Abstract
Although many bacteria are known to be naturally competent for DNA uptake, this ability varies dramatically between species and even within a single species, some isolates display high levels of competence while others seem to be completely nontransformable. Surprisingly, many nontransformable bacterial strains appear to encode components necessary for DNA uptake. We believe that many such strains are actually competent but that this ability has been overlooked because standard laboratory conditions are inappropriate for competence induction. For example, most strains of the gram-negative bacterium Legionella pneumophila are not competent under normal laboratory conditions of aerobic growth at 37°C. However, it was previously reported that microaerophilic growth at 37°C allows L. pneumophila serogroup 1 strain AA100 to be naturally transformed. Here we report that another L. pneumophila serogroup 1 strain, Lp02, can also be transformed under these conditions. Moreover, Lp02 can be induced to high levels of competence by a second set of conditions, aerobic growth at 30°C. In contrast to Lp02, AA100 is only minimally transformable at 30°C, indicating that Lp02 is hypercompetent under these conditions. To identify potential causes of hypercompetence, we isolated mutants of AA100 that exhibited enhanced DNA uptake. Characterization of these mutants revealed two genes, proQ and comR, that are involved in regulating competence in L. pneumophila. This approach, involving the isolation of hypercompetent mutants, shows great promise as a method for identifying natural transformation in bacterial species previously thought to be nontransformable.
The phenomenon of natural transformation, also known as competence, is defined as the ability of bacteria to take up and stably maintain exogenous DNA. This ability is prevalent in nature, as evidenced by the description of natural competence in over 40 different bacterial species, which are widely distributed among taxonomic and trophic groups (reviewed in reference 31). Although the purpose of natural transformation in nature remains unknown, it has been speculated to provide a means of genetic exchange, DNA repair, and/or nutrient acquisition (31).
The numerous examples of naturally transformable bacteria in the environment suggest that competence is a widely conserved trait, and yet, surprisingly, many bacterial species seem to lack this ability. Even closely related strains within a competent species can exhibit profound differences in the competence phenotype. For example, examination of a worldwide collection of Pseudomonas stutzeri strains revealed that less than one half were competent (45). In a similar analysis of Actinobacillus actinomycetemcomitans strains, only 1 of 17 was found to be transformable (52). Curiously, many bacteria that do not appear to be competent are known to encode components of DNA uptake machinery (9). It is possible that some of these strains contain only remnants of a once-functional uptake apparatus. Alternatively, they may actually be competent in the environment but have lost this ability due to passage in the laboratory. Finally, bacterial strains fully capable of natural transformation may only appear to be noncompetent due to the use of inappropriate assay conditions.
Consistent with the latter possibility, the ability to take up DNA is generally not constitutive but requires the development of a specific, genetically programmed physiological state (13). Moreover, development of the competent state varies greatly between organisms, making prediction of competence-inducing conditions difficult. For example, in Streptococcus pneumoniae competence is expressed transiently during the exponential phase of growth, when nutrients are plentiful, and is repressed in stationary phase (38, 48). In contrast, competence in Bacillus subtilis occurs in response to starvation and does not become apparent until the late exponential phase of growth (29). In each of these gram-positive species, the regulation of competence is mediated by quorum sensing (21, 46). Gram-negative species such as Neisseria gonorrhoeae, Haemophilus influenzae, and Acinetobacter calcoaceticus also display varied relationships between transformation and growth phase but do not use quorum sensing to induce the competent state (5, 17, 31, 40). From these studies, it is clear that induction of competence is highly variable, and for this reason many bacterial species capable of natural transformation may have yet to be recognized as such.
The gram-negative bacterium Legionella pneumophila provides an example of a competent species in which the transformable phenotype remained undiscovered due to a requirement for unusual inducing conditions. For over 20 years, L. pneumophila was not believed to be naturally competent. Recently, however, it was discovered that the L. pneumophila serogroup 1 strain AA100 can be transformed using microaerophilic growth at 37°C (47). In contrast, AA100 cannot be transformed using aerobic growth at 37°C, conditions normally used to culture L. pneumophila.
While investigating methods to induce competence in the laboratory setting, we discovered that another L. pneumophila serogroup 1 strain, Lp02, displays an unusually high level of competence at 30°C. The phenotype of enhanced competence, termed hypercompetence, has been described previously for bacterial mutants with defects in regulatory factors that increase or deregulate expression of the competence regulon (24, 28, 33, 49, 53). Hypercompetence can also result from mutations in components of the uptake machinery, as seen in P. stutzeri (18). Finally, hypercompetence can be induced indirectly: for example, an H. influenzae mutant with an altered peptidoglycan biosynthesis gene causes induction of the normal competence pathway (32).
We demonstrate here that Lp02 exhibits a hypercompetence phenotype at 30°C that is both growth phase and temperature regulated. In addition, we were able to recapitulate the hypercompetent phenotype in AA100 using mutagenesis and selection. Examination of AA100 hypercompetent mutant strains revealed two genes, proQ and comR, that normally repress natural transformation in L. pneumophila. The identification of highly transformable L. pneumophila strains could provide a useful tool for rapid and efficient genetic manipulation of this pathogen.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains are listed in Table 1. Strain AA100 is a streptomycin-resistant derivative of an L. pneumophila serogroup 1 clinical isolate (1). Strain Lp02 (thyA hsdR rpsL) is a derivative of the L. pneumophila serogroup 1 clinical isolate Philadelphia-1 (3). All L. pneumophila strains were cultured on N-2-acetamido-2-aminoethanesulfonic acid (ACES)-buffered charcoal yeast extract agar (CYE) or in ACES-buffered yeast extract broth (AYE) as described previously (15, 16). Lp02 and Lp02 derivatives were cultured on CYE or AYE supplemented with 100 μg of thymidine/ml. Kanamycin, chloramphenicol, and gentamicin were used at 30, 5, and 6.5 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant property(ies) | Reference or source |
|---|---|---|
| L. pneumophila | ||
| Philadelphia-1 | Clinical isolate, serogroup 1 | CDCa (Atlanta, Ga.) |
| AA100 | Clinical isolate, serogroup 1 | 1 |
| JR32 | Philadelphia-1 Smr | 41 |
| Lp01 | Philadelphia-1 rpsL hsdR | 3 |
| Lp02 | Lp01 thyA | 3 |
| JV1103 | Lp02 Cmr | This study |
| JV1160 | Lp02 Kanr | This study |
| JV1727 | AA100 comR::mini-Tn10kan | This study |
| JV1729 | AA100 proQ::mini-Tn10kan | This study |
| JV1763 | JV1727 + vector | This study |
| JV1766 | JV1727 + Lp02 comR | This study |
| JV1767 | JV1727 + AA100 comR | This study |
| JV1768 | JV1729 + vector | This study |
| JV1770 | JV1729 + Lp02 proQ | This study |
| JV1769 | JV1729 + AA100 proQ | This study |
| E. coli | ||
| DH5α::λpir | DH5α(λpir) tet::Mu | 23 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB laclqZΔM15 Tn10 (Tetr)] | Stratagene (La Jolla, Calif.) |
| Plasmids | ||
| pJK211-2 | Contains mini-Tn10 (Kanr/R6K ori) | James Kirby |
| pBluescript II KS+ | ColE1 cloning vector | Stratagene |
| pWSK29 | pSC101 cloning vector | 51 |
| pJB955 | pBluescript II KS+, 3.2-kb Lp02 insert | This study |
| pJB957 | pJB955, Kanr in center of insert | This study |
| pJB964 | pJB955, Cmr in center of insert | This study |
| pJB1389 | pJB955, Gentr in center of insert | This study |
| pJB1199 | pBluescript II KS+, 1-kb Lp02 insert, Kanr in center of insert | This study |
| pJB1517 | pWSK29, 500-bp Lp02 insert, Kanr in center of insert | This study |
| pJB1519 | pWSK29, 160-bp Lp02 insert, Kanr in center of insert | This study |
| pJB908 | RSF1010 cloning vector | 43 |
| pJB1301 | pJB908 derivative | This study |
| p34S-Gm | Cloning vector with Gentr cassette | 12 |
| pJB1653 | RSF1010 cloning vector, Gentr | This study |
| pJB1659 | pJB1653, proQ+ (from strain AA100) | This study |
| pJB1661 | pJB1653, proQ+ (from strain Lp02) | This study |
| pJB1663 | pJB1653, comR+ (from strain Lp02) | This study |
| pJB1665 | pJB1653, comR+ (from strain AA100) | This study |
CDC, Centers for Disease Control and Prevention.
