Abstract
In the Bateson-Dobzhansky-Muller (BDM) model of speciation, incompatibilities emerge from the deleterious interactions between alleles that are neutral or advantageous in the original genetic backgrounds, i.e., negative epistatic effects. Within species such interactions are responsible for outbreeding depression and F2 (hybrid) breakdown. We sought to identify BDM incompatibilities in the nematode Caenorhabditis elegans by looking for genomic regions that disrupt egg laying; a complex, highly regulated, and coordinated phenotype. Investigation of introgression lines and recombinant inbred lines derived from the isolates CB4856 and N2 uncovered multiple incompatibility quantitative trait loci (QTL). These QTL produce a synthetic egg-laying defective phenotype not seen in CB4856 and N2 nor in other wild isolates. For two of the QTL regions, results are inconsistent with a model of pairwise interaction between two loci, suggesting that the incompatibilities are a consequence of complex interactions between multiple loci. Analysis of additional life history traits indicates that the QTL regions identified in these screens are associated with effects on other traits such as lifespan and reproduction, suggesting that the incompatibilities are likely to be deleterious. Taken together, these results indicate that numerous BDM incompatibilities that could contribute to reproductive isolation can be detected and mapped within C. elegans.
Keywords: C. elegans, Bateson-Dobzhansky-Muller, reproductive isolation, negative epistasis, QTL, introgression line
To understand the mechanisms that lead to speciation, insight is required into the genetic basis of reproductive isolation. The most widely accepted explanation for the genetic basis of intrinsic, postzygotic reproductive isolation between species is the Bateson-Dobzhansky-Muller (BDM) model (Bateson 1909; Dobzhansky 1936; Muller 1942). This relies on negative epistasis between alleles and normally considers the case of alleles that have been fixed in different lineages. In hybrids, negative epistasis between alleles that have not been tested together by natural selection result in reduced hybrid fitness (Phillips 2008). Such epistatic interactions have been shown to be involved in, for instance, hybrid male sterility in Drosophila (Perez and Wu 1995; Orr and Irving 2001; Tao et al. 2003) and are also important in human disease and in complex traits more generally (see Phillips 2008 and Mackay 2014 for review). In recent years, the causal polymorphisms underlying BDM incompatibilities have been identified in a limited number of species, with divergence in both coding sequence and in regulatory elements producing incompatibilities (see Presgraves 2010 for review).
BDM incompatibilities will, however, also arise within species (see Cutter 2012 for review) and theoretical analyses suggest that interactions between synthetic deleterious loci are common (Phillips and Johnson 1998; Lachance et al. 2011). This is supported by the widespread observation of outbreeding depression in hybrids between divergent populations (e.g., Templeton 1986; Edmands 1999; Dolgin et al. 2007; Drury et al. 2013; Gimond et al. 2013). A small number of BDM incompatibilities have now been identified within species, mostly producing major effects (e.g., Seidel et al. 2008; Bikard et al. 2009; Drury et al. 2011; Baird and Stonesifer 2012). More recently, a genome-wide screen in D. melanogaster recombinant inbred lines (RILs) identified many epistatic interactions, two of which were shown to have major effects on fecundity (Corbett-Detig et al. 2013). It is, however, likely that the alleles and regions that have been found to date represent only a subset of the polymorphic incompatibilities within a species, i.e., the major effects identified to date represent those that are easy to detect (see Rockman 2012 for a general discussion of this issue).
As outbreeding depression has been documented between isolates of the free-living nematode Caenorhabditis elegans (Dolgin et al. 2007) it is likely that a range of potential incompatibilities exists between isolates. We therefore sought to identify small-effect incompatibilities between the isolates CB4856 and N2. We sought these by looking at the disruption of a complex, highly regulated and coordinated, phenotype, egg-laying, and undertook screens for genomic regions that disrupt this process. At 20°, C. elegans N2 eggs are normally laid about 3 hr after fertilization at around the 30-cell stage (Hirsh et al. 1976), with hatching occurring approximately 14 hr later (Sulston et al. 1983). Disruption of the egg-laying process produces an egl (egg laying abnormal) phenotype, with one class of egl mutation characterized by an increase in the number of fertilized eggs retained within the body and eggs being laid at a much later stage of development. Mutations producing this egl phenotype have been identified in genes that affect chemosensation, muscle development, the cell lineage, sex determination and dauer larvae development (Greenwald and Horvitz 1980; Horvitz and Sulston 1980; Waterston et al. 1980; Trent et al. 1983; WormBase [www.wormbase.org]). We therefore considered that this phenotype represented a suitably large target for the development of incompatibilities. Screens were undertaken using C. elegans RILs and introgression lines (ILs) produced from the isolates CB4856 and N2 (see the section Materials and Methods for details of these lines) and identified multiple quantitative trait loci (QTL) that result in a synthetic egl phenotype. For two of the QTL regions identified, analysis of the ILs indicates that the incompatibilities are a consequence of complex interactions between multiple loci. Incompatibility regions identified in these screens are also shown to be associated with negative effects on lifespan and on reproduction, suggesting that the incompatibilities are likely to be deleterious. In combination, these results indicate that numerous BDM incompatibilities that could lead to reproductive isolation can be detected within C. elegans.
Materials and Methods
Worms
Experiments were performed using the N2 (Bristol) isolate (obtained from the Caenorhabditis genetics center), wild isolates of C. elegans (obtained from Marie-Anne Félix, IBENS, Paris, France, and from the CGC), RILs produced from crosses between CB4856 and N2 (see, for details, Li et al. 2006; Kammenga et al. 2007, 2008; Li et al. 2010; Viñuela et al. 2010; Elvin et al. 2011; Rodriguez et al. 2012; Viñuela et al. 2012), and a panel of CB4856/N2 ILs derived from these RILs in which regions of the CB4846 genome have been introgressed into an N2 background (see, for details, Doroszuk et al. 2009; Green et al. 2013). Briefly, the RILs were created from crosses between N2 and CB4856, with the F1 progeny subsequently inbred, by transfer of single animals at each generation, for 20 generations. RILs were then genotyped at 121 markers across the genome (20 each on chromosomes I, II, III, IV and X, and 21 on V). The ILs were produced from specific RILs, chosen based on the CB4856 regions they contain, these RILs were back-crossed to N2, genotyped, further back-crossed as appropriate, and then genotyped at the same markers as the RILs and at two additional markers on chromosome IV (for a total of 123 markers). This resulted in the production of a panel of ILs, each containing a single segment of the CB4856 genome in an N2 background.
Worms were maintained using standard methods and fed on the OP50 strain of Escherichia coli (Stiernagle 2006). All experiments were undertaken at 20° and were initiated from synchronized populations of L1s produced by allowing eggs isolated from hypochlorite treated adults (Stiernagle 2006) to hatch on plates without food and to develop for 24 hr. Within assays, genotypes were randomized and plates blind coded, with plates that became infected by fungi excluded from analyses.
