Abstract
Arginine metabolism in pseudomonads with multiple catabolic pathways for its utilization as carbon and nitrogen sources is of particular interest as the model system to study control of metabolic integration. We performed transcriptome analyses to identify genes controlled by the arginine regulatory protein ArgR and to better understand arginine metabolic pathways of P. aeruginosa. We compared gene expression in wild-type strain PAO1 with that in argR mutant strain PAO501 grown in glutamate minimal medium in the presence and absence of arginine. Ten putative transcriptional units of 28 genes were inducible by ArgR and arginine, including all known ArgR-regulated operons under aerobic conditions. The newly identified genes include the putative adcAB operon, which encodes a catabolic arginine decarboxylase and an antiporter protein, and PA0328, which encodes a hypothetical fusion protein of a peptidase and a type IV autotransporter. Also identified as members of the arginine network are the following solute transport systems: PA1971 (braZ) for branched-chain amino acids permease; PA2042 for a putative sodium:serine symporter; PA3934, which belongs to the family of small oligopeptide transporters; and PA5152-5155, which encodes components of an ABC transporter for a putative opine uptake system. The effect of arginine on the expression of these genes was confirmed by lacZ fusion studies and by DNA binding studies with purified ArgR. Only five transcriptional units of nine genes were qualified as repressible by ArgR and arginine, with three operons (argF, carAB, and argG) in arginine biosynthesis and two operons (gltBD and gdhA) in glutamate biosynthesis. These results indicate that ArgR is important in control of arginine and glutamate metabolism and that arginine and ArgR may have a redundant effect in inducing the uptake systems of certain compounds.
Pseudomonas aeruginosa possesses four different catabolic pathways for utilization of arginine (11): the arginine deiminase (ADI) pathway, the arginine succinyltransferase (AST) pathway, the arginine decarboxylase (ADC) pathway, and the arginine dehydrogenase (ADH) pathway (Fig. 1). Under aerobic conditions, Haas and coworkers have established that arginine utilization occurs mainly through the AST pathway, which converts arginine to glutamate (20, 43). Recent studies in the laboratories of Lu and Abdelal have shown that the aru operon, which encodes the AST pathway, and the gdhB gene, which encodes a catabolic glutamate dehydrogenase, are inducible by arginine and that this effect is mediated by ArgR (18, 24). In P. aeruginosa, ArgR, the arginine-responsive regulator protein, is autoinduced from the aot-argR operon for arginine uptake and regulation (35). The ArgR protein of P. aeruginosa belongs to the AraC/XylS family of transcriptional regulators (7) and is thus quite different in structure and function from the ArgR proteins of enteric bacteria and Bacillus subtilis (3, 4, 6, 23, 25), which have a high degree of similarity in their three-dimensional structures and DNA-binding properties (48).
FIG. 1.
Arginine metabolic pathways in P. aeruginosa PAO1. Only key intermediates and known genes in the metabolic pathways are indicated. Arginine-repressible genes are shown with gray arrows, and arginine-inducible genes are depicted with dashed-line arrows. TCA, tricarboxylic acid.
Under anaerobic conditions, arginine can be used as a direct source of ATP via the ADI pathway (11, 47). While Anr, the anaerobic regulatory protein (8), is essential for induction of the arcDABC operon, which encodes an arginine:ornithine antiporter and enzymes of the ADI pathway, exogenous arginine can further induce its expression level through the interactions of Anr and ArgR (28, 38).
Elucidation of the functions of the ADC and ADH pathways in arginine utilization was hindered by the lack of genetic information about the missing genes. For the ADC pathway, although the presence of arginine-inducible ADC activity in the conversion of arginine into agmatine has been reported in an early study (29), the corresponding gene has not been identified. Recent studies in this laboratory have reported the characterization of the aguBA and spuC genes, which encode enzymes for the conversion of agmatine into 4-aminobutyrate of the ADC pathway (26, 34). Exogenous agmatine but not arginine induced these genes (14, 29). Very little is known about the enzymes or genes of the ADH pathway; only the gbuA gene and the bifunctional kauB gene have been characterized (32).
For arginine biosynthesis, only the argF gene and the carAB operon, which encode ornithine carbamoyltransferase and carbamoylphosphate synthetase, respectively, have been reported to be repressible by arginine (1, 17, 19, 49).
With the completion of the Pseudomonas Genome Project and the use of innovative DNA microarray technology (42), it has become feasible to identify and characterize genes of metabolic pathways in a very efficient and systematic way. Considering the relatively large size of the P. aeruginosa genome and its reputation as a metabolically versatile organism, it is very likely that many “hypothetical” or “unknown” genes encode catabolic enzymes for the utilization of different nutrients that this organism encounters in its varied habitats. The complex arginine metabolic network that enables P. aeruginosa to utilize arginine as a source of carbon, energy, and nitrogen is an excellent model system for a detailed investigation of the diversity of metabolic pathways and the associated regulatory mechanisms in this organism. This paper reports the results of transcriptome analysis, which confirm the previously reported ArgR-responsive regulation of certain operons but also reveal a wider regulatory network than previously recognized.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and chemicals.
