Abstract
The SecYEG heterotrimeric membrane protein complex functions as a channel for protein translocation across the Escherichia coli cytoplasmic membrane. SecY is the central subunit of the SecYEG complex and contains 10 transmembrane segments (TM1 to TM10). Previous mutation studies suggested that TM3 and TM4 are particularly important for SecY function. To further characterize TM3 and TM4, we introduced a series of cysteine-scanning mutations into these segments. With one exception (an unstable product), all the mutant proteins complemented the cold-sensitive growth defect of the secY39 mutant. A combination of this secY mutation and the secG deletion resulted in synthetic lethality, and the TM3 and TM4 SecY cysteine substitution mutations were examined for their ability to complement this lethality. Although they were all positive for complementation, some of the complemented cells exhibited significant retardation of protein export. The substitution-sensitive residues in TM3 can be aligned to one side of the alpha-helix, and those in TM4 revealed a tendency for residues closer to the cytosolic side of the membrane to be more severely affected. Disulfide cross-linking experiments identified a specific contact point for TM3 and SecG TM2 as well as for TM4 and SecG TM1. Thus, although TM3 and TM4 do not contain any single residue that is absolutely required, they include functionally important helix surfaces and specific contact points with SecG. These results are discussed in light of the structural information available for the SecY complex.
A major pathway of protein translocation across the Escherichia coli plasma membrane is mediated by Sec translocase. Among the membrane-embedded Sec components, SecY and SecE are the principal factors that are thought to constitute a channel-like pathway in the membrane for transport of preproteins (15). These proteins are essential for viability of E. coli. In addition, SecG takes part in the fully active SecYEG membrane-integrated complex. SecY has 10 transmembrane segments (TM1 to TM10), 6 cytoplasmic regions (C1 to C6), and 5 periplasmic regions (P1 to P5) (1). It interacts independently with SecE and SecG (7). SecE has three TMs with the cytoplasmic amino terminus and the periplasmic carboxy-terminal tail (21). SecG has two TMs (TM1 and TM2) with both termini facing the periplasm (18) and assists in the SecA insertion-deinsertion reaction cycles (9, 24) presumably through its remarkable property of undergoing topology inversion (18). Although SecG is dispensable for cell growth at least at higher temperatures, it is required for maximum translocase function in vivo and in vitro, especially at low temperatures or in the absence of the proton motive force (6).
Subunit interactions within the SecYEG complex have been studied in considerable detail. Genetic studies by Baba et al. (3) indicated that the C4 region of SecY is important for SecY-SecE interaction, and cross-linking studies showed that some C4 and C5 residues are indeed physically close to the conserved cytosolic region of SecE (20). In addition, SecY and SecE interact via their transmembrane regions as well (8, 30, 31). Site-specific cross-linking experiments identified a C2-TM3-P2-TM4-C3 segment of SecY as an interface with SecG (19, 29), a conclusion that was also supported by genetic suppression analysis (19).
Although TM regions might be important for transmembrane substrate transport and its regulation in SecYEG, their roles have not been sufficiently studied. We replaced the central six amino acid residues of each TM of SecY with unrelated sequences and suggested that the TM3 and TM4 regions are particularly important for SecY function along with TM2, TM7, and TM10 (22). In the present study, we attempted to elucidate further the roles of TM3 and TM4 amino acid residues in the translocation-facilitating function of SecY, with special reference to the previous implication that they are involved in interaction with SecG. We thus constructed a series of SecY derivatives having a single cysteine substitution in the TM3 and TM4 segments. Their complementation phenotypes and abilities to form a disulfide-bonded complex with appropriately engineered SecG proteins are described. In this way, we identified functionally important helix surfaces of TM3 and TM4 and specific contact points between the SecY and SecG TM regions.
MATERIALS AND METHODS
Media and transduction.
Minimal medium M9 was prepared as described previously (23). L medium contained 10 g of Bacto Tryptone per liter, 5 g of yeast extract per liter, and 5 g of NaCl per liter. P1 transduction was performed by the standard procedures (11). Introduction of ΔsecG was confirmed by PCR amplification with appropriate primers.
Bacterial strains.
E. coli K-12 strain MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR] was described by Silhavy et al. (23). Its derivative, GN31 (secY39) (10) carrying pSTD343 (lacIq; constructed and provided by Y. Akiyama) was used for complementation tests of SecY derivatives. KN370 (ΔsecG::kan) was described previously (17). THE453 (C600 recD1009 ΔsecG::kan zha203::Tn10; our laboratory stock) was used as a donor strain in P1 transduction of the zha203::Tn10 and ΔsecG markers. Strain SA101 (MC4100 ompT::kan ΔsecG::kan zha203::Tn10) was described previously (19).
Construction of plasmids encoding SecY variants having single-cysteine substitutions in TM3 and TM4.
We first constructed a plasmid encoding cysteine-less SecY-His6-Myc. A 1.1-kbp DNA fragment corresponding to the C-terminal ∼80% of cysteine-less SecY was amplified from pAJ51 (19) with a primer, 5′-TTCTATCTTTGCTCTGG-3′, corresponding to nucleotides 281 to 297 of secY and the primer 5′-ACTTCTCGGGCGACGGATCCTCGGCCGTAGCC-3′ (the underlined BamHI recognition sequence was designed such that a Ser codon substituted for the secY stop codon to allow continued translation into the his6-myc tag sequence). The amplified fragment was introduced into pNA3 (12, 22), carrying secY+-his6-myc, after digestion with SmaI and BamHI. This plasmid was named pHM459, into which two unique restriction sites, KpnI and NheI, were introduced at the positions corresponding to Gly58-Thr59 and Ala75-Ser76 of SecY, respectively, without amino acid substitutions, using the QuikChange method (Stratagene). The resulting plasmid, named pHM462, was used as template for QuikChange introduction of the TM3 and TM4 cysteine substitutions. To avoid fortuitous mutations, the NheI-SmaI TM3 fragment and the SmaI-HindIII TM4 fragment were confirmed for their sequences and cloned back into unmutagenized pHM462. The final plasmids are summarized in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmida | Encoded protein(s) |
