Abstract
Small noncoding RNAs play several roles in regulating gene expression. In the nucleus, small RNA-Argonaute complexes recruit epigenetic modifying activities to genomic sites. This pathway has been described in mammals primarily for the germline; however, its role in somatic cells is less characterized. Here, we describe in human somatic cells a potential link between the expression of small RNAs from the macrosatellite DXZ4 and Argonaute-dependent DNA methylation of this locus. DXZ4 was found to express a wide range of small RNAs potentially representing several classes of small RNAs. A subpopulation of these RNAs is bound by Argonaute. Moreover, we show AGO association with DXZ4 and that the Argonaute proteins AGO-1 and PIWIL4 may play a role in DNA methylation of DXZ4. We hypothesize that the RNAs are involved in Argonaute-dependent methylation of DXZ4 DNA.
Keywords: small noncoding RNA, somatic cells, DNA methylation, Argonaute, PIWI
At least half of the human genome is characterized as repetitive sequence that does not code for proteins. One of the two main families of DNA repeats are tandem repeats, including satellite DNA. DXZ4 is one of several macrosatellites identified in humans and consists of 50−100 copies of a 3-kb monomer (Giacalone et al. 1992). By virtue of its location on the X chromosome, DXZ4 is subject to X chromosome inactivation. However, contrary to what is expected, DXZ4 on the inactive X chromosome is highlighted by a euchromatic conformation consisting of dimethylated histone H3 lysine-4 (H3K4me2) and acetylated H3 lysine-9 (H3K9Ac) (Boggs et al. 2002; Chadwick 2008). In contrast, on the active X chromosome in both female and male cells, DXZ4 chromatin contains the heterochromatin-associated H3K9me3 modification (Chadwick 2008). Moreover on the active X, DXZ4 DNA is hypermethylated, also a characteristic of the repressed transcription state, and in contrast, hypomethylated on the inactive X (Giacalone et al. 1992; Chadwick 2008).
Studies in the mouse germline have demonstrated that Piwi-interacting RNAs (piRNAs) re-establish de novo DNA methylation in males (Carmell et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). This methylation is important for restricting transposon mobility. It has also been demonstrated that piRNA-mediated targeting induces allele-specific Rasgrf1 transcriptional silencing by de novo DNA methylation, thereby resulting in germline genomic imprinting of this locus (Watanabe et al. 2011). Recently, piRNA-like molecules have been identified in somatic tissues from multiple organisms (Ro et al. 2007; Ghildiyal et al. 2008; Li et al. 2009; Yan et al. 2011). A particularly convincing example is the induction of piRNAs in the central nervous system of the sea slug Aplysia by the neurotransmitter serotonin (Rajasethupathy et al. 2012). In neurons, a specific piRNA is induced by serotonin and inhibition of the Piwi protein abolished the serotonin-dependent methylation increase of a CpG island within the promoter of the CREB2 gene whose protein product is important for the persistence of memory. In human tissue culture cells, the PIWI protein PIWIL4 can induce histone H3 lysine 9 dimethylation (H3K9me2) at the p16Ink4a locus, resulting in down-regulation of the gene (Sugimoto et al. 2007). Also in tissue culture cells, microRNAs (miRNAs) have been found to interact with promoters to induce transcriptional gene silencing (Huang and Li 2012). Furthermore, endogenous small interfering RNAs (siRNAs) have been shown to be involved in H3K9me2 deposition at several genomic repeats to regulate genome stability in fruit fly somatic cells (Peng and Karpen 2007; Fagegaltier et al. 2009). Taken together, this evidence suggests that small RNA pathways can regulate gene expression by chromatin modification in somatic cells.
Because small RNAs are known to act on repetitive sequences (Peng and Karpen 2007; Fagegaltier et al. 2009), together with the fact that DXZ4 expresses ~85 nucleotide long RNAs (Chadwick 2008), we investigated whether small RNAs are expressed from DXZ4. In somatic cells, we detected the expression of a wide range of small RNAs from this locus with a subpopulation associating with Argonaute. Based on this finding, we further investigated whether the small RNA pathway plays a role in establishing epigenetic modifications at DXZ4 and found that Argonaute proteins are required for DNA methylation and that they bind DXZ4 chromatin. We speculate that the small RNAs are involved in the establishment of epigenetic modifications at this region.
Materials and Methods
Small RNA Northern hybridization
A total of 30 μg of total RNA was isolated using TriZol (Life Technologies). The RNA population <200 nucleotides was isolated from 5.5 × 106 cells (PureLink miRNA isolation kit; Life Technologies). Extracting the chromatin fraction from 5.5 × 106 cells was published elsewhere (Kugler et al. 1995). However, total RNA from the chromatin fraction was isolated using TriZol. RNA was separated on a denaturing 15% polyacrylamide gel, transferred to a HyBond-N membrane (GE Healthcare), ultraviolet crosslinked, and probed with 5′ 32P-phosphorylated oligonucleotides in ExpressHyb solution (Clontech) at 37°. Probes: DXZ4 1536-bp tgacgactcgtgtgtgccgtgg, DXZ4 2352-bp acacctatccccctggctcg, DXZ4 2942-bp ccccgggcccccttagccgatg. Probes for 3′ probing: DXZ4 cgcccccacgggaccgctctcgagg, cacacctatccccctggctcgctct, gcgagagcggtccgccgtgcccaag; miR-15a cacaaaccattatgtgctgcta.
Small RNA detection by quantitative reverse-transcription polymerase chain reaction (RT-PCR)
Custom TaqMan small RNA assays (Life Technologies) were performed according to the manufacture’s protocol using 500 ng of RNA <200 nucleotides. A U6 snoRNA probe (Life Technologies) was used as internal standard. Probed small RNAs included the following: DXZ4-2183as tcaccttggcttgggggacctcgagagcggtcccgt, DXZ4-2259as cgtcaacgcacctttaagggcgagagcggtccgccg, and DXZ4-2355as cctatccccctggctcgctctc.
