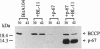Abstract
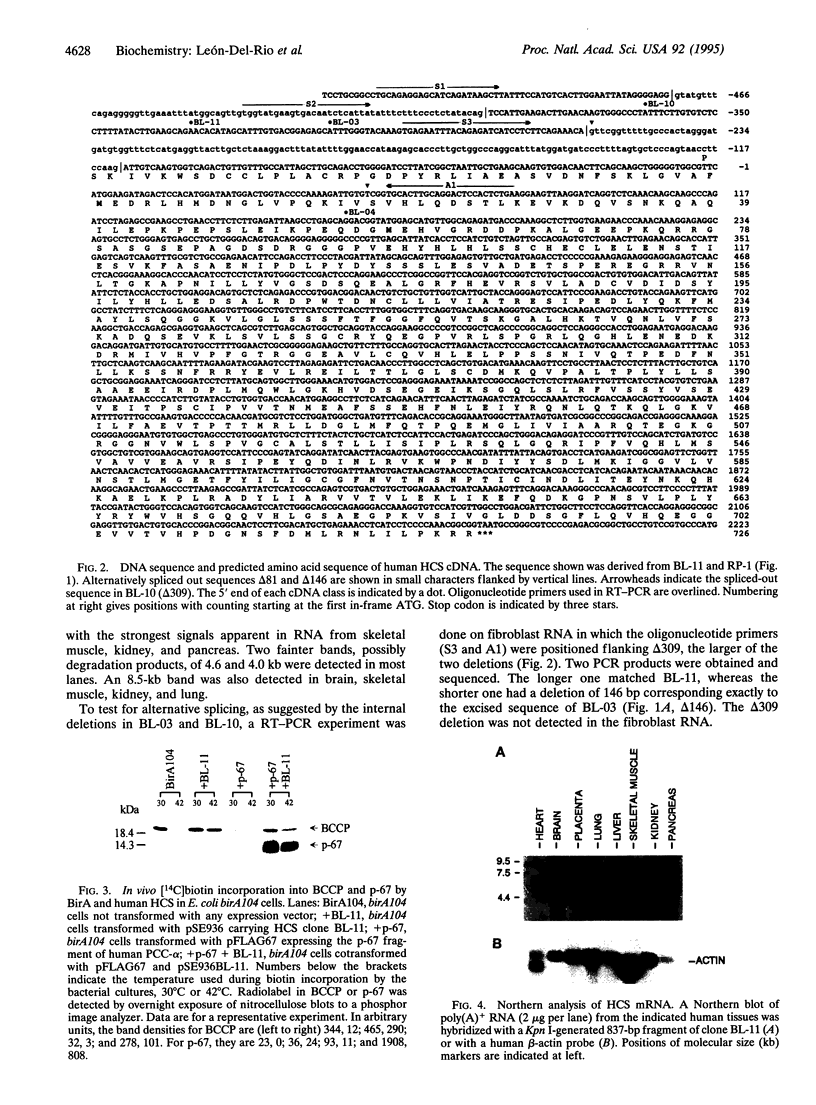
Holocarboxylase synthetase (HCS) catalyzes the biotinylation of the four biotin-dependent carboxylases in human cells. Patients with HCS deficiency lack activity of all four carboxylases, indicating that a single HCS is targeted to the mitochondria and cytoplasm. We isolated 21 human HCS cDNA clones, in four size classes of 2.0-4.0 kb, by complementation of an Escherichia coli birA mutant defective in biotin ligase. Expression of the cDNA clones promoted biotinylation of the bacterial biotinyl carboxyl carrier protein as well as a carboxyl-terminal fragment of the alpha subunit of human propionyl-CoA carboxylase expressed from a plasmid. The open reading frame encodes a predicted protein of 726 aa and M(r) 80,759. Northern blot analysis revealed the presence of a 5.8-kb major species and 4.0-, 4.5-, and 8.5-kb minor species of poly(A)+ RNA in human tissues. Human HCS shows specific regions of homology with the BirA protein of E. coli and the presumptive biotin ligase of Paracoccus denitrificans. Several forms of HCS mRNA are generated by alternative splicing, and as a result, two mRNA molecules bear different putative translation initiation sites. A sequence upstream of the first translation initiation site encodes a peptide structurally similar to mitochondrial presequences, but it lacks an in-frame ATG codon to direct its translation. We anticipate that alternative splicing most likely mediates the mitochondrial versus cytoplasmic expression, although the elements required for directing the enzyme to the mitochondria remain to be confirmed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker D. F., Campbell A. M. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J Mol Biol. 1981 Mar 15;146(4):469–492. doi: 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- Barker D. F., Campbell A. M. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J Mol Biol. 1981 Mar 15;146(4):451–467. doi: 10.1016/0022-2836(81)90042-5. [DOI] [PubMed] [Google Scholar]
- Barker D. F., Campbell A. M. Use of bio-lac fusion strains to study regulation of biotin biosynthesis in Escherichia coli. J Bacteriol. 1980 Aug;143(2):789–800. doi: 10.1128/jb.143.2.789-800.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buoncristiani M. R., Howard P. K., Otsuka A. J. DNA-binding and enzymatic domains of the bifunctional biotin operon repressor (BirA) of Escherichia coli. Gene. 1986;44(2-3):255–261. doi: 10.1016/0378-1119(86)90189-7. [DOI] [PubMed] [Google Scholar]
- Burri B. J., Sweetman L., Nyhan W. L. Mutant holocarboxylase synthetase: evidence for the enzyme defect in early infantile biotin-responsive multiple carboxylase deficiency. J Clin Invest. 1981 Dec;68(6):1491–1495. doi: 10.1172/JCI110402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. I., Cohen N. D. Regulation and intracellular localization of the biotin holocarboxylase synthetase of 3T3-L1 cells. Arch Biochem Biophys. 1983 Aug;225(1):237–247. doi: 10.1016/0003-9861(83)90026-7. [DOI] [PubMed] [Google Scholar]
