Abstract
The Lyme disease bacterium Borrelia burgdorferi is a motile spirochete with a flat-wave morphology. The periplasmic flagella, which are situated between the outer membrane sheath and cell cylinder, are essential for both the cell's wavy shape and motility. Here we focus on the structure and regulation of its periplasmic flagella. Previous studies have suggested that the periplasmic flagella consist of a polymer of the major filament protein FlaB and a minor protein, FlaA. We used immunoprecipitation methodology to present further evidence that FlaA is indeed a flagellar protein. In addition, in contrast to FlaA of the spirochete Brachyspira hyodysenteriae, B. burgdorferi FlaA did not impact the overall helical shape of the periplasmic flagella. We have previously shown that B. burgdorferi lacks the sigma factor-dependent cascade control of motility gene transcription found in other bacteria. To begin to understand motility gene regulation in B. burgdorferi, we examined the effects of an insertion mutation in flaB on the amounts of proteins encoded by motility genes. Of several motility gene-encoded proteins examined, only the amount of FlaA was decreased in the flaB mutant; it was 13% compared to the wild-type amount. Real-time reverse transcriptase PCR analysis indicated that this inhibition was not the result of a decrease in flaA mRNA. In addition, protein stability analysis suggested that FlaA was turned over in the flaB mutant. Our results indicate that the lack of FlaB negatively influences the amount of FlaA found in the cell and that this effect is at the level of either translational control or protein turnover.
Borrelia burgdorferi is a motile spirochete that is the causative agent of Lyme disease. The organelles for motility, the periplasmic flagella (PFs), reside between the outer membrane sheath and protoplasmic cell cylinder. Approximately 7 to 11 PFs are subterminally attached at each end of the cell, and these filaments overlap in the center of the cell (4, 27, 31). Motility is likely to be an important virulence factor for these spirochetes, as B. burgdorferi penetrates into tissues where other organisms fail to invade (11, 34). Moreover, motility has been postulated to be essential for the life cycle of these spirochetes in both the tick and the mammalian hosts (45, 55).
We are beginning to understand the dynamics of B. burgdorferi motility (see references 11, 42, and 45 for recent reviews). Recent results indicate that the PFs have both skeletal and motility functions (50, 61). Cells of mutants that are targeted in flaB, which encodes the major PF protein, are nonmotile and are rod shaped instead of being a flat wave as are wild-type cells (50, 61). During translational motility, backward moving waves are generated that are responsible for cell displacement (28). These waves result from the rotation of the relatively rigid left-handed helically shaped PFs closely juxtaposed to and interacting with the flexible protoplasmic cell cylinder: the PFs act as a gear generating waves along the cell cylinder from the anterior to the posterior end of the cell (11, 27, 28, 42). The bundles of PFs rotate asymmetrically, with the anterior bundle rotating counter-clockwise and the posterior one clockwise as viewed along the length of the PFs from their distal ends towards their insertion points on the cell (11, 28, 40).
The PFs of B. burgdorferi have been analyzed in some detail. These organelles have been shown to be left-handed, with a defined helix pitch and diameter (12). Their major constituent is a polymer of FlaB, which is similar in sequence at the N- and C-terminal domains to PFs of other spirochete species as well as to flagellin of other bacteria (54, 63). Based on purification of PFs, FlaB was considered for several years to be the only filament protein. However, DNA sequence analysis indicated the presence of a flaA homolog (17, 20). In other spirochete species, FlaA forms a sheath around the FlaB core. In addition, FlaA markedly affects the shape of the PFs in Brachyspira hyodysenteriae with respect to helix pitch and helix diameter (9, 38, 41, 42, 54). We found that flaA was expressed in B. burgdorferi, and when a gentler method of isolation was employed FlaA copurified with the PFs (23). However, in contrast to results seen with other spirochete species (9, 38, 41, 42, 54, 59) considerably less FlaA relative to FlaB was present in these purified PFs. Using immunoprecipitation, we provide further evidence that FlaA is associated with FlaB. In addition, we find that in contrast to other spirochetes there is considerably less FlaA relative to FlaB not only in purified PFs but also within the whole cell. Finally, we show that FlaA does not affect the PF shape for B. burgdorferi as it does for B. hyodysenteriae.
Bacterial flagella are complex organelles that often comprise a disproportionately large amount of protein in a motile cell. As a result, bacteria have evolved elegant mechanisms of flagellar assembly and control. Specifically, motile bacteria possess a cascade system of transcriptional control (1-3, 14, 48, 49). For example, in Escherichia coli and Salmonella enterica serovar Typhimurium the cell senses specific environmental signals to commence expression of class 1 genes flhC and flhD. FlhC and FlhD then promote the initiation of transcription of class 2 motility genes (2, 14). These genes include, among others, those that encode the proteins that comprise the basal body apparatus. Two of the class 2 genes, fliA and flgM, play critical roles in regulating class 3 gene transcription. Specifically, fliA encodes sigma28 that promotes transcription of class 3 genes, including the flagellar filament gene fliC. flgM encodes an anti-sigma28 factor, FlgM (2, 14), which is excreted into the medium when the flagellar hook-basal body is complete. FlgM insures that the class 3 genes are not transcribed until the initial flagellar structure is intact, thus providing the cell with a mechanism to sense the state of flagellar synthesis and adjust its transcription accordingly (2, 14).
The regulation of motility gene expression in B. burgdorferi is unique, as it lacks the cascade control of flagellar gene transcription found in other bacteria. The results of several studies indicate that gene transcription of the numerous motility operons is initiated by the housekeeping transcription factor sigma70 (11, 17, 20, 22, 24, 25, 42). Furthermore, no fliA homolog has been identified in its genome (17, 42). To begin to understand how the motility genes are regulated in B. burgdorferi, we are examining the effects of specific PF mutations on the synthesis of several motility and chemotaxis gene products. In this communication, we report our results determined with a previously described insertion mutant in flaB (50, 61).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
High-passage B. burgdorferi sensu stricto strain B31A, the flaB mutant MC-1, and the complemented flaB mutant MS17 have been previously described (8, 50, 61). Briefly, strain MC-1 has a flgB promoter-kanamycin cassette inserted into flaB of B. burgdorferi. This mutant is nonmotile and fails to synthesize PFs (50). Strain MS17 is a derivative of MC-1 containing a wild-type copy of flaB in the plasmid pED3. pED3 complements the flaB mutation in cis, i.e., only when the plasmid is integrated into the chromosome by a Campbell-type recombination event (61). Cells were grown in Barbour-Stoenner-Kelly II (BSK-II) broth or on plates at 32 to 34°C as previously described (50). S. enterica serovar Typhimurium strain SJW1103 is wild type for chemotaxis and flagellation, and strain SJW1368 Δ(cheW-flhD) is a master operon mutant which is aflagellate (18). These strains were the kind gift of the late R. Macnab, Yale University.