Embryo stage analysis in the RILs and ILs
The various stages of embryo morphogenesis are well defined in C. elegans (Von Ehrenstein and Schierenberg 1980) and can be identified with a dissecting microscope. Most screens undertaken for mutations producing an egl phenotype relied on screening worms early in the reproductive period to identify hermaphrodites that had died by internal hatching of progeny (bagging) or that were bloated with late-stage eggs (Greenwald and Horvitz 1980; Horvitz and Sulston 1980; Waterston et al. 1980; Trent et al. 1983). Subsequent analysis of these mutants showed that most worms capable of releasing eggs tended to lay them at a much later stage of development than the wild-type (Trent et al. 1983). As we aimed to identify genomic regions that, when in a different genetic background, produced an egl phenotype, we determined the stages of eggs laid by worms late in the reproductive period. Our reasoning for screening late in the reproductive life is that this would allow the identification of differences reliant on age-related loss-of-function. Preliminary experiments (data not shown, but see Figure 4 and Figure 5) indicated that both N2 and CB4856 continue to lay almost all eggs at very early stages of development (Supporting Information, Figure S4) throughout the reproductive period. We therefore considered that laying eggs at a late stage of development could be considered a consequence of an incompatibility between N2 and CB4856 alleles.
Figure 4.
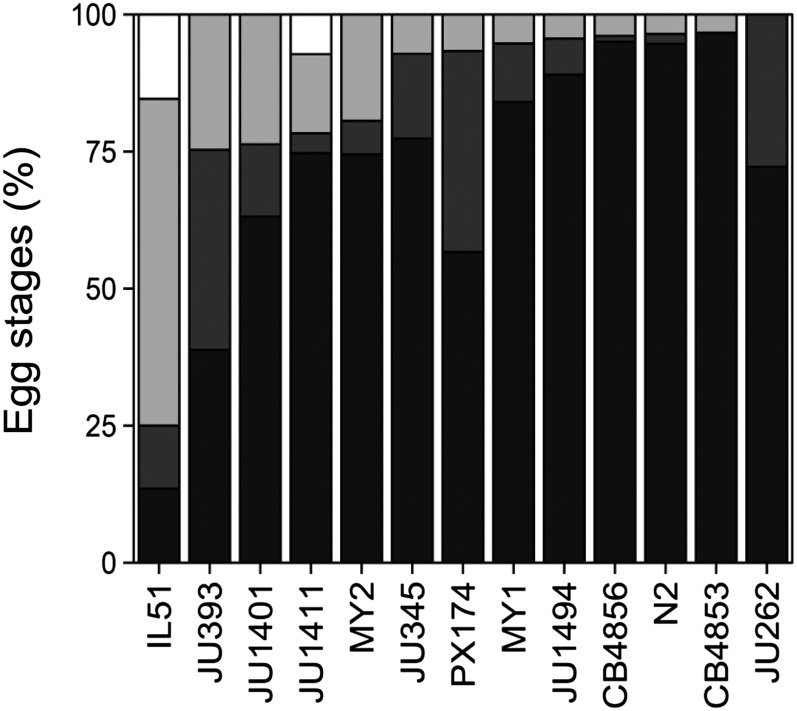
Embryo stages of wild-isolates. Embryo stage distribution shown as cumulative percentage of total progeny. From dark to light: Stage I, II, III, and L1 (in white). CB4856 scores from different experiments (n = 281). (N = IL51 (ewIR51): 52; JU393: 85; JU1401: 76; JU1411: 83; MY2: 98; JU345: 84; PX174: 30; MY1: 94; JU1494: 91; CB4856: 103; N2: 112; CB4853: 60; JU262: 18). For > stage II eggs all the wild isolates are significantly different from ewIR51 (P < 0.01, two-sided t-test on plate averages). None of the > stage II differences between the wild-isolated were significantly different (P > 0.05, two-sided t-test on plate averages).
Figure 5.
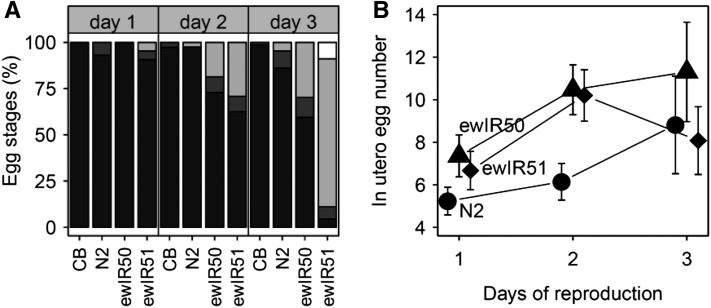
Variation across the reproductive period. (A) Embryo stage distribution across the reproductive period shown as cumulative percentage of total progeny. From dark to light: Stage I, II, III, and L1 (in white). (B) Number of eggs in utero across the reproductive period.
For embryo stage analysis, we classified progeny into four stages: stage I from fertilization to the end of gastrulation; stage II from ‘lima bean’ to ‘comma’ stage embryos; stage III ‘tadpole’ to ‘pretzel’ stage; and L1 (stages as described by Von Ehrenstein and Schierenberg 1980; see also Trent et al. 1983). Unless otherwise noted, embryo stages were assayed on the third day of reproduction, 6 d after recovery from L1 arrest, with adults transferred to fresh NGM plates 5 d after feeding to allow progeny to be discarded. On the day of assay, for each genotype, 5−10 worms were moved to a fresh NGM plate for 2 hr and then discarded. Eggs laid within this 2-hr window were then observed and the developmental stage classified. For the RIL and IL assays, lines were randomized across experimental blocks and N2 and CB4856 wild types were included as controls in each block. Other assays were conducted in the same manner. Analysis of embryo staging for each experimental block took less than an hour, and rescoring of plates during initial experiments indicated that this time did not affect embryo stage data.
All analyses were conducted in custom written scripts in “R” version 2.13.1 × 64. To analyze these data, the effect of genotype on the stage at which the eggs were deposited was tested by analysis of variance, with all the individual egg stage scores used as input “egg-stage~genotype+e.” This was only used to determine the effect of the genotype on the variation in egg-stage. For the IL and RIL data, the mean square of the trait and the residuals were then used to determine the heritability of the trait in each panel. To find genomic regions associated with the control of egg stage, we used QTL mapping. For QTL mapping in the RILs, we used a single marker model, with the percentage of total progeny at a certain stage used as a phenotype. In the RILs, the percentage of progeny at > stage II also was mapped. Genome-wide thresholds were determined by 1000 permutations. In each permutation round, the phenotypic scores were distributed randomly over the RILs after which genome-wide QTL were mapped. The most significant linkage was recorded for each permutation round. The 95% highest –log10(p) value was taken as the 0.05 genome-wide threshold. A similar method was used to determine the threshold for multiple QTL mapping (MQM).
Bin mapping
Bin mapping in the ILs was done as described by Doroszuk et al. (2009) and Green et al. (2013), with the exception that a χ2 test was used as a statistical test. The percentages of eggs per stage of N2 was used as expected distribution and tested against the distribution per bin. Threshold was determined by 10,000 permutations. Each permutation picked the egg-stage scores of two groups of three randomly selected dishes. These two groups were then used in a χ2 test. The 95% highest –log10(p) value was taken as the 0.05 FDR threshold. This method was also used to determine the threshold in IL vs. IL mapping.
MQM method
A forward marker selection was used as MQM method. The mapping was initiated by single marker mapping. The marker with most significant linkage was added to the mapping model as a cofactor. The cofactor was excluded from the model when markers closer than five markers from the cofactor were considered or when the significance of the cofactor was > 0.05. This process was repeated until no new QTL/cofactors could be added.
