Abstract
The arginine regulatory protein of Pseudomonas aeruginosa, ArgR, is essential for induction of operons that encode enzymes of the arginine succinyltransferase (AST) pathway, which is the primary route for arginine utilization by this organism under aerobic conditions. ArgR also induces the operon that encodes a catabolic NAD+-dependent glutamate dehydrogenase (GDH), which converts l-glutamate, the product of the AST pathway, in α-ketoglutarate. The studies reported here show that ArgR also participates in the regulation of other enzymes of glutamate metabolism. Exogenous arginine repressed the specific activities of glutamate synthase (GltBD) and anabolic NADP-dependent GDH (GdhA) in cell extracts of strain PAO1, and this repression was abolished in an argR mutant. The promoter regions of the gltBD operon, which encodes GltBD, and the gdhA gene, which encodes GdhA, were identified by primer extension experiments. Measurements of β-galactosidase expression from gltB::lacZ and gdhA::lacZ translational fusions confirmed the role of ArgR in mediating arginine repression. Gel retardation assays demonstrated the binding of homogeneous ArgR to DNA fragments carrying the regulatory regions for the gltBD and gdhA genes. DNase I footprinting experiments showed that ArgR protects DNA sequences in the control regions for these genes that are homologous to the consensus sequence of the ArgR binding site. In silica analysis of genomic information for P. fluorescens, P. putida, and P. stutzeri suggests that the findings reported here regarding ArgR regulation of operons that encode enzymes of glutamate biosynthesis in P. aeruginosa likely apply to other pseudomonads.
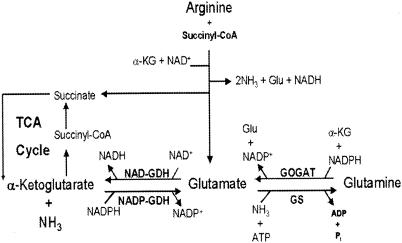
The arginine succinyltransferase (AST) pathway (Fig. 1) is the major route for arginine catabolism under aerobic conditions in Pseudomonas aeruginosa, This pathway converts l-arginine into l-glutamate with the concomitant release of three nitrogen moieties (11, 13, 14). Utilization of arginine as a carbon source entails deamination of glutamate to α-ketoglutarate, which is then channeled into the tricarboxylic acid (TCA) cycle. We have recently reported (18) the cloning and characterization of gdhB, which encodes a novel NAD+-dependent glutamate dehydrogenase (NAD-GDH; GdhB). The expression of gdhB was shown to be inducible by exogenous arginine, and this induction was mediated by ArgR, the arginine regulatory protein. The activity of GdhB, a tetramer of equal 180-kDa subunits, was also found to be subject to allosteric activation by arginine. The induction of gdhB expression and the activation by arginine of the encoded enzyme clearly serve as mechanisms that coordinate aerobic utilization of arginine as a carbon source with glutamate utilization via the TCA cycle.
FIG. 1.
The AST pathway and glutamate biosynthesis in P. aeruginosa PAO1. Only key intermediates and enzymes related to this report are shown. NADP-GDH (GdhA), anabolic NADP-dependent GDH; NAD-GDH (GdhB), catabolic NAD-dependent GDH; GOGAT, GltBD; Glu, glutamate, α-KG, α-ketoglutarate; CoA, coenzyme A.
The ArgR protein of P. aeruginosa does not exhibit any sequence homology to the arginine regulatory proteins from enteric bacteria (17, 19) or Bacillus subtilis (5). Rather, ArgR of P. aeruginosa is a member of the AraC/XylS family of transcriptional regulators (27) and functions like other members of this family (8), both as a transcriptional repressor and as an activator in control of operons responsible for arginine uptake and metabolism (20, 23, 24, 27). The operon that encodes ArgR of P. aeruginosa is autoinduced in the presence of exogenous arginine and is subject to carbon catabolite repression (23).
As a nitrogen source, the nitrogen moieties released from arginine via the AST pathway are used either by transamination into glutamate or by ammonia assimilation. Similar to enteric bacteria (28), ammonia assimilation in P. aeruginosa is catalyzed by an NADP-dependent GDH (NADP-GDH; GdhA) when the ammonia supply is high and by the combined actions of glutamine synthetase (GS) and glutamate synthase (GOGAT; GltBD) when the ammonia supply is limited. The presence of these three enzymes for ammonia assimilation has been demonstrated in P. aeruginosa (4, 15). While the corresponding genes have not been characterized, they are annotated in the finished genome project on the basis of sequence homology of the derived amino acid sequences (PAO1 Genome Annotation Project; www.pseudomonas.com).
