Abstract
In a process known as quorum sensing, bacteria communicate with one another by producing, releasing, detecting, and responding to signal molecules called autoinducers. Vibrio harveyi, a marine pathogen, uses two parallel quorum-sensing circuits, each consisting of an autoinducer-sensor pair, to control the expression of genes required for bioluminescence and a number of other target genes. Genetic screens designed to discover autoinducer-regulated targets in V. harveyi have revealed genes encoding components of a putative type III secretion (TTS) system. Using transcriptional reporter fusions and TTS protein localization studies, we show that the TTS system is indeed functional in V. harveyi and that expression of the genes encoding the secretion machinery requires an intact quorum-sensing signal transduction cascade. The newly completed genome of the closely related marine bacterium Vibrio parahaemolyticus, which is a human pathogen, shows that it possesses the genes encoding both of the V. harveyi-like quorum-sensing signaling circuits and that it also has a TTS system similar to that of V. harveyi. We show that quorum sensing regulates TTS in V. parahaemolyticus. Previous reports connecting quorum sensing to TTS in enterohemorrhagic and enteropathogenic Escherichia coli show that quorum sensing activates TTS at high cell density. Surprisingly, we find that at high cell density (in the presence of autoinducers), quorum sensing represses TTS in V. harveyi and V. parahaemolyticus.
Many bacteria utilize a cell-cell communication process termed quorum sensing to monitor cell population density and to regulate gene expression in response to fluctuations in cell numbers (39). Quorum-sensing bacteria make, release, and respond to signaling molecules called autoinducers (43). Specifically, as a population of autoinducer-producing bacteria grows in number the extracellular autoinducer concentration increases proportionally. When a critical threshold autoinducer concentration is reached, the bacteria detect the signal and respond as a group by coordinately altering gene expression. Behaviors controlled by quorum sensing are typically ones that to be successful require the synchronized activity of many cells. For example, quorum sensing regulates functions such as conjugation, secretion of virulence factors, antibiotic production, biofilm formation, and bioluminescence (for reviews, see references 18 and 39).
The marine bacterium Vibrio harveyi uses quorum sensing to control the expression of bioluminescence and other target genes (31, 42). This bacterium is found swimming freely in the seawater, attached to abiotic surfaces, and as a constituent of biofilm consortia in marine animals (43). V. harveyi is a significant marine pathogen and is known to secrete cysteine proteases and a variety of other exotoxins that contribute to virulence in aquatic animals, including juvenile shrimp, fish, and lobsters (16, 32, 33, 58, 62, 63).
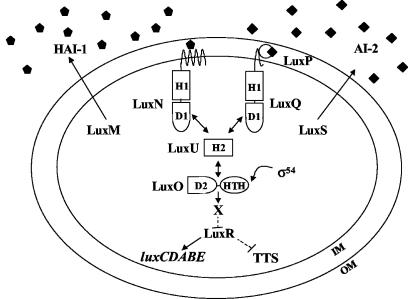
The V. harveyi quorum-sensing system is composed of two parallel two-component sensory circuits that converge to control targets such as luxCDABE, the luciferase operon (Fig. 1) (2, 3). System 1 of V. harveyi is comprised of the autoinducer signal HAI-1 (for V. harveyi autoinducer 1) and its two-component sensor LuxN. HAI-1 is the homoserine lactone (HSL) N-(3-hydroxybutanoyl)-HSL, and its production requires the LuxM synthase (formerly called LuxLM) (2, 10). System 2 is made up of the signal AI-2, which is the furanosyl borate diester 3A-methyl-5,6-dihydro-furo [2,3-d][1,3,2] dioxaborole-2,2,6,6A tetraol, and its cognate two-component sensor LuxQ (3, 12). AI-2 detection also requires the periplasmic AI-2 binding protein LuxP, which is proposed to interact with LuxQ (3, 12). AI-2 is produced by the LuxS synthase (48, 55). Sensory information from systems 1 and 2 converges at the phosphotransferase LuxU, and LuxU transmits the signal to the response regulator LuxO (19, 20). An unidentified repressor (X) and LuxR, a transcriptional activator, are also required for quorum-sensing regulation of luxCDABE (31, 35, 49).
FIG. 1.
Quorum sensing in V. harveyi. V. harveyi has two parallel quorum-sensing systems that regulate bioluminescence (luxCDABE) and other target genes. System 1 is composed of an autoinducer, HAI-1 (pentagons), and its cognate sensor LuxN. System 2 includes the autoinducer AI-2 (diamonds) and the sensor LuxPQ. The HAI-1 and AI-2 synthases are LuxM and LuxS, respectively. Information from both sensors is transduced to LuxU, and LuxU transmits the signal to LuxO. LuxO indirectly represses luxCDABE expression through a putative repressor we call X that appears to act at the level of luxR expression. LuxR is a transcriptional activator required for expression of luxCDABE. LuxR also controls TTS gene expression; however, in this case the action of LuxR is negative. We do not know whether this activity is direct or indirect. Details of the phosphorelay mechanism are given in the text. H and D stand for histidine and aspartate, which are the sites of phosphorylation. HTH denotes helix-turn-helix.
Genetic analyses have indicated that the V. harveyi quorum-sensing circuit functions as follows (Fig. 1). At low cell density, when the concentrations of HAI-1 and AI-2 are low the sensors LuxN and LuxQ act as kinases (19). They transfer phosphate to the shared phosphotransferase, LuxU, and LuxU subsequently transmits the phosphate to the downstream response regulator, LuxO. Phospho-LuxO is active and is proposed to work in conjunction with σ54 to activate the expression of an as-yet-unidentified repressor of Lux designated X (31). Phospho-LuxO through an unknown mechanism (that presumably involves X) represses expression of luxR (40). Because LuxR is required for transcription of luxCDABE, under low-cell-density conditions no light is produced. At high cell densities, interaction of the sensors LuxN and LuxPQ with their respective ligands, HAI-1 and AI-2, switches LuxN and LuxQ from kinase mode to phosphatase mode (21). The flow of phosphate in the system is reversed, which ultimately results in dephosphorylation of LuxO (19). In this state LuxO is inactive, which terminates the expression of X, the putative repressor of luxR expression. luxR is transcribed, and LuxR binds at the luxCDABE promoter and induces its expression (35, 40, 41). Thus, under high-cell-density conditions, light is produced.
Lux is not the exclusive target of quorum-sensing regulation in V. harveyi, as an earlier genetic screen for AI-2-regulated genes revealed 10 additional targets (42). Analysis of the regulation of these 10 targets showed that the autoinducers HAI-1 and AI-2 act synergistically and, furthermore, that both signals are required simultaneously for proper control of the target genes. This earlier work focused on information flow through the quorum-sensing circuit and not on the functions of the target genes. However, the analysis did reveal two genes with homology to components of type III secretion (TTS) systems, suggesting that V. harveyi could possess a quorum-sensing-regulated TTS system.
TTS systems are specialized secretion apparatuses used by many gram-negative plant and animal pathogens (for reviews, see references 13, 14, 15, and 50). These pathogens use TTS systems to inject effector virulence factors directly into the cytoplasm of eukaryotic host cells with which they are associated. TTS was first discovered and studied in Yersinia species. The Yersinia TTS channel is assembled from so-called Ysc (Yersinia secretion) and Lcr (low calcium response) proteins (38). This apparatus injects effector proteins known as Yops (Yersinia outer proteins) across the bacterial inner and outer membrane, past the eukaryotic cell membrane, and directly into the eukaryotic cell cytosol (8). Once inside the host cell, these effector proteins perform a range of functions that contribute to the propagation of the bacteria. TTS systems have subsequently been identified in numerous gram-negative bacterial pathogens, including enteropathogenic Escherichia coli and Vibrio parahaemolyticus (27, 34).
Over 20 proteins are generally required to form TTS channels (14). These proteins are highly conserved between pathogens that use TTS systems for virulence. In contrast, the effector proteins (Yops in Yersinia spp.) are not conserved between species. Unlike the proteins required for the channel, the effector proteins from different bacteria perform distinct functions. Presumably, the arsenal of effector proteins used by each bacterial pathogen has been optimized to aid in persistence in a specific host (reviewed in reference 26). Genes encoding TTS system components are generally clustered in pathogenicity islands or on virulence plasmids, although the organization of the clusters often differs between species (59).