P. aeruginosa PAO1 (12) and an isogenic argR::Gmr mutant (38) were used for expression analysis. Escherichia coli DH5α was used as the host for plasmid cloning. Luria-Bertani (LB) medium was used for strain maintenance (41). Minimal medium P (12) was supplied with 20 mM l-glutamate or l-arginine if indicated as the source of carbon or nitrogen. Cultures were grown aerobically at 37°C for all experiments. Where needed, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; carbenicillin, 200 μg/ml; gentamicin, 50 μg/ml.
RNA isolation, generation of cDNA probes, and data analysis.
Total RNA was isolated by the hot phenol method (27), followed by DNase I treatment and column RNA purification (Qiagen). Labeled cDNA probes were prepared in accordance with the protocol provided by the manufacturer (Affymetrix). cDNA was synthesized by annealing random primers (Invitrogen) to purified total RNA and subsequent extension with reverse transcriptase (SuperScript II; Invitrogen). Spike RNAs corresponding to B. subtilis genes dap, thr, phe, lys, and trp were included in the cDNA synthesis reaction mixtures as an internal control to monitor the processes of labeling, hybridization, and scanning efficiency (courtesy of Stephen Lory, Harvard Medical School).
The results of two independent experiments were merged for each of the four growth conditions: PAO1-glutamate (1E), PAO1-glutamate-arginine (1ER), PAO501-glutamate (5E), and PAO501-glutamate-arginine (5ER). The merged data were used for subsequent comparisons and assessed with Microarray Suite software (Affymetrix). All data were globally scaled to a target intensity of 500 to generate the absolute expression levels of transcripts for each chip. We performed pairwise comparison of 1E and 1ER, applied a twofold cutoff value, and eliminated transcripts with the absence call (P > 0.04) or with a signal level below 100. The following additional criteria were imposed in the analysis of transcript levels: 5ER = 5E < 1E < 1ER for ArgR activation genes and 5ER = 5E > 1E > 1ER for ArgR repression genes.
Construction of lacZ fusions.
DNA fragments containing the regulatory regions of interest (Fig. 2) were amplified by PCR from the genomic DNA of PAO1 with the following synthetic oligonucleotides designed to generate HindIII restriction sites on the forward primers: for PA1971 (braZ), 5′-CCAAGCTTTCGACATGGGCACGGATCT-3′ and 5′-GTTCATGCTGGAGAGGTACCGCGCT-3′; for PA1818 (adcA), 5′-CGCAAGCTTAGGCGCCGGTCGGCG-3′ and 5′-GGGAAATTTGAGGTCTTT-3′; for PA2041 (ygjU), 5′-CCAAGCTTGAGCCCGACCCAGTGAGG-3′ and 5′-TGTCATGCAGATTTCTACTCTTATAG-3′; for PA3934, 5′-CCAAGCTTCGACAAGCCCTTCTGACGAC and 5′-TTGCATGGATGAAAACTCTCGAAAC; for PA5152, 5′-CCAAGCTTCGGCGGCTCCATAGGCGGTCCCGC-3′ and 5′-GGCCATGGATTTTCCTCTTGTTAT-3′. The PCR products were purified from a 1% (wt/vol) agarose gel, digested by restriction endonuclease HindIII, and ligated to the HindIII and SmaI sites of the translational fusion vector pQF52 (39). The resulting plasmids contain the entire upstream intergenic sequences of the corresponding genes and the 5′ ends of their coding sequences fused in frame to the eighth codon of the lacZ gene in the vector. The nucleotide sequences of the resulting constructs were verified by nucleotide sequence determination.
FIG. 2.
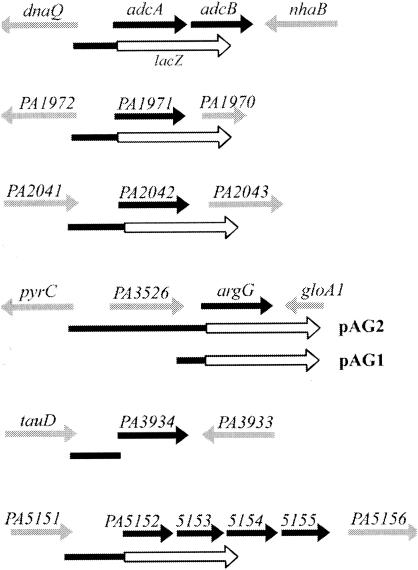
Schematic representations of chromosomal loci of ArgR-regulated genes. Only new genes of the ArgR regulon identified and subjected to further analysis in this study are shown as black arrows, and genes in their flanking regions are shown as gray arrows. The DNA fragments used in the construction of translational fusions of lacZ (white arrows) or in gel retardation assays are depicted as filled bars. The nomenclature and genetic organization of these genes are based on the current version of the Pseudomonas Genome Project (www.pseudomonas.com).
For construction of argG::lacZ translational fusions, two different DNA fragments were generated by PCR from either one of the two forward primers, 5′-GCCAAGCTTCGGGGCGCAGGAGGG-3′ or 5′-GCCAAGCTTCAGGGAAAACCCACG-3′, and the same reverse primer, 5′-CACATCCGCCATGCCATCACTCCA-3′. Following the cloning strategy described above, the resulting two argG::lacZ fusion plasmids were designated pAG1 and pAG2, respectively (Fig. 2), which contain either just the PA3526-argG intergenic region or an extension to the entire PA3526 gene and its putative regulatory region.
