Abstract
Protein inhibitor of activated STAT3 (PIAS3) is an endogenous inhibitor of STAT3 that negatively regulates STAT3 transcriptional activity and cell growth and demonstrates limited expression in the majority of human squamous cell carcinomas of the lung. In the present study we sought to determine if PIAS3 inhibits cell growth in non-small cell lung cancer (NSCLC) cell lines by inducing apoptosis. Our results demonstrate that over-expression of PIAS3 promotes mitochondrial depolarization, leading to cytochrome c release, caspase 9 and 3 activation and PARP cleavage. This intrinsic pathway activation was associated with decreased Bcl-xL expression and increased Noxa expression and was independent of p53 status. Furthermore, PIAS3 inhibition of STAT3 activity was also p53 independent. Microarray experiments were performed to discover STAT3-independent mediators of PIAS3-induced apoptosis by comparing the apoptotic gene expression signature induced by PIAS3 over-expression with that induced by STAT3 siRNA. The results showed that a subset of apoptotic genes was uniquely expressed only after PIAS3 expression. Thus, PIAS3 may represent a promising lung cancer therapeutic target because of its p53-independent efficacy as well as its potential to synergize with Bcl-2 targeted inhibitors.
Keywords: PIAS3, p53, STAT3, Noxa, apoptosis, lung cancer
Introduction
The American Cancer Society estimates that approximately 228,000 people in the United States will be newly diagnosed with cancer of the lung in 2013.1 Given the inherent resistance of lung cancer to cytotoxic agents, identifying molecules that drive lung cancer growth, survival, and metastasis is critical to the development of novel therapeutics. In this regard, members of the signal transducer and activator of transcription (STAT) family of transcription factors, in particular STAT3, are potential targets in lung cancer as well as other cancers.2
Blocking STAT3 signaling either directly or indirectly has been shown to affect tumor formation through inhibition of cell growth, induction of apoptosis, and/or inhibition of tumor angiogenesis. A number of recent studies have found STAT3 activation in lung cancer cell lines and tissues, suggesting a functional role for this target in lung cancer.3–5 Although there are several cellular inhibitors of STAT3 signaling6, interest in PIAS3 was increased by the recent discovery of its lack of expression in glioblastoma multiforme.7 Furthermore, ectopic expression of PIAS3 in a glioblastoma cell line caused inhibition of the transcriptional activity of STAT3 and cell growth. We have also identified a lack of PIAS3 protein expression in the majority of human squamous cell carcinomas of the lung8 and demonstrated that PIAS3 negatively regulates STAT3 transcriptional activity and cell growth in a concentration dependent manner.9 Taken together, these studies show that PIAS3 can inhibit the function of constitutively active STAT3 present in lung cancer. What remains unclear, however, is whether PIAS3-induced growth inhibition is caused by an activation of apoptosis.
Mutation of the p53 gene is a frequent event in carcinogenesis, occurring in about 50% of human tumors, including lung.10,11 The p53 protein is a potent inhibitor of cell growth, arresting cell cycle progression at several points and inducing apoptosis of cells undergoing uncontrolled growth.12 Lin et al have shown that expression of wild-type p53, but not mutant p53, significantly reduced STAT3 activation and DNA binding in a number of prostate cancer cell lines. Furthermore, they showed that cells with activated STAT3 from a variety of malignancies only harbor mutated or deleted p53, suggesting that p53 plays a major role in STAT3 activation and transcriptional activity.13 On the basis of this data we hypothesized that PIAS3 might functionally interact with STAT3 via p53 and sought to investigate this possibility.
In the present study, we demonstrate that PIAS3 inhibits cell growth in non-small cell lung cancer (NSCLC) cell lines by activating the intrinsic apoptosis pathway via altered expression of Bcl-2 family members. Furthermore, PIAS3-induced apoptosis and STAT3 inhibition were independent of p53 status.
Material and Methods
Cell culture and transient transfection
Human lung cancer cell lines A549, H1666, H358 and H1299 were purchased from American Type Culture Collection and maintained in DMEM/Ham’s F-12 medium supplemented with 10% (v/v) FBS (Hyclone) in a 5% CO2 humidified incubator at 37°C. Cells were transfected with either pCMV5 (mock) or pCMV5-mouse PIAS3 using Lipofectamine 2000 in Opti-MEM (InVitrogen/Life Technologies). After 5 h media was replaced with DMEM/F12 media containing fetal bovine serum (10%). Following 24 h of incubation, cells were collected for further analysis. In some experiments ABT-263 was added after the initial 5 h transfection.
Cell growth analysis
Cell growth and viability were assessed by trypan blue dye exclusion of manually counted cells, as well as the MTS assay, as described previously.9 Sub-G1 analysis was examined by flow cytometry using the propidium iodide (PI) DNA staining method. Samples were analyzed on a tri-laser FACSCalibur flow cytometer (Becton Dickinson) using CellQuest software (Becton Dickinson).
TUNEL assay
Apoptosis was detected by terminal deoxyribonucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining with an in situ cell death detection kit (ScienCell Research Laboratories) following the manufacturer’s directions. The nuclei of apoptotic cells were visualized by staining with DAB and counterstaining was performed using hematoxylin. Primary antibody and labeling mix were omitted in control sections. The results were examined under the light microscope at 400 × 3 magnification. The number of apoptotic cells over several random fields was counted out of a total of 100 and the percent TUNEL-positive cells was calculated. Each condition was performed in triplicate.
Mitochondrial depolarization assay
Mitochondrial membrane potential (Δψ) was assayed by flow cytometry following 24 h PIAS or mock transfection of A549 and H1299 cells. After 30 min loading with 50 μM TMRM, the cells were resuspended in flow buffer, which contained 5% FBS and 2 mM EDTA in 1x PBS. 50,000 events were collected from each sample on an Accuri C6 flow cytometer (Becton Dickinson). As a positive control a mitochondrial uncoupler, carbonyl cyanide p-chlorophenylhydrazone (CCCP), was used at 5 μm.