In this report we identify and characterize three TTS gene clusters in V. harveyi and show that they are similar in sequence, organization, and regulation to the recently reported system of V. parahaemolyticus (34). We show that the V. harveyi and V. parahaemolyticus TTS systems are functional and capable of secretion only under conditions that simulate low cell density and that transcription of both the V. harveyi and V. parahaemolyticus TTS system genes is repressed by autoinducers at high cell density. To our knowledge, this is the first report of TTS systems that are negatively regulated by quorum sensing.
MATERIALS AND METHODS
Bacterial strains and media.
The relevant genotypes of all V. harveyi strains and plasmids used in this study are listed in Table 1. E. coli JM109 {e14-(McrA−) recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15]} (Stratagene) was used for cloning of HAI-1-regulated target genes, and BL21(DE3) [F− ompT hsdSB (rB− mB−) gal dcm (DE3)] (Novagen) was used for VopD protein expression. E. coli strain JMH1123 is BL21(DE3) containing the protein expression vector pET-21b (Sigma). E. coli strains were grown at 37°C with aeration in Luria-Bertani broth (LB). V. harveyi was grown to high cell density (early stationary phase; optical density at 600 nm [OD600], ∼ 1) in autoinducer bioassay (AB) medium (24) at 30°C with aeration for β-galactosidase (β-Gal) assays. AB medium supplemented with 5 mM EGTA was used for secretion assays. AB HAI-1+ medium contained 10% (vol/vol) cell-free culture fluids from the V. harveyi luxS::Tn5 strain MM30 (i.e., HAI-1+ AI-2−). AB HAI-1− medium contained 10% V. harveyi luxM::Tn5 luxS::Cmr strain MM77 (i.e., HAI-1−, AI-2−) cell-free culture fluids. Cell-free culture fluids were prepared as described previously (2). V. harveyi was grown in heart infusion medium for all other experiments (19). Antibiotics (Sigma) were used at the following concentrations (in milligrams per liter) unless otherwise specified: ampicillin (Amp), gentamicin (Gm), kanamycin (Kan), and rifampin, 100; chloramphenicol (Cm) and tetracycline (Tet), 10. Polymyxin B was used at 50 U/liter. X-Gal (5-bromo-4-chloro-indoyl-β-galactopyranoside) (USB) was used at 40 mg/liter. The methods used to introduce lux mutations onto the chromosomes of V. harveyi strains have been described previously (2). Cosmids pJAF822 and pJAF783 were used for constructions involving luxO D47E and luxN D771A, respectively (19, 21). Cosmid pBB154 was used for restoration of the wild-type luxMN region (2).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| Strains | ||
| V. harveyi | ||
| BB120 | Wild type | 1 |
| BB721 | luxO::Tn5 | 4 |
| JAF548 | luxO D47E linked to Kanr | 19 |
| JAF633 | ΔluxM linked to Kanr | 21 |
| MM30 | luxS::Tn5 | 55 |
| MM77 | luxM::Tn5, luxS::Cmr | 42 |
| KM357 | luxR::Tn5 | This study |
| KM199 | vscP::mini-mulacZ Cmr | 42 |
| KM201 | vopB::mini-mulacZ Cmr | 42 |
| JMH385 | vopN::mini-mulacZ Cmr | This study |
| KM470 | vscP::mini-mulacZ Cmr, luxO D47E linked to Kanr | This study |
| KM476 | vopB::mini-mulacZ Cmr, luxO D47E linked to Kanr | This study |
| JMH401 | vopN::mini-mulacZ Cmr, luxOD47E linked to Kanr | This study |
| JMH70 | ΔluxM linked to Kanr, mini- mulacZ Cmr | This study |
| JMH97 | JMH70 with pLAFR2 | This study |
| JMH127 | JMH70 with pJAF822 | This study |
| JMH276 | JMH70 with pJAF783 | This study |
| V. parahaemo- lyticus | ||
| LM5312 | Wild type; opaque phenotype; BB22OP | 36 |
| LM4437 | Translucent phenotype; BB22TR opaR::Tn5 | 36 |
| Plasmids | ||
| pBR322 | Ampr | 9 |
| pRK2013 | Broad host range, tra Kanr | 17 |
| pPH1JI | Broad host range, tra, mob Gmr | 6 |
| pUC18 | NEB cloning vector, MCS in frame with lacZα | 61 |
| pET21-b | Protein expression vector, C-terminal six-His tag | Novagen |
| pLAFR2 | Broad-host-range cosmid; mob, Tetr | 22 |
| pJAF822 | pLAFR2 with luxO D47E linked to Kanr | 19 |
| pJAF783 | pLAFR2 with luxN D771A linked to Kanr | 21 |
| pBB154 | pLAFR2 with luxMN region | 2 |
| pJMH113 | VopD-His6 fusion in pET21-b | This study |
| pJMH25 | pLAFR2 with TTS system region | This study |
| pJMH33 | pLAFR2 with TTS system region | This study |
DNA manipulations.
DNA manipulations were performed as previously described (47). PCRs were conducted using Ex-Taq (Takera) polymerase according to the manufacturer's specifications. PCR products were purified using a PCR cleanup kit (Qiagen). Calf alkaline phosphatase was purchased from Roche. T4 ligase and all restriction endonucleases were purchased from New England Biolabs. Plasmid isolations from E. coli were performed using a Quick Spin kit (Qiagen) according to the manufacturer's protocol. All oligonucleotides were obtained from the Princeton University Syn/Seq facility, which also performed all sequencing reactions. Probes for Southern blot experiments were labeled with [α-32P]dATP (Amersham Biosciences) using an Amersham multiprime DNA labeling kit.
Genetic screen for genes regulated by HAI-1 in V. harveyi.
V. harveyi strain JAF633 (ΔluxM-Kanr) was mutagenized with the transposon mini-MulacZ (Cmr) as described previously (21, 35). Cmr colonies were arrayed on heart infusion Cm agar plates and subsequently stamped to AB HAI-1+ and AB HAI-1− plates containing the chromogen X-Gal to visualize β-Gal activity. Colonies were compared after 2 days of growth at 30°C. Those transposon insertion mutants exhibiting differential β-Gal activity under the plus and minus HAI-1 conditions were further analyzed. Transcription of reporter fusions was quantitated by performing β-Gal assays as previously described (42, 51), and the β-Gal activity of strains grown to early stationary phase (OD600 ∼ 1) in AB HAI-1+ liquid medium was compared with the activity of the strains when grown in AB HAI-1− liquid medium. These β-Gal units were used to calculate the HAI-1 severalfold induction or repression for each lacZ fusion strain. Genomic DNA adjacent to the transposon insertions was amplified by a two-round PCR procedure or by direct cloning of the transposon fusion junctions. These procedures have been described previously (42, 56). The amplified or cloned DNA fragments were sequenced to determine their identities.
Identification and sequencing of the V. harveyi TTS system locus.
PCR-amplified DNA adjacent to the vopN::mini-MulacZ transposon in strain JMH70 was used to probe a V. harveyi genomic cosmid library (2, 56). Cosmids that hybridized to the probe were isolated and digested with either EcoRI or HindIII, and the digests were probed with the same vopN-containing fragment from JMH70. Hybridizing fragments were subcloned and sequenced. To extend the DNA sequence, regions at the termini of our sequenced subclones were used to reprobe the genomic DNA contained in the cosmids following digestion with EcoRV, SaeI, or XbaI. All hybridizing DNA fragments were subcloned into the pUC18 cloning vector (New England Biolabs) for sequencing. Overlapping fragments of V. harveyi genomic sequence were assembled and analyzed using Sequencher version 4.1 (Gene Codes) and NCBI Blast (http://www.ncbi.nlm.nih.gov/BLAST/) and the BLASTX program. Protein molecular masses were calculated with Protparam (http://us.expasy.org/tools/protparam.html). The following primers were used to amplify V. harveyi vscF: 5′-CGA TGC GAC CAA CAG TG-3′ and 5′-CAC CTT CTG CAG GAT CG-3′.
