Abstract
Background
Projectin is a giant modular protein of Drosophila muscles and a key component of the elastic connecting filaments (C-filaments), which are involved in stretch activation in insect Indirect Flight Muscles. It is comparable in its structure to titin, which has been implicated as a scaffold during vertebrate myofibrillogenesis.
Methods
We performed immunofluorescence studies on Drosophila pupal tissue squashes and isolated myofibrils to identify the pattern of appearance and assembly for projectin and several other myofibrillar proteins, using both wild type and mutant fly stocks.
Results and conclusions
In the first step of assembly, projectin immunolocalization appears as random aggregates colocalizing with α-actinin, kettin and Z(210), as well as, F-actin. In the second step of assembly, all these proteins become localized within discrete bands, leading ultimately to the regularly spaced I-Z-I regions of myofibrils. This assembly process is not affected in myosin heavy chain mutants, indicating that the anchoring of projectin to the thick filament is not essential for the assembly of projectin into the developing myofibrils. In the actin null mutation, KM88, the early step involving the formation of the aggregates takes place despite the absence of the thin filaments. All tested Z-band proteins including projectin are present and are colocalized over the aggregates. This supports the idea that interactions of projectin with other Z-band associated proteins are sufficient for its initial assembly into the forming myofibrils. In KM88, though, mature Z-bands never form and projectin I-Z-I localization is lost at a later stage during pupal development. In contrast, treatment of adult myofibrils with calpain, which removes the Z-bands, does not lead to the release of projectin. This suggests that after the initial assembly with the Z-bands, projectin also establishes additional anchoring points along the thick and/or thin filaments. In conclusion, during pupation the initial assembly of projectin into the developing myofibril relies on early association with Z-band proteins, but in the mature myofibrils, projectin is also held in position by interactions with the thick and/or the thin filaments.
Keywords: Drosophila, muscles, IFM, myofibrillogenesis, Z-band, C-filaments, titin
Background
The giant protein, projectin, is found in all Drosophila muscle types, including the asynchronous Indirect Flight Muscles (abbreviated as IFMs) [1-8]. Projectin belongs to a protein family, which was originally based on the characteristics of the Caenorhabditis elegans protein, twitchin, and now also includes the Drosophila proteins D-titin, stretchin and the vertebrate protein, titin. The above are all giant, modular proteins composed predominantly of two repeated domains, called Ig (Immunoglobulin-C2) and Fn3 (Fibronectin III) motifs [9-16]. In IFMs, immunofluorescence data indicate that projectin is localized within the sarcomeric region encompassing the Z band and the two adjacent I band regions (called the I-Z-I region). Based on its estimated length of 0.1 μm [6], one molecule of projectin is long enough to be anchored within the Z-band, to extend over the I region and to overlap with the A-band [17,18]. The orientation of the molecule, however, is still unknown, even though it has been proposed, by analogy with titin's orientation, that projectin NH2-terminus is most likely embedded within the Z-band.
During myofibrillogenesis, specific protein interactions lead to the formation of the thin and thick filaments, the Z bands, as well as, their organization into a highly structured sarcomere. This process has been well studied in Drosophila melanogaster IFMs using combined electron microscopy, molecular and genetic approaches (reviewed in [19,20]). A time frame of thick and thin filaments, as well as Z band assembly is relatively well established [19,21]. At the start of pupation, most of the Drosophila larval muscles are histolyzed and new adult muscles need to be formed. In particular, the dorsal-ventral set of IFMs form de novo by fusions of myoblast from imaginal disks. On the other hand, the dorsal-longitudinal set of IFMs is build through the fusion of myoblasts with larval templates, which are the remnants of not-fully histolyzed larval muscles [22,23]. In the IFMs, microtubules and "fine filaments" appear by 32 hours After the Start of Pupation (abbreviated as ASP) [21]. By 42 hours ASP, initial myofibrils occur inside sleeves of microtubules and dense Z bodies are present, although still irregular. Thin and thick filaments are found interdigitated between the Z bodies but with still no true striation. Around 50 hours ASP, striated narrow myofibrils can be found throughout the muscle [21]. Within this time frame, the steps of projectin assembly are, however, still unknown.
In this paper, we describe the steps of projectin assembly into the developing myofibrils during the pupation period. The presented data are also correlated to the corresponding behavior of several other myofibrillar proteins, including actin, α-actinin, Z(210), kettin, and myosin heavy chain. The effects of IFM-specific null mutations affecting myosin heavy chain and actin proteins on projectin assembly into the myofibrils are also presented. The data are further correlated with the effects of calpain treatments on isolated adult IFM myofibrils. The implications on the assembly and interactions of the projectin protein are discussed.
Results
Pattern of myofibrillar proteins assembly during pupation
Pupal squashes were performed in approximately 6-hour intervals starting at 24 h ASP (see Methods [24]). The typical cell distribution and cell morphology obtained with the squash technique for the period from 30 hours to 48 hours ASP is presented in Figure 1 panels A, C and E. The corresponding immunofluorescence indicates that cells immunostaining for muscle proteins are large rounded cells (myosin, Z(210) and projectin in panels B, D and F respectively). This pattern is similar to the images obtained from frozen cryostat sections of pupae at the same stage (data not shown). Because many muscles besides IFMs are also forming at the same time, the identity of specific cells as early IFM myotubes was evaluated by immunolabeling for the Z(210) protein, which is only found in IFMs and the large cells of the Tergal depressor of trochanter (TDT or jump muscles) [3]. Cells expressing Z(210) were identified as early as 40 hours ASP. Many, but clearly not all, cells within a pupal squash stain positive for the Z(210) antibody (compare Figure 1C and 1D and arrows indicating Z(210) negative cells). As seen at higher magnification, the cells staining for the Z(210) protein are also multinucleated as shown by DAPI staining, (Figure 1G and 1I). Because of both the presence of the Z(210) protein and the multinucleated pattern, we consider these early cells as precursors for the IFMs or the large cells of the TDT.
Figure 1.