|---|---|
| pHM462 | Cys-less SecY-His6-Myc |
| pHM464 | SecY(A136C)-His6-Myc |
| pHM465 | SecY(Q131C)-His6-Myc |
| pHM466 | SecY(I129C)-His6-Myc |
| pHM467 | SecY(Y122C)-His6-Myc |
| pHM468 | SecY(S132C)-His6-Myc |
| pHM469 | SecY(A128C)-His6-Myc |
| pHM470 | SecY(Q118C)-His6-Myc |
| pHM471 | SecY(T120C)-His6-Myc |
| pHM472 | SecY(T137C)-His6-Myc |
| pHM473 | SecY(P140C)-His6-Myc |
| pHM474 | SecY(Y119C)-His6-Myc |
| pHM475 | SecY(G123C)-His6-Myc |
| pHM476 | SecY(T124C)-His6-Myc |
| pHM477 | SecY(L125C)-His6-Myc |
| pHM478 | SecY(L127C)-His6-Myc |
| pHM479 | SecY(F130C)-His6-Myc |
| pHM480 | SecY(I135C)-His6-Myc |
| pHM481 | SecY(V126C)-His6-Myc |
| pHM482 | SecY(I133C)-His6-Myc |
| pHM483 | SecY(G134C)-His6-Myc |
| pHM484 | SecY(G138C)-His6-Myc |
| pHM485 | SecY(L139C)-His6-Myc |
| pHM486 | SecY(N141C)-His6-Myc |
| pHM487 | SecY(R121C)-His6-Myc |
| pHM488 | SecY(G123C I135C)-His6-Myc |
| pNA3 | SecY-His6-Myc |
| pNA61 | SecY(Y157C)-His6-Myc |
| pNA62 | SecY(F158C)-His6-Myc |
| pNA63 | SecY(T159C)-His6-Myc |
| pNA64 | SecY(A160C)-His6-Myc |
| pNA65 | SecY(V161C)-His6-Myc |
| pNA66 | SecY(V162C)-His6-Myc |
| pNA67 | SecY(S163C)-His6-Myc |
| pNA68 | SecY(L164C)-His6-Myc |
| pNA69 | SecY(V165C)-His6-Myc |
| pNA70 | SecY(T166C)-His6-Myc |
| pNA71 | SecY(G167C)-His6-Myc |
| pNA72 | SecY(T168C)-His6-Myc |
| pNA73 | SecY(M169C)-His6-Myc |
| pNA74 | SecY(F170C)-His6-Myc |
| pNA75 | SecY(L171C)-His6-Myc |
| pNA76 | SecY(M172C)-His6-Myc |
| pNA77 | SecY(W173C)-His6-Myc |
| pNA78 | SecY(L174C)-His6-Myc |
| pNA79 | SecY(G175C)-His6-Myc |
| pNA80 | SecY(E176C)-His6-Myc |
| pSA61 | SecE SecG(L10C) |
| pSA62 | SecE SecG(I11C) |
| pSA63 | SecE SecG(V12C) |
| pSA64 | SecE SecG(A13C) |
| pSA65 | SecE SecG(I14C) |
| pSA66 | SecE SecG(G15C) |
| pSA67 | SecE SecG(L16C) |
| pSA68 | SecE SecG(V17C) |
| pSA69 | SecE SecG(T61C) |
| pSA70 | SecE SecG(L62C) |
| pSA71 | SecE SecG(F63C) |
| pSA72 | SecE SecG(F64C) |
| pSA73 | SecE SecG(I65C) |
| pSA74 | SecE SecG(I66C) |
| pSA75 | SecE SecG(S67C) |
| pSA76 | SecE SecG(L68C) |
| pSTD343 | LacIq |
| pTYE100 | SecE SecG |
| pTWV228 | Vector |
| pSTV28 | Vector |
pHM462 to pHM488 and pNA3 to pNA80 are pBR-based plasmids. pSA61 to pSA76 are pACYC184-based plasmids. pHM462 to pHM488, pNA61 to pNA80, and pSA61 to pSA76 were constructed in this study. pNA3 (12) and pTYE100 (13) were described previously.
Construction of plasmids encoding SecG variants having single-cysteine substitutions in TM1 and TM2.
Plasmid pTYE100 encoded SecE and SecG under the control of the tac promoter (13). QuikChange site-directed mutations were introduced into its secG region to construct plasmids encoding SecG variants having single-cysteine substitutions in the TM regions, as listed in Table 1.
Protein export measurement.
Export of a periplasmic protein, maltose-binding protein (MBP), was examined by pulse-labeling cells with [35S]methionine and immunoprecipitating labeled MBP species, which were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) into the precursor and mature forms (13). Radioactive proteins were visualized and quantified by a Fuji BAS1800 phosphorimager. Export efficiency was assessed from the proportion of the processed mature form in the labeled MBP molecules.
Immunoblotting.
Intracellular accumulation of SecY and SecG was examined by subjecting whole-cell proteins from a fixed number of cells, as estimated from the culture turbidity, to SDS-PAGE and visualization by immunoblotting using appropriate antibodies and Fuji LAS1000 luminescent image analyzer. All the details were described previously (13).
AMS modification.
Cells of GN31 (secY39) carrying pSTD343 (lacIq) and an additional single-cysteine SecY plasmid were grown at 37°C in M9 minimal medium containing glycerol and amino acids supplemented with maltose (0.4%) and isopropyl-β-d-thiogalactoside (IPTG, 1 mM). A portion of an early-exponential-phase culture was then treated with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) (3 mM) at 37°C for 30 min or not treated with AMS. Subsequently, samples were divided into two portions, one of which was pulse-labeled for 30 s and processed for MBP export assessment as described above. Another portion was used to determine whether the SecY derivative was susceptible to further modification with O-(2-maleimidoethyl)-O′-methyl-polyethylene glycol 5,000 (Mal-PEG). For the latter assay, cultures were directly treated with trichloroacetic acid (final concentration, 5%). After acetone washing, protein precipitates were solubilized with 1% SDS containing 2 mM Tris (2-carboxyethyl) phosphine hydrochloride without or with 5 mM Mal-PEG. Samples were incubated at room temperature for 30 min and then at 37°C for 30 min. Proteins were separated by SDS-PAGE and visualized by anti-Myc immunoblotting as described above.
Disulfide cross-linking.
Preparations of crude membranes were used for disulfide cross-linking experiments. Cells of SA101 carrying an appropriate combination of SecY and SecEG plasmids were grown at 37°C in L medium containing appropriate antibiotics, harvested, and converted into spheroplasts as described previously (19). They were resuspended in a solution containing 10% (wt/vol) sucrose, 3 mM EDTA (pH 7.0), 1 mM dithiothreitol, and 0.1 mM phenylmethylsulfonyl fluoride and disrupted by sonication at 4°C. After clarification of unbroken cells by low-speed centrifugation (18,500 × g for 10 min at 4°C), crude membrane materials containing the outer and inner membranes were isolated by ultracentrifugation (125,000 × g for 30 min at 4°C) and resuspended in 50 mM HEPES-KOH (pH 7.2)-20% glycerol. Disulfide oxidation was performed essentially by the method of Veenendaal et al. (30). Membrane suspensions in 50 mM HEPES-KOH (pH 7.2)-20% glycerol were mixed with 1/10 volume of a solution containing 10 mM CuSO4 and 30 mM o-phenanthroline and incubated for 30 min at the indicated temperature. The reaction was terminated with 10 mM neocuproine. Subsequently, trichloroacetic acid (final concentration, 5%) was added to precipitate proteins, which were collected by centrifugation, washed with acetone, and dissolved in 1.5% SDS-100 mM Tris-HCl (pH 9.0)-5 mM EDTA-15 mM iodoacetamide by agitating at room temperature for 30 min. Samples were subjected to SDS-PAGE, under reducing or nonreducing conditions. SecY and SecG, as well as their covalent complexes, were detected by immunoblotting using antibodies against the Myc epitope and against SecG.