Expression, purification, and analysis of VopD.
vopD was amplified from the genome of V. harveyi with the following primers: 5′-GCG AAG CTT ACT GGA CGC GCT GAC CTT TAC-3′ and 5′-GCG CTC GAG TAC CGT AGG GAT AGA GGC-3′. These primers introduce upstream and downstream HindIII and XhoI sites, respectively, and were used for cloning into the expression vector pET-21b to generate a C-terminal VopD-6His protein fusion. The cloned product was sequenced to verify its correct construction. The VopD-6His recombinant vector (pJMH113) was maintained in E. coli BL21(DE3). This strain, JMH1113, was grown overnight in LB-Amp (200 mg/liter) at 37°C with aeration. The culture was diluted 1:100 into fresh LB-Amp (200 mg/liter) supplemented with 0.2% glucose and grown at 30°C with aeration to an OD600 of 0.8, whereupon 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma) was added and the culture was incubated for an additional 7 h. The cells were harvested by centrifugation, resuspended at 1:100 volume in 10 mM β-mercaptoethanol-20 mM Tris-HCl (pH 7.5)-20 mM NaCl-5 μg of DNAse I/ml, and lysed via a French press. The sample was subjected to centrifugation (Eppendorf model 5415C) at maximum speed at 4°C for 1 h to pellet the lysed cells. The supernatants were discarded, and the insoluble fraction was resuspended in 8 M urea for 10 min at 25°C. The sample was subjected to centrifugation as described above to remove any remaining insoluble material, and the cleared supernatant was applied to a His-affinity nickel resin column (Sigma). The VopD-6His fusion protein was purified according to HIS-Select HC nickel affinity gel denaturing conditions protocols (Sigma). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to confirm the size of the protein fusion. To prepare polyclonal antibodies against VopD, 400 μg of purified VopD-6His protein was resuspended in a 1:1 volume of Freund's complete adjuvant (Sigma) and injected into a New Zealand White rabbit. At 2-week intervals, rabbits were given booster injections with 200 μg of VopD-His in a 1:1 volume of Freund's incomplete adjuvant (Sigma). Polyclonal sera were adsorbed to JMH1123 E. coli lysates prior to performing Western blot analyses. Purified LuxS-6His was kindly provided by F. Hughson. Anti-LuxS polyclonal antibodies were prepared as described above. The Princeton University Animal Facility generated all polyclonal antibodies used in this study.
VopD secretion assays.
V. harveyi cultures were grown for 14 h in AB medium with aeration at 30°C. The cultures were diluted 1:10 in fresh AB medium and supplemented with 5 mM EGTA. Cultures were incubated at 30°C with shaking for an additional 6 to 7 h. Cultures were normalized to the same OD600, and approximately 2 × 108 cells of each strain were pelleted by centrifugation (Eppendorf Model 5415C) at maximum speed, resuspended in 75 μl of SDS-PAGE sample buffer, and boiled for 10 min (whole-cell lysate). The remainder of each culture was subjected to centrifugation for 5 min at 4,000 × g (Sorvall GLC-2B). The supernatants were removed and filtered through 0.22-μm-pore-diameter syringe filters (Millipore). A total of 33 μg of bovine serum albumin (BSA) (New England Biolabs) was added to a volume of cell-free culture fluid containing 5 × 108 cells. Proteins were concentrated approximately 20-fold by trichloroacetic acid (TCA) precipitation. Protein pellets were resuspended in 50 μl of SDS-PAGE sample buffer and boiled for 10 min (concentrated cell-free culture fluid). Two identical 12.5% PAGE gels were prepared; one was used for protein visualization by Coomassie staining, and the other was used for Western blot analysis. Following Western transfer, the nitrocellulose membranes were divided in half; the top portion was probed with antibody against VopD, and the lower portion was probed with anti-LuxS antibody. Specifically, 1:10,000 dilutions of the VopD and LuxS primary antisera and a 1:5,000 dilution of anti-rabbit immunoglobulin G (heavy plus light chains)-horseradish peroxidase conjugate (Promega) were incubated with the Western blot membranes. Enhanced-chemiluminescence ECL Western blotting substrate and protocol were utilized for VopD and LuxS detection (Amersham). The BSA protein band was visualized by high-sensitivity Coomassie staining as described previously (60).
Nucleotide sequence accession number.
The nucleotide sequence of the V. harveyi TTS system locus and the translated protein sequences have been deposited into the GenBank database under the accession number AY524044.
RESULTS
Identification of quorum-sensing-regulated genes in V. harveyi.
The autoinducers HAI-1 and AI-2 are hypothesized to be used for intracellular and intercellular communication, respectively (1), and our initial investigations have focused on determining whether the different autoinducer input states (no autoinducer, HAI-1 only, AI-2 only, and HAI-1 plus AI-2) regulate identical or distinct subsets of genes. To that end, we previously performed a genetic screen in V. harveyi to identify genes that are controlled by the exogenous addition of AI-2 in an HAI-1-producing strain and discovered 10 genes, including two putative TTS homologues (42). Interestingly, these genes only displayed significant induction/repression in the simultaneous presence of both HAI-1 and AI-2. This initial finding led us to hypothesize that the V. harveyi quorum-sensing circuit may act as a coincidence detector that distinguishes the presence of both autoinducers from the other three potential input states. The requirement for both autoinducers for the regulation of these targets was not surprising, because they were isolated on the basis of differential expression under HAI-1+, AI-2+conditions compared to HAI-1+, AI-2− conditions. Here we report the results of the reciprocal screen, that is, a screen for differential gene expression in an HAI-1−, AI-2+ V. harveyi strain following the addition of HAI-1. We are currently performing a screen for genes regulated by either or both autoinducers in an HAI-1−, AI-2− V. harveyi strain.
To identify genes regulated by HAI-1, the ΔluxM (i.e., HAI-1−) strain JAF633 was mutated with the transposon mini-MulacZ (Cmr) (11). Insertion mutants were arrayed on replicate agar grids containing 10% cell-free culture fluids prepared from the luxS::Tn5 strain (i.e., HAI-1+, AI-2−) strain MM30 or from the luxM::Tn5 luxS::Cmr (i.e., HAI-1−, AI-2−) strain MM77. Of 10,000 fusions assayed, 36 exhibited differential lacZ expression in the presence and absence of HAI-1. As mentioned previously, in the screen by Mok et al. for AI-2-regulated targets all of the targets identified that depended on AI-2 also required the simultaneous presence of HAI-1 for their regulation (42). This was also the case in the screen conducted in the present study; that is, all of the HAI-1 regulated targets required the simultaneous presence of AI-2 for their control. While the screens had the potential to reveal classes of genes regulated only by HAI-1 or only by AI-2, they were not performed to saturation and such genes were not identified. Importantly, these screens could not identify genes that respond to the presence of one autoinducer in the absence of the other.
As reported previously, inverted repeats at the termini of the mini-MulacZ transposon prevent PCR amplification of the transposon-genome fusion junction (11, 37, 42). Using restriction digestion of the chromosome, we successfully cloned and sequenced eight of the HAI-1-controlled targets. The genes encode proteins with a variety of predicted functions, including a TTS system component, an enolase, a σ54-dependent response regulator, a hybrid sensor histidine kinase-response regulator, a methyl-accepting chemotaxis protein, and an uncharacterized protein containing a type 3 chitin-binding domain.
To us, the most interesting genes found in these screens are the ones encoding putative TTS proteins because these are potential virulence determinants. The HAI-1 screen revealed a gene with homology to yopN and popN of Yersinia species and Pseudomonas aeruginosa, respectively. We name this gene vopN. In the AI-2 screen we found genes that we call vscP and vopB. Presumably, TTS genes came out of both screens, because, as mentioned previously, all of the genes identified to date require both HAI-1 and AI-2 for regulation. Here we present a characterization of the TTS system in V. harveyi. The remainder of the genes identified in the autoinducer screens await further analysis.
Quorum-sensing regulation of vopN.