Strain and plasmid construction.
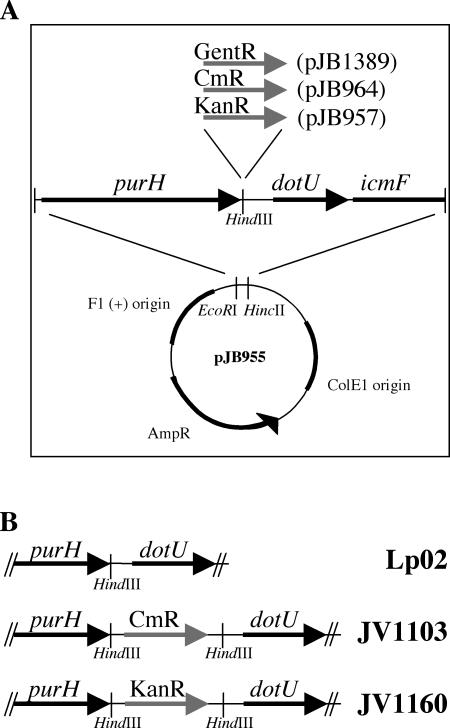
Plasmids for natural competence reporter systems were built by first cloning a region of the Lp02 chromosome into pBluescript II KS+ to make pJB955, then introducing drug resistance cassettes into the center of the pJB955 insert. Plasmid pJB955 contains a 3.2-kb HincII/EcoRI fragment of DNA adjacent to the dot/icm region II cloned into the HincII/EcoRI sites of pBluescript II KS+. Natural competence reporter constructs pJB957, pJB964, and pJB1389 are pJB955 derivatives with Kanr, Cmr, or Gentr cassettes (respectively) cloned into a HindIII site in the pJB955 insert. Complementary strains were constructed by naturally transforming the wild-type L. pneumophila strain Lp02 with reporter plasmid pJB957 or pJB964. This resulted in strains with a Kanr or Cmr cassette inserted on the chromosome in the HindIII site adjacent to dot/icm region II. The Kanr and Cmr strains created were designated JV1103 and JV1160, respectively, and their phenotypes were confirmed by Southern blotting.
The cloning vector pJB1653, used to create proQ and comR complementing clones, is a Gentr derivative of the RSF1010 cloning vector pJB908 (43). First, the pJB908 polylinker HindIII site was replaced with a NotI site, creating plasmid pJB1301. Plasmid pJB1301 was then digested with NotI, the cohesive ends were filled in using Klenow polymerase, and the Gentr cassette from plasmid p34S-Gm (12) was cloned into this site on a SmaI fragment to make plasmid pJB1653. pJB1659, pJB1661, pJB1663, and pJB1665 complementing clones were created as follows: the proQ or comR open reading frames were PCR amplified from chromosomal DNA of the appropriate strain with primers containing BamHI or SalI restriction sites. Primers were as follows: proQ, 5′-CCCGGATCCGACTAAACACAATAAGGGTACC; proQ, 3′-CCCGTCGACGCATTTACTCTGTTGTTTCC; comR, 5′-CCCGGATCCCTTTGCTACACTTTTGCC; and comR, 3′-CCCGTCGACTTACTCTACGTTCCTTGCG. The resulting PCR product was digested with BamHI and SalI, cloned into the BamHI/SalI sites of pJB1653, and confirmed by sequencing.
Plasmids used to assay the length of DNA required for homologous recombination, pJB1199, pJB1517, and pJB1519, were created in a similar fashion as those described above. BamHI and SalI engineered primers were used to PCR amplify the Kanr cassette from pJB957 with 80, 250, or 500 bp of L. pneumophila chromosomal DNA flanking either side. The resulting PCR product was digested with BamHI and SalI and cloned into the BamHI/SalI sites of the vector pBluescript II KS+ or pWSK29 (51). This produced a series of plasmids containing a Kanr cassette flanked by variable lengths of L. pneumophila Lp02 DNA.
Plate transformation assays.
A single colony was patched onto CYE and grown at 37°C for 2 days. Bacteria from this patch were used to make a second, dime-sized patch on CYE, to which DNA (typically 1 μg) resuspended in a 10-μl volume of sterile water was immediately added. DNA was carefully spread over the entire surface of the patch, and it was incubated at 30°C for 48 h. The entire patch was then resuspended in 1 ml of sterile water, and dilutions were plated on media selective for transformants versus nonselective media to obtain counts of transformed and total viable cells. The transformation frequency represents the total number of natural competence transformants divided by the total number of viable cells in a given transformation patch.
Where the experimental temperature was varied, secondary patches were incubated in the presence of 200 ng of plasmid DNA at 30, 34, and 37°C for 2 days or at 26°C for 5 days and cells were then resuspended and plated as described above. For the plate photograph, cell density was normalized to an optical density at 600 nm (OD600) of 1.0 and 10 μl of a 1/10 dilution was plated on selective media. Where the length of flanking DNA was varied, cells were transformed with 1 μg of DNA fragment alone or 1 μg of fragment contained in a plasmid.
Liquid transformation assays.
To assess transformation frequency versus growth phase, a 25-ml starter culture was grown to stationary phase at 30°C with low-speed shaking (100 rpm) and then back-diluted into 500 ml of AYE to an OD600 of 0.02. Aliquots from this culture were periodically assayed for transformation frequency as follows: 1 ml of culture was removed, transferred to a test tube containing 1 μg of plasmid DNA, and incubated at 30°C with shaking for 2 h. Dilutions were then plated on media with and without antibiotics to obtain counts of transformed and total viable cells. Transformation frequency represents the total number of natural competence transformants divided by the total number of viable cells in a given test tube.
To assess the kinetics of DNA uptake, 1-ml aliquots were removed from an exponential-phase culture (grown at 30°C with shaking), exposed to 1 μg of DNA for various lengths of time, and then incubated with DNase I (to 0.1 mg/ml) and MgCl2 (10 mM final concentration) at 37°C for 15 min and plated as described above. This amount of DNase was 100-fold in excess of what was sufficient to degrade 1 μg of plasmid DNA in the absence of cells.
To assess the effects of conditioned media, nutrient availability, and pH on transformation frequency, 1-ml aliquots were removed from an exponential-phase culture (grown at 30°C with shaking), gently pelleted, resuspended in various media, and further incubated for 2 h at 30°C with shaking. One microgram of DNA was added, and cells were incubated for an additional 2 h (30°C with shaking) and plated as described above. Addition of nutrients to conditioned media was accomplished by supplementing them with 1/10 volume of 100-mg/ml yeast extract.
To compare transformation frequencies between L. pneumophila strains, cells from a 2-day 37°C patch were inoculated into 25 ml of AYE, to an OD600 of 0.02. One microgram of DNA was added, and cells were cultured at 30°C with low-speed shaking (100 rpm) to the late stationary phase of growth (68 h) and then plated to obtain counts of transformants and of viable cells. Transformation frequencies were calculated as described above.
Isolation of hypercompetent AA100 mutants.
Transposon mutagenesis of strain AA100 was accomplished using plasmid pJK211-2, which has a temperature-sensitive origin of replication and harbors a mini-Tn10 transposon containing a Kanr cassette and an R6K origin of replication. Kanr colonies (16,000) from a single transposition reaction were collected into 10 pools. Cells from each pool were resuspended and mixed with 12 μg of pJB964 (Cmr) DNA, spotted onto CYE, and incubated at 30°C for 48 h. Bacteria were then resuspended in sterile water and plated to select for Cmr transformants.
After isolation and confirmation of hypercompetent mutants, the location of the mini-Tn10 insertion was determined by recovering the transposon and its flanking chromosomal DNA as a plasmid (as follows) and then sequencing the flanking regions. Chromosomal DNA from a transposon mutant was digested with restriction endonucleases that do not recognize sites within the transposon. Digested DNA was then circularized via ligation and electroporated into Escherichia coli strain DH5α::λpir. Transformed bacteria containing DNA with the mini-Tn10 transposon were selected for, as the R6K origin contained within the transposon allowed replication of these circularized fragments as plasmids, and the kanamycin resistance marker on the transposon allowed a direct selection for such plasmids. Recovered plasmids were sequenced using primers that hybridized to the ends of the transposon.
