Abstract
Aer is a membrane-associated protein that mediates aerotactic responses in Escherichia coli. Its C-terminal half closely resembles the signaling domains of methyl-accepting chemotaxis proteins (MCPs), which undergo reversible methylation at specific glutamic acid residues to adapt their signaling outputs to homogeneous chemical environments. MCP-mediated behaviors are dependent on two specific enzymes, CheR (methyltransferase) and CheB (methylesterase). The Aer signaling domain contains unorthodox methylation sites that do not conform to the consensus motif for CheR or CheB substrates, suggesting that Aer, unlike conventional MCPs, might be a methylation-independent transducer. Several lines of evidence supported this possibility. (i) The Aer protein was not detectably modified by either CheR or CheB. (ii) Amino acid replacements at the putative Aer methylation sites generally had no deleterious effect on Aer function. (iii) Aer promoted aerotactic migrations on semisolid media in strains that lacked all four of the E. coli MCPs. CheR and CheB function had no influence on the rate of aerotactic movements in those strains. Thus, Aer senses and signals efficiently in the absence of deamidation or methylation, methylation changes, methylation enzymes, and methyl-accepting chemotaxis proteins. We also found that chimeric transducers containing the PAS-HAMP sensing domain of Aer joined to the signaling domain and methylation sites of Tar, an orthodox MCP, exhibited both methylation-dependent and methylation-independent aerotactic behavior. The hybrid Aear transducers demonstrate that methylation independence does not emanate from the Aer signaling domain but rather may be due to transience of the cellular redox changes that are thought to trigger Aer-mediated behavioral responses.
Methyl-accepting chemotaxis proteins (MCPs) mediate many of the chemotactic behaviors of bacteria and archaea (see references 6 and 20 for recent reviews). MCPs typically possess an extracellular ligand-binding domain for monitoring environmental chemoeffector levels and an intracellular signaling domain that controls motor responses and undergoes reversible methylation at multiple sites. Methylation enables MCPs to detect chemical changes over time by comparing current chemoeffector levels, reflected in the fraction of ligand-occupied molecules, with chemoeffector levels during the past few seconds, represented by the average methylation state of the molecules. Whenever current conditions differ from those of the recent past, MCPs produce a feedforward excitation signal that modulates the organism's motility and a feedback adaptation signal that updates MCP methylation state to correspond to the new chemical environment (see references 12 and 33 for recent reviews). The ability of MCPs to adapt to homogeneous chemical environments via methylation changes endows these chemoreceptors with a wide dynamic range for stimulus detection and signaling (45).
Escherichia coli has four transmembrane MCPs that share a highly conserved signaling and methylation site domain: Tsr (serine chemoreceptor), Tar (aspartate and maltose chemoreceptor), Trg (ribose and galactose chemoreceptor), and Tap (dipeptide chemoreceptor). The recently discovered aerotaxis transducer, Aer, also has a C-terminal MCP-like signaling domain, but it lacks a periplasmic sensing domain (9, 37). Instead, Aer has a central hydrophobic segment that anchors it to the inner face of the cytoplasmic membrane and an N-terminal cytoplasmic PAS domain that binds flavin adenine dinucleotide (FAD) (8, 38). Aer is thought to produce aerotactic responses by detecting oxygen-related cellular redox changes with its FAD prosthetic group (reviewed in reference 50).
E. coli strains defective in CheR (MCP methyltransferase) and/or CheB (MCP methylesterase) function exhibit severely impaired chemotactic responses to stimuli sensed by Tar, Tap, Trg, and Tsr (35, 36, 56), demonstrating that the ability to modulate methylation state is crucial to those MCP functions. In contrast, sensory adaptation to aerotactic stimuli appears to be a methylation-independent behavior in E. coli (30). Niwano and Taylor (30) found that cheR mutants, or wild-type cells depleted of AdoMet by methionine starvation or cycloleucine treatment, had essentially unaltered response times for oxygen stimuli. Tar-, Trg-, and Tsr-mediated responses in the same cells were prolonged, indicating that methylation-dependent sensory adaptation had been compromised. These findings suggest that Aer, whose MCP-like signaling domain lacks conventional methylation site motifs, might be a methylation-independent transducer.
To determine whether aerotaxis mediated by Aer is a methylation-independent behavior, we examined Aer for CheR- or CheB-dependent covalent modifications, assessed Aer function in cheR and cheB mutants, and examined the behavior of methylation site mutants of Aer. These studies demonstrated that Aer-mediated aerotaxis was not dependent on CheR or CheB, on methylatable sites in the signaling domain, or on any of the conventional MCPs in E. coli. Moreover, a chimeric transducer containing the Aer sensing domain joined to the signaling domain and methylation sites of an orthodox MCP exhibited both methylation-dependent and methylation-independent aerotactic behavior. These findings indicate that sensory adaptation can occur at more than one step, and by more than one mechanism, in the Aer signaling pathway.
MATERIALS AND METHODS
Bacterial strains.