RESULTS
Cysteine-scanning mutation analysis of TM3 and TM4.
Results of our TM substitution experiments suggested that TM3 and TM4 regions of SecY are important for the function of SecY (22), whereas site-specific cross-linking identified the C2-TM3-P2-TM4-C3 region of SecY as a part of the interface with SecG (18). To elucidate the functional importance of each amino acid residue of TM3 and TM4, we introduced cysteine substitution mutations into these regions (Fig. 1). The parental plasmid for mutagenesis carried a cysteine-less version of SecY-His6-Myc, which accumulated stably in the cell (data not shown) and retained full biological activity (Fig. 2, column 16). Altogether, 44 single-cysteine variants (24 TM3 substitutions and 20 TM4 substitutions) were constructed (Fig. 1 and Table 1).
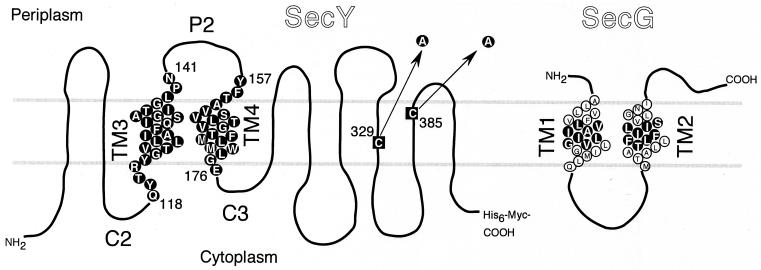
FIG. 1.
SecY and SecG residues that were subjected to cysteine substitution mutation. SecY and SecG amino acid sequences are shown according to their topology models (1, 18). The two cysteine residues of SecY (black squares) were converted to alanine as indicated by the arrows. Single-cysteine mutations were then introduced into residues shown by circles with a black background. SecG residues that were converted to cysteine are similarly shown.
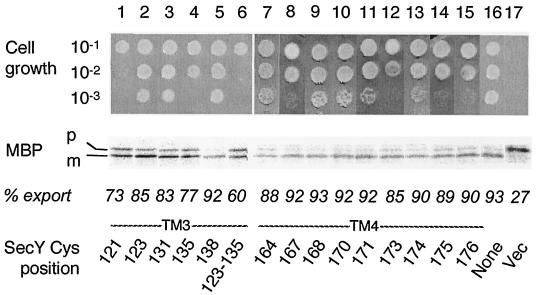
FIG. 2.
Abilities of single-cysteine SecY derivatives to complement the secY39 defect. Plasmids encoding single-cysteine SecY derivatives as specified by the positions (indicated by residue numbers) of substitution were introduced into strain GN31 (secY39). For growth tests (top), cells were grown in L broth at 37°C until mid-log phase and diluted serially (10−1 to 10−3 as indicated) with 0.9% NaCl; 2-μl portions of each suspension were spotted onto L-agar plates, which were incubated at 37°C for 12 h (data not shown) as well as at 20°C for 48 h (photographs). For protein export assay, the same set of bacterial cells was grown at 37°C in M9 minimal medium containing glycerol and amino acids supplemented with maltose (0.4%) and IPTG (1 mM) until early log phase. Thirty minutes after the temperature was shifted to 20°C, cells were pulse-labeled with [35S]methionine for 1 min followed by immunoprecipitation of MBP, SDS-PAGE, and phosphorimaging visualization of the labeled MBP species. The positions of the precursor (p) and mature (m) forms of MBP are indicated. The proportions (% export) of the mature form are shown below the electrophoretic patterns. The Cys position mutated in SecY is shown at the bottom of the figure. Vec, vector.
The mutant secY plasmids thus constructed were introduced into a secY39 mutant, a cold-sensitive mutant (2, 13), to test their ability to complement this SecY defect (2, 14). Immunoblotting experiments demonstrated that all the single-cysteine SecY variants accumulated stably in the cell, except for SecY(Arg121Cys) that had a decreased (by ∼20%) cellular abundance (data not shown). Whereas the plating efficiency of the secY39 mutants decreased more than 1,000-fold at 20°C, the nonpermissive temperature (Fig. 2, column 17) (22), 38 of the mutant forms of SecY fully rescued the growth defect (the data for only some of the mutants are shown in Fig. 2). The plating efficiencies of variants with Ile135Cys (column 4), Gly167Cys (column 8), Trp173Cys (column 12), Gly175Cys (column 14), and Glu176Cys (column 15) substitutions were slightly lower than normal (∼1/10). The plasmid expressing the unstable Arg121Cys variant was still less potent in complementation (Fig. 2, column 1).
The complemented mutant cells were examined for their protein export activities by pulse-labeling MBP. Without a complementing secY plasmid, only 20 to 30% of MBP was labeled as the mature form at 20°C by a 1-min pulse-labeling with [35S]methionine (Fig. 2, lane 17). This value was increased to more than 80% by introduction of any plasmid with a single cysteine substitution of SecY, except for Arg121Cys (73% mature form [Fig. 2, lane 1]) and Leu135Cys (77% mature form [Fig. 2, lane 4]).
Complementation tests in the absence of SecG.
Although the above complementation tests indicated that none of the TM3-TM4 cysteine substitution mutants lost SecY function, these tests may not have been sensitive enough to report a relatively minor defect. Since a secY deletion mutant is not available and introducing the mutations from plasmids into the chromosome could be time-consuming and/or impossible (22), we used a more convenient but sensitive genetic test. SecG is required for optimal functioning of the SecYE complex but is actually dispensable, its deletion being associated with a cold-sensitive phenotype in certain genetic backgrounds.
We found that the secY39 and ΔsecG::Kanr mutations are synthetically lethal. Thus, in P1 transduction experiments using a donor strain having two closely linked mutations (zha203::Tn10 and ΔsecG::Kanr) and a recipient secY39 strain, no Kanr marker was cotransduced with the selective marker, Tetr (Tn10). In contrast, about 80% of Tetr transductants received the Kanr marker when the recipient strain had been complemented with a secY+ or secG+ plasmid. Indeed, the normal cotransduction frequency between these markers is ∼80%.