Enzyme assays.
For the measurements of β-galactosidase activities, cells were grown in glutamate minimal medium in the presence or absence of arginine. Cell cultures in the logarithmic phase were collected by centrifugation, and the cell pellets were suspended in 50 mM potassium phosphate buffer, pH 7.0. Cells were broken with a French pressure cell at 8,000 lb/in2, and soluble cell extracts were prepared for the measurements of β-galactosidase activity with o-nitrophenyl-β-d-galactopyranoside as the substrate (30). Protein concentration was determined by the method of Bradford (2) with bovine serum albumin as the standard.
Gel retardation assay.
DNA probes containing the regulatory regions of interest were prepared by labeling with [γ-32P]ATP by T4 polynucleotide kinase (New England BioLabs). The radioactively labeled DNA probe (0.1 nM) was allowed to interact with the purified ArgR protein in 20 μl of a mixture containing 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM EDTA, 5% (vol/vol) glycerol, bovine serum albumin (20 μg/ml), and 10 ng of sheared salmon sperm DNA. Reaction mixtures were incubated for 10 min at room temperature and applied to a 5% polyacrylamide gel in Tris-acetate-EDTA running buffer. After being dried, the gel was autoradiographed by exposure to a phosphorimager plate (Fuji).
RESULTS
Identification of ArgR-regulated genes.
For GeneChip experiments, four RNA samples were prepared from wild-type strain PAO1 or its argR derivative PAO501 grown in glutamate (E) minimal medium in the presence or absence of arginine (R) under aerobic conditions (PAO1-E, PAO1-ER, PAO501-E, and PAO501-ER). Depending on the location of its binding sites, ArgR can serve as either a repressor or an activator (39). Since ArgR expression itself is subject to carbon catabolite repression by tricarboxylic acid cycle intermediates in P. aeruginosa (35), glutamate was used as the source of carbon and nitrogen in the reference minimal medium in order not to obscure the effect of arginine and ArgR. In analysis of the data, if the gene is inducible by arginine, its expression levels among the RNA samples would be PAO1-ER > PAO1-E > PAO501-E = PAO501-ER; the reverse order is expected for arginine-repressible genes. By applying such criteria in data analysis in addition to the global scaling approach proposed by the manufacturer (Microarray Suite 5.0; Affymetrix), candidate genes of the ArgR regulon were identified (Table 1). The number of genes under control of ArgR was 37; of these, 28 are inducible and 9 are repressible.
TABLE 1.
Microarray analysis of genes controlled by ArgR and l-arginine under aerobic growth conditions in P. aeruginosa strains PAO1 and PAO501 (argR::GmΩ)
| Gene no.a | Gene name | Maximum changeb | Protein description or function |
|---|---|---|---|
| PA0328 | 3.7 | Unknown | |
| PA0888 | aotJ | 6.0 | Arginine:ornithine ABC transporter |
| PA0889 | aotQ | 8.7 | Arginine:ornithine ABC transporter |
| PA0890 | aotM | 7.4 | Arginine:ornithine ABC transporter |
| PA0891 | 4.9 | Unknown | |
| PA0892 | aotP | 3.2 | Arginine:ornithine ABC transporter |
| PA0893 | argR | 6.2 | Arginine-responsive transcriptional regulator |
| PA0895 | aruC | 26.1 | AST pathway for arginine utilization |
| PA0896 | aruF | 55.0 | AST pathway for arginine utilization |
| PA0897 | aruG | 25.7 | AST pathway for arginine utilization |
| PA0898 | aruD | 30.3 | AST pathway for arginine utilization |
| PA0899 | aruB | 29.5 | AST pathway for arginine utilization |
| PA0900 | 4.8 | Unknown | |
| PA0901 | aruE | 7.7 | AST pathway for arginine utilization |
| PA1818 | adcA | 19.8 | Putative catabolic ADC |
| PA1819 | adcB | 6.2 | Putative arginine:agmatine antiporter |
| PA1971 | braZ | 18.2 | Permease for branched-chain amino acids |
| PA2042 | 4.3 | Sodium:serine symporter; ygjU | |
| PA3068 | gdhB | 19.9 | Catabolic NAD-dependent glutamate dehydrogenase |
| PA3934 | 8.9 | Putative short oligopeptide transporter | |
| PA5152 | 5.7 | ABC transporter | |
| PA5153 | 3.2 | ABC transporter | |
| PA5154 | 5.0 | ABC transporter | |
| PA5155 | 4.1 | ABC transporter | |
| PA5170 | arcD | 7.5 | Arginine:ornithine antiporter |
| PA5171 | arcA | 5.0 | ADI pathway for arginine utilization |
| PA5172 | arcB | 3.8 | ADI pathway for arginine utilization |
| PA5173 | arcC | 3.7 | ADI pathway for arginine utilization |
| PA3525 | argG | −6.7 | Arginine biosynthesis; argininosuccinase |
| PA3537 | argF | −12.2 | Arginine biosynthesis; omithine carbamoyltransferase |
| PA3538 | −3.9 | Transport of small molecules | |
| PA4588 | gdhA | −3.5 | Anabolic NADP-dependent glutamate dehydrogenase |
| PA4756 | carB | −3.3 | Carbamoylphosphate synthetase, heavy subunit |
| PA4757 | −2.1 | Unknown | |
| PA4758 | carA | −3.2 | Carbamoylphosphate synthetase, light subunit |
| PA5035 | gltD | −2.2 | Glutamate synthase, light subunit |
| PA5036 | gltB | −4.1 | Glutamate synthase, heavy subunit |
Gene numbers are from the Pseudomonas Genome Project (www.pseudomona.scom). Boldface type indicates genes or gene products previously reported to be controlled by arginine and ArgR.