Fixed locus mapping
To investigate the effect of the major QTL of the left of chr IV on QTL mapping, we fixed the locus by splitting the RILs into two groups. One group with an N2 allele at the left of chr IV and one group with a CB4856 allele at the left of chr IV. Single marker mapping using linear regression was subsequently used to find QTL in these two groups of RILs.
Sub-IL generation
To further investigate the effects of introgressions on chromosome IV on the control of egg stage, we also analyzed an additional set of sub-ILs (ewIR4001-4011). These were generated by crossing ewIR052 with N2 and selecting for new recombinants in the F4 offspring. F4 offspring were obtained by single worm decent. Restriction fragment length polymorphism markers described in Li et al. (2006) and Doroszuk et al. (2009) spanning the original ewIR052 introgression were used for recombination detection.
IL vs. IL mapping
To test whether the egg-stage distribution between each IL-pair were different, a χ2 test was used. The percentages of eggs per stage of one IL was used as expected distribution and tested against the distribution of the other IL. Pairs were then compared as described by Shao et al. (2010) and Green et al. (2013) to find QTL.
Embryo stage analysis in wild isolates
Preliminary experiments and the RIL and IL analyses indicated that N2 and CB4856 lay the majority of their eggs at very early stages of development. To investigate natural variation in this trait more broadly, we assayed, as described previously, a range of wild isolates. The IL ewIR51, which contains a CB4856 introgression on chromosome IV that results in the production of large numbers of late stage progeny (see Figure 2 and Figure S2), was included in these assays as a control.
Figure 2.
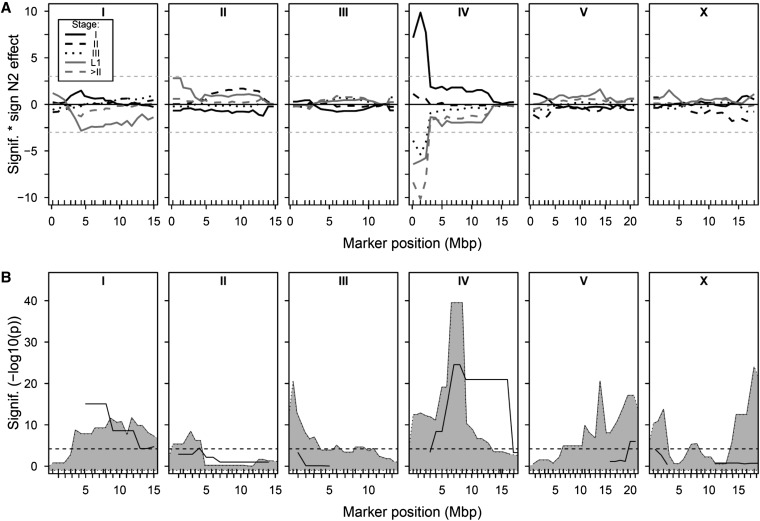
QTL mapping in the RILs and ILs. (A) Mapping of embryo stage in the RILs, with the significance (−log10(p)) multiplied by the sign of the effect of the N2 allele plotted against the marker positions in mega base pairs for the percentage of total eggs in stage I eggs (black solid line), stage II eggs (black dashed line), stage III eggs (black dotted line), stage IV eggs (gray solid line), L1s (gray dashed line), and the proportion of progeny > stage II (gray dotted line). (B) Genome-wide bin mapping of late-stage embryo production (proportion > stage II), showing the significance (−log10(p)) by chi-square test of ILs sharing a certain genomic part against N2. RILs, recombinant inbred lines; ILs, introgression lines.
Analysis of the chromosome IV QTL
To determine how the embryo stage of progeny changed across the reproductive period, we compared ewIR51, ewIR52 (another IL containing the chromosome IV QTL), CB4856, and N2. Here the embryo stages of progeny were determined, as described previously, daily for the first 3 d of the reproductive period. To determine whether the production of large numbers of late stage embryos was associated with an increase in the number of fertilized eggs in utero, as seen in many egl mutants (Trent et al. 1983), we compared ewIR51, ewIR52, and N2. To do this, individual hermaphrodites were transferred to a drop of hypochlorite solution (Stiernagle 2006) on an NGM plate with food. Plates were then incubated at 20° for 2 d when the number of progeny that had developed was determined. Again, these assays were undertaken daily for the first 3 d of the reproductive period.
Relationship to other traits
To determine how variation in other life history traits relates to the synthetic egl effects observed in the RIL and IL lines, all ILs containing introgressions on chromosomes II and IV were assayed for body length, lifetime fecundity and lifespan. These analyses also identified any animals that died by bagging. These assays used standard methods for the analysis of reproductive traits in C. elegans (Hodgkin and Doniach 1997). Body length was determined as described by Harvey and Orbidans (2011) for worms 2 d after recovery from L1 arrest, with individuals photographed using a Moticam 2000 video camera (Motic, Wetzlar, Germany) and the length from the mouth to the base of the tail, determined in ImageJ (http://rsb.info.nih.gov/ij/). Worms were considered to have died if they were not moving and failed to respond to touch.
Data storage
All data were stored in WormQTL (www.wormqtl.org; Snoek et al. 2013, 2014b; Van Der Velde et al. 2014).
Results
Analyses of the RILs (101 lines) and the ILs (87 lines) indicated that genotype significantly affected the stage at which eggs were laid (P < 1e-15 in both cases). The heritability of the egg stage was also very high (estimated as 96.1% in the RILs and 92.9% in the ILs based on individual egg measurements and 74.8% on multiple population averages per genotype in the ILs), although variability between replicates suggests that the heritability based on the individual egg measurements is most likely an overestimation. In both sets of lines, the N2 and CB4856 controls are not significantly different (χ2 test; p~1), with both lines laying mostly stage I eggs (~96% and ~90%, respectively, for N2 and CB4856).
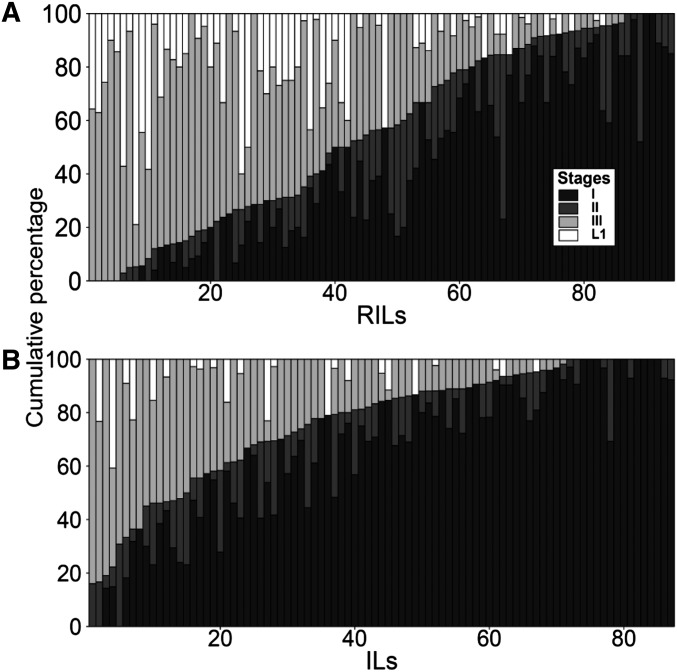
The phenotypic distribution in both the RILs and ILs shows a one-sided transgression, with many genotypes laying large proportions of their eggs at much later stages (Figure 1 and Figure S1) than either of the parental isolates. About half of the RILs laid 50% or more eggs at stage III or later, with about 20% of the ILs displaying such extreme phenotypes (Figure 1). These lines therefore phenocopy mild egl mutations, i.e., they would be classified as M/E, most/early, with all or most progeny released, a few early-stage eggs, and many late-stage eggs observed on the plate (Trent et al. 1983). The observed transgression therefore provides evidence that the stage at which an egg is deposited is a polygenic trait. Moreover, it suggests that either N2 or CB4856 each carry positive and negative allele(s) of the genes involved that are acting additively, or that the observed effects are a consequence of incompatibilities between diverged N2 and CB4856 alleles at different loci, i.e., negative epistatic effects, or a combination of both of these. That more RILs than ILs show an egl phenotype, suggests that multiple regions of the genome and interactions between those contribute to the laying of late stage eggs (comparison between Figure 1, A and B).
