Abstract
Microchimerism represents a condition where one individual harbors genetically distinct cell populations, and the chimeric population constitutes <1% of the total number of cells. The most common natural source of microchimerism is pregnancy. The reciprocal cell exchange between a mother and her child often leads to the stable engraftment of hematopoietic and non-hematopoietic stem cells in both parties. Interaction between cells from the mother and those from the child may result in maternal immune cells becoming sensitized to inherited paternal alloantigens of the child, which are not expressed by the mother herself. Vice versa, immune cells of the child may become sensitized toward the non-inherited maternal alloantigens of the mother. The extent of microchimerism, its anatomical location, and the sensitivity of the techniques used for detecting its presence collectively determine whether microchimerism can be detected in an individual. In this review, we focus on the clinical consequences of microchimerism in solid organ and hematopoietic stem cell transplantation, and propose concepts derived from data of epidemiologic studies. Next, we elaborate on the latest molecular methodology, including digital PCR, for determining in a reliable and sensitive way the extent of microchimerism. For the first time, tools have become available to isolate viable chimeric cells from a host background, so that the challenges of establishing the biologic mechanisms and function of these cells may finally be tackled.
Keywords: pregnancy, transplantation, graft-versus-host disease, maternal antigen, paternal antigen, mixed chimerism, microchimerism, FACS sorting, qPCR, digital PCR, single cell analysis, monoclonal antibodies
History of chimerism
The very first observation of chimerism was reported in 1945 by Ray Owen1 (reviewed in ref. 2). A cow had given birth to twin calves, which turned out to be derived from two different bulls. This form of fraternal twinning is relatively common in cattle. Furthermore, each of these genetically dissimilar twins carried blood antigens from the mother as well as from both sires. Owen went on to systematically study the blood type in 80 pairs of bovine heterozygotic twin calves, and he found that it was identical between the dissimilar twins in the majority of cases.1 He attributed this result to vascular anastomosis between the placentas of bovine twins. The individual calves displayed a situation, which is nowadays called “mixed chimerism,” where cells from two distinct zygote lineages coexist in one organism. In rare human cases, blood from healthy donors, who have a twin sibling, was found to be a mixture of two kinds of blood cells.3-5 Studies by Billingham, Brent, Medawar, and Hasek provided support for the principle that mammals and birds immunologically react only to a limited extent to foreign tissue cells to which they have been previously exposed in fetal/neonatal life.6-8 In neonatal mice artificially-induced “macrochimerism” by blood cell transfusion led to antigen-specific tolerance and the acceptance of a skin graft given later in life.6 Owen and colleagues later provided evidence for the existence of actively acquired tolerance to rhesus (Rh) blood group antigens: Rh-negative children of Rh-positive mothers acquire persistent tolerance toward the Rh antigen,9 possibly as a result of exposure to the antigen in the uterus. The concept, that the degree of the mother’s tolerance toward the child’s RhD antigen is related to the RhD status of the grandmother,9 and the role of chimerism therein, has recently been debated.10
Mixed Chimerism (Macrochimerism) as a Result of Hematopoietic Stem Cell Transplantation (HSCT)
The balance between immunity and immune regulatory mechanisms will determine the degree of alloimmune responses induced in transplant recipients, and may thereby have significant clinical implications in relation to overall survival. However, immune regulation does not occur without immunity. This concept also applies to conditions of donor-specific immunologic tolerance. Tolerance to particular antigens probably arises from the fact that the effect of regulatory immune cells dominates over mechanisms exploited by effector type immune cells. To understand the biologic mechanisms of microchimerism, as discussed below, it is helpful to first evaluate the underlying immune modulatory mechanisms of mixed chimerism (macrochimerism) and their impact on clinical outcome.
Mixed chimerism: the ultimate platform to induce immunological tolerance to solid allografts?
The feasibility of inducing long-lasting donor-specific tolerance through the establishment of mixed or even full donor chimerism has been studied in animal models and in humans.11-16 This type of immune intervention may form the ultimate clinical platform for obtaining sustained allograft function without the need for life-long immunosuppressive medication. The immunological concept of this approach is simple: together with active removal of pre-existing host T cells by conditioning therapy, infusion of hematopoietic stem cells obtained from the same donor as the one who will donate the kidney allograft introduces new donor-derived antigen-presenting cells (APC) along the host-derived APC that were already present. Both types of APC will, among other locations, end up in the thymus where they facilitate the deletion of high avidity T cells that are specific for donor- or host-specific alloantigens. Consequently, the new peripheral alloreactive T cell pool generated after the HSCT procedure will be grossly devoid of cells that recognize donor-specific human leukocyte antigen (HLA) class I or II molecules expressed on cells present in the subsequently transplanted kidney allograft. This so called deletional tolerance occurs through positive and negative selection of immature host T cells facilitated by thymus-residing APC (central tolerance), as well as through deletion of host alloreactive T cells upon interaction with donor HLA-expressing leukocytes present in the periphery (peripheral tolerance).
