Abstract
Bats comprise 20% of all mammalian species and display a number of characteristics, including true flight, echolocation, and a heightened ability to resist viral load that uniquely position this group for comparative genomic studies. Here we searched for evidence of genomic variation consistent with sensory rewiring through bat evolution. We focused on two species with divergent sensory preferences. Myotis davidii is a bat species that echolocates and possesses dim- but not daylight-adapted vision whereas the black flying fox (Pteropus alecto) has highly developed day vision but does not echolocate. Using the naked mole rat as a reference, we found five functional genes (CYP1A2, RBP3, GUCY2F, CRYBB1, and GRK7) encoding visual proteins that have degenerated into pseudogenes in M. davidii but not P. alecto. In a second approach genome-wide codon usage bias (CUB) was compared between the two bat species. This CUB ranking systematically enriched for vision-related (CLN8, RD3, IKZF1, LAMC3, CRX, SOX8, VAX2, HPS1, RHO, PRPH2, and SOX9) and hearing-related (TPRN, TMIE, SLC52A3, OTOF, WFS1, SOD1, TBX18, MAP1A, OTOS, GPX1, and USH1G) machinery in M. davidii but not P. alecto. All vision and hearing genes selectively enriched in M. davidii for which orthologs could be identified also were more biased in the echolocating M. lucifugus than the nonecholocating P. vampyrus. We suggest that the existence of codon bias in vision- and hearing-related genes in a species that has evolved echolocation implies CUB is part of evolution’s toolkit to rewire sensory systems. We propose that the two genetic changes (pseudogene formation and CUB) collectively paint a picture of that incorporates a combination of destruction and gain-of-function. Together, they help explain how natural selection has reduced physiological costs associated with the development of a smaller eye poorly adapted to day vision but that also contribute to enhanced dim light vision and the hearing adaptations consonant with echolocation.
Keywords: evolution, vision, codon usage bias, pseudogene, bat
As pointed out previously (Jones et al. 2013), bats perceive the world using a range of sensory mechanisms, some of which have undergone specialization (Altringham and Fenton 2003). Myotis davidii and Pteropus alecto are Chiropterans that use different habitats, prefer different diets, and occupy different sensory niches (Zhang et al. 2013). Previous work has compared and contrasted their genomes to shed light on the genetic basis on a host of their phenotypic similarities and differences. Recently, a new connection was drawn between bat immunity and bioenergetic demands based on gene family expansions and contractions and other lines of genetic evidence (Zhang et al. 2013). In taking a sensory perspective of the two species, P. alecto forages through vision and smell but cannot echolocate. In contrast, M. davidii is an echolocator thought to have relatively poor day vision. It exhibits the loss of function often observed when biological systems avoid paying the cost of unnecessary capacity (Weibel et al. 1991).
Basic morphological data in echolocating bats—such as differences in the lens, cornea, vitreous body shape and proportion of rods and cones—are consistent with reduced visual acuity (Bhatnagar 1975; Pettigrew 1986; Fuzessery et al. 1993; Kalko et al. 1996). A complication to the sensory ecology of myotid bats is that although they possess a smaller eye, this does not exclusively imply degradation. Recent work in M. brandtii suggests this echolocator may be adapted to dim light conditions such as found at dawn and dusk and also ultraviolet light (Seim et al. 2013). These visual adaptations are related to photopigment opsins, rhodopsin, and downstream signaling components (Seim et al. 2013). The molecular basis of visual adaptation in M. davidii per se is unknown.
Taking a broader sensory perspective, a very recently published analysis based on the ratio of nonsynonymous to synonymous substitutions common to both echolocating dolphins and echolocating bats identified genes encoding sensory proteins, particularly vision and hearing, as striking signatures of convergent evolution (Parker et al. 2013). The hearing adaptation relates to the echolocation facility, a multimodal capability comprising both specialized vocalization on the one hand and signal reception on the other. A recent review of the genetic basis of the sensory ecology of echolocating bats can be found here in Jones et al. (2013). Genes thought to be of particular interest for echolocation per se include those encoding proteins related to vocalization (FOXP2) and enhanced hearing (SLC26A5).
There is some debate over whether bat echolocation is (1) a derived characteristic not present in the common ancestor that has subsequently evolved in multiple echolocating lineages by convergent evolution, or (2) was present in the ancestral species then selectively lost (Teeling 2009). Either way, our aim is to contribute to the elucidation of the genetic basis of sensory rewiring, with a focus on these two species. We compare the output to two other bat species for whom genome-wide data already exist, the echolocator M. lucifugus and the nonecholocator P. vampyrus. Pteropodids diverged from other bats 55 mya, whereas the myotid bats are considered by some authorities to be a more recent lineage that split 15 mya. Assembled genomes for these four bats provide great scope for comparative genomics. Indeed, it has been pointed out that the future for understanding the molecular basis of sensory biology is promising, with comparative genomics having great potential (Jones et al. 2013). Here, we find a combination of hypothesis-driven and hypothesis-free approaches to be mutually reinforcing, shedding molecular insight into the apparent rewiring of M. davidii’s visual modality, and also complementary changes in the hearing modality that we suggest likely relate to echolocation.
Our motivation for exploring genome-wide patterns of codon usage bias (CUB) in the context of comparative mammalian evolution was our recent finding that pathway-level alterations in translation efficiency appear to have an underappreciated role in driving evolutionary change through gain of function, even in complex eukaryotes such as mammals and birds (Hudson et al. 2011). We previously observed systematic CUB in genes fundamental to important characteristics of each lineage, such as cell wall and chloroplast physiology in plants, mitochondrial function in birds, and hair formation in mammals (Hudson et al. 2011). Other recent work also has reinforced a likely role for CUB in human evolution. For example, a large scale tissue-specific analysis in humans previously found that the tRNA adapatation index significantly correlated with expression levels both globally and in a range of tissues taken in isolation (Waldman et al. 2010). These lines of evidence support a role for CUB in mammalian evolution, but genome-wide CUB analysis has never previously been applied to any bat species.
Here, we apply a very sensitive CUB detection tool that, when applied genome-wide, finds a coordinated pattern of enrichment for genes encoding vision and hearing-related machinery in the echolocating M. davidii but not the nonechlocating fruitbat P. alecto. Orthologs of those vision and hearing genes also were more biased in M. lucifugus than P. vampyrus, although the effect was not so strong. The outlier visual proteins tend to be highly expressed in human eye tissue but this is not always the case, implying no simple relationship to abundance. Also based on comparative human data, some of these genes are expected to have highly restricted tissue-specific expression patterns, whereas others are widely expressed. Given observations we have made applying the same method in a number of other genomes, we submit that the most likely explanation to be selection on translation efficiency that manifests at the pathway level. We propose that the coordinate impact of these changes (pseudogenes and CUB) is sensory rewiring, presumably helping to manipulate the Myotis retinogenic pathway toward the development of a smaller, low light-adapted eye with additional changes relating to the enhanced hearing that contributes to the echolocation facility.