To examine whether the mutant SecY plasmids can complement the secY39 ΔsecG lethality, they were introduced into the secY39 strain, and the resulting plasmid-bearing mutant cells were used as recipients for Tetr transductant selection at 37°C. Although the SecY(Arg121Cys) plasmid did not allow cotransduction of ΔsecG::kan, all the others did so. The complemented double mutants were still cold sensitive due to the ΔsecG::kan mutation. Pulse-labeling at 20°C showed that export of MBP was retarded more in the double mutant cells that were complemented with some of the mutant SecYs than those complemented with others. The less active SecY variants contained one of the Gly123Cys, Gln131Cys, and Ile135Cys alterations in TM3 and one of the Leu164Cys, Thr168Cys, Phe170Cys, Leu171Cys, Leu174Cys, and Gly175Cys alterations in TM4 (Fig. 3).
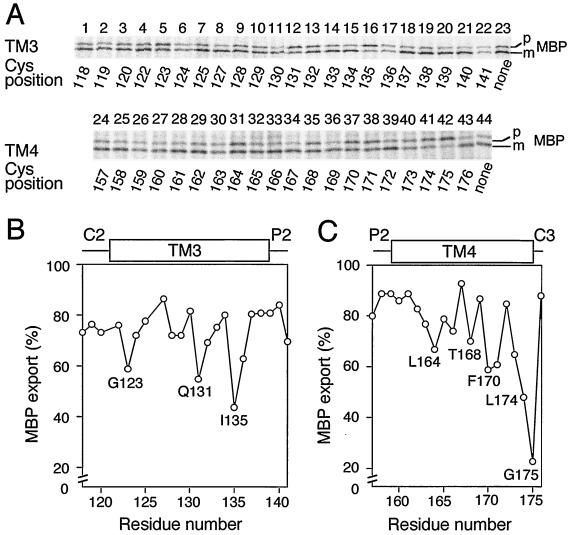
FIG. 3.
Protein export activities of the secY39 ΔsecG cells complemented with single-cysteine SecY derivatives. (A) Profiles of pulse-labeled MBP. The ΔsecG::kan mutation was introduced by P1 transduction into strain GN31 (secY39) carrying plasmid pSTD343 (lacIq) as well as a plasmid encoding the cysteine-less SecY-His6-Myc (lanes 23 and 44) or one of the single-cysteine SecY derivatives with substitutions in TM3 and TM4 at the residues indicated below the blots by residue numbers. Cells were grown at 37°C in M9 minimal medium containing glycerol, amino acids, maltose (0.4%), and IPTG (1 mM) until early log phase. The temperature of a portion of the culture was then shifted to 20°C for 30 min, and then the culture was pulse-labeled with [35S]methionine for 1 min. MBP was immunoprecipitated and separated into the precursor (p) and mature (m) forms. Labeled MBP molecules were visualized by phosphorimaging. (B and C) Graphical representations of the MBP export activities as a function of the mutation locations. The proportions of the labeled mature form of MBP (MBP export) are plotted against the mutation positions as indicated by amino acid residue numbers in TM3 (B) and TM4 (C).
The extents of MBP export in the complemented double mutant strains, as indicated by signal sequence processing, were plotted against the positions of the cysteine substitutions for TM3 (Fig. 3B) and TM4 (Fig. 3C). Interestingly, periodic changes in export proficiency were noted for both sets of the mutations. The pattern obtained for TM3 showed several minima that were separated by three or four amino acid residues (Fig. 3B). For TM4, protein export proficiency appeared to decrease gradually as the alterations approached the cytosolic side of the TM segment. The cysteine substitution-sensitive residues in TM3 and TM4 tended to be localized to one side of each alpha-helix (28) (see Discussion). These helix surfaces might be functionally important, especially in the absence of SecG.
The importance of residues Gly123, Gln131, and Ile135 was also studied in the secG+ cells by combining two of them (Table 1). It was noted that a Gly123Cys-Ile135Cys combination resulted in poor growth (Fig. 2, column 6) and a retardation of MBP export (Fig. 2, lane 6). Another double mutant with the Gly123Cys-Gln131Cys combination also exhibited a slight export defect (data not shown). Thus, the residues that face the “functional” side of the TM3 helix are important even in the presence of SecG.
Extracellular effects of a sulfhydryl alkylating agent on protein export.
Only very limited information is available about the functioning of the SecY amino acid residues located in the periplasmic side. We made use of the TM3 single-cysteine derivatives to partially address this question. Mutant cells with an engineered cysteine residue in the region between residues 134 to 141, the periplasmic half of TM3, were treated with 3 mM AMS, a membrane-impermeable thiol modifier (27), at 37°C for 30 min. Subsequently, cells were pulse-labeled with [35S]methionine for 30 s, and MBP was immunoprecipitated (Fig. 4A; data not shown for Gly134Cys, Ile135Cys, and Ala136Cys). Among those tested, SecY(Gly138Cys) and SecY(Leu139Cys) were inhibited by AMS (Fig. 4A, lanes 2 and 3). The MBP processing efficiencies of SecY(Gly138Cys) and SecY(Leu139Cys) were decreased from more than 90% to 50 and 73%, respectively.
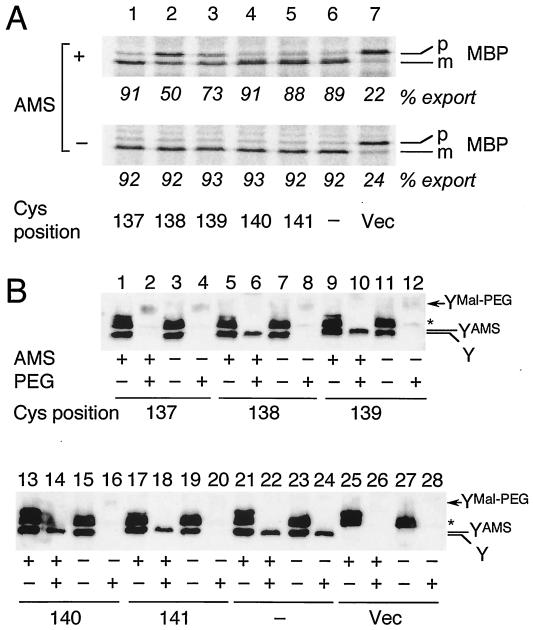
FIG. 4.
(A) Effects of AMS treatment of intact cells on protein export activity. Cells of strain GN31 (secY39) carrying plasmid pSTD343 (lacIq) as well as a plasmid encoding a TM3 single-cysteine SecY derivative (lanes 1 to 5 [Cys substitution positions shown at the bottom of panel A]), cysteine-less SecY-His6-Myc (lane 6), or vector (Vec) (lane 7) were grown at 37°C in M9 minimal medium containing glycerol, amino acids, maltose (0.4%), and IPTG (1 mM) until early log phase. Each culture was divided into two portions, one of which was treated with 3 mM AMS at 37°C for 30 min (+) and not treated with AMS (−). Cells were then pulse-labeled with [35S]methionine for 30 s at 37°C and processed for MBP immunoprecipitation for visualization of labeled MBP species. The positions of the precursor (p) and mature (m) forms of MBP are indicated. The percent export values represent proportions of the mature form. (B) Verification of AMS modification by the blockage of subsequent Mal-PEG modification. Cells expressing SecY with cysteine substitutions, as indicated by the residue numbers, were grown as described above for panel A and treated with AMS as described above (+) or not treated with AMS (−). The samples were precipitated with 5% (final concentration) trichloroacetic acid and solubilized with 1% SDS supplemented with (+) or without (−) 5 mM Mal-PEG, as described in Materials and Methods. After SDS-PAGE, the SecY-His6-Myc proteins were detected by anti-Myc immunoblotting. The positions of SecY-His6-Myc (Y), its AMS-modified form (YAMS), and its Mal-PEG-modified form (YMal-PEG) are indicated. The asterisk marks a nonspecific background protein.