Optimized protocol for transforming L. pneumophila strain Lp02.
Based on the conditions tested in this paper, we recommend the following protocol when introducing DNA into Lp02 by natural transformation. First, make a dime-sized patch on a CYE-thymidine plate from a single fresh colony of Lp02. Add 1 μg of plasmid DNA or 1 μg of chromosomal DNA in 10 μl of water or Tris-EDTA to the freshly patched strain. The transforming plasmids should contain at least 500 bp of Lp02 DNA flanking a selectable marker (1 kb total). Incubate the transformation reaction mixture at 30°C for 48 h and then plate it on selective media to isolate transformed cells. Typically, one can obtain approximately 105 transformants per μg of plasmid DNA (using 500 bp of flanking DNA) and 104 transformants per μg of chromosomal DNA.
RESULTS
Competence in L. pneumophila strain Lp02 is temperature dependent.
L. pneumophila was not believed to be naturally competent until recently, when transformation was described for the serogroup 1 strain AA100 (47). Transformation of this strain was found to be dependent upon microaerophilic growth, a condition not normally used to culture L. pneumophila, which may explain why competence went undetected for over 20 years (47). In addition to AA100, a number of laboratories use derivatives of L. pneumophila Philadelphia-1, a strain not previously described as competent. While determining whether Philadelphia-1 derivatives could be transformed using the conditions described for AA100, we discovered that the wild-type laboratory strain Lp02 was able to incorporate a marker at efficiencies comparable to those previously reported for AA100 (data not shown).
With the goals of facilitating genetic screens and enabling high-throughput genomic analysis using natural transformation, we reexamined the published transformation protocol in order to potentially improve its efficiency. First, we generated a set of reporter plasmids and Lp02-derived strains with which to easily assay transformation. The reporter plasmids contained several kilobases of Lp02 DNA interrupted with a drug resistance marker that could be selected for in L. pneumophila (Fig. 1A). The reporter strains contained an insertion with a different drug resistance marker in the corresponding region of the chromosome (Fig. 1B). The site of insertion is in a presumably neutral location immediately downstream of the housekeeping gene purH, well upstream of the virulence gene dotU (44, 50). This system of reporters allowed DNA transformation to be assayed not only by uptake of the marker on the plasmid but also by the concomitant loss of the linked chromosomal drug marker.
FIG. 1.
Reporter plasmids and strains used to assay Lp02 competence. (A) Reporter plasmids used for transforming L. pneumophila were constructed by first cloning a 3-kb piece of L. pneumophila DNA into pBluescript II KS+ to generate plasmid pJB955. Three pJB955 derivatives (pJB1389, pJB964, and pJB957) were constructed by cloning a gentamicin, chloramphenicol, or kanamycin resistance cassette, respectively, into the unique HindIII site in the 3-kb insert. (B) Two Lp02-derived reporter strains were used to assay natural competence. JV1103, a Cmr-marked version of Lp02, was constructed by natural transformation using plasmid pJB964. JV1160, a Kanr-marked version of Lp02, was constructed by natural transformation using plasmid pJB957. With these strains and plasmids, successful recombination can be assessed not only by gain of a drug resistance marker from the transforming reporter plasmid but also by loss of a drug resistance marker from the reporter strain.
Using these reporter plasmids and strains, we attempted to develop an assay that would be easy to perform and would potentially yield a higher overall frequency of transformation. We made several changes from the published protocol based on the following observations from the literature. First, maximal expression of the L. pneumophila type IV pilus biogenesis genes, which are required for natural competence, occurs at 30°C (30). Second, many competent species can be transformed on a solid surface rather than in liquid broth, making manipulations simpler and less prone to contamination. Using these observations, we developed a plate assay for Lp02 transformation wherein the Kanr reporter plasmid pJB957 was mixed with a small amount of the Cmr-marked Lp02 strain JV1103 and incubated for 2 days before transformants were selected for on media containing kanamycin.
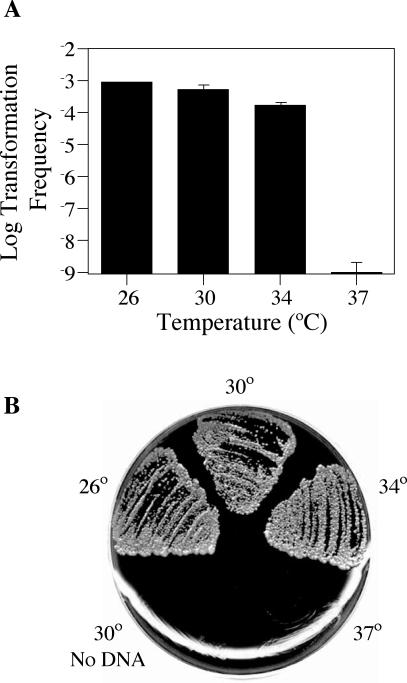
To optimize this protocol, identical transformation reactions were performed at various temperatures for several days until a patch of growth was apparent. The cells were swabbed into water, serially diluted, and plated on media selective for the reporter plasmid as well as nonselective media to determine total numbers of bacteria. We found that the Cmr Lp02 derivative strain JV1103 could be transformed with 200 ng of the Kanr reporter plasmid pJB957 at rates approaching 1 transformant per 1,000 total cells (Fig. 2), which was equivalent to ∼107 transformants per μg of DNA. This level of transformation was approximately 100-fold higher than what Stone and Abu Kwaik found for strain AA100 (47) or what we obtained for strain Lp02 using their published protocol (data not shown). We found that transformation of Lp02 required a temperature between 26 and 34°C, with maximal transformation occurring at 26°C (Fig. 2). Reactions at 37°C yielded no Kanr transformants, similar to assays performed in the absence of transforming DNA. Because L. pneumophila growth at 30°C is more than twice as fast as growth at 26°C, assays were henceforth performed at 30°C. Transformation at this temperature occurred with high fidelity, since 97% of the transformants were the result of homologous recombination at the intended locus resulting in gene replacement. These results suggest that transformation of the L. pneumophila strain Lp02 occurs with high fidelity and is regulated by temperature, consistent with the known temperature regulation of L. pneumophila type IV pilus expression.
FIG. 2.
Natural transformation of Lp02 is temperature dependent. (A) Transformation frequency as a function of growth temperature. L. pneumophila reporter strain JV1103 was grown on solid medium at 26, 30, 34, and 37°C in the presence of 200 ng of DNA from the reporter construct pJB957 (Kanr). After 48 h for the 30, 34, and 37°C samples or 120 h for the 26°C sample, bacteria were plated on selective media versus nonselective media in order to determine the number of Kanr transformants in the total cell population. The limit of detection for transformation frequency was 10−9; error bars indicate standard deviations. (B) Plate photograph of Kanr L. pneumophila transformants. Approximately 106 cells from each transformation reaction represented in panel A were plated on media containing kanamycin.
Characterization of competence in strain Lp02: DNA source and minimum length of homology.
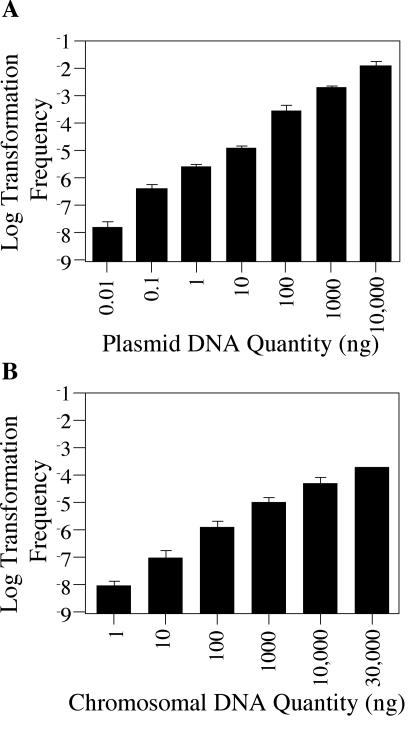
Previously it was shown that AA100 could be transformed both by L. pneumophila chromosomal DNA and by L. pneumophila DNA contained on a plasmid (47). To determine if the 30°C plate transformation assay was comparable in this regard, the effect of DNA source and quantity on the transformation frequency of Lp02 was examined. Similar to AA100, Lp02 could be transformed with either chromosomal DNA or plasmid DNA. For both plasmid and chromosomal DNA, a linear relationship between DNA quantity and transformation frequency was observed (Fig. 3), with average values of 3 × 107 transformants per μg of plasmid DNA and 4 × 104 transformants per μg of chromosomal DNA. Transformants could be detected using as little as 10 pg of plasmid pJB957, and frequencies of up to 1 in 100 were obtainable with 10 μg of plasmid DNA (Fig. 3A). Interestingly, this amount of plasmid DNA did not appear to be saturating. Similarly, as much as 30 μg of chromosomal DNA from the Kanr strain JV1160 was not sufficient to saturate the reaction (Fig. 3B). Since comparison of the same mass of plasmid and chromosomal DNA was equivalent to using a 1,000-fold molar excess of plasmid DNA, the overall transformation frequencies for plasmid and chromosomal DNA are actually quite similar. Therefore, L. pneumophila is able to take up the two substrates with approximately equal efficiencies.