Materials and Methods
Hypothesis-driven pseudogene analysis
A list of vision-related genes obtained from the gene ontology database (GO:0007601: visual perception) and visual system from the QIAGEN pathway library (http://www.qiagen.com/Products/Genes%20and%20Pathways/) defined presence, copy number, and nature of corresponding genes in the M. davidii and P. alecto genomes. Proteins from human vision-related genes and their neighboring genes were downloaded from Ensembl release 64. The longest transcript was chosen to represent each gene in cases when alternative splicing variants have been shown to exist. We then subjected all proteins to tBlastn analysis against the bat genomes with the similarity cutoff threshold of e-value = 1e−5. With the human protein set as a reference, we found the best hit for each human protein in the two bat genomes by using the criteria that more than 30% of the aligned sequence showed an identity above 30%. We used GeneWise algorithm (Birney et al. 2004) (with parameters -genesf -for -quiet) to define the detailed exon-intron structure of each bat gene and to identify potential pseudogenes among the bat visual genes.
Initially, we targeted the examination of the bat orthologs for vision-related pseudogenes identified in the naked mole rat Heterocephalus glaber (Kim et al. 2011), a species with poor vision adapted to life in the dark. Genes containing frame shifts or premature stop codons were considered candidates. We filtered as follows: (1) To avoid reported frame shifts or premature stop codons that were due to a flaw in the GeneWise algorithm, we aligned all human proteins to their corresponding loci in the human genome, and genes with frame shifts or premature stop codons in human-to-human alignments were filtered; (2) Using the results of the human-to-human alignment from GeneWise, candidate pseudogenes with obvious splicing errors near their frame shifts or premature stop codons were filtered; (3) Candidate pseudogenes with a low number of reads covering their frame shift or premature stop codon sites were considered assembly errors. Genes with a considerable number of reads due to genotype variation at these sites were treated as heterozygous and filtered.
Hypothesis-free genome-wide CUB analysis
A number of CUB statistics have been proposed, and there is little consensus on the optimal approach. A very recent review outlining the pros and cons of some commonly applied CUB quantification approaches can be found here in Behura and Severson (2013). Our CUB statistic uses the concept of entropy, which estimates data regularities based on a combination of order and proportion. Its strength lies in its broad applicability and sensitivity. Because it is not systematically influenced by gene length and accounts for amino acid composition, it can be fairly applied within and between genomes. It does not account, however, for background GC content, nor does it determine whether bias is toward preferred or nonpreferred codons. For this reason, background GC content and a more detailed analysis of codon usage must be retrospectively assessed on a case-by-case basis (i.e., in those genes whose entropic properties have been determined as extreme and therefore worthy of deeper investigation).
To summarize, we determined the extent of CUB for every coding sequence using an information theory-based statistic as previously described (Hudson et al. 2011). For each observed coding sequence, we computed the entropy of the nucleic acid and compared it against the average entropy of 20 random sequences. The random sequences were generated in a way that they would encode the same amino acid sequence, but where the codons were selected at random. Because entropy provides a measure of data regularity, the differential entropy between the observed sequence and the random corresponds to the extent of regularity attributable to CUB.
We first ranked all coding sequences on extent of CUB on a within-genome basis. To ascertain the extent of functional enrichment analysis the ranked lists for Myotis and Pteropus were subjected to the GOrilla webtool (Eden et al. 2009). GOrilla uses hypergeometric statistics to determine whether particular biological processes are enriched within any ranked input set of genes. We used the single ranked list option based on human functional annotation, and imported SWISSPROT IDs, which is one of the designated preferred input identifiers for GOrilla analysis. We also compared relative codon bias in the orthologs in common between the two species (of which we identified 6748). This secondary analysis helps identify genes that may not be extreme on a within genome basis but still possess markedly different properties between the two species.
Because the GOrilla enrichment relies on incomplete annotation of functional processes and can overlook functional connections even in well-annotated genes, we used the top enrichment as a focus for further manual curation, based on mining the PubMed literature database. A subset of outlier CUB genes were retrospectively assessed for regional GC content. This was computed from the gene itself, plus 10 kb upstream from the start codon and 10 kb downstream from the stop codon.
Independent validation of divergent CUB:
The differential entropy statistic is one of many ways of measuring CUB. In an effort to provide an independent line of evidence for differential CUB in the two bat species and subsequent differential functional enrichment, we also ran the previously published effective numbers of codons (ENC) (Wright 1990) through the CodonW website: http://codonw.sourceforge.net/. According to Behura and Severson (2013), ENC is used to measure how far the codon usage of a gene departs from equal usage of synonymous codons. EC values range from 20 (extreme codon bias when one codon is used exclusively for each amino acid) to 61 (in cases of no codon bias where synonymous codons are equally likely to code the amino acid).
CUB outlier genes quality checking:
Because it is possible that gaps in the genome assembly data can result in artifacts with the cds sequences, the cds sequence data for all M. davidii CUB outlier genes were confirmed by comparison with cds sequences from other species. We used translation BLAST to identify potential assembly problems, including gaps in our cds sequences. The bats M. brandtii and M. lucifugus plus other mammalian species available in Genbank were used for sequence alignment. In the event of potential missing exons we cross-referenced back to the M. davidii genomic assembly to confirm the quality of genomic sequence data.
CUB analysis in two additional bat species: M. lucifugus and P. vampyrus:
To detect comparative evidence for CUB in the sensory apparatus in echolocating bats, we computed CUB in an additional echolocator (M. lucifugus) and nonecholocator (P. vampyrus). The current genome assemblies from the Ensemble database for P. vampyrus (pteVam1) and M. lucifugus (Myoluc2.0) were used for this analysis. For these two comparison species, we obtained coding sequence data from Biomart, yielding 14,141 annotated cds for M. lucifugus and 8, 845 for P. vampyrus. CUB was estimated using the entropy statistic as previously described.
Results
Hypothesis-driven pseudogene analysis
Of the 19 visual perception genes identified as inactivated or missing in the naked mole rat, we identified five that have also become pseudogenes in M. davidii but not P. alecto (CYP1A2, RBP3, GUCY2F, CRYBB1, and GRK7; Figure 1). Taxonomically the naked mole rat H. glaber is placed within rodents. Despite some exceptional traits the overall properties of the H. glaber genome are similar to other mammals (Kim et al. 2011) making the comparison to the two bat species reasonable.
Figure 1.
Gene structure, partial amino acid ,and nucleotide alignments for five M. davidii pseudogenes. Red triangles illustrate the location of stop codons compared with P. alecto. Amino acid alignment surrounding the stop codon region in P. alecto and M. davidii. Asterisks represent stop codons. Nucleotide alignment between P. alecto and M. davidii corresponding to the same region in amino acid alignment. Gray nucleotides represent the region after a frame shift in M. davidii that leads to a premature stop codon (red).
For CYP1A2, CRYBB1, and RBP3 single or multiple nucleotide deletions were responsible for frame-shift mutations leading to premature stop codons in M. davidii. For CRYBB1, the deletion was 31 nucleotides in the first exon compared with P. alecto. In contrast, the GRK7 and GUCY2F pseudogenes were caused by single-point mutations that created single stop codons. In the case of GUCY2F two point mutations had created two stop codons in close vicinity to each other. The similarity between the intact P. alecto genes compared to their corresponding regions in the M. davidii genome, ranged from 85 to 90%.
Hypothesis-free genome-wide CUB analysis
We calculated codon usage preferences for the two species genome-wide (Table 2 and Table 3). These genome-wide figures can be used to help establish preferred and nonpreferred codons for that particular species in concert with the available tRNA pool. In turn this helps shed light on the translational implications of CUB. Namely, is CUB likely to increase or decrease the energetic cost and efficiency of translation for the gene in question?