We then examined the actual abilities of the cysteine residues to receive the chemical modification by externally added AMS. Thus, the AMS-treated and untreated samples were subjected to SDS denaturation and to the second modification by using Mal-PEG (see Materials and Methods). When SecY was derivatized with Mal-PEG, its SDS-PAGE mobility was retarded due to the added mass of this reagent. This resulted in the disappearance of the normal-mobility band (Fig. 4B, compare the Y bands in lanes 3 and 4) with the concomitant appearance of a new, slowly migrating and broad band (YMal-PEG in Fig. 4B, lane 4). The Mal-PEG-modified SecY band was faint, presumably due to its broadening and low-efficiency blotting. The band indicated by the asterisk in Fig. 4B was due to a background protein, which we believe contained some periplasmic and cytosolic cysteine residues, the latter of which remained reactive with Mal-PEG irrespective of the previous treatment of the intact cells with AMS (Fig. 4B, lanes 2, 6, 10, 14, 18, 22, and 26).
The cysteine introduced at position 137 was not blocked by AMS treatment; therefore, this SecY derivative disappeared after denaturation and Mal-PEG modification, whether or not the cells had been treated with AMS (Fig. 4B, lanes 2 and 4). In contrast, all the other cysteine variants tested were significantly affected by AMS treatment, such that large fractions of the Y band became refractory to the Mal-PEG modification (Fig. 4B, lanes 6, 10, 14, and 18). They were all fully modified by Mal-PEG without pretreatment with AMS (Fig. 4B, lanes 8, 12, 16, and 20). Close inspections indicated that the mobility of the Y band was slightly shifted upward in the AMS-treated samples, presumably representing its AMS-modified form (YAMS observed in Fig. 4B, lanes 5, 6, 9, 10, 13, 14, 17, and 18). The cysteine-less SecY was not affected by AMS or Mal-PEG (Fig. 4B, lanes 21 to 24). These results demonstrate that cysteine residues introduced between Gly138 and Asn141 were accessible by AMS from the periplasmic side of the membrane. Our results indicate that AMS can pass through the outer membrane, probably through the porins. This bulky moiety interfered with the translocase function when it modified the side chain at position 138 or 139 but not at position 140 or 141. Interestingly, Gly138 resides on the implicated functional side of the TM3 helix (see below).
Disulfide cross-linking between the TM segments of SecY and SecG.
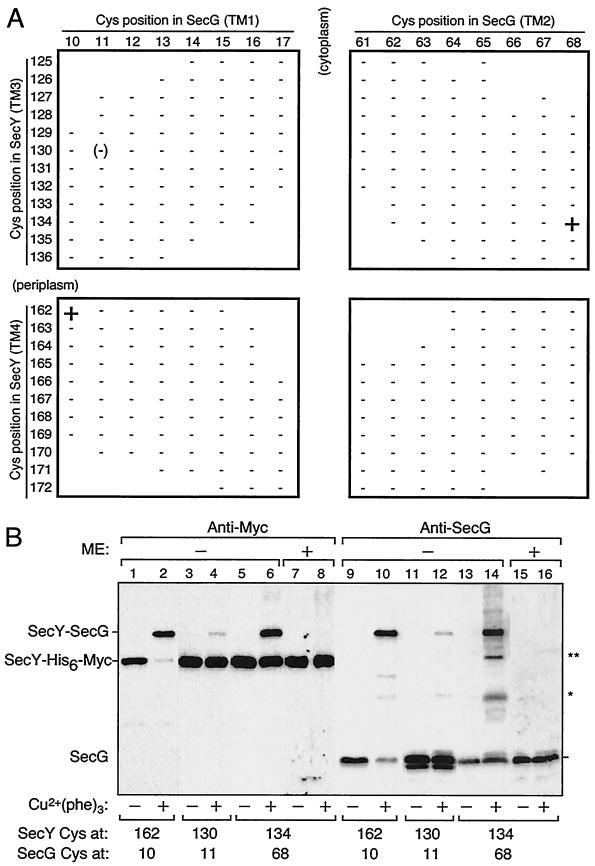
The cross-linking results of Satoh et al. showed that the C2-TM3-P2-TM4-C3 region of SecY is located close to SecG (19). To obtain further information on the SecY-SecG interaction within their TM domains, we examined whether an intermolecular disulfide bond can be formed between combinations of SecY and SecG single-cysteine variants. We examined 12 cysteine substitutions in TM3 (Leu125 to Ala136) and 11 cysteine substitutions in TM4 (Val162 to Met172) (Fig. 1) of SecY. As possible cross-linking partners, we constructed a total of 16 SecG single-cysteine variants with substitutions at the central eight residues of each TM (Leu10 to Val17 in TM1 and Thr61 to Leu68 in TM2; Fig. 5). Two compatible plasmids, one coding for a SecY mutant and the other coding for a SecG mutant, were introduced into the ΔsecG strain. On the basis of the topology models of SecY and SecG proteins (Fig. 1) (1, 17), we omitted some combinations with extreme topological discrepancies (blanks in Fig. 5A).
FIG. 5.
SecY-SecG disulfide cross-linking involving SecY TM3 and TM4. (A) SecY-SecG combinations tested. Membrane fractions prepared from cells carrying two plasmids, one expressing single-cysteine SecY-His6-Myc and the other expressing single-cysteine SecG, were subjected to Cu2+(phenanthroline)3 oxidation. The SecY-SecG combinations are indicated by the residue numbers where cysteine substitutions had been introduced. Samples were analyzed by nonreducing SDS-PAGE and immunostaining using anti-Myc and anti-SecG antibodies. Symbols: +, significant formation of a cross-linked product; −, absence of cross-linking. The 130-11 combination gave very faint cross-linked product and was scored as (−). Combinations without a plus or minus have not been tested. (B) Electrophoretic evidence for cross-linking. Membranes were incubated in the presence (+) or absence (−) of Cu2+-phenanthroline conjugate [Cu2+(phe)3] at 37°C and then electrophoresed after treatment with (+) or without (−) 5% β-mercaptoethanol (ME). Proteins were detected by anti-Myc (lanes 1 to 8) and anti-SecG (lanes 9 to 16) immunoblotting. The SecY-SecG combinations are shown at the bottom by the residues of cysteine substitutions. The identities of the products indicated by asterisks have not been established.
