory. Cheese whey phage titers (y axis) observed on the starter strains B to P indicated on the z axis on a given day (x axis) before (negative days) and after the introduction of the new starter strain. Reprinted from reference 14 with permission. (B) Phages in a European lake. Chlorophyll a, bacterial abundance, bacteria with intracellular mature phage particles, free phages <100 nm in diameter, phage production, and burst size in the Lake of Constance during different seasons between late March and August of 1992. Error bars indicate standard deviations. Reprinted from reference 32 with permission.
Cyclic phage development was also seen in other environments. Two types of Serratia phages were associated with the soil surrounding sugar beets: temperate Siphoviridae with a long latent period and big burst size and virulent Podoviridae with a short latent period and a small burst size. Over a 6-month observation period, the initially predominant siphovirus population changed to one dominated by the podovirus. Apparently, the two phages are adapted to two quite different niches in the rhizosphere, where the availability and physiological status of bacteria and plants changes with time (2). Some reports indicate that phage-host interaction can be quite complicated in the soil. In streptomycetes, spatial heterogeneity in phage-host interaction and temporal changes in phage susceptibility defined bacterial escape strategies from phage lysis. It was revealed that germinating spores were more susceptible to phage infection than hyphae of developed mycelia. Mature, resistant mycelia adsorb most of the Streptomyces-specific soil phages and thus protect younger, susceptible hyphae from infection (15).
Fluctuating phage titers are also a common observation in the dairy factory. This reflects the buildup and ensuing disappearance of phages specific for a starter strain imposed by the starter strain rotation system. Phage dynamics were also documented in a large intervention trial in a cheese factory (14) (Fig. 1A). One starter combination was replaced by a second that was insensitive to the resident phages of the factory. The intervention resulted in a nearly immediate disappearance of the resident phages. However, 5 to 7 days after the intervention, the first phages infecting the new starters were detected. Restriction enzyme analysis of the phage DNA traced the origin of the new phages to the rare phages in the raw milk samples delivered to the factory during the intervention period. Phage titers increased rapidly to the former levels, and starter rotation had to be reintroduced. Persistence of the phage in the absence of a propagating starter strain was not observed in the factory (unpublished observations).
In the human gut, it was reported that phage types and titers differed when healthy adults and patients suffering from traveler's diarrhea were compared (28). The former excreted low titers of lambda-like phages, while the latter yielded higher numbers of T4-like phages. This change in the phage population was suggested to reflect disturbances of the intestinal microflora during a bout of diarrhea.
COMPLEXITY: PHAGE GENOMICS
Are phages genetically as diverse as they are numerous in the biosphere? Only a few phage groups have been intensively studied by sequencing projects (coliphages, mycobacteriophages, and dairy phages). These investigations demonstrated that the genetic relatedness between phages reflected more the phylogenetic relationships between their host bacteria than the possibilities of current gene transfer in a given ecological setting. For example, phages from dairy streptococci were closely related to phages from pathogenic streptococci living on the skin. Lactococcus phages are the prime cause of cheese fermentation failures. Due to this economic interest, their genetic diversity was intensively explored. Different types of prolate-headed and large and small isometric-headed Siphoviridae (phages with long noncontractile tails) and rare Podoviridae (phages with short tails) were described. Interlaboratory phage comparisons led to the definition of 12 lactococcal phage species. In meat fermentation, you also find Myoviridae (phages with contractile tails) infecting Lactobacillus starters (see Fig. 2 for the basic tailed phage types). Mycobacteria are hosts to an amazingly diverse group of phages (51). Their morphology stimulated the researchers to use such fanciful names as Corndog phage. As variable as the morphologies are, their genomes are just as variable. Within the 14 sequenced mycobacteriophages, 10 lacked DNA sequence similarity with the other mycobacteriophages, and nearly half of the predicted proteins lacked any database matches. The workhorses of molecular bacteriology, Escherichia coli and Bacillus subtilis, are also hosts to about 10 phages with fundamentally different genome organization, not to mention substantial differences at the DNA sequence level within each group of phages. If these observations of 10 or more phages per bacterial species can be generalized, phages might indeed represent the largest unexplored reservoir of sequence information in the biosphere. Random sequencing efforts of viral DNA from two uncultured marine water samples (10) combined with a statistical analysis revealed between 400 and 7,000 different viral types in the two investigated 100-liter samples, with the most abundant type representing 3% of the total viral population. However, a different picture was obtained by the few complete marine phage genomes. This analysis led to the description of a new phage family (Corticoviridae), while other marine phages shared protein sequence similarity with coliphages T7, T4, lambda, P2, and dairy siphophages, suggesting a limited number of prototype phage genomes (18, 72) (Fig. 3).