The finding that gdhB, which is required for utilization of arginine (and glutamate) as a carbon source, is a member of the ArgR regulon in P. aeruginosa raised the intriguing question of whether ArgR also plays a role in controlling the expression of genes responsible for utilization of arginine as a nitrogen source. This paper reports studies that demonstrate that ArgR indeed mediates repression by arginine of the gltBD and gdhA genes. In addition, gltBD and gdhA are shown to be subject to regulation by the availability of glutamate and ammonia.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) enriched medium (30) was used with the following supplements as required: ampicillin at 50 μg/ml (Escherichia coli); carbenicillin at 200 μg/ml (P. aeruginosa); gentamicin at 100 μg/ml; streptomycin at 500 μg/ml; and 5-bromo-3-indolyl-β-d-galactopyranoside (X-Gal) at 0.03% (wt/vol). Minimal medium P, described by Haas et al. (12), and minimal medium E (30) were used for the growth of P. aeruginosa and E. coli, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Wild type | 12 |
| PAO1-Sm | Spontaneous Smr mutant of PAO1 | 26 |
| PAO501 | argR::Gmr | 26 |
| PAO502 | gltD::Gmr | This study |
| E. coli | ||
| DH5α | F−/endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA relA1 Δ(lacIZYA-argF) U169 deoR [φ80dlacΔ(lacZ)M15] | Bethesda Research Laboratories |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Kmr) | 9 |
| K-12 | Wild type | Laboratory collection |
| Plasmids | ||
| pQF52 | Apr; lacZ translational fusion vector derived from broad-host-range plasmid pQF50 | 27 |
| pKA58 | carA promoter region in pUC19 | 26 |
| pKB39 | gltB::lacZ fusion in pQF52 | This study |
| pSH1 | gdhA::lacZ fusion in pQF52 | This study |
| pKB41 | gltBD operon in pUC19 | 16 |
| pRTP1-M | Apr; conjugation vector | 32 |
| pGMΩ1 | Gmr cassette | 31 |
Antibiotic resistance phenotypes: Apr, ampicillin for E. coli and carbenicillin for P. aeruginosa; Gmr, gentamicin; Kmr, kanamycin; Smr, streptomycin; Tcr, tetracycline.
Gel retardation assays.
Homogeneous ArgR purified as previously described (31) was mixed at various concentrations with an end-labeled DNA fragment (10−11 M) in a reaction mixture (total volume, 20 μl) containing 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM EDTA, 50 μg of bovine serum albumin per ml, and 10 ng of sheared salmon sperm DNA. The reaction mixture was incubated at room temperature for 20 min and applied to a 5% (wt/vol) polyacrylamide gel in Tris-acetate-EDTA buffer.
DNA probes containing the regulatory regions of gltBD and gdhA (Fig. 2) were amplified by PCR from pKB41 or genomic DNA of PAO1 with the following synthetic primers designed to generate HindIII or SmaI restriction sites: for gltBD, 5′-TCGGCCAGGCGCATTGATC-3′ and 5′-CTGCCCGGGGGCGATCAGGCCAAATCC-3′; for gdhA, 5′-GCGAAGCTTAGACCCGGCCGTAGGTA-3′ and 5′-GAAAGCGTCGACGGATTTGCGT-3′. The PCR products were purified from a 1% (wt/vol) agarose gel and labeled at the 5′ end with either [α-32P]ATP by T4 nucleotide kinase or [γ-32P]dATP by Klenow fragment.
FIG. 2.
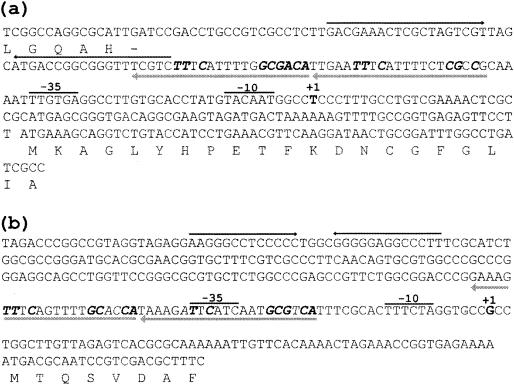
Nucleotide sequences of P. aeruginosa promoters and flanking regions for gltBD (a) and gdhA (b). The transcriptional initiation sites are indicated by +1 above the nucleotides, and the −10 and −35 regions of each promoter are labeled accordingly. The hypersymmetric sequences marked by convergent arrows are possible rho-independent transcriptional terminators. The ArgR-binding sites determined in this study are underlined, and the bases identical to the consensus sequence 5′-TGTCGCN6GNAAN5-3′ on the complementary strand are shown by bold italics.
Enzyme assays.
GOGAT and anabolic GDH activities were assayed at 37°C by measuring the initial rates of NADPH oxidation at 340 nm (21). The reaction mixture (2 ml) contained 0.35 mM NADPH, 5 mM sodium α-ketoglutarate, 100 mM Tris-HCl (pH 7.5), and 20 mM l-glutamine for GltBD or 20 mM NH4Cl for GdhA. Reactions were started by addition of glutamate. The specific activities are expressed as nanomoles of NADPH oxidized per minute per milligram of protein. Protein concentration was determined by the method of Bradford (3) with bovine serum albumin as the standard. β-Galactosidase activity was determined by the method of Miller (22).
Purification of GOGAT.