Immunoblotting and antibodies
Whole cell lysates were prepared in RIPA buffer. Cytosols were prepared as described previously.14 Protein concentrations were estimated using Bradford reagent (BioRad). Equal amounts of protein were resolved by SDS-PAGE and electroblotted onto PVDF membrane. The membrane was blocked in TBS containing 0.1% Tween-20 (TBS-T) and 5% dry milk powder then probed with primary antibody in TBS-T/5% milk overnight at 4°C. Secondary antibody in TBS-T/5% milk was incubated for 1 h, followed by chemiluminescence detection. The antibodies used in western blots were obtained from Santa Cruz Biotechnology (Bcl-2 sc492/7392, p21 sc397, p53 sc71817), BD Biosciences (cytochrome c #556433, Bak #556382), Novus (Cidec NBP2-15902, Noxa NB600-1159), Millipore (Dapk2 07-1229), Cell Signaling (Puma #4976, Bcl-xL #2764, Bim #2933, caspase 9 #9502, caspase 3 #9665), Calbiochem (VDAC/Anti-porin #529536) and Sigma (actin A5441).
Retrovirus production and infection
Phoenix-Ampho packaging cells were transfected with an ‘all-in-one’ retroviral construct (pL6N2-RHS3H/ZF2-PL) (Ariad Pharmaceuticals) with or without a mouse PIAS3 insert using HD FuGene reagent (Roche). Retroviral supernatants were harvested 48 h after transfection, filtered and the supernatants (1 ml) combined with 3 ml of medium containing polybrene (4 μg/ml final) before adding to A549 cells. The MOI was 10. Cells were centrifuged at room temperature at 650 × g for 90 min before incubation at 37°C. After 48 h of infection, cells were placed in selection medium containing G418 (850 μg/ml) for one week to isolate stable cells. The resulting stable cells infected with the PIAS3-containing retrovirus were called ‘A549P’ cells. This retroviral construct allows for the simultaneous delivery of a transcription factor cassette and a regulatable target gene in the same retroviral vector. The transcription factor cassette is activated by a cell-permeable ‘dimerizing’ agent, in this case the rapamycin analog AP21967, to induce transcription of the target gene, in this case mPIAS3.
Quantitative real-time PCR
RNA was extracted from cells using the RNeasy Mini Kit (Qiagen), digested with DNase I (Promega) and reverse transcribed using the Superscript III First Strand synthesis kit for RT-PCR (Invitrogen) to synthesize cDNA. Real time PCR was performed using TaqMan Fast Universal Master Mix and the TaqMan probe for human PIAS3 (Hs00966025_g1) on an Applied Biosystems 7500 Fast Sequence Detection System according to the manufacturer’s instructions. Samples were run in triplicate and standardized against endogenous β-actin (Human ACTB Endogenous Control, Applied Biosystems). The resulting relative PIAS3 mRNA amounts in each sample (ΔCT = PIAS3 CT − β-actin CT) were normalized to control values (ΔΔCT) to yield fold changes. A similar method was used to measure CIDEC (Hs01032998 probe) and DAPK2 (Hs00204888 probe) transcripts.
Luciferase assay
Cells were co-transfected with luciferase reporter construct pSTAT3-Luc or pTA-Luc alone as a nonspecific control together with pCMV5-PIAS3 or pCMV5 alone using HD FuGENE. The cells were incubated in DMEM/F12 medium for 48 h, treated or left untreated with 20 ng/mL EGF for 15 min, then lysed with passive lysis buffer (Promega). The supernatant was assessed for luciferase activity using a Berthold luminometer (Lumat LB9507) after briefly mixing the supernatant (20 μl) with 100 μl of firefly luciferase assay substrate solution. The activity of luciferase was normalized to protein concentrations in the lysate. Transfections were repeated at least three times and the relative changes are presented as mean ± SE.
p53 knockdown with shRNA lentivirus
The p53 shRNA lentivirus was produced similar to the ‘all-in-one’ retrovirus except using human embryonic kidney 293 (HEK293) packaging cells.15 A549, H1666, and H358 cells were infected with lentivirus for 48 h before beginning transfection experiments, as described above. Cells were incubated with or without EGF then analyzed for STAT3 luciferase as described above.
Microarray analysis
A549 cells were left untreated or transfected separately with either pCMV5 vector encoding human PIAS3 or siRNA against STAT3. After 36 h of growth in complete medium, all cells were switched to serum-free medium for the last 12 h and stimulated with epidermal growth factor (EGF) at 50 ng/mL for the final 15 min. Cells were harvested and cRNA was synthesized using the GeneChip IVT Labeling Kit (Affymetrix) using biotinylated ribonucleotides (Enzo Diagnostics) and hybridized to human U133A GeneChip micro-arrays (Affymetrix). Staining was performed on an Affymetrix fluidics station and micro-arrays scanned on a Hewlett Packard GeneArray Scanner. Data was compiled with Affymetrix Microarray Suite 5.0 software. Each experimental condition was repeated in two independent cultures. Expander was used to analyze the intensity data from each of the micro-arrays.16 Raw data was normalized using Robust Multichip Analysis and the hierarchical clustering method implemented in Expander was used to cluster genes based on their expression profiles. Standard t-tests were used for differential expression analysis (p < 0.05, absolute fold change greater than 1.2). The gene expression data from six samples have been deposited in the GEO database (GSE42979).