FIG. 2.
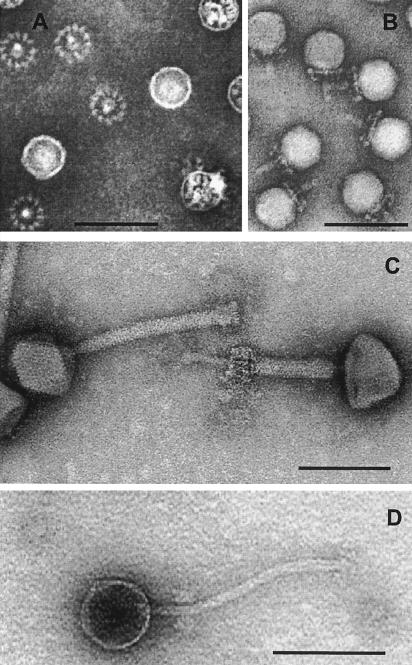
Morphological diversity of phages isolated from food fermentation. The negative-stain electron microscopic pictures show a Lactobacillus plantarum myovirus LP65 (note the contracted tail in the right phage in panel C), a L. plantarum siphovirus LP45 (panel D), and a Staphylococcus carnosus stc1 or -2 podovirus (panel B, side view of stc2; panel A, stc1 [from beneath, the phage baseplate becomes visible as a wheel-like structure]). All phages were isolated from meat (salami) fermentation. The bars are 100 nm.
FIG. 3.
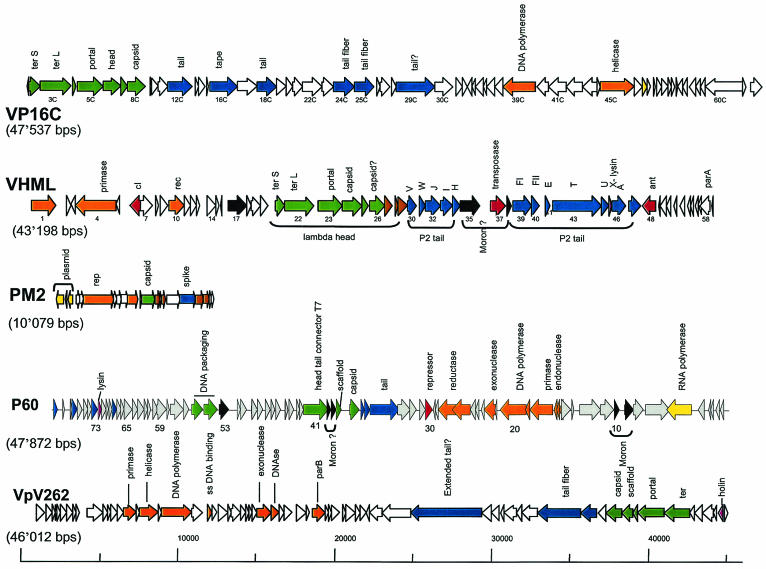
Genomic diversity of phages isolated from the ocean. The genome maps of the currently sequenced marine phages are shown. From top to bottom: Vibrio parahaemolyticus phage VP16C resembling dairy Siphoviridae over the structural genes (GenBank accession no. AY328853; AY328852 for the related phage VP16T); Vibrio harveyi phage VHML resembling E. coli siphophage lambda over the head and myophage P2 over the tail genes (no. AY133112); Alteromonas phage PM2, the first lipid-containing bacterial virus attributed to the new phage family Corticoviridae (no. NC_000867); cyanophage P60 infecting Synechococcus sp., which shows a distant E. coli phage T7-like genome organization (no. NC_003390); V. parahaemolyticus phage VpV262, another marine phage with a T7-like genome organization (no. NC_003907). Not shown are a Roseobacter phage SIO1 (no. NC_002519), since it only distantly resembled T7 phages and otherwise nothing else in the database. Also not shown is the broad-host-range vibriophage KVP40 (no. NC_005083) because its 244,835-bp genome is larger than all depicted phage genomes together. KVP40 resembles E. coli myovirus T4 closely over the structural and DNA replication modules but differs by large insertions of DNA lacking database matches. The likely functions of the genes are color coded: green, head; blue, tail; brown, head-to-tail genes (in VP16C) and transmembrane structural genes (in PM2); orange, DNA replication; yellow, transcription; red, lysogeny and recombination; mauve, lysis; black, lysogenic conversion genes.