GOGAT of P. aeruginosa was purified from a strain of E. coli DH5α harboring a high-copy-number plasmid, pKB41 (Table 1), carrying the gltBD genes (16). Cultures were grown in LB medium (2 liters) for maximal repression of chromosomally encoded E. coli GltBD (34). Cells were suspended in 20 mM potassium phosphate buffer (PPB), pH 7.6, containing 1 mM EDTA at a ratio of 1 g of cells to 2 ml of buffer. Phenylmethylsulfonyl fluoride was added to a final concentration of 1 mM immediately prior to the passage of cells through an AMINCO French pressure cell at 800 lb/in2. The cell debris was removed by centrifugation at 48,000 × g for 30 min. Streptomycin sulfate (1 g/100 ml) was added to the supernatant at 0°C with stirring and equilibrated for 30 min. After centrifugation at 48,000 × g for 30 min, the supernatant was subjected to ammonium sulfate fractionation. The fraction precipitating between 30 and 45% saturation was dissolved in 50 ml of 20 mM PPB. This solution was filtered through a Millipore membrane (0.2 μm pore size) and applied to a Pharmacia Mono Q column (HR 10/10) previously equilibrated with 20 mM PPB. GltBD was eluted with a linear gradient of KCl. Fractions containing GltBD activity (0.33 to 0.35 M KCl) were combined, concentrated by precipitation with 70% ammonium sulfate, and dissolved in a minimal volume of 0.1 M potassium phosphate (pH 7.6). The solution was applied to a Pharmacia Superose 6 gel filtration column (H/R 10/30) equilibrated with 0.1 M PPB. Fractions containing GltBD were combined, diluted to 20 mM PPB, and then applied to a Pharmacia HiTrap heparin column equilibrated with 20 mM PPB. Following gradient elution with KCl, fractions containing GltBD activity were combined and dialyzed against buffer containing 20 mM potassium-HEPES (pH 7.2), 2 mM α-ketoglutarate, and 1 mM EDTA.
Determination of amino-terminal amino acid sequence.
Purified GltBD (2 μg) was applied to a (wt/vol) sodium dodecyl sulfate (SDS)-5% polyacrylamide gel, and the constituent subunits were separated by electrophoresis at 100 V for 2 h. After electrophoresis, the large and small subunits were transferred to a polyvinylidene difluoride membrane with the LKB 2051 Midget MultiBlot electrophoretic transfer unit. The amino-terminal amino acid sequences of the separated subunits were determined by Edman degradation with a protein sequencer at the Molecular Genetics Facility of Georgia State University.
Construction of a gltB::lacZ and gdhA::lacZ translational fusions.
DNA fragments containing the regulatory regions of gltBD and gdhA (Fig. 2) were amplified by PCR from pKB41 or genomic DNA of PAO1 with the following synthetic oligonucleotides designed to generate HindIII or SmaI restriction sites: for gltBD, 5′-TCGGCCAGGCGCATTGATC-3′ and 5′-CTGCCCGGGGGCGATCAGGCCAAATCC-3′; for gdhA, 5′-GCGAAGCTTAGACCCGGCCGTAGGTA-3′ and 5′-GAAAGCGTCGACGGATTTGCGT-3′. The PCR products were purified from a 1% (wt/vol) agarose gel, digested by restriction endonucleases HindIII and SmaI, and ligated to the corresponding sites of lacZ translational fusion vector pQF52 (Table 1). The nucleotide sequences of the resulting constructs, pKB39 and pSH1 for gltBD and gdhA, respectively, were verified by nucleotide sequence determination.
Gene replacement.
A 1.6-kb EcoRI fragment containing the gentamicin resistance (Gmr) cassette was isolated from plasmid pGMΩ1 (31) by agarose gel electrophoresis and cloned into the EcoRI site of gltD on pKB41 containing the entire gltBD operon. The resulting plasmid containing the gltD::Gmr region was digested by KpnI and cloned into the KpnI site of a conjugation vector, pRTP1-M (32). The resulting gene replacement plasmid was mobilized into a spontaneous streptomycin-resistant P. aeruginosa strain, PAO1-Sm, by biparental plate mating with E. coli SM10 as described by Gambello and Iglewski (9). Following incubation at 37°C for 16 h, transconjugants were selected on LB plates supplemented with gentamicin (250 μg/ml) and streptomycin (500 μg/ml).
Nucleotide sequence accession number.
The sequences of gltBD and the flanking regions from P. aeruginosa have been deposited in the GenBank database under accession number U81261.
RESULTS
The gltBD operon encodes the two unequal subunits of GOGAT. The gltBD operon of PAO1 has been cloned and sequenced previously by Kwon and Abdelal (16). The open reading frames of the gltB and gltD genes have coding capacities for polypeptides of 161.6 and 52.6 kDa, respectively. The derived amino acid sequences of GltB and GltD of PAO1 exhibit 75 and 80% sequence similarity (data not shown), respectively, to the large and small subunits of GOGAT of E. coli (25). The gltBD operon of PAO1 is separated by two putative rho-independent terminators from an upstream PA5037 gene, which encodes a hypothetical protein, and from the downstream hemE gene, which encodes uroporphyrinogen decarboxylase (PAO1 genome annotation project; www.pseudomonas.com).