Quorum-sensing control of the transcription of the putative TTS gene vopN is shown in Fig. 2. In this experiment, we quantitated the change in expression of the vopN::mini-MulacZ fusion in response to the presence of HAI-1. The parent strain lacks luxM, so it does not produce endogenous HAI-1 but does produce AI-2. In the absence of HAI-1, the fusion is highly expressed (Fig. 2, leftmost black bar). Addition of exogenous HAI-1 causes a 74-fold reduction in lacZ expression (leftmost white bar). This result shows that HAI-1 represses the expression of vopN.
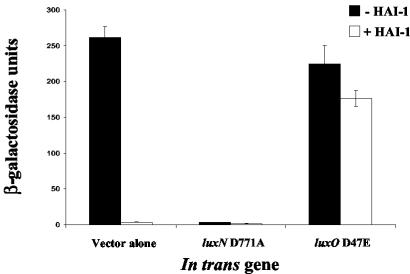
FIG. 2.
The LuxN-LuxO circuit controls the HAI-1-regulated targets. β-Gal assays were performed to measure the activity from the fusion in JMH70 (vopN::mini-MulacZ) with the following in trans alleles: vector alone (JMH97), luxN D771A (JMH276), and luxO D47E (JMH127). Cultures were supplemented with 10% V. harveyi MM77 (HAI-1−, AI-2−) cell-free culture fluids (black bars) or V. harveyi MM30 (HAI-1+, AI-2−) cell-free culture fluids (white bars). All experiments were performed in triplicate. JMH70 produces wild-type levels of AI-2.
We wondered whether the components of Lux quorum-sensing system 1 are required for transducing the HAI-1 signal to the vopN target. To investigate this, we introduced dominant alleles of luxN and luxO into the above-descrived V. harveyi vopN::mini-MulacZ transcriptional fusion strain and determined whether these alleles had any effect on lacZ reporter expression. To test the role of LuxN in HAI-1-mediated repression of the target, we introduced a luxN null mutation (luxN D771A) in trans that eliminates both the kinase and phosphatase activity of LuxN. The luxN D771A allele, when carried in trans in a strain containing wild-type luxN, confers the luxN null phenotype to the strain, presumably because it forms nonfunctional heterodimers with wild-type LuxN. Thus, this allele of LuxN makes the cells unresponsive to HAI-1 (21). Following introduction of luxN D771A, the level of activity of the vopN-lacZ fusion remains low (<3 U) in the absence and presence of HAI-1 (Fig. 2, middle bars). Therefore, LuxN is required to transduce the HAI-1 signal to the target gene. This result shows that when LuxN does not transmit information through the system, the fusion remains in a state mimicking high cell density. We suggest that the input from AI-2 via system 2 is responsible for this effect. We provide a fuller interpretation of these results in the Discussion.
To test whether LuxO is required for regulation of the vopN-lacZ fusion, we introduced the luxO D47E allele and assayed β-Gal activity in the absence and presence of exogenous HAI-1 (Fig. 2, rightmost black and white bars, respectively). This dominant LuxO allele mimics phospho-LuxO, and its presence “locks” the cells into the low-cell-density (i.e., no autoinducer) condition (19). When LuxO D47E is present, we observe high-level LacZ activity in the absence and presence of HAI-1, indicating that the fusion is not responsive to HAI-1 and, furthermore, that it is locked in a state mimicking the low-cell-density condition (compare to vector-alone control). This result demonstrates that like LuxN, LuxO is an essential component of the signaling pathway coupling HAI-1 to expression of vopN.
Identification and organization of a TTS system in V. harveyi.
Genes encoding TTS proteins are organized into clusters in many bacterial species. Therefore, we predicted that genes specifying additional TTS components in V. harveyi could be located in close proximity to the yopN-like vopN gene. We used PCR to amplify and clone ∼200 bp of V. harveyi genomic DNA adjacent to the vopN::mini-MulacZ transposon junction in JMH70, and this PCR product was used to probe a V. harveyi genomic cosmid library (2, 56). Cosmids from our library that hybridized to the vopN-containing probe from JMH70 were isolated and sequenced (see Materials and Methods). This analysis revealed many more putative TTS genes. The map of this region is shown in Fig. 3.
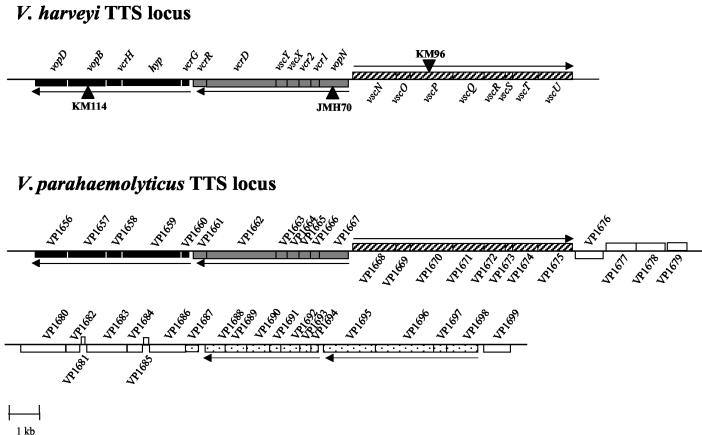
FIG. 3.
Organization of the TTS loci in V. harveyi and V. parahaemolyticus. The organizations of the loci encoding components of TTS systems in V. harveyi (accession number AY524044) and V. parahaemolyticus are shown. Transposon insertions obtained in V. harveyi in TTS system genes are denoted by black triangles and labeled as follows: KM114 (vopB::mini-MulacZ), JMH70 (vopN::mini-MulacZ), and KM96 (vscP::mini-MulacZ). Three homologous TTS system gene clusters found in both Vibrio species are indicated with black and gray shading and stripes and are aligned in the figure. Adjacent genes in V. parahaemolyticus that do not appear to encode TTS system components are shown with white boxes. Boxes indicating additional V. parahaemolyticus genes that encode proteins with homology to TTS functions that have not been identified in V. harveyi are marked with black dots. A horizontal arrow indicates the predicted direction of transcription of each operon.
We find three TTS system gene clusters in the vicinity of the yopN-like gene in the V. harveyi genome. These gene clusters are organized identically to the recently discovered TTS system locus of V. parahaemolyticus (34). Table 2 lists the putative TTS system components, which we have designated Vop, Vsc, or Vcr for Vibrio outer protein, Vibrio secretion, or Vibrio calcium response to correspond to their closest homologues based on TTS systems in other bacteria, most notably Yersinia and Pseudomonas spp. We identified 20 putative V. harveyi TTS system proteins. Of note are the gene we have called vscP on the basis of its location between vscO and vscQ and the gene we have called hyp (for hypothetical TSS protein) located between vcrG and vcrH. While we predict on the basis of their locations that these proteins are involved in TTS, apparent homologues are only found in V. parahaemolyticus and not in any other sequenced bacterium with a TTS system.
TABLE 2.
V. harveyi TTS proteins
| V. harveyi protein | No. of aaa residues | Molecular mass | Locus of highest aa homology | Predicted function | % aa identity |
|---|---|---|---|---|---|
| VscU | 349 | 38.7 | VP1675 | Translocation protein in TTS | 89 |
| VscT | 260 | 29.0 | VP1674 | Translocation protein in TTS | 87 |
| VscS | 88 | 9.5 | VP1673 | Translocation protein in TTS | 93 |
| VscR | 216 | 24.2 | VP1672 | Translocation protein in TTS | 96 |
| VscQ | 321 | 36.1 | VP1671 | Putative translocation protein in TTS | 70 |
| VscP | 431 | 47.7 | VP1670 | Putative translocation protein in TTS | 60 |
| VscO | 153 | 18.4 | VP1669 | Putative TTS protein | 76 |
| VscN | 440 | 48.0 | VP1668 | ATP synthase in TTS | 94 |
| VopN | 297 | 33.1 | VP1667 | Putative outer membrane protein | 84 |
| Vcr1 | 94 | 11.1 | VP1666 | Hypothetical protein in TTS | 74 |
| Vcr2 | 123 | 13.8 | VP1665 | Putative PopN chaperone TTS | 78 |
| VscX | 125 | 14.5 | VP1664 | Putative TTS protein | 74 |
| VscY | 112 | 12.9 | VP1663 | Putative TTS protein | 68 |
| VcrD | 705 | 77.9 | VP1662 | Low calcium response protein | 85 |
| VcrR | 137 | 15.9 | VP1661 | Putative regulator of TTS | 80 |
| VcrG | 96 | 10.9 | VP1660 | Putative regulator of TTS | 65 |
| Hyp | 605 | 66.6 | VP1659 | Putative TTS protein | 55 |
| VcrH | 163 | 18.2 | VP1658 | Low calcium response protein | 96 |
| VopB | 399 | 41.5 | VP1657 | Putative translocator protein | 70 |
| VopD | 334 | 35.6 | VP1656 | Putative translocator protein | 75 |
aa, amino acids.