FIG. 3.
The frequency of Lp02 transformation is dependent on DNA source and quantity. (A) Transformation frequency as a function of plasmid DNA quantity. Reporter strain JV1103 was grown on solid media at 30°C in the presence of various quantities of pJB957 (Kanr) plasmid DNA. After 48 h, bacteria were plated on selective media versus nonselective media in order to determine the number of kanamycin-resistant transformants in the total cell population. (B) Transformation frequency as a function of chromosomal DNA quantity. Transformation was assayed as in panel A using JV1160 (Kanr) chromosomal DNA instead of plasmid DNA. The limit of detection for transformation frequency was 10−9; error bars indicate standard deviations.
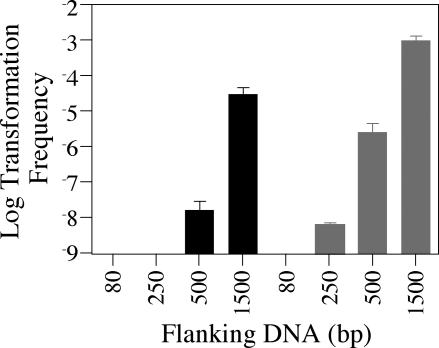
A second predicted parameter of transformation is the length of DNA available for homologous recombination. In order to identify the minimum length of homologous DNA sufficient for transformation via natural competence, a series of constructs was made that contained various amounts of L. pneumophila DNA flanking a selectable marker (Kanr). We were unable to detect transformation with linear DNA fragments containing 80 or 250 bp of flanking homologous sequence but could detect transformation with linear fragments containing 500 or 1,500 bp of flanking sequence (Fig. 4). When the DNA fragments were present on a closed circular plasmid, transformants were then obtainable with only 250 bp of flanking sequence. In addition, the use of circular rather than linear DNA increased the overall frequency of transformation 10- to 100-fold when 500 or 1,000 bp of flanking sequence was used (Fig. 4), presumably due to increased protection of the circular form from degradation.
FIG. 4.
The frequency of Lp02 transformation is dependent on the amount of homologous DNA available for recombination. The figure shows transformation frequency as a function of the length of homologous DNA flanking a selectable marker. Reporter strain JV1103 was grown on solid media at 30°C in the presence of 1 μg of a DNA fragment consisting of a Kanr marker flanked by 80, 250, 500, or 1,500 bp of L. pneumophila DNA on either side. Fragments were used as linear molecules (black bars) or were present as part of a plasmid (gray bars). After 48 h, bacteria were plated on selective media versus nonselective media in order to determine the number of kanamycin-resistant transformants in the total cell population. The limit of detection for transformation frequency was 10−9; error bars indicate standard deviations.
Characterization of competence in strain Lp02: kinetics of DNA uptake.
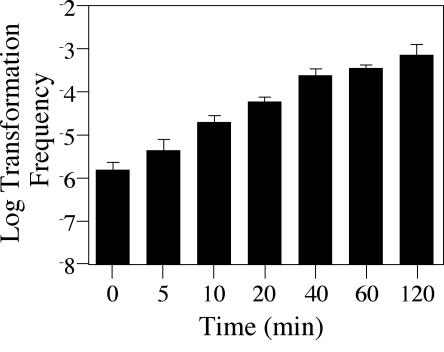
Another characteristic of natural competence is that uptake of DNA is thought to occur rapidly. DNA is first bound to the bacterial cell and then transported across the cell wall into the cytoplasm, where one strand is integrated onto the bacterial chromosome (20, 25). Because incoming DNA is converted into a DNase-protected state prior to integration, the rate of uptake can be easily measured. To assess the rapidity with which Lp02 can bind and protect transforming DNA, we switched to a 30°C liquid assay where a broth-grown exponential JV1103 culture was incubated with the reporter plasmid pJB957 for various amounts of time. The reaction was terminated by addition of DNase I, which cleaves any free, unprotected DNA (see Materials and Methods). DNA uptake into JV1103 was extremely rapid since addition of DNase immediately after exposure to DNA still resulted in detectable transformants (Fig. 5). As expected, levels of transformation increased linearly with time of exposure to DNA.
FIG. 5.
Kinetics of transformation in a broth assay. The figure shows transformation frequency as a function of time. Reporter strain JV1103 was grown in AYE at 30°C with low-speed shaking to early exponential phase. One-milliliter aliquots were removed, and 1 μg of reporter construct pJB957 (Kanr) DNA was added for 0, 5, 10, 20, 40, 60, or 120 min, followed by a 15-min incubation at 37°C with DNase I at 0.1 mg/ml. Bacteria were plated on selective media in order to determine the number of kanamycin-resistant transformants in the total cell population. The limit of detection for transformation frequency was 10−8; error bars indicate standard deviations.
Characterization of competence in strain Lp02: growth phase regulation.
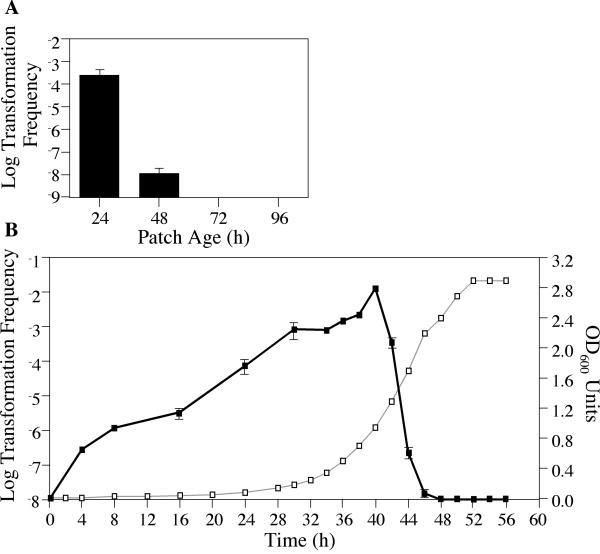
To further understand the requirements for transformation of Lp02 at 30°C, we tested whether the growth phase of the bacteria was important. A relationship between transformation frequency and growth phase has been described for many naturally competent bacteria (4, 37, 39, 48). To broadly assess whether such a relationship exists in L. pneumophila, we performed a variation on the standard plate assay, letting the bacteria grow at 30°C for 24 to 96 h before adding pJB957 DNA for 4 h followed by plating on selective media. Typically, L. pneumophila streaked heavily onto a plate will form a light patch after 24 h of incubation that roughly corresponds to an exponentially growing culture. In contrast, 48 h of growth will result in a dense patch that is close to saturation. By varying the time prior to addition of DNA, we observed that a 24-h patch could be transformed several logs more efficiently than a 48-h patch. In contrast, transformation was not detectable when DNA was added to a 72- or a 96-h patch (Fig. 6A). Thus, competence for DNA uptake on plates appears to occur during early rather than late stages of L. pneumophila growth.
FIG. 6.