GOGAT (GltBD) of P. aeruginosa was purified from a recombinant E. coli strain carrying pKB41 (Table 1) grown under conditions (LB medium) in which the host GltBD is maximally repressed. Interestingly, GltBD from P. aeruginosa binds to heparin and such binding greatly facilitated its purification from this organism. The large and small subunits of GltBD were separated by SDS-polyacrylamide gel electrophoresis, and the first 17 amino acid residues of the amino termini were determined. The amino-terminal sequences of GltB and GltD were XGFGLIAHMQGEPSHQL and SERLNSDRLNNDFQFIE, respectively. Comparison of these sequences to the derived sequences in Fig. 2a indicated that the first 14 residues of the translated GltB sequence are absent and that the purified GltB protein has a terminal cysteine residue, which normally cannot be detected by a protein sequencer. Similar amino-terminal processing of GltB has been reported for E. coli (25). In the case of GltD, comparison of the derived sequence to the determined sequence indicated that the first methionine residue of GltD was removed. The molecular masses of GltB and GltD were estimated to be 162 and 54 kDa, respectively, from a plot of electrophoretic mobility in SDS-15% polyacrylamide gel against the logarithm of molecular masses of known polypeptides. These values are in good agreement with those calculated from the derived sequences. The molecular mass of GltBD of P. aeruginosa was estimated by molecular sieving to be 230 kDa, indicating that the native enzyme is a single heterodimer. In contrast, the native GltBD protein of E. coli self-associates into a tetrameric form (25). Furthermore, unpublished work done in this laboratory showed that GltBD of E. coli did not bind to heparin or to DNA, unlike GltBD of P. aeruginosa, which binds to heparin (this study) and has been found earlier to bind certain DNA fragments (16). The possible significance of these differences between the two enzymes from P. aeruginosa and E. coli are under investigation.
Identification of gltBD and gdhA promoters.
The gdhA gene of PAO1 (PA4588), which encodes the anabolic GDH, has been annotated on the basis of sequence similarity (80%) to the corresponding gene of E. coli (33). Like GDHs of other bacteria (18), the derived sequence of GdhA of PAO1 does not show significant sequence similarity to the catabolic GdhB protein, which was characterized earlier in this laboratory. The results of nucleotide sequence analysis indicated that the gdhA gene is separated by a putative rho-independent terminator from an upstream PA4589 gene and the downstream convergent ccpR gene, which encodes cytochrome peroxidase (7).
The transcriptional initiation sites of the gltBD and gdhA genes were determined by primer extension experiments (data not shown) and identified on the nucleotide sequences as shown in Fig. 2. In the case of gltBD, sequences resembling the −10 and −35 consensus sequences of the σ70 RNA polymerase of E. coli were identified in the proper positions relative to the transcriptional initiation site. For gdhA, the −10 and −35 sequences of the identified transcriptional initiation site were less homologous to the E. coli consensus sequences.
Expression of GOGAT and anabolic GDH is repressed by arginine.
P. aeruginosa PAO1 was grown in glutamate minimal medium in the absence or presence of exogenous arginine, and GltBD and GdhA activities were measured in cell extracts. The results (Table 2) showed that exogenous arginine represses the specific activities of each approximately fourfold. Low levels of both enzymes were observed in extracts of cells grown in minimal medium supplemented with arginine as the sole source of carbon, energy, and nitrogen.
TABLE 2.
Effects of l-arginine and ArgR on repression of gltBD and gdhA opereons
| Strain | Genotype | Mediuma | Sp actb
|
|||
|---|---|---|---|---|---|---|
| GOGAT | NADP- GDH | β-Galacto- sidasec
|
||||
| gltB | gdhA | |||||
| PAO1 | Wild type | Glu | 16 | 65 | 68 | 92 |
| Glu + Arg | 4 | 16 | 33 | 28 | ||
| Arg | 3 | 18 | NDd | ND | ||
| PAO501 | argR::Gmr | Glu | 36 | 114 | 154 | 115 |
| Glu + Arg | 26 | 95 | 159 | 127 | ||
Cells were grown at 37°C in minimal medium with the indicated supplements at 20 mM as the sources of carbon and nitrogen. Glu, glutamate; Arg, arginine.
Specific activities (nmoles per milligram per minute) are mean values from two measurements with standard errors below 15%.
For the measurement of β-galactosidase activities, the gltB::lacZ fusion (pKB39) and the gdhA::lacZ fusion (pSH1) were introduced into the host strain as indicated and the recombinant strains were grown under the conditions described in footnote a. The specific activities (nmoles per milligram per minute) are the average of two measurements with standard errors below 10%.
ND, not determined.
The specific activities of GltBD and GdhA were also determined in strain PAO501, an argR derivative of PAO1. As shown in Table 2, the specific activities of both enzymes were twofold higher in extracts of PAO501 cells grown in glutamate minimal medium, and the repression by exogenous arginine was essentially abolished. These results indicate that ArgR mediates the observed arginine repression in the parent strain, PAO1.