The human gastrointestinal tract is rich in commensal life forms. About 1011 bacteria per ml of colonic content are normally found. The number of bacterial cells in the gut exceeds the total number of cells constituting the human body. Gut bacteria contain about 400 bacterial species, but only 40 species account for 99% of the total population. Also, shotgun cloning of the uncultured virus fraction of human feces underlined the genetic diversity of this fraction (9). Statistical calculations suggested between 160 and 1,200 viral genotypes in the intestine.
NUTRITION: INFECTING THE STARVED CELL
Scientists in the laboratory generally work with media maintaining optimal growth of their bacteria. Bacteria in the marine environment have to live with much less nutrients.
Consequently, in situ burst size (the number of phages produced per infected cell) is generally smaller in the marine environment than in the laboratory, where bacteria grow to larger sizes. It was predicted that the normal state of a marine bacterium corresponds to the nutritional state of a laboratory bacterium under stationary phase. In the laboratory, bacteria cannot be productively infected with phages in the stationary phase. In general, infected stationary-phase cells release progeny phage only after resumption of cell growth. This observation could explain the persistence of virulent phages within populations of nongrowing cells. However, there are reports of Pseudomonas phages that productively infect host cells maintained under starvation conditions (57). In this case, the latent period was lengthened and the burst size greatly reduced when compared to those of logarithmic-phase infection (71). In fact, mycobacteriophages have found a way to replicate in slow-growing cells (51).
The nutritional considerations are also relevant for applied phage research like the therapeutic use of phages against pathogenic bacteria. Empirical phage therapy was conducted in the United States during the 1930s and in the Soviet Union until quite recently, where substantial success against bacterial diarrhea and skin infections was reported (59). Notably, oral T4-like phages survived the gastrointestinal passage in mice (19) and humans (unpublished results). However, the presence of a host cell and a corresponding phage is not sufficient to lead to a phage infection in the intestine. The host cell must be in an appropriate physiological state to allow productive infection, and the cell must be in an accessible anatomical location for the phage. Mouse experiments demonstrated that the resident E. coli flora was protected against oral phage exposure, although the majority of the fecally excreted E. coli strains were infected by the same phage in vitro (19). At first glance, one would expect that the gut is a nutritionally privileged anatomical site. However, E. coli within the lumen of the colon is nutritionally deprived and nonreplicating (53) and is thus a poor target for phages. The metabolically active intestinal E. coli cells are found in microcolonies associated with the mucus layer of the gut mucosa (37, 52), where they are probably physically shielded from luminal phages. In contrast, E. coli strains freshly introduced into the mouse infection model were fully susceptible to oral phage exposure (19). Apparently, these new E. coli cells were actively replicating but not yet associated with the mucus layer of the gut mucosa. This example illustrates that we need a detailed knowledge of the ecology of target bacteria and the therapeutic phages in the gut of the mammalian host if phage therapy is to be successful (38). Similar considerations apply for the application of phages to other body sites, e.g., the skin. The Soviet experience suggested the use of phages for the treatment of wound infections (59), and food microbiologists explore the use of phages to decontaminate the skin of slaughtered chicken from Salmonella and Campylobacter spp. (30) and E. coli O157 for beef (38). The potential medical and food safety applications of phages are favored by the availability of broad-host-range Myoviridae for Staphylococcus aureus (50) and Listeria monocytogenes (41).
POLYVALENCY: FINDING THE RIGHT CELL
In near coastal water, the bacterial population consists of 106 cells per ml distributed over 100 different bacterial host species (68). Less than 104 cells per ml therefore constitute the average target population for a species-specific phage. Streptococcus thermophilus phages are found with titers of 200 infectious phages per ml of raw milk (14), while its target cell is only detected in raw milk after enrichment. How can phage infection cycles be maintained under these conditions? Laboratory experiments with three phages, including T4, showed that phage production did not occur below 104 cells/ml (67). Statistical analysis and back-calculations in natural marine environments predicted 105 cells/ml as minimal bacterioplankton concentrations for successful virus production. However, some marine viruses replicated efficiently down to 103 specific host cells/ml (62). The determination of the variations of host cell concentration over time allowed the approximation that cyanophage replication still occurred when the host cell concentration fell to 102 cells/ml (65).
Broad-host-range (polyvalent) phages could be a solution to this dilemma. Indeed, some observations suggest that phages isolated from nutrient-poor marine environments showed a trend towards increased polyvalency, possibly representing an adaptation to low host cell concentrations.