RESULTS
PIAS3 expression induces apoptosis in both A549 and H1299 cells
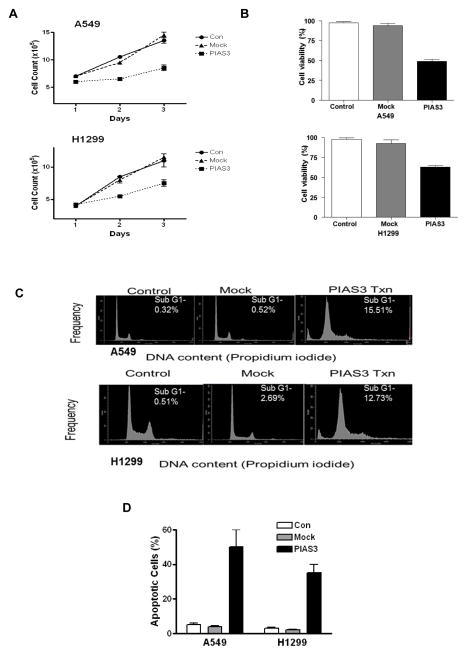
Although we have previously demonstrated that transient over-expression of PIAS3 inhibits cell growth in several lung cancer cell lines9 we have not yet determined if this inhibition results from apoptosis or if p53 status plays a role. Thus, here we used two different lung cancer cell lines of known p53 status, A549 (p53 wild-type) and H1299 (p53 null), to look for apoptosis induction by PIAS3. Initially, cells were transiently transfected with pCMV-5 containing full length PIAS3 to confirm its growth inhibitory effects. Cell number and viability were measured 48 h after transfection by trypan blue exclusion and the MTS assay. Mock transfection with empty vector or no transfection were used as controls. As shown in Figure 1, transfection with PIAS3 significantly inhibited the proliferation (panel A) and viability (panel B) of both A549 and H1299 cells. To determine whether the observed growth inhibition was due to apoptosis, we initially performed cell cycle analysis to determine the amount of sub-G1 DNA in A549 and H1299 cells after PIAS3 transfection. As shown in Figure 1C, PIAS3 increased the sub-G1 population by 30-fold in A549 cells and by 5-fold in H1299 cells, indicating a substantial increase in apoptotic cells. We confirmed that PIAS3 induced apoptosis using TUNEL assays. Transfection of PIAS3 clearly increased the percentage of apoptotic cells in both A549 and H1299 cells (Figure 1D). Thus, PIAS3 induces growth inhibition via an increase in apoptosis.
Figure 1. PIAS3 inhibits cell growth and induces apoptosis in non-small cell lung cancer cells.
A549 and H1299 cells were transfected with PIAS3 or empty plasmid (mock) or not transfected (control). In both cell lines, a significant decrease in proliferation was seen 24 h after the transfection by both cell count measurement (A) (p<0.01 for all groups) or MTS viability assay (B) (p<0.01 for all groups) compared to controls (N=4, data reported as mean ± SE). (C) Analysis of apoptosis by sub-G1 DNA content using flow cytometry of PIAS3 over-expressing cells. A549 and H1299 cell lines were transfected with empty plasmid or PIAS3. After 48 h, cells were stained by propidium iodide to analyze DNA content. The results shown are representative of at least three independent experiments. (D) Analysis of apoptosis by TUNEL staining in cells 48 h after transfection.
PIAS3 activates the intrinsic pathway of apoptosis
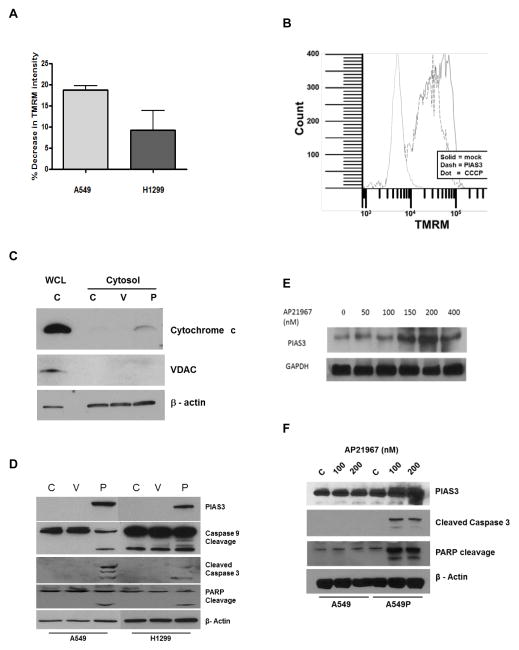
PIAS3 likely activates the intrinsic pathway of apoptosis. Therefore, we examined the effect of PIAS3 transfection on mitochondrial depolarization. As shown in Figure 2A, PIAS3 induced mitochondrial depolarization in both A549 and H1299 cells, as measured as a loss of TMRM fluorescence, relative to mock transfected cells, after 24 h in three independent experiments. A representative flow diagram is shown in Figure 2B. CCCP was used as a positive control. Furthermore, mitochondrial depolarization should result in cytochrome c release and apoptosome activation. Indeed, western blotting experiments demonstrated an increase in cytochrome c release into the cytosol after PIAS3 transfection (Figure 2C). The lack of VDAC in the cytosol demonstrated little mitochondrial contamination. Cytochrome c release was associated with an increase in cleaved caspase 9 and caspase 3, indicating activation (Figure 2D). Caspase 3 activation was confirmed by the appearance of cleaved poly (ADP-ribose) polymerase (PARP), a downstream substrate of caspase 3.17 Collectively, these results provide strong support for the idea that PIAS3 activates the intrinsic apoptotic pathway.
Figure 2. PIAS3 induces mitochondrial depolarization and apoptosome activation.