E. coli K-12 strains used in this work are listed in Table 1. All are derivatives of RP437, our reference wild type for chemotaxis studies (35).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant properties | Reference |
|---|---|---|
| Strains | ||
| RP437 | thr-1 leuB6 his-4 metF159 eda-50 (chemotaxis wild-type) | 35 |
| RP526 | mutD | 18 |
| RP9010 | recD::mini-Tn10-1903 | R. C. Stewart, personal communication |
| UU1117 | Δaer-1 | 9 |
| UU1247 | Δaer-1 ygjG::Gm | This work |
| UU1249 | Δaer-1 Δ(tar-cheR)2208 Δtsr-7028 Δtrg-100 ygjG::Gm zbd::Tn5 | This work |
| UU1250 | Δaer-1 Δ(tar-tap)5201 Δtsr-7028 Δtrg-100 ygjG::Gm zbd::Tn5 | This work |
| UU1535 | Δaer-1 Δ(tar-cheB)2234 Δtsr-7028 Δtrg-100 ygjG::Gm zbd::Tn5 | This work |
| Plasmids | ||
| pCJ30 | IPTG-inducible Ptac expression vector | 9 |
| pNP1 | pCJ30 with NdeI site at ATG start codon in polylinker | N. Pershing and P. Ames, unpublished results |
| pSB20 | pCJ30 aer+ (IPTG-inducible) | 9 |
| pSB137 | pSB20 Aer-D60N | 8 |
| pBSL142 | Carries Ω-Gm element | 1 |
| pRSETc | Cloning vector | 41 |
| pACYC184 | Cloning vector | 14 |
| pPA144 | pACYC184 tsr+ (native promoter) | 3 |
| pDM5 | pSB20 Aer[E482D/E483D] (Aer-4*) | This work |
| pDM6 | pSB20 Aer[Q286A/Q287A] (Aer-1*) | This work |
| pDM7 | pSB20 Aer[Q293A/Q294A] (Aer-2*) | This work |
| pDM8 | pSB20 Aer[N300A/Q301A] (Aer-3*) | This work |
| pDM9 | pSB20 Aer[K492A/H493A] (Aer-5*) | This work |
| pDM13 | pSB20 Aer[1*/2*/4*] | This work |
| pKG128 | pDM13 Aer[1/*2/*/4*/T431M] | This work |
| pSB90 | pSB20 Aer[C193A/C203A/C253A] (Aer-C3*) | 8 |
| pKG119 | pNP1 Aer-C3*[1-260]/Tar[269-553] (Aear-260) | This work |
| pKG122 | pNP1-Aer-C3*[1-269]/Tar[278-553] (Aear-269) | This work |
Plasmids.
Plasmids central to this work are listed in Table 1. Parental plasmids included pCJ30, an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Ptac expression vector (9); pSB20, a pCJ30 derivative that expresses wild-type aer (9); and pSB90, a pSB20 derivative that encodes a fully functional Aer protein with cysteine-to-alanine mutations at the three native cysteine residues in Aer (8).
Construction of ygjG::Gm.
A gentamicin resistance (Gm) insertion linked to the aer locus was constructed to facilitate introduction of chromosomal aer alleles into various strain backgrounds. The ygjG open reading frame is adjacent to the promoter-proximal end of aer (10). It encodes an ArgF-like protein that is functionally redundant in E. coli K-12 and causes no tight auxotrophy or other readily selectable phenotype (A. C. Miller and S. I. Bibikov, unpublished results). An internal portion of the ygjG coding region spanning a BssHI restriction site was PCR amplified from chromosomal DNA of RP437. The oligonucleotide primers introduced at the fragment ends a BamHI site and a PstI site, which were used to clone the ′ygjG′ fragment into the pRSETc vector. The gentamicin resistance gene in plasmid pBSL142 was excised with MluI and inserted into the compatible BssHI site of pRSETc-′ygjG′. A linear BamHI-PstI fragment carrying the Gm insertion and flanking segments of ygjG was transferred into RP9010 by electroporation, selecting for gentamicin-resistant transformants. The resulting ygjG::Gm insertion was 97% linked to the aer locus by P1 generalized transduction. A Δaer-1 ygjG::Gm donor strain (UU1247) was constructed in order to introduce the Δaer-1 mutation into other genetic backgrounds by P1 transduction.
Construction of Aer methylation site mutants.
Mutational changes at the putative methylation sites of Aer were constructed by oligonucleotide-directed mutagenesis of pSB20, using the QuikChange mutagenesis kit (Stratagene, La Jolla, Calif.). Candidate plasmid mutants were verified by sequencing the entire aer coding region and adjacent expression signals. Multiple mutants were constructed by successive rounds of site-directed mutagenesis.
Construction of Aer/Tar chimeras.
Aear hybrids were constructed by PCR amplification of the N-terminal aer coding segment (codons 1 to 260 and 1 to 269) from pSB90 using the KOD HiFi DNA polymerase (Novagen, Madison, Wis.). The PCR primers introduced an NdeI site at the aer start codon and either an AatII site (at codon 260) or a BglII site (at codon 269) at the other end of the coding fragment. The coding segment for the Tar signaling domain (tar codons 269 to 553 and 278 to 553) was amplified from plasmid pLC113 (4) with primers that introduced either an AatII site (at codon 269) or a BglII site (at codon 278) and a BamHI site at the other fragment end. The aer and tar PCR products were digested with AatII or BglII, ligated, and then used as a PCR template to amplify the hybrid gene. The amplified genes were gel isolated and inserted between the NdeI and BamHI sites of the expression vector pNP1. Both chimeric constructs (Aear-260 and Aear-269) were confirmed by sequencing the entire hybrid coding region.
Behavioral assays.
Chemotactic ability was assessed by rate of colony expansion on semisolid tryptone agar (34). Aerotaxis in UU1117 was assessed by colony migration on semisolid succinate minimal plates (9). Aerotaxis in UU1250, UU1249, and UU1535 was assessed by colony migration on semisolid tryptone agar. Plates were routinely incubated at 30°C.
Isolation of aerotactic pseudorevertants.
Plasmid pDM13 was mutagenized by passage through RP526 and then transformed into UU1250. Pooled transformants were spread in a line on the surface of a semisolid tryptone plate and incubated overnight. Aerotactic revertants that moved outward from the inoculum were picked for further analysis.
SDS-polyacrylamide gel electrophoresis (PAGE) analysis of Aer modification patterns.