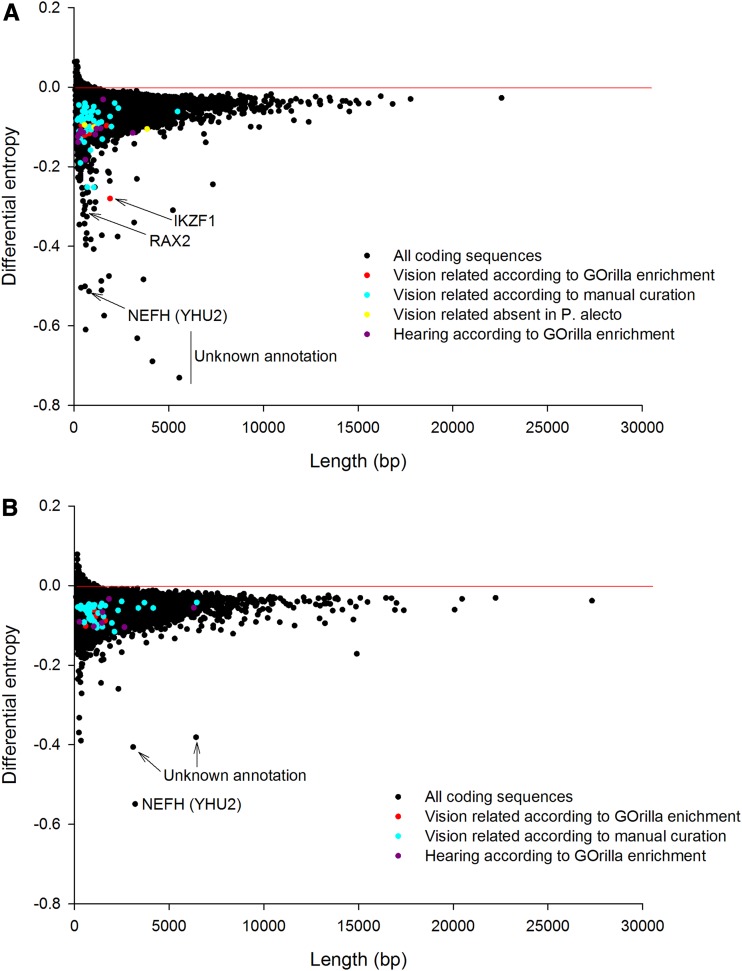
Figure 2.
Genes showing extreme codon bias (negative differential entropy) on a within-genome basis in (A) M. davidii and (B) P. alecto, with those encoding vision and hearing related proteins highlighted.
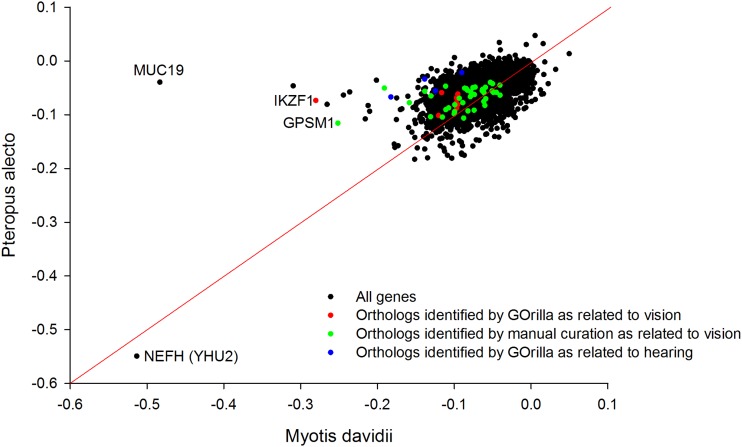
Figure 3.
Orthologs showing differential codon bias (distance from diagonal) between M. davidii and P. alecto on a between-genome basis. Genes encoding vision and hearing elated proteins are highlighted. Of the sensory genes IKZF1 is particularly prominent. The highly codon biased LAMC3, RHO, and SOX9 identified by GOrilla as vision-related in M. davidii appear to be absent in P. alecto. MUC19, highly biased in M. davidii, is a Mucin family member expressed in the inner ear. NEFH is equally highly biased in both bat species.
We next used a hypothesis-free screen, based on CUB, to test for pathway functional enrichment. Before functional enrichment, GOrilla screens the imported gene list, removes duplicates (keeping the only the most highly ranked candidate), then assigns GO characteristics where those exist. For M. davidii we imported 13,654 genes that could be assigned SWISS PROT identifiers, of which 37% were recognized. A total of 6951 were recognized by Gene Symbol, 1731 duplicates were removed, and of the remaining 5220, 5056 had a GO term assigned. For P. alecto we imported 12,188 genes that could be assigned to SWISS PROT identifiers of which 40% were recognized. A total of 6183 were recognized by Gene Symbol, 1056 duplicates were removed, and of the remaining 5127, 4956 had a GO term assigned.
NEFH (alias YHU2, encoding the neurofilament heavy polypeptide) possessed extreme CUB in both species. Consistent with previous research in other organisms, the translation initiation factor IF2 (PRP2) was among the most biased genes in both species.
Of the biased genes within M. davidii we found the top ‘Process’ enrichment to be “retina development of camera type eye” (hypergeometric test P-value = 0.0000775; False Discovery Rate q-value = 0.766) based on the prominent bias of the following genes: CLN8, RD3, IKZF1, LAMC3, CRX, SOX8, VAX2, HPS1, RHO, PRPH2, SOX9; Figure 2A and Supporting Information, File S1). The remaining functional enrichments reported in descending order by GOrilla for M. davidii were “O-glycan processing” (P-value = 0.000127), “protein O-linked glycosylation” (P-value = 0.000393), “adenylate cyclase-inhibiting G-protein coupled receptor signalling pathway” (P-value = 0.000508), “renal absorption” (P-value = 0.00051), “renal water transport” (P-value = 0.00051), “peripheral nervous system neuron axonogenesis” (P-value = 0.000593), “positive regulation of transcription from RNA polymerase II promoter in response to stress” (P-value = 0.000791), “sensory perception of light stimulus” (P-value = 0.000853), “sensory perception of sound” (P-value = 0.000932), and “negative regulation of neutrophil differentiation” (P-value = 0.000989).
The ‘sensory perception of light stimulus’ contained the following additional genes not recognized in the ‘retina development’ function, but which play a related physiological role: OPN4, NR2E3, CNGB1, RDH8, ROM1, CLN6, WFS1, KIFC3, RGR, RP1L1, RDH5, CABP4, AIPL1, RAX2, NRL and USH1G. The sensory perception of sound comprised the following genes (TPRN, TMIE, SLC52A3, OTOF, WFS1, SOD1, TBX18, MAP1A, OTOS, GPX1 and USH1G). Overall, we considered the collective presence of 4 functions in M. davidii (retina development, peripheral axonogenesis, light perception and sound perception) clearly associated with sensory rewiring to be noteworthy.