(A) Flow cytometric analysis of mitochondrial depolarization measured as loss of TMRM staining 24 h after PIAS3 or mock transfection. Bar graph shows results in A549 and H1299 cells representing the mean ± SE of three independent experiments. (B) Representative experiment showing the TMRM fluorescence intensity of 50,000 events in A549 cells averaged over three independent experiments. CCCP was the positive control. (C) and (D) Western blotting for downstream markers of mitochondrial depolarization in A549 and H1299 cells that were transfected with PIAS3 (P) or empty plasmid (V) or not transfected (C) for 24 h. Cytochrome c blots (C) used both A549 cytosolic extracts and whole cell lysates (WCL) for analyses, the blots in (D) all used whole cell lysates of A549 and H1299 cells. (E) Dose-response induction of PIAS3 expression by western blotting in A549P cells. PIAS3 expression was examined after 24 h incubation of cells with the indicated concentrations (nM) of AP21967. GAPDH was used as a loading control. (F) Western blotting for apoptotic markers after 24 h PIAS3 induction. Parental A549 and stably transfected A549P cells were incubated in the absence (C) or presence of two different concentrations of rapamycin (100 nm or 200 nm). β-actin was used as a loading control.
To confirm our transient transfection results we established a model system for stable but regulated expression of PIAS3 in cells. We infected A549 cells with a retroviral vector that allows PIAS3 expression only in the presence of the rapamycin analog (rapalog) AP21967. We called these cells A549P cells. Initial experiments demonstrated the ability of the rapalog to induce PIAS3 expression in a dose-dependent manner up to 200 nM AP21967 with no leakage of PIAS3 expression in the absence of AP21967 (Figure 2E). We then incubated A549P cells for 24 h with AP21967 (100 nm and 200 nm) and western blotted for apoptotic markers. As shown in Figure 2F, we observed a dose-dependent increase in cleaved caspase 3 and PARP in A549P lysates only in the presence of AP21967 and not in parental A549 cells, demonstrating that AP21967 lacks any cellular effect on its own, as previously reported.18 These results demonstrate that regulated expression of PIAS3 induces apoptosis.
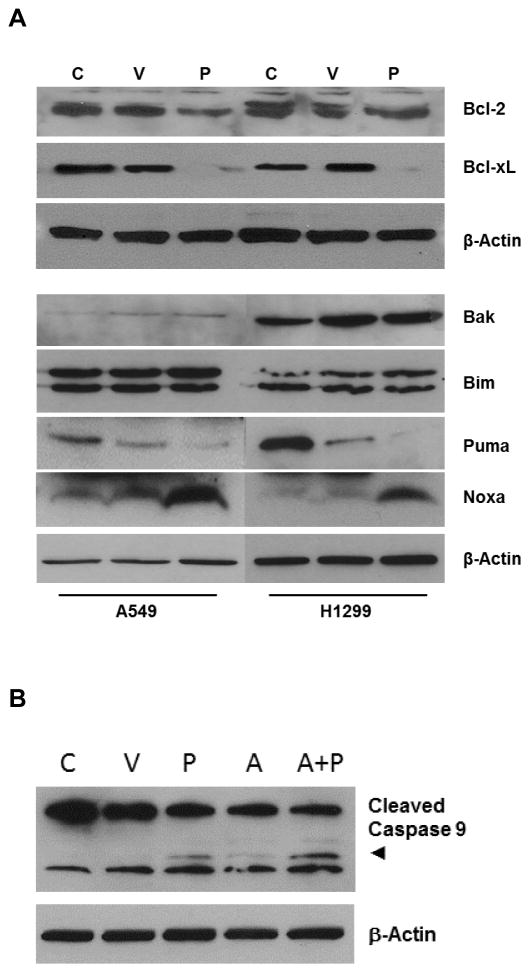
We next looked for changes in the expression of Bcl-2 family members induced by PIAS3 transfection. Two pro-survival family members, Bcl-xL and to a lesser extent Bcl-2, demonstrated reduced levels after PIAS3 transfection (Figure 3A). With regard to pro-apoptotic family members, Noxa was the only one tested that demonstrated an increase in expression after PIAS3 transfection. We next reasoned that if PIAS3 changes the expression of Bcl-2 proteins, then it should synergize with small molecule inhibitors of this protein family to induce apoptosis. Indeed, the results of Figure 3B demonstrate that incubation of PIAS3 transfected A549 cells with a low concentration of ABT-263 (A+P) results in enhanced cleavage, and therefore activation of caspase 9. ABT-263 by itself (A) had no effect. Taken together, PIAS3 induces mitochondrial depolarization by selectively down-regulating pro-survival Bcl-xL and up-regulating pro-apoptotic Noxa proteins in both p53 wild-type A549 and p53 null H1299 cells.
Figure 3. PIAS3 regulates Bcl-xL and Noxa expression.
(A) A549 and H1299 cells were transfected with PIAS3 (P) or empty plasmid (V) or not transfected (C) for 24 h. Protein lysates were analyzed by western blotting for Bcl-2 family members. Two sets of experiments were used for these studies, each with its own loading control. (B) Synergy of PIAS3 with ABT-263. A549 cells were transfected with PIAS3 (P) or mock transfected (C) then incubated for 24 h in the absence or presence of 10 μM ABT-263 (A). Protein lysates were western blotted for caspase 9 cleavage.