In addition to the three gene clusters corresponding to the ones we identified in V. harveyi, V. parahaemolyticus possesses a conserved cluster of 12 genes that are predicted to be required to form a functional TTS apparatus (black dots in Fig. 33) (34). This region contains genes homologous to a putative TTS chaperone and the yscLKJIHGF and yscDCB genes of Yersinia spp. (44). In V. parahaemolyticus, 11 open reading frames of mostly unknown functions separate the fourth gene cluster from the three contiguous clusters described above. We sequenced roughly 4 kb upstream and 2 kb downstream of the region shown for V. harveyi in Fig. 3, but we did not identify any genes encoding probable TTS system components. However, using PCR primers flanking an internal 48-codon region of VP1694 (the putative yscF homologue in the fourth V. parahaemolyticus gene cluster) we were able to amplify a DNA fragment from the V. harveyi genome. Sequencing shows that this fragment is 95% identical to the V. parahaemolyticus VP1694 sequence (data not shown). This result leads us to believe that additional TTS components exist in V. harveyi and are likely conserved with those in V. parahaemolyticus.
Quorum sensing controls the transcription of TTS genes in V. harveyi.
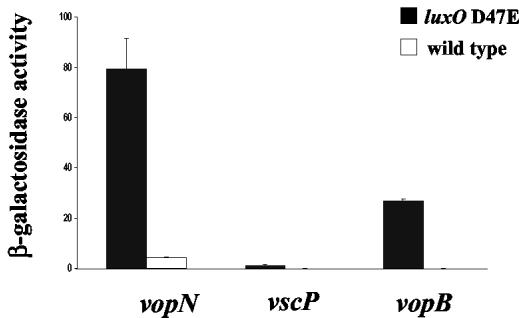
Our discovery that lacZ fusions to genes encoding putative TTS system components are regulated by autoinducers indicates that quorum sensing controls TTS. While not confirmed, sequence analysis reveals that each of the three fusions we obtained in our transposon mutageneses appears to reside in a different TTS system operon in V. harveyi (Fig. 3). Thus, investigation of the quorum-sensing regulation of vopB, vopN, and vscP gives us a means to examine quorum-sensing control of the expression of all three TTS system operons. For this analysis, we measured and compared the β-Gal activity produced by the three TTS system chromosomal fusions at high cell density in an otherwise wild-type V. harveyi strain and in a V. harveyi strain carrying the luxO D47E allele on the chromosome. As mentioned previously, LuxO D47E mimics phospho-LuxO, so the presence of this allele locks the cells into a state characteristic of low cell density (19). Therefore, this comparison provides us a measure of the expression of the operons in the presence and absence of autoinducer. We routinely use locked V. harveyi mutants that mimic the low-cell-density state to measure lacZ fusions to quorum-sensing controlled targets, because, unlike measurements of Lux which can be reliably carried out in both low- and high-cell-density cultures, accurately assaying LacZ activity in low-cell-density (<5 × 106 cells/ml) cultures is not possible. Figure 4 shows that transcription of all three TTS genes is higher in the luxO D47E strain (black bars) than in the wild-type strain (white bars). Thus, we infer that the three TTS system clusters are transcribed at low cell density in the absence of autoinducers and repressed at high cell density by the presence of autoinducers. The minimal and maximal activities of each fusion differ, which is expected since each fusion reports the expression of a distinct operon.
FIG. 4.
Quorum-sensing regulation of TTS system genes in V. harveyi. The activity levels of the vopN::mini-MulacZ, vscP::mini-MulacZ, and vopB::mini-MulacZ fusions were measured in V. harveyi containing luxO D47E on the chromosome to simulate low cell density (black bars) and in the wild-type V. harveyi background at high cell density (white bars). Strains were grown 14 h in AB medium. Experiments were performed in triplicate.
The TTS component VopD is produced and secreted at low cell density.
Although our limited sequencing allowed us to uncover many proteins required for TTS, we predict on the basis of analysis of other TTS systems that additional proteins must be required to assemble a functional channel. Additionally, our sequence analysis did not reveal genes encoding obvious TTS effectors. For these two reasons, we questioned whether the TTS system in V. harveyi is complete and functional or whether the locus we identified is simply a vestige of a virulence determinant that is no longer operational. To test whether the V. harveyi TTS system is intact and capable of secretion, we needed a method to monitor secretion through the channel. Importantly, our sequence analysis revealed vopD, encoding a protein similar to YopD and PopD of Yersinia and Pseudomonas spp., respectively. YopD and PopD are components of their respective TTS translocation channels, and under low-calcium conditions both are secreted into the extracellular environment (7, 23, 30, 45, 46, 57). We hypothesized that under low-calcium conditions, the V. harveyi protein VopD would be released into cell-free culture fluids. If so, we reasoned that monitoring VopD production and release would allow us to examine TTS gene expression and protein secretion in our V. harveyi quorum-sensing mutants. We purified VopD from recombinant E. coli and generated polyclonal antibodies to it to monitor its production and localization (intracellular versus extracellular).
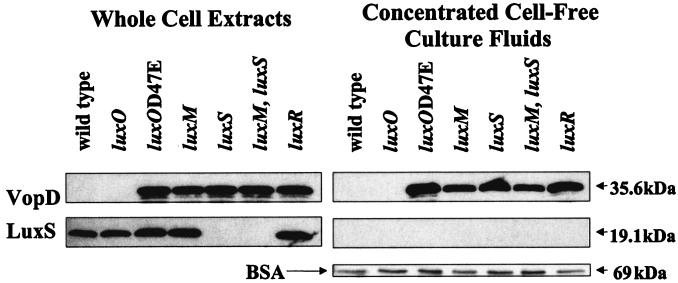
We grew various V. harveyi Lux regulatory mutants in low-calcium medium to induce secretion of VopD (see Materials and Methods). Western blotting was used to analyze intracellular VopD (whole-cell extracts) and secreted VopD (concentrated cell-free culture fluids). Figure 5 shows that at high cell density, VopD protein is not present in wild-type whole-cell extracts or in concentrated cell-free culture fluids. Based on Lux phenotypes, introduction of the luxO deletion into V. harveyi locks it into a state that constitutively mimics high cell density (4). Similar to the wild-type strain at high cell density, there is no detectable VopD in whole-cell extracts or in culture fluids prepared from the luxO null mutant. In contrast, VopD protein is present in cell extracts and in cell-free culture fluids prepared from all of the V. harveyi mutant strains that are locked into a form characteristic of low cell density (luxO D47E, luxM [HAI-1−], luxS [AI-2−] luxM, luxS [HAI-1−, AI-2−], and luxR). Taken together, the results in Fig. 5 show that the VopD protein is synthesized and secreted under low-cell-density but not high-cell-density conditions. These results are entirely consistent with those in Fig. 4 showing that the TTS system genes are only transcribed at low cell density.
FIG. 5.