Crude membranes were prepared from cells carrying a total of 307 combinations of plasmids (Fig. 5A). The membranes were then subjected to oxidation with Cu2+-o-phenanthroline and analyzed by SDS-PAGE under nonreducing conditions. SecG and SecY-His6-Myc were detected with anti-SecG and anti-Myc immunoblotting, respectively, whereas their covalent complex was detected by either antibody as a mobility-shifted band (Fig. 5B). As summarized in Fig. 5A, we identified two specific SecY-SecG combinations that gave positive cross-linking, residues 162 and 10 and residues 134 and 68 (Fig. 5B, lanes 2 and 6). The efficiency of cross-linking was almost quantitative for the 162-10 combination and somewhat lower for the 134-68 combination. Although a cross-linked product was slightly detected for a 130 (SecY)-11 (SecG) combination (Fig. 5B, lane 4), its significance is questionable. We did not observe any cross-linked product in other combinations of SecY and SecG derivatives. The cross-linking reactions depended on the oxidant (Fig. 5B). The SecY(Gly134Cys)-SecG(Leu68Cys) combination gave an additional two bands after oxidation (Fig. 5B, lane 14, asterisks). They did not seem to have the C-terminal (Fig. 5B, lane 6) or N-terminal (data not shown for anti-SecY immunoblotting) sequences of SecY. From the SDS-PAGE mobility, the lower band (indicated by a single asterisk) might have been a SecG dimer (16). The cross-linked band between SecY(Gly134Cys)-SecG(Leu68Cys) disappeared after treatment with β-mercaptoethanol, confirming that they were connected by a disulfide bond (Fig. 5B, lanes 8 and 16). The other products, the 162-10 and 130-11 combinations, were only partially reduced by treatments with reducing agents (data not shown). Although the reason for the latter observations is unknown (see also Discussion), the specific combinations of engineered cysteines were clearly required for the disulfide bond formation. These data suggest that SecY and SecG interact not only via their cytosolic and periplasmic regions (19) but also via their TM regions. Specifically, a TM3 residue Gly134 is in the vicinity of Leu68 of SecG TM2, and a TM4 residue Val162 is close to Leu10 of SecG TM1.
DISCUSSION
TM regions of membrane proteins can play at least two roles. Whereas some TM regions may only function to anchor the protein to the membrane with correct local orientations, others may have more direct functional roles. Functional importance of TM3 and TM4 of SecY was suggested previously from our mutation studies, in which six-amino-acid segments in the middle of each SecY TM were replaced by unrelated sequences from the LacY TM regions (22). TM3 (Leu127-Ser132) and TM4 (Leu164-Met169) substitutions severely compromised SecY function without destabilizing the protein (22). In contrast, none of the cysteine-scanning single-amino-acid substitutions in TM3 and TM4 created in this study impaired SecY function to the same extent as the six-amino-acid replacements did. These results do not necessarily dismiss the importance of the TM3 and TM4 residues. A severe loss of function by a single amino acid change can be expected for such a residue that directly contributes to an enzymatic active site. It is doubtful that SecY has such a specific “active site,” since it does not catalyze any chemical reaction. Each of amino acid residues in SecY TMs might have a more cumulative type of role.
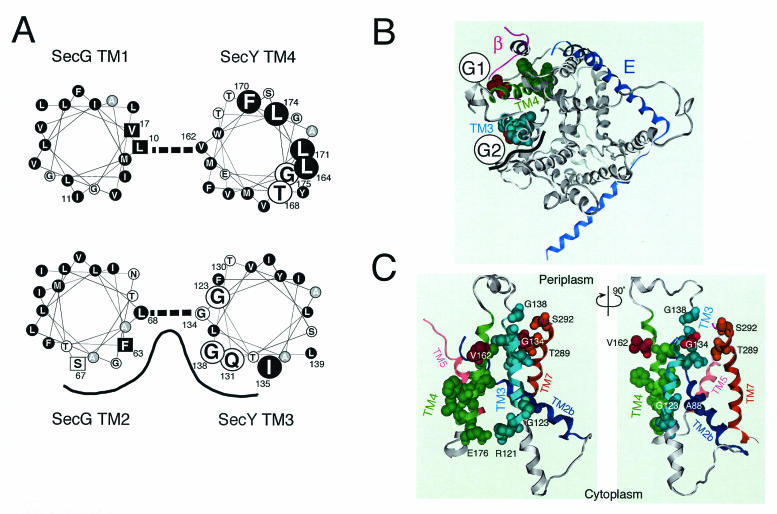
The sequence features and topology determination experiments had predicted that a SecYEG complex contains a total of 15 TM helices (1, 18, 21), which were indeed visualized by a two-dimensional crystal analysis (4). More recently, high-resolution determination of the X-ray structure of the SecYEβ from Methanococcus jannaschii (28) revealed clearly that TM3 and TM4, like other TM segments, form alpha-helices. van den Berg et al. (28) discussed the structure of E. coli SecYEG by integrating its two-dimensional crystal-based density map (4) and the structure of the M. jannaschii complex. We also modeled the E. coli SecY structure as shown in Fig. 6B and C from the published coordinates of the archaeal complex (PDB accession code, 1RHZ), using the Molecular Operating Environment program (Chemical Computing Group, Inc., Montreal, Quebec, Canada). The resulting top view (from the periplasmic side) and two side views are presented in Fig. 6B and C, respectively. For clarity, the SecE (blue) and the β (magenta) subunits from the M. jannaschii SecY complex are superimposed (Fig. 6B). These analyses justify the helical wheel representations of the E. coli SecY TM3 and TM4 (Fig. 6A). The TM3 (Arg121-Gly138) and TM4 (Tyr157-Glu176) regions are highlighted in teal and green, respectively, in Fig. 6B and C.
FIG. 6.