The ArgR-mediated arginine repression was further analyzed with lacZ fusions carried on a low-copy-number vector. Plasmids pKB39 and pSH1, carrying gltB::lacZ and gdhA::lacZ translational fusions, respectively, were introduced into PAO1 and PAO501. The effect of arginine on expression of β-galactosidase was examined in glutamate minimal medium in the absence or presence of arginine. The results (Table 2) showed that exogenous arginine represses lacZ expression from pKB39 and pSH1 in PAO1, but not in the argR derivative, PAO501. These results indicated the presence of arginine-repressible promoters in the upstream regions flanking the gltBD and gdhA genes.
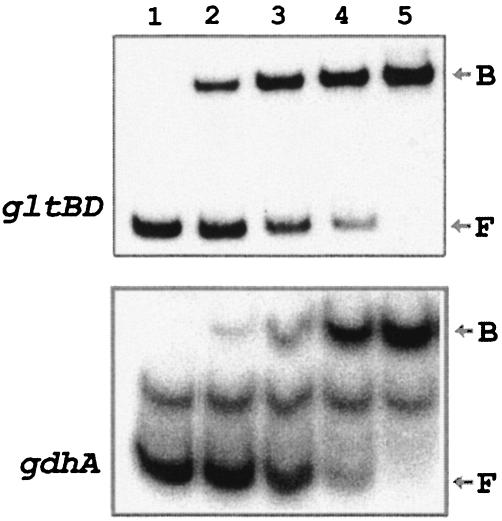
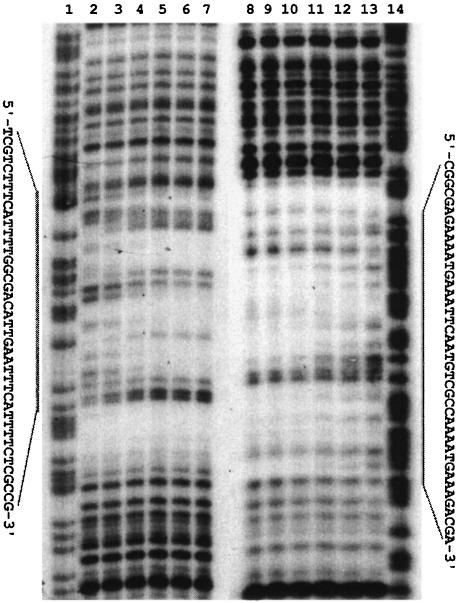
In vitro binding of ArgR to the promoter regions for gltBD and gdhA. Gel retardation experiments were carried out with homogeneous ArgR and a 304-bp DNA fragment carrying the control region for the gltBD operon. The results (Fig. 3) showed that the ArgR protein binds specifically to the gltBD regulatory region with an apparent dissociation constant of 20 pM as determined from a plot of the percentage of bound DNA against the concentration of ArgR (data not shown). DNase I footprinting analysis was used to define the ArgR binding site. Binding of ArgR protects a 42-bp region against nuclease digestion on both strands (Fig. 4), 40 bases upstream of the transcriptional initiation site of the gltB promoter (Fig. 2a).
FIG. 3.
Gel retardation experiments. The radioactive 32P-labeled gltBD and gdhA operator DNA fragments were incubated with the following concentrations of ArgR (from lanes 1 to 5): 0, 15, 30, 60, and 120 pM for gltBD and 0, 50, 100, 200, and 350 pM for gdhA. The reactions were performed as described in Materials and Methods. B, DNA-protein complex; F, free probe.
FIG. 4.
DNase I footprinting analysis of ArgR with the gltBD regulatory region. The DNA fragments used were labeled at the 3′ end of the sense strand (lanes 1 to 7) and its complementary strand (lanes 8 to 14). Lanes: 1 and 14, the corresponding G+A Maxam-Gilbert sequencing ladders; 2 to 7 and 13 to 8, DNase I footprinting with increasing concentrations of ArgR (0, 0.6, 1.2, 2.5, 5, and 10 nM). The protection regions are indicated by bars and the corresponding nucleotide sequences.
Similar experiments with gdhA demonstrated specific binding of ArgR to a 323-bp DNA fragment carrying the control region (Fig. 3). The location of this ArgR binding site was determined from DNase I footprinting analysis (data not shown). The protected region starts at 21 bases upstream of the transcriptional initiation site of the gdhA promoter (Fig. 2b).
Repression of GltBD and GdhA by glutamate and nitrogen limitation.
Since GltBD and GdhA play major physiological roles in glutamate biosynthesis and ammonia assimilation, the possible effects of glutamate and other nitrogen sources on their activities were investigated. To study the effect of exogenous glutamate, PAO1 and its argR derivative, PAO501, were grown in glucose-ammonium minimal medium in the presence or absence of glutamate. The results (Table 3) showed that exogenous glutamate represses GltBD and GdhA in wild-type PAO1 by 4.6- and 5.5-fold, respectively. The repression is retained in the argR derivative, thus precluding a role for ArgR in glutamate repression. It should be noted that the repression ratios for GltBD and GdhA in PAO501 are lower than those observed in PAO1 (2.7- and 2.5-fold, respectively).