Gel electrophoresis and Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with an enhanced chemiluminescent detection method (ECL; Amersham Pharmacia) were carried out as previously reported (23). Equal amounts (except as noted) of cellular protein in lysates, as determined by a Bio-Rad protein assay kit, were applied to each lane. Several monoclonal and polyclonal antibodies kindly provided by other investigators included the following: monoclonal anti-FlaA (B. Johnson, Centers for Disease Control and Prevention, Atlanta, Ga.), monoclonal anti-OspA and DnaK (J. Benach, State University of New York, Stony Brook), monoclonal anti-FlaB (H9724; A. Barbour, University of California, Irvine), rabbit anti-MotB (J. Carroll, Rocky Mountain Laboratories, Hamilton, Mt.), rabbit polyclonal anti-FlaA of Treponema pallidum (S. Norris, University of Texas, Houston), rabbit anti-CheA of E. coli (P. Matsumura, University of Illinois, Chicago), and rabbit anti-FliM of E. coli (D. Blair, University of Utah, Salt Lake City). Rabbit anti-CheY2 and anti-FlgE will be described elsewhere (M. Motaleb, M. Sal, and N. Charon, manuscript in preparation). Specific monoclonal or polyclonal reactivity to B. burgdorferi DnaK, FlaA, FlaB, FliI, MotB, CheA1, CheA2, and CheY3 has been previously reported (5, 10, 15, 25, 26, 40, 50, 53). Insertion mutations in flaA, flaB, flgE, motB, cheA1, cheA2, cheY2, and cheY3 verified antibody specificity by Western blotting (40, 50; M. Sal, M. A. Motaleb, and N. W. Charon, Abstr. Bacterial Locomotion and Signal Transduction Meeting VII, Cuernavaca, Mexico, p. 87, 2003; M. Motaleb, M. Sal, and N. Charon, unpublished data). Anti-FliM reactivity was verified using S. enterica serovar Typhimurium strains SJW1103 and SJW1368.
Quantitative immunoblot assays.
To quantify FlaA and FlaB in cell lysates, PFs were purified using the Triton X-100 method as previously described (23). PF proteins were separated on SDS-PAGE gels, and the bands corresponding to FlaA and FlaB were cut from the gels which were then electroeluted using ExElutor (Labnet International Inc., Woodbridge, N.J.). FlaA and FlaB were concentrated by centrifugation in Amicon Ultra concentrators (Millipore, Bedford, Mass.). Protein concentrations were determined using a Bio-Rad protein assay kit as well as SDS-PAGE gels stained with Coomassie brilliant blue R-250 with bovine serum albumin as a standard. Different amounts of purified FlaA, FlaB, and cell lysates were probed with monoclonal anti-FlaA and anti-FlaB by the use of Western blotting. Signals were quantified by FluorChem spot densitometry (Alpha Innotech, San Leandro, Calif.).
Immunoprecipitation and PF morphology.
Immunoprecipitation was carried out as previously described (43). Approximately 5 × 109 B. burgdorferi cells were washed twice with 150 mM phosphate-buffered saline (pH 7.5)-5 mM MgCl2 and were lysed by incubation at 37°C for 1 h in TSEA buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.05% sodium azide, pH 7.5) containing 1% Nonidet P-40 and 50 μg of phenylmethylsulfonyl fluoride/ml. The lysate was centrifuged (1,600 × g for 30 min, 25°C), and the pellet was resuspended in phosphate-buffered saline and sonicated for 1 min intermittently for 20 min in ice followed by centrifugation (14,800 × g for 30 min, 25°C). Approximately 200 μl of the supernatant fluid was incubated with 15 μl of the monoclonal anti-FlaB for 1 h at 25°C in the presence of 1% bovine serum albumin. Controls consisted of anti-FlaB incubated with no cell sonicate and the cell sonicate incubated with no antibody. After incubation, approximately 50 μl of protein A (Pansorbin [Staphylococcus aureus Cowan 1]; Calbiochem Behring, La Jolla, Calif.) was added to each sample and further incubated for 1 h at 25°C. The immunoprecipitates and controls were centrifuged at 1,600 × g at 25°C and washed three times with 1 ml of TSEA buffer containing 0.05% Tween-20. The final pellets were suspended in 60 μl of electrophoresis sample buffer, boiled for 5 min, and briefly centrifuged as before. For Western blotting, 10 μl of the supernatant fluids was applied to each lane of a SDS-PAGE gel. The left- or right-hand orientation, helical pitch, and helical diameter of the purified PFs were determined using dark-field microscopy as previously described (12, 23, 41). PFs containing both FlaA and FlaB were purified using the Triton X-100 method as previously described (23). PFs containing FlaB and no FlaA were purified with Sarkosyl as previously reported (12, 23).
Real-time quantitative RT-PCR and RNase protection assays.
Total cellular RNA was extracted using an RNeasy kit from QIAGEN and then digested with Turbo DNase I (Ambion) at 37°C for 2 h. Equal amounts of total RNA (approximately 0.3 μg) were used to generate cDNA according to the protocol of the manufacturer (Promega). To ensure that the RNA preparations were free from genomic DNA, reverse transcription was performed with or without reverse transcriptase. Quantitative PCR was carried out in duplicate on cDNA with SYBR Green master mix (QIAGEN) and a LightCycler (Roche Biochemicals). A standard curve was generated using 10-fold serial dilutions (1.0 pg to 0.1 fg) from the flaA reverse transcriptase PCR (RT-PCR) product. 16S rRNA was used as an internal control. PCR primers for flaA (BB0668; GenBank accession no. U62900) were the following: forward primer, 5′-CACTAGTTCTTTTTGCTCAA-3′; reverse primer, 5′-ACTGCAGTACCTTTTTGACTC-3′. For 16S rRNA (GenBank accession no. L40596), the primers were the following: forward primer, 5′-GTGGCGAACGGGTGAGTAAC-3′; reverse primer, 5′-CCGTCAGCTTTCGCCATTGC-3′. Results are expressed with respect to the threshold cycle at which the measured fluorescence increased above background (57).