Maximum changes (fold) in gene expression are indicated by the ratio of PAO1 (wild type) to PAO501 (argR) grown in glutamate minimal medium in the presence of arginine for arginine induction genes and the ratio of PAO501 to PAO1 with a minus sign for arginine repression genes.
Arginine induction.
Eighteen of the 28 arginine-inducible genes are in four transcriptional units that have been reported recently as members of the ArgR regulon: the aotJQMOP-argR (PA0888-0893) operon for arginine and ornithine uptake and regulation (35); the aruCFGDBE operon (PA0895-0901), which encodes enzymes of the AST pathway (18); the gdhB gene (PA3068), which encodes catabolic glutamate dehydrogenase (24); and the arcDABC operon (PA5170-5173), which encodes enzymes of the ADI pathway (28).
Genes that are inducible by arginine and ArgR but have not been reported previously as members of the ArgR regulon include PA1818 and PA1819. These two genes encode a putative arginine/ornithine/lysine decarboxylase and an amino acid/amine permease, respectively, according to current genomic annotations. The amino acid sequence of the putative decarboxylase exhibits 57% similarity to that of the catabolic ADC (AdiA) of E. coli (44), and the downstream permease sequence has 56% similarity to that of AdiC (YjdE) of the arginine:agmatine antiporter of E. coli (10). On the basis of sequence homology and the observed arginine activation effect, we propose that PA1818 and PA1819 be designated adcA and adcB for their possible roles in the ADC pathway of P. aeruginosa.
Interestingly, the remaining arginine-inducible genes, with the exception of PA0328, are all related to transport systems of small molecules. PA5152-5155 likely represents an operon that encodes components of an ABC transporter. This hypothesis is based on a high sequence similarity to the corresponding components of an octopine/nopaline transport system of Agrobacterium tumefaciens (46) and the Art system in arginine uptake of E. coli (50). PA1971 (braZ) has been reported as the gene for an Na+-coupled transporter of branched-chain amino acids in P. aeruginosa (16). PA2042 encodes a putative symporter protein showing 83% similarity to the SstT sodium:serine symporter of E. coli (37), and the hypothetical protein of PA3934 with 16 predicted transmembrane helices exhibited sequence similarity to the family of small oligopeptide transporters (Pfam 03169). PA0328 encodes a hypothetical outer membrane protein with a peptidase domain (Pfam 4386) at the N terminus and a type V autotransporter domain (Pfam 3797) at the C terminus.
Arginine repression.
As previously reported (39), argF, which encodes ornithine carbamoyltransferase, and the carA-orf-carB-greA operon, which encodes the small (CarA) and large (CarB) subunits of carbamylphosphate synthetase, are repressed by arginine and ArgR. In addition, the results show that the argG gene, which encodes argininosuccinate synthetase, which has not been previously reported to be under the control of ArgR, is indeed repressed 6.7-fold by arginine. Furthermore, PA3538, which encodes a putative ATP-binding component of ABC transporters, is also repressible by arginine. Since PA3538 is located only 5 bp downstream of argF in the same transcriptional orientation, it is very likely that argF and PA3538 belong to the same transcriptional unit.
Other arg genes that encode biosynthetic enzymes, such as those catalyzing the conversion of ornithine from glutamate (argA-E and argJ) and the last step of arginine biosynthesis (argH), did not qualify as ArgR-repressible genes in this analysis since their repression ratios were all less than twofold. The only exception is argD, which is the same gene as the arginine-inducible aruC gene of the AST pathway (11, 18). These results were consistent with the conclusions of earlier reports (12, 17, 49).
The gltBD operon and the gdhA gene, which encode glutamate synthase and anabolic glutamate dehydrogenase in glutamate biosynthesis, were found to be repressible by exogenous arginine. A detailed analysis of the role of ArgR in the control of these genes is presented in a separate report (15).
Data verification.
For candidate genes as new members of the ArgR regulon, LacZ translational fusions of these genes were constructed as described in Materials and Methods to validate the data of transcriptome analysis. These include the arginine-inducible genes PA1971, PA1818, PA2042, and PA5152. The resulting plasmids were introduced into wild-type strain PAO1 and argR mutant strain PAO501. The effect of exogenous arginine on the expression of these promoters was analyzed by measurement of the β-galactosidase activities of these recombinant strains grown in glutamate minimal medium in the presence or absence of arginine. As shown in Table 2, all of these fusions exhibit arginine-inducible promoter activities in wild-type strain PAO1, and the induction effect of arginine was abolished in argR mutant strain PAO501.
TABLE 2.