Strains containing two compatible plasmids (pPA144 [Tsr] or pACYC184 [vector] and pSB20 [Aer] or pCJ30 [vector]) were grown at 35°C to early log phase in H1 minimal glycerol medium (34) containing 1% Casamino Acids, 100 μg of ampicillin/ml, and 50 μg of chloramphenicol/ml. Cells were harvested by centrifugation and lysed by boiling in sample buffer (26), as described previously (4). Lysate proteins were analyzed on sodium dodecyl sulfate (SDS)-containing 10 to 20% polyacrylamide gradient gels (44:0.3, acrylamide/bisacrylamide) and visualized with an anti-Tsr antibody, essentially as described previously (4).
Aer methyl incorporation test.
Cells were grown in tryptone broth at 35°C to mid-log phase and harvested by centrifugation. Cell pellets were washed twice in motility medium (10 mM potassium phosphate buffer [pH 7.0], 0.1 mM EDTA, 20 mM sodium lactate) containing 200 μg of chloramphenicol/ml and resuspended in the same medium plus 30 μg of puromycin/ml to inhibit protein synthesis. After incubation for 10 min at 30°C, l-[methyl-3H]methionine (specific activity, 36 mCi/μmol) was added to a final concentration of 3 μM. After an additional 45-min incubation at 30°C, cold trichloroacetic acid was added to a final concentration of 20%, and the cells were pelleted by centrifugation and washed with 200 μM acetone at 4°C. Cells were lysed in sample buffer and subjected to SDS-PAGE, as described previously (19). Gels were treated with 2,5-diphenyloxazole solution in dimethyl sulfoxide and exposed for 30 days, using preflashed film and a fluorescent screen.
RESULTS
Aer is not detectably modified by CheR or CheB.
A wild-type Aer plasmid (pSB20) was expressed in strains with various combinations of cheR and cheB defects to look for evidence of CheR- or CheB-dependent modifications. To provide an internal control in those experiments, the strains also carried a second compatible plasmid expressing wild-type Tsr, an orthodox MCP. Tsr carries a pentapeptide sequence (NWETF) at its C terminus that binds to CheR and CheB molecules and enhances the efficiency of the methylation and demethylation reactions (7, 55). Tsr molecules with this CheR/CheB tether can assist the methylation reactions of other Tsr molecules in trans (27) and probably assist other MCP receptors, e.g., Tap and Trg, that do not have CheR/CheB tethering sites (21, 53). Thus, we reasoned that if Aer, which lacks the NWETF tail, was a substrate for either enzyme, Tsr might assist those reactions.
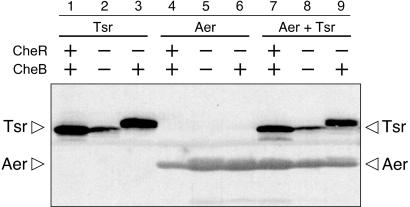
We first looked for Aer modifications in the form of CheR- or CheB-dependent band shifts in denaturing PAGE. For reasons not well understood, CheR-promoted methylation increases the SDS-PAGE mobility of MCP molecules (13, 15, 19), whereas CheB-promoted deamidation and demethylation reduce their mobility (24, 25, 40, 43). Although the actual mass changes are miniscule, the band shifts correspond in magnitude to apparent molecular mass differences of several thousand Daltons. Under these gel conditions, Tsr exhibited the expected CheR/CheB-dependent band shifts (Fig. 1). In a host with neither CheR nor CheB function, Tsr migrated as a single band of intermediate mobility corresponding to the unmethylated, undeamidated form (Fig. 1, lanes 2 and 8). In a host that lacked CheR function but had CheB function, Tsr migrated as a single band with slow mobility, corresponding to the unmethylated, but deamidated, form (Fig. 1, lanes 3 and 9). In a host with both CheR and CheB function, Tsr formed several faster-migrating bands representing different methylation states (Fig. 1, lanes 1 and 7). In contrast, Aer either alone or in the presence of Tsr migrated as a single major band at the same position in all three host strains (Fig. 1, lanes 4 to 9). (A minor, CheR/CheB-independent band apparent in some of the Aer lanes might represent a different posttranslational processing event.) These results suggest that Aer is not modified by CheR and CheB under physiological conditions where Tsr is modified. However, we cannot exclude the possibility that Aer undergoes CheR or CheB modifications that are not detectable as band shifts in the gel electrophoresis system we used, despite its suitability for other MCPs. To address this concern, we also attempted to incorporate tritium-labeled methyl groups (from 3H-methyl methionine via S-adenosyl methionine) into Aer, using the same strains as in the band shift experiments. Tsr readily incorporated the methyl label, but Aer did not (data not shown). We conclude that if Aer is capable of accepting methyl groups, it did so below the detection limits of our assay.
FIG. 1.
Absence of CheR- and CheB-dependent Aer band shifts in SDS-PAGE. Protein extracts of strains containing pPA144 (Tsr), pSB20 (Aer), or both plasmids were analyzed on 10 to 20% gradient gels, as described in Materials and Methods. Tsr and Aer were visualized by Western blotting with a polyclonal antiserum directed against residues 290 to 470 of the Tsr signaling domain (2). Host strains were UU1250 (CheR+ CheB+), UU1535 (CheR− CheB−), and UU1249 (CheR− CheB+).
CheR, CheB, and MCPs are not required for aerotaxis.