There was no formal vision related enrichment detected for P. alecto (Figure 2B) or indeed of any other sensory rewiring such as sound perception. The top ‘Biological Process’ enrichments in descending order were “cellular developmental process” (P-value = 0.0000178), “regulation of intrinsic apoptotic signalling pathway” (P-value = 0.000559), “G-protein coupled receptor signalling pathway” (P-value = 0.000634), “developmental process” (P-value = 0.000192), “regulation of interferon-gamma biosynthetic process” (P-value = 0.000267), “positive regulation of MAPK cascade” (P-value = 0.000354), “cell differentiation” (P-value = 0.000381), “cell surface receptor signalling pathway” (P-value = 0.00056),“regulation of MAPK cascade” (P-value = 0.000652), “regulation of interferon-gamma production” (P-value = 0.000703) and “blood vessel development” (P-value = 0.000813). In both species, there was some indication of enrichment relating to immune function.
When we compared these orthologs of the two genomes directly on a between-genome basis (Figure 3), IKZF1 (also identified in the “retina development of the camera eye”) displayed a prominent outlier value in M. davidii. LAMC3, RHO, and SOX9 were genes annotated as vision related that were absent in the P. alecto annotation and so orthologs could not be directly compared. A number of other orthologs were found to possess more bias in Myotis than in Pteropus, some of which were manually explored for their possible connection to the visual system. Although not identified as relating to hearing by GOrilla enrichment, the outlier position of MUC19 is of interest, given (1) the general role of mucin family members in signal transduction during hearing (Kerschner 2007) and (2) that MUC19 specifically encodes a major gel-forming mucin expressed in the middle ear (Kerschner et al. 2009). However, the mucins are known to bind pathogens, so an immune function or some other role cannot be discounted.
The overall distribution of these data was not symmetrical. There were more relatively biased orthologs in M. davidii than in P. alecto. This may reflect the fact that the Myotis bats are a much more recent lineage that, according to some sources, diverged approximately 15 mya from other bat species. Alternatively, the skewed data may reflect the fact that M. davidii possesses more derived phenotypic characteristics than P. alecto, resulting from the evolution of enhanced echolocation in this species. The genome-wide GC content is similar in the two bat species, with P. alecto (39.7%) slightly more biased than M. davidii (42.7%). This finding implies genome-wide GC content does not account for the slight skew in the between species distribution.
It is clear from scrutinizing the plots that the CUB statistic has the appealing feature of being independent of gene length because the mass of the data in Figure 1 follows a horizontal line in both species. It is also immediately apparent that, sitting close to the horizontal line, the vast majority of genes in both bat species do not exhibit extreme CUB. Nevertheless, in both cases there are a small subset of genes, many of which are short and some of which are long, that possess properties of relatively extreme CUB. Moreover, this subset enriches for a particular biological function, even though the physical location, gene length, and other molecular details vary. This observation can be generalized. We detected the exact same broad patterns previously identified in six other eukaryotic genomes: Arabidopsis, S. cerevisiae, C. elegans, chicken, chimpanzee, and human (Hudson et al. 2011) i.e., whereas most genes are unbiased, a small subset of variable length are biased and these enrich for particular functions that appear relevant to that lineage.
We calculated the GC content up and downstream of the differentially biased IKZF1, GPSM1, and SPRR1 (cornifin) and found them to be either almost identical, or more out of equilibrium in the Pteropodid genome. Table 1 summarizes these findings. Therefore, regional background GC content is unable to account for the greater CUB in these Myotid coding sequences. Gene length changes between species is also unable to account for the observation of differential CUB, as although it varies, the variation is not systematic. M. davidii IKZF1 is longer than in P. alecto, GPSM1 is shorter, and SPRR1 is almost identical, whereas CUB is substantially more extreme in M. davidii in all three cases.
Table 1. The genomic properties of three vision-related genes showing more CUB in M. davidii than P. alecto.
| Gene | GC% M. davidii | GC% P. alecto | Gene length M. davidii | Gene length P. alecto |
|---|---|---|---|---|
| IKZF1 | 49.6 | 53.2 / 51.5 | 1908 | 228/816 |
| GPSM1 | 49.1 | 65.4 | 705 | 2118 |
| SPRR1 | 44.0 | 47.1 | 339 | 342 |
Background GC% and gene length cannot account for the observation. CUB, codon usage bias.
Independent validation of divergent CUB:
The ENC measure of genome-wide CUB reinforced the change in the visual system of M. davidii and partially reinforced the change in its auditory system compared with P. alecto. However, the sensory enrichments observed for M. davidii were no longer ranked among the top ten on a within-genome basis.
The specific functional enrichments “retina development in camera-type eye” (based on CLN8, BAX, PRPH2, LAMC3, IKZF1, SOX9, NPHP4, PFDN5, PVRL1, SOX8, RD3) and “sensory perception of sound” (based on ASIC2, TPRN, MYO15A, WFS1, SOBP, ESPN, GPX1, TIMM13, GRM7, TMIE, SLC52A3, OTOF, SOD1, MAP1A, OTOS, SIX1, USH1G, MYO7A) were both significant in M. davidii (P = 0.000294 and P = 0.00066, respectively) but not P. alecto, broadly reinforcing the patterns identified by the original analysis performed using the entropy-based statistic. Interestingly, the Myotid bat also had the functional enrichment (P = 0.000232) “sensory perception of mechanical stimulus” (based on ASIC2, TPRN, MYO15A, TSHZ3, WFS1, SOBP, GPX1, TIMM13, GRM7, TMIE, SLC52A3, OTOF, SOD1, MAP1A, OTOS, SIX1, USH1G, MYO7A). The overlap in content with “sensory perception of sound” given genes such as TMIE (trans-membrane of inner ear), and components of the otolith (OTOF and OTOS) suggests this function is also essentially related to inner ear development.
Vision enrichments for P. alecto are present but to a lesser extent and do not convincingly highlight sensory rewiring. Although P. alecto does have vision-related enrichments given the presence of “closure of optic fissure” (P = 0.00026) “positive regulation of lens epithelial cell proliferation” (P = 0.00026), and “regulation of lens epithelial cell proliferation” (P = 0.00026), these were all based on the ranked position of a single gene, SOX11. Finally, “Inner ear development” (based on PDGFA, CCM2, CYTL1, HPCA, LHX3, GATA3, SHH, CEBPA, SOX2, and SIX1) was enriched in Pteropus (P = 0.000816). The genes comprising the hearing enrichment in P. alecto are different to those highlighted in M. davidii.