The concept outlined above was shown to be robust in small and large animal models.15,17,18 In the nineties, sporadic clinical cases have been described where immunologic tolerance toward the renal allograft was observed after a bone marrow transplant from the same donor, wherein no or minimal dosing of immunosuppressive medication was applied.19-21 In this new millennium, the potential applicability of combined HSCT and kidney transplantation procedures has been further explored in the clinic.13,14,22,23 From seven patients who underwent such combined treatment for multiple myeloma and end-stage renal failure, five are alive with no signs of a relapse of the malignancy, and three out of these five have normal to near-normal renal function in complete absence of immunosuppressive therapy.23 Although substantial co-morbidities were observed in this very small patient group, sustained mixed chimerism, particularly in the T cell population, is perhaps the most ideal setting to assure protection from allograft rejection by host T cells as well as from graft-vs.-host disease (GvHD) inflicted by donor T cells. Similarly, durable chimerism in transplant recipients was associated with tolerance toward the donor kidney allograft after a mobilized stem cell graft (enriched for hematopoietic stem cells and plasmacytoid precursor dendritic cells) and non-myeloablative conditioning.24 GvHD was not observed. Presence of sustained chimerism was used as an indicator for successfully weaning off immunosuppressive therapy by one year after transplantation in five transplant recipients.
Persistent mixed chimerism is indicative of the co-presence of host- and donor-derived regulatory T cells
Persistent mixed chimerism is frequently observed in Thalassemia major patients after they have undergone HLA-identical HSCT following myeloablative conditioning. In this setting, the proportion of residual host hematopoiesis may range from 10 to 70%.25 Although residual (host-derived) APC after HSCT are generally considered as a risk factor for graft rejection, Thalassemia major patients in whom host and donor cells co-persist for more than two years after graft infusion often remain blood transfusion-independent for prolonged periods of time.26,27 T cell cloning experiments showed that mixed chimerism in these patients is associated with the presence of both effector cells and regulatory T cells (Tregs) reactive against host or donor alloantigens.27 The Treg clones reciprocally inhibited proliferation and IFNγ release by effector type alloreactive T cells of host and donor origin, respectively. The immunesuppressive function of these Tregs depends, at least in part, on the cytokine IL-10. These observations point out that persistent clinical presentation of mixed chimerism after allogeneic HSCT is likely the result of both central tolerance and peripheral tolerance induced by host- and donor-derived alloreactive Tregs. The next challenge will be to find effective and safe conditioning regimens, which favor the induction of stable mixed chimerism without unwanted side effects, as myeloablative conditioning is often accompanied by toxicity issues that may cause severe transplantation-related morbidity and mortality.
Microchimerism as a Result of Solid Organ Transplantation
Donor microchimerism in the recipient
In contrast to hematopoietic macrochimerism (often >25% as observed by Owen, Medawar, and colleagues, and by the group of Sykes and Sachs), a much lower extent of chimerism, usually less than 0.1%, is frequently seen after solid organ transplantation. Low levels of donor chimerism, referred to as “microchimerism,” result from the migration of passenger hematopoietic (stem)cells from the allograft into the peripheral circulation and tissues of the recipient. While macrochimeric conditions appear to be related to tolerance mechanisms, the clinical impact of donor microchimerism in solid organ transplantation is less clear.28 In several studies it was shown that microchimerism in humans is associated with graft acceptance29,30 and a lower incidence of rejection31,32 after kidney, liver or small bowel transplantation. In one of those studies, transplant recipients, who displayed microchimerism at 2 mo after graft kidney implantation, had a significantly lower incidence of biopsy-proven acute rejection during the subsequent four years compared with patients who were microchimerism-negative.32 Whereas in a mouse model it was shown that microchimerism was the causal factor in maintaining deletion of donor-specific CD8+ T cells,33 microchimerism after human transplantation does not seem to be accompanied by diminished anti-donor T cell responses.31 In a girl who had received a liver allograft from a deceased, unrelated male donor, spontaneous donor chimerism (94–100%) was observed in T, B, and NK cells, and granulocytes at 13 mo post-transplantation.34 After reduction and complete withdrawal of all immunosuppressive medication the recipient showed immunological tolerance toward the graft.