For most phages investigated in the laboratory, host species specificity is the rule. The polyvalent phages infecting different genera in the Enterobacteriaceae must be regarded with some caution because this family is such a closely related bacterial group. Even data on marine phages indicate that most of them are host species specific, many even demonstrate strain specificity. Polyvalence was more prevalent in cyanophages, but fluorescence-labeled cyanophages (33) demonstrated that they attached specifically only to their host and not other bacteria of the natural consortium. Data from the ocean showed that polyvalency was correlated with phage morphology. Phages isolated from high-light-adapted Prochlorococcus hosts yielded exclusively Podoviridae that were strain specific (60). In contrast, low-light-adapted Prochlorococcus hosts yielded Myoviridae that also infected Synechococcus spp., a phylogenetically related cyanobacterium. Similarly, Synechococcus-infecting Myoviridae also cross-infected Prochlorococcus spp., lending some support to the polyvalency concept in the marine environment. Also, in other environments, Myoviridae showed a broader host range than Siphoviridae and Podoviridae.
Polyvalency was not described in dairy phages: the analysis of hundreds of phage isolates from cheese factories showed a very narrow host range. We are only aware of exceptions for Lactobacillus phages appearing in sauerkraut fermentation: one phage isolate could infect two ecologically related Lactobacillus species (42). Furthermore, about 30% of lactobacilli constituting the major commensals in the vagina of healthy women were lysogenic. Many lysogens could be induced by mitomycin C, and some phages could infect up to five different Lactobacillus species, which dominate the vaginal flora (36).
Even if polyvalency is not the rule, many phages have developed efficient methods to change their host range by elegant genetic tricks. A classical case is phage Mu, containing a recombinase that inverses the orientation of the receptor-interacting gene leading to the synthesis of a new receptor recognition specificity. A coliphage possessed two different tail fiber proteins and showed the combined host range of phages containing either one or the other tail gene (56). The similarities in the tail fiber genes of coliphages belonging to different phage families (P2, T4, lambda) provide evidence that illegitimate recombination resulting in domain exchanges occurs at previously unappreciated levels (31). Similarly, phages infecting lactic streptococci can alter their host range by exchanging variable domains flanked by conserved collagen-like repeats that serve as target sites for homologous recombination (23). A fundamentally different method of host range changes was recently described for Bordetella phages (40). At one genome end, the mtd (major tropism determinant) gene encodes the phage protein responsible for host cell recognition. Its C-terminal end varies between isolates differing in host range. Directly adjacent to mtd is a second nonidentical copy of the variable end of mtd called TR (for template repeat), followed by a reverse transcriptase gene. Tropism switching is the result of a TR-dependent reverse transcriptase-mediated process that introduces base pair substitutions leading to amino acid changes at about 20 defined positions in the variable part of mtd. The authors proposed a mechanism for this process analogous to the site-specific retrohoming ability of group II introns.
LYSOGENY: A SURVIVAL STRATEGY FOR BOTH PHAGE AND BACTERIA
Theory predicts that lysogeny becomes the preferred strategy when the cell density falls below the lower limit necessary for maintenance of the phage density by repeated cycles of lytic infections. The argument is that the production of temperate phages is independent of host cell density. Indeed, two marine surveys revealed 40% mitomycin C-inducible cells, and similar proportions of lysogens were identified in Pseudomonas colonies from lakes. In contrast, UV or sunlight was not a good inducer of prophages in water samples. The surveys showed a trend for lysogeny to be more prevalent in oligotrophic environments (35). This observation fits with theory, since this setting is dominated by the low density of slow-growing bacteria. Other data contradict this interpretation. Surveys in estuarine waters showed a seasonal development of lysogeny with highs in the summer months when eutrophic conditions were prevalent and lows in the winter months when cells were at their minimum (20, 44). There are further contradictions with expectations. First, spontaneous induction of prophages is generally low (10−2 to 10−5 phage per bacterium per generation). This release can only account for far less than 1% of the phage concentrations in the ocean (35). Second, large phage surveys in the North Sea revealed that only 10% of the phage isolates are temperate. In contrast, the genome maps of the major virulent S. thermophilus phages isolated from dairies still betray their origin from temperate parental phages. The preponderance of virulent phages in dairy collections might therefore represent a secondary character and an adaptation to the abundance of host cells in the dairy environment. In fact, serial passage of a temperate S. thermophilus phage resulted in its replacement by a virulent derivative deletion mutant after only a few days.