PIAS3 inhibits STAT3 activity independent of p53 status
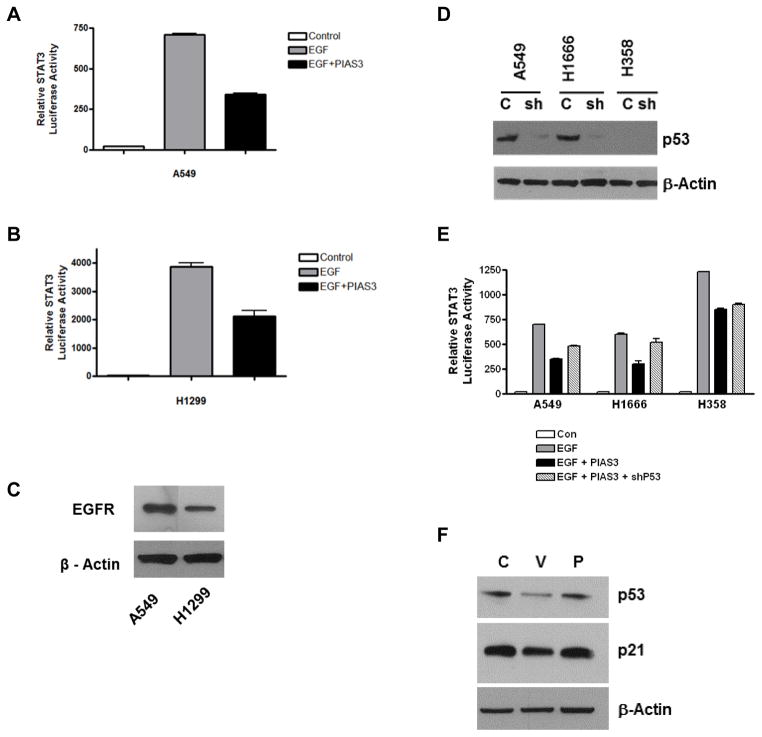
STAT3 activity promotes cell survival through many target proteins, including Bcl-xL expression, therefore PIAS3 inhibition of STAT3 activity likely contributes to PIAS3-induced apoptosis. However, because p53 is also known to inhibit STAT3 activity13, it was of interest to determine its role in PIAS3-induced STAT3 inhibition. In initial experiments we examined the effect of PIAS3 on EGF-stimulated STAT3 luciferase activity using A549 and H1299 cells. As shown in Figures 4A and 4B, a 60% reduction in EGF-stimulated STAT3 transcriptional activity was observed in A549 cells and a 50% reduction in activity was observed in H1299 cells after PIAS3 transfection, although stimulated STAT3 activity was much higher in H1299 cells. Control experiments showed that total EGFR expression in the two cell lines was similar (Figure 4C). Together, these data suggest that in an EGF-driven model, PIAS3 can inhibit STAT3 transcriptional activity irrespective of p53 status.
Figure 4. PIAS3 inhibits STAT3 activity independent of p53 status.
(A) A549 and (B) H1299 cells were transfected with STAT3 luciferase in the absence or presence of PIAS3. After 48 h, some cells were stimulated with EGF for 15 min after which lysates were prepared and analyzed. Results were normalized to protein concentration. (C) Western blot showing endogenous EGFR levels in A549 and H1299 cells. (D) Western blot of p53 levels. Protein lysates were prepared from A549, H1666 and H358 cells after 48 h incubation ± p53 shRNA lentivirus. (E) Combined effect of PIAS3 and p53 knockdown on EGF-stimulated STAT3 luciferase activity was done as described in Material and Methods. Each bar represents mean ± SD of three independent biological repeats. (F) Western blotting showing effects of PIAS3 transfection on p53 and p21 expression after 24 h in A549 cells.
To further investigate the role of p53 status, we examined if knockdown of p53 with shRNA would alter the PIAS3 inhibitory effect on STAT3 activity. To this end we utilized two p53 wild type cells, A549 and H1666, and one p53 null cell, H358. As shown in Figure 4D, the p53 shRNA effectively reduced endogenous p53 expression in both A549 and H1666 cells. When the same p53 shRNA was used in STAT3 luciferase experiments, we observed that the negative effect of PIAS3 on STAT3 transcriptional activity is only partially reversed in the p53 wild-type A549 and H1666 cells (Figure 4E). As expected, this was not observed in p53 null H358 cells.
Taken together, the above data indicate that the presence of p53 has little effect on the degree of PIAS3 inhibition of STAT3 luciferase activity. However, to examine the relationship of PIAS3 and p53 more directly, we determined the effect of PIAS3 transfection on p53 stability and transcriptional activity. As show in Figure 4F, PIAS3 transfection had no effect on p53 protein levels or the expression of a downstream transcriptional target of p53, p21.
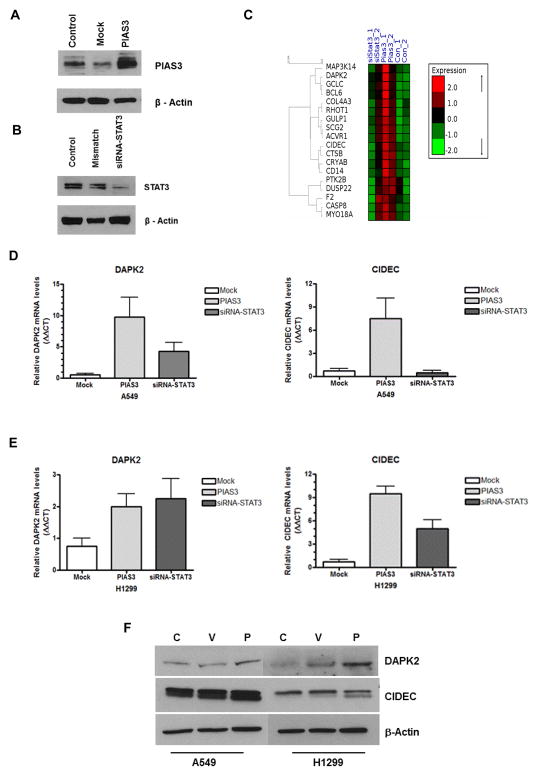
Overexpression of PIAS3 up-regulates STAT3-independent, apoptosis promoting genes CIDEC and DAPK2
Our data show that both PIAS3-induced apoptosis and STAT3 inhibition are p53-independent. It remains less clear, however, if PIAS3-induced apoptosis occurs exclusively via a STAT3-dependent mechanism. In order to clarify this, we used microarray analysis of A549 cells to examine changes in apoptotic gene expression profiles induced by overexpression of PIAS3 versus changes induced by siRNA knockdown of STAT3. PIAS3 overexpression and STAT3 knockdown were confirmed by immunoblotting (Figures 5A and 5B). We were able to achieve a high degree of PIAS3 overexpression in PIAS3 transfected cells (transfection efficiency 30–40%) and were also able to achieve a significant reduction in STAT3 with targeted siRNA (>80% knockdown).