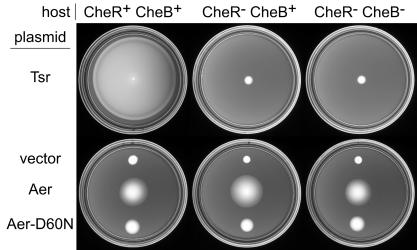
We constructed three strains with the aer locus as well as the four conventional MCP loci deleted. One had functional cheR and cheB genes (UU1250); one contained a deletion of cheR (UU1249); in one both cheR and cheB (UU1535) were deleted. A plasmid expressing wild-type Aer under IPTG-inducible control (pSB20) allowed all three strains to form large colonies on tryptone semisolid agar containing 50 μM IPTG, the optimal inducer concentration (Fig. 2). At lower IPTG levels, colony size was considerably reduced, whereas at higher inducer concentrations, colony size remained close to the maximum (data not shown). Three lines of evidence argue that colony expansion under these conditions is driven by aerotactic behavior. (i) The colony morphology is typical of aerotactic swarms on other media. The colonies have very sharp borders, indicative of directed movements rather than random swimming. Moreover, the colonies are dome shaped, indicating that cells move fastest at the bottom of the agar where the oxygen gradient, established through cell metabolism, should be steepest. (ii) Cells expressing mutant forms of Aer, for example, Aer-D60N, which has an FAD-binding defect in the PAS domain (8), failed to form large colonies under these conditions (Fig. 2). This indicates that Aer sensing and signaling functions contribute to colony expansion. (iii) Chemotactic responses promoted by conventional MCPs were abrogated in the cheR- and cheB-defective hosts. For example, pJC3, a wild-type Tsr plasmid, failed to mediate serine chemotaxis in the UU1249 and UU1535 backgrounds (Fig. 2).
FIG. 2.
Methylation-independent aerotaxis on tryptone semisolid agar plates. Host strains were UU1250 (CheR+ CheB+), UU1535 (CheR− CheB−), and UU1249 (CheR− CheB+); plasmids were pJC3 (Tsr), pCJ30 (vector), pSB20 (Aer), and pSB137 (Aer-D60N). Plates in the upper row contained 20 μM IPTG and were incubated at 30°C for 15 h. Plates in the lower row contained 50 μM IPTG and were incubated at 30°C for 17 h. All plates contained 50 μg of ampicillin/ml.
These results show that Aer-mediated aerotaxis can be monitored on tryptone soft agar in strains that contain no other MCPs. Aer is evidently able to assemble ternary signaling complexes and modulate CheA activity with no assistance from other MCP receptors. Moreover, Aer functions quite well in cheR and cheR cheB hosts, demonstrating that the MCP deamidation and methylation-demethylation systems are not crucial to Aer signaling. (The slight differences in colony size evident in Fig. 2 are probably due to differential polarity effects of the three tar-tap deletions used to construct the host strains, but this proposition has not yet been experimentally tested.)
Aer methylation sites are not required for aerotaxis.
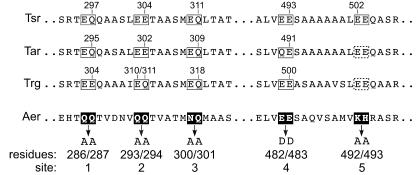
The signaling domain of Aer is similar to that of other MCPs except for its unorthodox methylation sites (Fig. 3). The methylation sites identified in Tar, Tsr, and Trg are typically the second residue of EE or EQ pairs embedded in characteristic amino acid tracts; the overall nine-residue methylation site motif is (A/S)-X-X-E-(E/Q)-X-(A/T/S)-A-(A/S/T) (24, 32, 52). The few known methylation sites that deviate from this consensus (e.g., Tsr-502, Tar-491, Trg-310) are less efficiently methylated (32, 39, 51). The corresponding positions in the Aer signaling domain share none of these features: The first two sites are QQ pairs with atypical neighboring residues; the third is an NQ pair with orthodox neighbors; the fourth is an EE pair, but with atypical neighbors; and the fifth is a KH pair with atypical neighbors.
FIG. 3.
Methylation regions of Tsr, Tar, Trg, and Aer. Known methylation sites in Tsr, Tar, and Trg are shown in open boxes marked with their residue number(s). The Tar and Trg sites shown in dashed boxes correspond in position and in sequence motif to the fifth methylation site of Tsr but are not known to be methylated. Black boxes and white residue letters indicate the corresponding sites in Aer, below which are shown the Aer residue numbers, the Aer methylation site designations, and the mutational replacements made at each site.
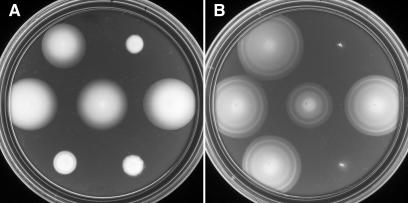
If CheR and CheB are not needed for Aer function, then the methylation sites in Aer should also be dispensable. To explore this proposition, we created pairs of mutations (designated 1* to 5*) at each of the five methylation sites of Aer: Aer site 4 was converted to a DD pair; the others were converted to AA pairs (Fig. 3). These changes are known to eliminate methylation and deamidation when made at conventional MCP methylation sites (29, 31, 42). In UU1250, the 1*, 2*, and 3* mutants retained aerotactic ability, whereas the 4* and 5* mutants did not (Fig. 4). Clearly, the first three methylation sites are not individually crucial to Aer function. In UU1117, an aerΔ strain that has the other MCPs, all but the 5* mutant mediated a robust aerotactic response (Fig. 4). The ability of the 4* mutant to function in the presence of other MCPs suggests that its principal defect is in setting an appropriate swimming pattern. In UU1117, the high-abundance Tar and Tsr transducers will establish the swimming pattern, which the Aer 4* mutant can evidently modulate in response to aerotactic stimuli. The Aer mutant at site 5, the least orthodox of the methylation sites, is not assisted by other MCPs, indicating that its defect is more profound. The 5* mutant protein could be unstable or simply unable to signal properly. Perhaps wild-type Aer must have basic residues at this position. In any event, it is highly unlikely that either of the wild-type residues (K and H) could serve as a substrate site for CheR or CheB. In contrast, site 3, whose wild-type residues are similarly unorthodox, tolerated alanine substitutions with no loss in Aer stability or function.
FIG. 4.