However, lysogeny is a survival strategy for phage as well as for bacteria (17). Lysogens frequently outcompete the nonlysogenic congeners, possibly due to the selective advantage conferred by lysogenic conversion genes of the prophages. Some of them are relatively universal, such as immunity functions and superinfection exclusion genes. Other prophages contribute genes that make the lysogen competitive under special ecological situations (serum resistance conferred to the lysogen by the phage lambda bor gene during blood growth of E. coli [3]). This phenomenon seems to be widespread in bacterial pathogens where many virulence factors are encoded by prophages. However, even the laboratory phages P1, P2, lambda, and Mu confer to the E. coli lysogen a higher metabolic activity and faster and longer growth than the nonlysogens (25, 39).
Shiga-toxin producing E. coli (STEC) strains represent a spectacular case of lysogeny. These strains are commonly found in the intestines of asymptomatic cattle, while in humans the STEC O157:H7 strains are dangerous food pathogens. The available evidence suggests that they are derived over the last 50 years from the enteropathogenic E. coli strain O55:H7 by the acquisition of two prophages encoding the Shiga toxins Stx1 and Stx2, the major pathogenicity factor of STEC (58). In fact, the two sequenced O157:H7 isolates contained 16 to 18 prophages, including many closely related lambda-like prophages. Whole-genome PCR scanning of eight distinct O157 strains revealed a high degree of genomic diversity, mainly due to extensive structural and positional diversity of the prophages, implying that prophages are the major factor in generating genomic diversity in the O157 lineage (49).
Toxin production differed in human and bovine STEC isolates (54). The lysogen expresses Stx in case of low iron concentration (a typical growth-limiting factor for intestinal bacteria) (64), leading to intestinal hemorrhage (liberating iron from red blood cells leading to resumed bacterial growth and Stx downregulation). Notably, stx is under the control of the Fur repressor and thus part of a large bacterial iron-controlled regulon. Similarly, the diphtheria toxin encoded by a corynephage is under the control of the DTxR, the master repressor of an iron regulon in this gram-positive bacterium. Stx has no physiological secretion pathway and is only released by lysing cells. As antibiotics induce the prophage, chemotherapy can result in an aggravation of the clinical condition. In a fascinating illustration of the selfish gene concept, STEC recruits bystander intestinal E. coli cells via infection with the released Stx phage for an amplification of the suicidal Stx production (29). Experiments with mice demonstrated that the resistance or susceptibility pattern of the intestinal flora towards the released Stx phage exerts either a protective or an enhancing effect on the severity of STEC infections. Clinically, this observation could explain the variability of the disease symptoms in different O157-infected patients.
The observation that pathogenic bacteria convert environmental and commensal bystander bacteria via lysogenization into toxin-producing bacteria is not restricted to STEC (55) but was also found in Vibrio spp. (26) and Streptococcus pyogenes (11).
PHAGE RESISTANCE
Phages are a nuisance in the dairy industry, where they interfere with industrial milk fermentation when they attack starter bacteria. Therefore, food microbiologists have intensively studied phage-host interactions. Lactic acid bacteria were under substantial evolutionary pressure to develop antiphage strategies, as demonstrated by numerous phage resistance systems naturally found in lactococci. These mechanisms range from the blocking of phage DNA injection (45) to abortive infection systems (abiA to abiU) interfering with phage DNA replication, RNA transcription, phage development, and morphogenesis. The evolutionary arms race between bacteria and their phages has been well documented by dairy microbiologists. For example, against bacterial restriction systems, phages used different escape strategies covering either the loss of restriction sites (47) or the gain of a methylase gene (34). Acquisition of chromosomal DNA by lactococcal phages confronted with abi mechanisms has been repetitively demonstrated (6, 24). The high number of prophages in Lactococcus lactis offers in fact a large supply of new genetic material to superinfecting phages which is accessible via homologous recombination. Interestingly, the rich knowledge on phage resistance mechanisms acquired in the dairy field could also contribute to the understanding of the coexistence of phages and bacteria in the marine environment or the management of phage infections in humans (e.g., the cigarette smoke-induced prophage propagation purportedly linked to a decline of the protective Lactobacillus flora in the vagina which might open the way to vaginosis and candida superinfections) (36).