The frequency of Lp02 transformation is dependent on bacterial growth phase. (A) Transformation frequency as a function of patch age. Reporter strain JV1103 was grown on solid media at 30°C for 24, 48, 72, or 96 h. One microgram of reporter construct pJB957 (Kanr) DNA was added and incubated for an additional 4 h, and then cells were plated on selective versus nonselective media in order to determine the number of kanamycin-resistant transformants in the total cell population. The limit of detection for transformation frequency was 10−9; error bars indicate standard deviations. (B) Transformation frequency and growth phase as a function of time. Reporter strain JV1103 was grown to stationary phase, back-diluted into fresh AYET medium, and cultured at 30°C with gentle shaking to stationary phase. Periodically, 1-ml aliquots were removed, the optical density was measured, and a portion of the cells were incubated for 2 h with 1 μg of reporter construct pJB957 DNA. These bacteria were then plated on selective versus nonselective media in order to determine the number of transformants in the total cell population. The optical density of the culture at a given time is indicated by open squares connected by a gray line, and the scale is shown on the right-hand y axis. The log transformation frequency of the culture at a given time is indicated by filled squares connected by a black line, and the scale is shown on the left-hand y axis. The limit of detection for transformation frequency was 10−8; error bars indicate standard deviations.
To gain a more precise representation of the relationship between growth phase and transformation frequency, assays were performed using a liquid culture. Fresh medium was inoculated with cells from a stationary-phase culture of JV1103 and incubated at 30°C with gentle shaking. Periodically, cells were removed, the optical density of the culture was measured, and 1 μg of DNA was added. The cells and DNA were incubated for 2 h at 30°C with shaking prior to plating on selective medium to assay competence. Consistent with the previous experiment, the frequency of transformation steadily increased during the early exponential phase of growth, peaking at mid-exponential phase. It then dropped precipitously, decreasing to below detectable limits upon entrance into stationary phase (Fig. 6B). Thus, Lp02 competence is not only temperature but also growth phase regulated in both the plate and liquid assays.
Growth phase regulation of natural competence in Lp02 is not due to quorum sensing.
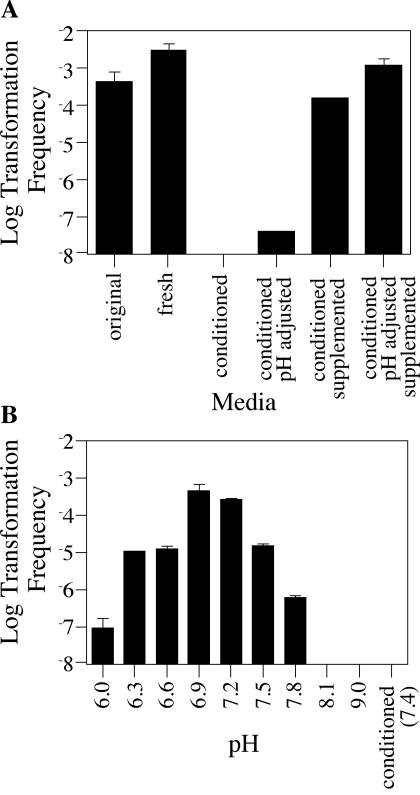
Lp02 is maximally competent during early exponential phase and completely nontransformable in late exponential and stationary phase. This growth phase regulation would be consistent with a quorum sensing mechanism, which is commonly used by microorganisms. For example, B. subtilis and S. pneumoniae regulate competence in a growth phase-dependent manner by sensing the presence of secreted peptide pheromones in the medium (21, 46). To determine if L. pneumophila is regulating competence via quorum sensing, cells were removed from an exponential-phase liquid culture, gently pelleted, resuspended in fresh medium or in conditioned medium from a stationary-phase JV1103 culture, and exposed to pJB957 DNA and the transformation frequency was determined. Addition of fresh medium had no effect on the transformability of exponential cells, whereas addition of conditioned medium completely abolished transformation, consistent with the presence of a factor used for quorum sensing (Fig. 7A).
FIG. 7.
Frequency of transformation versus pH and nutrient availability. (A) Transformation frequency as a function of medium source. Repression of competence by conditioned media can be reversed by addition of nutrients and adjustment of pH. Reporter strain JV1103 was grown in AYET at 30°C with shaking to early exponential phase. One-milliliter aliquots were removed, and cells were pelleted and resuspended in 1 ml of the original medium (pH 6.8), fresh medium (pH 6.8), conditioned medium (pH 7.2), conditioned medium with the pH adjusted (pH 6.8), conditioned medium supplemented with nutrients (pH 7.2), or conditioned medium with the pH adjusted and supplemented with nutrients (pH 6.8) and incubated at 30°C with shaking for 2 h. One microgram of pJB957 (Kanr) DNA was added, and cells were further incubated for 2 h. Bacteria were plated on selective media in order to determine the number of transformants in the total cell population. (B) Transformation frequency as a function of medium pH. Strain JV1103 was grown in AYE at 30°C with low-speed shaking to early exponential phase. One-milliliter aliquots were removed, and cells were gently pelleted and resuspended in 1 ml of AYET at pH 6.0, 6.3, 6.6, 6.9, 7.2, 7.5, 7.8, 8.1, or 9.0 or in conditioned AYET (pH 7.4) and incubated at 30°C with shaking for 2 h. One microgram of reporter construct pJB957 (Kanr) DNA was added, and cells were further incubated for 2 h. Bacteria were plated on selective media in order to determine the number of kanamycin-resistant transformants in the total cell population. The limit of detection for transformation frequency was 10−8; error bars indicate standard deviations.
However, the inhibitory effect of conditioned medium could also be due to changes caused by bacterial growth. To examine whether pH changes in the medium could alter the transformability of L. pneumophila, cells were removed from an exponential-phase liquid culture, gently pelleted, resuspended in fresh medium varying from pH 6.0 to 9.0 for 2 h, and then assayed for competence. Normal AYE-thymidine (AYET) is buffered to a pH of 6.9, and Lp02 was seen to be maximally transformable at this pH (Fig. 7B). Not surprisingly, extreme pHs of above 8 or below 6.3 had a pronounced inhibitory effect on transformation. However, conditioned medium had an average pH of 7.4, and this pH was not significantly inhibitory when fresh medium was used (Fig. 7B), suggesting that changes in medium pH due to bacterial growth were not primarily responsible for the inhibitory effect of conditioned medium. Moreover, adjusting the pH of conditioned medium from 7.4 to 6.9 was not sufficient to eliminate competence repression (Fig. 7A), suggesting the presence of another factor in conditioned medium which regulates competence.
In addition to changes in pH, conditioned medium is significantly different from fresh medium due to a depletion of nutrients. To determine whether a decrease in nutrient availability might also contribute to competence regulation, we exposed exponential-phase bacteria to conditioned medium versus conditioned medium supplemented with yeast extract (see Materials and Methods). The addition of nutrients greatly reduced the effects of conditioned medium on competence, though it did not completely eliminate them (Fig. 7A). However, when the pH of conditioned medium was also adjusted, the frequency of transformation for bacteria exposed to this medium was very similar to that of bacteria exposed to fresh medium. Thus, growth phase-dependent regulation of competence in L. pneumophila strain Lp02 is not due to the presence of a secreted factor detected by quorum sensing. Instead, it is most likely due to changes in nutrient availability and pH.
Lp02 is unique among several serogroup 1 strains in its ability to be transformed on solid medium.
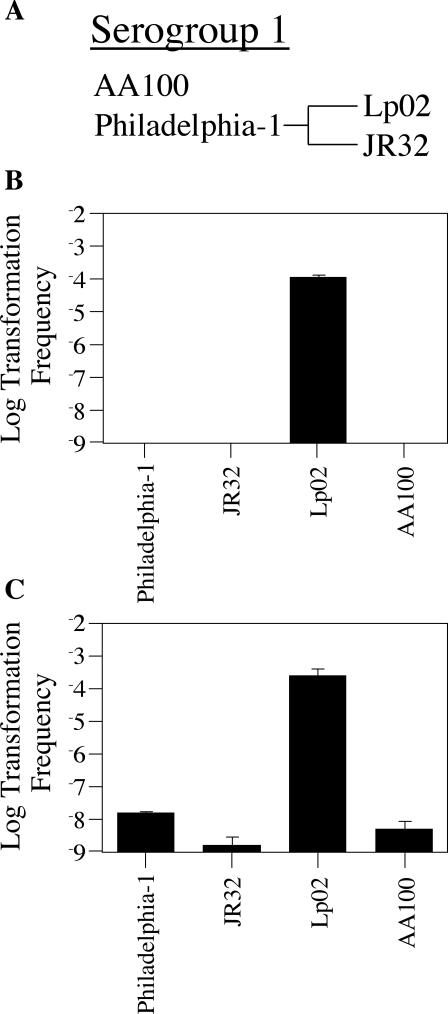
To determine if the ability to be transformed on plates at 30°C was specific to Lp02, a number of commonly used L. pneumophila strains were examined under these conditions. In contrast to our findings with strain Lp02, no transformants could be detected with the serogroup 1 strain AA100 (Fig. 8B), even though it was previously shown to be competent at 37°C under microaerophilic conditions (47). JR32, a strain closely related to Lp02, also did not appear to be competent by the plate assay. Even L. pneumophila Philadelphia-1, the progenitor strain for both Lp02 and JR32, was not transformed via the plate assay (Fig. 8B). Nevertheless, each of these strains was capable of being transformed when assayed for competence by a 30°C liquid assay (Fig. 8C), although Philadelphia-1, JR32, and AA100 were transformed at frequencies reduced by at least 10,000-fold compared to that for Lp02. Thus, strain Lp02 possesses enhanced transformability in these assays and can be described as hypercompetent.