Aerotaxis promoted by Aer methylation-site mutants. (A) Plasmid-containing UU1250 colonies on tryptone soft agar plates containing 50 μM IPTG and incubated for 16 h at 30°C. (B) Plasmid-containing UU1117 colonies on minimal succinate soft agar (with no IPTG) and incubated for 22 h at 30°C. Both plates contained 50 μg of ampicillin/ml. Plasmids in both strain backgrounds were as follows (reading from left to right): top row, pSB20 (Aer) and pCJ30 (vector control); middle row, pDM6 (Aer-1*), pDM7 (Aer-2*), and pDM8 (Aer-3*); bottom row, pDM5 (Aer-4*) and pDM9 (Aer-5*).
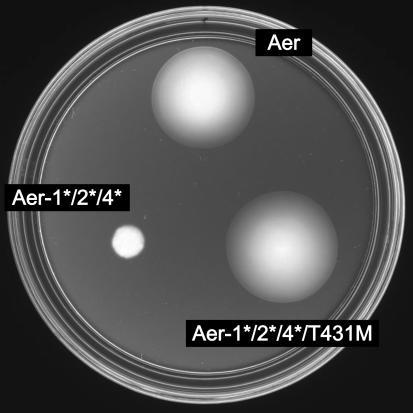
To determine whether Aer methylation sites 1, 2, and 4 are collectively essential for aerotactic signaling, we constructed a 1*/2*/4* triple mutant. The triple mutant, like the 4* mutant alone, failed to mediate aerotaxis in UU1250 (Fig. 5) but functioned normally in UU1117 (data not shown), where other MCPs can set the cell's steady-state swimming pattern. This behavior suggests that the 1*/2*/4* triple mutant is fundamentally competent for stimulus detection and flagellar signaling. To test this idea, we sought Aer mutations that would allow the Aer 1*/2*/4* mutant to function in UU1250. One such phenotypic revertant is shown in Fig. 5. It retained all three methylation-site lesions but had acquired a new mutation in the Aer signaling domain (T431M). Thus, when assisted by MCPs or by a second-site suppressor mutation in its own signaling domain, the Aer methylation site triple mutant exhibits normal aerotactic sensing and signaling ability.
FIG. 5.
Aerotaxis promoted by a pseudorevertant of the Aer 1*/2*/4* triple methylation site mutant. Plasmid-containing UU1535 (CheR− CheB−) colonies were on a tryptone soft agar plate containing 50 μM IPTG and 50 μg of ampicillin/ml. The plate was incubated at 30°C for 17 h. Plasmids were pSB20 (Aer+), pDM13 (Aer-1*/2*/4*), and pKG128 (Aer-1*/2*/4*/T431 M).
Aer-MCP chimeras also exhibit methylation-independent signaling behavior.
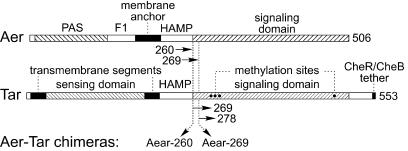
To determine whether the methylation-independent signaling behavior of Aer was correlated with its atypical signaling domain, we constructed two Aer chimeras that had the signaling domain from Tar, a methylation-dependent MCP (Fig. 6). Similar Aer-Tsr chimeras have been reported (8, 38), but we chose the Tar signaling domain to avoid potential complications of Tsr, which also plays a role in aerosensing behavior (23, 37, 57). The Aer and Tar proteins were joined at two homologous positions near the junction of their HAMP and signaling domains, creating chimeras with the PAS-HAMP portion of Aer and the signaling domain of Tar, including its methylation sites and C-terminal CheR/CheB tethering site (Fig. 6).
FIG. 6.
Domain organization and functional features of Aer and Tar and the positions of join points in Aer-Tar chimeras. The primary structures of the molecules are drawn to the same scale and aligned at the chimera join points. In the Aear-260 chimera, Aer residues 1 to 260 were joined to Tar residues 269 to 553. In the Aear-269 chimera, Aer residues 1 to 269 were joined to Tar residues 278 to 553.
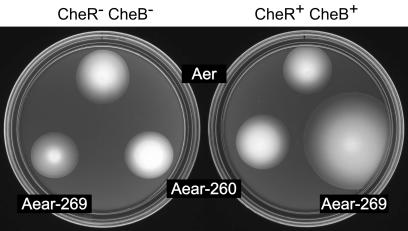
Both Aear-260 and Aear-269 supported aerotaxis in UU1535, which lacks CheR and CheB function, demonstrating that methylation-independent signaling may emanate from the PAS-HAMP portion of Aer (Fig. 7). Moreover, Aer and both Aear chimeras produced comparable migration rates (Fig. 7) at the same optimal expression level (data not shown), indicating that the Aer and Tar signaling domains have similar signaling properties in the absence of CheR and CheB modifications. However, in UU1250, which has CheR and CheB function, the Aear-269 chimera produced dramatically faster aerotactic migrations (Fig. 7). The improved behavior presumably reflects a contribution from the methylation-dependent signaling system of Tar. This Aear hybrid may use the feedback-controlled methylation system to optimize a steady-state swimming pattern and to assist in sensory adaptation to aerotactic stimuli. The failure of the Aear-260 chimera to benefit from CheR and CheB function indicates that the region between Aer residues 260 and 269 is somehow crucial for methylation-dependent signaling in the hybrid transducers. Paradoxically, this region must come from Aer in order to confer methylation-dependent signaling behavior on the Aear hybrid.
FIG. 7.
Aerotactic signaling by Aer-Tar chimeras. Plasmid-containing colonies were on tryptone soft agar plates containing 50 μM IPTG and 50 μg of ampicillin/ml. Plates were incubated at 30°C for 15 h. Strains were UU1535 (CheR− CheB−) and UU1250 (CheR+ CheB+). Plasmids were pSB20 (Aer), pKG121 (Aear-269), and pKG119 (Aear-260).