Verification of arginine- and ArgR-dependent genes by promoter-lacZ fusions
| Gene no. (name)a | Host strainb | Sp act (nmol/min/mg)c
|
|
|---|---|---|---|
| Glu | Glu + Arg | ||
| PA1971 (braZ) | PAO1 | 57 | 251 |
| PAO501 | 9 | 17 | |
| PA1818 (adcA) | PAO1 | 233 | 1,214 |
| PAO501 | 58 | 90 | |
| PA2042 (ygjU) | PAO1 | 91 | 422 |
| PAO501 | 91 | 79 | |
| PA5152 | PAO1 | 123 | 677 |
| PAO501 | 44 | 56 | |
| PA3525 (argG) | PAO1 | 52 | 20 |
| PAO501 | 106 | 102 | |
Gene numbers and names are from the Pseudomonas Genome Project (www.pseudomonas.com).
PAO1 is the wild-type strain of P. aeruginosa, and PAO501 is the argR mutant.
Specific activities are averages of two measurements for each growth condition, with standard errors of less than 5%. Cells were grown in minimal medium with the indicated supplements at 20 mM as the sources of carbon and nitrogen. Glu, glutamate; Arg, arginine.
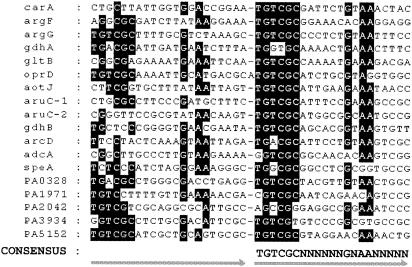
Binding of homogeneous ArgR to the promoter regions of these putative arginine-inducible genes was confirmed by gel retardation assays (Fig. 3), indicating the presence of ArgR binding sites in regulatory regions of these genes. The putative ArgR binding sites were identified by sequence alignment as shown in Fig. 4. Sequence alignment identified putative ArgR binding sites that exhibit similarity to the consensus ArgR binding site reported in previous work from this laboratory (24). The consensus ArgR binding site consists of two half-sites in a direct-repeat arrangement with the consensus sequence 5′-TGTCGCN6GNAAN5-3′. In most cases, the second half-site sequences are relatively more conserved than the first half-site sequences.
FIG. 3.
Gel retardation experiments. The radioactively labeled operator DNA fragments of the genes indicated were incubated with increasing concentrations of ArgR from lane 1 to lane 4: 0 (lane 1), 7.0 × 10−10 M (lane 2), 1.4 × 10−9 M (lane 3), and 2.8 × 10−9 M (lane 4). B, ArgR-DNA complex; C, negative control probe; F, free probe.
FIG. 4.
Sequence alignment of ArgR binding sites. The sequences were obtained from the results of DNase I footprinting of known ArgR-regulated genes or by sequence analysis of genes identified in this study. The consensus sequence was deduced from the second half-sites, which are more conserved in most cases. Nucleotides identical to those of the consensus site are shaded.
For arginine-repressible genes, argG is the only new candidate among enzymes of arginine biosynthesis. An argG::lacZ fusion, pAG1 (Fig. 2), that covers the intergenic region between argG and the upstream PA3526 gene, was constructed and introduced into PAO1 and PAO501 to test its repression by arginine. This fusion exhibited a negligible level of promoter activity even in the absence of exogenous arginine. Accordingly, another argG::lacZ fusion, pAG2, covering the entire PA3526 gene and its putative promoter region (Fig. 2) was constructed. The PA3526 gene encodes a probable peptidoglycan-associated outer membrane protein with no apparent association with arginine metabolism. As shown in Table 2, the expression of lacZ from pAG2 was repressible by arginine in PAO1 and exhibited no arginine repression in PAO501. Analysis of the PA3526-argG intergenic sequence (pAG1; Fig. 2) revealed a putative ArgR binding site, and the results of gel retardation assays with the purified ArgR protein confirmed the binding of ArgR to this region. The combined results strongly support the hypothesis that transcription of argG is initiated either from the promoter of PA3526 or from an internal promoter in the coding sequence of PA3526, which is differentially repressed by the binding of ArgR to the intergenic region.
DISCUSSION
With one exception, genes of the ArgR regulon in P. aeruginosa identified by transcriptome analysis in this study include all known members as reported previously. The data reported here also identified 15 new candidate members of the ArgR regulon. For most of these new candidates, the identification was supported by data from studies of promoter fusions and gel retardation assays. Furthermore, sequence alignment identified putative ArgR binding sites in the new candidate operons based on homology with the previously reported (24) consensus sequence of well-characterized ArgR binding sites.
The single member of the ArgR regulon that was not identified by GeneChip analysis is oprD, which encodes outer membrane porin D. While the interaction of ArgR and the oprD regulatory region has been demonstrated in vitro by gel retardation assays and DNase I footprinting, only marginal induction by arginine was observed by Western analysis (36). The growth conditions used in this study might be suboptimal for expression of oprD, thus obscuring the marginal effect of arginine and ArgR on expression of OprD. Therefore, in those cases in which ArgR is not the major transcriptional regulator, it is conceivable that such genes will be difficult to detect in DNA microarray experiments under conditions in which growth variation is limited. Similar conditions might contribute to the low repression ratio of a number of the putative ArgR-regulated genes identified in this study.