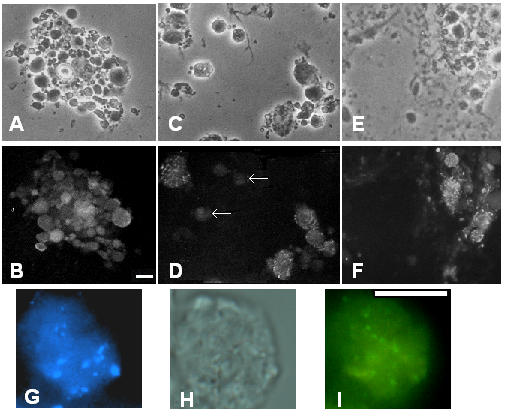
Early forming IFMs are multinucleated and expressed the Z(210) protein. Pupae were aged and after dissection, the tissues were spread and processed for immunofluorescence as described in Methods. Panels A-F: low magnification phase images (A, C and E) and corresponding immunofluorescence microscopy, panel B: myosin heavy chain, panel D: Z(210), and panel F: projectin. Arrows in panel D indicate cells negative for Z(210) staining. Panels G-I: Higher magnification image of a single cell. Panel G: DAPI nuclear staining; panel H: phase contrast; panel I: Z(210) immunostaining. DAPI indicates the presence of several nuclei within one cell. The apparent size difference for the various nuclei can be attributed to their different positions within the vertical focal plan of a single cell. As the same cell is represented in all three panels, the Z(210) staining insures that the cell shown is a multinucleated IFM precursor. Scale bars in B and I = 10 μm
At each stage tested, the presence and pattern for the following myofibrillar proteins were assessed: projectin, myosin heavy chain, α-actinin, kettin, Z(210) [3] and F-actin. Staining with projectin antibodies appears, by 30 hours, as few randomly dispersed aggregates throughout the cells' cytoplasm (data not shown). By 36–40 hours ASP, the projectin staining becomes brighter and is found within a greater number of aggregates (Figure 2A). The identity of the cells showing projectin in this aggregate pattern as early IFM myotubes was evaluated by immunolabeling for the Z(210) protein. As shown in Figure 2B, the cells where projectin is found in an aggregate pattern also show immunostaining for Z(210) which colocalizes over the same aggregates. Double immunofluorescence staining, between projectin and either α-actinin or kettin demonstrates that staining for the other Z-band-associated proteins can be aligned over the projectin's aggregates (Compare Figure 2E and 2F for projectin and kettin). Colocalization of other Z band proteins with the Z(210) protein was also obtained (Figure 2G and 2H for α-actinin and Z(210) respectively), again identifying these myotubes as the precursors of the IFMs or large cells of the TDT. The staining for F-actin obtained with FITC-labeled phalloidin shows that actin also colocalizes with these aggregates (compare Figure 2C and 2D for projectin and F-actin) consistent with the early association of growing thin filaments with the forming I-Z-I complex, which has been reported before [21]. These results are, therefore, consistent with the early anchoring of projectin to the I-Z-I complex during its assembly within the forming myofibrils.
Figure 2.
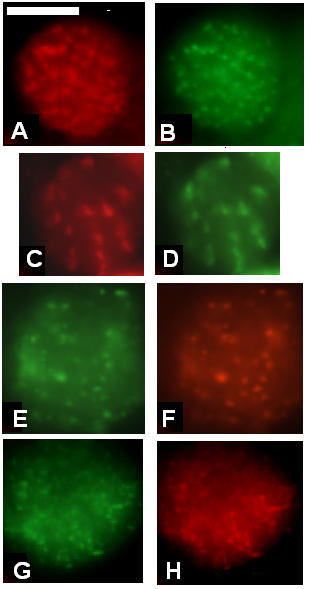
Projectin and other Z-band associated proteins show early colocalization during pupal myofibrillogenesis. Immunofluorescence microscopy images representing the staining pattern obtained at 36–40 hours ASP for projectin, several Z-band proteins, as well as F-actin. Pupae were aged and after dissection, the tissues were spread and processed for immunofluorescence as described in Methods. Corresponding left and right panels represent images from double immunostaining on the same cell. Panels A, C and E: projectin, panel B: Z(210), panel D: F-actin, panel F: kettin; panel G: α-actinin and panel H: Z(210). The five proteins follow a similar pattern of aggregates where I-Z-I proteins colocalize. Scale bar in A is for panels A through H and represents 10 μm.
For each protein tested, the same immunofluorescence pattern was observed when the different antibodies available against a particular protein were used (see Methods for list of antibodies). That the colocalizations are not an artifact of bleed through of the secondary antibodies between the two filters is supported by the data from single immunofluorescence where the aggregate pattern was observed individually for all proteins tested (data not shown). Also, when FITC conjugated secondary antibody is used alone, there is no fluorescence in the rhodamine channel and vice-versa and staining with either of the secondary antibodies alone is always negative (data not shown).
By 48 hours ASP, the staining for projectin, as well as kettin, α-actinin and Z(210) has changed to a pattern of ordered bands where all tested Z-band-associated proteins are colocalized (compare Figure 3A and 3B for projectin and α-actinin respectively). The identity of these cells as IFM was ensured by double staining with Z(210) as shown in Figure 3C and 3D (projectin and Z(210) respectively), as well as 3E and 3F (α-actinin and Z(210) respectively).
Figure 3.
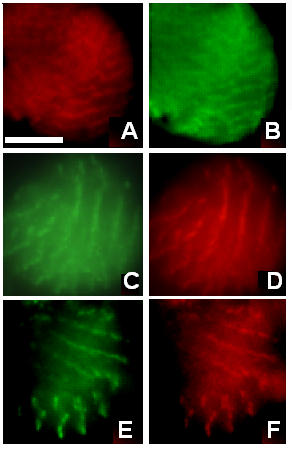
Projectin assembles within early Z-bands together with other I-Z-I associated proteins. Immunofluorescence microscopy images representing the staining pattern obtained at 48 hour ASP for several Z-band proteins. Pupae were aged and after dissection, the tissues were spread and processed for immunofluorescence as described in Methods. Corresponding left and right panels represent images from double immunostaining on the same cell. Panel A: projectin; panel B: α-actinin; panel C: projectin; panel D: Z(210); panel E: α-actinin, and panel F: Z(210). All tested Z-band proteins, including projectin, colocalize over discrete bands together with F-actin. Scale bar in A is for panels A through F and represents 10 μm.