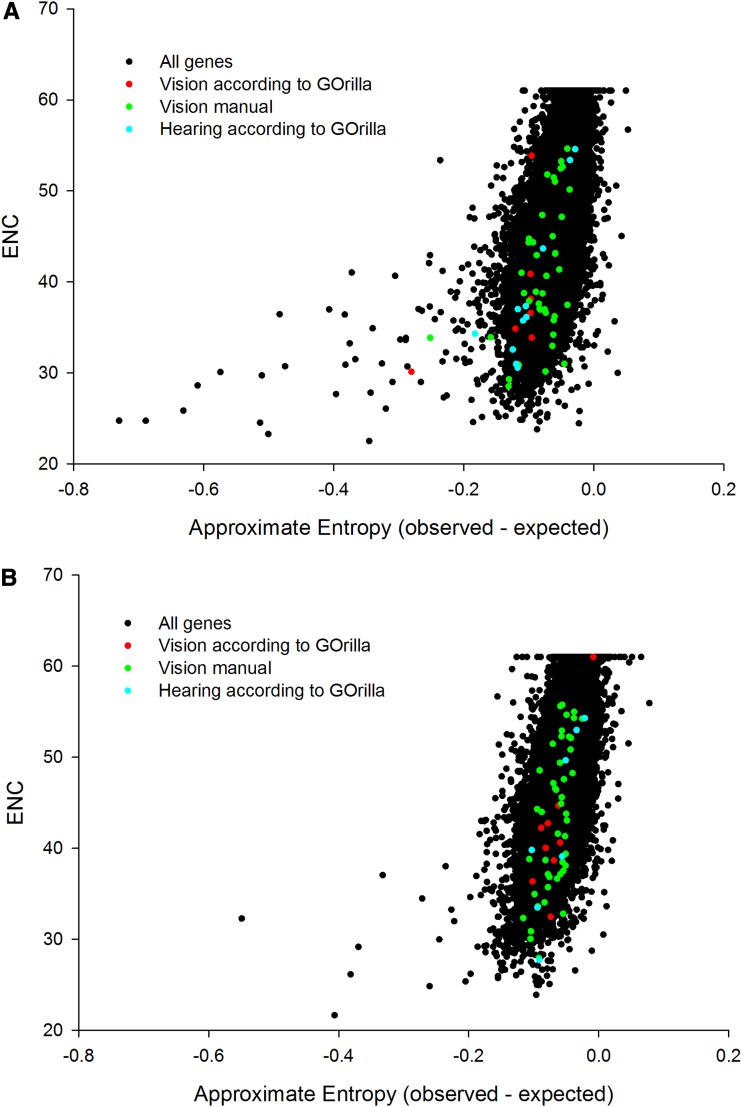
The genome-wide relationship between the two different CUB statistics (entropy vs. ENC) was plotted (Figure 4). We discovered a positive relationship (correlations of 0.619 and 0.672 in M. davidii and P. alecto, respectively). The entropy CUB statistic appears to discriminate the highly biased genes more sensitively, as evidenced by the skew at the bottom left of both species. This enhanced discrimination of entropy as one assesses CUB in the more biased genes appears a possible driver of any observed discrepancies in functional enrichment.
Figure 4.
The relationship between ENC and entropy based CUB estimation for (A) M. davidii and (B) P. alecto.
CUB outlier genes quality checking:
M. davidii LAMC3, CLN8, and SOX9 contained apparent gaps in the cds after translation BLAST and comparison with other bats and mammals. IKZF1, WFS1, and SOX8 were in the expected reading frame but not contiguous. Collectively, these genes possess low-quality genomic sequence (string of Ns) in predicted exonic regions that might explain any missing cds sequence compared with other mammals. In contrast, alignment of M. lucifugus sequence for IKZF1, LAMC3, SOX9, and SOX8 to other mammals is more conserved. This finding implies these genes may contain genomic assembly errors peculiar to M. davidii rather than being indicative of true evolutionary change. SOD1 appears to be partial sequence only.
CUB enrichment across two additional bat species: M. lucifugus and P. vampyrus:
In an effort to find independent comparative evidence for CUB in genes encoding proteins related to the sensory apparatus of echolocating bats, we computed the entropy based CUB in an additional echolocator (M. lucifugus) and a nonecholocator (P. vampyrus) species (File S2 and Figure 5). Sensory perception was not enriched in either species according to GOrilla analysis. The top three functional enrichments for M. lucifugus were G-protein−coupled receptor signaling pathway (P = 0.000000137), G-protein−coupled receptor signaling pathway coupled to cyclic nucleotide messenger (P = 0.00000667), and negative regulation of blood pressure (P = 0.0000295). For P. vampyrus they were G-protein−coupled receptor signaling pathway (P = 0.0000000196), adenylate cyclize-activating G-protein−coupled receptor signaling pathway (P = 0.0000296), and feeding behavior (P = 0.0000315).
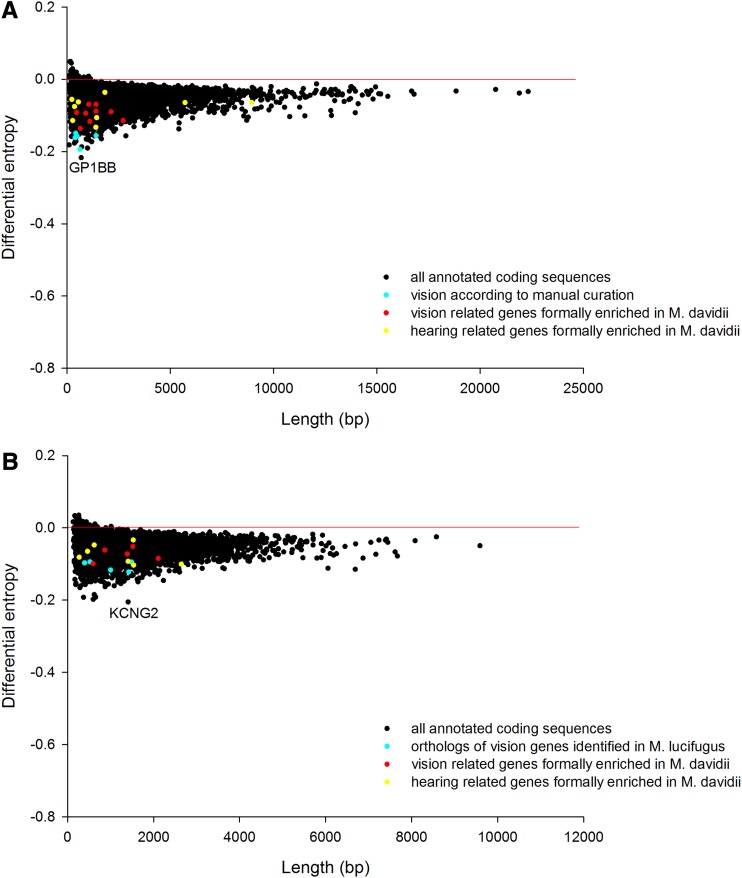
Figure 5.
Genes showing extreme codon bias (negative differential entropy) on a within-genome basis in (A) M. lucifugus and (B) P. vampyrus, with those encoding vision and hearing related proteins highlighted.
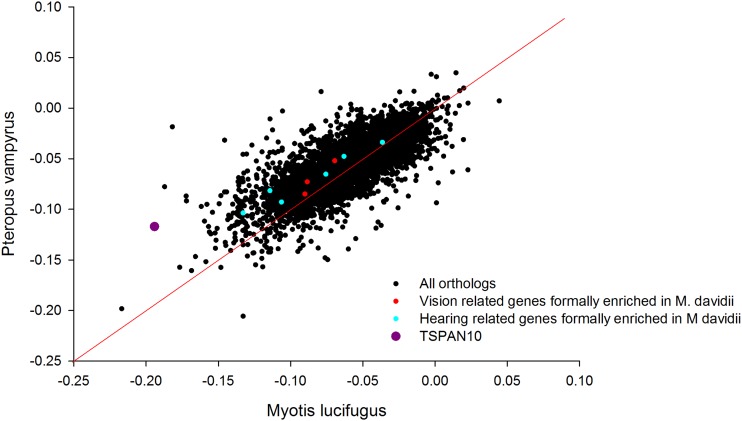
Next, we identified the orthologs of those vision and hearing sensory genes previously enriched in M. davidii. In the three vision (SOX8, HPS1, IKZF1) and six hearing (TMIE, SLC52A3, TBX18, OTOS, GPX1 and USH1G) instances in which both M. lucifugus and P. vampyrus orthologs could be identified, they were always more biased in M. lucifugus than P. vampyrus (Figure 6).