For arginine catabolic pathways, a likely candidate for the gene that encodes the first enzyme of the ADC pathway (PA1818; adcA) was identified in this study. Similar to the case in enteric bacteria, the adcA gene is likely to form an operon with the following adcB gene, which encodes a putative arginine:agmatine antiporter (10). The presence of an arginine-inducible ADC in P. aeruginosa was reported from an early study conducted by Stalon and coworkers (43). However, a recent report by Nakada and Itoh (33) reported that the biosynthetic ADC encoded by the speA gene contributes more than 95% of the ADC activity in cell extracts. While this study tentatively identified PA1818 as the operon that encodes the catabolic ADC on the basis of its induction by exogenous arginine and the amino acid sequence homology, the encoded protein and its possible physiological function in arginine utilization or polyamine synthesis are currently under investigation in this laboratory.
Consistent with earlier reports (11, 20, 32), other known genes of the ADC and ADH pathways were not identified as ArgR-inducible genes by transcriptome analysis. This argues against the function of ArgR in the control of the ADC and ADH pathways for arginine utilization in P. aeruginosa. Furthermore, although not shown here, a separate set of genes was categorized as arginine inducible in the argR mutant but not in the wild type. The implications of these genes in the ADH and ADC pathways and in polyamine metabolism will be the subject of a separate report. In P. putida, the ADH and AST pathways are equally important for arginine utilization (45). Therefore, a comparative genomic approach might be helpful in revealing the difference in arginine utilization among pseudomonads at the genetic level.
In P. aeruginosa, the arginine biosynthetic genes are completely scattered on the chromosome. In contrast, the arginine biosynthetic genes of enteric bacteria and bacilli form multigene operons and are tightly regulated at the transcriptional level by repression in response to the concentration of arginine (9, 31, 40, 51). In the presence of exogenous arginine, the ArgR protein of PAO1 seems to repress the expression of only 3 out of 10 enzymes of the arginine biosynthetic pathway encoded by argF, carAB, and argG. The involvement of ArgR in the control of argG is the new discovery of this study, while repression of argF and carAB by ArgR has already been documented. The repression ratio of carAB (3.0) is significantly lower than those of argF (12.2) and argG (6.7). This could be related to the fact that carAB expression is also subject to pyrimidine control via the attenuation mechanism (22). For genes involved in ornithine biosynthesis, virtually no repression effect by ArgR and arginine was observed in this study. In addition, no obvious sequence resembling the ArgR binding sites can be found in the putative regulatory region of these genes. However, arginine could exert feedback control on biosynthesis by allosteric inhibition. For example, N-acetylglutamate synthetase, which catalyzes the first step of arginine biosynthesis, is subject to inhibition by arginine (13).
The unique features of the ArgR regulon in P. aeruginosa extend into glutamate metabolism. Lu and Abdelal (24) have previously reported that gdhB, which encodes a catabolic glutamate dehydrogenase is induced by ArgR and arginine. The encoded enzyme is also subject to allosteric activation by arginine and inhibition by citrate. These results highlighted the role of the catabolic glutamate dehydrogenase in linking the product of the AST pathway with the tricarboxylate cycle. The identification of the gltBD and gdhA operons, which encode two major enzymes of glutamate biosynthesis, as ArgR-repressed genes revealed a higher level of coordination of the regulatory networks that govern arginine and glutamate metabolism in P. aeruginosa (15).
Efficient uptake is the essential first step in the utilization of any compound. We have shown previously the importance of an ABC transporter encoded by the aot operon in arginine uptake (35). However, the presence of additional arginine transport systems was evidenced by the reduced, but still inducible, arginine uptake of the aot mutant (35). The arcD gene, which encodes an arginine:ornithine antiporter, is induced under anaerobic conditions from the arcDABC operon (8). Many new members of the ArgR regulon that encode polypeptides for solute transport are inducible by arginine: the PA5152-5155 operon, PA3934, PA2042, PA1971, and AdcB for a putative arginine:agmatine antiporter. In addition, PA3538, which encodes the ATP-binding component of ABC transporters, is repressible by arginine and likely forms an operon with the upstream argF gene. The induction effect of arginine on these genes in vivo and the binding of ArgR to the cognate regulatory regions in vitro were confirmed by lacZ fusions and gel retardation experiments. However, the results of sequence analysis have suggested their putative functions in the uptake of octopine:nopaline (PA5152-5155), short oligopeptides (PA3934), or l-serine (PA2042). Furthermore PA1971 (braZ) has been reported to function in the transport of branched-chain amino acids (16). Therefore, it is likely that arginine and ArgR may exert a redundant effect in inducing the uptake of these compounds.
The availability of highly organized and diverse transport systems for arginine uptake in P. aeruginosa reflects the importance of this amino acid as a nutrient for this organism. These systems enable P. aeruginosa to be an effective scavenger of l-arginine, which can serve as a source of carbon, energy, or nitrogen or indeed as a sole source of all three. It is intriguing that arginine serves as the substrate of nitric oxide synthetase for the synthesis of an important second messenger molecule, NO (21). Perhaps maintaining such a sophisticated arginine metabolic network in P. aeruginosa provides this opportunistic human pathogen an advantage in establishing infections, such as in cystic fibrosis patients (5).