RNA interference
We transiently transfected 8 × 104–3.5 × 105 HEK293T cells or 2 × 105 MRC-5 cells per 12-well dish with 5–20 nM siRNA the following day after seeding using Lipofectamine RNAiMax (Life Technologies). Cells were analyzed 48–72 hr posttransfection. Silencer Select Validated siRNAs (Life Technologies) were as follows: negative control no. 1 4390843; AGO-1 s25500, s25501; PIWIL4 s44573; Dicer s23756.
In vitro Drosha cleavage assay
In vitro Drosha processing was performed essentially as described (Zeng and Cullen 2006), except that nuclear extracts were prepared from HEK293T cells transfected with expression plasmids for Drosha and DGCR8 and used directly in the assay instead of using purified recombinant proteins.
Probing of 3′ RNA ends
NaIO4 treatment and β-elimination was performed as described (Vagin et al. 2006).
Chromatin immunoprecipitation (ChIP)
ChIP was performed essentially as described with minor adaptations (Kim and Rossi 2009). A total of 5 µg of anti-pan-AGO (Z. Mourelatos and EMD Millipore MABE56) was preconjugated to 25 µL of protein A/G Plus-agarose-beads (sc-2003; Santa Cruz Biotechnology) in 0.5% bovine serum albumin/phosphate-buffered saline (PBS) for at least 6 hr. Trypsinized HEK293T cells were crosslinked with 1% formaldehyde for 10 min at room temperature in Dulbecco’s Modified Eagle Medium and 10% serum, followed by a 5-min quench with 125 mM glycine. Cell pellet was resuspended in ChIP lysis buffer (50 mM HEPES at pH 7.5, 140 mM NaCl, 10% Triton X-100, 0.1% sodium deoxycholate, protease inhibitors) and incubated on ice. After 10 min, samples were centrifuged, pellets resuspended again in ChIP lysis buffer, and incubated for 10 min on ice. Extracts were then sonicated to generate ~200- to 500-bp DNA fragments and cleared via centrifugation for 10 min. Then, 300 µg of chromatin was added to the antibody-coated beads and incubated overnight. Beads were washed at 4° 1× with 1 mL of ChIP lysis buffer, 2× with 1 mL of ChIP lysis buffer high salt (50 mM HEPES at pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate), followed by two washes with 1 mL of ChIP wash buffer (10 mM Tris-HCl at pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA). Antibody complexes were eluted 2× for 10 min at 65° in (50 mM Tris pH 8.0, 10mM EDTA, and 1% sodium dodecyl sulfate), crosslinks were reversed at 65° for 4 hr. Eluates were incubated with Proteinase K, and DNA was extracted with phenol/chloroform and ethanol-precipitated.
Semiquantitative detection of PIWIL4 mRNA expression:
For RT-PCR, 1 µg of total RNA was reverse transcribed using oligo d(T) primers. PIWIL4 cDNA was amplified for 30 cycles with the following primer pair: PIWIL4s CCTGATGGTGGTCGGTATTGA and PIWIL4as ACACAATTATCCGTGCTGGC.
DNA methylation analysis
Part of the siRNA-transfected cell population was used to assess knockdown efficiency by quantitative RT-PCR. Genomic DNA from the remaining cells was isolated and bisulfite modified with the EpiTect Plus DNA Bisulfite kit (QIAGEN). Modified DNA was amplified, subcloned, and sequenced. DXZ4 and H19 primers specific to bisulfite-modified DNA have been published elsewhere (Borghol et al. 2006; Chadwick 2008). Sequence reads were analyzed with the QUMA online tool (Kumaki et al. 2008).
Fluorescence in situ hybridization (FISH)
For the DXZ4 FISH probe, a full-length DXZ4 monomer was PCR-amplified from human fibroblast genomic DNA (5′-gctttgccaccgaactcatcg, 5′-aagcttgagaaatggagactc) and gel eluted. For the XIST FISH probe, a bacterial artificial chromosome plasmid (RP11-13M9, CHORI BPRC) was used. Then, 25 ng of probe DNA was labeled with the BioPrime DNA labeling kit (Life Technologies), using Cy3- or Cy5-conjugated dCTP (GE Healthcare), and stored in 70% ethanol at −20° until further use. To prepare FISH probes for hybridization, probes were precipitated with yeast transfer RNA (tRNA) and Salmon Sperm DNA (both Life Technologies). After washes with 75% and 100% ethanol, probes were air-dried and denatured for 10 min in 50–100 µL of 100% formamide at 85°. An equal volume of 2× hybridization buffer (25% dextran sulfate/4× saline sodium citrate [SSC]) was then added, and probes were prehybridized for 90 min at 37°.
DNA and RNA FISH experiments were performed essentially as described (Calabrese et al. 2012). Cells were grown on coverslips, fixed for 10 min in 2% paraformaldehyde/PBS at room temperature, and permeabilized for 10 min on ice in 0.5% Triton X-100/PBS. Cells were then dehydrated by serial 3-min incubations with 75%, 85%, 95%, and 100% ethanol and air-dried for 5 min. For combined DNA/RNA FISH experiments, cells were heat denatured at 80° for 9 min in 70% formamide/2× SSC followed by two washes in cold 2× SSC. Probes were jointly hybridized overnight at 37°. Coverslips were washed 2× for 5 min in 50% formamide/2× SSC at 37°, then 2× for 5 min in 2× SSC at 37°. DAPI was added at 100 ng/mL to one of the wash buffers.
