INTRODUCTION
While tobacco smoking remains the most important risk factor for the development of small cell lung cancer (SCLC), an epidemiological study reported that 3% of patients with SCLCs are never-smokers.1 Several reports describe the rare, but well-documented, phenomenon of transformation to SCLC as a mechanism of acquired resistance (AR) to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in 4–14% of patients initially treated for EGFR-mutant lung adenocarcinomas, including in some patients who are never-smokers.2–5
Scant literature exists describing de novo SCLCs among patients who are never-smokers. We report on 23 patients with SCLCs who classify themselves as never-smokers.
MATERIALS AND METHODS
Patient identification and smoking history documentation
We performed a retrospective review of 1040 SCLC patients evaluated at Memorial Sloan-Kettering Cancer Center (MSKCC) between 2005 and 2012. Patients with lung cancers assessed at MSKCC complete a prospective, self-administered smoking questionnaire at the initial visit. Never-smokers are defined as those patients who report having smoked ≤ 100 cigarettes in their lifetime.
Pathologic review of small cell lung cancer
The diagnosis of SCLC was confirmed through histologic and immunohistochemical (IHC) testing (chromogranin, synaptophysin, CD56, and MIB1) by a thoracic pathologist.
KRAS, EGFR, ALK, and RB testing
Patients with sufficient tissue underwent testing for EGFR and KRAS mutations and ALK rearrangements. EGFR mutations (exon 19 deletions and exon 21 L858R amino acid substitutions) were identified by mutation-specific PCR-based methods.6–8 KRAS codon 12 and 13 mutation identification was performed by both mass-spectrometry (Sequenom, Inc., San Diego, CA)-based genotyping or direct sequencing. ALK rearrangements were tested using either fluorescence in situ hybridization (dual-color break-apart ALK probe, Abbott Molecular) or IHC (ALK-01 Ventana 790-2918). RB expression was analyzed using IHC (clone 1F8, Leica Biosystems).
Comprehensive, integrated mutation analysis of actionable cancer genes using next generation sequencing (NGS)
DNA was extracted from biopsied tissue and cytology specimens (and patient-matched normal tissue) using Qiagen nucleic acid extraction kits. Using our MSK-IMPACT assay (Integrated Mutation Profiling of Actionable Cancer Targets), bar-coded sequence libraries were prepared (Illumina TruSeq), and exon capture was performed on bar-coded pools by hybridization (Agilent SureSelect Target Enrichment) using custom oligonucleotides to capture all exons and select introns of 279 cancer genes. DNA was sequenced on an Illumina HiSeq 2000 to maximize sensitivity for detecting mutations. This strategy enables the identification of mutations, indels, and copy number alterations involving these 279 genes.
RESULTS
Incidence
Two percent of patients (23/1040) with SCLCs were never-smokers. Among never-smokers with SCLCs, 83% (19/23) had de novo SCLCs, and 17% (4/23) of patients had small cell transformation as a mechanism of AR to EGFR TKIs in EGFR-mutant lung adenocarcinoma. Baseline characteristics are listed in Table 1. Clinical and pathologic characteristics of each patient are listed in Table 2.
TABLE 1.
CLINICAL CHARACTERISTICS OF PATIENTS WITH SMALL CELL LUNG CANCER WHO ARE NEVER SMOKERS
| SCLC as Acquired Resistance (n = 4)‡ | De novo SCLC (n = 19) | |
|---|---|---|
| Sex | ||
| Male | 0 | 9 |
| Female | 4 | 10 |
| Median age at diagnosis(Range) | 48 (42–75)* | 65 (35–94) |
| Race | ||
| White | 2 | 13 |
| Black | 0 | 3 |
| Asian | 2 | 3 |
| Stage at diagnosis | ||
| Limited | 0 | 5 |
| Extensive | 4 | 14 |
| Brain Metastases at Presentation | ||
| Yes | 2 | 4 |
| No | 2 | 15 |
All four of these patients had been treated with erlotinib.
For patients with SCLC transformation as a mechanism of acquired resistance to EGFR TKIs, median age at diagnosis is the age at which SCLC transformation was diagnosed.