DISCUSSION
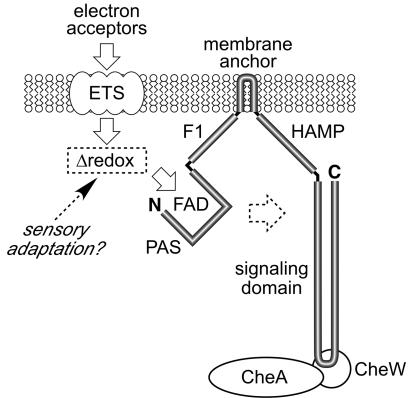
Our current view of Aer signaling, which incorporates the findings of the present study, is summarized in Fig. 8. Aer is thought to use the FAD bound to its PAS domain as a redox sensor for detecting changes in environmental levels of oxygen and other electron acceptors (50). The PAS domain in turn modulates the activity of the Aer signaling domain, either directly or through the intervening F1, membrane anchor, and/or HAMP domains. The Aer signaling domain resembles that of conventional methyl-accepting receptors and, like MCPs, appears to be capable of regulating CheA autophosphorylation, presumably in ternary complexes with the CheW coupling protein. However, unlike MCPs, Aer signaling and sensory adaptation are not dependent on methylation changes. Instead, we suggest that the redox signal sensed by Aer may be the site of sensory adaptation for aerotaxis, as discussed below.
FIG. 8.
Current working model of Aer signal transduction. Aer is anchored to the cytoplasmic side of the inner membrane and probably monitors respiratory status via redox changes in a component of the electron transport system (ETS). Transience of the redox signal could account for methylation-independent sensory adaptation during aerotaxis, as discussed in the text.
Aer-mediated aerotaxis is methylation independent.
Three lines of evidence from this study demonstrate that Aer is a methylation-independent transducer. (i) The Aer protein was not detectably modified by either CheR or CheB, the MCP-specific methyltransferase and methylesterase, respectively. The possibility remains that Aer carries an undetected modification, perhaps promoted by a different enzyme. (ii) Aer contains unorthodox methylation sites that do not conform to the consensus motif for CheR or CheB substrates. Moreover, replacements at those sites with amino acids that cannot be deamidated by CheB or methylated by CheR generally had no deleterious effect on Aer function. Although the site 5 alteration abolished Aer function, the wild-type residues at that site are the least likely ones to be CheR/CheB substrates and are probably important in some other aspect of Aer signaling. (iii) Aer promoted aerotactic migrations in a strain that lacked MCPs, CheR, and CheB. Thus, Aer senses and signals efficiently in the absence of methylation, methylation changes, methylation enzymes, and methyl-accepting chemotaxis proteins.
Methylation-independent sensory adaptation.
These findings are consistent with the work of Niwano and Taylor, who demonstrated methylation-independent sensory adaptation to oxygen stimuli (30). Although our study did not directly examine adaptation behavior, it seems likely that Aer-mediated cell migrations in soft agar media require sensory adaptation ability. Thus, the methylation-independent signaling properties of Aer probably underlie the methylation-independent adaptive responses observed by Niwano and Taylor. Because chimeric Aer molecules with the signaling domain of Tar also mediated aerotaxis in strains lacking CheR and CheB, the PAS-HAMP portion of Aer may be responsible for methylation-independent behavior. For example, if the redox signal detected by Aer were transient, this would provide a simple mechanism for methylation-independent sensory adaptation in aerotaxis. The origin of that signal is unknown but most likely involves interaction between Aer and a component of the electron transport system. ArcB, a membrane-anchored respiratory monitor that regulates gene expression, obtains its sensory information from oxidized quinone electron carriers (22).
A more general mechanism of methylation-independent sensory adaptation may also contribute to Aer signaling behavior. Mutants defective in CheR or CheB function detect and respond to changing chemoeffector levels, but their responses are prolonged, consistent with impaired sensory adaptation. However, cheR and cheB mutants do exhibit slow, partial recoveries from MCP-mediated responses that imply existence of an inefficient methylation-independent mechanism of sensory adaptation (11, 36, 46, 48, 56). The nature of this secondary adaptation system is unknown. Conceivably, it could be the primary adaptation system for Aer signaling.
Steady-state swimming pattern influences Aer function.
In the presence of MCPs, Aer, a low-abundance receptor, has little control over the cell's steady-state swimming pattern. Methylation enables the MCPs to adjust the cell's swimming pattern for optimal chemotactic responsiveness. In cheR mutants, unmethylated MCPs produce low CheA kinase activity and counterclockwise-biased flagellar rotation. In cheB mutants, overmethylated MCPs produce high CheA activity and clockwise-biased rotation. Consequently, although Aer is a methylation-independent transducer, cheR and cheB mutants that contain MCPs and Aer at their normal expression levels do not show aerotactic behavior in soft agar media (S. I. Bibikov and K. Hyland, unpublished results). In contrast, CheR and CheB cannot influence the swimming pattern of strains lacking MCPs, and in such strains Aer alone can establish a swimming pattern suitable for aerotactic responses. However, proficient behavior requires Aer expression above normal levels, presumably to compensate for the lack of other receptors.
The Aer-4* methylation site mutant could not carry out aerotaxis in UU1250, an MCP-less strain, but exhibited normal aerotaxis in UU1117, which has other MCPs. This MCP helping effect most likely reflects the ability, particularly of Tar and Tsr, to set the cell's swimming pattern via methylation-dependent sensory adaptation. We designate such Aer mutants bias dependent because they can be seen to sense and signal normally when the cells are provided with an appropriate (i.e., roughly wild-type) steady-state flagellar rotational bias and random-walk swimming pattern. Roughly one-third of Aer mutants isolated on the basis of aerotaxis defects in UU1250 have proven to be bias dependent (Bibikov and Hyland, unpublished; M. Burón and J. S. Parkinson, unpublished results).