Results and Discussion
DXZ4 expresses a wide range of chromatin-associated small RNAs
Northern blots with RNA from the cell lines HEK293T and human umbilical vein endothelial cell were probed for regions that express 85-nucleotide DXZ4 RNAs detecting multiple transcripts including small RNAs between 20 and 40 nucleotides long (Figure 1A). This size range corresponds to several classes of small RNAs known to mediate epigenetic modifications (Kim et al. 2009). In addition to these small RNAs, the probes detected several longer transcripts. Because of their design, the probes are predicted to detect the 85-nucleotide transcripts as well as any larger transcripts (Chadwick 2008), providing a putative explanation for the detection of multiple signals per probe. However, although we used stringent hybridization and washing conditions, we cannot exclude the presence of unintended cross hybridization signals.
Figure 1.
Small RNA expression from DXZ4. (A) Northern hybridization detecting small RNAs expressed from DXZ4 in the cell lines HEK293T and human umbilical vein endothelial cell (HUVEC). The numbers below correlate with the start position of the Northern probes in DXZ4, which colocalize with DXZ4 regions associated with characteristic histone modifications. (B) Distribution of the lengths of DXZ4-matching unique small RNA reads obtained from merged high-throughput sequencing data from K562, HEK293, MCF10a, H1, and IMR-90 cell lines (n = 97). Coloring corresponds to size range of known small RNA classes. (C) Alignment of DXZ4 small RNAs from (B) and from the K562 chromatin fraction only (n = 47), respectively. Refer to the text for details. The colors of the dots reflect the colors used in (B) indicating small RNA size. (D) Relative expression of DXZ4 small RNAs in HEK293T detected by quantitative reverse-transcription polymerase chain reaction. cDNA synthesis was performed in the presence or absence of reverse transcriptase. Each reaction was performed n ≥ 6. (E) Alignment of sequenced DXZ4 small RNAs coprecipitated in AGO immunoprecipitations in HeLa cells. Alignment includes multiple RNA copies.
Further, we identified DXZ4-matching small RNAs from several published high-throughput small RNA sequencing studies from a total of 3.2 × 107 reads (Fejes-Toth et al. 2009; Mayr and Bartel 2009; Bernstein et al. 2010). Up to three mismatches were tolerated to factor in single-nucleotide polymorphisms between DXZ4 monomers, potential posttranscriptional editing, and sequencing errors (Tremblay et al. 2011). More specifically, we considered only reads that did not coalign to other genomic sites with the same or a greater stringency than to DXZ4 (Figure 1, B and C). This filtration led to the exclusion, among others, of all small RNAs aligning to a continuous stretch of 775 nucleotides in the center of the DXZ4 monomer representing 38% of highly stringent DXZ4 reads. The region includes 664 nucleotides with more than 99% identity to an intronic sequence of ARID5b and is followed by a CT repeat. A total of 55% of the remaining 97 unique DXZ4 small RNA sequences matched perfectly. The sequence data confirmed the specificity of our small RNA Northern, because many of the probes we used were identical or partially overlapped with sequenced small RNAs, for example the probes employed generating the data in Figure 1A. The relative small number of identified unique reads indicates a low expression level DXZ4 small RNAs. Approximately 25% of the small RNA sequences were in the sense direction and 75% in the antisense direction. Interestingly, almost 22% of the sequences were 35, 36, or 37 nucleotides long (Figure 1B). Although piRNAs do not typically exceed 32 nucleotides, those with 36 nucleotides have been found (Ro et al. 2007). Alternatively, the DXZ4 complementary sequences could be derived from other classes of small RNAs, for example, tRNA-derived small RNAs, which can reach a length of 38 nucleotides (Lee et al. 2009; Peng et al. 2012). However, the DXZ4 sequence did not reveal any annotated transfer-RNA genes, nor could we identify tRNA-like sequences using tRNA search programs (Laslett and Canback 2004; Schattner et al. 2005).
Because DXZ4 expresses 85-nucleotide RNAs from regions characterized by specific histone modifications (Chadwick 2008), we plotted the position and size of the DXZ4 small RNAs identified by high-throughput sequencing relative to the DXZ4 monomer (Figure 1C). Sequenced RNAs associated with chromatin also were analyzed individually to reveal any potential link between the region of specific histone modifications and the genomic origin of the small RNAs. In general, the results show that the DXZ4 small RNA expression was not restricted to regions previously shown to harbor specific histone modifications. However, a number of DXZ4 sites, in particular around nucleotide 800, were associated with an increased number of small RNAs. This enrichment was mostly not generated by multiple copies of the same sequences but instead by slightly different RNAs. We did not recognize a correlation between the lengths of the small RNAs and their origins (Figure 1C).
To validate the expression of DXZ4 small RNAs via an alternate method, we performed quantitative RT-PCR with RNA from HEK293T, MRC-5, and IMR-90 cells. We chose RNA species that were identified in the high-throughput sequencing datasets. The detection of expression of all three small RNAs in HEK293T cells validated the existence of DXZ4 small RNAs by an additional independent method (Figure 1D). The late amplification signals corroborate a low expression level of these small RNAs as indicated in the sequencing data analysis earlier. One of the small RNA assays also detected expression in MRC-5 cells. However, the small RNAs were barely or not detectable in IMR-90, respectively. This differential small RNA expression correlates with our observations identifying these RNAs by Northern hybridization and in the sequencing datasets in some cell lines but not in all.
The presence of DXZ4 small RNAs in the K562 chromatin fraction prompted us to investigate chromatin-associated RNAs in MRC-5, IMR-90, and HEK293T cells. We explored their presence in a 300-base pair DXZ4 window by a Northern hybridization-based scanning approach using consecutive oligonucleotide probes. This sequence window encompasses one of the known regions containing histone modifications (Chadwick 2008). Many small RNAs in both orientations were detected, including some that were specific to one or two of the cell lines corroborating results described previously (Supporting Information, Figure S1A and B). In agreement with the sequencing data, small RNAs detected by Northern hybridization from the chromatin fraction originated from sites dispersed along the entire monomer sequence.