FIG. 8.
Lp02 is significantly more competent than other L. pneumophila serogroup 1 strains. (A) Relationship among the three commonly studied L. pneumophila serogroup 1 strains Lp02, JR32, and AA100. Strains Lp02 and JR32 are derivatives of the same parent strain, L. pneumophila Philadelphia-1. AA100 is a derivative of an unrelated serogroup 1 strain, L. pneumophila Wadsworth. (B) Transformation frequency (plate assay) as a function of strain origin. Four L. pneumophila serogroup 1 strains were assayed using the 30°C plate assay. Strains were grown on solid media at 30°C in the presence of 1 μg of reporter construct pJB957 (Kanr) plasmid DNA. After 48 h, bacteria were plated on selective versus nonselective media in order to determine the number of kanamycin-resistant transformants in the total cell population. The limit of detection for transformation frequency was 10−9; error bars indicate standard deviations. (C) Transformation frequency (broth assay) as a function of strain origin. Four L. pneumophila serogroup 1 strains were assayed using the 30°C liquid assay. The four strains were inoculated into AYET broth to an OD600 of 0.02 and cultured at 30°C with low-speed shaking (100 rpm) in the presence of 1 μg of reporter construct pJB957 (Kanr) plasmid DNA. After 72 h, bacteria were plated on selective versus nonselective media in order to determine the number of kanamycin-resistant transformants in the total cell population. The limit of detection for transformation frequency was 10−9; error bars indicate standard deviations.
Identification of regulators of competence.
Since Lp02 originated from L. pneumophila Philadelphia-1, and neither Philadelphia-1 nor its derivative JR32 displays the level of competence that Lp02 does, we reasoned that the hypercompetence phenotype might have been fortuitously acquired during the derivation of Lp02 from Philadelphia-1. Lp02 was generated by isolating three independent mutations sequentially in L. pneumophila Philadelphia-1. These include spontaneous mutations conferring resistance to streptomycin, a dependence on thymidine supplementation, and a lack of a functional restriction-modification system (3). It is possible that Lp02 acquired mutations in addition to the desired ones while the strain was being passaged. For instance, Lp02 might have lost a factor that normally regulates competence under laboratory conditions, resulting in the hypercompetence phenotype.
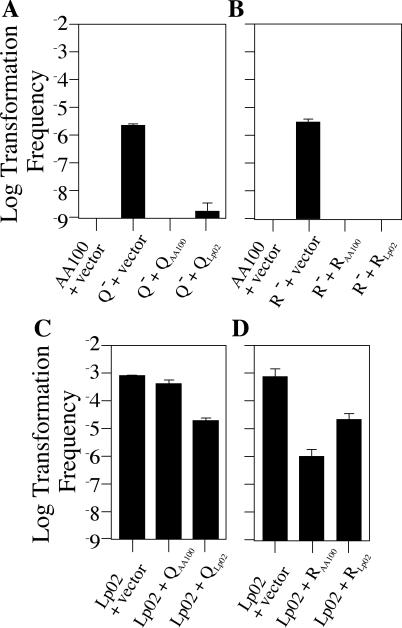
To test this possibility, we attempted to identify an inhibitor of natural competence by mutagenizing AA100 with a mini-Tn10 transposon and then selecting for mutant strains that exhibited competence at 30°C on plates (see Materials and Methods). By this approach, two independent mutants, JV1729 and JV1727, were isolated that rendered AA100 amenable to plate transformation although at frequencies 100-fold lower than that of Lp02 (Fig. 9). The first mutant, JV1729, was found to contain an insertion in a gene with homology to the proQ gene of E. coli. E. coli ProQ has been proposed to be a regulator of the osmoprotectant pump ProP (11, 26). The second mutant, JV1727, contained an insertion in a gene which encodes a protein predicted to contain a helix-turn-helix, raising the possibility that it might be a transcriptional regulator. We have named this second gene comR, for competence regulator. In both cases, the transposon insertions were solely responsible for the hypercompetence phenotype, since movement of the insertion into an unmutagenized version of AA100 recapitulated the hypercompetence phenotype in the original strain (data not shown).
FIG. 9.
Inactivation of proQ or comR induces competence of strain AA100. (A) Transformation frequency and complementation of strain AA100 containing a proQ mutation. AA100 containing the empty vector pJB1653 (AA100 + vector), AA100 proQ::mini-Tn10kan (JV1729) containing the empty vector pJB1653 (Q− + vector), JV1729 containing the AA100 proQ complementing clone pJB1659 (Q− + QAA100), and JV1729 containing the Lp02 proQ complementing clone pJB1661 (Q− + QLp02) were assayed for the ability to take up pJB964 (Cmr) DNA by the 30°C plate assay. (B) Transformation frequency and complementation of strain AA100 containing a comR mutation. AA100 containing the empty vector pJB1653 (AA100 + vector), AA100 comR::mini-Tn10kan (JV1727) containing the empty vector pJB1653 (R− + vector), JV1727 containing the AA100 comR complementing clone pJB1665 (R− + RAA100), and JV1727 containing the Lp02 comR complementing clone pJB1663 (R− + RLp02), were assayed for the ability to take up pJB964 DNA by the 30°C plate assay. (C) Transformation frequency of strain Lp02 in the presence of proQ complementing clones. Lp02 containing either the empty vector pJB1653 (Lp02 + vector), the AA100 proQ complementing clone pJB1659 (Lp02 + QAA100), or the Lp02 proQ complementing clone pJB1661 (Lp02 + QLp02) was assayed for the ability to take up pJB964 DNA by the 30°C plate assay. (D) Transformation frequency of strain Lp02 in the presence of comR complementing clones. Lp02 containing either the empty vector pJB1653 (Lp02 + vector), the AA100 comR complementing clone pJB1665 (Lp02 + RAA100), or the Lp02 comR complementing clone pJB1663 (Lp02 + RLp02) was assayed for the ability to take up pJB964. In each transformation reaction, bacteria were exposed to 1 μg of reporter plasmid DNA for 48 h and then plated on selective versus nonselective media in order to determine the number of chloramphenicol-resistant transformants in the total cell population. The limit of detection for transformation frequency was 10−9; error bars indicate standard deviations.
Comparison of AA100 and Lp02 proQ.
As proQ appears to be a repressor of the competent state in AA100, we reasoned that Lp02 hypercompetence might be due to its inactivation. To test whether the Lp02 proQ gene is functional, we constructed proQ complementing clones from both Lp02 and AA100 DNA, where proQ gene expression is driven by an exogenous promoter. Each clone was transformed into strain JV1729 (AA100 proQ::mini-Tn10kan) in order to check its ability to repress competence. We found that the presence of either Lp02 or AA100 proQ resulted in nearly full repression of JV1729 competence, indicating that each gene could complement the loss of proQ and restore competence inhibition (Fig. 9A). Thus, both Lp02 and AA100 proQ complementing clones appear to encode functional proteins.
Although the Lp02 proQ gene can encode a functional protein in our complementation studies, it is possible that Lp02 does not express sufficient levels of ProQ. For example, Lp02 may contain a proQ promoter mutation or may have lost a positive regulatory factor. To determine if the presence of the proQ complementing clones inhibits Lp02 hypercompetence, we introduced them into this strain and assayed competence. In contrast to JV1729, complete loss of Lp02 hypercompetence in the presence of either clone was not observed (Fig. 9C), suggesting that hypercompetence is not simply due to loss of ProQ activity.
Comparison of AA100 and Lp02 comR.