Important helix surfaces of TM3 and TM4. (A) Helical wheel representations and SecY-SecG contact sites. Amino acid residues in TM3 and TM4 of SecY and TM1 and TM2 of SecG are shown in helical wheel representations viewed from the periplasmic side. TM3 and TM2 as well as TM4 and TM1 are paired as shown by thick broken lines on the basis of our disulfide cross-linking results. The functionally important SecY residues identified in this study are shown by large letters. Hydrophobic amino acids are shown by circles with a black background. Alanine is shown by circles with a gray background. Residues in SecG at which suppressor mutations against the secY104 mutation have been isolated (19) are shown by squares. The proposed TM3-TM2 helix surface with relatively hydrophilic amino acids is indicated by a black line. (B) A periplasmic top view of SecY on the basis of the three-dimensional structure of the M. jannaschii SecY complex. The coordinates for the M. jannaschii SecY complex (28) were used to model E. coli SecY according to the Molecular Operating Environment program. A top view of SecY from the periplasmic side is shown in gray, with TM3 and TM4 highlighted in teal and green, respectively, and with the M. jannaschii SecE (E) (blue) and β (magenta) subunits superimposed. The functionally important TM3 and TM4 residues identified in this study are indicated by space-filled side chains, in which the residues of contact with SecG (Gly134 and Val162) are shown in red. Schematic images for the locations of the TM segments of SecG are shown by G1 and G2. The black wavy line indicates the proposed hydrophilic surface formed by SecYTM3 and SecG TM2. (C) Side views around TM3 and TM4 of SecY. A view from the putative SecG side (G1-G2 mid point in panel B) is shown on the left, whereas its ∼90°-rotated version is shown on the right. TM2b (Ser76-Pro100), TM3, TM4, TM5 (Ile187-His205), and TM7 (Asn270-Ser292) are shown in dark blue, teal, green, pink, and brown, respectively. The important TM3 and TM4 residues, including the Arg121-Glu176 pair (see the text), are space filled for their side chains, along with residues in other TMs that are discussed in the text.
We examined the abilities of the TM3 and TM4 cysteine substitution variants to correct the lethal export defect caused by the secY39 ΔsecG double mutation. This strict complementation test identified several residues that contribute to the full functionality of SecY (shown by space-fill representations in Fig. 6B and C). It should be noted that their importance might not be restricted to the SecG-deficient conditions, since some double substitutions compromised SecY function in the secG+ background (Fig. 2). At any rate, it is remarkable that residues assigned as functionally important exhibited specific features in their alignments across the membrane (Fig. 6A). The important TM3 residues are distributed in a periodical fashion along the polypeptide, a feature that might define an important helix surface of this TM segment. Similarly, the important TM4 residues tended to exhibit one-sided distribution axially, as well as a vertical polarity in that its cytosolic side is enriched with them. In earlier studies by Taura et al., one such residue, Gly175, was characterized by a Gly175Asp secY104 mutation that caused cold-sensitive growth and protein export (25, 26). It is imaginable that TM4 has an important role in its cytosolic side.
Our disulfide cross-linking experiments identified two putative SecY-SecG contact points involving their TMs. One involves Gly134 located within the functional surface of TM3 helix of SecY and Leu68 within TM2 of SecG. The other involves Val162 located in the opposite side of the SecY TM4 helix surface that was assigned as functionally important and Leu10 of SecG TM1. The SecY side chains involved in the contact are shown in red in Fig. 6B and C. These results led us to locate SecG TM2 close to SecY TM3 and SecG TM1 close to SecY TM4, as shown schematically in Fig. 6B. Since each TM pair contains only a unique contact site (Fig. 5A), the SecY and SecG TMs may cross each other rather than running in parallel. Indeed, SecY TM4 is tilted in the bilayer (28). The above locations of the SecG TMs are different from the SecYEG arrangement proposed by van den Berg et al. (28) on the basis of the similar orientations of SecG TM2 and the β chain of M. jannaschii. It should be pointed out in this respect that SecG in SecA-containing organisms and β in SecA-lacking organisms might be totally different in their functions. In addition, SecG was reported to invert its orientation of membrane integration during function, in conjunction with conformational changes in SecA (9, 18, 24). Thus, the orientation in the “resting state” alone cannot be taken as a strong guide to formulate a structural model. We propose the arrangement shown in Fig. 6B as an alternative disposition of SecG TMs in the SecYEG complex.
The SecY TM3 helix and SecG TM2 helix show some amphiphilicity (Fig. 6A). The cross-linking results indicate that their hydrophilic surfaces face each other if not running in parallel. Also, the hydrophilic side of SecY TM3 coincides with the functionally important helix surface identified above. This relatively hydrophilic helix surface also includes residue 138 that was shown to be accessible from the periplasmic side by the sulfhydryl alkylating agent that inactivated the translocase function by modifying the engineered cysteine at this position. Thus, this relatively hydrophilic part of the SecY-SecG TM segments (shown by wavy black line in Fig. 6B) is partially open to the periplasmic side and functionally important. While residue 137 is no longer accessible by AMS, the residues between positions 138 and 141 are all periplasmically exposed and capable of being modified by AMS. However, AMS modification at position 140 or 141 had no effect on protein export. Thus, residues 138 and 139 may occupy a critical region of the channel such that a bulky moiety at these positions is interfering, although a tryptophan substitution for Gly138 did not inactivate the channel (H. Mori, unpublished results).
We note that Thr289 and Ser292 in TM7 face the functionally important part of TM3. A TM7 region near the periplasm has been discussed by van den Berg et al. to have a role in channel gating and lateral movement of a signal sequence (28). SecY TM3 and SecG TM2 might contribute to such a process. It should also be noted that Phe63 and Ser67 of SecG, which are located within the TM3-TM2 interface region (Fig. 6A) are known as sites of mutations that suppress the secY104 defect, consistent with their contribution to the gate control of the channel.
Although the important TM3 residues are mostly located toward the periplasmic side of the membrane, Gly123 is located near the cytoplasmic surface and facing Ala88 in TM2b. Thus, TM3's role is not confined to a periplasmic event. We believe that TM3 and TM4 interact, particularly at their cytosolic ends, where Arg121 of TM3 is located close to Glu176 of TM4. Their estimated distance of 3.8 Å suggests that they form a salt bridge. Indeed, the Arg121Cys and Glu176Cys variants showed reduced functionality (Fig. 2). Also, cytosolic regions before TM3 and after TM4 include residues in close proximity to the cytosolic region of SecG (19).
Although uneven localization of the important residues in TM4 to one side of the helix is evident, neither this helix nor interacting TM1 helix of SecG is particularly amphiphilic (Fig. 6A). Overall, they are very hydrophobic. The contact site with SecG TM1 is on the opposite side of the functional TM4 surface and of the secY104 alteration site. The contact point on SecG, Leu10, and its “neighbor” Val17 are also known as sites of suppressor alterations (Leu10Arg, Leu10His, and Val17Asp) against the secY104 defect (19). The high efficiency of cross-linking between cysteines introduced into position 162 (SecY) and position 10 (SecG) suggests that these residues are very close to each other. In fact, we observed that this intermolecular cross-linkage was not readily broken by a reducing agent even in the presence of SDS. Although this phenomenon is not fully understood, the extreme hydrophobicity of this region might have precluded the access of the reducing agent to this disulfide bond. In any case, the charge-introducing suppressor mutations seem to indicate that weakening of this hydrophobic interaction acts to shift the channel conformation to a more open state.
The importance of the cytosolic side of TM4 is corroborated by the discussion of van den Berg et al. that the following flexible loop containing some conserved glycine residues is involved in the widening of the hourglass-shaped main conduit (28). We speculate that such a regulatory function can be assisted by SecG that interacts with the C2-TM3-P2-TM4-C3 region of SecY in manners partially resolved by our present genetic information and our previous cross-linking studies (19).