Association of DXZ4 small RNAs with Argonaute proteins would strongly support a function of these small RNAs. We therefore searched for DXZ4 small RNAs in co-precipitated RNA populations from Argonaute immunoprecipitations (Dueck et al. 2012). Matching sequences were filtered by the same parameters as specified previously. AGO-1, AGO-2, and AGO-3 are expressed in HeLa cells and were associated with DXZ4 small RNAs in both orientations (Figure 1E). A total of 55% of the incorporated reads matched perfectly to DXZ4. In contrast, the IgG control and the coprecipitated material from AGO-4, which is not expressed in these cells, did not reveal any DXZ4 sequences. Aligning the identified sequences to the DXZ4 monomer showed a concentration in the 5′ region that includes but also extends beyond the promoter (Figure 1E).
Characterization of DXZ4 small RNAs
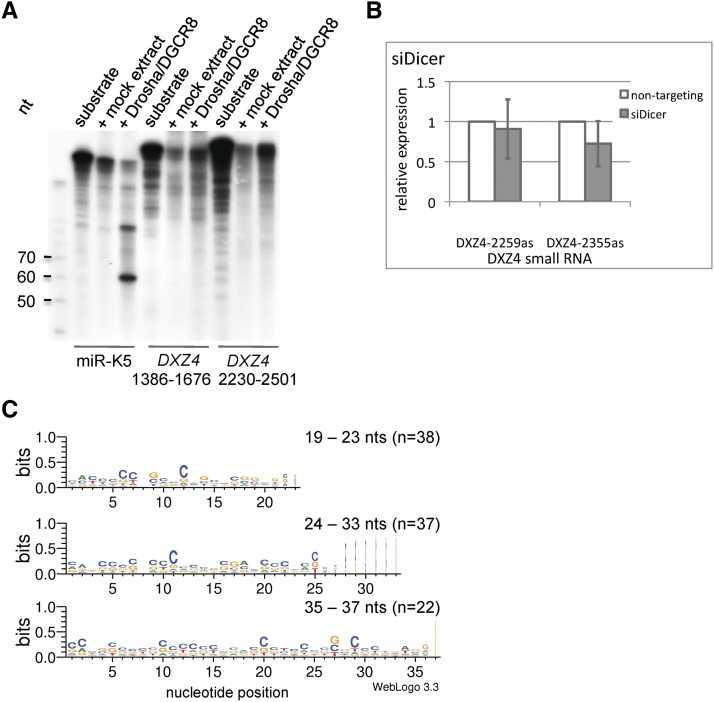
The size range of DXZ4 small RNAs corresponds to several classes of small RNAs, including siRNAs, miRNAs, and piRNAs (Kim et al. 2009). To characterize these RNAs, we tested processing of potential precursor-miRNAs by the Drosha/DGCR8 complex (Lee et al. 2003; Denli et al. 2004; Gregory et al. 2004). Because processing requires folding of the primary miRNA into a characteristic hairpin stem loop structure (Zeng et al. 2005), approximately 65-nucleotide long DXZ4 RNA fragments were tested with a secondary structure algorithm (Zuker 2003). For several fragments, one or several thermodynamically stable hairpin structures were identified, implying DXZ4 transcripts could resemble precursor-miRNAs. Two of those potential precursors were analyzed in a Drosha cleavage assay (Zeng and Cullen 2006). The Kaposi’s sarcoma-associated herpesvirus miR-K5 precursor that previously had been identified as a substrate (Gottwein et al. 2006) yielded the expected ~65 nucleotide cleavage product (Figure 2A). In contrast, both DXZ4 transcripts were not cleaved, suggesting they are not proper Drosha substrates. It has been shown that subtle differences in the stem-loop structure can severely impair its ability to be processed (Gottwein et al. 2006). Although those DXZ4 fragments could potentially fold into stem loops, we conclude that the limited number of DXZ4 RNA fragments tested is not regular precursor-miRNAs. It is known that some miRNAs are generated by Drosha-independent noncanonical pathways (Okamura et al. 2007; Ruby et al. 2007; Bogerd et al. 2010; Valen et al. 2011).
Figure 2.
Characterization of DXZ4 small RNAs. (A) In vitro Drosha assay with extracts from HEK293T cells either mock transfected or cotransfected with expression plasmids for Drosha and DGCR8. Radiolabeled in vitro transcripts from two DXZ4 regions characterized by specific histone modifications and the KSHV precursor-miRNA miR-K5 served as substrates for Drosha/DGCR8 processing and were separated by polyacrylamide gel electrophoresis. (B) Effect of Dicer depletion on DXZ4 small RNA expression in HEK293T cells. Relative expression of two small RNAs was determined by quantitative reverse-transcription polymerase chain reaction (n = 5). The difference with DXZ4-2355as is not statistically significant (P-value = 0.089, t-test). (C) Sequence logos for DXZ4 small RNAs grouped according to their lengths.
Dicer is another RNaseIII endoribonuclease that plays a central role in the processing of certain classes of small RNAs; it is required for the maturation of endogenous siRNAs and miRNAs but not of piRNAs (Bernstein et al. 2001; Grishok et al. 2001; Vagin et al. 2006; Houwing et al. 2007). We tested a requirement of Dicer in DXZ4 small RNA expression by transiently depleting Dicer expression in HEK293T cells (Figure S3B). DXZ4 small RNA expression was analyzed by determining the expression of two DXZ4 small RNAs of different size by quantitative RT-PCR: a 36-nucleotide RNA (DXZ4-2259as) and a 22-nucleotide RNA (DXZ4-2355as). Dicer depletion had no significant effect on the abundance of both small RNAs (Figure 2B).