Acknowledgments
We thank Steve Lory for array processing.
This work was supported by grant NSF9985660 from the National Science Foundation, the Georgia Research Alliance, and the Research Program Enhance Grant of the Georgia State University Research Foundation. We gratefully acknowledge Cystic Fibrosis Foundation Therapeutics, Inc., for subsidizing the P. aeruginosa Affymetrix GeneChip arrays.
REFERENCES
- 1.Abdelal, A. T., L. Bussey, and L. Vickers. 1983. Carbamoylphosphate synthetase from Pseudomonas aeruginosa: subunit composition, kinetic analysis and regulation. Eur. J. Biochem. 129:697-702. [PubMed] [Google Scholar]
- 2.Bradford, M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Charlier, D., M. Roovers, F. Van Vliet, A. Boyen, R. Cunin, Y. Nakamura, N. Glansdorff, and A. Pierard. 1992. Arginine regulon of Escherichia coli K-12: a study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J. Mol. Biol. 226:367-386. [DOI] [PubMed] [Google Scholar]
- 4.Czaplewski, L. G., A. K. North, M. C. Smith, S. Baumberg, and P. G. Stockley. 1992. Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol. Microbiol. 6:267-275. [DOI] [PubMed] [Google Scholar]
- 5.Darling, K., and T. Evans. 2003. Effects of nitric oxide on Pseudomonas aeruginosa infection of epithelial cells from a human respiratory cell line derived from a patient with cystic fibrosis. Infect. Immun. 71:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dion, M., D. Charlier, H. Wang, D. Gigot, A. Savchenko, J. Hakket, N. Glansforff, and V. Sakanyan. 1997. The highly thermostable arginine repressor of Bacillus stearothermophilus: gene cloning and repressor-operator interactions. Mol. Microbiol. 25:385-398. [DOI] [PubMed] [Google Scholar]
- 7.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamper, M., A. Zimmermann, and D. Haas. 1991. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 173:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glansdorff, N. 1996. Biosynthesis of arginine and polyamines, p. 408-433. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 10.Gong, S., H. Richard, and J. W. Foster. 2003. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas, D., M. Galimands, M. Gamper, and A. Zimmermann. 1990. Arginine network of Pseudomonas aeruginosa: specific and global controls, p. 303-316. In S. Silver, A.-M. Chakrabarty, B. Iglewski, and S. Kaplan (ed.), Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. American Society for Microbiology, Washington, D.C.
- 12.Haas, D., B. W. Holloway, A. Schambock, and T. Leisinger. 1977. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol. Gen. Genet. 154:7-22. [DOI] [PubMed] [Google Scholar]
- 13.Haas, D., V. Kurer, and T. Leisinger. 1972. N-Acetylglutamate synthetase of Pseudomonas aeruginosa: an assay in vitro and feedback inhibition by arginine. Eur. J. Biochem. 31:290-295. [DOI] [PubMed] [Google Scholar]
- 14.Haas, D., H. Matsumoto, P. Moretti, V. Stalon, and A. Mercenier. 1984. Arginine degradation in Pseudomonas aeruginosa mutants blocked in two arginine catabolic pathways. Mol. Gen. Genet. 193:437-444. [DOI] [PubMed] [Google Scholar]
- 15.Hashim, S., D.-H. Kwon, A. Abdelal, and C.-D. Lu. 2004. The arginine regulatory protein mediates repression by arginine of the operons encoding glutamate synthase and anabolic glutamate dehydrogenase in Pseudomonas aeruginosa. J. Bacteriol. 186:3848-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hishino, T., K. Kose-Terain, and Y. Uratani. 1991. Isolation of the braZ gene encoding the carrier for a novel branched-chain amino acid transport system in Pseudomonas aeruginosa PAO. J. Bacteriol. 173:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isaac, J. H., and B. W. Holloway. 1972. Control of arginine biosynthesis in Pseudomonas aeruginosa. J. Gen. Microbiol. 73:427-438. [DOI] [PubMed] [Google Scholar]
- 18.Itoh, Y. 1997. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J. Bacteriol. 179:7280-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh, Y., L. Soldati, V. Stalon, P. Falmagne, Y. Terawaki, T. Leisinger, and D. Haas. 1988. Anabolic ornithine carbamoyltransferase of Pseudomonas aeruginosa: nucleotide sequence and transcriptional control of the argF structural gene. J. Bacteriol. 170:2725-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jann, A., H. Matsumoto, and D. Haas. 1988. The fourth arginine catabolic pathway of Pseudomonas aeruginosa. J. Gen. Microbiol. 134:1043-1053. [DOI] [PubMed] [Google Scholar]
- 21.Jorens, P. G., P. A. Vermeire, and A. G. Herman. 1993. l-Arginine-dependent nitric oxide synthase: a new metabolic pathway in the lung and airways. Eur. Respir. J. 6:258-266. [PubMed] [Google Scholar]
- 22.Kwon, D.-H. 1995. Ph.D. thesis. Georgia State University, Atlanta.