THE LOOK AHEAD: INTEGRATIVE BIOLOGY WITH PHAGES
Over the last 10 years, genome sequencing has fundamentally changed microbiology. However, a genomic sequence is fundamentally static. True biological processes are defined by the dynamic interaction of biological systems. Phage and bacterium interaction is conceptually the simplest case of such genome interaction. The infection process provides a precise kinetics for this interaction. Microarray analysis of phage transcription throughout the infection cycle is still limited to phage T4-infected E. coli cells (43). In the next logical step, microarray analysis from the invading phage must be integrated with that of the infected bacterium, especially when it concerns such a well-investigated organism as E. coli.
Phage infection is a highly dynamic process, probably inducing bacterial gene expression. Some of these events might even be visible in thin-section electron microscopy (Fig. 4). If the genomes of both the phage and the bacterium were sequenced, transcription of both genomes could be analyzed on microarrays at different time points after phage infection. If this analysis is combined with thin-section immune electron microscopy and proteomics, one could combine cell biology with gene and protein expression studies to examine the consequences of two genomes confronting each other in a hostile encounter.
FIG. 4.
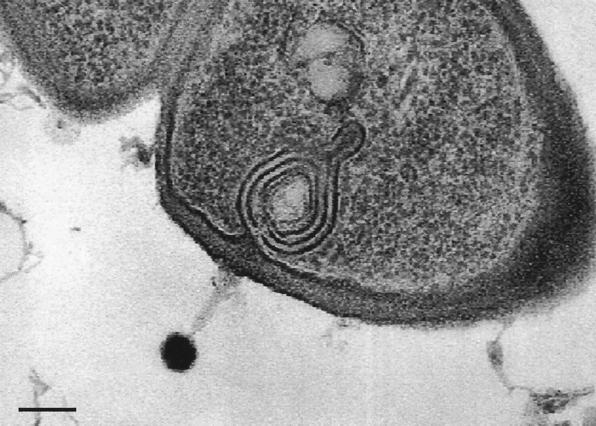
Phage-host interaction. Thin-section electron microscopy of myophage LP65 infecting an oral commensal Lactobacillus plantarum strain. Note the conspicuous membrane system underlying the phage adsorption site. The phage is the same as the myovirus shown in a negative-staining preparation in the central panel of Fig. 2. The bar is 100 nm.
However, the analysis must not stop here, since this corresponds still to the test tube situation that has dominated phage biology in the past. The phage research community should immediately envision the next level of complexity with interacting systems. An attractive test system is provided by streptococcal lysogens. Many prophages from low-GC-content, gram-positive bacteria encode candidate lysogenic conversion genes located between the phage lysin gene and the right attachment site (4). In lactic streptococci and in lactobacillus commensals, these prophage genes are expressed during broth culture (63). In contrast, the expression level of the corresponding prophage genes in pathogenic streptococci is low. Interestingly, when Streptococcus pyogenes comes in contact with pharyngeal cells (the natural target cells in streptococcal angina), the expression of the lysogenic conversion gene is upregulated (12). Broudy and coworkers have identified a low-molecular-weight compound released by the pharyngeal cell that induces the expression of this prophage gene (13). In this cell culture system, one has the chance to study by microarray analysis the interaction of a phage, a bacterium, and a human cell in a system that is directly relevant for the disease process. The mammalian host not only promotes prophage induction in S. pyogenes, but it also favors the lysogenization of nontoxigenic bystander bacteria resulting in the generation of new toxin-producing cells (11). Within the mammalian host, bacteria apparently alter not only their gene expression (46) but their genomes themselves.
There are still other suitable model systems to study phage-host interaction in a relevant ecological context. A lysogenic avian E. coli pathogen, when injected into a chicken, showed an upregulation of prophage gene expression (22). This and similar whole-animal experimental systems allow the analysis of genome interactions at the next complexity level. As the data analysis for such complex genome interactions will represent a substantial challenge, the old virtue of phages, i.e., “small is beautiful,” will again become an asset in the postgenomic era.
Acknowledgments
We thank Anne Constable for reading the review and Barry Dowsett (Salisbury, United Kingdom) for electron microscopy of the Staphylococcus phage.
We thank the Swiss National Science Foundation for the support of Sandra Chibani-Chennoufi (grant 5002-057832).