Figure 5. PIAS3 induces STAT3-independent apoptotic gene expression.
Confirmation of PIAS3 overexpression (A) and STAT3 knockdown with siRNA (B) in A549 cells after 48 h treatment by western blotting. No treatment (Control) and either mock empty vector transfection (A) or mismatched siRNA (B) were used as experimental controls. (C) Heat map of microarray data of apoptotic genes from A549 cells treated as described above. The mRNA signals of hybridization experiments were clustered based on significance. Green indicates negative values and red indicates positive values. Samples were clustered using a hierarchical clustering program to show sample-to-sample relationships (brackets). The full list of genes is in the supplementary figure. (D) and (E) Validation of DAPK2 and CIDEC mRNA expression by real-time qPCR analysis after 48 h of indicated treatment in A549 (D) and H1299 (E) cells. Samples were normalized against β-actin and results expressed as ΔΔCT. (F) Western blotting of CIDEC and DAPK2 protein expression 24 h after PIAS3 transfection.
We initially analyzed 500 genes annotated as apoptotic genes in the microarray (supplementary figure). From these 500 genes a group of 18 genes clustered whose expression was specifically increased by PIAS3 overexpression relative to control and siRNA knockdown cells (Figure 5C). Among them, we selected two genes for validation by real-time qPCR, CIDEC and DAPK2, due to their strong association with apoptosis. CIDEC is a member of the cell death- inducing DNA fragmentation factor α--like effector (CIDE) family that was initially identified as a group of pro-apoptotic proteins.19 DAPK2 (death-associated protein kinase 2) is a member of the serine/threonine protein kinase family whose overexpression induces cell death,20 and restoration of down-regulated DAPK2 expression in Hodgkin lymphoma induces apoptosis.21 After PIAS3 transfection, there was a 9-fold increase in DAPK2 mRNA expression and more than a 6-fold increase in CIDEC mRNA expression in A549 cells compared to mock controls (Figure 5D). siRNA knockdown of STAT3 was less effective, confirming the array results. Identical experiments in H1299 cells produced similar increases in CIDEC mRNA expression, but little change in DAPK2 expression (Figure 5E). Increased DAPK2 protein expression could be detected by western blotting after PIAS3 transfection in both cell types, however this was less apparent for CIDEC expression (Figure 5F). Taken together, these results support our idea that STAT3-independent pathways likely exist for PIAS3-induced apoptosis.
Discussion
Apoptosis is a tightly regulated process and plays an important role in development, maintenance of homeostasis, and elimination of damaged cells.22,23 However, most cancer cells harbor mutations that allow them to evade apoptosis, such as amplification of Bcl-2 or the mutation/loss of tumor suppressors such as TP53 and PTEN. Therefore, overcoming this intrinsic resistance to apoptotic cell death is an important therapeutic goal of many anti-cancer drugs.
PIAS3 is thought to induce growth inhibition through its ability to inhibit STAT3 transcriptional activity.2,6 However, given the constitutive activation of STAT3 in a number of solid tumors, including lung, PIAS3 inhibition of this signal transduction protein also likely promotes apoptosis.24 Here we show that PIAS3 can induce apoptosis in human lung carcinoma cells and does so by stimulating mitochondrial depolarization. This leads to cytochrome c release, apoptosome formation and caspase 3 activation. The mitochondrial depolarization appears to be triggered by a simultaneous loss of Bcl-xL and increased Noxa expression. The loss of Bcl-xL expression likely results from PIAS3 inhibition of STAT3 activity, as Bcl-xL is an established transcriptional target of STAT3.25 It is presently unclear how PIAS3 induces Noxa.
These experimental findings are significant for several reasons. First, PIAS3 induction of apoptosis was p53 independent. This was shown by the ability of PIAS3 to induce apoptosis in H1299 cells, which are p53 null, as well as by its inability to upregulate the expression of several p53 responsive proteins such as Bak, Puma (Figure 3A) and p21 (Figure 4F) in A549 cells, which harbor wild-type p53. PIAS3 also retained its ability to inhibit STAT3 activity in p53 null cells, even though these cells demonstrated higher basal activity (Figure 4B and 4E). This is likely related to the known negative regulatory effect of p53 on STAT3 transcriptional activity13 and may explain the greater aggressiveness, as well as worse prognosis, of lung cancer in patients with p53 abnormalities.26 Because about 49% of NSCLC tumors are p53 mutant, our results indicate that finding ways to up-regulate PIAS3 may promote apoptosis in both p53 mutant and p53 wild-type tumors. In this regard, it has recently been shown that curcumin has the ability to increase endogenous PIAS3 levels in ovarian cancer cells.27 This finding indicates that it may be possible to therapeutically increase PIAS3 expression with small molecules.
Second, PIAS3-induced apoptosis was associated with increased Noxa expression. Noxa, unlike many pro-apoptotic BH3-only proteins, demonstrates selective binding to only one Bcl-2 pro-survival family member, Mcl-1.28 Mcl-1 is critical for survival of many NSCLC cell lines29 and demonstrates limited expression in NSCLC tumors as well.30 Thus, increasing PIAS3 expression may not only represent a new strategy to induce apoptosis in NSCLC, but may also be synergistic with current Bcl-2 directed therapies.31,32 For example, ABT-737 is often ineffective in many lung cancer cells because it has no binding affinity for Mcl-1, so ‘executioner’ binding partners of Bcl-2 proteins, such as Bax, that are displaced by this drug are simply scavenged by Mcl-1, limiting the amount of cell death produced by ABT-737. Thus, PIAS3-induced increases in Noxa will limit the ability of Mcl-1 to scavage Bax and thereby free Bax to oligomerize and induce mitochondrial depolarization and apoptosis. Indeed, we provide proof-of-principal for this synergy in Figure 3B using ABT-263. Noxa expression can also be increased by the proteasome inhibitor bortezomib and the HDAC inhibitor vorinostat,33 expanding the number of potential drugs that could synergize with increased PIAS3.