- 23.Lim, D. B., J. D. Oppenheim, T. Eckhardt, and W. K. Maas. 1987. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc. Natl. Acad. Sci. USA 84:6697-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, C.-D., and A. T. Abdelal. 2001. The gdhB of Pseudomonas aeruginosa encodes an arginine-inducible NAD dependent glutamate dehydrogenase which is subject to allosteric regulation. J. Bacteriol. 183:490-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, C. D., J. E. Houghton, and A. T. Abdelal. 1992. Characterization of the arginine repressor from Salmonella typhimurium and its interactions with the carAB operator. J. Mol. Biol. 225:11-24. [DOI] [PubMed] [Google Scholar]
- 26.Lu, C.-D., Y. Itoh, Y. Nakada, and Y. Jiang. 2002. Functional analysis and regulation of the divergent spuABCDEFGH-spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 184:3766-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, C. D., M. Kilstrup, J. Neuhard, and A. Abdelal. 1989. Pyrimidine regulation of tandem promoters for carAB in Salmonella typhimurium. J. Bacteriol. 171:5436-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, C.-D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercenier, A., J. P. Simon, D. Haas, and V. Stalon. 1980. Catabolism of l-arginine by Pseudomonas aeruginosa. J. Gen. Microbiol. 116:381-389. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Mountain, A., N. M. Mann, R. N. Munton, and S. Baumberg. 1984. Cloning of a Bacillus subtilis restriction fragment complementing auxotrophic mutants of eight Escherichia coli genes of arginine biosynthesis. Mol. Gen. Genet. 197:82-89. [DOI] [PubMed] [Google Scholar]
- 32.Nakada, Y., and Y. Itoh. 2002. Characterization and regulation of the gbuA gene, encoding guanidinobutyrase in the arginine dehydrogenase pathway of Pseudomonas aeruginosa PAO1. J. Bacteriol. 184:3377-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakada, Y., and Y. Itoh. 2003. Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 149:707-714. [DOI] [PubMed] [Google Scholar]
- 34.Nakada, Y., Y. Jiang, T. Nishijyo, Y. Itoh, and C.-D. Lu. 2001. Molecular characterization and regulation of the aguBA operon, responsible for agmatine utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6517-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishijyo, T., S. M. Park, C. D. Lu, Y. Itoh, and A. T. Abdelal. 1998. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J. Bacteriol. 180:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochs, M. M., C.-D. Lu, R. E. Hancock, and A. T. Abdelal. 1999. Amino acid mediated induction of the basic amino acid-specific outer membrane porin OprD from Pseudomonas aeruginosa. J. Bacteriol. 181:5426-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa, W., Y.-M. Kim, T. Mizushima, and T. Tsuchiya. 1998. Cloning and expression of the gene for the Na+-coupled serine transporter from Escherichia coli and characteristics of the transporter. J. Bacteriol. 180:6749-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, S.-M., C.-D. Lu, and A. T. Abdelal. 1997. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 179:5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, S.-M., C.-D. Lu, and A. T. Abdelal. 1997. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J. Bacteriol. 179:5309-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakanyan, V., D. Charlier, C. Legrain, A. Kochikyan, I. Mett, A. Pierard, and N. Glansdorff. 1993. Primary structure, partial purification and regulation of key enzymes of the acetyl cycle of arginine biosynthesis in Bacillus stearothermophilus: dual function of ornithine acetyltransferase J. Gen. Microbiol. 139:393-402. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 43.Stalon, V., C. Vander Wauven, P. Momin, and C. Legrain. 1987. Catabolism of arginine, citrulline and ornithine by Pseudomonas and related bacteria. J. Gen. Microbiol. 133:2487-2495. [DOI] [PubMed] [Google Scholar]
- 44.Stim, K. P., and B. N. Bennett. 1993. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-inducible arginine decarboxylase of Escherichia coli. J. Bacteriol. 175:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tricot, C., V. Stalon, and C. Legrain. 1991. Isolation and characterization of Pseudomonas putida mutants affected in arginine, ornithine and citrulline catabolism: function of the arginine oxidase and arginine succinyltransferase pathways. J. Gen. Microbiol. 137:2911-2918. [DOI] [PubMed] [Google Scholar]
- 46.Valdivia, R., L. Wang, and S. Winans. 1991. Characterization of a putative periplasmic transport system for octopine accumulation encoded by Agrobacterium tumefaciens Ti plasmid pTiA6. J. Bacteriol. 173:841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vander Wauven, C., A. Pierard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Duyne, G. D., G. Ghosh, W. K. Maas, and P. B. Sigler. 1996. Structure of the oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol. 256:377-391. [DOI] [PubMed] [Google Scholar]
- 49.Voellmy, R., and T. Leisinger. 1978. Regulation of enzyme synthesis in the arginine biosynthetic pathway of Pseudomonas aeruginosa. J. Gen. Microbiol. 109:25-35. [DOI] [PubMed] [Google Scholar]
- 50.Wissenbach, U., S. Six, J. Bongaerts, D. Ternes, S. Steinwachs, and G. Unden. 1995. A third periplasmic transport system for l-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol. Microbiol. 17:675-686. [DOI] [PubMed] [Google Scholar]
- 51.Yang, H., S.-M. Park, W. G. Nolan, C.-D. Lu, and A. T. Abdelal. 1997. Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus stearothermophilus. Eur. J. Biochem. 249:443-449. [DOI] [PubMed] [Google Scholar]