Figure 1.
Embryo stage distribution in the RILs and ILs. The cumulative percentage of embryo stages per RIL (A) and IL (B). Lines were sorted by the percentage of embryos > stage II. RILs, recombinant inbred lines; ILs, introgression lines.
QTL mapping in the RILs and ILs
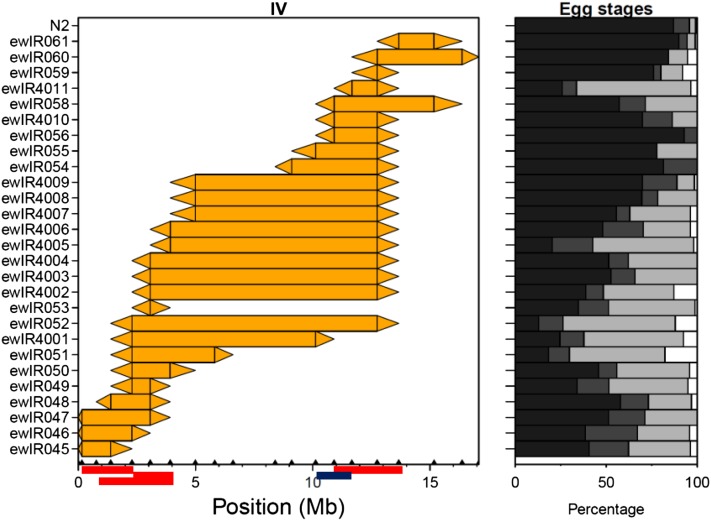
QTL mapping in the RILs identified one highly significant locus at the left of chromosome IV (Figure 2A). This locus can be found for the percentage of progeny at stage I, stage III, L1, and > stage II, with the CB4856 allele at this locus increased the proportions of late stage progeny. These analyses also identified minor QTL for the proportion of L1s on both chromosomes I and II. MQM analysis indicated that additional QTL can be detected on chromosomes I, III, and IV (Table 1 and Figure S2). A two-locus scan for epistatic interactions suggested that there were interactions between many of these QTL, but, due to limited power, these were not significant after correction for multiple testing.
Table 1. Locations and effect of QTL detected for egg-stages.
| Chr | N2L | CBL | CBR | N2R | CB Effect | Detected by |
|---|---|---|---|---|---|---|
| I | 2818974 | 3502476 | 3502476 | 4338254 | + | MQM, (BIN), Single IL, IL vs. IL |
| I | 9569913 | 10259909 | 10259909 | 11085295 | + | Single IL, IL vs. IL |
| I | 11085295 | 11085295 | 11085295 | 11760179 | − | IL vs. IL |
| II | 2755074 | 3403575 | 4147051 | 4800868 | + | (BIN), Single IL, IL vs. IL |
| II | 4147051 | 4800868 | 10414073 | 11180836 | − | IL vs. IL |
| III | 5925983 | 6847169 | 7998164 | 8318553 | − | IL vs. IL |
| III | 10027496 | 10613119 | 10613119 | 11341120 | + | MQM, Single IL, IL vs. IL |
| III | 10613119 | 11341120 | 11341120 | 12301725 | − | IL vs. IL |
| IV | Not applicable | 151889 | 1381409 | 2288742 | + | SM, MQM, Single IL, IL vs. IL |
| IV | 2288742 | 3067374 | 3067374 | 3920366 | + | SM, MQM, Single IL, IL vs. IL |
| IV | 10122930 | 10909560 | 10909560 | 11668242 | − | IL vs. IL |
| IV | 10909560 | 11668242 | 11668242 | 12748880 | + | SM, MQM, Single IL, IL vs. IL |
| V | 10368660 | 10912994 | 16008404 | 17377158 | + | Single IL, IL vs. IL |
| V | 17377158 | 18574593 | 18574593 | 19525561 | − | IL vs. IL |
| V | 18574593 | 19525561 | 20758352 | 20893784 | + | Single IL, IL vs. IL |
| X | 5010049 | 5770179 | 5770179 | 7067019 | − | IL vs. IL |
| X | 5770179 | 7067019 | 7982354 | 8691677 | + | Single IL, IL vs. IL |
The column label Chr show the chromosome on which the QTL was found. N2L, CBL, CBR, and N2R show the position of the left N2, left CB, right CB, and right N2 boundaries of the QTL. The “Detected by” indicates the methods by which the QTL were found/supported. QTL, quantitative trait loci.
Bin mapping in the ILs using the data from the initial genome-wide screen identified a total of 8 QTL where the CB4856 introgression increased the production of late-stage eggs (Figure 2B). ILs with introgressions harboring one of these QTL were retested in a separate experiment and this analysis resulted in the confirmation of four of the eight QTL (Table 1 and Figure 2B), with three of these QTL overlapping the major QTL and minor QTL identified in the RILs. The IL analysis also suggests the presence of additional QTL on chromosome V and on the X chromosome. In combination, the RIL and IL analyses therefore reproducibly identify regions of chromosomes I, II and IV where introgression of the CB4856 region into an N2 background results in an increased production of late-stage eggs.
The QTL identified by bin mapping span very large regions of the genome, up to almost a whole chromosome in the case of chromosomes I and IV (Table 1 and Figure 2B). Because of this, we investigated the individual ILs for clues on the number of alleles/QTL present. This was done by using a χ2 test to test for a difference in stage numbers between N2 and the individual ILs (Table 1 and Figure S3). These analyses detect and confirm the stage increasing CB4856 QTL on chromosomes I, II, III, and IV. Given that ~90% of progeny in N2 and CB4856 are stage I eggs, comparison of the ILs and N2 will only detect CB4856 alleles that increase progeny stage. Such analyses suggest that many regions of the genome disrupt the normal process of egg-laying. For example, on chromosome I this suggests the presence of at least three separate QTL as three nonoverlapping ILs are different from N2 (Table 1, Table S1 and Figure S3). In contrast to such analyses, comparison of overlapping ILs allows the identification of regions that contain CB4856 alleles that decrease progeny stage (Table 1 and Figure 3). These comparisons support the conclusions that the QTL detected here can be separated into multiple factors.
Figure 3.