RNase protection assays were carried out using a Hybspeed RPA kit from Ambion according to standard procedures (23). Briefly, primers for the B. burgdorferi flaA and 16S rRNA were used to amplify specific regions by PCR. The amplified regions (305-bp flaA and 296-bp 16S rRNA) were cloned into pGEM-T Easy vector (Promega), and the resultant plasmids were linearized with SpeI (flaA) or NcoI (16S rRNA). Antisense riboprobe was synthesized with [α-32P]UTP (Amersham Pharmacia) and a transcription kit (Ambion) and purified using a 5% polyacrylamide-8 M urea gel. Hybridization and digestion were done as described previously (23). Transcript levels were quantified using a Molecular Dynamics PhosphorImager.
Protein turnover.
Techniques previously employed for other bacteria (33) were used to determine the stability of FlaA in growing cells. Wild-type and MC-1 strains were grown in BSK-II medium at 33°C until cells reached a density of 5 × 108 cells per ml. Cells (2 ml) were added to 40 ml of BSK-II medium containing spectinomycin (final concentration, 100 μg/ml) and incubated at 33°C. Samples were removed and processed for Western blotting at indicated time points.
RESULTS
Association of FlaA with FlaB.
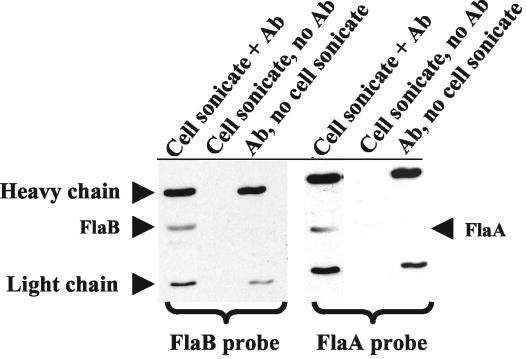
Ge et al. previously found that FlaA and FlaB copurified when the Triton X-100 procedure was used for PF isolation (23). To confirm that these two proteins were actually associated with one another, FlaB was immunoprecipitated from cell lysates of B. burgdorferi and the resulting immunoprecipitate was probed with anti-FlaB (Fig. 1, left panel) and anti-FlaA (Fig. 1, right panel) in Western blotting. We found that FlaA coprecipitated with FlaB (Fig. 1, right panel), while no coprecipitation occurred with another abundant B. burgdorferi protein, OspA (data not shown). These results suggest that there is a specific association of FlaA with FlaB in situ.
FIG. 1.
Immunoprecipitation of FlaA with monoclonal H9724 anti-FlaB. Wild-type-cell sonicates were immunoprecipitated with FlaB antibody (Ab) and then probed with monoclonal antibody to FlaB (left panel) and FlaA (right panel) by Western blotting. The upper and lower bands of heavy and light chains of the primary antibody are identified. As controls without FlaB antibody, neither FlaB nor FlaA was precipitated (middle lanes).
FlaA is a minor PF protein.
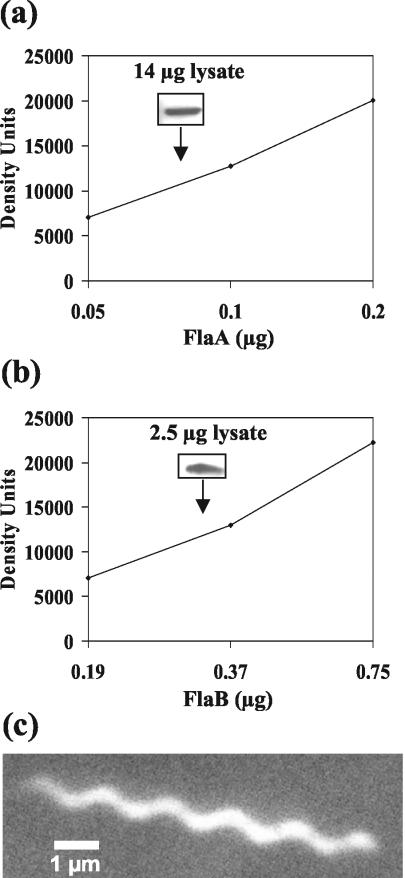
SDS-PAGE of purified PFs was previously used to estimate the relative amounts of FlaA and FlaB in B. burgdorferi. In these studies, FlaA was found to be a minor component (23). These results are in contrast to those obtained with other spirochete species, for which the amount of FlaA is approximately equal to that of FlaB (9, 41, 53, 54, 59). One concern is that perhaps much of the B. burgdorferi FlaA was lost during the purification of PFs. Accordingly, we determined the amounts of both FlaA and FlaB in total cell lysates by the use of quantitative immunoblotting. Western blot results indicated that FlaA contributed less than 0.5% to the total protein of the cell (Fig. 2a). In contrast, FlaB was found to contribute 10 to 14% of the total cell protein (Fig. 2b). This relatively large amount of FlaB is in agreement with the results seen with other multiflagellated bacteria. In S. enterica serovar Typhimurium, which has approximately eight flagella per cell, the filament protein FliC contributes 8% of the total cell protein (51). B. hyodysenteriae has approximately 10 to 13 PFs per bundle, and these PFs have been estimated to contribute 5 to 10% of the total protein (39). Our results are consistent with the PFs being a major constituent of B. burgdorferi and with these organisms accumulating considerably less FlaA than FlaB.
FIG. 2.
Quantitative Western blot analysis of periplasmic flagellar protein levels in wild-type B. burgdorferi. (a and b) Different amounts of gel-purified FlaA, FlaB, and cell lysates were separated and probed with monoclonal anti-FlaA (a) and anti-FlaB (b) antibodies. The amounts of FlaA and FlaB in the cell lysates were calculated from the standard curves obtained from purified FlaA (a) and FlaB (b), respectively. Inserts illustrate Western blot reactivity at a specific amount of protein lysate. (c) Dark-field micrograph of a PF bundle containing FlaA and FlaB, which were isolated using the Triton X-100 method (23).
Influence of FlaA on PF structure.
FlaA has been shown to affect the PF structure of B. hyodysenteriae. Specifically, PFs from mutants that lack FlaA have a helix pitch and helix diameter markedly different from those of the wild type (41). B. burgdorferi PFs were previously shown to be left-handed and have a helix pitch of 1.48 μm ± 0.03 μm and helix diameter of 0.28 μm ± 0.03 μm. These measurements were made with PFs isolated using Sarkosyl; such PFs lack FlaA (12). To examine whether FlaA influenced the PF shape of B. burgdorferi, we isolated PFs containing both FlaA and FlaB by the use of the gentler technique that replaces Sarkosyl with Triton X-100 (23). We found that PFs with both FlaA and FlaB were identical in shape to those isolated without FlaA (left-handed; helix pitch, 1.5 μm ± 0.028 μm; helix diameter, 0.33 μm ± 0.027 μm) (Fig. 2c). The results indicate that in contrast to the results seen with B. hyodysenteriae, FlaA does not influence the helical shape of the PFs.