Figure 6.
Orthologs showing differential codon bias (distance from diagonal) between M. lucifugus and P. vampyrus on a between-genome basis. Genes encoding vision and hearing elated proteins are highlighted. Of the sensory genes, TSPAN10, which has a highly restricted expression pattern in retina, is particularly prominent. All the sensory genes prioritized by CUB in M. davidii are more biased in M. lucifugus than P. alecto.
Finally, we manually explored the M. lucifugus and P. vampyrus CUB outliers by using the GO information in the Gene function of the National Center for Biotechnology Information database and searching the literature in PubMed. In M. lucifugus on a within-genome basis the second (TSPAN10, alias OCSP) (Wistow et al. 2002), 15th (HCRT) (Liu et al. 2011), 20th (FIZ1) (Mali et al. 2008), 25th (RTN4R) (Wang et al. 2008), and 34th (TRAPPC6A) (Gwynn et al. 2006) were all vision-related (Figure 5A). TSPAN10 has a very strong tissue-restricted expression in retina based on the BioGPS normalized tissue expression library (Wu et al. 2009). In fact, retina is the only tissue of the 84 assayed in which TSPAN10 has an expression level greater than threefold of the mean (File S3). FIZ1 is a transcriptional co-factor that interacts with NRL and CRX to drive expression of rod photoreceptor-specific genes (Mali et al. 2008). Intriguingly, one of these partners (CRX) was actually prioritized by CUB in M. davidii. TRAPPC6A contributes to melanosome formation, and when mutated in mouse yields a loss of pigmentation phenotype in the retinal epithelial layer (Gwynn et al. 2006).
Although there was no formal enrichment for sensory perception in M. lucifugus, the CUB data obtained for the two myotid bats is broadly comparable at a pathway level (vision and hearing). Although some details differ, this is highly suggestive of a role for CUB in echolocating bat sensory evolution.
Discussion
In this study we performed a comparative genomic assessment of Myotid and Pteropodid bats by using multiple approaches, and the results provide information on the genetic origins of sensory rewiring in an echolocating bat. We began by identifying pseudogenes using the naked mole rat as a comparative species with poor vision. Pseudogenes are relatives of genes that have subsequently lost their protein coding function, usually by disabling mutations such as premature stop codons and frameshifts. The observed correspondence between the naked mole rat and the echolocating bat (CYP1A2, RBP3, GUCY2F, CRYBB1, and GRK7) points to the general importance of these particular genes in controlling mammalian vision and also to pseudogenes in particular as a genetic signature after loss of physiological function (Newman and Robinson 2005; Meredith et al. 2013).
The next analysis was a hypothesis-free screen based on the genome-wide extent of CUB. In simple organisms, CUB is known to alter translation efficiency and subsequent protein abundance, but the potential role in governing translation efficiency during mammalian evolution is less clear (Duret 2002). Supporting the concept it is known that protein expressed from a foreign gene in human cells can be increased by two orders of magnitude if the sequence is engineered to contain the most commonly used human codons (Gustafsson et al. 2004). However, there is an apparent lack of correlation between codon bias and protein abundance, and it is possible for codon-usage studies to be systematically influenced by amino acid content, gene length, and other potential artifacts (Chamary et al. 2006).
Despite these variable findings, all these studies have been impacted by an inability to exhaustively interrogate an entire and well-annotated genome (Behura and Severson 2013). For this reason, recent work that focuses on complete genome screens as well as other factors such as accounting for tissue and developmental stage, along with this study, may provide the first complete evidence for correlation between CUB and protein abundance in humans and other mammals (Waldman et al. 2010). A recent study concluded that although variation in mutational bias is the dominant force influencing CUB, translational selection acts as a weak additional factor and that this is widely true across the vertebrates (Doherty and McInerney 2013).
On a within-genome basis, we were struck with the finding that the top functional enrichment for the echolocating M. davidii was retina development, a process not enriched at all on a within-genome basis in P. alecto. The additional M. davidii enrichments for ‘axonogenesis’, ‘light perception,’ and ‘sound perception’ that were absent in P. alecto reinforced evidence for sensory rewiring in the echolocating species. Equivalent analyses run on the additional echolocator M. lucifugus and the additional nonecholocator P. vampyrus did not receive formal enrichment scores for vision and hearing. However, manual exploration of the outliers found intriguing support for vision, particularly through TSPAN10, which has a very prominent tissue specific expression in retina, and the transcriptional co-activator FIZ1, which interacts with CRX to drive the expression of rod photoreceptor-specific genes.
To complement our CUB within-genome analysis (Figure 2), we also compared the orthologs of M. davidii and P. alecto directly (Figure 3). Orthologs identified as substantially more biased in M. davidii (but not previously identified by the within-genome GOrilla analysis) were explored manually for possible connections to the visual and hearing systems through targeted PubMed literature searches. Although these genes are bound together by their shared role in the biological processes of vision and hearing, they are somewhat diverse in terms of other more detailed molecular and functional properties. In regard to the visual proteins, some are transcription factors (e.g., CRX), whereas others are visual pigment proteins (e.g., RHO). Based on the TIGER database (Liu et al. 2008), which provides normalized tissue mRNA comparisons in humans, many of the vision genes are abundantly expressed in eye, and some are not. Furthermore, some are expressed highly across a wide range of tissues whereas others have an extremely restricted expression profile.
Clearly, this range of tissue-level expression profiles renders any simple correlation of CUB to protein abundance unlikely. However, this does not mean one should discount CUB as a driver of eukaryotic evolution, as previously argued (Waldman et al. 2010). Waldman et al. (2010) found significantly greater correlations observed in adult tissues than the fetus, pointing to the importance of developmental stage in these kind of assessments (Waldman et al. 2010). In that work, related sets of functional gene groups were found to be translated efficiently in each tissue, pointing to the importance of viewing groups of genes as components in a broader biological process or pathway (Waldman et al. 2010). In the future, comparison of biased genes to actual expression level in components of the bats sensory system would be of interest, but unfortunately these data are not presently available.
To allow the CUB outliers to be placed in context, we calculated the genome-wide preferred codons for M. davidii and P. alecto based on all the available coding sequences available from the genome sequencing efforts (Table 2 and Table 3). These data can be exploited to make predictions of bias toward or against preferred codons. In the context of explaining the low light−adapted vision and small eyes of M. davidii, we would predict enhanced translation efficiency (bias toward preferred codons) in negative regulators of eye size and visual pigments adapted for low light, and the converse (nonpreferred codons) for positive regulators of eye size and pigments adapted for day vision. However, the developmental pathways controlling retinogenesis are incomplete, and the genetic changes in the highlighted genes need to be assessed on a case-by-case basis and await future functional validation. Of the vision related genes highly biased in M. davidii IKZF1 encodes a transcription factor called Ikaros family zinc finger 1, an ortholog of which confers competence to mouse retinal progenitor cells (Elliott et al. 2008). Manual exploration identified additional highly differentially biased genes encoding proteins expressed in eye components—GPSM1 (Kerov et al. 2005) and SPRR1 (Tong et al. 2006). Enrichments for light and sound perception include numerous additional sensory genes not included in the specific retinogenic enrichment. The observation of extreme CUB in CRX in M. davidii and its binding co-factor FIZ1 in M. lucifugus supports this molecular pathway as a likely hotspot in vision-related adaptations that have evolved in nocturnal echolocating bats.