Comparison between ILs of chromosome IV. The CB4856 introgression per IL is shown by the colored rectangle. Triangles join adjacent CB4856 and N2 markers. Embryo stage distribution is shown as cumulative percentage of total progeny. From dark to light: Stage I, II, III, and L1 (in white). QTL are indicated on the X axis by red (+) or blue (−) boxes (denoting that the CB4856 allele increases or decreases the proportion of late stage embryos, respectively). ILs, introgression lines; QTL, quantitative trait loci.
Embryo stage analysis in wild isolates
To determine whether late-stage egg production was seen in wild isolates of C. elegans, the embryo stage of hermaphrodites from a range of wild isolates on the third day of reproduction was tested. These analyses indicated that there are differences between lines, but that wild isolates all lay eggs at a predominantly early stage of development (Figure 4). This further supports our classification of the late-stage embryo production trait as an incompatibility.
Analysis of the chromosome IV QTL
Analysis of embryo stage across the reproductive period indicates that the trait is age-related, such that the proportion of embryos laid at later stages of development increases throughout the reproductive period (Figure 5A). This finding suggests that it may represent a change in the rates at which the worms are senescing. Previously identified differences in developmental speed between RILs derived from crosses between the isolates N2 and CB4856 only span a few hours (Francesconi and Lehner 2014; Snoek et al. 2014a) and cannot therefore cause the (large) differences in egg-stages between lines.
Many egl mutations cause worms to retain large numbers of eggs in utero, with young adults displaying a slightly bloated phenotype and older worms often containing many times the normal number of fertilized embryos. Comparison of two ILs containing the major chromosome IV QTL to N2 (Figure 5B) indicated that the number of eggs in utero is slightly increased during the first two days of reproduction, but that there is no increase seen on the third day of reproduction.
Relationship to other traits
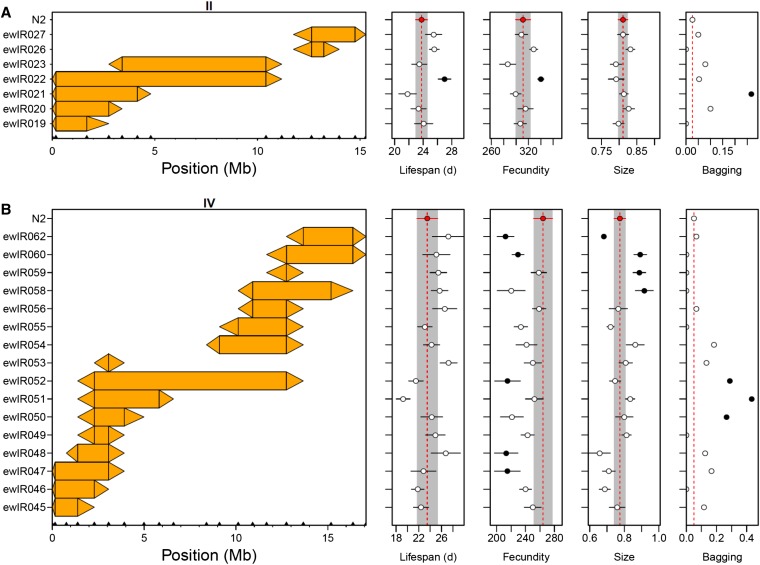
Analysis of all ILs containing introgressions on chromosomes II and IV indicated that all traits were variable (Figure 6, A and B), with these analyses defining QTL for all traits (Table 2 and Table S2). Comparison of these QTL to those found in previous analyses indicates that many QTL are found in multiple studies. For instance, variation in body size between N2 and BO has previously been mapped to chromosome IV (Knight et al. 2001), and one of the chromosome IV body size QTL identified here (Table 2) contains tra-3, a gene polymorphic between CB4856 and N2 that affects how body size changes across temperatures (Kammenga et al. 2007). Similarly, previous comparisons using CB4856 and N2 RILs identified a fecundity QTL on chromosome IV (Gutteling et al. 2007), although this was found at 12° and not at 24°. The patterns of variation identified here do however indicate that the control of these traits is complex, with chromosome IV containing five separate QTL affecting body size (Table 2).
Figure 6.
Incompatibility QTL are associated with variation in other traits. Average lifespan, lifetime fecundity, body size at L4 and proportion of worms that die by bagging for ILs containing introgressions on chromosome II (A) and IV (B). The CB4856 introgression per IL is shown by the colored rectangle. Triangles join adjacent CB4856 and N2 markers. Error bars represent ± 1 SE, dashed lines and shaded bars represent trait values in N2 and ILs significantly different from N2 (P < 0.05) are shown in black. QTL, quantitative trait loci.
Table 2. Locations and effect of QTL detected for body length, lifetime fecundity, lifespan, and bagging.
| Trait | Chr | N2L | CBL | CBR | N2R | CB effect | Detected by |
|---|---|---|---|---|---|---|---|
| Size | IV | 766649 | 1381409 | 3067374 | 3920366 | − | Single IL |
| IV | 5819735 | 6599685 | 12748880 | 13667267 | − | IL vs. IL | |
| IV | 8397264 | 9102404 | 9102404 | 10122930 | + | IL vs. IL | |
| IV | 11668242 | 12748880 | 12748880 | 13667267 | + | Single IL | |
| IV | 12748880 | 13667267 | 16371991 | 17084259 | − | Single IL | |
| Lifespan | II | Not applicable | 176721 | 2755074 | 3403575 | + | IL vs. IL |
| II | 4147051 | 4800868 | 10414073 | 11180836 | + | IL vs. IL | |
| IV | 3920366 | 4991858 | 5819735 | 6599685 | − | IL vs. IL | |
| Fecundity | II | Not applicable | 176721 | 2755074 | 3403575 | + | IL vs. IL |
| II | 4147051 | 4800868 | 10414073 | 11180836 | + | IL vs. IL | |
| IV | 12748880 | 13667267 | 16371991 | 17084259 | − | Single IL, IL vs. IL | |
| Bagging | II | Not applicable | 176721 | 2755074 | 3403575 | + | IL vs. IL |
| II | 4147051 | 4800868 | 10414073 | 11180836 | + | IL vs. IL | |
| IV | 3067374 | 3920366 | 3920366 | 4991858 | + | IL vs. IL |
QTL limits are shown by the locations of the flanking markers with N2 genotype and the adjacent markers with a CB4856 genotype. QTL marked as Single IL were detected in comparisons between ILs and N2, those marked IL vs. IL were detected in comparisons between ILs. Only ILs on chromosome II and IV were tested. QTL, quantitative trait loci.
There was no overall correlation between the traits assayed, showing that multiple independent functional allelic differences exist between N2 and CB4856 (on chromosomes II and IV). These analyses do however indicate that QTL affecting bagging, lifetime fecundity and lifespan can be identified in regions associated with the production of late stage progeny (Figure 6). These data also provide direct evidence for epistatic interactions affecting both lifespan and fecundity on chromosome II, with the IL vs. IL analyses of ewlR021-23 indicating an epistatic interaction between the CB4856 region in ewlR021 and the region in ewlR023 (Figure 6 and Table 2).