- Chiba Y., Suzuki Y., Aoki Y., Ishida Y., Narisawa K. Purification and properties of bovine liver holocarboxylase synthetase. Arch Biochem Biophys. 1994 Aug 15;313(1):8–14. doi: 10.1006/abbi.1994.1351. [DOI] [PubMed] [Google Scholar]
- Cleary P. P., Campbell A., Chang R. Location of promoter and operator sites in the biotin gene cluster of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2219–2223. doi: 10.1073/pnas.69.8.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne-Smith C., Jang S. H., Shi Q., DeWille J., Sasaki G., Kolattukudy P. E. Cytoplasmic accumulation of a normally mitochondrial malonyl-CoA decarboxylase by the use of an alternate transcription start site. Arch Biochem Biophys. 1992 Nov 1;298(2):576–586. doi: 10.1016/0003-9861(92)90452-3. [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Jr The E. coli bio operon: transcriptional repression by an essential protein modification enzyme. Cell. 1989 Aug 11;58(3):427–429. doi: 10.1016/0092-8674(89)90421-2. [DOI] [PubMed] [Google Scholar]
- Eisenberg M. A., Prakash O., Hsiung S. C. Purification and properties of the biotin repressor. A bifunctional protein. J Biol Chem. 1982 Dec 25;257(24):15167–15173. [PubMed] [Google Scholar]
- Elledge S. J., Mulligan J. T., Ramer S. W., Spottswood M., Davis R. W. Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman G. L., Wolf B. Deficient acetyl CoA carboxylase activity in multiple carboxylase deficiency. Clin Chim Acta. 1981 Apr 9;111(2-3):147–151. doi: 10.1016/0009-8981(81)90181-9. [DOI] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990 Oct;4(1):33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- Ghneim H. K., Bartlett K. Mechanism of biotin-responsive combined carboxylase deficiency. Lancet. 1982 May 22;1(8282):1187–1188. doi: 10.1016/s0140-6736(82)92253-x. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Pfanner N., Nicholson D. W., Neupert W. Mitochondrial protein import. Biochim Biophys Acta. 1989 Jan 18;988(1):1–45. doi: 10.1016/0304-4157(89)90002-6. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Salmond G. P., Gill D. R., Downie J. A., Evans I. J., Holland I. B., Gray L., Buckel S. D., Bell A. W. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986 Oct 2;323(6087):448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- Howard P. K., Shaw J., Otsuka A. J. Nucleotide sequence of the birA gene encoding the biotin operon repressor and biotin holoenzyme synthetase functions of Escherichia coli. Gene. 1985;35(3):321–331. doi: 10.1016/0378-1119(85)90011-3. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski D. J., Mehl R., Yin H. L. Genomic organization and biosynthesis of secreted and cytoplasmic forms of gelsolin. J Cell Biol. 1988 Feb;106(2):375–384. doi: 10.1083/jcb.106.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Del-Rio A., Gravel R. A. Sequence requirements for the biotinylation of carboxyl-terminal fragments of human propionyl-CoA carboxylase alpha subunit expressed in Escherichia coli. J Biol Chem. 1994 Sep 16;269(37):22964–22968. [PubMed] [Google Scholar]