However, several clinical studies did not show an effect of microchimerism on acceptance of heart, liver, and kidney grafts 35–37. Microchimerism after liver transplantation has even been observed coinciding with graft rejection.38,39 Discrepancies in the clinical impact of microchimerism may be the result of inter-patient differences in the level or subtypes of microchimeric cells. Indeed, the type of microchimeric cell has been advocated to be the underlying factor determining whether microchimerism is related to tolerance or immunity toward host-derived foreign cells.
Recipient chimerism in the donor organ
Recipient chimerism in transplants probably results from migration of precursor- and/or stem cells from the recipient into the transplanted donor organ. Recipient chimerism was observed in endothelial cells of kidney grafts.40 The highest frequencies of chimerism were seen during the most severe types of rejection, i.e., vascular rejection. In another study, 88% of kidney transplant patients had chimerism in 2.4 to 6.6% of tubular epithelial cells in their donor graft.41 This tubular chimerism was not related to clinical outcome. Renal allografts undergoing chronic dysfunction often contain mesenchymal cells from recipient origin in vascular and interstitial compartments.42 In female donor heart transplants, Y-positive chimeric cells of male recipients were observed in 14 to 20% of myocytes, coronary arterioles, and capillaries.43 Chimeric cells were positive for stem cell markers and early heart cell differentiation markers. Recipient chimerism was, however, not related to clinical outcome.
The results outlined above suggest that precursor cells migrate from the recipient into the graft, a process possibly triggered or enhanced by donor tissue damage. However, in many of those cases studied solely by fluorescent in situ hybridization (FISH), it is plausible that the male cells found in female donor hearts may in fact be derived pre-implantation, i.e., via male offspring of female allograft donors.44
Pregnancy-Induced Immunization and Fetal-Maternal Microchimerism
The most common natural source of microchimerism is pregnancy. The maternal-fetal interface in the placenta harbors both cells from the mother and from the fetus. Progenitor cell trafficking over the placenta may induce microchimerism for a prolonged period of time after birth in the mother and the child.45-47 Maternal immune cells may become sensitized to mismatched inherited paternal antigens (IPA) expressed by cells or cellular debris acquired from the developing fetus. This is reflected by the formation of HLA alloantibodies to IPA in a significant proportion of parous women.48-50 These antibodies are often directed against mismatched HLA-A, -B and –DR antigens, which are not expressed on trophoblast cells, the fetal cell layer which forms the border between mother and child. Therefore it is most likely that fetus-derived chimeric leukocytes are responsible for the induction of these alloantibodies. In addition, the continuous release until birth of placental antigens into the maternal circulation facilitates the systemic induction of maternal effector type as well as regulatory type T cells.
Human fetal or maternal microchimeric cells can be found in circulating blood cells such as granulocytes,51 lymphocytes,52 monocytes,53 and myeloid dendritic cells54,55 as well as in organs and tissues56 (Table 1). While blood-borne CD45+ chimeric cells are obviously derived from engrafted hematopoietic stem cells,57,58 the origin and characteristics of the stem cell giving rise to tissue-resident microchimerism, as found in bone marrow, endocrine organs, and other tissues (reviewed in ref. 59) is still unclear. Three candidates have been suggested in this context: mesenchymal stromal cells,60 pregnancy-associated progenitor cells,61 and transdifferentiating hematopoietic stem cells.62
Table 1. Naturally acquired, circulating, or tissue-resident microchimeric cell types identified in humans and mice.
| Hematopoietic cell types | References |
|---|---|
| CD34+ / c-kit+ stem cells | 57,58,68 |
| CD66b+ granulocytes | 51,86 |
| CD3+ T cells | 53,86 |
| CD56+ CD16+ natural killer cells | 53 |
| CD20+ B cells | 53,86 |
| CD14+ monocytes | 53,86 |
| CD11b+ macrophages | 68 |
| CD11c+ myeloid dendritic cells | 55,86,157 |
| Non-hematopoietic cell types | References |
| CD45- / c-kit- bone marrow mesenchymal stromal cells | 60,68 |
| Cardiomyocytes | 44,158 |
| Kidney tubular epithelial cells | 44 |
| Hepatocytes | 44,56 |
| Lung epithelial cells | 56 |
| Pancreatic insulin-producing β cells | 56,159 |
Using quantitative PCR technology for the detection of nonshared polymorphic HLA genes unique to mother- and fetus-derived cells, it was shown that adult women often carry multiple types of naturally acquired chimeric cells derived from different sources, i.e., from their mother and from their offspring. Addressing the prevalence of these two types of chimeric cells specifically within short-lived and continuously replenished cells of myeloid origin, Nelson and colleagues demonstrated that maternal microchimerism is actually more common in granulocytes obtained from adult females than fetal microchimerism.51 Neither the prevalence nor concentration of fetal cells in the maternal circulation seems to be affected by the number of pregnancies. In contrast, the prevalence and level of maternal cells was found to be significantly lower in multiparous women.46 This unexplained decline in maternal microchimeric cells, ultimately resulting in the dominance of fetal microchimerism in time, could be the result of reciprocal alloimmune reactions between fetus- or mother-derived immune cells (graft-graft interactions), resulting in a survival benefit for one of the two parties as is often seen in the setting of double umbilical cord blood (UCB) transplantation.63,64 Alternatively, host alloimmune responses directed against circulating fetal chimeric cells may additionally lead to a specific loss of tolerance to non-inherited maternal antigens (NIMA) expressed by the maternal grandmother; the latter situation could eventually lead to specific eradication of HLA class Ilow hematopoietic stem cells of grandmother origin by host HLA class I-restricted effector cells, while leaving NIMAneg fetal stem cells untouched.