Similar to the case for proQ complementation, the comR insertion strain JV1727 (AA100 comR::mini-Tn10kan) transformed with comR from either Lp02 or AA100 exhibited full competence repression, indicating that both comR complementing clones are functional (Fig. 9B). The presence of comR in strain Lp02 did not fully repress its ability to take up DNA, indicating that Lp02 hypercompetence is not solely due to loss of comR (Fig. 9D). However, a partial decrease in Lp02 transformation frequency was observed in the presence of both comR complementing clones, consistent with a regulatory role for this gene in both AA100 and Lp02 strains.
DISCUSSION
While examining the competence phenotype of the Philadelphia-1 derivative strain Lp02, we discovered that it displays hypercompetence under certain laboratory conditions. Transformation experiments were initially based on previous work demonstrating that a different L. pneumophila serogroup 1 strain, AA100, was able to take up DNA at 37°C under microaerophilic conditions (47). By varying the published protocol, we discovered that Lp02, in contrast to AA100, was highly competent at 30°C with aerobic growth. In an attempt to identify the genetic basis of hypercompetence, we isolated mutant strains of AA100 that exhibit enhanced DNA uptake. Characterization of these mutants revealed two genes, comR and proQ, that repress competence in L. pneumophila.
One striking characteristic of Lp02 hypercompetence is the effect of temperature. Transformation occurs during aerobic growth on agar plates at 30°C, whereas no transformation is observed when the same assay is performed at 37°C. This temperature dependence is consistent with expression studies showing that genes encoding components of the L. pneumophila type IV pilus, which is required for transformation, are transcribed at 30 and not at 37°C (30, 47). It is curious that the transformation assays described by Stone and Abu Kwaik were performed at 37°C but clearly depended on the presence of type IV pili (47). A likely explanation for this is that the microaerophilic conditions of their assay induced pilus expression sufficient for transformation. Thus, oxygen availability and temperature are two factors that appear to regulate competence in L. pneumophila.
A linear relationship between plasmid DNA quantity and transformation frequency was observed for both Lp02 and AA100 (47). Whereas we were unable to identify saturating amounts of DNA for Lp02 transformation, Stone and Abu Kwaik found that 8 μg or more was sufficient to saturate AA100 transformation (47). In addition, we noted a correlation between transformation frequency and the length of the DNA substrate. Successful Lp02 transformation depended on the presence of at least 250 bp of flanking DNA and increased proportionally with the length of DNA.
We also discovered that Lp02 competence appears to be controlled by growth phase, with transformation being maximal during exponential growth and completely absent by the beginning of stationary phase. A number of other bacteria exhibit competence during exponential growth, including N. gonorrhoeae, Deinococcus radiodurans, Synechococcus species, and Chlorobium species (31). However, growth phase regulation of Lp02 competence most closely resembles that of A. calcoaceticus, which is maximally transformable in exponential phase and loses competence upon entry into stationary phase (39).
Repression of Lp02 competence during stationary phase results from nutrient depletion and an increase in pH, as the inhibitory effect of conditioned medium can be totally abolished by the addition of yeast extract and lowering of the pH. These data suggest that L. pneumophila does not rely on an extracellular signal molecule such as a quorum sensing autoinducer to regulate the competent state. Alternatively, as proposed for A. calcoaceticus, control of L. pneumophila competence may be under a growth phase-regulated promoter. Another striking example of growth phase regulation in L. pneumophila is the induction of virulence by entry into stationary phase (7, 19). Hammer and Swanson have shown that the stationary-phase induction of virulence traits can be mimicked by artificial expression of the stringent response gene ppGpp synthetase (relA) during exponential phase (19). Furthermore, they have shown that ppGpp production in L. pneumophila is stimulated by conditions of nutrient depletion—the same conditions seen to repress natural competence in this work. It will be important to determine if relA expression might repress competence and/or expression of the type IV pilus while inducing virulence.
An initial concern in using natural competence as a genetic tool was the fidelity of the recombination event, particularly because the transformation substrate was often uncut plasmid DNA. In B. subtilis and S. pneumoniae, incoming substrate DNA is cleaved, on average, into ∼13.5- or 6-kb fragments (14, 36) of which a single strand is transported across the bacterial membrane while its complement is degraded (14, 27, 34). Similar degradation is thought to occur in the gram-negative species N. gonorrhoeae and H. influenzae (8, 22), and it is likely that L. pneumophila modifies incoming DNA in the same fashion. Despite these modifications, however, reconstitution of plasmids taken up by competence machinery has been shown to occur (2). Though none of the plasmid substrates used in these studies can replicate in L. pneumophila, the possibility of generating plasmid integrants was a concern. Another concern was the possibility of spontaneous mutation resulting in a strain that might be falsely classified as a transformant. To address these issues, we developed a reporter system with which transformants could be quickly and easily checked for the appropriate recombination event. By this system, the percentage of true transformation for a range of experiments was calculated and found to average 97%. These findings are consistent with those of Stone and Abu Kwaik, who observed a high percentage of homologous transformation with strain AA100 (47). Judging from these results, it is reasonable to conclude that the fidelity of L. pneumophila transformation reactions is high and that the vast majority of natural competence transformants are legitimate.
The discovery that Lp02 is hypercompetent at 30°C compared to its progenitor strain Philadelphia-1 was surprising. The most likely explanation for this observation is that Lp02 was altered during its derivation and rendered hypercompetent. For example, it could have sustained a mutation in a competence regulatory gene resulting in up-regulation of an inducer of competence or loss of a repressor of competence. We favor the latter model since it is more likely and since Lp02 is already known to have sustained at least one deletion during its derivation, resulting in loss of the lvh locus (6, 42).
Based on this idea that competence is normally repressed in L. pneumophila, we attempted to mimic the hypercompetence phenotype of Lp02 in AA100 via gene disruption. The strain was mutagenized, and hypercompetent mutants were selected for, resulting in the identification of two potential L. pneumophila competence regulator genes, comR and proQ. The comR gene is predicted to encode a novel protein that contains a putative helix-turn-helix motif but has no significant homologies by BLAST search. The presence of a DNA binding motif suggests that this protein may be a novel transcriptional regulator controlling competence in L. pneumophila. The proQ gene encodes a protein with homology to the E. coli ProQ protein (26, 35), which functions as a positive regulator on the solute transporter protein ProP (10, 11, 26, 35). E. coli ProP functions in osmoprotection by transporting certain organic solutes such as proline and glycine betaine into cells to maintain a balance of osmotic pressure (reviewed in reference 54). In E. coli, proline uptake by ProP in response to a hypotonic environment is greatly impaired in a proQ mutant (10, 11, 26, 35). The fact that disruption of the L. pneumophila AA100 proQ homologue results in increased transformation frequencies suggests a possible relationship between osmolarity and competence regulation.
In order to determine if Lp02 hypercompetence was due to inactivation of proQ, we first tested whether the Lp02 version of this gene could complement the AA100 proQ mutant. The fact that the Lp02 proQ gene could complement indicates that it does not contain a mutation that destroys its activity. Furthermore, expression of proQ from either strain in Lp02 did not fully repress competence, suggesting that Lp02 is not a proQ mutant. Similar to the case with proQ, the comR gene from Lp02 could complement the corresponding AA100 mutant but could not fully repress Lp02 competence, indicating that Lp02 is also not a comR mutant. However, the presence of comR in Lp02 partially inhibited transformation, consistent with its functioning as a competence repressor. Thus, the hypercompetence phenotype of Lp02 does not appear to be due solely to a lesion in one of these regulatory factors and is instead likely caused by a mutation in some third, as yet unidentified, factor.
Natural competence for DNA transformation is an intriguing phenomenon which also has useful genetic applications. Considering that no transducing phage have been discovered for Legionella species, the addition of natural transformation to the genetic armament has been very beneficial. The wild-type laboratory strain Lp02 can be transformed very simply and efficiently, at rates of 107 transformants per μg of DNA. Although other commonly used L. pneumophila strains are not naturally competent under the conditions described here, inactivation of the proQ or comR gene could provide an easy method for increasing the competence of AA100, and possibly that of other L. pneumophila species. Further study of strain Lp02 and proQ and comR is likely to lend insight into the complex control pathways for expression of the competent state. Finally, our findings provide further evidence that the competent state is highly regulated and demonstrate that hypercompetence can be easily induced as the result of a single genetic lesion.