- Li S. J., Cronan J. E., Jr The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992 Jan 15;267(2):855–863. [PubMed] [Google Scholar]
- Purdue P. E., Lumb M. J., Danpure C. J. Molecular evolution of alanine/glyoxylate aminotransferase 1 intracellular targeting. Analysis of the marmoset and rabbit genes. Eur J Biochem. 1992 Jul 15;207(2):757–766. doi: 10.1111/j.1432-1033.1992.tb17106.x. [DOI] [PubMed] [Google Scholar]
- Samols D., Thornton C. G., Murtif V. L., Kumar G. K., Haase F. C., Wood H. G. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988 May 15;263(14):6461–6464. [PubMed] [Google Scholar]
- Saunders M. E., Sherwood W. G., Duthie M., Surh L., Gravel R. A. Evidence for a defect of holocarboxylase synthetase activity in cultured lymphoblasts from a patient with biotin-responsive multiple carboxylase deficiency. Am J Hum Genet. 1982 Jul;34(4):590–601. [PMC free article] [PubMed] [Google Scholar]
- Saunders M., Sweetman L., Robinson B., Roth K., Cohn R., Gravel R. A. Biotin-response organicaciduria. Multiple carboxylase defects and complementation studies with propionicacidemia in cultured fibroblasts. J Clin Invest. 1979 Dec;64(6):1695–1702. doi: 10.1172/JCI109632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusher L. B., Gillman E. C., Martin N. C., Hopper A. K. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9789–9793. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella M. C., Schauerte H., Straub K. L., Leptin M. Identification of secreted and cytosolic gelsolin in Drosophila. J Cell Biol. 1994 May;125(3):607–616. doi: 10.1083/jcb.125.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Yoshida T., Tuboi S. Evidence that rat liver mitochondrial and cytosolic fumarases are synthesized from one species of mRNA by alternative translational initiation at two in-phase AUG codons. Eur J Biochem. 1992 Jul 15;207(2):767–772. doi: 10.1111/j.1432-1033.1992.tb17107.x. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Aoki Y., Ishida Y., Chiba Y., Iwamatsu A., Kishino T., Niikawa N., Matsubara Y., Narisawa K. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat Genet. 1994 Oct;8(2):122–128. doi: 10.1038/ng1094-122. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. P., Shewchuk L. M., Brennan R. G., Otsuka A. J., Matthews B. W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H. G., Barden R. E. Biotin enzymes. Annu Rev Biochem. 1977;46:385–413. doi: 10.1146/annurev.bi.46.070177.002125. [DOI] [PubMed] [Google Scholar]
- Xu X., Matsuno-Yagi A., Yagi T. DNA sequencing of the seven remaining structural genes of the gene cluster encoding the energy-transducing NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochemistry. 1993 Jan 26;32(3):968–981. doi: 10.1021/bi00054a030. [DOI] [PubMed] [Google Scholar]