Only RNAs with both the 2′ and 3′ hydroxy termini such as animal siRNAs and miRNAs react with sodium periodate (NaIO4) (Elbashir et al. 2001; Hutvagner et al. 2001). In contrast, piRNAs are modified by 2′-O-methylation blocking the NaIO4 reaction. NaIO4-reacted RNAs migrate faster than unreacted RNA in denaturing gel electrophoresis, generating a signal shift in subsequent Northern hybridizations. This difference provides a direct way to distinguish endogenous siRNAs and miRNAs from piRNAs (Vagin et al. 2006; Kirino and Mourelatos 2007). We used this chemical probing to characterize DXZ4 small RNAs which, regarding their spectrum of sizes, potentially belong to more than one class of small RNA (Figure S2). miR-15a, which was expected to be unmodified at its 3′ end, was sensitive to the treatment and showed a ~2 nucleotide shift. We used two Northern probes to detect DXZ4 small RNAs in both orientations. Individual RNAs in both the <200 nucleotide population and the chromatin-associated RNA fraction shifted after treatment whereas other RNAs did not. This discriminative behavior of individual small RNAs adds to previous lines of evidence indicating the expression of small RNAs from DXZ4 with different characteristics.
Inert RNAs are indicative of a block of the NaIO4 reaction suggesting a 3′ modification of certain DXZ4 small RNAs, potentially analogous to as previously documented for piRNAs. Primary piRNAs from Drosophila and mouse testis have a strong preference for uridine at their 5′ position (Girard et al. 2006; Aravin et al. 2007; Brennecke et al. 2007). In this regard, and in reference to their “ping-pong” amplification biogenesis mechanism, secondary piRNAs have an adenine bias at nucleotide position ten (Brennecke et al. 2007). In contrast, DXZ4 small RNAs did not display any nucleotide preference suggesting that they are generated by a ping-pong unrelated mechanism (Figure 2C). Primary piRNAs are generated by mechanisms that are not well understood, particularly those piRNAs generated outside the mammalian germline (Ishizu et al. 2012). In certain cell types, such as somatic follicle cells in flies or mouse spermatocytes, piRNAs are only generated by the primary biogenesis pathway (Siomi et al. 2011; Yan et al. 2011). The absence of multiple copies of the same RNA in the sequencing data suggests that DXZ4 RNAs are not generated by amplification. However, nucleotide preferences are not prevalent in siRNAs and miRNAs.
In summary, our characterization of DXZ4 small RNAs suggests that this RNA population likely represents a noncanonical group of RNAs. Although DXZ4 small RNAs share certain characteristics with known small RNA classes, all together our assays did not lead to a clear assignment to a previously described RNA class. Further characterization will require deep-sequencing approaches to capture the entire population.
Argonaute proteins associate with DXZ4 chromatin
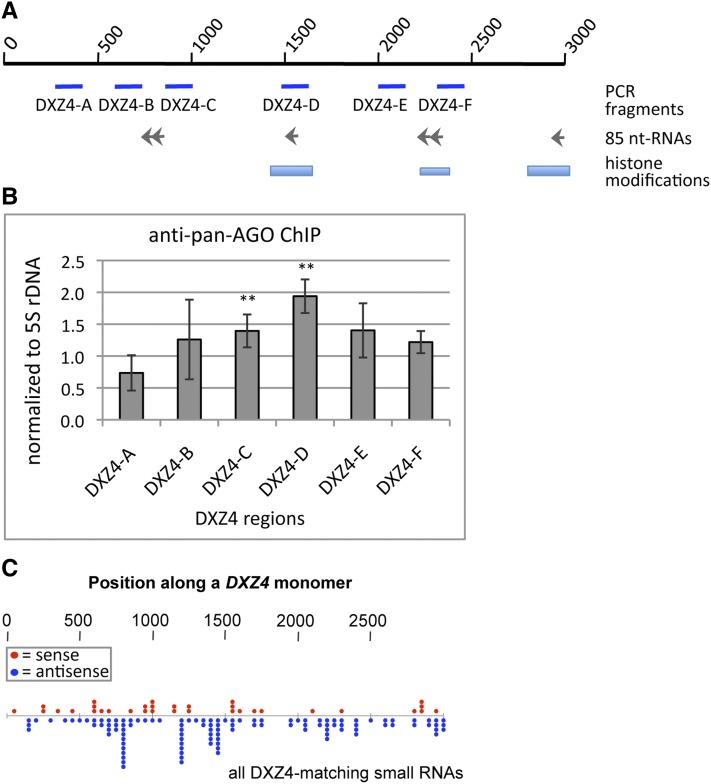
Small RNAs associate with Argonaute proteins to guide them to nascent transcripts in a sequence-specific manner (Grewal and Jia 2007). Based on our finding that several AGO proteins bound a substantial number of DXZ4 small RNAs (Figure 1E), we asked whether AGO directly associates with DXZ4. To address this question, we performed ChIP using a pan-AGO antibody specific for AGO-1 through AGO-4 in HEK293T cells (Nelson et al. 2007; Schwartz et al. 2008). Coprecipitated DNA was detected in immunoprecipitated chromatin by quantitative PCR and normalized relative to a species-matched IgG control. DXZ4 was analyzed using a set of PCR fragments dispersed across a DXZ4 monomer (Figure 3A). We used the 5S rDNA repeat cluster, which only showed a modest enrichment compared with IgG as a negative control to copy number-match the DXZ4 array and further normalized DXZ4 enrichments relative to it. DXZ4 amplification between base pairs 820 and 1600 of the monomer was significantly enriched relative to the IgG control (Figure 3B).
Figure 3.