TABLE 2.
CLINICAL AND PATHOLOGIC CHARACTERISTICS OF SMALL CELL LUNG CANCERS AMONG NEVER SMOKERS
| PATIENT # | SEX | DE NOVO OR ACQUIRED RESISTANCE | STAGE | PATHOLOGY | MUTATIONS IDENTIFIED |
|---|---|---|---|---|---|
| 1† | M | DE NOVO | EXTENSIVE | SCLC | |
| 2 | F | DE NOVO | LIMITED | SCLC / SPINDLE CELL | |
| 3† | F | DE NOVO | LIMITED | SCLC | |
| 4 | F | DE NOVO | LIMITED | SCLC | EGFR WT / KRAS WT |
| 5 | F | DE NOVO | EXTENSIVE | SCLC | EGFR WT / KRAS WT |
| 6 | F | DE NOVO | EXTENSIVE | SCLC | EGFR WT / KRAS WT |
| 7 | M | DE NOVO | EXTENSIVE | SCLC | EGFR WT / KRAS WT |
| 8 | F | DE NOVO | EXTENSIVE | SCLC | EGFR WT / KRAS WT |
| 9 | M | DE NOVO | EXTENSIVE | SCLC / ADC | EGFR L858R / KRAS WT |
| 10 | M | DE NOVO | LIMITED | SCLC / ADC | |
| 11 | F | DE NOVO | EXTENSIVE | SCLC / Large Cell neuroendocrine carcinoma | |
| 12 | M | DE NOVO | EXTENSIVE | SCLC | |
| 13 | M | DE NOVO | EXTENSIVE | SCLC | EGFR Exon 19 del* |
| 14 | F | AR | EXTENSIVE | SCLC | EGFR Exon 19 del |
| 15 | F | AR | EXTENSIVE | SCLC / NEUROENDOCR INE CA | EGFR Exon 19 del |
| 16 | F | AR | EXTENSIVE | SCLC / ADC | EGFR Exon 19 del |
| 17 | M | DE NOVO | EXTENSIVE | SCLC | |
| 18 | F | DE NOVO | LIMITED | SCLC | |
| 19 | F | DE NOVO | EXTENSIVE | SCLC | |
| 20 | F | DE NOVO | EXTENSIVE | SCLC | |
| 21 | M | DE NOVO | EXTENSIVE | SCLC | |
| 22 | F | AR | EXTENSIVE | SCLC | EGFR Exon 19 del |
| 23 | M | DE NOVO | EXTENSIVE | SCLC | EGFR WT / KRAS WT |
This patient’s tumor also harbored a PIK3CA E545K mutation.
Please see Table 4 for next generation sequencing results.
M – male
F – female
AR – acquired resistance
SCLC – small cell lung cancer
ADC – adenocarcinoma
NSCLC – non-small cell lung cancer
De novo Small Cell Lung Cancers
Pathologic Characteristics
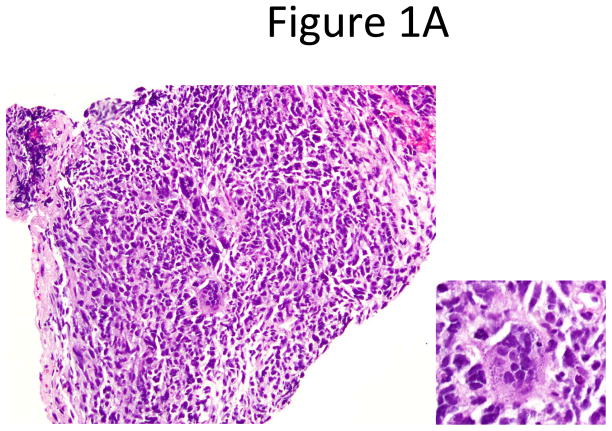
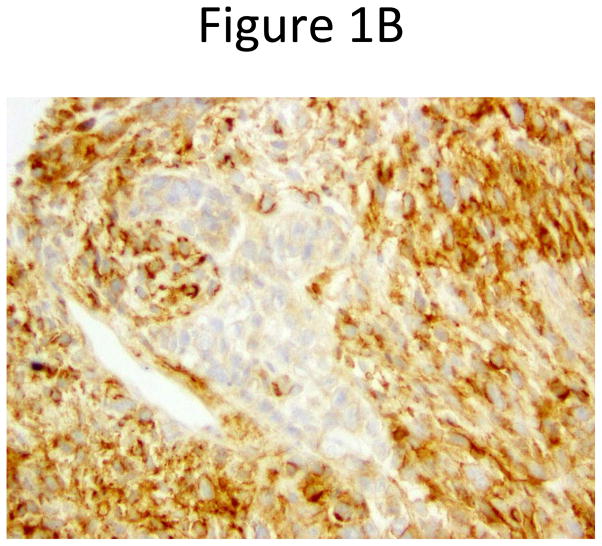
Pathologic re-review confirmed the following: 15 pure SCLC; one mixed SCLC and large cell neuroendocrine carcinoma; one SCLC and spindle and giant cell carcinoma (Figures 1A and 1B); and two mixed SCLC and adenocarcinoma.