The Aear-269 chimeric transducer also exhibited bias-dependent signaling behavior. In the presence of CheR and CheB, Aear-269 produced rapid aerotactic migrations, whereas in the absence of those functions, Aear-269 functioned identically to Aear-260 and to Aer. The CheR/CheB-dependent behavioral enhancement of the Aear-269 chimera could be due to an improved swimming pattern but might also reflect more efficient gradient tracking via the methylation-dependent adaptation system. This hybrid transducer may actually use both methylation-dependent and methylation-independent modes of adaptation, working in additive or synergistic fashion.
Aerotaxis might be responsible for methylation-independent chemotaxis.
Mutants defective in CheR or CheB function readily acquire mutations in other components of the chemotaxis machinery that restore a pseudo-wild swimming pattern and enhance the rate of migration in soft agar (16, 44, 47). Part of the speed-up effect is due to the phenomenon of pseudotaxis, the ability of cells that both run and tumble to percolate through an agar matrix faster than cells that incessantly run or tumble (5, 54). However, the faster-spreading pseudorevertant colonies also have a sharp ring at their perimeter suggestive of a chemotactic response (44, 46-48, 54). Stock and colleagues (46, 47) have argued that cheR cheB double mutants, which have a wild-type swimming pattern, use a methylation-independent adaptation mechanism to move away from repellent compounds produced by the growing colony. Although controversial, this methylation-independent chemotaxis appears to be a more purposeful behavior than pseudotaxis (cf. reference 54) and has never been satisfactorily explained. Perhaps it is Aer-mediated aerotaxis. As explained above, aerotaxis can occur in cells lacking CheR and/or CheB, but only if they have a roughly wild-type swimming pattern, as the chemotactic pseudorevertants do.
Evolving Aer.
The Aer transducer in E. coli functions best in the presence of MCPs and a methylation-dependent adaptation system. In the absence of MCPs, Aer mediates much slower aerotactic migrations than those promoted by the Aear-269 chimera, which has a methylation-dependent signaling domain. Did the Aer signaling domain originate from an MCP and subsequently lose its methylation faculties? Or is Aer a proto-MCP that has not yet acquired methylation competence? An initial analysis of MCP-like transducers in microbial genomes indicates that Aer is more likely to be a young transducer rather than the progenitor of present-day MCPs (I. B. Zhulin, personal communication).
Whether the Aear-269 chimera resembles the ancestral Aer transducer or the next step in Aer evolution, it would seem to be an ideal subject for studying many Aer-specific signaling questions. For example: how does Aer monitor the cell's electron acceptor status? How are Aer input signals relayed to the output domain? How is Aer signal output modulated by stimulus input? With Aear-269 it should be possible to avoid bias-dependent signaling lesions in future aerotaxis mutant hunts, because its methylation-dependent adaptation system should compensate for most such defects. Alternatively, it should be possible to isolate Aear-269 mutants in hosts that lack CheR/CheB function and then test them in other hosts to distinguish bias-dependent lesions from signal transduction lesions. Finally, Aear-269 may prove tractable for biochemical studies of the Aer sensing mechanism, because the Tar signaling domain functions well in vitro (17, 28, 49).
Acknowledgments
We thank Mark Johnson (Loma Linda University) and Igor Zhulin (Georgia Institute of Technology) for helpful comments on the manuscript.
This work was supported by research grants GM19559 and GM62904 from the National Institute of General Medical Sciences and by the University of Utah Research Foundation Funding Incentive Seed Grant Program. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute. A.C.M. received support from the Bioscience Undergraduate Research Program, Department of Biology, University of Utah.
REFERENCES
- 1.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160:63-67. [DOI] [PubMed] [Google Scholar]
- 2.Ames, P., and J. S. Parkinson. 1994. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 176:6340-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames, P., and J. S. Parkinson. 1988. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell 55:817-826. [DOI] [PubMed] [Google Scholar]
- 4.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ames, P., Y. A. Yu, and J. S. Parkinson. 1996. Methylation segments are not required for chemotactic signalling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol. Microbiol. 19:737-746. [DOI] [PubMed] [Google Scholar]
- 6.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 7.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1999. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc. Natl. Acad. Sci. USA 96:10667-10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 11.Block, S. M., J. E. Segall, and H. C. Berg. 1982. Impulse responses in bacterial chemotaxis. Cell 31:215-226. [DOI] [PubMed] [Google Scholar]
- 12.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 13.Boyd, A., and M. I. Simon. 1980. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J. Bacteriol. 143:809-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, A. C. Y., H. A. Erlich, R. P. Gunsalus, J. H. Nunberg, R. J. Kaufman, R. T. Schimke, and S. N. Cohen. 1980. Initiation of protein synthesis in bacteria at a translational start codon of mammalian cDNA: Effects of the preceding nucleotide sequence. Proc. Natl. Acad. Sci. USA 77:1442-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chelsky, D., and F. W. Dahlquist. 1981. Multiple sites of methylation in the methyl accepting chemotaxis proteins of Escherichia coli. Prog. Clin. Biol. Res. 63:371-381. [PubMed] [Google Scholar]
- 16.Chen, J. 1992. Genetic studies of transmembrane and intracellular signaling by a bacterial chemoreceptor. Ph.D. thesis. University of Utah, Salt Lake City, Utah.