Acknowledgments
We thank Petra Levin, Patrick Bardill, and Carr Vincent for critical analysis of the manuscript; Steve DeLira and Jennifer Miller for construction of several plasmids and strains used in this study; and James Kirby for the generous gift of the mini-Tn10 plasmid pJK211-2.
J. A. Sexton was supported by the Washington University, Department of Internal Medicine, Infectious Diseases Training Grant #5 T32 AI07172-22. J. P. Vogel was supported by the Whittaker Foundation, the American Lung Association, and NIH grant AI48052-02.
REFERENCES
- 1.Abu Kwaik, Y., B. I. Eisenstein, and N. C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behnke, D. 1981. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol. Gen. Genet. 182:490-497. [DOI] [PubMed] [Google Scholar]
- 3.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 4.Bertolla, F., F. Van Gijsegem, X. Nesme, and P. Simonet. 1997. Conditions for natural transformation of Ralstonia solanacearum. Appl. Environ. Microbiol. 63:4965-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, G. D., T. Sox, E. Blackman, and P. F. Sparling. 1977. Factors affecting genetic transformation of Neisseria gonorrhoeae. J. Bacteriol. 129:983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brassinga, A. K., M. F. Hiltz, G. R. Sisson, M. G. Morash, N. Hill, E. Garduno, P. H. Edelstein, R. A. Garduno, and P. S. Hoffman. 2003. A 65-kilobase pathogenicity island is unique to Philadelphia-1 strains of Legionella pneumophila. J. Bacteriol. 185:4630-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., and S. A. Hill. 1998. Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae. J. Bacteriol. 180:5117-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claverys, J. P., and B. Martin. 2003. Bacterial “competence” genes: signatures of active transformation, or only remnants? Trends Microbiol. 11:161-165. [DOI] [PubMed] [Google Scholar]
- 10.Culham, D. E., J. Henderson, R. A. Crane, and J. M. Wood. 2003. Osmosensor ProP of Escherichia coli responds to the concentration, chemistry, and molecular size of osmolytes in the proteoliposome lumen. Biochemistry 42:410-420. [DOI] [PubMed] [Google Scholar]
- 11.Culham, D. E., B. Lasby, A. G. Marangoni, J. L. Milner, B. A. Steer, R. W. van Nues, and J. M. Wood. 1993. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J. Mol. Biol. 229:268-276. [DOI] [PubMed] [Google Scholar]
- 12.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 14.Dubnau, D., and C. Cirigliano. 1972. Fate of transforming DNA following uptake by competent Bacillus subtilis. Formation and properties of products isolated from transformed cells which are derived entirely from donor DNA. J. Mol. Biol. 64:9-29. [DOI] [PubMed] [Google Scholar]
- 15.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabay, J. E., and M. A. Horwitz. 1985. Isolation and characterization of the cytoplasmic and outer membranes of the Legionnaires' disease bacterium (Legionella pneumophila). J. Exp. Med. 161:409-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodgal, S. H., and R. M. Herriott. 1961. Studies on transformations of Haemophilus influenzae. J. Gen. Physiol. 44:1201-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graupner, S., and W. Wackernagel. 2001. Pseudomonas stutzeri has two closely related pilA genes (type IV pilus structural protein) with opposite influences on natural genetic transformation. J. Bacteriol. 183:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 20.Havarstein, L. S. 1998. Bacterial gene transfer by natural genetic transformation. APMIS Suppl. 84:43-46. [DOI] [PubMed] [Google Scholar]
- 21.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahn, M. E., and H. O. Smith. 1984. Transformation in Haemophilus: a problem in membrane biology. J. Membr. Biol. 81:89-103. [DOI] [PubMed] [Google Scholar]
- 23.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 24.Kong, L., K. J. Siranosian, A. D. Grossman, and D. Dubnau. 1993. Sequence and properties of mecA, a negative regulator of genetic competence in Bacillus subtilis. Mol. Microbiol. 9:365-373. [DOI] [PubMed] [Google Scholar]
- 25.Koomey, M. 1998. Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl. 84:56-61. [DOI] [PubMed] [Google Scholar]
- 26.Kunte, H. J., R. A. Crane, D. E. Culham, D. Richmond, and J. M. Wood. 1999. Protein ProQ influences osmotic activation of compatible solute transporter ProP in Escherichia coli K-12. J. Bacteriol. 181:1537-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacks, S., B. Greenberg, and M. Neuberger. 1974. Role of a deoxyribonuclease in the genetic transformation of Diplococcus pneumoniae. Proc. Natl. Acad. Sci. USA 71:2305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacks, S. A., and B. Greenberg. 2001. Constitutive competence for genetic transformation in Streptococcus pneumoniae caused by mutation of a transmembrane histidine kinase. Mol. Microbiol. 42:1035-1045. [DOI] [PubMed] [Google Scholar]
- 29.Lazazzera, B. A. 2000. Quorum sensing and starvation: signals for entry into stationary phase. Curr. Opin. Microbiol. 3:177-182. [DOI] [PubMed] [Google Scholar]
- 30.Liles, M. R., V. K. Viswanathan, and N. P. Cianciotto. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, C., and R. J. Redfield. 2000. Point mutations in a peptidoglycan biosynthesis gene cause competence induction in Haemophilus influenzae. J. Bacteriol. 182:3323-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, B., M. Prudhomme, G. Alloing, C. Granadel, and J. P. Claverys. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867-878. [DOI] [PubMed] [Google Scholar]
- 34.Mejean, V., and J. P. Claverys. 1984. Use of a cloned DNA fragment to analyze the fate of donor DNA in transformation of Streptococcus pneumoniae. J. Bacteriol. 158:1175-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milner, J. L., and J. M. Wood. 1989. Insertion proQ220::Tn5 alters regulation of proline porter II, a transporter of proline and glycine betaine in Escherichia coli. J. Bacteriol. 171:947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison, D. A., and W. R. Guild. 1973. Structure of deoxyribonucleic acid on the cell surface during uptake by pneumococcus. J. Bacteriol. 115:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nester, E. W., and B. A. D. Stocker. 1963. Biosynthetic latency in early stages of deoxyribonucleic acid transformation in Bacillus subtilis. J. Bacteriol. 86:785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pakula, R., and W. Walczak. 1963. On the nature of competence of transformable streptococci. J. Gen. Microbiol. 31:125-133. [DOI] [PubMed] [Google Scholar]
- 39.Palmen, R., P. Buijsman, and K. J. Hellingwerf. 1994. Physiological regulation of competence induction for natural transformation in Acinetobacter calcoaceticus. Arch. Microbiol. 162:344-351. [Google Scholar]
- 40.Palmen, R., A. J. Driessen, and K. J. Hellingwerf. 1994. Bioenergetic aspects of the translocation of macromolecules across bacterial membranes. Biochim. Biophys. Acta 1183:417-451. [DOI] [PubMed] [Google Scholar]
- 41.Purcell, M., and H. A. Shuman. 1998. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66:2245-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samrakandi, M. M., S. L. Cirillo, D. A. Ridenour, L. E. Bermudez, and J. D. Cirillo. 2002. Genetic and phenotypic differences between Legionella pneumophila strains. J. Clin. Microbiol. 40:1352-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sexton, J. A., J. S. Pinkner, R. Roth, J. E. Heuser, S. J. Hultgren, and J. P. Vogel. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186:1658-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3:178-185. [DOI] [PubMed] [Google Scholar]
- 45.Sikorski, J., N. Teschner, and W. Wackernagel. 2002. Highly different levels of natural transformation are associated with genomic subgroups within a local population of Pseudomonas stutzeri from soil. Appl. Environ. Microbiol. 68:865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solomon, J. M., R. Magnuson, A. Srivastava, and A. D. Grossman. 1995. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 9:547-558. [DOI] [PubMed] [Google Scholar]
- 47.Stone, B. J., and Y. Abu Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomasz, A., and R. D. Hotchkiss. 1964. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc. Natl. Acad. Sci. USA 51:480-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 51.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 52.Wang, Y., S. D. Goodman, R. J. Redfield, and C. Chen. 2002. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol. 184:3442-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ween, O., S. Teigen, P. Gaustad, M. Kilian, and L. S. Havarstein. 2002. Competence without a competence pheromone in a natural isolate of Streptococcus infantis. J. Bacteriol. 184:3426-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood, J. M., E. Bremer, L. N. Csonka, R. Kraemer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437-460. [DOI] [PubMed] [Google Scholar]