Table 2. Patterns of genome-wide codon usage in Myotis davidii.
| UUU | 15.7 (141141) | UCU | 14.4 (128939) | UAU | 10.7 (96582) | UGU | 9.7 (87355) |
| UUC | 20.5 (183882) | UCC | 19.1 (171243) | UAC | 15.5 (139000) | UGC | 12.6 (112906) |
| UUA | 7.2 (64716) | UCA | 11.3 (101170) | UAA | 0.6 (5059) | UGA | 1.2 (10439) |
| UUG | 12.5 (112218) | UCG | 5.0 (45139) | UAG | 0.5 (4775) | UGG | 12.4 (111457) |
| CUU | 12.1 (108263) | CCU | 16.9 (151920) | CAU | 10.0 (90245) | CGU | 4.2 (37777) |
| CUC | 20.4 (183451) | CCC | 21.6 (194040) | CAC | 16.6 (148955) | CGC | 9.9 (89098) |
| CUA | 6.4 (57262) | CCA | 15.9 (143248) | CAA | 11.8 (106136) | CGA | 6.0 (54257) |
| CUG | 41.3 (371130) | CCG | 7.6 (68087) | CAG | 36.1 (323855) | CGG | 12.0 (108187) |
| AUU | 14.6 (131554) | ACU | 12.0 (108140) | AAU | 15.5 (139605) | AGU | 12.1 (108595) |
| AUC | 21.5 (193277) | ACC | 19.8 (177653) | AAC | 20.0 (179409) | AGC | 20.5 (183722) |
| AUA | 6.9 (62193) | ACA | 13.9 (125180) | AAA | 23.6 (211742) | AGA | 11.5 (103418) |
| AUG | 22.1 (198262) | ACG | 7.2 (65088) | AAG | 33.3 (298847) | AGG | 12.9 (115789) |
| GUU | 10.3 (92630) | GCU | 17.9 (161068) | GAU | 20.8 (186580) | GGU | 9.9 (89712) |
| GUC | 15.4 (137894) | GCC | 29.3 (263071) | GAC | 27.4 (246001) | GGC | 22.2 (199804) |
| GUA | 6.4 (57396) | GCA | 15.2 (136268) | GAA | 28.7 (25792) | GGA | 15.5 (138999) |
| GUG | 29.2 (261930) | GCG | 7.6 (68234) | GAG | 41.6 (374083) | GGG | 17.3 (155758) |
These data help set the expectation for whether a given M. davidii codon is preferred or nonpreferred. Observed bias in any particular gene can therefore be interpreted as likely to increase or decrease the cost and efficiency of translation
Table 3. Patterns of genome-wide codon usage in Pteropus alecto.
| UUU | 16.3 (156476) | UCU | 15.2 (146151) | UAU | 11.3 (108424) | UGU | 10.0 (96432) |
| UUC | 20.0 (192106) | UCC | 17.7 (170229) | UAC | 14.8 (141890) | UGC | 12.2 (116902) |
| UUA | 7.7 (74291) | UCA | 12.0 (114942) | UAA | 0.5 (5244) | UGA | 1.1 (10325) |
| UUG | 13.1 (125571) | UCG | 5.2 (49969) | UAG | 0.5 (4808) | UGG | 12.3 (117792) |
| CUU | 13.1 (125218) | CCU | 17.6 (168660) | CAU | 10.4 (100155) | CGU | 4.9 (46662) |
| CUC | 19.8 (190319) | CCC | 20.7 (198601) | CAC | 15.7 (150619) | CGC | 10.6 (101489) |
| CUA | 7.3 (69639) | CCA | 16.7 (160345) | CAA | 12.2 (116897) | CGA | 6.4 (61551) |
| CUG | 39.5 (379104) | CCG | 7.7 (74204) | CAG | 35.0 (336173) | CGG | 12.0 (114783) |
| AUU | 15.1 (144545) | ACU | 12.9 (124104) | AAU | 16.3 (156140) | AGU | 12.5 (119863) |
| AUC | 20.4 (196052) | ACC | 18.5 (177795) | AAC | 18.9 (181664) | AGC | 20.1 (192594) |
| AUA | 7.3 (70411) | ACA | 14.6 (139725) | AAA | 24.3 (233289) | AGA | 11.8 (112920) |
| AUG | 21.5 (206522) | ACG | 6.9 (65967) | AAG | 31.9 (305590) | AGG | 12.1 (116001) |
| GUU | 10.6 (101643) | GCU | 18.5 (177510) | GAU | 21.4 (205693) | GGU | 10.5 (100839) |
| GUC | 15.2 (146033) | GCC | 28.8 (275932) | GAC | 26.3 (253066) | GGC | 22.5 (215905) |
| GUA | 7.2 (69462) | GCA | 15.7 (150363) | GAA | 29.6 (284341) | GGA | 15.8 (151181) |
| GUG | 27.9 (267333) | GCG | 8.0 (76818) | GAG | 40.3 (387415) | GGG | 16.6 (159580) |
These data help set the expectation for whether a given P. alecto codon is preferred or nonpreferred. Observed bias in any particular gene can therefore be interpreted as likely to increase or decrease the cost and efficiency of translation.
Overall, our findings across two echolocating (M. davidii and M. lucifugus) and two nonecholocating (P. alecto and P. vampyrus) bats strongly support a recent observation made across echolocating mammals (bats and bottlenose dolphins) using a different comparative genomics approach. In this independent study, it was found that patterns of convergent protein evolution (based on nonsynonymous vs. synonymous amino acid substitutions) identify pathways contributing to hearing and vision as among the strongest signals (Parker et al. 2013). The exact overlap between our differential CUB analysis vs. the protein evolution study of (Parker et al. 2013) included the genes RHO and OTOF, which are involved in light and sound perception, respectively. The fact that not all genes exist in both analyses suggests that multiple regulatory mechanisms are likely to contribute to change of function.
NEFH (alias YHU2; encoding the neurofilament heavy polypeptide) possessed extreme CUB in both M. davidii and P. alecto, implying it was either present in the ancestor or has subsequently converged due to a fundamental role in both bat lineages. A broader comparative analysis is needed to shed light on this gene, but unfortunately an annotated ortholog has not been identified in either the M. lucifugus or P. vampyrus sequences. One could sequence NEFH in a range of bats and one or more Laurasiatherian outgroups. NEFH is most highly expressed in the peripheral nervous system (Liu et al. 2008), so a generic sensory explanation seems plausible i.e., one not specific to echolocation. Finally, a number of unannotated highly biased genes in both M. davidii and P. alecto were detected. These may represent targets for future functional studies.
Overall, our findings provide insight into two genetic mechanisms contributing to evolution’s toolkit for adapting phenotypes. The deterioration of a number of core visual genes in M. davidii is consistent with genomic evolution in the naked mole rat (Kim et al. 2011) and the poor day vision and small eyes of this species. Pseudogene formation is clearly a loss-of-function decay process. Disabling mutations are allowed to persist because of relaxation of selection pressure on that particular protein. Because mutation is random, if unchecked it will cause DNA sequence to tend toward (random) equilibrium. This finding contrasts with extreme CUB, which enforces structure and regularity on DNA sequence. This enforced regularity is strongly indicative of an applied evolutionary force, and tends to be associated with gain-of-function.