The TTS protein VopD is produced and secreted under low-cell-density conditions. VopD present in whole-cell extracts (left panels) and concentrated cell-free culture fluids (right panels) was assayed by Western blotting using polyclonal antibodies directed against VopD. The strains of V. harveyi used in this analysis were grown in low-calcium, EGTA-containing medium and were as follows: BB120 (wild type), BB721 (luxO), JAF548 (luxO D47E), JAF633 (luxM), MM30 (luxS), MM77 (luxM, luxS), and KM357 (luxR). The nitrocellulose membranes were probed with anti-LuxS antibody as a control for leakage of cytoplasmic protein into the cell-free culture fluids. Gels containing identical samples of concentrated cell-free culture fluids were stained with Coomassie blue to visualize the BSA protein precipitation control.
To demonstrate that the released VopD is a result of secretion through a functional secretion apparatus and not due to leakage as a consequence of cell lysis, we monitored the constitutively produced, cytoplasmic protein LuxS in the same preparations used for the VopD analysis. The left panels of Fig. 5 show that the antibody directed against V. harveyi LuxS detects LuxS protein in all the whole-cell extracts except those made from strains lacking luxS (lanes 5 and 6 from the left). The right panels show that there is no LuxS in any of the concentrated cell-free culture fluid preparations, confirming that the presence of VopD in cell-free culture fluids is not due to problems with the integrity of the cells. To ensure that the absence of VopD in the cell-free culture fluids of high-cell-density wild-type and luxO strains was not due to improper TCA protein precipitation, we added BSA to the culture fluid samples prior to TCA precipitation. The right panels of Fig. 5 show these controls and that the differences in VopD observed in the culture fluids were not a consequence of differences in the amount of protein precipitated or loaded onto the gels.
A functional TTS apparatus is required for secretion but not for production of the TTS proteins.
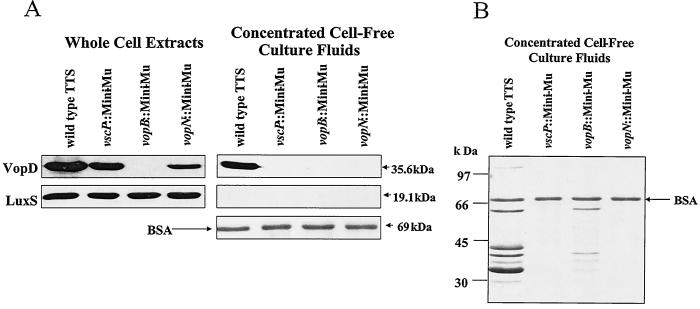
To demonstrate that VopD is secreted through the channel we have identified and not by some other unidentified mechanism or secretion system, we assayed secretion in mutant strains harboring insertions in components of the TTS apparatus. The luxO D47E mutation was transferred to the chromosomes of the V. harveyi strains carrying the mini-MulacZ insertions in vscP, vopB, and vopN. As described previously (Fig. 4 and 5), the luxO D47E strain constitutively expresses the three TTS system gene clusters containing these genes. VopD protein is produced in the luxO D47E strain that is wild type for the TTS locus and in the vscP::mini-MulacZ and vopN::mini-MulacZ mutants but not in the vopB::mini-MulacZ insertion mutant, presumably because the transposon in vopB is polar on vopD (Fig. 6A [whole-cell extracts] and Fig. 3). The VopD protein is only secreted in the strain that is wild type for all three TTS system operons (Fig. 6A [concentrated cell-free culture fluids]). We conclude that a functional TTS channel is not required for production of VopD. However, because VopD is not secreted in the vscP and vopN insertion mutant strains, which are presumably null for vscP and vscN and polar on downstream genes in their respective operons, we conclude that the TTS channel we have identified is required for VopD secretion. While we have only monitored VopD, we predict that this channel is capable of secretion of additional proteins because the proteins contained in extracellular fluids prepared from strains that are wild type for the TTS system are not identical to fluids made from TTS mutants (Fig. 6B). Specifically, extracellular fluid of the luxO D47E strain possessing a functional TTS apparatus contains seven bands that are not present in fluids from the luxO D47E vscP::mini-Mu and luxO D47E vopN::mini-Mu mutants. The luxO D47E vopB::mini-Mu mutant shows residual secretion of some of these proteins. Importantly, this insertion is near the end of one of the TTS operons (Fig. 3) and it is only polar on yopD. Some secretion occurs presumably because, as in other TTS systems, YopB and YopD are required for protein translocation but not for secretion per se (5, 25, 52). Fewer bands are detected because those corresponding to YopB (41 kDa) and YopD (36 kDa) are absent.
FIG. 6.
Transposon insertions in genes encoding the TTS system abolish secretion but not production of VopD in V. harveyi. (A) Whole-cell extracts (left panels) and concentrated cell-free culture fluids (right panels) of V. harveyi TTS fusions in the luxO D47E strain background were analyzed by Western blotting to visualize VopD and LuxS protein. The strain designated the wild-type TTS system is JAF548 (luxO D47E). The mini-MulacZ (Mini-Mu) insertion strains in vscP, vopB, and vopN in the luxO D47E background are KM470, KM476, and JMH401, respectively. The Coomassie-stained portion of the gel containing BSA is shown as a control for protein loading and precipitation. (B) Coomassie-stained gel containing cell-free culture fluids used for the Western blot shown in panel A.
Quorum sensing controls the expression of the TTS system of V. parahaemolyticus.
It is intriguing that we have found a functional TTS system in V. harveyi, because while several strains of V. harveyi are potent marine pathogens our laboratory wild-type strain appears to be avirulent in preliminary shrimp virulence assays (K. Burnett, unpublished data). The recently completed genome of V. parahaemolyticus predicts a TTS system very similar to the one we have identified in V. harveyi (34). V. parahaemolyticus is a marine bacterium, strains of which are pathogenic to humans, and TTS is proposed to be involved in virulence (T. Iida, personal communication). Interestingly, V. parahaemolyticus possesses all of the Lux regulators present in V. harveyi (i.e., LuxM [VP1967], LuxN [VP1968], LuxS [VP1967], LuxPQ [VPA1221 and VPA1220, respectively], LuxU [VP2098], LuxO [VP2099], and LuxR [called OpaR {VP2516} in V. parahaemolyticus]) but lacks the luciferase operon (34). Because of the apparent similarities between the quorum-sensing and TTS systems of V. parahaemolyticus and V. harveyi, we tested whether quorum sensing could also control TTS in V. parahaemolyticus.
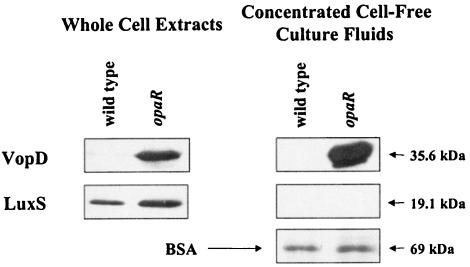
The VopDs and LuxSs of V. harveyi and V. parahaemolyticus share 75 and 94% amino acid identity levels, respectively. We reasoned, therefore, that we could use our antibodies to V. harveyi VopD and LuxS to detect V. parahaemolyticus VopD and LuxS. High-cell-density whole-cell extracts and concentrated cell-free culture fluids were prepared from wild-type V. parahaemolyticus and from a V. parahaemolyticus opaR transposon insertion mutant. As mentioned previously, OpaR is homologous to LuxR, and OpaR and LuxR are functionally interchangeable because both are capable of proper luxCDABE regulation (36). On the basis of these results, we presume that OpaR functions at the bottom of the quorum-sensing regulatory cascade in V. parahaemolyticus. Consistent with this hypothesis, in V. parahaemolyticus, as in V. harveyi, VopD is not made or secreted by the wild-type strain at high cell density (Fig. 7). However, Fig. 7 also shows that like the V. harveyi locked low-cell-density luxR mutant, the V. parahaemolyticus opaR mutant produces and secretes VopD. As in Fig. 5 and 6, the LuxS and BSA controls are shown in Fig. 7 to confirm that the V. parahaemolyticus VopD protein present in cell-free culture fluids is a result of secretion and not cell lysis or a problem with TCA precipitation. These results strongly suggest that the V. harveyi-like quorum-sensing circuit present in V. parahaemolyticus regulates TTS. While V. harveyi and V. parahaemolyticus are closely related evolutionarily, V. harveyi infects marine organisms whereas V. parahaemolyticus is capable of colonization of the human gastrointestinal tract. These differences make it critical to establish whether there is a requirement for TTS and quorum sensing in the virulence of both V. parahaemolyticus and V. harveyi.