Discussion
Within the Caenorhabditis species, there is a continuum between distinct, reproductively isolated, species and species where isolates are at the very earliest stages of speciation (Baird and Stonesifer 2012; Kozlowska et al. 2012; Gimond et al. 2013). The polymorphisms that result in outbreeding depression and hybrid breakdown within species underlie developmental transitions that can ultimately lead to speciation. Our analyses of ILs and RILs derived from the isolates CB4856 and N2 indicate that many of these lines phenocopy mild egl mutations, laying progeny at an advanced stage of development (Figure 1). Genetic analyses of these data revealed multiple QTL affecting egg-laying (Figure 2 and Table 1). These data indicate that the stage at which an egg is deposited is a polygenic trait. However, it is not clear from this analysis if this is a consequence of the additive action of positive and negative allele(s) from CB4856, of epistatic interactions between loci, or a combination of both. The observation that all of the wild isolates lay very early stage eggs (Figure 4) and that the QTL are associated with increased bagging does however argue that laying late stage eggs is deleterious and therefore that selection will be acting to minimize this.
The other phenotypes linked to these egl effects involve fitness traits (Table 2). The clearest association is with bagging, with ILs underlying the QTL on both chromosomes showing increased bagging (Figure 6 and Table 2). This association between laying late stage progeny and an increased rate of bagging is unsurprising given that this is a common phenotype in egl mutants (Trent et al. 1983). The patterns of bagging observed on both chromosome II and IV indicates that these do not represent simple interactions between two loci. For example, comparison of ILs ewIR21-23 (Figure 6) suggest the presence of interactions with other loci on the same chromosome (e.g., between alleles present in ewLR21 and those in ewlR23). As this trait is, like production of late-stage embryo trait, based on the proportion of the population showing the trait, it is not possible to use these comparisons to distinguish between QTL acting additively and those that are a function of epistatic interactions. This is not the case for the lifespan and fecundity QTL that we detect in the two incompatibility regions (Table 2), as positive effect QTL would be detected in comparisons between ILs and N2. Here, both regions support the interpretation of the QTL as epistatic interactions. For instance, comparisons between ILs on chromosome II define two positive effect QTL for both fecundity and lifespan (Table 2), but the introgressions in this region are not consistent with this, as it would imply two positive effect QTL in ewlR22 and one each in ewlR21 and 23 (Figure 6). Because ewlR21 and 23 are not different to N2, a more parsimonious explanation would be that the increased lifespan and fecundity seen in ewlR22 is a consequence of an interaction between CB4856 alleles that are separated in ewlR21 and 23. In this context, it noteworthy that ewlR21 has a slightly reduced lifespan in this assay and has been previously shown, using these ILs, to contain a CB4856 allele that reduces lifespan (Doroszuk et al. 2009). A similar case for a complex interaction can be made for the lifespan QTL identified on chromosome IV (Table 2), a QTL also found by Doroszuk et al. (2009). Given that fecundity QTL are detected at both ends of chromosome IV (Figure 6 and Table 2), it is not clear if a model of additive QTL is more consistent with these data than one reliant on epistatic interactions.
Given the detrimental effects of the QTL we have detected, it is likely that they would represent weak postzygotic barriers. Conceptually, the effects we have detected can be viewed in a number of differing ways. They could be the consequence of transgressive segregation, although in this case this is unlikely as the trait mapped is essentially synthetic and not seen in either parent or in other wild isolates. Alternatively, the trait could be the result of a disruption of canalization and represent the exposure of cryptic genetic variation. In general, canalization acts to limit trait sensitivity to changes in the environment and/or the genetic background (Waddington 1942; Schmalhausen 1949; Lerner 1954). Within species, such incompatibilities will appear similar to cryptic variation, a situation where genetic or environmental perturbation is required to reveal otherwise hidden genetic variation (Gibson and Dworkin 2004; Li et al. 2006; Masel and Siegal 2009; Snoek et al. 2012; Paaby and Rockman 2014). Here, the origin of cryptic variation may represent the evolution of epistatic correction of deleterious effects of a particular mutation (that may or may not also produce adaptive changes). Such changes would be analogous to the local compensatory mutations that occur both between and within species to correct structural changes in proteins (Long et al. 2013)
The life history of C. elegans may facilitate the build-up of such deleterious mutations. For example, fixation within a line of adaptive mutations that have pleiotropic deleterious effects, or mildly deleterious mutations (as aided by the extensive selfing and the bottlenecking resulting from the C. elegans life-history) would allow the subsequent selection for compensatory mutations. As compensatory mutations appear commonly in C. elegans, as shown by experiments that have reimposed selection on mutation accumulation lines (Estes and Lynch 2003; Denver et al. 2010; Estes et al. 2011), this could result in a negative interaction between the compensatory mutation and the original allele. This would produce a situation where local adaptation (first mutation advantageous) or cryptic genetic variation (first mutation deleterious and now associated with a compensatory mutation) would produce, at least, a pair of coadapted genes. In making the RILs and the ILs the links between coadapted genes might be broken up and cryptic genetic variation that only exists to correct otherwise deleterious polymorphisms is revealed. It is clear that there is significant genotypic and phenotypic variation between C. elegans wild isolates (Hodgkin and Doniach 1997; Viney et al. 2003; Barrière and Félix 2005; Barrière and Félix 2007; Harvey et al. 2008, 2009; Maydan et al. 2010; Andersen et al. 2012; Green et al. 2013; Thompson et al. 2013; Volkers et al. 2013; Snoek et al. 2014b). Large-scale analysis of C. elegans isolates reveals little grouping by isolation environment or by country of origin on a global scale (Andersen et al. 2012), although there is evidence at smaller scales that suggests local adaptation (Volkers et al. 2013). Hence, there is much potential for local adaptation to produce the kinds of interactions proposed here.
The mapping resolution of the QTL identified here precludes a detailed search for candidate genes. However, comparison of the locations of the QTL identified here to the results of expression QTL (eQTL) studies of lines produced from crosses between N2 and CB4856 (Li et al. 2006; Rockman et al. 2010; Viñuela et al. 2010, 2012; Snoek et al. 2013; Van Der Velde et al. 2014) suggest that a number of the genome hotspots for trans acting eQTL do co-localize with incompatibility QTL. This is particularly the case with the incompatibility QTL on the top of chromosome IV (Figure 2), where a very strong eQTL hotspot has been identified under a range of conditions (Rockman et al. 2010; Viñuela et al. 2010, 2012). This part of chromosome IV also contains multiple QTL affecting dauer larvae development in growing populations (Green et al. 2013) and a large number of separate QTL affecting olfactory preference between Serratia marcescens, a bacterium pathogenic to C. elegans, and E. coli (Glater et al. 2014). The large number of phenotypes now known to be linked this region and the observed complexity of their regulation, as implied by the number of separable QTL in the region (Green et al. 2013; Glater et al. 2014) (Table 1 and Figure 5), mean that determining how these variants are related will be interesting for their potential role in speciation. More generally, given the extensive lab adaptation observed in the N2 isolate (McGrath et al. 2009; Weber et al. 2010; Duveau and Félix 2012) it would be informative to investigate the role of these changes in the incompatibilities observed here as such alleles are known to be of recent origin. This would therefore demonstrate that short periods of strong selection can rapidly produce incompatibilities.