The CUB evidence we have gathered for manipulation of translation efficiency at the biological pathway level has not previously been reported in the context of bat sensory ecology and warrants further investigation. The CUB data collectively imply a suite of adaptations to both the visual system (perhaps positively impacting on adaptation to dim light) and the enhancement of hearing (which presumably relates to echolocation). Overall, the general prioritization of sensory rewiring in the echolocator is highly resonant with the recent findings of Parker et al. (2013), despite a different line of genetic evidence and indeed different choice of echolocating species.
Supplementary Material
Acknowledgments
We wish to acknowledge fruitful discussions with Dr. James Kijas and Dr. Chris Elvin. This research was facilitated by the CSIRO Animal, Food, and Health Sciences post-genomics workshop, held at Coffs Harbor during April 2013 and was supported by CSIRO. Q.G. was supported by in part by the Shanghai Postdoctoral Sustention fund (no. 12R21410200). M.L.B. was supported by an Australian Research Council Future Fellowship (FT110100234).
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.011262/-/DC1
Communicating editor: D. J. de Koning
Literature Cited
- Altringham J. D., Fenton M. B., 2003. Sensory ecology and communication in the Chiroptera, pp. 90–127 in Bat Ecology, edited by Kunz T. H., Fenton M. B.. University of Chicago Press, Chicago [Google Scholar]
- Behura S. K., Severson D. W., 2013. Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes. Biol. Rev. Camb. Philos. Soc. 88: 49–61 [DOI] [PubMed] [Google Scholar]
- Bhatnagar K. P., 1975. Olfactory in Artibeus jamaicensis and Myotis lucifugus in the context of vision and echolocation. Experientia 31: 856. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., 2004. GeneWise and Genomewise. Genome Res. 14: 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary J. V., Parmley J. L., Hurst L. D., 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7: 98–108 [DOI] [PubMed] [Google Scholar]
- Doherty A., McInerney J. O., 2013. Translational selection frequently overcomes genetic drift in shaping synonymous codon usage patterns in vertebrates. Mol. Biol. Evol. 30: 2263–2267 [DOI] [PubMed] [Google Scholar]
- Duret L., 2002. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 12: 640–649 [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J., Jolicoeur C., Ramamurthy V., Cayouette M., 2008. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 60: 26–39 [DOI] [PubMed] [Google Scholar]
- Fuzessery Z. M., Buttenhoff P., Andrews B., Kennedy J. M., 1993. Passive sound localization of prey by the pallid bat (Antrozous p. pallidus). J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 171: 767–777 [DOI] [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S., Minshull J., 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22: 346–353 [DOI] [PubMed] [Google Scholar]
- Gwynn B., Smith R. S., Rowe L. B., Taylor B. A., Peters L. L., 2006. A mouse TRAPP-related protein is involved in pigmentation. Genomics 88: 196–203 [DOI] [PubMed] [Google Scholar]
- Hudson N. J., Gu Q., Nagaraj S. H., Ding Y. S., Dalrymple B. P., et al. , 2011. Eukaryotic evolutionary transitions are associated with extreme codon bias in functionally-related proteins. PLoS ONE 6: e25457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G., Teeling E. C., Rossiter S. J., 2013. From the ultrasonic to the infrared: molecular evolution and the sensory biology of bats. Front Physiol 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalko E. K. V., Herre E. A., Handley C. O., Jr., 1996. Relation of fig fruit characteristics to fruit-eating bats in the New and Old World tropics. J. Biogeogr. 23: 565–576 [Google Scholar]
- Kerov V. S., Natochin M., Artemyev N. O., 2005. Interaction of transducin-alpha with LGN, a G-protein modulator expressed in photoreceptor cells. Mol. Cell. Neurosci. 28: 485–495 [DOI] [PubMed] [Google Scholar]
- Kerschner J. E., 2007. Mucin gene expression in human middle ear epithelium. Laryngoscope 117: 1666–1676 [DOI] [PubMed] [Google Scholar]
- Kerschner J. E., Khampang P., Erbe C. B., Kolker A., Cioffi J. A., 2009. Mucin gene 19 (MUC19) expression and response to inflammatory cytokines in middle ear epithelium. Glycoconj. J. 26: 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. B., Fang X., Fushan A. A., Huang Z., Lobanov A. V., et al. , 2011. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Xu G. Z., Wang L., Jiang S. X., Yang X. L., et al. , 2011. Gene expression and protein distribution of orexins and orexin receptors in rat retina. Neuroscience 189: 146–155 [DOI] [PubMed] [Google Scholar]
- Liu X., Yu X., Zack D. J., Zhu H., Qian J., 2008. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali R. S., Peng G. H., Zhang X., Dang L., Chen S., et al. , 2008. FIZ1 is part of the regulatory protein complex on active photoreceptor-specific gene promoters in vivo. BMC Mol. Biol. 9: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith R. W., Gatesy J., Emerling C. A., York V. M., Springer M. S., 2013. Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9: e1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman L. A., Robinson P. R., 2005. Cone visual pigments of aquatic mammals. Vis. Neurosci. 22: 873–879 [DOI] [PubMed] [Google Scholar]
- Parker J., Tsagkogeorga G., Cotton J. A., Liu Y., Provero P., et al. , 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502: 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettigrew J. D., 1986. Flying primates? Megabats have the advanced pathway from eye to midbrain. Science 231: 1304–1306 [DOI] [PubMed] [Google Scholar]
- Seim I., Fang X., Xiong Z., Lobanov A. V., Huang Z., et al. , 2013. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat. Commun. 4: 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E. C., 2009. Hear, hear: the convergent evolution of echolocation in bats? Trends Ecol. Evol. 24: 351–354 [DOI] [PubMed] [Google Scholar]
- Tong L., Corrales R. M., Chen Z., Villarreal A. L., De Paiva C. S., et al. , 2006. Expression and regulation of cornified envelope proteins in human corneal epithelium. Invest. Ophthalmol. Vis. Sci. 47: 1938–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman Y. Y., Tuller T., Shlomi T., Sharan R., Ruppin E., 2010. Translation efficiency in humans: tissue specificity, global optimization and differences between developmental stages. Nucleic Acids Res. 38: 2964–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chan C. K., Taylor J. S., Chan S. O., 2008. Localization of Nogo and its receptor in the optic pathway of mouse embryos. J. Neurosci. Res. 86: 1721–1733 [DOI] [PubMed] [Google Scholar]
- Weibel E. R., Taylor C. R., Hoppeler H., 1991. The concept of symmorphosis: a testable hypothesis of structure-function relationship. Proc. Natl. Acad. Sci. USA 88: 10357–10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistow G., Bernstein S. L., Wyatt M. K., Fariss R. N., Behal A., et al. , 2002. Expressed sequence tag analysis of human RPE/choroid for the NEIBank Project: over 6000 non-redundant transcripts, novel genes and splice variants. Mol. Vis. 8: 205–220 [PubMed] [Google Scholar]
- Wright F., 1990. The ’effective number of codons’ used in a gene. Gene 87: 23–29 [DOI] [PubMed] [Google Scholar]
- Wu C., Orozco C., Boyer J., Leglise M., Goodale J., et al. , 2009. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K. A., et al. , 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339: 456–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.