Influence of flaB::kan on the synthesis of select gene products in motility operons.
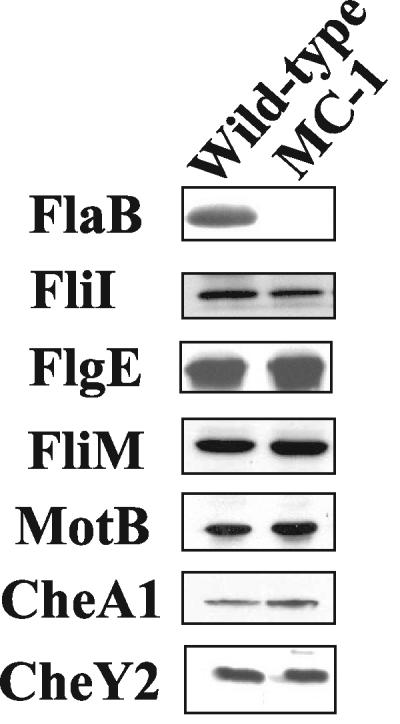
Previous results indicated that B. burgdorferi lacks the typical cascade control of flagellar gene regulation seen in other bacteria (11, 17, 20, 22, 24, 25, 42). To begin to understand how motility genes are regulated in B. burgdorferi, we examined the effects of a flaB::kan mutation (mutant MC-1) on the synthesis of other flagella and chemotaxis proteins. The flagellar filament gene flaB maps far to the left on the linear B. burgdorferi chromosome (17) and is transcribed as a monocistronic mRNA (19, 60). We first examined protein synthesis with respect to select genes in the large 21-kb flgB operon that encodes most of the hook, basal body, and motility genes. This operon maps approximately 180 kb away from flaB (17). Those genes chosen for analysis were fliI, flgE, fliM, and motB; each maps several open reading frames (ORFs) away from the others. FliI in other bacteria is associated with both basal body protein and flagellin export. FlgE is the flagellar hook structural protein which serves as a universal joint between the flagellar basal body and the filament (6). FliM is part of the rotary apparatus of the motor, and MotB is the stator involved in energy transduction to the motor (6). Insertion mutations in flgE and motB led to the expected nonmotile phenotypes (Sal et al., Abstr. Bacterial Locomotion and Signal Transduction Meeting VII; Motaleb, Sal, and Charon, unpublished), indicating that these genes function in motility as predicted from their sequence. Western blot analysis indicated that although the synthesis of FlaB was inhibited in MC-1 as reported previously (50), no decrease in the amount of protein is evident with respect to FlgE, FliI, FliM, and MotB (Fig. 3). These results suggest that flaB::kan did not alter the synthesis of the encoded proteins for the select motility genes assayed in this operon.
FIG. 3.
Effect of flaB::kan on other motility and chemotaxis proteins. Equal amounts (10 μg) of cell lysates from wild-type and the flaB::kan mutant MC-1 strains were probed by Western blotting with antibodies specific for the indicated proteins. DnaK was used as an internal control as shown in Fig. 4b. The apparent sizes (in kilodaltons) of the proteins are as follows: FlaB, 41; FliI, 48; FlgE, 40; FliM, 37; MotB, 24; CheA1, 80; CheY2, 12.
We examined the effect of the flaB mutation on a putative chemotaxis operon, which is approximately 400 kb away from flaB. This operon consists of cheW2, orf566, cheA1, cheB2, orf569, and cheY2. Preliminary results indicate that mutations in cheW2, orf569, and cheY2 negatively impact chemotaxis (C. Li, M. Motaleb, and N. Charon, unpublished data). However, although Western blot analysis indicated that CheA1 was produced it was found not to be essential for chemotaxis (40). We tested whether flaB::kan affected the expression of CheA1 and CheY2. Western blot analysis indicated that the levels of CheA1 and CheY2 in the MC-1 mutant were equal to those in the wild type (Fig. 3). These results indicate that flaB::kan did not influence the amounts of CheA1 and CheY2.
Protein synthesis of select genes in the flaA operon.
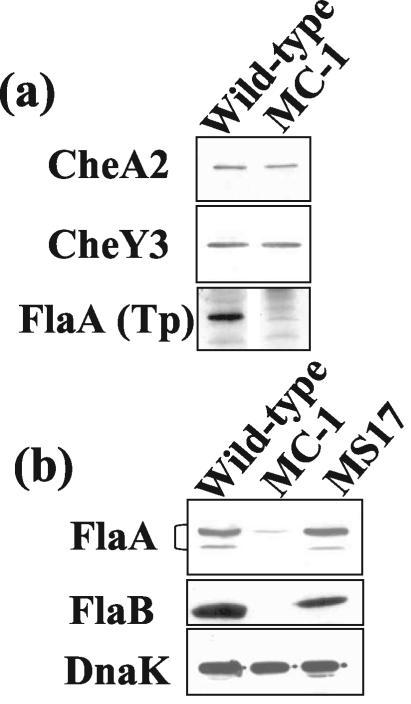
Another relatively large operon consists of flaA-cheA2-cheW3-cheX-cheY3 (20, 22, 23). It maps approximately 550 kb away from flaB. Targeted mutations in cheA2, cheX, and cheY3 markedly affect chemotaxis in B. burgdorferi (40; M. A. Motaleb and N. W. Charon, unpublished data). We tested the effect of the flaB::kan mutation on the synthesis of FlaA, CheA2, and CheY3. Western blot analysis indicated that both CheA2 and CheY3 in strain MC-1 were synthesized at levels equivalent to that for the wild type (Fig. 4a). However, the results obtained with Western blots probed with either a heterologous polyclonal FlaA antiserum (Fig. 4a) or a monoclonal antibody directed to B. burgdorferi FlaA (Fig. 4b) indicated that the level of FlaA in MC-1 was markedly decreased. When the monoclonal antibody was used, FlaA often formed a doublet, with the major band migrating to approximately 38 kDa and the minor band at 37 kDa. With the monoclonal antibody, approximately 15 μg of protein lysate loaded in the gel lane was necessary for detection of FlaA in strain MC-1. No FlaA in strain MC-1 was seen with the polyclonal antiserum under the same conditions. Quantitative immunoblotting analysis indicated that the amount of FlaA in MC-1 was approximately 13% of the amount seen with the wild-type strain (Fig. 4b). To further test whether this inhibition of FlaA accumulation was specifically related to flaB::kan, we tested the flaB::kan-complemented strain MS17. This strain is wavy and motile and has PFs (61). We found that the level of FlaA in strain MS17 was equivalent to that seen with the wild-type strain (Fig. 4b). These results suggest that the flaB::kan mutation specifically decreases FlaA accumulation in B. burgdorferi.