FIG. 7.
Quorum sensing controls TTS in V. parahaemolyticus. Whole-cell extracts (left panels) and concentrated cell-free culture fluids (right panels) prepared from the V. parahaemolyticus wild type (strain LM5312) and the opaR null mutant (LM4437) were analyzed by Western blotting and probed with antibodies to V. harveyi VopD and LuxS. As described for Fig. 5 and 6A, the BSA control is shown.
DISCUSSION
This investigation focuses on the identification, organization, and quorum-sensing regulation of a TTS system in V. harveyi. In V. harveyi, quorum sensing regulates the expression of at least three gene clusters encoding a TTS system (Fig. 3 and 4), and we know this secretion channel is operational because it secretes at least the VopD protein (Fig. 5 and 6). Several independent methods were used to show that secretion occurs at low cell density and not at high cell density (Fig. 2 and 4 to 6). Additionally, a closely related marine bacterium, V. parahaemolyticus, a human pathogen, appears similar with respect to regulation of its TTS system (Fig. 7). The V. harveyi TTS is regulated via the known Lux circuitry (specifically HAI-1, LuxN, LuxO, and LuxR), and it is repressed by the presence of autoinducers (Fig. 2 and 5).
The above-described results, together with the knowledge that two quorum-sensing circuits function in parallel to control target gene expression in V. harveyi (42), allow us to interpret the results of the regulation experiment shown in Fig. 2. We suggest that the vopN fusion in JMH70 is expressed at low cell density in the absence of autoinducers and is repressed at high cell density by autoinducers (leftmost bars). Because HAI-1 and AI-2 act synergistically, both autoinducers are required simultaneously to mimic high-cell-density conditions (42). In this control experiment, the strain produces AI-2, which induces the phosphatase activity of LuxQ. However, LuxN is locked as a kinase in the absence of HAI-1; this competing activity results in a net phosphorylation of LuxO. This activity is sufficient to simulate low-cell-density conditions, and so the target fusion is highly expressed. Addition of HAI-1 converts LuxN to a phosphatase. When both LuxN and LuxQ are phosphatases, they promote the complete dephosphorylation of LuxO and the transition from low- to high-cell-density mode (19). Under this condition vopN is repressed. When the LuxN D771A null allele is present (middle bars), there is neither kinase nor phosphatase input from LuxN (21). However, the AI-2-dependent phosphatase activity of LuxQ leads to dephosphorylation of LuxO, which locks the cells into a state mimicking high cell density. Under these conditions the vopN target is repressed. Finally, the LuxO D47E allele constitutively mimics phospho-LuxO (right-hand bars) and locks the cell into low-cell-density mode regardless of the activity of the sensors (19). Under this condition vopN is highly expressed.
If regulatory functions other than LuxN and LuxO were responsible for controlling transcription of the vopN::mini-MulacZ fusion in JMH70, we would expect that the dominant in trans luxN D771A and luxO D47E alleles would have no impact on its expression. However, our experiments show that both LuxN and LuxO do regulate the lacZ fusion in JMH70. As mentioned previously, all 36 of the target genes we identified in this screen require LuxN and LuxO for HAI-1-mediated regulation. Because we know that LuxU is obligatory for the interaction between LuxN and LuxO, we infer that LuxU is also necessary for HAI-1-regulation of lacZ expression in JMH70 and the other fusion strains, although we did not explicitly test this assumption (20).
Our sequence analysis revealed 20 genes encoding putative TTS proteins. While we have yet to identify all of the components required for the assembly of the TTS channel, we assume these proteins are present in V. harveyi because the channel is capable of secretion. Clues to their location and identities come from the newly finished genome sequence of V. parahaemolyticus. As in V. harveyi, three adjacent operons on the V. parahaemolyticus large chromosome encode many of the proteins required for assembly of the secretion channel. In V. parahaemolyticus, another two gene clusters encoding an additional 12 TTS system proteins are separated by roughly 9.5 kb from the three contiguous TTS system operons (Fig. 3) (34). We suspect that an analogous region exists in V. harveyi, because oligonucleotide primers made for one member of this gene cluster in V. parahaemolyticus (the yscF homologue) were successfully used to amplify a genomic fragment from V. harveyi. The cosmids we isolated and used for analyzing the V. harveyi TTS system locus do not span the intervening sequence shown in Fig. 3. However, the vscF fragment itself can be used to probe our genomic library for cosmids that contain this predicted TTS system genomic region.
The TTS system region of V. parahaemolyticus depicted in Fig. 3 resides on the larger of the two V. parahaemolyticus chromosomes. In addition to this region, the small chromosome of V. parahaemolyticus contains a pathogenicity island that encodes several putative effector proteins, such as a YopP homologue (VPA1346) (34). DNA probes revealed that all virulent clinical strains of V. parahaemolyticus but not environmental strains possess the TTS system effectors on the small chromosome. The TTS system genes on the large chromosome of V. parahaemolyticus were present in every environmental and virulent clinical strain tested (34). We currently do not know whether V. harveyi possesses genes encoding effector proteins on its small chromosome. Because our wild-type laboratory strain of V. harveyi has a functional TTS system yet is avirulent in a shrimp model infection, it is possible that the strain lacks effectors. Analysis of this region of the genome of V. harveyi, when the sequence becomes available, could prove informative if, as in V. parahaemolyticus, possession of effectors correlates with ability to colonize a host and/or pathogenicity.
As a preliminary test for the presence of effectors in V. harveyi, we used Coomassie-stained SDS gels to analyze cell-free culture fluids prepared from various V. harveyi strains grown under low-calcium conditions (i.e., conditions promoting TTS). In comparison to wild-type cells grown to high cell density and cells with transposon insertions in vscP and vopN, cells of strains locked in low-cell-density mode that are wild type for the TTS system secrete at least seven additional proteins (Fig. 6B). We suspect that these proteins are TTS effectors and/or proteins like VopD and VopB, which are released by the channel under low-calcium conditions. We do not exclude the possibility that there are more than seven effectors-TTS components released because of the insensitivity of the Coomassie staining procedure.
Quorum sensing has previously been implicated in the control of TTS. Sperandio et al. (53, 54) reported that induction of expression of the TTS systems of both enterohemorrhagic and enteropathogenic E. coli requires luxS or intestinal hormones such as epinephrine for full activation. In both V. harveyi and V. parahaemolyticus, quorum sensing regulates TTS in a manner that is opposite to that of E. coli. The unexpected result that these marine bacteria secrete putative effectors at low cell density when they are presumably not in a community and terminate secretion when they are at high cell density could be important for understanding pathogenicity in clinically relevant marine bacteria such as V. parahaemolyticus. Importantly, while it has been established that TTS is necessary for virulence in many gram-negative bacterial species (13) and that TTS mutants of V. parahaemolyticus display reduced virulence (T. Iida, personal communication), TTS has not yet been shown to be required for pathogenicity in V. harveyi. To our knowledge, the data in the present manuscript provide the first evidence showing that quorum sensing negatively regulates TTS. Presumably, the differences in how E. coli and Vibrio spp. use quorum sensing to regulate TTS reflects optimization of a common phenomenon (cell-cell communication) for use in distinct niches.
Interestingly, we find that all the TTS phenotypes of V. harveyi are reciprocal to their bioluminescence phenotypes (Fig. 1). It has previously been observed that the V. harveyi quorum sensory system is capable of dual positive and negative regulation (31, 42). Reciprocal regulation of gene expression by the same sensory cascade is highly economical, and it likely benefits V. harveyi by facilitating the simultaneous activation and repression of distinct sets of genes. We do not yet understand why it is beneficial for V. harveyi to express particular traits such as bioluminescence at high cell density and other traits such as a TTS system at low cell density.