REFERENCES
- 1.Ashelford, K. E., M. J. Day, and J. C. Fry. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., J. C. Fry, M. J. Bailey, A. R. Jeffries, and M. J. Day. 1999. Characterization of six bacteriophages of Serratia liquefaciens CP6 isolated from the sugar beet phytosphere. Appl. Environ. Microbiol. 65:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barondess, J. J., and J. Beckwith. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346:871-874. [DOI] [PubMed] [Google Scholar]
- 4.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 8.Bratbak, G., M. Heldal, T. F. Thingstad, and P. Tuomi. 1996. Dynamics of virus abundance in coastal seawater. FEMS Microbiol. Ecol. 19:263-269. [Google Scholar]
- 9.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broudy, T. B., and V. A. Fischetti. 2003. In vivo lysogenic conversion of Tox− Streptococcus pyogenes to Tox+ with lysogenic streptococci or free phage. Infect. Immun. 71:3782-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broudy, T. B., V. Pancholi, and V. A. Fischetti. 2001. Induction of lysogenic bacteriophage and phage-associated toxin from group A streptococci during coculture with human pharyngeal cells. Infect. Immun. 69:1440-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broudy, T. B., V. Pancholi, and V. A. Fischetti. 2002. The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase. Infect. Immun. 70:2805-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruttin, A., F. Desiere, N. d'Amico, J. P. Guerin, J. Sidoti, B. Huni, S. Lucchini, and H. Brussow. 1997. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 63:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burroughs, N. J., P. Marsh, and E. M. Wellington. 2000. Mathematical analysis of growth and interaction dynamics of streptomycetes and a bacteriophage in soil. Appl. Environ. Microbiol. 66:3868-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushman, F. 2002. Lateral DNA transfer. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chibani-Chennoufi, S., J. Sidoti, A. Bruttin, E. Kutter, S. A. Sarker, and H. Brussow. 2004. Comparison of in vitro and in vivo bacteriolytic activity of Escherichia coli phages: implication for phage therapy. Antimicrob. Agents Chemother. [DOI] [PMC free article] [PubMed]
- 20.Cochran, P. K., and J. H. Paul. 1998. Seasonal abundance of lysogenic bacteria in a subtropical estuary. Appl. Environ. Microbiol. 64:2308-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danovaro, R., E. Manini, and A. Dell'Anno. 2002. Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 68:1468-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 24.Durmaz, E., and T. R. Klaenhammer. 2000. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl. Environ. Microbiol. 66:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edlin, G., L. Lin, and R. Kudrna. 1975. Lambda lysogens of E. coli reproduce more rapidly than non-lysogens. Nature 255:735-737. [DOI] [PubMed] [Google Scholar]
- 26.Faruque, S. M., M. M. Rahman, Asadulghani, K. M. Nasirul Islam, and J. J. Mekalanos. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXΦ. Infect. Immun. 67:5723-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 28.Furuse, K. 1987. Distribution of coliphages in the general environment: general considerations, p. 87-124. In S. M. Goyal, C. Gerba, and G. Bitton (ed.), Phage ecology. John Wiley & Sons, New York, N.Y.
- 29.Gamage, S. D., J. E. Strasser, C. L. Chalk, and A. A. Weiss. 2003. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect. Immun. 71:3107-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goode, D., V. M. Allen, and P. A. Barrow. 2003. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microbiol. 69:5032-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haggard-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennes, K. P., and M. Simon. 1995. Significance of bacteriophages for controlling bacterioplankton growth in a mesotrophic lake. Appl. Environ. Microbiol. 61:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hennes, K. P., C. A. Suttle, and A. M. Chan. 1995. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl. Environ. Microbiol. 61:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill, C., L. A. Miller, and T. R. Klaenhammer. 1991. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 173:4363-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang, S. C., and J. H. Paul. 1997. Significance of lysogeny in the marine environment: studies with isolates and a model of lysogenic phage production. Microb. Ecol. 35:235-243. [DOI] [PubMed] [Google Scholar]
- 36.Kilic, A. O., S. I. Pavlova, S. Alpay, S. S. Kilic, and L. Tao. 2001. Comparative study of vaginal Lactobacillus phages isolated from women in the United States and Turkey: prevalence, morphology, host range, and DNA homology. Clin. Diagn. Lab. Immunol. 8:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogfelt, K. A., L. K. Poulsen, and S. Molin. 1993. Identification of coccoid Escherichia coli BJ4 cells in the large intestine of streptomycin-treated mice. Infect. Immun. 61:5029-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kudva, I. T., S. Jelacic, P. I. Tarr, P. Youderian, and C. J. Hovde. 1999. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl. Environ. Microbiol. 65:3767-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, L., R. Bitner, and G. Edlin. 1977. Increased reproductive fitness of Escherichia coli lambda lysogens. J. Virol. 21:554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, M., R. Deora, S. R. Doulatov, M. Gingery, F. A. Eiserling, A. Preston, D. J. Maskell, R. W. Simons, P. A. Cotter, J. Parkhill, and J. F. Miller. 2002. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295:2091-2094. [DOI] [PubMed] [Google Scholar]