FIG. 4.
Effect of flaB::kan on proteins encoded by genes in the flaA operon. (a) Western blot analysis of wild-type and MC-1 strains. FlaA was probed with polyclonal T. pallidum FlaA antiserum (Tp). (b) Western blot probed with monoclonal antibodies anti-FlaA, FlaB, and DnaK to the wild-type strain, strain MC-1, and the complemented strain MS-17. DnaK was used as an internal control. For FlaA, FlaB, and DnaK detection in the wild-type and MS-17 strains, approximately 10 μg of cell lysate was loaded into each lane. For strain MC-1, 20 μg of cell lysate was loaded into the lane for FlaA and 10 μg was loaded for DnaK. The apparent sizes (in kilodaltons) of the proteins are as follows: CheA2, 98; CheY3, 10; FlaA, 38; DnaK, 72.
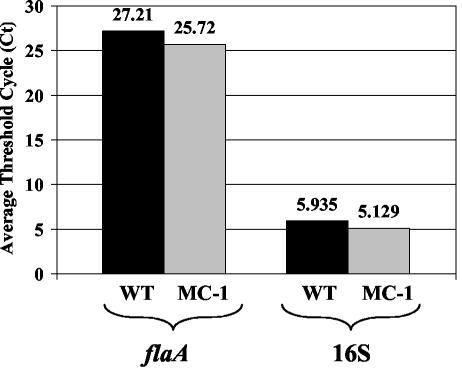
Transcription of flaA in strain MC-1.
One mechanism that could explain the reduced FlaA protein levels in the flaB::kan mutant is inhibition of flaA transcription. To test for this possibility, mRNA levels were measured for the wild-type strain and flaB::kan mutant by both quantitative RT-PCR and RNase protection assays. 16S rRNA was used as a control in both assays. Quantitative RT-PCR results indicated that the level of flaA transcript in MC-1 was similar to or slightly greater than that seen with the wild-type strain (Fig. 5). Furthermore, RNase protection assay results yielded PhosphorImager units for the wild-type and MC-1 strains of 2,150 and 2,020 for flaA and 1,175 and 1,015 for 16S rRNA, respectively. These results taken together reveal that the flaB::kan mutation did not negatively influence flaA mRNA synthesis. The results are also consistent with the lack of inhibition of downstream gene expression in this operon in strain MC-1, as exemplified by the results seen with CheA2 and CheY3 (Fig. 4a).
FIG. 5.
Detection and quantification of relative amounts of flaA transcript in the wild-type (WT) and MC-1 strains using real-time RT-PCR. Average threshold cycle (Ct) values were calculated from duplicate samples in each experiment.
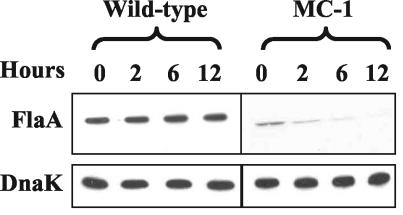
Posttranscriptional control of FlaA.
Two explanations are evident with respect to the lack of FlaA protein detected in MC-1. One hypothesis states that FlaA synthesis is controlled at the translational level. For example, perhaps FlaB is an activator for translation of FlaA. Alternatively, perhaps FlaA is rapidly degraded without FlaB. To test for possible turnover, spectinomycin was added to growing cells and the stability of FlaA was monitored by Western blot analysis (Fig. 6). Spectinomycin at 100 μg/ml was found to rapidly inhibit protein synthesis and growth, and the cells remained intact for several hours. DnaK was used as a control, as transcription of this protein has been previously shown to be relatively constant under different growth conditions (56). We found that the level of DnaK in spectinomycin-treated cells was stable in both the wild-type and MC-1 strains for as long as 12 h (Fig. 6). Similar results were found with respect to FlaA in the wild-type strain. However, the level of FlaA in strain MC-1 showed an approximately 50% decrease by 2 h. By 12 h, there still was a trace of FlaA present. These results indicate that FlaA is degraded in the MC-1 strain, with a half-life of approximately 2 h.
FIG. 6.
Degradation of FlaA. Stability of FlaA was determined by adding spectinomycin (100 μg/ml of culture) to growing B. burgdorferi cells. At different time points, samples were subjected to Western blotting with monoclonal anti-FlaA. DnaK was used as an internal control. Approximately 5 μg of lysate was loaded into each lane for the wild-type strain. For strain MC-1, approximately 20 μg was loaded for detection of FlaA and 5 μg was loaded for detection of DnaK.
DISCUSSION
Motility is energetically costly to B. burgdorferi. We found that the contribution of the major PF filament protein FlaB to the organism is substantial; it constitutes between 10 and 14% of the cellular protein total. Furthermore, at least 5% of the genome is dedicated to motility and chemotaxis (17). Specifically, approximately 42 ORFs homologous to motility genes of other bacteria have been identified; this estimate includes the multiple homologs of certain chemotaxis (cheY, cheA, cheW, cheR, and cheB) and flagellar genes (fliG). Targeted mutagenesis of many of these genes indicates that they function in motility and chemotaxis (40; M. Motaleb, C. Li, M. Sal, R. Bakker, and N. Charon, unpublished data). In addition, eight more ORFs of undetermined function map within the major motility operons. Because of their location, these ORFs are also likely to be involved in motility and chemotaxis. Thus, we estimate that approximately 50 of the total of 1,000 ORFs encoded by the B. burgdorferi chromosome are involved in these functions.