The myosin staining was also evaluated in these two pupal stages and compared to the aggregate pattern obtained for the Z-band-associated proteins. At 40 hours ASP, myosin staining appears diffuse throughout the entire cytoplasm (Figure 4B). Double staining between projectin and myosin shows the presence in the same cells of the aggregate staining for projectin and a diffuse cytoplasmic distribution for myosin (compare Figure 4A and 4B). Even at a later stage (48 hours ASP), myosin is still found in a diffuse, random pattern with a stronger signal located around the plasma membrane (Figure 4D) whereas the double immunofluorescence with projectin shows the I-Z-I band localization (Figure 4C and 4D).
Figure 4.
Projectin assembles independently from myosin heavy chain. Immunofluorescence microscopy images representing the staining pattern obtained at 36–40 hours and 48 hours ASP for projectin and myosin heavy chain. Panels A and B represent the 36–40 hours ASP stage and panels C and D the 48 hours ASP stage. Panels A and C are stained for projectin and panels B and D for myosin. Myosin staining is still diffuse at 48 hours ASP whereas projectin shows the band pattern already described in Figure 3. Scale bar in B represents 10 μm for panels A through D.
By 60 hours ASP, a fully organized striated pattern is recognizable for all the tested proteins including myosin (data not shown).
Effects of mutations in myosin and actin on projectin assembly
Pupal squashes of several mutant stocks were performed at two different times during pupal development: approximately 40 hours and 48 hours asp. At both stages the presence and localization of several myofibrillar proteins were assessed as described above and compared to the wild type assembly pattern. These two stages were specifically chosen as they represent respectively the time point when aggregates are the most abundant and easier to visualize and the time point for early Z-band assembly as described above.
All three myosin mutations, Mhc7, Mhc10 and Mhc11, are IFM-specific null mutations and there is no staining with any anti-myosin antibody available to us, even when the images were purposefully overexposed (Figure 5B). In the same myosin-negative cell, projectin clearly shows a regular banding pattern (Figure 5A) similar to the one observed in wild type pupae (Figure 3A for example). As shown in Figure 5 for the Mhc7 allele, F-actin and projectin assemble following a pattern similar to the wild type, first as aggregates around 40 hour ASP (Figures 5C and 5D for projectin and F-actin respectively) and then as regularly spaced Z-bands after 48 hours ASP (Figures 5E and 5F for projectin and F-actin respectively). α-actinin and kettin also assemble following the wild type pattern (data not shown). The assembly data obtained for the Mhc10 and Mhc11 alleles are identical to the data presented in Figure 5 for Mhc7 (data not shown).
Figure 5.
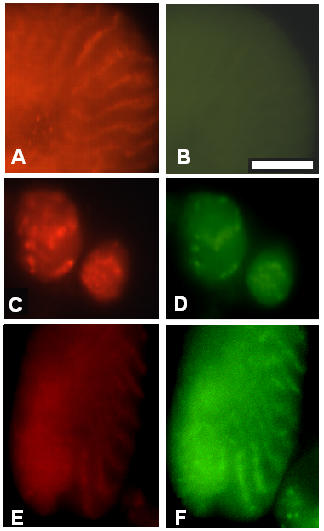
The assembly of I-Z-I associated proteins is not affected in myosin heavy chain Mhc7 mutant. Immunofluorescence microscopy images representing the staining pattern obtained in Mhc7 mutants at 36–40 hours ASP and the 48 hours ASP. Pupae from the Mhc7 mutant stock were aged and after dissection, the tissues were spread and processed for immunofluorescence microscopy as described in Methods. All left panels are stained for projectin, and all right panels correspond to the double staining on the same cell for one of the other tested proteins. Panel A, C and E: projectin; panel B: myosin heavy chain; panels D and F: F-actin (FITC-labeled phalloidin). Panels C and D are at 36–40 hours ASP stage; panels A, B, E and F are at the 48 hours ASP stage. The image in panel B is purposefully overexposed to demonstrate the absence of myosin staining. Projectin forms Z-bands together with the other I-Z-I associated proteins, even in the complete absence of myosin. Scale bar in B is for panels A through F and represents 10 μm.
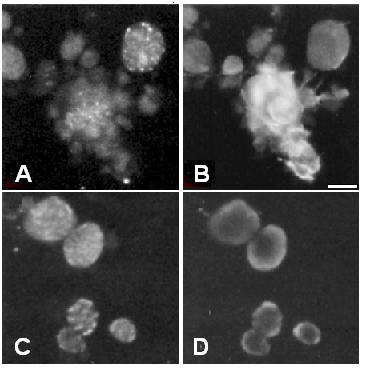
Two actin mutations were also selected, KM88 and E93K, which are both mutant alleles of the IFM-specific Act88F gene [25,26]. In the E93K mutant, actin still assembles in thin filaments but in the adult IFMs, the Z-bands are degenerated [26]. In this mutant allele, projectin assembles properly through the expected steps. At the 40-hour time point co-staining over aggregates is visible between projectin and actin (Figure 6A and 6B), as well as with kettin and α -actinin (data not shown). The staining changes by 48 hours ASP toward a Z-band pattern indistinguishable from the pupal wild type (Figure 6C and 6D). Therefore, the E93K actin mutation does not disrupt the early formation of Z-bands and the assembly of projectin during pupation. In adult E93K flies, IFM myofibrils can be prepared and immunofluorescence staining indicates that projectin is still present within the myofibrils (Figure 6E). By comparison to the immunostaining obtained for myosin (FITC/green staining in Figure 6F), projectin's localization over the I-Z-I region is further confirmed, as the two fluorescence patterns do not overlap. Therefore, even though Z-bands are degenerated in the E93K adult IFM myofibrils, the localization of projectin over the I-Z-I region is maintained.
Figure 6.