- 41.Loessner, M. J., M. Rudolf, and S. Scherer. 1997. Evaluation of luciferase reporter bacteriophage A511::luxAB for detection of Listeria monocytogenes in contaminated foods. Appl. Environ. Microbiol. 63:2961-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, Z., F. Breidt, V. Plengvidhya, and H. P. Fleming. 2003. Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 69:3192-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luke, K., A. Radek, X. Liu, J. Campbell, M. Uzan, R. Haselkorn, and Y. Kogan. 2002. Microarray analysis of gene expression during bacteriophage T4 infection. Virology 299:182-191. [DOI] [PubMed] [Google Scholar]
- 44.McDaniel, L., L. A. Houchin, S. J. Williamson, and J. H. Paul. 2002. Lysogeny in marine Synechococcus. Nature 415:496. [DOI] [PubMed] [Google Scholar]
- 45.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2002. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 43:509-520. [DOI] [PubMed] [Google Scholar]
- 46.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1993. Restriction/modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the dairy industry. Appl. Environ. Microbiol. 59:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neve, H., U. Kemper, A. Geis, and K. J. Heller. 1994. Monitoring and characterization of lactococcal bacteriophage in a dairy plant. Kiel. Milchwirtsch. Forschungsber. 46:167-178. [Google Scholar]
- 49.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pantucek, R., A. Rosypalova, J. Doskar, J. Kailerova, V. Ruzickova, P. Borecka, S. Snopkova, R. Horvath, F. Gotz, and S. Rosypal. 1998. The polyvalent staphylococcal phage phi 812: its host-range mutants and related phages. Virology 246:241-252. [DOI] [PubMed] [Google Scholar]
- 51.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 52.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect. Immun. 62:5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poulsen, L. K., T. R. Licht, C. Rang, K. A. Krogfelt, and S. Molin. 1995. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J. Bacteriol. 177:5840-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie, J. M., P. L. Wagner, D. W. Acheson, and M. K. Waldor. 2003. Comparison of Shiga toxin production by hemolytic-uremic syndrome-associated and bovine-associated Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 69:1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage Φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scholl, D., S. Rogers, S. Adhya, and C. R. Merril. 2001. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schrader, H. S., J. O. Schrader, J. J. Walker, T. A. Wolf, K. W. Nickerson, and T. A. Kokjohn. 1997. Bacteriophage infection and multiplication occur in Pseudomonas aeruginosa starved for 5 years. Can. J. Microbiol. 43:1157-1163. [DOI] [PubMed] [Google Scholar]
- 58.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1051. [DOI] [PubMed] [Google Scholar]
- 61.Suttle, C. A. 1994. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28:237-243. [DOI] [PubMed] [Google Scholar]
- 62.Suttle, C. A., and A. M. Chan. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ventura, M., C. Canchaya, M. Kleerebezem, W. M. de Vos, R. J. Siezen, and H. Brussow. 2003. The prophage sequences of Lactobacilllus plantarum strain WCFS1. Virology 316:245-255. [DOI] [PubMed] [Google Scholar]
- 64.Wagner, P. L., J. Livny, M. N. Neely, D. W. Acheson, D. I. Friedman, and M. K. Waldor. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957-970. [DOI] [PubMed] [Google Scholar]
- 65.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinbauer, M. G., and M. G. Hofle. 1998. Size-specific mortality of lake bacterioplankton by natural virus communities. Aquat. Microb. Ecol. 15:103-113. [Google Scholar]
- 67.Wiggins, B. A., and M. Alexander. 1985. Minimum bacterial density for bacteriophage replication: implications for significance of bacteriophages in natural ecosystems. Appl. Environ. Microbiol. 49:19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wommack, K. E., R. T. Hill, T. A. Muller, and R. R. Colwell. 1996. Effects of sunlight on bacteriophage viability and structure. Appl. Environ. Microbiol. 62:1336-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wommack, K. E., J. Ravel, R. T. Hill, and R. R. Colwell. 1999. Hybridization analysis of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 65:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woods, D. R. 1976. Bacteriophage growth on stationary phase Achromobacter cells. J. Gen. Virol. 32:45-50. [DOI] [PubMed] [Google Scholar]
- 72.Zhong, Y., F. Chen, S. W. Wilhelm, L. Poorvin, and R. E. Hodson. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]