We found that there was considerably less FlaA than FlaB in B. burgdorferi, with FlaA contributing less than 0.5% of the total cellular protein. The function of FlaA in spirochete motility is presently unknown. In several spirochete species, FlaA forms a protein sheath around the FlaB core (9, 11, 38, 41). Null mutants in flaA in B. hyodysenteriae have been characterized (41, 58). These mutants lack the flagellar protein sheath and have an altered PF helical shape, but they still retain motility (41). Microscopic observations and swarm plate assays indicate that these flaA mutants of B. hyodysenteriae are less motile than the wild type (41, 58). In B. burgdorferi, no sheath is evident in electron microscopic observation (Motaleb and Charon, unpublished). As shown by immunoprecipitation, FlaA associates with FlaB, confirming that it is a flagellar protein. In contrast to the results seen with B. hyodysenteriae (41), FlaA did not influence the helical shape of the PFs from B. burgdorferi. Preliminary results with a flaA::kan mutant yielded a PF shape identical to that seen with the wild type (Motaleb and Charon, unpublished). Future experiments will focus on localizing FlaA on the PF filament by immunogold technology.
We found by Western blot analysis that FlaA in B. burgdorferi often migrated as a doublet (Fig. 4b). These doublets were most commonly observed when large amounts of protein were loaded in the gels. The major protein was approximately 38 kDa in size, which is consistent with our published results obtained with strain 212 and the results of others (23, 26). The minor protein of approximately 37 kDa was often not seen (Fig. 1, 2, and 6), and we could only detect it by Western blotting. The minor 37-kDa band has not been previously reported (23, 26). Several possible explanations could account for the lack of previously detection of the minor FlaA protein. One possibility is differences with anti-FlaA reagents. In our previous studies, we used a heterologous polyclonal antibody to T. pallidum FlaA (23). As shown in Fig. 4, the minor protein was not detected using this antiserum; it was only detected using the monoclonal anti-FlaA of B. burgdorferi. Furthermore, when we used the monoclonal antibody to test strain 212, the Western blot pattern was identical to that seen with strain B31 (Motaleb and Charon, unpublished). Thus, the 37-kDa band is not strain specific. In addition, this minor protein was not detected by Gilmore et al., who first obtained and analyzed the FlaA monoclonal antibody (26). One possible explanation is that these investigators used the less-sensitive colorimetric assay for immunostaining. Because the 37-kDa protein was present in relatively small amounts, it may have gone undetected (26). Motaleb and Charon found that prolonged incubation of cell lysates at 23 or 37°C did not change the ratio of the two FlaA bands (Motaleb and Charon, unpublished); thus, it is unlikely that the 37-kDa protein is the result of protein degradation. Both proteins are likely to be encoded by flaA, as a flaA::kan mutant lacks both proteins but still synthesizes FlaB at a level similar to that seen with the wild-type strain (Motaleb and Charon, unpublished). Our preliminary experiments performed to characterize the minor band by mass spectroscopy have been unsuccessful. It will be interesting in future experiments to sort out the relationship of the two proteins. Several lines of evidence suggest that the N terminus of the FlaA is cleaved by signal peptidase I (20, 21, 23, 26) and that this protein is also glycosylated (23). Perhaps the lower-molecular-weight form is unglycosylated FlaA. Alternatively, others have shown that the carboxy-terminal end of certain outer membrane proteins of B. burgdorferi is cleaved (52); thus, the low-molecular-weight protein could be the result of specific protease digestion.
We examined the effects of an insertion mutation in flaB on the synthesis of other motility proteins. Because B. burgdorferi lacks sigma-dependent cascade control of flagellar gene regulation (11, 17, 20, 22, 24, 25, 42), we could not predict how a flaB mutation would influence the expression of other motility genes. Western blot analysis indicates that eight of nine putative chemotaxis and motility proteins tested had unaltered levels of accumulation compared to the wild-type results. Of those tested, the only gene product found to be significantly reduced was FlaA at a level of approximately 13% of the wild-type level (Fig. 4b). In E. coli and S. enterica serovar Typhimurium, mutations in fliC, which encodes the only protein found in the filament, do not effect the expression of other flagellar genes (14). Thus, the results obtained are similar in some respects to those obtained with E. coli. However, several species of bacteria have multiple filament proteins. For example, Caulobacter crescentus and Vibrio parahaemolyticus have six (16, 47-49), B. hyodysenteriae has four (41, 58), and Helicobacter pylori, H. mustelae, and Campylobacter jejuni have two (35, 62, 65). In these species, mutants with mutations in the genes that encode specific filament proteins still synthesize the other filament proteins at levels close to those seen with the wild-type strain (35, 41, 49, 62). Thus, B. burgdorferi is different from these other species of bacteria, as flaB::kan did decrease the amount of FlaA.
We investigated the basis of FlaA inhibition in the flaB::kan mutant strain. The results indicated that there was no change in the amount of flaA message. These results suggest that the inhibition of FlaA in the MC-1 is posttranscriptional. Posttranscriptional control of flagellar gene expression in other species has been noted. For example, in S. enterica serovar Typhimurium this control is coupled to the secretion and assembly of basal body proteins and flagellin (2, 7, 36, 37). In addition, the membrane protein FlK regulates translation of the flagellum-specific anti-sigma factor FlgM in response to flagellar ring assembly (37). In C. crescentus, synthesis of both FljK and FljL flagellar filament proteins have been shown to be under translational control at specific checkpoints in flagellar assembly (3, 49). Recently, Limberger et al. isolated a T. denticola mutant with a mutation in tap1 (44). tap1 is a homolog to fliK, the flagellar hook assembly gene found in other bacteria. This mutant failed to make flagellar filament FlaB proteins, but RT-PCR results indicated that flaB continued to be transcribed. Because the relative amount of flaB mRNA in tap1 was not determined, it is unclear whether the flaB mRNA in the mutant achieved wild-type levels.
There are two likely explanations for the inhibition of FlaA accumulation in the FlaB mutant. First, FlaA could be degraded by cell proteases. Spectinomycin inhibition results indicate that whereas this protein is stable in the wild-type strain, the small amount detected in the flaB::kan mutant was degraded, with a half-life of approximately 2 h. This turnover rate relative to the 8- to 10-h generation time would readily explain the small amount of FlaA detected in the flaB mutant. To our knowledge, these results are the first evidence for protein turnover in B. burgdorferi. Degradation of FlaA in this mutant could be related to the complicated assembly of spirochete flagellar filaments. Specifically, FlaB has no signal sequence and is excreted into the periplasmic space by a type III secretory pathway in a manner similar to that of other bacterial flagellin (11, 13, 20, 46). On the other hand, FlaA of B. burgdorferi and other spirochetes contains a signal peptidase I cleavage site near its N terminus (20). Several lines of evidence indicate that it reaches the periplasmic space by the type II pathway (13, 20, 21, 23, 26, 42). Both proteins evidently form a complex in the periplasmic space for filament formation. Perhaps in the absence of FlaB, FlaA is more vulnerable to protease digestion. In other systems, proteins that comprise a complex but fail to pair with their partners, e.g., RcsA and RcsB in E. coli, are often unstable and readily degradable (29). This hypothesis is further supported by our finding that among the downstream genes in the flaA operon, the amounts of CheA2 and CheY3 were unaltered in strain MC-1 compared to the results seen with the wild-type strain (Fig. 4a).