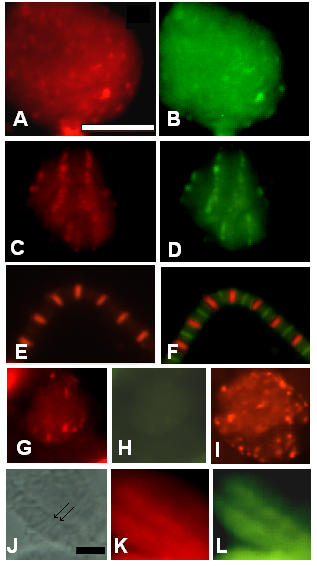
The assembly of all I-Z-I associated proteins is affected in the KM88 actin mutant. Immunofluorescence microscopy images representing the staining pattern obtained in the E93K and KM88 actin mutants at 36–40 hours and 48 hours ASP as well as in purified adult myofibrils. Pupae from the E93K and KM88 mutant stocks were aged and after dissection, the tissues were spread and processed for immunofluorescence microscopy as described in Methods. Myofibrils were prepared from adult E93K and processed for immunofluorescence microscopy as described in Methods. Panels A through F represent data for the E93K mutant. Panels A and B: 36–40 hours ASP pupae stained respectively for projectin and F-actin; panels C and D: 48 hours ASP pupae stained respectively for projectin and F-actin; panels E and F: E93K adult myofibrils stained respectively for projectin and myosin + projectin. Panels G through L show data for the KM88 mutant. Panels G and H: 36–40 hours ASP pupae stained respectively for projectin and F-actin; panel I: 36–40 hours ASP pupae stained for kettin. Panel J-L: 60 hours ASP pupae, with J showing the phase contrast image and panels K and L stained respectively for projectin and kettin. Arrows in J indicate two visible Z-bands. Projectin forms Z-bands together with the other I-Z-I associated proteins in the E93K mutant, but not in the KM88 mutant pupae. Scale bar in A is for panels A through I and represents 10 μm. Scale bar in J is for panels J through L and represents 10 μm.
In KM88 mutant flies, no actin is present in the IFMs, and in the adults both the thin filaments and the Z bands are absent [25,27]. As shown in Figure 6G for projectin, the early step involving the formation of the aggregates takes place in the KM88 mutants, whereas F-actin is not detectable even with an overexposed image (Figure 6H). The early colocalization is also observed for kettin (Figure 6I) and α-actinin (data not shown). The identity of these cells as IFM precursors was confirmed by the staining with an anti-Z(210) antibody, such that Z(210)-positive cells show aggregates but are negative for actin staining (data not shown). So, even in the absence of thin filaments, early aggregation of Z-band associated proteins takes place, and projectin is recruited within these aggregates. The next step involving the formation of early Z-bands is, however, not observed. In late pupae (50 hours and older) a striated pattern can be observed in some fibers under the phase contrast setting (Figure 6J). As shown in Figure 6K and 6L, these same muscles do not exhibit a recognizable striated Z-band pattern when stained with either projectin or kettin. Therefore, the final Z-band assembly for projectin, kettin and α-actinin does not take place in the KM88 mutant. Since IFM myofibrils cannot be prepared from adult KM88, we were not able to further assess by immunofluorescence the presence and localization of projectin in adult IFMs.
Analysis of calpain-digested myofibrils
Lakey et al. [28] demonstrated that, when purified IFM myofibrils are incubated in the presence of the protease, calpain, the Z-bands are preferentially digested as shown by the specific loss of α-actinin and kettin (see also Figure 7). We performed a similar analysis to assess the behavior of projectin in response to calpain treatment and its attachment sites within the myofibrils.
Figure 7.
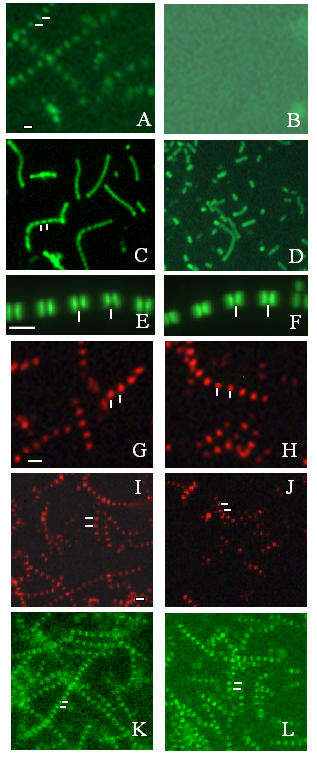
Projectin is not released from calpain-digested myofibrils. Immunofluorescence microscopy images representing the staining pattern obtained on purified adult IFM myofibrils with or without calpain digestion. All left panels represent untreated myofibrils, and all right panels correspond to myofibrils treated with 72 μg of calpain/slide. Myofibrils were processed for immunofluorescence microscopy as described in Methods. Panels A and B: kettin (MAC155); panels C and D: myosin (3E8); panels E and F: F-actin (FITC-labeled phalloidin); panels G and H: projectin (Core 1P); panels I and J: projectin (3b11); and panels K and L: projectin (P5). White lines mark the position of the Z-bands. projectin is not released upon calpain digestion of the Z-bands. Scale bar in A is for panels A through D and I through L and represents 10 μm; scale bar in E is for panels E and F and represents 10 μm, scale bar in G is for panels G and H and represents 10 μm.
Calpain-digested myofibrils were analyzed by immunofluorescence microscopy as described in Methods. Non-treated myofibrils were incubated in calpain activation buffer but without calpain for the same amount of time and condition as the treated myofibrils. When no calpain was added, all proteins tested (kettin, α-actinin, actin, myosin and projectin) are detected within their respective unchanged sarcomeric domains (Figure 7 panels A, C and E for kettin, myosin and F-actin respectively; Figure 7 panels G, I and K for projectin). Upon addition of calpain, all staining is lost for both kettin (Figure 7, Panel B) and α-actinin (not shown), and this result is consistent with previously published data [28]. Myofibrils, however, could still be detected under the phase contrast setting indicating that myofibrils devoid of Z bands are still present (not shown). Accordingly, A-band staining using a myosin antibody was still observed in calpain-treated myofibrils (Figure 7D). Similarly, F-actin visualization using FITC-labeled phalloidin staining does not give indication of any visible loss of F-actin upon calpain digestion (Figure 7F). Therefore, the data indicate that under the conditions used, detectable calpain digestion is limited to the Z band and that thin and thick filaments remain relatively untouched and in position. Another difference observed sometimes between the no calpain and the calpain-treated slides was that myofibrils tended to be smaller after calpain treatment (Figure 7C and 7D). These data were usually obtained using the protocol with a centrifugation step after the calpain treatment (see Methods). Later in the analysis, this step was omitted, and no significant decrease in myofibril length was observed after digestion. It is probable that the fibrils are more fragile after the calpain treatment and, therefore, tend to break apart more easily during the centrifugation step.