To date, the mechanisms that isolate four Caenorhabditis species, C. elegans, C. briggsae, C. remanei, and C. sp. strain CB5161, now named C. brenneri (Sudhaus and Kiontke 2007), have been described (Baird et al. 1992). Work on more recently isolated Caenorhabditis species, which can form viable, and in some cases fertile, hybrids, has also started to address the genetic bases of speciation in this group (Baird and Stonesifer 2012; Kozlowska et al. 2012; Gimond et al. 2013). Because outbreeding depression is also observed in the other predominantly self-fertilizing Caenorhabditis species (Ross et al. 2011; Baird and Stonesifer 2012; Kozlowska et al. 2012; Gimond et al. 2013) it is likely that BDM incompatibilities will also be detectable within these species. Over the longer term, the identification of the causative loci for the QTL identified here would allow comparison with the changes that produce more extreme reproductive isolation and the alleles involved in the very early stages of speciation that have been detected in other Caenorhabditis species (Dey et al. 2012; Kozlowska et al. 2012). This suggests that the Caenorhabditis species have the potential to be hugely informative about the genetics of speciation and more generally about the role of epistatic interactions in the control of complex traits.
Supplementary Material
Acknowledgments
We thank Wormbase (www.wormbase.org) for being a rich and versatile source of information. We thank Morris Swertz and Joeri van der Velde for their help with making the data accessible through WormQTL. Strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). Elements of the work reported here were undertaken by H.E.O. while supported by a Nuffield Foundation Undergraduate Research Bursary (URB/39836). S.C.H. was supported by a Research Grant from the Royal Society and L.B.S. and J.E.K. were funded by the ERASysbio-plus ZonMW project GRAPPLE-Iterative modeling of gene regulatory interactions underlying stress, disease, and ageing in C. elegans (project nr. 90201066) and NEMADAPT (Molecular architecture of environmental adaptation in natural populations of the nematode Caenorhabditis elegans), NWO-ALW (Nederlandse Organisatie voor Wetenschappelijk Onderzoek. Aard- en Levenswetenschappen project 855.01.151).
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.013151/-/DC1
Data are archived in WormQTL (http://www.wormqtl.org).
Communicating editor: B. J. Andrews
Literature Cited
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S. E., Stonesifer R., 2012. Reproductive isolation in Caenorhabditis briggsae: dysgenic interactions between maternal- and zygotic-effect loci result in a delayed development phenotype. Worm 1: 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S. E., Sutherlin M. E., Emmons S. W., 1992. Reproductive isolation in Rhabditidae (Nematoda, Secernentea)—mechanisms that isolate 6 species of 3 genera. Evolution 46: 585–594 [DOI] [PubMed] [Google Scholar]
- Barrière A., Félix M. A., 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15: 1176–1184 [DOI] [PubMed] [Google Scholar]
- Barrière A., Félix M. A., 2007. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics 176: 999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W., 1909. Heredity and variation in modern lights, pp. 85–101 in Darwin and Modern Science, edited by Seward A. C.. Cambridge University Press, Cambridge [Google Scholar]
- Bikard D., Patel D., Le Mette C., Giorgi V., Camilleri C., et al. , 2009. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323: 623–626 [DOI] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Zhou J., Clark A. G., Hartl D. L., Ayroles J. F., 2013. Genetic incompatibilities are widespread within species. Nature 504: 135–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., 2012. The polymorphic prelude to Bateson-Dobzhansky-Muller incompatibilities. Trends Ecol. Evol. 27: 209–218 [DOI] [PubMed] [Google Scholar]
- Denver D. R., Howe D. K., Wilhelm L. J., Palmer C. A., Anderson J. L., et al. , 2010. Selective sweeps and parallel mutation in the adaptive recovery from deleterious mutation in Caenorhabditis elegans. Genome Res. 20: 1663–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A., Jeon Y., Wang G. X., Cutter A. D., 2012. Global population genetic structure of Caenorhabditis remanei reveals incipient speciation. Genetics 191: 1257–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. S., Charlesworth B., Baird S. E., Cutter A. D., 2007. Inbreeding and outbreeding depression in Caenorhabditis nematodes. Evolution 61: 1339–1352 [DOI] [PubMed] [Google Scholar]
- Doroszuk A., Snoek L. B., Fradin E., Riksen J., Kammenga J., 2009. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res. 37: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury D. W., Jideonwo V. N., Ehmke R. C., Wade M. J., 2011. An unusual barrier to gene flow: perpetually immature larvae from inter-population crosses in the flour beetle, Tribolium castaneum. J. Evol. Biol. 24: 2678–2686 [DOI] [PubMed] [Google Scholar]
- Drury D. W., Ehmke R. C., Jideonwo V. N., Wade M. J., 2013. Developmental trajectories and breakdown in F1 interpopulation hybrids of Tribolium castaneum. Ecol. Evol. 3: 1992–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duveau F., Félix M. A., 2012. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 10: e1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands S., 1999. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution 53: 1757–1768 [DOI] [PubMed] [Google Scholar]
- Elvin M., Snoek L. B., Frejno M., Klemstein U., Kammenga J. E., et al. , 2011. A fitness assay for comparing RNAi effects across multiple C. elegans genotypes. BMC Genomics 12: 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes S., Lynch M., 2003. Rapid fitness recovery in mutationally degraded lines of Caenorhabditis elegans. Evolution 57: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Estes S., Phillips P. C., Denver D. R., 2011. Fitness recovery and compensatory evolution in natural mutant lines of C. elegans. Evolution 65: 2335–2344 [DOI] [PubMed] [Google Scholar]
- Francesconi M., Lehner B., 2014. The effects of genetic variation on gene expression dynamics during development. Nature 505: 208–211 [DOI] [PubMed] [Google Scholar]
- Gibson G., Dworkin I., 2004. Uncovering cryptic genetic variation. Nat. Rev. Genet. 5: 681–690 [DOI] [PubMed] [Google Scholar]
- Gimond C., Jovelin R., Han S., Ferrari C., Cutter A. D., et al. , 2013. Outbreeding depression with low genetic variation in selfing Caenorhabditis nematodes. Evolution 67: 3087–3101 [DOI] [PubMed] [Google Scholar]
- Glater E. E., Rockman M. V., Bargmann C. I., 2014. Multigenic natural variation underlies Caenorhabditis elegans olfactory preference for the bacterial pathogen Serratia marcescens. G3 (Bethesda) 4: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. W., Snoek L. B., Kammenga J. E., Harvey S. C., 2013. Genetic mapping of variation in dauer larvae development in growing populations of Caenorhabditis elegans. Heredity (Edinb) 111: 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. S., Horvitz H. R., 1980. unc-93(e1500): A behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics 96: 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteling E. W., Riksen J. A., Bakker J., Kammenga J. E., 2007. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity (Edinb) 98: 28–37 [DOI] [PubMed] [Google Scholar]
- Harvey S. C., 2009. Non-dauer larval dispersal in Caenorhabditis elegans. J. Exp. Zool. B Mol. Dev. Evol. 312B: 224–230 [DOI] [PubMed] [Google Scholar]
- Harvey S. C., Orbidans H. E., 2011. All eggs are not equal: the maternal environment affects progeny reproduction and developmental fate in Caenorhabditis elegans. PLoS ONE 6: e25840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S. C., Shorto A., Viney M. E., 2008. Quantitative genetic analysis of life-history traits of Caenorhabditis elegans in stressful environments. BMC Evol. Biol. 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D., Oppenheim D., Klass M., 1976. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 49: 200–219 [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Doniach T., 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146: 149–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R., Sulston J. E., 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96: 435–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga J. E., Doroszuk A., Riksen J. A., Hazendonk E., Spiridon L., et al. , 2007. A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 3: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga J. E., Phillips P. C., De Bono M., Doroszuk A., 2008. Beyond induced mutants: using worms to study natural variation in genetic pathways. Trends Genet. 24: 178–185 [DOI] [PubMed] [Google Scholar]