AGO associated with DXZ4 chromatin. (A) Schematic representation of a DXZ4 monomer showing fragments amplified to analyze coprecipitated DXZ4 DNA. Locations of RNAs and histone modifications were adopted from Chadwick (2008). (B) Chromatin immunoprecipitation (ChIP) in HEK293T cells using a pan-AGO antibody. Coprecipitated DNA was quantified by real-time polymerase chain reaction. Relative DXZ4 enrichment over host matched IgG was normalized to the repetitive 5S rDNA locus (n = 4–5). Double asterisks indicate significant enrichment (P < 0.05) calculated by using the t-test: DXZ4-C P-value = 0.029, DXZ4-D P-value = 0.006. (C) Alignment of DXZ4 matching small RNA reads including alignment to non-DXZ4 genomic loci with the same or higher stringency. Data obtained from merged high-throughput sequencing data from K562, HEK293, MCF10a, H1, and IMR-90 cell lines (n = 154).
DXZ4-matching RNAs that align to a different genomic locus with the same or a higher stringency irrespective of their origin have an equivalent potential in guiding Argonaute to DXZ4. When we compared the ChIP data with the distribution of this comprehensive small RNA population, it became obvious that the fragments with significant ChIP enrichment coincided with sites of mounded small RNA alignment to DXZ4 (Figure 3C). In summary, these data indicate that one or several members of the AGO subfamily are part of DXZ4 chromatin.
Argonaute proteins possibly play a role in DNA methylation at DXZ4
The detection of AGO in DXZ4 chromatin prompted us to investigate a potential functional consequence of AGO association with DXZ4. One of the characteristics of DXZ4 is it’s hypermethylation on the active X chromosome (Giacalone et al. 1992; Chadwick 2008). To investigate whether Argonaute proteins are essential in establishing or maintaining DXZ4 methylation, we determined the effects of AGO-1 depletion on CpG methylation. Furthermore, because it is known that the piRNA pathway is involved in DNA methylation of protein-coding genes and transposons in mouse male germ cells (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008; Watanabe et al. 2011), we included the PIWI protein PIWIL4, which is expressed in somatic cells (Sugimoto et al. 2007). Using RT-PCR, we could confirm PIWIL4 expression in MRC-5, IMR-90, and HEK293T fibroblast lines (Figure S3A). RNAi-mediated knockdown of AGO-1 and PIWIL4 reduced the mRNA of these genes by at least 70% (Figure S3B).
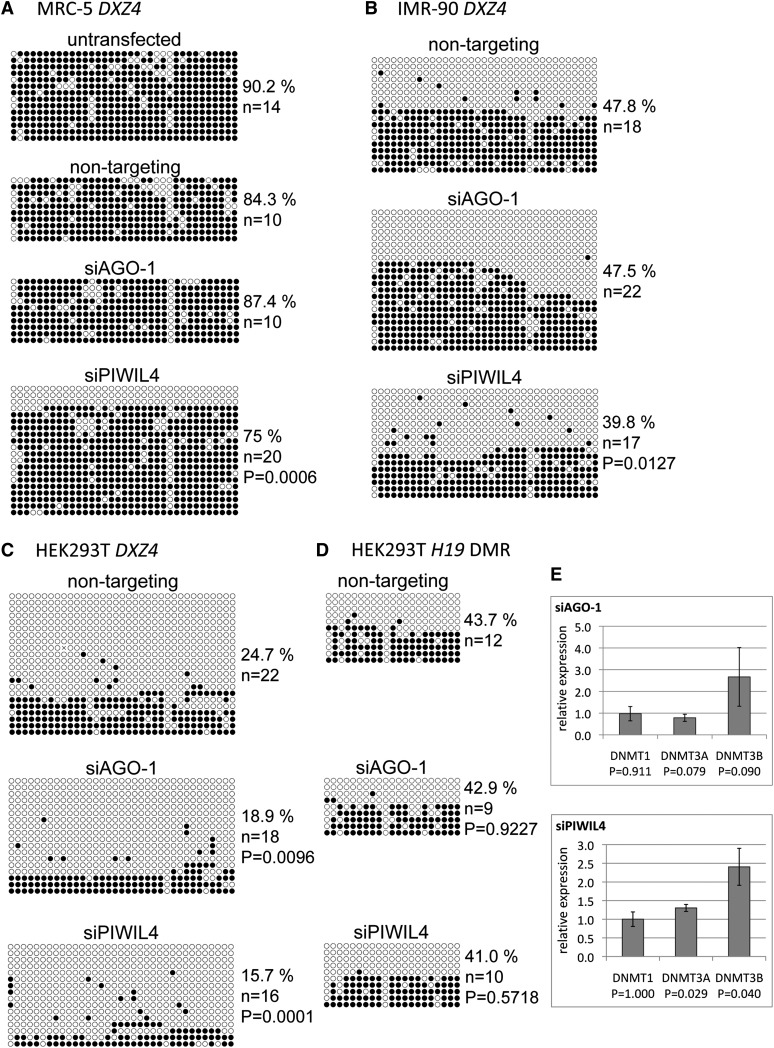
Sequencing of bisulfite-treated genomic DNA isolated from siRNA-transfected cells was used to determine DNA methylation levels. A DXZ4 fragment was analyzed that covers 35 CpG dinucleotides and contains the internal promoter and the CTCF binding region (Chadwick 2008). This DXZ4 fragment was nearly completely methylated in the male primary fibroblasts MRC-5, which is analogous to previously published data (Chadwick 2008). Although AGO-1 knockdown did not affect DXZ4 methylation, depletion of PIWIL4 reduced the number of methylated CpGs statistically significant compared with levels found in mock transfected cells (Figure 4A). We further analyzed DXZ4 methylation in IMR-90 fibroblasts, which are a female counterpart of MRC-5. As expected, DXZ4 methylation in these cells was approximately 50% because only DXZ4 on the active X chromosome is methylated. Argonaute knockdown confirmed the result observed with the MRC-5 cells with a statistically significant reduction of DXZ4 methylation in PIWIL4 depleted IMR-90 cells (Figure 4B).
Figure 4.