Calpain-digested myofibrils were then analyzed using five different projectin antibodies, namely MAC150, P5, Core1P, 3b11 and kinase (see Methods for description and origin of these antibodies). All preparations were double stained with an anti-kettin antibody to insure that calpain digestion has taken place (not shown). Projectin staining after calpain treatment is still present when projectin is detected using any of the five antibodies, as presented in Figure 7G,7H,7I,7J,7K,7L for P5, Core1P and 3b11 respectively. This indicates that the projectin molecule is not being released from the adult IFM myofibrils upon digestion of the Z-band. So, even at a concentration when all other tested Z-band proteins have been removed as shown by the immunofluorescence data and by gel analysis (data not shown), projectin is still within the myofibrils at its proper localization. These data are consistent with the existence of protein-protein interactions between projectin and the thick and/or thin filaments within the mature myofibrils, insuring physical integrity and strength.
Discussion
The pattern of Drosophila projectin appearance, localization and assembly was investigated during the early pupation period and compared to that of others Z-band associated proteins in both wild type and several mutant backgrounds.
By 36–40 hours ASP, double immunofluorescence staining for projectin and all Z-band proteins tested (kettin, α-actinin and Z(210)) shows colocalization over abundant, randomly distributed aggregates. The identity of these early myotubes as early IFMs was confirmed by the immunostaining for the Z(210) protein, which is an IFM/jump muscle-specific protein and can serve as a marker. DAPI staining also indicates that by 36–40 hours the cells are multinucleated, which is consistent with previous data indicating that fusion starts to occur around 24–30 hours into pupation [21]. This aggregate pattern, where all the Z-band proteins are colocalized, can be corroborated with the electron microscopy studies by Reedy and Beall [21] where they reported the presence of Z bodies by 42 hours ASP. The FITC-labeled phalloidin staining for F-actin is consistent with the previously described early association of growing thin filaments with forming Z-bands. By 48 hours ASP, the colocalization of projectin with α-actinin and kettin has evolved to a visible pattern of ordered lines or bands. The assembly data described here are, therefore, compatible with the localization of projectin within the I-Z-I region being driven during early myofibril formation by protein-protein interactions between projectin and the previously suggested I-Z-I complex [19,27]. Similarly, in some studies, vertebrate titin was found in periodically aligned spots together with α-actinin [29]. The interactions involved for Z-band targeting and the regions of projectin responsible for these bindings are still unknown, but an important role for projectin's terminal Ig domains can be suggested because the equivalent titin Ig domains have been implicated in the in vitro binding between titin and α-actinin [30,31] as well as the anchoring of titin to the Z-band [32,33]. Similarly, projectin PEVK-like domain [16] might be important for binding projectin to the thin filaments, as described for titin PEVK domain [34].
Double staining between projectin and myosin demonstrates the presence in the same cells of projectin's aggregates together with a diffuse cytoplasmic distribution for myosin. As reported by Reedy and Beall [21], myosin filaments appear within microtubule sleeves by 42 hours ASP, but these filaments do not show a characteristic sarcomeric pattern until later. It appears, therefore, that projectin's and myosin heavy chain's patterns of assembly differ markedly during pupation. The separate pathway identified for the myosin heavy chain protein is in agreement with the proposed scheme where thick filaments assemble somewhat independently from the I-Z-I complex and only later form the proper interactions to establish the final sarcomeric architecture [19,21,27]. This leaves open the question of whether projectin establishes interactions with the myosin thick filaments only later in development.
In light of this possibility, we follow projectin assembly in both actin and myosin heavy chain mutants [[14,26,27,35,36], reviewed in [20]]. In all three IFM-specific null Mhc mutants tested, all Z-band proteins, including projectin, as well as F-actin first assemble as aggregates and then later as regularly spaced Z-bands, indistinguishable from the wild type pattern. In the KM88 null actin mutant, no thin filaments assemble due to the lack of the IFM-specific actin isoform Act88F. In this mutant, α-actinin, kettin and projectin, however, still colocalize during early pupal development into aggregates similar to those observed in the wild type. The possibility exists that initial myofibrils may form on a scaffold of cytoskeletal actin. As this form of actin would still be present in the KM88 mutant, projectin assembly over the aggregates could depend on interactions with cytoskeletal actin. However, FITC-labeled phalloidin should also label cytoskeletal actin and the staining would appear in our KM88 preparations as either aggregates or a filamentous staining; we have never observed such a pattern. Therefore, in the absence of the F-actin, early aggregation of Z-band associated proteins takes place, including the assembly of projectin within those early structures. This indicates that, at an early stage, interactions of projectin with other Z-band-associated proteins are sufficient for its targeting to the I-Z-I region. Interactions with F-actin, which may normally take place, are not essential at least for the initial recruiting and anchoring of projectin into the I-Z-I complex. The next step involving the formation of banded structures is, however, not observed in the KM88 pupae. This probably reflects the need for F-actin in order to proceed with and maintain the formation of Z-bodies/bands. The mutant analysis data are, therefore, consistent with the initial localization of projectin being dependent primarily on its interactions with other Z-band associated proteins. The possibility of interactions with F-actin as part of the I-Z-I complex cannot be excluded, but the KM88 mutant data suggest that they are not essential for the association of projectin with other Z-band associated proteins. The association with the thick filaments would be a subsequent process independent from the assembly of the early I-Z-I complex.
To further characterize other anchoring points for the projectin protein, we carried out studies on adult IFM myofibrils treated with calpain. Data indicated that under the conditions used, detectable calpain digestion is limited to the Z-bands and that F-actin and thick filaments remain relatively untouched and in their correct position. Immunolocalization studies showed that, following calpain treatment, staining for projectin was still apparent, indicating that even at concentrations when all other tested Z-band proteins have been removed, projectin is not being released from the myofibrils and is still found at its proper localization. Earlier studies using Western analysis of calpain-digested myofibrils indicated that projectin appears smaller [28]. This observed truncation of projectin suggested that the Z-band domain of projectin is cleaved during calpain treatment. The fact that projectin is still present and correctly localized within the myofibrils following calpain digestion strongly suggests that projectin must be held in place by interactions with other components besides the Z band proteins, such as the thick and thin filaments. Binding of vertebrate titin to myosin is well established [37-40]. Data are also available to support titin/actin interactions [34,39,41,42]. The existence of the same kind of interactions between projectin and actin or myosin could explain how projectin is not released from the myofibrils even though the Z-bands have been degraded by the calpain treatment.