- Knight C. G., Azevedo R. B., Leroi A. M., 2001. Testing life-history pleiotropy in Caenorhabditis elegans. Evolution 55: 1795–1804 [DOI] [PubMed] [Google Scholar]
- Kozlowska J. L., Ahmad A. R., Jahesh E., Cutter A. D., 2012. Genetic variation for postzygotic reproductive isolation between Caenorhabditis briggsae and Caenorhabditis Sp 9. Evolution 66: 1180–1195 [DOI] [PubMed] [Google Scholar]
- Lachance J. L., Johnson N. A., True J. R., 2011. The population genetics of X-autosome synthetic lethals and steriles. Genetics 189: 1011–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner I. M., 1954. Genetic Gomeostasis. Wiley, New York [Google Scholar]
- Li Y., Alvarez O. A., Gutteling E. W., Tijsterman M., Fu J., et al. , 2006. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Breitling R., Snoek L. B., van der Velde K. J., Swertz M. A., et al. , 2010. Global genetic robustness of the alternative splicing machinery in Caenorhabditis elegans. Genetics 186: 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., Rabanal F. A., Meng D., Huber C. D., Farlow A., et al. , 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., 2014. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J., Siegal M. L., 2009. Robustness: mechanisms and consequences. Trends Genet. 25: 395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydan J. S., Lorch A., Edgley M. L., Flibotte S., Moerman D. G., 2010. Copy number variation in the genomes of twelve natural isolates of Caenorhabditis elegans. BMC Genomics 11: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. T., Rockman M. V., Zimmer M., Jang H., Macosko E. Z., et al. , 2009. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61: 692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1942 Isolating mechanisms, evolution and temperature, pp. 71–125 in Temperature, Evolution, Development, Vol. 6, edited by T. Dobzhansky. Biological Symposiua: A Series of Volumes Devoted to Current Symposia in the Field of Biology. Jaques Cattell Press, Lancaster, PA. [Google Scholar]
- Orr H. A., Irving S., 2001. Complex epistasis and the genetic basis of hybrid sterility in the Drosophila pseudoobscura Bogota-USA hybridization. Genetics 158: 1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Rockman M. V., 2014. Cryptic genetic variation: evolution’s hidden substrate. Nat. Rev. Genet. 15: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez D. E., Wu C. I., 1995. Further characterization of the Odysseus locus of hybrid sterility in Drosophila: one gene is not enough. Genetics 140: 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P. C., 2008. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9: 855–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P. C., Johnson N. A., 1998. The population genetics of synthetic lethals. Genetics 150: 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180 [DOI] [PubMed] [Google Scholar]
- Rockman M. V., 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Skrovanek S. S., Kruglyak L., 2010. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330: 372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M., Snoek L. B., Riksen J. A., Bevers R. P., Kammenga J. E., 2012. Genetic variation for stress-response hormesis in C. elegans lifespan. Exp. Gerontol. 47: 581–587 [DOI] [PubMed] [Google Scholar]
- Ross J. A., Koboldt D. C., Staisch J. E., Chamberlin H. M., Gupta B. P., et al. , 2011. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 7: e1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalhausen I. I., 1949. Factors of Evolution: The Theory of Stabilizing Selection. Blakiston, Philadelphia [Google Scholar]
- Seidel H. S., Rockman M. V., Kruglyak L., 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Sinasac D. S., Burrage L. C., Hodges C. A., Supelak P. J., et al. , 2010. Analyzing complex traits with congenic strains. Mamm. Genome 21: 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek L. B., Terpstra I. R., Dekter R., Van den Ackerveken G., Peeters A. J., 2012. Genetical genomics reveals large scale genotype-by-environment interactions in Arabidopsis thaliana. Front. Genet. 3: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek L. B., Van der Velde K. J., Arends D., Li Y., Beyer A., et al. , 2013. WormQTL–public archive and analysis web portal for natural variation data in Caenorhabditis spp. Nucleic Acids Res. 41: D738–D743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek L. B., Sterken M. G., Volkers R. J., Klatter M., Bosman K. J., et al. , 2014a A rapid and massive gene expression shift marking adolescent transition in C. elegans. Sci. Rep. 4: 3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek L. B., van der Velde K. J., Li Y., Jansen R. C., Swertz M. A., et al. , 2014b Worm variation made accessible: take your shopping cart to store, link, and investigate! Worm 3: e28357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans (February 11, 2006), WormBook, The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1,, http//www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhaus W., Kiontke K., 2007. Comparison of the cryptic nematode species Caenorhabditis brenneri sp n. and C remanei (Nematoda: Rhabditidae) with the stem species pattern of the Caenorhabditis elegans group. Zootaxa 1456: 45–62 [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic-cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119 [DOI] [PubMed] [Google Scholar]
- Tao Y., Zeng Z. B., Li J., Hartl D. L., Laurie C. C., 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. II. Mapping hybrid male sterility loci on the third chromosome. Genetics 164: 1399–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A. R., 1986. Coadaptation and outbreeding depression, pp. 33–59 in Conservation Biology: The Science of Scarcity and Diversity, edited by Soulé M.. Sinauer Associates, Sunderland, MA [Google Scholar]
- Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., et al. , 2013. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent C., Tsuing N., Horvitz H. R., 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104: 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velde K. J., de Haan M., Zych K., Arends D., Snoek L. B., et al. , 2014. WormQTLHD–a web database for linking human disease to natural variation data in C. elegans. Nucleic Acids Res. 42: D794–D801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney M. E., Gardner M. P., Jackson J. A., 2003. Variation in Caenorhabditis elegans dauer larva formation. Dev. Growth Differ. 45: 389–396 [DOI] [PubMed] [Google Scholar]
- Viñuela A., Snoek L. B., Riksen J. A., Kammenga J. E., 2010. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 20: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuela A., Snoek L. B., Riksen J. A., Kammenga J. E., 2012. Aging uncouples heritability and expression-QTL in Caenorhabditis elegans. G3 (Bethesda) 2: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkers R. J., Snoek L. B., Hubar C. J., Coopman R., Chen W., et al. , 2013. Gene-environment and protein-degradation signatures characterize genomic and phenotypic diversity in wild Caenorhabditis elegans populations. BMC Biol. 11: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Ehrenstein G., Schierenberg E., 1980, Cell lineages and development of Caenorhabditis elegans and other nematodes, pp. 1–71 in Nematodes as Biological Models, edited by Zuckerman B. M.. Academic Press, New York [Google Scholar]
- Waddington C. H., 1942. The canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [DOI] [PubMed] [Google Scholar]
- Waterston R. H., Thomson J. N., Brenner S., 1980. Mutants with altered muscle structure in Caenorhabditis elegans. Dev. Biol. 77: 271–302 [DOI] [PubMed] [Google Scholar]
- Weber K. P., De S., Kozarewa I., Turner D. J., Babu M. M., et al. , 2010. Whole genome sequencing highlights genetic changes associated with laboratory domestication of C. elegans. PLoS One 5: e13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.