Figure 1.
Clinical Characteristics
Five of 19 patients with de novo SCLCs were either lost to followup or had inadequate information available regarding treatment course and response. Of the 14 cases with available treatment history, all received first-line etoposide/platinum doublets. Fifty-seven percent of these patients (8/14) had a response to chemotherapy that lasted ≥3 months from completion of first-line therapy. Median time to progression was 11 months (95%CI: 5–13 months). Median overall survival (OS) from the time of SCLC diagnosis was 23 months (95%CI: 10–27 months).
EGFR, KRAS, ALK, and RB testing
Eight of the 19 de novo SCLC cases underwent testing for EGFR Exon 19 deletion and Exon 21 L858R mutation. An EGFR L858R mutation was found in one patient, whose tumor contained SCLC and adenocarcinoma components. This patient initially received erlotinib, carboplatin, and etoposide, with progression of disease within three months of completing chemotherapy. While he continued erlotinib throughout his subsequent chemotherapeutic regimens, his disease rapidly progressed. The tumor of one patient with pure SCLC harbored an EGFR Exon 19 deletion and PIK3CA E545K mutation. This patient had progressive disease after four cycles of etoposide/platinum. He was then initiated on erlotinib, with progression of disease after just four weeks of therapy.
There were no KRAS mutations or ALK rearrangements identified in the cases tested for these alterations. Six of seven cases tested for RB expression demonstrated RB loss (Table 3).
TABLE 3.
PATHOLOGIC CHARACTERISTICS OF SMALL CELL LUNG CANCERS AMONG NEVER SMOKERS
| Pathologic confirmation of SCLC | SCLC as Acquired Resistance (n = 4) | De novo SCLC(n = 19) |
|---|---|---|
| Pure SCLC | 2 | 15 |
| Mixed Histology | 2 | 4 |
| EGFR mutations found / EGFR testing performed | 4*/4 | 2/8 |
| KRAS mutations found / KRAS testing performed | 0/2 | 0/8 |
| ALK rearrangements found / ALK testing performed | 0/0 | 0/5 |
| RB loss found / RB testing performed | 0/0 | 6/7 |
All 4 patients with EGFR mutations had EGFR Exon 19 deletions present at biopsies taken at baseline and at the time of acquired resistance to EGFR TKIs.
Acquired Resistance Small Cell Lung Cancer
Pathologic characteristics
Of the four patients with SCLC as a mechanism of AR to EGFR TKIs, two had pure SCLC, one had mixed histology of SCLC and adenocarcinoma, and one had SCLC, adenocarcinoma, and large cell neuroendocrine carcinoma components.