TABLE 3.
Effect of glutamate on repression of gltBD and gdhA operons
| Strain | Genotype | Supplementa | Sp act (nmol/min/mg)b
|
|
|---|---|---|---|---|
| GOGAT | NADP-GDH | |||
| PAO1 | Wild type | None | 73 | 589 |
| Glu | 16 | 106 | ||
| PAO501 | argR::Gmr | None | 88 | 345 |
| Glu | 33 | 140 | ||
Cells were grown at 37°C in minimal medium in the presence of 0.2% glucose and 10 mM NH4Cl in the absence or presence 20 mM glutamate (Glu).
Specific activities are mean values from two measurements with standard errors below 15%.
To analyze the effect of other nitrogen sources, PAO1 was grown in succinate minimal medium supplemented with ammonium, nitrate, proline, or serine as the sole source of nitrogen. The results (Table 4) showed that the level of GdhA specific activity generally correlates with the effectiveness of these nitrogen sources in supporting growth of P. aeruginosa, Thus, the highest specific activity was observed with ammonium (doubling time of 47 min) and the lowest was observed with nitrate and serine (doubling times of 75 and 154 min, respectively).
TABLE 4.
Effects of nitrogen sources on expression of gltBD and gdhA operons in P. aeruginosa PAO1
| Nitrogen | Doubling time (min) | Sp act (nmol/min/mg)a
|
|
|---|---|---|---|
| GOGAT | NADP-GDH | ||
| Ammonium | 47 | 46 | 418 |
| Proline | 58 | 8 | 116 |
| Glutamate | 69 | 14 | 84 |
| Nitrate | 75 | 12 | 17 |
| Serine | 154 | 10 | 25 |
Cells were grown at 37°C in succinate minimal medium supplemented with the indicated nitrogen sources at 20 mM. Specific activities are mean values from two measurements with standard errors below 15%.
It is worth noting that the specific activities of GltBD and GdhA in glucose-ammonium minimal medium are about 60 and 40% higher, respectively, than those measured in succinate-ammonium minimal medium (compare enzyme activities in Tables 3 and 4).
Growth phenotype of gltD::Gmr derivative on different nitrogen sources.
Reitzer (28) has proposed that in enteric bacteria, reduction of glutamine biosynthesis resulting from a defect in GOGAT shuts off the Ntr system and, hence, the catabolic genes for nitrogen utilization. To investigate if such a hypothesis is applicable to P. aeruginosa, a gltD::Gmr derivative, PAO502, was constructed and its ability to utilize various amino acids as sole nitrogen sources was examined in succinate minimal medium. PAO502 was capable of utilizing 20 mM l-arginine, l-proline, l-glutamate, or l-glutamine as a sole nitrogen source as effectively as the parent strain, PAO1. However, unlike the parent strain, PAO502 did not utilize 20 mM l-serine or 20 mM nitrate as a sole nitrogen source.
DISCUSSION
Glutamate is the end product of the AST pathway for arginine utilization (Fig. 1). In P. aeruginosa, an arginine-inducible catabolic GDH (GdhB), encoded by the gdhB gene, catalyzes further breakdown of glutamate into α-ketoglutarate, which is then utilized through the TCA cycle (18). Induction of the expression of gdhB and activation of the encoded dehydrogenase by arginine serve to direct the flow of glutamate into the TCA cycle. The repression by exogenous arginine of the gdhA gene reported here could serve to minimize the operation of an energy-consuming futile cycle involving the simultaneous function of gdhA and gdhB when P. aeruginosa uses arginine as a carbon source (Fig. 1). Similarly, arginine repression of the gltBD operon minimizes loss of energy when glutamate is readily available as the product of the AST pathway. Thus, the overall physiological significance of the observed role of arginine in controlling expression of gdhA, gdhB, and gltBD, which encode three key enzymes of glutamate metabolism, is conservation of energy while ensuring flow of the carbon skeletons of arginine into the TCA cycle (for use of arginine as an energy source) and flow of ammonia into glutamine (for use of arginine as a nitrogen source).
Several lines of evidence support the conclusion that ArgR mediates repression by exogenous arginine of GltBD and GdhA. (i) Repression by arginine is abolished in an argR derivative of PAO1 in which argR was inactivated by gene replacement (Table 2). (ii) Gel retardation assays showed that homogeneous ArgR binds specifically to DNA fragments carrying the control regions for the gltBD and gdhA genes (Fig. 3). (iii) DNase I footprinting experiments (Fig. 4) showed that ArgR protects regions homologous to the ArgR binding site and centered at positions −60 (for the gltB promoter) and −41 (for the gdhA promoter) relative to the transcription start site.