The effects of PIAS3 on Noxa expression are in addition to its well-known role as an inhibitor of STAT3 activity, which leads to decreased Bcl-xL expression and apoptosis. STAT3 has been a target for drug development for years, with little success due to its lack of intrinsic activity. Other experimental methods for targeting STAT3 have included targeting upstream tyrosine kinases responsible for STAT3 activation34,35 and a variety of agents for which their effect on STAT3 is most likely indirect.36,37 Thus targeting PIAS3 represents a novel strategy to inhibit STAT3 activity. We also investigated the possibility of STAT3-independent mediators of PIAS3-induced apoptosis in our study, and introduced two candidate genes with promising specificity for PIAS3, CIDEC and DAPK2, but these require further validation.
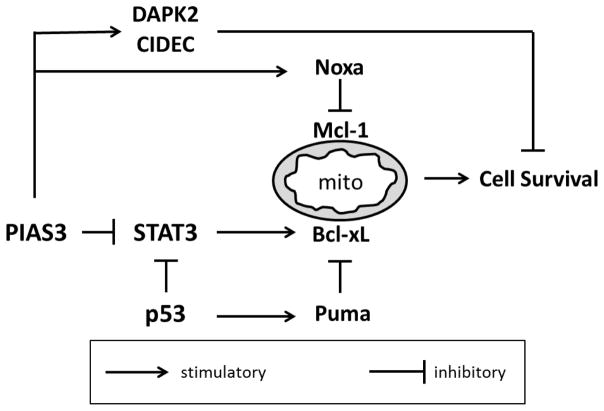
In summary, our results indicate that PIAS3 strongly suppresses the proliferation of NSCLC cells by inducing apoptosis via the intrinsic pathway (see Figure 6). Upstream events leading to apoptosis include decreased STAT3 transcriptional activity with concomitant loss of Bcl-xL expression, or may involve STAT3-independent mechanisms, such as increased DAPK2 and CIDEC expression. Future studies on the mechanism of PIAS3-induced Noxa expression may provide insight into novel therapies targeting the Bcl-2 family. We believe that PIAS3 is a promising candidate around which to build lung cancer strategies because of its efficacy in p53 mutant cancer cells as well as its potential to synergize with existing therapeutic modalities.
Figure 6. Proposed mechanism(s) of PIAS3-induced apoptosis in lung cancer cells.
PIAS3 acts through STAT3 dependent and independent pathways to disrupt Bcl-2 protein family binding equilibrium at the mitochondria (mito) and promote apoptosis. These effects are p53 independent. Puma is shown as one example of how p53 regulates mitochondrial apoptotic pathways.
Supplementary Material
Novelty/Impact.
PIAS3 inhibits cell growth in several lung cancer cell lines although the exact mechanism remains unclear. Our results indicate that PIAS3 activates the intrinsic apoptotic pathway and includes p53- and STAT3-independent mechanisms. We believe this makes PIAS3 a promising candidate around which to build cancer therapeutic strategies.
Acknowledgments
We thank Dr. George Stark for providing the retroviral Phoenix-Ampho packaging cells and Dr. Mark Jackson for providing the p53 shRNA lentiviral construct and HEK293 packaging cells. Supported by grant K23 CA109348 from the National Institutes of Health (AD), a support grant from the University Hospitals Seidman Cancer Center (AD) and the Gene Expression & Genotyping Core Facility of the Case Comprehensive Cancer Center (P30 CA43703). The flow cytometry facility in the Department of Dermatology’s Skin Diseases Research Center (SDRC) is supported in part by the National Institutes of Health Grant (5P30AR039750) and the Ohio Department of Development - Center for Innovative Immunosuppressive Therapeutics (TECH 09-023).
References
- 1.http://www.cancer.org/cancer/lungcancer-non-smallcell/detailedguide/non-small-cell-lung-cancer-key-statistics
- 2.Lai SY, Johnson FM. Defining the role of the JAK-STAT pathway in head and neck and thoracic malignancies: Implications for future therapeutic approaches. Drug Resist Updates. 2010;13(3):67–78. doi: 10.1016/j.drup.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, Bromberg JF. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117(12):3846–56. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. 2005;11(23):8288–94. doi: 10.1158/1078-0432.CCR-05-0827. [DOI] [PubMed] [Google Scholar]
- 5.Cortas T, Eisenberg R, Fu P, Kern J, Patrick L, Dowlati A. Activation state EGFR and STAT-3 as prognostic markers in resected non-small cell lung cancer. Lung Cancer. 2007;55(3):349–55. doi: 10.1016/j.lungcan.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5(8):593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 7.Brantley EC, Nabors LB, Gillespie GY, Choi YH, Palmer CA, Harrison K, Roarty K, Benveniste EN. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res. 2008;14(15):4694–704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kluge A, Dabir S, Vlassenbroeck I, Eisenberg R, Dowlati A. Protein inhibitor of activated STAT3 expression in lung cancer. Mol Oncol. 20115(3):256–64. doi: 10.1016/j.molonc.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dabir S, Kluge A, Dowlati A. The association and nuclear translocation of the PIAS3-STAT3 complex is ligand and time dependent. Mol Cancer Res. 2009;7(11):1854–60. doi: 10.1158/1541-7786.MCR-09-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 11.Harris CC, Hollstein M. Clinical implications of the p53 tumor-suppressor gene. N Engl J Med. 1993;329(18):1318–27. doi: 10.1056/NEJM199310283291807. [DOI] [PubMed] [Google Scholar]
- 12.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351(6326):453–6. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Tang H, Jin X, Jia G, Hsieh JT. p53 regulates Stat3 phosphorylation and DNA binding activity in human prostate cancer cells expressing constitutively active Stat3. Oncogene. 2002;21(19):3082–8. doi: 10.1038/sj.onc.1205426. [DOI] [PubMed] [Google Scholar]