Clinical Characteristics
All four patients were women [median 48 years; range 40–75 years] at the diagnosis of SCLC transformation from EGFR-mutant lung adenocarcinoma. Once the diagnosis of SCLC was made, two patients received platinum/etoposide. One of these patients underwent local therapy with surgical resection of a lung nodule that demonstrated recurrent growth while all other sites of disease had resolved.9 Pathology review of this specimen with acquired resistance demonstrated SCLC, adenocarcinoma, and large cell neuroendocrine carcinoma components. After receiving 6 cycles of adjuvant carboplatin, etoposide, and erlotinib therapy, this patient had a nine month disease-free interval after treatment. The two patients who did not receive platinum/etoposide therapy had varied treatments and clinical courses.
EGFR, KRAS, ALK, and Rb testing
The patients with SCLC as a mechanism of AR to EGFR TKI had persistence of the original EGFR mutation in their tumors confirmed on biopsy taken at the time of resistance. Of three cases also tested for EGFR Exon 20 T790M mutation at the time of AR, two had the second-site EGFR Exon 20 T790M mutation, in addition to SCLC. Insufficient tissue was available for RB testing in these samples.
Comprehensive mutation analysis of actionable cancer genes using NGS
Two patients had adequate tissue available for NGS. In one patient, analysis revealed five mutations in four genes: PHOX2B, NOTCH1, RB1, and TP53. Additionally, amplification was seen in TERT. NGS analysis of the second patient’s sample revealed two mutations in the two genes CBL and GNAS, with amplification seen in MYCL1. Neither patients’ samples had smoking-associated G -->T transversions (Table 4).
TABLE 4.
NEXT GENERATION SEQUENCING OF ALL EXONS AND SELECTED INTRONS OF 279 CANCER TARGET GENES
| Sample | Gene altered | Alteration present | Protein alteration | Base Pair alteration |
|---|---|---|---|---|
| Pt 1 | PHOX2B | Missense Mutation | P82L | G → A |
| NOTCH1 | Frame Shift Insertion | P2485fs | ||
| RB1 | Splice Site | R500_splice | G → A | |
| TP53 | Frame Shift Deletion | V218fs | ||
| TP53 | Frame Shift Deletion | V73fs | ||
| TERT | Amplification | |||
| Pt 2 | CBL | Missense Mutation | C401S | G → C |
| GNAS | Missense Mutation | M102V | A → G | |
| MYCL1 | Amplification | |||
DISCUSSION
Two percent of SCLCs at our institution occurred in never-smokers. This represents the largest cohort of clinically and pathologically described never-smokers with SCLC. 83% of these diagnoses were made de novo.
Pathologic review of these SCLC cases confirmed the diagnosis. Immunohistochemistry for RB loss was performed on seven samples, and six of these revealed Rb loss, consistent with lung cancer of SCLC lineage.10 Of eight patients with de novo SCLCs tested for EGFR mutations, one patient with mixed SCLC and adenocarcinoma had an EGFR L858R mutation, and a second patient with pure SCLC had a tumor with mutations in both EGFR (exon 19 deletion) and PIK3CA (E545K). Unlike patients with EGFR-mutant lung adenocarcinomas and the majority of patients with SCLC, these patients suffered rapid progression of disease that did not respond to therapy with EGFR TKIs or platinum/etoposide. Median OS in SCLC never-smokers was 23 months, which is longer than generally seen in SCLC.11
Of the four cases with SCLC as a mechanism of AR to EGFR TKIs in EGFR-mutant lung adenocarcinoma, two received cisplatin and etoposide. One patient had a partial response with a disease-free survival of 9 months, and the other did not respond. This small group represents a heterogeneous population in which the disease biology and clinical course can mirror both EGFR-mutant lung adenocarcinoma and SCLC.
The presence of EGFR mutations in tumors from patients with SCLCs remains an area of controversy. While an EGFR mutation in the setting of a mixed diagnosis of SCLC and adenocarcinoma can be seen with the EGFR mutation arising from the adenocarcinoma component, the etiology of the EGFR mutations in a single biopsy of SCLC is unclear. Prior work from our group has demonstrated that EGFR mutations do not exist in pure SCLC or squamous cell lung cancer.12,13 One interpretation is that this finding of pure SCLC represents sampling error in the setting of tumor heterogeneity rather than two oncogenic mechanisms in a single cell.