Drosophila projecti, as a key component of the elastic C-filaments in Insect Indirect Flight Muscles, becomes associated very early with the building blocks of the I-Z-I complex and these interactions are critical for the initial and correct assembly of projectin into the developing myofibril. Projectin's interactions with F-actin and the thick filaments are not, at least initially, essential for the association of projectin with the I-Z-I complex and its assembly into the myofibrils. These interactions are established later during pupal myofibrillogenesis, and in adult myofibrils they are strong enough to keep projectin localized within the myofibrils upon removal of Z bands.
Methods
Staged pupae collection and squashes
Pupae were staged according to the protocol of Bainbridges and Bownes [24]. Pupae were collected at the immobile white prepupae stage with everted spiracles. The pupal case becomes brown within an hour of immobility and marks the beginning of pupation [24]. The pupae were then aged for the appropriate time at 22°C, and the timing used in this study is based on hours after the start of pupation (abbreviated as ASP). Developmental landmarks were also used to confirm staging [24]. Because of the asynchrony of pupal development, the chronology indicated in this paper represents the times/stages where any particular pattern is predominant. Staged pupae were hand-dissected from their pupal case using fine glass needles. Either the whole pupa or the thoracic area (for pupae aged 40 hours ASP and later stages) was used and the tissues were teased apart. The tissues were spread on poly-L-lysine coated slides by applying hand pressure with a coverslip and processed for immunofluorescence as described below.
Drosophila mutants
Three myosin mutants were obtained from Dr. Charles Emerson's laboratory (U. Penn. Medical School): Mhc7, Mhc10 and Mhc11. All three are null mutations, which specifically disrupt the myosin heavy chain protein only in IFMs. These stocks are, therefore, homozygous viable, though flightless. The two actin mutations, E93K and KM88, were obtained from Dr. John Sparrow's laboratory (York University, UK). They are IFM-specific as they are mutated alleles of the gene coding for the IFM-specific actin isoform (Act88F), and are homozygous viable flightless.
Myofibril extraction procedure
Myofibrils were extracted from adult wild type Oregon R or from E93K flies using a procedure, which yields primarily asynchronous myofibrils [3]. Approximately 0.5 grams of flies were homogenized in Hodges/EGTA buffer (0.1 M KCl, 1 mM MgCl2, 2.5 mM EGTA, pH 7) containing the following protease inhibitors: 0.5 mM phenylmethanesulfonylfluoride (PMSF), 0.1 mg/ml soybean trypsin inhibitor (STI), 5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 0.2 mg o-phenanthroline (OPT). This mixture was filtered and submitted to several sucrose gradient centrifugations as described [3]. In the last gradient, the band at the 1.25 M/2.5 M sucrose interface was collected as this fraction is highly enriched in IFM myofibrils. The preparation was kept at 4°C for no more than a week before being used for immunofluorescence.
Calpain digestion of myofibrils
Myofibrils were digested with calpain using the procedure of Lakey et al. [28]. Myofibrils were typically washed once with Hodges/EGTA buffer to remove the sucrose. Pelleted myofibrils were further washed 2 times before being resuspended in calpain activating solution (CAS: 0.1 M NaCl, 20 mM Tris-HCl pH 7, 2 mM MgCl2, 10 mM β-mercaptoethanol, 2 mM NaN3, 5 mM CaCl2). In early studies, myofibrils were treated with calpain in solution, and then pelleted by centrifugation before being deposited on a slide for analysis. In later studies, a drop of untreated fibrils was first deposited on poly-L-lysine coated slides and left to settle for approximately 10 minutes to facilitate myofibril absorption to the slide. Various amounts of calpain (Sigma) in calpain activating solution were then added directly onto the myofibrils. In both protocols, calpain amounts typically ranged from 0 μg (in controls) to 72 μg of calpain per slide. After addition of the calpain, myofibrils were incubated at 25°C for 90 minutes. Calpain-treated myofibrils were then fixed with 4% formaldehyde in 1X PBS immediately at the end of the incubation period and processed for immunofluorescence microscopy as described below.
Immunofluorescence
The immunofluorescence procedure was as described before [7]. For double labeling, the two primary antibodies were applied together, as well as, the two secondaries. Primary antibody combinations were selected such that each primary would be recognized by a different secondary (see below). Both rhodamine and fluorescein tagged-secondary antibodies (Pierce) were used for identification at a 1:200 dilution. F-actin was detected using FITC-labeled phalloidin (Sigma) at a dilution of 1:100, which was routinely added with the secondary antibody. Slides were mounted in mounting medium with DAPI (Vectorshield Inc.). In all control immunofluorescence experiments, reacting the preparations with the secondary antibodies alone indicated no background staining. Slides were examined by epifluorescence microscopy at either 80× or 200× magnification. Early images were recorded on Kodachrome color slides, which were subsequently digitized using a scanner. In later stages, fluorescent images were directly recorded using a microscope-mounted digital camera. All images were processed and figures assembled using Abobe PhotoshopR software.
Projectin fusion protein and antibody production
A cDNA fragment representing a portion of the central core region of the projectin protein [13] with 2 Ig and 6 Fn3 domains was amplified by RT-PCR. The DNA fragment was then gel-purified and subcloned into a pQE vector (Qiagen Inc.), which provides an NH2-terminal 6X His tag. The bacterial fusion protein was induced and purified on Ni-NTA columns from bacterial cultures, as recommended by the manufacturer (Qiagen Inc.). The Core1P purified fusion protein was run on SDS-polyacrylamide gels followed by transfer onto nitrocellulose membranes. The area of the membrane containing the protein was cut out and used for rabbit immunization (Pocono Rabbit Farm). The Core1P antibody specificity was assessed by standard Western analysis of total adult proteins as described before [7] (data not shown).
Other antibodies
The following antibodies were used in the immunofluorescence studies.