Reduction in CpG methylation at DXZ4 after Argonaute depletion. (A–C) CpG methylation of a 494-base pair DXZ4 region was assessed by bisulfite sequencing. Each row of circles represents an individual sequenced clone. Open circles indicate unmethylated and filled circles indicate methylated cytosine residues of a CpG. Percent methylated CpGs and number of sequenced clones are indicated. P-values were calculated by via the Fisher’s exact test for data pairs from cell populations treated with the nontargeting siRNA or with an Argonaute siRNA. (D) Analysis of CpG methylation of the differential methylated region (DMR) of the H19 locus. (E) mRNA expression levels of DNA methyltransferases in Argonaute-depleted cells relative to nontargeting siRNA-transfected cells determined by quantitative RT-PCR (n = 3–4). P-values were calculated using the t-test.
We included a second female cell line, HEK293T, in our analysis. In these cells, DXZ4 DNA methylation in mock transfected HEK293T was at 25% instead of the expected 50%. Because the HEK293T cell line is a derivative of the experimentally transformed female HEK293 line that has an aberrant chromosome content (Graham et al. 1977), we determined the active and inactive X chromosome ratio by in situ hybridizations. X chromosomes were labeled using a probe of the DXZ4 monomer in DNA-FISH. Four signals per nucleus were observed in most of the cells, indicating the presence of four X chromosomes (Figure S4). Four X chromosomes also were seen by using a bacterial artificial chromosome clone encompassing the X inactive-specific transcript (XIST) locus as a probe (data not shown). To detect the inactive X chromosomes, DXZ4 DNA-FISH was combined with XIST RNA-FISH. The noncoding XIST is expressed from and preferentially coats the inactive X, generating a diffuse fluorescence signal marking the X chromosome (Clemson et al. 1996). Two XIST signals colocalized with DXZ4 signals, suggesting two of the four X chromosomes were silenced and two should be active (Figure S4). Thus, our data detecting 25% DNA hypermethylation of DXZ4 in these cells suggest that only one of the two active X chromosomes is expressed from this region. Moreover, it is also possible that DXZ4 is methylated on only one of the active X chromosomes or the DXZ4 arrays on both active X chromosomes are incompletely methylated containing methylated and unmethylated monomers accumulating at 25% methylation.
In HEK293T cells, depletion of PIWIL4 again significantly reduced DXZ4 methylation? (Figure 4C). Moreover, AGO-1 depletion resulted in a statistically significant reduction of DXZ4 CpG methylation as well. In the context of the association of DXZ4 small RNAs with AGO-1 (Figure 1E), this observation suggests a guiding role of these RNAs for Argonaute in cis. The cause of the different AGO-1−dependent outcomes in the different cell types is unknown. A possible explanation could be the presence of compensation mechanisms that are present in primary cells but are lacking in the transformed HEK293T cell line.
To investigate whether Argonaute knockdown induced methylation changes at other loci, we examined CpG methylation at the H19 locus. We chose H19 because disruption of the piRNA pathway does not affect H19 DNA methylation (Watanabe et al. 2011). As expected, knockdown of AGO-1 or PIWIL4 did not change DNA methylation globally (Figure 4D).
To consider a potential indirect effect of Argonaute depletion on DNA methylation levels, we determined the effect on the expression of DNA methyltransferases, enzymes that catalyze the addition of methyl groups to DNA. The three known mammalian DNA methyltransferases are expressed in HEK293T cells (Choi et al. 2011). Argonaute knockdown did not result in a reduction of DNMT levels, indicating that the reduced DXZ4 methylation was not due to an effect on DNMT expression (Figure 4E). Overall, these findings support a role for Argonaute proteins in establishing or maintaining DNA methylation at DXZ4 in somatic cells. A requirement for PIWIL4 in somatic cell DNA methylation extends previous reports showing a role for components of the piRNA pathway outside of the germline. Although the use of a pan-AGO antibody in our ChIP experiments does not allow us to specify the AGO protein, based on its role in DXZ4 methylation, we speculate that AGO-1 is likely among the precipitated AGO members.
It has previously been shown that DXZ4 is expressed from both the active and the inactive X chromosome (Chadwick 2008). Considering the mainly suppressive nature of DNA methylation, a reduction could potentially have a positive effect on DXZ4 expression. We therefore investigated the effect of AGO-1 and PIWIL4 depletion on DXZ4 transcription in male MRC-5 cells, which are normally hypermethylated. DXZ4 expression was analyzed at three regions of the DXZ4 monomer by quantitative RT-PCR (Figure S5). Only AGO-1 knockdown generated a slight increase in DXZ4 expression at one of the three regions tested. Taken together, neither AGO-1 nor PIWIL4-mediated reduction in DNA methylation caused a clear change in DXZ4 transcription output. Because the data presented here indicate that Argonaute and Dicer are not required for DXZ4 small RNA expression, it is not known by which means these small RNAs are generated. However, it seems plausible that DXZ4 small RNA expression is generally linked to the transcription of long DXZ4 RNAs that is controlled by the bidirectional promoter residing within each monomer (Chadwick 2008). How the small RNAs possibly mature from the longer transcripts requires further investigation.
Characterization of the physiological functions of diverse small RNAs has lagged behind their discovery (Valen et al. 2011). Here, we describe a potential link between the expression of small RNAs, members of the Argonaute family, and DNA methylation of the macrosatellite DXZ4 in human somatic cells. Furthermore, we provide additional evidence that the piRNA pathway is active in nongermline cells. In the germline, Piwi functions in maintaining structural integrity of the genome (Mani and Juliano 2013). We speculate that Argonaute/macrosatellite interactions in somatic cells may have similar functions.