Another possible explanation is translational control of FlaA. For example, perhaps FlaB or another factor acts as a positive effector for translation of the flaA message. This type of control has been noted in other systems, such as the Com protein stimulating translation of mom mRNA in bacteriophage Mu (64). Genomic analysis of B. burgdorferi indicates that it has only a few two-component signal transduction systems, and obvious operator-repressor systems are not evident (17). Recent evidence indicates that RpoN and RpoS are both involved in the regulation of gene expression in tick and mammalian hosts (32). Perhaps B. burgdorferi evolved specific adaptive strategies for these hosts and consequently shed many of the control mechanisms found in other bacteria. Along these lines, several other genes (e.g., genes Bbk2.10 and elpA.1 and genes flaA and flaB in a flgE mutant) have been identified in B. burgdorferi for which there was abundant mRNA present but the level of the encoded protein was undetectable or markedly decreased (30; Sal et al., Abstr. Bacterial Locomotion and Signal Transduction Meeting VII). The results presented here and elsewhere do not allow us to differentiate which type of mechanism (translational control versus degradation) is occurring with respect to FlaA regulation and these other proteins; perhaps both are occurring simultaneously. To precisely sort this out, rapid pulse-labeling experiments would be helpful. However, because of the rich nutrient requirements of B. burgdorferi our attempts to use this approach have so far been unsuccessful.
Acknowledgments
We thank A. Barbour, D. Blair, J. Benach, F. Cabello, J. Carroll, B. Johnson, P. Matsumura, S. Norris, and the late R. Macnab for kindly sharing strains, monoclonal antibodies, and antisera. We also thank T. Elliot, M. Hirsch, S. Minnich, J. Olson, E. Rucks, R. Bakker, C. Li, and D. Yelton for suggestions and critical comments on the manuscript.
This research was supported by U.S. Public Health Service grant AI29743 to N.W.C. and American Heart Association grant 0365225B to M.A.M.
REFERENCES
- 1.Aizawa, S.-I., I. B. Zhulin, L. M. Márquez-Magaña, and G. W. Ordal. 2001. Chemotaxis and motility, p. 437-452. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 2.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. K., and A. Newton. 1997. Posttranscriptional regulation of Caulobacter flagellin genes by a late flagellum assembly checkpoint. J. Bacteriol. 179:2281-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, A. G., and S. F. Hayes. 1986. Biology of Borrelia species. Microbiol. Rev. 50:381-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour, A. G., S. F. Hayes, R. A. Heiland, and M. E. Schrumpf. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 52:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 7.Bonifield, H. R., S. Yamaguchi, and K. T. Hughes. 2000. The flagellar hook protein, FlgE, of Salmonella enterica serovar Typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J. Bacteriol. 182:4044-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brahamsha, B., and E. P. Greenberg. 1988. A biochemical and cytological analysis of the complex periplasmic flagella from Spirochaeta aurantia. J. Bacteriol. 170:4023-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll, J. A., N. El Hage, J. C. Miller, K. Babb, and B. Stevenson. 2001. Borrelia burgdorferi RevA antigen is a surface-exposed outer membrane protein whose expression is regulated in response to environmental temperature and pH. Infect. Immun. 69:5286-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charon, N. W., and S. F. Goldstein. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 36:47-73. [DOI] [PubMed] [Google Scholar]
- 12.Charon, N. W., S. F. Goldstein, S. M. Block, K. Curci, J. D. Ruby, J. A. Kreiling, and R. J. Limberger. 1992. Morphology and dynamics of protruding spirochete periplasmic flagella. J. Bacteriol. 174:832-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charon, N. W., E. P. Greenberg, M. B. H. Koopman, and R. J. Limberger. 1992. Spirochete chemotaxis, motility, and the structure of the spirochetal periplasmic flagella. Res. Microbiol. 143:597-603. [DOI] [PubMed] [Google Scholar]
- 14.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman, J. L., and J. L. Benach. 1992. Characterization of antigenic determinants of Borrelia burgdorferi shared by other bacteria. J. Infect. Dis. 165:658-666. [DOI] [PubMed] [Google Scholar]
- 16.Ely, B., T. W. Ely, W. B. Crymes, Jr., and S. A. Minnich. 2000. A family of six flagellin genes contributes to the Caulobacter crescentus flagellar filament. J. Bacteriol. 182:5001-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, G. M., T. Hirano, H. U. Ferris, L. L. Devgan, M. Kihara, and R. M. Macnab. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 48:1043-1057. [DOI] [PubMed] [Google Scholar]
- 19.Gassmann, G. S., E. Jacobs, R. Deutzmann, and U. B. Gobel. 1991. Analysis of the Borrelia burgdorferi GeHo fla gene and antigenic characterization of its gene product. J. Bacteriol. 173:1452-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge, Y., and N. W. Charon. 1997. An unexpected flaA homolog is present and expressed in Borrelia burgdorferi. J. Bacteriol. 179:552-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge, Y., and N. W. Charon. 1997. FlaA, a putative flagellar outer sheath protein, is not an immunodominant antigen associated with Lyme disease. Infect. Immun. 65:2992-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge, Y., and N. W. Charon. 1997. Molecular characterization of a flagellar/chemotaxis operon in the spirochete Borrelia burgdorferi. FEMS Microbiol. Lett. 153:425-431. [DOI] [PubMed] [Google Scholar]
- 23.Ge, Y., C. Li, L. Corum, C. A. Slaughter, and N. W. Charon. 1998. Structure and expression of the FlaA periplasmic flagellar protein of Borrelia burgdorferi. J. Bacteriol. 180:2418-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge, Y., I. Old, I. S. Girons, and N. W. Charon. 1997. The flgK motility operon of Borrelia burgdorferi is initiated by a σ70-like promoter. Microbiology 143:1681-1690. [DOI] [PubMed] [Google Scholar]