Projectin: MAC150, Core1P, 3b11, P5 and kinase; Myosin heavy chain: 3e8 and 722; α-actinin: 4g6 and MAC276; Kettin: MAC155; Z(210): 3d10. The three "MAC" antibodies are rat monoclonals (generous gifts from Dr. Belinda Bullard, EMBL, Heidelberg, [4]). Antibodies 3e8, 4g6, 3b11, P5 and 3d10 are mouse monoclonals (generous gifts from Dr. Judith Saide, Boston University School of Medicine, [3]). 722 is a rabbit polyclonal (a generous gift from Dr. Dan Kiehart, [43]). Kinase is a rabbit polyclonal directed against twitchin kinase, which cross-reacts with projectin kinase domain (generous gift of Dr. G. Benian, Emory University). Core1P antibody is described above.
List of abbreviations
ASP: after start of pupation
IFM: indirect flight muscle
Mhc: myosin heavy chain
TDT: Tergal depressor of the trochanter
CAS: calpain activating solution
Authors' contributions
AS and RS carried out the time points and mutant analysis and drafted the manuscript. CB and TR performed the calpain analysis.
Acknowledgments
Acknowledgements
We want to thank Drs. J. Saide, B. Bullard, C. Emerson and D. Kiehart for providing antibodies against various myofibrillar proteins. This research was supported by an NSF grant # MCB-9996318 to AAS. The material presented is also based upon work jointly supported by the National Science Foundation/EPSCoR under Grant No. EPS-0132573 and National Institutes of Health/BRIN under Grant No. 8-P0RR16461A.
Contributor Information
Agnes Ayme-Southgate, Email: southgatea@cofc.edu.
Christophe Bounaix, Email: christophe.bounaix-du-puch@laposte.net.
Theresa E Riebe, Email: hemitoy@aol.com.
Richard Southgate, Email: southgater@cofc.edu.
References
- Bullard B, Hammond KS, Luke BM. The site of paramyosin in insect flight muscle and the presence of an unidentified protein between myosin filaments and Z line. J Mol Biol. 1977;115:417–440. doi: 10.1016/0022-2836(77)90163-2. [DOI] [PubMed] [Google Scholar]
- Saide JD. Identification of a connecting filament protein in insect fibrillar flight muscle. J Mol Biol. 1981;153:661–679. doi: 10.1016/0022-2836(81)90412-5. [DOI] [PubMed] [Google Scholar]
- Saide JD, Chin-Bow S, Hogan-Sheldon J, Busquets-Turner L, Vigoreaux JO, Valgeirsdottir K, Pardue ML. Characterization of components of Z-bands in the fibrillar flight muscle of Drosophila melanogaster . J Cell Biol. 1989;109:2157–2167. doi: 10.1083/jcb.109.5.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey A, Ferguson C, Labeit S, Reedy M, Larkins A, Butcher G, Leonard K, Bullard B. Identification and localization of high molecular weight proteins in insect flight and leg muscles. EMBO J. 1990;9:3459–3467. doi: 10.1002/j.1460-2075.1990.tb07554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DH, Matsuno A, Terakado K, Matsuura T, Kimura S, Maruyama K. Projectin is an invertebrate connectin (titin): isolation from crayfish claw muscle and localization in crayfish claw muscle and insect flight muscle. J Muscle Res Cell Motil. 1990;11:497–511. doi: 10.1007/BF01745217. [DOI] [PubMed] [Google Scholar]
- Nave R, Weber K. A Myofibrillar protein of insect muscle related to vertebrate titin connects Z band and A band: purification and molecular characterization of invertebrate mini-titin. J Cell Sci. 1990;95:535–544. doi: 10.1242/jcs.95.4.535. [DOI] [PubMed] [Google Scholar]
- Vigoreaux JO, Saide JD, Pardue M-L. Structurally different Drosophila striated muscles utilize distinct variants of Z-band associated proteins. J Muscle Res Cell Motil. 1991;12:340–354. doi: 10.1007/BF01738589. [DOI] [PubMed] [Google Scholar]
- Maroto M, Vinos J, Marco R, Cervera M. Autophosphorylating protein kinase activity of titin-like arthropod projectin. J Mol Biol. 1992;224:287–291. doi: 10.1016/0022-2836(92)90994-u. [DOI] [PubMed] [Google Scholar]
- Benian GM, Kiff JE, Neckelmann N, Moerman DG, Waterston RH. Sequence of an Unusually Large Protein Implicated in Regulation of Myosin Activity in C. elegans . Nature. 1989;342:45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- Benian GM, L'Hernault SW, Morris ME. Additional sequence complexity in the muscle gene, unc22 and its encoded protein twitchin of Caenorhabditis elegans. Genetics. 1993;134:1097–1104. doi: 10.1093/genetics/134.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Ayme-Southgate A, Vigoreaux JO, Benian GM, Pardue ML. Drosophila has a twitchin/titin-related gene that appears to encode projectin. Proc Natl Acad Sci USA. 1991;88:7973–7977. doi: 10.1073/pnas.88.18.7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayme-Southgate A, Southgate R, Saide J, Benian G, Pardue ML. Both synchronous and asynchronous muscle isoforms of projectin (the Drosophila bent locus product) contain functional kinase domains. J Cell Biol. 1995;128:393–403. doi: 10.1083/jcb.128.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg CC, Labeit S, Bullard B, Leonard K, Fyrberg EA. Drosophila projectin: relatedness to titin and twitchin and correlation with lethal (4) 102CDa and bent-dominant mutants. Proc R Soc Lond B. 1992;249:33–40. doi: 10.1098/rspb.1992.0080. [DOI] [PubMed] [Google Scholar]
- Daley J, Southgate R, Ayme-Southgate A. Structure of the projectin isoforms and implications for projectin assembly and functions. J Mol Biol. 1998;279:201–210. doi: 10.1006/jmbi.1998.1756. [DOI] [PubMed] [Google Scholar]
- Southgate R, Ayme-Southgate A. Drosophila projectin contains a spring-like PEVK region which is alternatively spliced. J Mol Biol. 2001;313:1037–1045. doi: 10.1006/jmbi.2001.5115. [DOI] [PubMed] [Google Scholar]
- Crossley AC. The morphology and development of the Drosophila muscular system. In: Asburner M, Wright TRF, editor. In The genetics and biology of Drosophila. 2B. London: Academic Press; 1978. pp. 499–559. [Google Scholar]
- Trombitas K. Connecting filaments: a historical perspective. In: Pollack GH, Granzier H, editor. In Proceedings: Elastic filaments of the cell. Kluwer Academic/Plenum Publishers; 2000. pp. 1–23. [Google Scholar]