Undoubtedly, a signal peptide and SecA will be additional factors that interact with this region of the complex. These elements should function with remarkable mobility (5, 18). Further exploitation of genetics and structural biology will be promising for our deeper understanding of the dynamic functioning of protein translocase.
Acknowledgments
We thank Yoshinori Akiyama for stimulating discussions; Tomoya Tsukazaki for help in the SecY structure modeling; and Michiyo Sano, Kunihiko Yoshikaie, and Kiyoko Mochizuki for technical support.
This work was supported in part by CREST, JST (Japan Science and Technology Agency) (to K.I.), by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.M. and K.I.), and by National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Akiyama, Y., and K. Ito. 1987. Topology analysis of the SecY protein, an integral membrane protein involved in protein export in Escherichia coli. EMBO J. 6:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., A. Jacq, E. Brickman, J. Beckwith, T. Taura, C. Ueguchi, Y. Akiyama, and K. Ito. 1990. Characterization of cold-sensitive secY mutants of Escherichia coli. J. Bacteriol. 172:7005-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T., T. Taura, T. Shimoile, Y. Akiyama, T. Yoshihisa, and K. Ito. 1994. A cytoplasmic domain is important for the formation of a SecY-SecE translocator complex. Proc. Natl. Acad. Sci. USA 91:4539-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breyton, C., W. Haase, T. A. Rapoport, W. Kuhlbrandt, and I. Collinson. 2002. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418:662-665. [DOI] [PubMed] [Google Scholar]
- 5.Economou, A., and W. Wickner. 1994. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78:835-843. [DOI] [PubMed] [Google Scholar]
- 6.Hanada, M., K. Nishiyama, and H. Tokuda. 1996. SecG plays a critical role in protein translocation in the absence of the proton motive force as well as at low temperature. FEBS Lett. 381:25-28. [DOI] [PubMed] [Google Scholar]
- 7.Homma, T., T. Yoshihisa, and K. Ito. 1997. Subunit interactions in the Escherichia coli protein translocase: SecE and SecG associate independently with SecY. FEBS Lett. 408:11-15. [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann, A., E. H. Manting, A. K. Veenendaal, A. J. Driessen, and C. van der Does. 1999. Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry 38:9115-9125. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto, G., H. Mori, and K. Ito. 1998. Roles of SecG in ATP- and SecA-dependent protein translocation. Proc. Natl. Acad. Sci. USA 95:13567-13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto, G., T. Yoshihisa, and K. Ito. 1997. SecY and SecA interact to allow SecA insertion and protein translocation across the Escherichia coli plasma membrane. EMBO J. 16:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, J. H. 1972. Experiments in molecular genetics, p. 431. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Mori, H., Y. Akiyama, and K. Ito. 2003. A SecE mutation that modulates SecY-SecE translocase assembly, identified as a specific suppressor of SecY defects. J. Bacteriol. 185:948-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori, H., and K. Ito. 2001. An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc. Natl. Acad. Sci. USA 98:5128-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori, H., and K. Ito. 2003. Biochemical characterization of a mutationally altered protein translocase: proton motive force stimulation of the initiation phase of translocation. J. Bacteriol. 185:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori, H., and K. Ito. 2001. The Sec protein-translocation pathway. Trends Microbiol. 9:494-500. [DOI] [PubMed] [Google Scholar]
- 16.Nagamori, S., K. Nishiyama, and H. Tokuda. 2002. Membrane topology inversion of SecG detected by labeling with a membrane-impermeable sulfhydryl reagent that causes a close association of SecG with SecA. J. Biochem. 132:629-634. [DOI] [PubMed] [Google Scholar]
- 17.Nishiyama, K., M. Hanada, and H. Tokuda. 1994. Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 13:3272-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama, K., T. Suzuki, and H. Tokuda. 1996. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell 85:71-81. [DOI] [PubMed] [Google Scholar]
- 19.Satoh, Y., G. Matsumoto, H. Mori, and K. Ito. 2003. Nearest neighbor analysis of the SecYEG complex. 1. Identification of a SecY-SecG interface. Biochemistry 42:7434-7441. [DOI] [PubMed] [Google Scholar]
- 20.Satoh, Y., H. Mori, and K. Ito. 2003. Nearest neighbor analysis of the SecYEG complex. 2. Identification of a SecY-SecE cytosolic interface. Biochemistry 42:7442-7447. [DOI] [PubMed] [Google Scholar]
- 21.Schatz, P. J., K. L. Bieker, K. M. Ottemann, T. J. Silhavy, and J. Beckwith. 1991. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 10:1749-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimokawa, N., H. Mori, and K. Ito. 2003. Importance of transmembrane segments in Escherichia coli SecY. Mol. Gen. Genomics 269:180-187. [DOI] [PubMed] [Google Scholar]
- 23.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Suzuki, H., K. Nishiyama, and H. Tokuda. 1998. Coupled structure changes of SecA and SecG revealed by the synthetic lethality of the secAcsR11 and ΔsecG::kan double mutant. Mol. Microbiol. 29:1013-1021. [DOI] [PubMed] [Google Scholar]
- 25.Taura, T., T. Yoshihisa, and K. Ito. 1997. Protein translocation functions of Escherichia coli SecY: in vitro characterization of cold-sensitive secY mutants. Biochimie 79:517-521. [DOI] [PubMed] [Google Scholar]
- 26.Taura, T., Y. Akiyama, and K. Ito. 1994. Genetic analysis of SecY: additional export-defective mutations and factors affecting their phenotypes. Mol. Gen. Genet. 243:261-269. [DOI] [PubMed] [Google Scholar]
- 27.Uchida, K., H. Mori, and S. Mizushima. 1995. Stepwise movement of preproteins in the process of translocation across the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 270:30862-30868. [DOI] [PubMed] [Google Scholar]
- 28.van den Berg, B., W. M. Clemons, Jr., I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature 427:36-44. [DOI] [PubMed] [Google Scholar]
- 29.van der Sluis, E. O., N. Nouwen, and A. J. Driessen. 2002. SecY-SecY and SecY-SecG contacts revealed by site-specific crosslinking. FEBS Lett. 527:159-165. [DOI] [PubMed] [Google Scholar]
- 30.Veenendaal, A. K., C. van der Does, and A. J. Driessen. 2001. Mapping the sites of interaction between SecY and SecE by cysteine scanning mutagenesis. J. Biol. Chem. 276:32559-32566. [DOI] [PubMed] [Google Scholar]
- 31.Veenendaal, A. K., C. van der Does, and A. J. Driessen. 2002. The core of the bacterial translocase harbors a tilted transmembrane segment 3 of SecE. J. Biol. Chem. 277:36640-36645. [DOI] [PubMed] [Google Scholar]