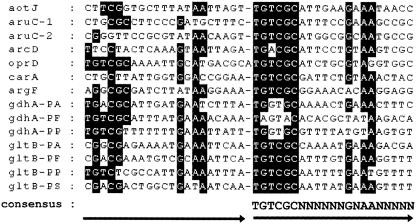
The ArgR binding sites for the gltBD and gdhA genes were compared with the well-characterized ArgR binding sites for the aot, aru, argF, car, arc, and oprD promoters (Fig. 5), revealing that the gltB and gdhA promoters comprise two tandem repeats with a consensus sequence of 5′-TGTCGCN6GNAAN5-3′. In the ArgR-repressible operons argF and carAB, the binding sites completely overlap the target promoters for these operons, indicating that in these cases ArgR exerts its effect by steric hindrance (27). This appears to be the case for gdhA. However, in the case of gltBD, the ArgR binding site is located 7 bases upstream from the −35 region of the promoter and in the reverse orientation relative to the direction of transcription. It is possible that binding of ArgR in such a spatial arrangement could inhibit the binding of the alpha subunit of RNA polymerase to an UP element in the −40 to −60 region of the promoter (29).
FIG. 5.
Sequence alignment of ArgR binding sites. All of the ArgR binding sites of P. aeruginosa shown have been characterized by DNase I footprinting experiments. The first and second halves of the binding sites are depicted by arrows. The consensus sequence was deduced from the more conserved second half-sites. Nucleotides identical to those of the consensus are shaded. PA, P. aeruginosa; PF, P. fluorescens; PP, P. putida; PS, P. stutzeri.
Expression of gltBD and gdhA was found to be also subject to repression by glutamate and nitrogen limitation. Mechanisms for these controls have not been reported in P. aeruginosa. In E. coli, the leucine-responsive regulatory protein Lrp is required for gltBD expression (6) and the Nac protein represses gltBD under nitrogen limitation but has no significant effect on gdhA (10). In contrast, both gltBD and gdhA of Klebsiella aerogenes are repressed by Nac (10). In B. subtilis, the gltAB operon, which encodes GOGAT, requires GltC for its activation (1) and is repressed by the global nitrogen regulator TnrA under nitrogen limitation (2, 35). No Lrp or Nac functional homologues have been hitherto identified for P. aeruginosa.
The gltD knockout mutant constructed in this study exhibited a growth defect in the utilization of nitrate and serine as nitrogen sources. However, the gltD mutant, like the parent PAO1, utilized glutamate, glutamine, arginine, or proline as a sole source of carbon and nitrogen. The growth phenotype of this mutant is consistent with a previous report by Brown and Tata (4). Reitzer has proposed that in enteric bacteria, a defect in GOGAT results in the accumulation of glutamine under nitrogen limitation, which, in turn, shuts off the Ntr system and the catabolic genes for nitrogen utilization (28). Bender et al. have reported evidence that supports the alternative hypothesis that the inability of GOGAT mutants to utilize alternative nitrogen sources is the result of glutamate starvation rather than an effect of the Ntr system (10). In P. aeruginosa, while the latter explanation could account for the inability of the gltD mutant to utilize nitrate or serine, the normal growth phenotype on other amino acids indicates that the Ntr system is less important in this organism. Thus, induction of catabolic pathways for utilization of compounds as both carbon and nitrogen sources might involve mechanisms that are different from the Ntr system.
The findings reported here regarding regulation by arginine of glutamate biosynthesis in P. aeruginosa likely have physiological relevance to other pseudomonads that are proficient in utilizing arginine as a source of carbon, energy, and nitrogen. This hypothesis is supported by in silica analyses of the published genome sequences of P. putida, P. fluorescens, and P. stutzeri at the National Center for Biotechnology Information. In particular, these three pseudomonads have operon structures that are homologous to the aot-arg, aru, and gdhB operons, which function in arginine uptake, utilization, and regulation in P. aeruginosa PAO1 (18, 23, 26). Furthermore, sequences that are homologous to the ArgR-binding sites reported here for gdhA and gltBD in PAO1 were found to be conserved in the corresponding regions in P. putida, P. fluorescens, and P. stutzeri (Fig. 5), except gdhA of P. stutzeri, which does not have this gene.
Acknowledgments
This work was supported by grant MCB-9985660 from the National Science Foundation.