- 17.Cochran, A., and P. Kim. 1996. Imitation of Escherichia coli aspartate receptor signaling in engineered dimers of the cytoplasmic domain. Science 271:1113-1116. [DOI] [PubMed] [Google Scholar]
- 18.Cox, E. C., and D. L. Horner. 1986. DNA sequence and coding properties of mutD(dnaQ) a dominant Escherichia coli mutator gene. J. Mol. Biol. 190:113-117. [DOI] [PubMed] [Google Scholar]
- 19.Engstrom, P., and G. L. Hazelbauer. 1980. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell 20:165-171. [DOI] [PubMed] [Google Scholar]
- 20.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng, X., A. A. Lilly, and G. L. Hazelbauer. 1999. Enhanced function conferred on low-abundance chemoreceptor Trg by a methyltransferase-docking site. J. Bacteriol. 181:3164-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 23.Greer-Phillips, S. E., G. Alexandre, B. L. Taylor, and I. B. Zhulin. 2003. Aer and Tsr guide Escherichia coli in spatial gradients of oxidizable substrates. Microbiology 149:2661-2667. [DOI] [PubMed] [Google Scholar]
- 24.Kehry, M. R., M. W. Bond, M. W. Hunkapiller, and F. W. Dahlquist. 1983. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc. Natl. Acad. Sci. USA 80:3599-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehry, M. R., and F. W. Dahlquist. 1982. Adaptation in bacterial chemotaxis: CheB-dependent modification permits additional methylations of sensory transducer proteins. Cell 29:761-772. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Li, J., G. Li, and R. M. Weis. 1997. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry 36:11851-11857. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y., M. Levit, R. Lurz, M. G. Surette, and J. B. Stock. 1997. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 16:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiyama, S., T. Nara, M. Homma, Y. Imae, and I. Kawagishi. 1997. Thermosensing properties of mutant aspartate chemoreceptors with methyl-accepting sites replaced singly or multiply by alanine. J. Bacteriol. 179:6573-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niwano, M., and B. L. Taylor. 1982. Novel sensory adaptation mechanism in bacterial chemotaxis to oxygen and phosphotransferase substrates. Proc. Natl. Acad. Sci. USA 79:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowlin, D. M., J. Bollinger, and G. L. Hazelbauer. 1988. Site-directed mutations altering methyl-accepting residues of a sensory transducer protein. Proteins 3:102-112. [DOI] [PubMed] [Google Scholar]
- 32.Nowlin, D. M., J. Bollinger, and G. L. Hazelbauer. 1987. Sites of covalent modification in Trg, a sensory transducer of Escherichia coli. J. Biol. Chem. 262:6039-6045. [PubMed] [Google Scholar]
- 33.Parkinson, J. S. 2003. Bacterial chemotaxis: a new player in response regulator dephosphorylation. J. Bacteriol. 185:1492-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson, J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkinson, J. S., and P. T. Revello. 1978. Sensory adaptation mutants of E. coli. Cell 15:1221-1230. [DOI] [PubMed] [Google Scholar]
- 37.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, M. S., and F. W. Dahlquist. 1991. Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J. Biol. Chem. 266:9746-9753. [PubMed] [Google Scholar]
- 40.Rollins, C., and F. W. Dahlquist. 1981. The methyl-accepting chemotaxis proteins of Escherichia coli: a repellent-stimulated, covalent modification, distinct from methylation. Cell 25:333-340. [DOI] [PubMed] [Google Scholar]
- 41.Schoepfer, R. 1993. The pRSET family of T7 promoter expression vectors for Escherichia coli. Gene 124:83-85. [DOI] [PubMed] [Google Scholar]
- 42.Shapiro, M. J., and D. E. Koshland, Jr. 1994. Mutagenic studies of the interaction between the aspartate receptor and methyltransferase from Escherichia coli. J. Biol. Chem. 269:11054-11059. [PubMed] [Google Scholar]
- 43.Sherris, D., and J. S. Parkinson. 1981. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 78:6051-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherris, D. I. 1984. Protein methylation in bacterial chemotaxis. Ph.D. thesis. University of Utah, Salt Lake City, Utah.
- 45.Sourjik, V., and H. C. Berg. 2002. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 99:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stock, J., G. Kersulis, and D. E. Koshland, Jr. 1985. Neither methylating nor demethylating enzymes are required for bacterial chemotaxis. Cell 42:683-690. [DOI] [PubMed] [Google Scholar]
- 47.Stock, J. B., A. Borczuk, F. Chiou, and J. E. Burchenal. 1985. Compensatory mutations in receptor function: a reevaluation of the role of methylation in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 82:8364-8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stock, J. B., A. M. Maderis, and D. E. Koshland, Jr. 1981. Bacterial chemotaxis in the absence of receptor carboxylmethylation. Cell 27:37-44. [DOI] [PubMed] [Google Scholar]
- 49.Surette, M., and J. Stock. 1996. Role of α-helical coiled-coil interactions in receptor dimerization, signaling, and adaptation during bacterial chemotaxis. J. Biol. Chem. 271:17966-17973. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, B. L., I. B. Zhulin, and M. S. Johnson. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 51.Terwilliger, T. C., J. Y. Wang, and D. E. Koshland, Jr. 1986. Kinetics of receptor modification. The multiply methylated aspartate receptors involved in bacterial chemotaxis. J. Biol. Chem. 261:10814-10820. [PubMed] [Google Scholar]
- 52.Terwilliger, T. C., J. Y. Wang, and D. E. Koshland, Jr. 1986. Surface structure recognized for covalent modification of the aspartate receptor in chemotaxis. Proc. Natl. Acad. Sci. USA 83:6707-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 86:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]
- 56.Yonekawa, H., H. Hayashi, and J. S. Parkinson. 1983. Requirement of the cheB function for sensory adaptation in Escherichia coli. J. Bacteriol. 156:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, H. S., J. H. Saw, S. Hou, R. W. Larsen, K. J. Watts, M. S. Johnson, M. A. Zimmer, G. W. Ordal, B. L. Taylor, and M. Alam. 2002. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol. Lett. 217:237-242. [DOI] [PubMed] [Google Scholar]