Supplementary Material
Acknowledgments
We thank the Magnuson lab members for constant input, Joshua Starmer for visualizing data and statistical advice, and the Praveen Sethupathy lab for generous help with the TaqMan qPCR. We are also thankful to Zissimos Mourelatos for kindly providing the pan-AGO antibody and to Eva Gottwein for the miR-K5 construct. This work was supported by National Institutes of Health grant RO1 GM101974.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.012260/-/DC1
Communicating editor: J. K. Kim
Literature Cited
- Aravin A. A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G. J., 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Sachidanandam R., Bourc’his D., Schaefer C., Pezic D., et al. , 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Stamatoyannopoulos J. A., Costello J. F., Ren B., Milosavljevic A., et al. , 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28: 1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J., 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bogerd H. P., Karnowski H. W., Cai X., Shin J., Pohlers M., et al. , 2010. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol. Cell 37: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs B. A., Cheung P., Heard E., Spector D. L., Chinault A. C., et al. , 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30: 73–76 [DOI] [PubMed] [Google Scholar]
- Borghol N., Lornage J., Blachere T., Sophie Garret A., Lefevre A., 2006. Epigenetic status of the H19 locus in human oocytes following in vitro maturation. Genomics 87: 417–426 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., et al. , 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Calabrese J. M., Sun W., Song L., Mugford J. W., Williams L., et al. , 2012. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M. A., Girard A., van de Kant H. J., Bourc’his D., Bestor T. H., et al. , 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chadwick B. P., 2008. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 18: 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. H., Heo K., Byun H. M., An W., Lu W., et al. , 2011. Identification of preferential target sites for human DNA methyltransferases. Nucleic Acids Res. 39: 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C. M., McNeil J. A., Willard H. F., Lawrence J. B., 1996. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A. M., Tops B. B., Plasterk R. H., Ketting R. F., Hannon G. J., 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Dueck A., Ziegler C., Eichner A., Berezikov E., Meister G., 2012. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 40: 9850–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S. M., Lendeckel W., Tuschl T., 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D., Bouge A. L., Berry B., Poisot E., Sismeiro O., et al. , 2009. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc. Natl. Acad. Sci. USA 106: 21258–21263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth K., Sotirova V., Sachidanandam R., Assaf G., Hannon G. J. et al. Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project , 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M., Seitz H., Horwich M. D., Li C., Du T., et al. , 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacalone J., Friedes J., Francke U., 1992. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet. 1: 137–143 [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G. J., Carmell M. A., 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Gottwein E., Cai X., Cullen B. R., 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol. 80: 5321–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R., 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36: 59–74 [DOI] [PubMed] [Google Scholar]
- Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., et al. , 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Jia S., 2007. Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., et al. , 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Houwing S., Kamminga L. M., Berezikov E., Cronembold D., Girard A., et al. , 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Huang V., Li L. C., 2012. miRNA goes nuclear. RNA 9: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan J., Pasquinelli A. E., Balint E., Tuschl T., et al. , 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Ishizu H., Siomi H., Siomi M. C., 2012. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 26: 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Rossi J. J., 2009. Transcriptional gene silencing using small RNAs. Methods Mol. Biol. 555: 119–125 [DOI] [PubMed] [Google Scholar]
- Kim V. N., Han J., Siomi M. C., 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kirino Y., Mourelatos Z., 2007. Mouse Piwi-interacting RNAs are 2’-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 14: 347–348 [DOI] [PubMed] [Google Scholar]
- Kugler W., Enssle J., Hentze M. W., Kulozik A. E., 1995. Nuclear degradation of nonsense mutated beta-globin mRNA: a post-transcriptional mechanism to protect heterozygotes from severe clinical manifestations of beta-thalassemia? Nucleic Acids Res. 23: 413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y., Oda M., Okano M., 2008. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 36: W170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., et al. , 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D., Canback B., 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., et al. , 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Shibata Y., Malhotra A., Dutta A., 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Vagin V. V., Lee S., Xu J., Ma S., et al. , 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S. R., Juliano C. E., 2013. Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev. 80: 632–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Bartel D. P., 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. T., De Planell-Saguer M., Lamprinaki S., Kiriakidou M., Zhang P., et al. , 2007. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA 13: 1787–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Hagen J. W., Duan H., Tyler D. M., Lai E. C., 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Shi J., Zhang Y., Zhang H., Liao S., et al. , 2012. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 22: 1609–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. C., Karpen G. H., 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 9: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P., Antonov I., Sheridan R., Frey S., Sander C., et al. , 2012. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 149: 693–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S., Park C., Song R., Nguyen D., Jin J., et al. , 2007. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA 13: 1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Bartel D. P., 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P., Brooks A. N., Lowe T. M., 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33: W686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J. C., Younger S. T., Nguyen N. B., Hardy D. B., Monia B. P., et al. , 2008. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 15: 842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M. C., Sato K., Pezic D., Aravin A. A., 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nature 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Kage H., Aki N., Sano A., Kitagawa H., et al. , 2007. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem. Biophys. Res. Commun. 359: 497–502 [DOI] [PubMed] [Google Scholar]
- Tremblay D. C., Moseley S., Chadwick B. P., 2011. Variation in array size, monomer composition and expression of the macrosatellite DXZ4. PLoS 6: e18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V. V., Sigova A., Li C., Seitz H., Gvozdev V., et al. , 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Valen E., Preker P., Andersen P. R., Zhao X., Chen Y., et al. , 2011. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat. Struct. Mol. Biol. 18: 1075–1082 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Tomizawa S., Mitsuya K., Totoki Y., Yamamoto Y., et al. , 2011. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332: 848–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Hu H. Y., Jiang X., Maierhofer V., Neb E., et al. , 2011. Widespread expression of piRNA-like molecules in somatic tissues. NAR 39: 6596–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Cullen B. R., 2006. Recognition and cleavage of primary microRNA transcripts. Methods Mol. Biol. 342: 49–56 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi R., Cullen B. R., 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.