- 25.Ge, Y., I. G. Old, I. Saint Girons, and N. W. Charon. 1997. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus σ70 promoter. J. Bacteriol. 179:2289-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmore, R. D., Jr., R. L. Murphree, A. M. James, S. A. Sullivan, and B. J. Johnson. 1999. The Borrelia burgdorferi 37-kilodalton immunoblot band (P37) used in serodiagnosis of early Lyme disease is the flaA gene product. J. Clin. Microbiol. 37:548-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein, S. F., K. F. Buttle, and N. W. Charon. 1996. Structural analysis of Leptospiraceae and Borrelia burgdorferi by high-voltage electron microscopy. J. Bacteriol. 178:6539-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein, S. F., N. W. Charon, and J. A. Kreiling. 1994. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc. Natl. Acad. Sci. USA 91:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottesman, S., and M. R. Maurizi. 1992. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 56:592-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hovind Hougen, K. 1984. Ultrastructure of spirochetes isolated from Ixodes ricinus and Ixodes dammini. Yale J. Biol. Med. 57:543-548. [PMC free article] [PubMed] [Google Scholar]
- 32.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, R. C., N. Marek, and C. Kodner. 1984. Infection of Syrian hamsters with Lyme disease spirochetes. J. Clin. Microbiol. 20:1099-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177:3010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlinsey, J. E., J. Lonner, K. L. Brown, and K. T. Hughes. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487-497. [DOI] [PubMed] [Google Scholar]
- 37.Karlinsey, J. E., H. C. T. Tsui, M. E. Winkler, and K. T. Hughes. 1998. Flk couples flgM translation to flagellar ring assembly in Salmonella typhimurium. J. Bacteriol. 180:5384-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koopman, M. B., E. Baats, C. J. van Vorstenbosch, B. A. Van der Zeijst, and J. G. Kusters. 1992. The periplasmic flagella of Serpulina (Treponema) hyodysenteriae are composed of two sheath proteins and three core proteins. J. Gen. Microbiol. 138:2697-2706. [DOI] [PubMed] [Google Scholar]
- 39.Koopman, M. B., O. S. de Leeuw, B. M. Van der Zeijst, and J. G. Kusters. 1992. Cloning and DNA sequence analysis of a Serpulina (Treponema) hyodysenteriae gene encoding a periplasmic flagellar sheath protein. Infect. Immun. 60:2920-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, C., R. G. Bakker, M. A. Motaleb, M. L. Sartakova, F. C. Cabello, and N. W. Charon. 2002. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc. Natl. Acad. Sci. USA 99:6169-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, C., L. Corum, D. Morgan, E. L. Rosey, T. B. Stanton, and N. W. Charon. 2000. The spirochete FlaA periplasmic flagellar sheath protein impacts flagellar helicity. J. Bacteriol. 182:6698-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, C., M. A. Motaleb, M. Sal, S. F. Goldstein, and N. W. Charon. 2000. Spirochete periplasmic flagella and motility. J. Mol. Microbiol. Biotechnol. 2:345-354. [PubMed] [Google Scholar]
- 43.Limberger, R. J., and N. W. Charon. 1986. Treponema phagedenis has at least two proteins residing together on its periplasmic flagella. J. Bacteriol. 166:105-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Limberger, R. J., L. L. Slivienski, J. Izard, and W. A. Samsonoff. 1999. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J. Bacteriol. 181:3743-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lux, R., A. Moter, and W. Shi. 2000. Chemotaxis in pathogenic spirochetes: directed movement toward targeting tissues? J. Mol. Microbiol. Biotechnol. 2:355-364. [PubMed] [Google Scholar]
- 46.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 47.McCarter, L. L. 1995. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J. Bacteriol. 177:1595-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minnich, S. A., N. Ohta, N. Taylor, and A. Newton. 1988. Role of the 25-, 27-, and 29-kilodalton flagellins in Caulobacter crescentus cell motility: method for construction of deletion and Tn5 insertion mutants by gene replacement. J. Bacteriol. 170:3953-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motaleb, M. A., L. Corum, J. L. Bono, A. F. Elias, P. Rosa, D. S. Samuels, and N. W. Charon. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. USA 97:10899-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neidhardt, F. C. 1987. Chemical composition of Escherichia coli, p. 3-6. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 52.Noppa, L., Y. Ostberg, M. Lavrinovicha, and S. Bergstrom. 2001. P13, an integral membrane protein of Borrelia burgdorferi, is C-terminally processed and contains surface-exposed domains. Infect. Immun. 69:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norris, S. J., N. W. Charon, R. G. Cook, M. D. Fuentes, and R. J. Limberger. 1988. Antigenic relatedness and N-terminal sequence homology define two classes of major periplasmic flagellar proteins of Treponema pallidum subsp. pallidum and Treponema phagedenis. J. Bacteriol. 170:4072-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norris, S. J., and Treponema pallidum Polypeptide Research Group. 1993. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Microbiol. Rev. 57:750-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nuttall, P. A., G. C. Paesen, C. H. Lawrie, and H. Wang. 2000. Vector-host interactions in disease transmission. J. Mol. Microbiol. Biotechnol. 2:381-386. [PubMed] [Google Scholar]
- 56.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:2003-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosey, E. L., M. J. Kennedy, D. K. Petrella, R. G. Ulrich, and R. J. Yancey, Jr. 1995. Inactivation of Serpulina hyodysenteriae flaA1 and flaB1 periplasmic flagellar genes by electroporation-mediated allelic exchange. J. Bacteriol. 177:5959-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruby, J. D., H. Li, H. Kuramitsu, S. J. Norris, S. F. Goldstein, K. F. Buttle, and N. W. Charon. 1997. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J. Bacteriol. 179:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadziene, A., D. D. Thomas, V. G. Bundoc, S. C. Holt, and A. G. Barbour. 1991. A flagella-less mutant of Borrelia burgdorferi. Structural, molecular, and in vitro functional characterization. J. Clin. Investig. 88:82-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sartakova, M. L., E. Y. Dobrikova, M. A. Motaleb, H. P. Godfrey, N. W. Charon, and F. C. Cabello. 2001. Complementation of a nonmotile flaB mutant of Borrelia burgdorferi by chromosomal integration of a plasmid containing a wild-type flaB allele. J. Bacteriol. 183:6558-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wassenaar, T. M., N. M. Bleumink-Pluym, and B. A. Van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson, D. R., and T. J. Beveridge. 1993. Bacterial flagellar filaments and their component flagellins. Can. J. Microbiol. 39:451-472. [DOI] [PubMed] [Google Scholar]
- 64.Wulczyn, F. G., and R. Kahmann. 1991. Translational stimulation: RNA sequence and structure requirements for binding of Com protein. Cell 65:259-269. [DOI] [PubMed] [Google Scholar]
- 65.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and nonmotile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]