- 14.Unkila M, McColl KS, Thomenius MJ, Heiskanen K, Distelhorst CW. Unreliability of the cytochrome c-enhanced green fluorescent fusion protein as a marker of cytochrome c release in cells that overexpress Bcl-2. J Biol Chem. 2001;276(42):39132–7. doi: 10.1074/jbc.M104986200. [DOI] [PubMed] [Google Scholar]
- 15.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 16.Ulitsky I, Maron-Katz A, Shavit S, Sagir D, Linhart C, Elkon R, Tanay A, Sharan R, Shiloh Y, Shamir R. Expander: from expression microarrays to networks and functions. Nature Protoc. 2010;5(2):303–22. doi: 10.1038/nprot.2009.230. [DOI] [PubMed] [Google Scholar]
- 17.Lazebni A, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371(6495):346–7. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 18.Bayle JH, Grimley JS, Stankunas K, Gestwicki JE, Wandless TJ, Crabtree GR. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol. 2006;13(1):99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Inohara N, Koseki T, Chen S, Wu X, Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17(9):2526–33. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai T, Nomura F, Hoshino K, Copeland NG, Gilbert DJ, Jenkins NA, Akira S. Death-associated protein kinase 2 is a new calcium/calmodulin-dependent protein kinase that signals apoptosis through its catalytic activity. Oncogene. 1999;18(23):3471–80. doi: 10.1038/sj.onc.1202701. [DOI] [PubMed] [Google Scholar]
- 21.Tur MK, Neef I, Jost E, Galm O, Jäger G, Stöcker M, Ribbert M, Osieka R, Klinge U, Barth S. Targeted restoration of down-regulated DAPK2 tumor suppressor activity induces apoptosis in Hodgkin lymphoma cells. J Immunother. 2009;32(5):431–41. doi: 10.1097/CJI.0b013e31819f1cb6. [DOI] [PubMed] [Google Scholar]
- 22.Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 23.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407(6805):770–6. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 24.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8(4):945–54. [PubMed] [Google Scholar]
- 25.Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene. 2000;19(56):6613–26. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 26.Campling BG, El-Deiry WS. Clinical implication of p53 mutation in lung cancer. Mol Biotechnol. 2003;24(2):141–56. doi: 10.1385/MB:24:2:141. [DOI] [PubMed] [Google Scholar]
- 27.Saydmohammed M, Joseph D, Syed V. Curcumin suppresses constitutive activation of STAT-3 by up-regulating protein inhibitor of activated STAT-3 (PIAS3) in ovarian and endometrial cancer cells. J Cell Biochem. 2010;110(2):447–56. doi: 10.1002/jcb.22558. [DOI] [PubMed] [Google Scholar]
- 28.Ploner C, Kofler R, Villunger A. Noxa: at the tip of the balance between life and death. Oncogene. 2009;27(Suppl 1):S84–S92. doi: 10.1038/onc.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Guttikonda S, Roberts L, Uziel T, Semizarov D, Elmore SW, Leverson JD, Lam LT. Mcl-1 is critical for survival in a subgroup of non-small-cell lung cancer cell lines. Oncogene. 2011;30(16):1963–8. doi: 10.1038/onc.2010.559. [DOI] [PubMed] [Google Scholar]
- 30.Sakakibara-Konishi J, Oizumi S, Kikuchi J, Kikuchi E, Mizugaki H, Kinoshita I, Dosaka-Akita H, Nishimura M. Expression of Bim, Noxa, and Puma in non-small cell lung cancer. BMC Cancer. 2012;12(286):286. doi: 10.1186/1471-2407-12-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogler M, Dinsdale D, Dyer MJS, Cohen GM. Bcl-2 inhibitors: small molecules with a big impact on cancer therapy. Cell Death Differ. 2009;16(3):360–67. doi: 10.1038/cdd.2008.137. [DOI] [PubMed] [Google Scholar]
- 32.Hauck P, Chao BH, Litz J, Krystal GW. Alterations in the Noxa/Mcl-1 axis determine sensitivity of small cell lung cancer to the BH3 mimetic ABT-737. Mol Cancer Ther. 2009;8(4):883–92. doi: 10.1158/1535-7163.MCT-08-1118. [DOI] [PubMed] [Google Scholar]
- 33.Barbone D, Cheung P, Battula S, Busacca S, Gray SG, Longley DB, Bueno R, Sugarbaker DJ, Fennell DA, Broaddus VC. Vorinostat eliminates multicellular resistance of mesothelioma 3D spheroids via restoration of Noxa expression. PLoS One. 2012;7(12):e52753. doi: 10.1371/journal.pone.0052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dowlati A, Nethery D, Kern JA. Combined inhibition of epidermal growth factor receptor and JAK/STAT pathways results in greater growth inhibition in vitro than single agent therapy. Mol Cancer Ther. 2004;3(4):459–63. [PubMed] [Google Scholar]
- 35.Quintas-Cardama A, Verstovsek S. Molecular pathways: JAK/STAT pathway: mutations, inhibitors, and resistance. Clin Can Res. 2013;19(8):1933–40. doi: 10.1158/1078-0432.CCR-12-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dowlati A, Kluge A, Nethery D, Halmos B, Kern JA. SCH66336, inhibitor of protein farnesylation, blocks signal transducer and activators of transcription 3 signaling in lung cancer and interacts with a small molecule inhibitor of epidermal growth factor receptor/human epidermal growth factor receptor 2. Anticancer Drugs. 2008;19(1):9–16. doi: 10.1097/CAD.0b013e3282f1a908. [DOI] [PubMed] [Google Scholar]
- 37.Turkson J, Zhang S, Mora LB, Burns A, Sebti S, Jove R. A novel platinum compound inhibits constitutive Stat3 signaling and induces cell cycle arrest and apoptosis of malignant cells. J Biol Chem. 2005;280(38):32979–88. doi: 10.1074/jbc.M502694200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.