In V. harveyi, all the quorum-sensing-regulated targets we have studied are jointly regulated by HAI-1 and AI-2 and control is mediated by the known Lux circuit: LuxN or LuxPQ, together with LuxU, LuxO, and, importantly, LuxR (J. M. Henke and K. C. Mok, unpublished data). It is intriguing that LuxR is involved in control of all of the targets we have identified, because LuxR is known to be a transcriptional activator of luxCDABE and we observe both positive and negative regulation of the different targets. Perhaps LuxR acts as both an activator and a repressor, similar to what has been shown for its counterpart, HapR, in V. cholerae (28, 29). Alternatively, LuxR-mediated repression could be indirect and could proceed via the activation of a downstream repressor. Experiments are under way to determine whether, at high cell density, LuxR directly binds to the promoters of repressed genes, such as the TTS loci identified here.
The screens we have performed thus far to identify components of the quorum-sensing regulon have not been saturated, and additional classes of target genes, such as genes regulated exclusively by HAI-1 or AI-2, may exist. However, our investigations have begun to provide insight into the types of physiological processes that are under the control of cell-to-cell communication. Our identification of putative virulence factors such as TTS proteins that are controlled by quorum sensing, and, importantly, the unexpected nature of this control, could have significant consequences in the application of antimicrobial therapies aimed at interfering with cell-cell communication.
Acknowledgments
This work was supported by National Institutes of Health grant 1 R01 GM 065859 and National Science Foundation grant MCB004-94447.
We thank Fred Hughson and Stephan Schauder for the purified LuxS protein and LuxS polyclonal antibodies. Linda McCarter kindly supplied us with the V. parahaemolyticus strains LM5312 and LM4437. We are grateful to Karen Burnett for performing the V. harveyi shrimp virulence assays. We appreciate the Princeton Syn/Seq facility for sequencing and the Princeton Animal Facility for the VopD antibody preparation. We thank Karina Xavier, Michiko Taga, and Kenny Mok for insightful discussions.
REFERENCES
- 1.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403-412. [DOI] [PubMed] [Google Scholar]
- 5.Bergman, T., S. Håkansson, A. Forsberg, L. Norlander, A. Macellaro, A. Bäckman, I. Bölin, and H. Wolf-Watz. 1991. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role of LcrH and LcrV. J. Bacteriol. 173:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beringer, J. E., J. L. Beynon, A. V. Buchanan-Wollaston, and A. W. B. Johnston. 1978. Transfer of the drug resistance transposon Tn5 to Rhizobium. Nature 276:633-634. [Google Scholar]
- 7.Boland, A., M. P. Sory, M. Iriarte, C. Kerbourch, P. Wattiau, and G. R. Cornelis. 1996. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 15:5191-5201. [PMC free article] [PubMed] [Google Scholar]
- 8.Bolin, I., D. A. Portnoy, and H. Wolf-Watz. 1985. Expression of the temperature-inducible outer membrane proteins of yersiniae. Infect. Immun. 48:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolivar, F., R. L. Rodriquez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 10.Cao, J. G., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264:21670-21676. [PubMed] [Google Scholar]
- 11.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis, G. R. 2002. The Yersinia Ysc-Yop 'type III' weaponry. Nat. Rev. Mol. Cell Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 14.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 15.Cornelis, G. R., and H. Wolf-Watz. 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23:861-867. [DOI] [PubMed] [Google Scholar]
- 16.Diggles, B. K., G. A. Moss, J. Carson, and C. D. Anderson. 2000. Luminous vibriosis in rock lobster Jasus verreauxi (Decapoda: Palinuridae) phyllosoma larvae associated with infection by Vibrio harveyi. Dis. Aquat. Organ. 43:127-137. [DOI] [PubMed] [Google Scholar]
- 17.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donabedian, H. 2003. Quorum sensing and its relevance to infectious diseases. J. Infect. 46:207-214. [DOI] [PubMed] [Google Scholar]
- 19.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 20.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman, J. A., B. N. Lilley, and B. L. Bassler. 2000. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol. Microbiol. 35:139-149. [DOI] [PubMed] [Google Scholar]
- 22.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 23.Frithz-Lindsten, E., A. Holmstrom, L. Jacobsson, M. Soltani, J. Olsson, R. Rosqvist, and A. Forsberg. 1998. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol. Microbiol. 29:1155-1165. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg, E. P., J. W. Hastings, and S. Ulitzer. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 179:87-91. [Google Scholar]
- 25.Hakansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 26.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 29.Kovacikova, G., W. Lin, and K. Skorupski. 2003. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J. Bacteriol. 185:4825-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, V. T., and O. Schneewind. 1999. Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu. Mol. Microbiol. 31:1619-1629. [DOI] [PubMed] [Google Scholar]
- 31.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 32.Liu, P. C., and K. K. Lee. 1999. Cysteine protease is a major exotoxin of pathogenic luminous Vibrio harveyi in the tiger prawn, Penaeus monodon. Lett. Appl. Microbiol. 28:428-430. [DOI] [PubMed] [Google Scholar]
- 33.Liu, P. C., K. K. Lee, C. C. Tu, and S. N. Chen. 1997. Purification and characterization of a cysteine protease produced by pathogenic luminous Vibrio harveyi. Curr. Microbiol. 35:32-39. [DOI] [PubMed] [Google Scholar]
- 34.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 35.Martin, M., R. Showalter, and M. Silverman. 1989. Identification of a locus controlling expression of luminescence genes in Vibrio harveyi. J. Bacteriol. 171:2406-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarter, L. L. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180:3166-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metcalf, W. W., P. M. Steed, and B. L. Wanner. 1990. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J. Bacteriol. 172:3191-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michiels, T., J.-C. Vanooteghem, C. Lambert de Rouvroit, B. China, A. Gustin, P. Boudry, and G. R. Cornelis. 1991. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J. Bacteriol. 173:4994-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto, C. M., P. V. Dunlap, E. G. Ruby, and E. A. Meighen. 2003. LuxO controls luxR expression in Vibrio harveyi: evidence for a common regulatory mechanism in Vibrio. Mol. Microbiol. 48:537-548. [DOI] [PubMed] [Google Scholar]
- 41.Miyamoto, C. M., E. E. Smith, E. Swartzman, J. G. Cao, A. F. Graham, and E. A. Meighen. 1994. Proximal and distal sites bind LuxR independently and activate expression of the Vibrio harveyi lux operon. Mol. Microbiol. 14:255-262. [DOI] [PubMed] [Google Scholar]
- 42.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 45.Rosqvist, R., K. E. Magnusson, and H. Wolf-Watz. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosqvist, R., C. Persson, S. Hakansson, R. Nordfeldt, and H. Wolf-Watz. 1995. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib. Microbiol. Immunol. 13:230-234. [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 49.Showalter, R. E., M. O. Martin, and M. R. Silverman. 1990. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 172:2946-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silhavy, T. J. 1997. Death by lethal injection. Science 278:1085-1086. [DOI] [PubMed] [Google Scholar]
- 51.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 53.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 57.Tardy, F., F. Homble, C. Neyt, R. Wattiez, G. R. Cornelis, J. M. Ruysschaert, and V. Cabiaux. 1999. Yersinia enterocolitica type III secretion-translocation system: channel formation by secreted Yops. EMBO J. 18:6793-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teo, J. W., L. H. Zhang, and C. L. Poh. 2003. Cloning and characterization of a metalloprotease from Vibrio harveyi strain AP6. Gene 303:147-156. [DOI] [PubMed] [Google Scholar]
- 59.Winstanley, C., and C. A. Hart. 2001. Type III secretion systems and pathogenicity islands. J. Med. Microbiol. 50:116-126. [DOI] [PubMed] [Google Scholar]
- 60.Wong, C., S. Sridhara, J. C. Bardwell, and U. Jakob. 2000. Heating greatly speeds Coomassie blue staining and destaining. BioTechniques 28:426-428, 430, 432. [DOI] [PubMed] [Google Scholar]
- 61.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, X.-H., P. G. Meaden, and B. Austin. 2001. Duplication of hemolysin genes in a virulent isolate of Vibrio harveyi. Appl. Environ. Microbiol. 67:3161-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, X.-H., and B. Austin. 2000. Pathogenicity of Vibrio harveyi to salmonids. J. Fish Dis. 23:93-102. [Google Scholar]