REFERENCES
- 1.Belitsky, B. R., P. J. Janssen, and A. L. Sonenshein. 1995. Site required for GltC-dependent regulation of Bacillus subtilis glutamate synthase expression. J. Bacteriol. 177:5686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky, B. R., L. V. J. Wray, S. H. Fisher, D. E. Bohannon, and A. L. Sonenshein. 2000. Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J. Bacteriol. 182:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. R., and R. Tata. 1981. Growth of Pseudomonas aeruginosa mutants lacking glutamate synthase activity. J. Bacteriol. 147:193-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czaplewski, L. G., A. K. North, M. C. Smith, S. Baumberg, and P. G. Stockley. 1992. Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis, Mol. Microbiol. 6:267-275. [DOI] [PubMed] [Google Scholar]
- 6.Ernsting, B. R., J. W. Denninger, R. M. Blumenthal, and R. G. Matthews. 1993. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J. Bacteriol. 175:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fulop, V., C. J. Ridout, C. Greenwood, and J. Hajdu. 1995. Crystal structure of the di-haem cytochrome c peroxidase from Pseudomonas aeruginosa, Structure 3:1225-1233. [DOI] [PubMed] [Google Scholar]
- 8.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambello, J. R., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goss, T. J., A. Perez-Matos, and R. A. Bender. 2001. Roles of glutamate synthase, gltBD, and gltF in nitrogen metabolism of Escherichia coli and Klebsiella aerogenes, J. Bacteriol. 183:6607-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas, D., M. Galimands, M. Gamper, and A. Zimmermann. 1990. Arginine network of Pseudomonas aeruginosa: specific and global controls, p. 303-316. In S. Silver, A.-M. Chakrabarty, B. Iglewski, and S. Kaplan (ed.), Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. American Society for Microbiology, Washington, D.C.
- 12.Haas, D., B. W. Holloway, A. Schambock, and T. Leisinger. 1977. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa, Mol. Gen. Genet. 154:7-22. [DOI] [PubMed] [Google Scholar]
- 13.Itoh, Y. 1997. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa, J. Bacteriol. 179:7280-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jann, A., V. Stalon, C. Vander Wauven, T. Leisinger, and D. Haas. 1986. N2-succinylated intermediates in an arginine catabolic pathway of Pseudomonas aeruginosa, Proc. Natl. Acad. Sci. USA 83:4937-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen, D. B., H. J. op den Camp, P. J. Leenen, and C. van der Drift. 1980. The enzymes of the ammonia assimilation in Pseudomonas aeruginosa, Arch. Microbiol. 124:197-203. [DOI] [PubMed] [Google Scholar]
- 16.Kwon, D.-H. 1995. Ph.D. thesis. Georgia State University, Atlanta.
- 17.Lim, D. B., J. D. Oppenheim, T. Eckhardt, and W. K. Maas. 1987. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc. Natl. Acad. Sci. USA 84:6697-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, C.-D., and A. T. Abdelal. 2001. The gdhB of Pseudomonas aeruginosa encodes an arginine-inducible NAD-dependent glutamate dehydrogenase which is subject to allosteric regulation. J. Bacteriol. 183:490-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, C. D., J. E. Houghton, and A. T. Abdelal. 1992. Characterization of the arginine repressor from Salmonella typhimurium and its interactions with the carAB operator. J. Mol. Biol. 225:11-24. [DOI] [PubMed] [Google Scholar]
- 20.Lu, C.-D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa, J. Bacteriol. 181:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meers, J. L., D. W. Tempest, and C. M. Brown. 1970. Glutamine (amide):2-oxoglutarate aminotransferase oxidoreductase (NADP), an enzyme involved in the synthesis of glutamate by some bacteria. J. Gen. Microbiol. 64:187-194. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Nishijyo, T., S. M. Park, C. D. Lu, Y. Itoh, and A. T. Abdelal. 1998. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa, J. Bacteriol. 180:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochs, M. M., C.-D. Lu, R. E. Hancock, and A. T. Abdelal. 1999. Amino acid mediated induction of the basic amino acid-specific outer membrane porin OprD from Pseudomonas aeruginosa, J. Bacteriol. 181:5426-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver, G., G. Gosset, R. Sanchez-Pescador, E. Lozoya, L. M. Ku, N. Flores, B. Becerril, F. Valle, and F. Bolivar. 1987. Determination of the nucleotide sequence for the glutamate synthase structure genes of Escherichia coli K-12. Gene 60:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Park, S.-M., C.-D. Lu, and A. T. Abdelal. 1997. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 179:5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, S.-M., C.-D. Lu, and A. T. Abdelal. 1997. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J. Bacteriol. 179:5309-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reitzer, L. J. 1996. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine, p. 391-407. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 29.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 262:1407-1412. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-833. [PubMed] [Google Scholar]
- 32.Stibitz, S., W. Black, and S. Falkow. 1986. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene 50:133-140. [DOI] [PubMed] [Google Scholar]
- 33.Valle, F., B. Becerril, E. Chen, P. Seeburg, H. Heyneker, and F. Bolivar. 1984. Complete nucleotide sequence of the glutamate dehydrogenase gene from Escherichia coli, Gene 27:193-199. [DOI] [PubMed] [Google Scholar]
- 34.Velazquez, L., L. Camarena, J. L. Reyes, and F. Bastarrachea. 1991. Mutations affecting the Shine-Dalgarno sequences of the untranslated region of the Escherichia coli gltBDF operon. J. Bacteriol. 173:3261-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wray, L. V. J., A. E. Ferson, K. Rohrer, and S. H. Fisher. 1996. TnrA, a transcriptional factor required for global nitrogen regulation in Bacillus subtilis, Proc. Natl. Acad. Sci. USA 93:8841-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]