- Fyrberg EA, Beall C. Genetic approaches to myofibril form and function in Drosophila. Trends in Genet. 1990;6:126–131. doi: 10.1016/0168-9525(90)90127-R. [DOI] [PubMed] [Google Scholar]
- Bernstein SI, O'Donnell PT, Cripps RM. Molecular genetics analysis of muscle development, structure and function in Drosophila. Int Rev Cytol. 1993;143:63–152. doi: 10.1016/s0074-7696(08)61874-4. [DOI] [PubMed] [Google Scholar]
- Reedy MC, Beall C. Ultrastructure of developing flight muscle in Drosophila. I. assembly of myofibrils. Dev Biol. 1993;160:443–465. doi: 10.1006/dbio.1993.1320. [DOI] [PubMed] [Google Scholar]
- Fernandes J, Bate M, VijayRaghavan K. Development of the indirect flight muscles of Drosophila. Development. 1991;113:67–77. doi: 10.1242/dev.113.1.67. [DOI] [PubMed] [Google Scholar]
- Roy S, VijayRhagavan K. Patterning muscles using organizers: larval muscle templates and adult myoblasts actively interact to pattern the dorsal longitudinal flight muscles of Drosophila. J Cell Biol. 1998;141:1135–1145. doi: 10.1083/jcb.141.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge P, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- Karlik CC, Coutu MD, Fyrberg EA. A nonsense mutation within the Act88F actin gene disrupts myofibril formation in Drosophila indirect flight muscles. Cell. 1984;38:711–719. doi: 10.1016/0092-8674(84)90266-6. [DOI] [PubMed] [Google Scholar]
- Sparrow J, Reedy M, Ball E, Kyrtatas V, Molloy J, Durston J, Hennessey E, White D. Functional and ultrastructural effects of a missense mutation in the indirect flight muscle-specific actin gene of Drosophila melanogaster. J Mol Biol. 1991;222:963–982. doi: 10.1016/0022-2836(91)90588-w. [DOI] [PubMed] [Google Scholar]
- Beall CJ, Sepanski MA, Fyrberg EA. Genetic dissection of Drosophila myofibril formation: effects of actin and myosin heavy chain null alleles. Genes and Dev. 1989;3:131–140. doi: 10.1101/gad.3.2.131. [DOI] [PubMed] [Google Scholar]
- Lakey A, Ferguson C, Labeit S, Reedy M, Larkins A, Butcher G, Leonard K, Bullard B. Kettin, a large modular protein in the Z-Disc of insect muscles. EMBO J. 1993;12:2863–2871. doi: 10.1002/j.1460-2075.1993.tb05948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT, Maher PA. Immunocytochemical studies of cardiac myofibrillogenesis in early chick embryos. I: Presence of immunofluorescent titin spots in premyofibril stages. J Cell Biol. 1987;105:2781–2793. doi: 10.1083/jcb.105.6.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-M, Jeng C-J. Interactions between titin and alpha actinin. Biomed Res. 1992;13:197–202. doi: 10.1016/0167-9473(92)90007-3. [DOI] [Google Scholar]
- Ohtsuka H, Yajima H, Maruyama K, Kimura S. Binding of the N-terminal 63 kDa portion of connectin/titin to alpha-actinin as revealed by the yeast two-hybrid system. FEBS Letters. 1997;401:65–67. doi: 10.1016/S0014-5793(96)01432-9. [DOI] [PubMed] [Google Scholar]
- Turnacioglu KK, Mittal B, Sanger JM, Sanger WJ. Partial characterization of zeugmatin indicates that it is part of the Z-band region of titin. Cell Motil Cytoskeleton. 1996;34:108–121. doi: 10.1002/(SICI)1097-0169(1996)34:2<108::AID-CM3>3.3.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Turnacioglu KK, Mittal B, Dabiri GA, Sanger JM, Sanger WJ. An N-terminal fragment of titin coupled to green fluorescent protein localizes to the Z-bands in living muscle cells: overexpression leads to myofibril disassembly. Mol Biol Cell. 1997;8:705–717. doi: 10.1091/mbc.8.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierez-Cru zG, van Heerden A, Wang K. Modular Motifs, Structural Folds and Affinity Profiles of PEVK Segments of Human Fetal Skeletal Muscle Titin. J Biol Chem. 2001;10:7442–7449. doi: 10.1074/jbc.M008851200. [DOI] [PubMed] [Google Scholar]
- Chun M, Falkenthal S. Ifm (2)2 is a myosin heavy chain allele that disrupts myofibrillar assembly only in the indirect flight muscle of Drosophila melanogaster . J Cell Biol. 1988;107:2613–2621. doi: 10.1083/jcb.107.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell PT, Bernstein SI. Molecular and ultrastructure defects in a Drosophila myosin heavy chain mutant: Differential effects on muscle function produced by similar thick filament abnormalities. J Cell Biol. 1988;107:2601–2612. doi: 10.1083/jcb.107.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst DO, Osborn M, Weber K. Myogenesis in the mouse embryo: differential onset of expression of myogenic proteins and the involvement of titin in myofibril assembly. J Cell Biol. 1989;109:517–527. doi: 10.1083/jcb.109.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S, Gautel M, Lakey A, Trinick J. Towards a molecular understanding of titin. EMBO J. 1992;11:1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soteriou A, Gamage M, Trinick J. A survey of interactions made by the giant protein titin. J Cell Sci. 1993;104:119–123. doi: 10.1242/jcs.104.1.119. [DOI] [PubMed] [Google Scholar]
- Houmeida A, Holt J, Tskhovrebova L, Trinick J. Studies of the interactions between titin and myosin. J Cell Biol. 1995;131:1471–1481. doi: 10.1083/jcb.131.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J-P. Cloned rat cardiac titin class I and class II motifs: expression, purification, characterization and interaction with F-actin. J Biol Chem. 1995;270:6908–6916. [PubMed] [Google Scholar]
- Linke WA, Rudy DE, Centner T, Gautel M, Witt CC, Labeit S, Gregorio CC. I-band titin in cardiac muscle is a three-element molecular spring and is critical for maintaining thin filament structure. J Cell Biol. 1999;246:631–644. doi: 10.1083/jcb.146.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Feghali R. Cytoplasmic myosin from Drosophila melanogaster. J Cell Biol. 1986;103:1517–1525. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]