Further molecular analysis using NGS of exons of 279 cancer genes revealed mutations in TP53, Notch, and RB1, consistent with prior reports regarding the molecular basis of SCLC.14,15, 16 Interestingly, no smoking-associated G -->T transversions were observed in the two samples tested.
Our analysis has several limitations. This is a retrospective study. Given the rarity of patients with SCLCs who are never-smokers, prospective identification and analysis of these patients is difficult. The available tissue for molecular analysis was limited, and molecular testing could only be performed on a minority of the patients identified. The patients in this series were identified as never-smokers based on self-report and direct questioning by the medical staff.
Patients with SCLCs who are never-smokers represent a clinically and pathologically distinct population because they bear little similarity to non-SCLCs among never-smokers. Their molecular characteristics are unique compared to never-smoker patients with adenocarcinomas. The transformation of EGFR-mutant lung adenocarcinomas to a SCLC phenotype co-incident with the emergence of EGFR TKI AR is an area of much interest and research relative to its rarity. This study further supports the need for comprehensive, multiplexed genotyping to improve our ability to provide optimal care and facilitate research in these unique populations.
Acknowledgments
This research was supported in part by NIH 2P01 CA129243 (PI: Dr. Mark Kris, Targets for therapy for carcinomas of the lung).
Footnotes
Presented, in part, at the American Society of Clinical Oncology Annual Meeting in Chicago, Illinois, May 31 – June 4, 2013.
Conflicts of Interest and Source of Funding: The authors have no direct financial disclosures or conflicts with this research.
References
- 1.Ou SH, Ziogas A, Zell JA. Prognostic factors for survival in extensive stage small cell lung cancer (ED-SCLC): the importance of smoking history, socioeconomic and marital statuses, and ethnicity. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2009;4:37–43. doi: 10.1097/JTO.0b013e31819140fb. [DOI] [PubMed] [Google Scholar]
- 2.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morinaga R, Okamoto I, Furuta K, et al. Sequential occurrence of non-small cell and small cell lung cancer with the same EGFR mutation. Lung cancer (Amsterdam, Netherlands) 2007;58:411–413. doi: 10.1016/j.lungcan.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355:213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 5.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li AR, Chitale D, Riely GJ, et al. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J Mol Diagn. 2008;10:242–248. doi: 10.2353/jmoldx.2008.070178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu HA, Sima CS, Huang J, et al. Local therapy with continued EGFR tyrosine kinase inhibitor therapy as a treatment strategy in EGFR-mutant advanced lung cancers that have developed acquired resistance to EGFR tyrosine kinase inhibitors. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2013;8:346–351. doi: 10.1097/JTO.0b013e31827e1f83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modi S, Kubo A, Oie H, et al. Protein expression of the RB-related gene family and SV40 large T antigen in mesothelioma and lung cancer. Oncogene. 2000;19:4632–4639. doi: 10.1038/sj.onc.1203815. [DOI] [PubMed] [Google Scholar]
- 11.Vallieres E, Shepherd FA, Crowley J, et al. The IASLC Lung Cancer Staging Project: proposals regarding the relevance of TNM in the pathologic staging of small cell lung cancer in the forthcoming (seventh) edition of the TNM classification for lung cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2009;4:1049–1059. doi: 10.1097/JTO.0b013e3181b27799. [DOI] [PubMed] [Google Scholar]
- 12.Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1167–1176. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rekhtman NAM, Lau C, He M, Moreira AL, Travis WD, Zakowski MF, Pietanza MC, Riely GJ, Kris MG, Ladanyi M. Analysis of EGFR and KRAS Mutations in Small Cell Carcinoma and Large Cell Neuroendocrine Carcinoma of Lung. 14th Annual World Conference on Lung Cancer; Amsterdam, The Netherlands. 2011. [Google Scholar]
- 14.D'Angelo SP, Pietanza MC. The molecular pathogenesis of small cell lung cancer. Cancer biology & therapy. 2010;10:1–10. doi: 10.4161/cbt.10.1.12045. [DOI] [PubMed] [Google Scholar]
- 15.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]