Maternal microchimerism and tolerance to NIMA in the offspring
In 1953, Billingham and colleagues stated that they “are inquiring into the possibility that [actively acquired tolerance] may occur naturally by the accidental incorporation of maternal cells into the fetus during normal development.”6 Fifty-five years later, Mold and colleagues showed that maternal cells engraft in fetal lymph nodes after having crossed the placenta,65 leading to 0.0035–0.83% of cells in mesenteric lymph nodes being of maternal origin. Specific tolerance of the child’s immune system to non-shared NIMA is maintained long after birth.65,66 Noteworthy, mesenteric lymph nodes belong to a set of organs and tissues collectively termed the mucosa. Natural exposure of naïve T cells to (allo)antigens encountered at the mucosa or in mucosa-draining lymph nodes generally leads to the induction of immunological tolerance. This may explain why successful establishment of NIMA-specific tolerance in newborn mice depends on maternal cells acquired both through placental exchange during fetal development and through nursing after birth.67,68
The processes underlying neonatal tolerance induction to NIMA in humans are poorly understood.69 Mold and colleagues demonstrated that chimeric maternal lymphocytes detectable in fetal mesenteric lymph nodes are protected from immune-mediated destruction by the presence of high numbers of fetal Tregs.65 Whereas fetal T cells show a proliferative response when combined in vitro with APC from the blood of unrelated adult donors, they do not respond against the NIMA expressed by maternal cells.65 In the latter case, fetal alloreactive T cell expansion is suppressed by fetal CD4+CD25highFoxp3+ Tregs. In addition to the site where microchimeric cells are encountered, also the type of chimeric cells could be instrumental for the induction of NIMA-specific tolerance. Burlingham and coworkers proposed that maternal MHC class IIpos cells from hematopoietic origin such as dendritic cells and macrophages are instrumental for presentation of non-shared maternal antigens to NIMA-specific fetal CD4+ Treg.68 The potent suppressive function of these NIMA-specific CD4+ Treg was elegantly demonstrated in transplant models wherein NIMA-expressing heart allografts were protected from rejection when implanted in offspring mice, which had been exposed prior to transplantation to the same NIMA antigen as present on the heart allograft during pregnancy and neonatal life.70,71 Besides dendritic cells, also circulating T cells have been associated with allograft tolerance induction in mice and humans.54,72,73 Whether single-cell type chimerism or multilineage chimerism, resulting in co-existing myeloid and lymphoid chimeric cells, is required for successful induction and life-long maintenance of alloantigen-specific tolerance74 remains to be studied in more detail. Based on the identification of CD8+ T regulator cells specific for NIMA presented in the context of maternal MHC class I alleles,66 it should be noted that also maternal chimeric cells lacking MHC class II expression, i.e., tissue-resident somatic cells, may be involved in the induction of Treg.
In a more recent study Mold and colleagues showed that fetal naïve CD4+ T cells are much more responsive to stimulation with allogeneic cells, but more prone to develop into CD25+Foxp3+ Tregs, compared with adult CD4+ T cells.75 The findings in humans described above have been confirmed in mice. While fetal tolerance to NIMA seems to be grossly mediated by the effects of alloantigen-specific Tregs,67 also B cells may become unresponsive to NIMA during fetal life: in 26 highly-sensitized patients awaiting a kidney transplant, a preferential non-responsiveness of alloantibodies to NIMA was found.76 Thus, several studies have provided evidence that natural exposure to maternal microchimeric cells favors the induction of NIMA-specific tolerance in the offspring.
Priming to NIMA
In contrast to development of NIMA-specific tolerance, there is also evidence that exposure to NIMA can lead to alloantigen-specific priming.66 It has been proposed that the quality and quantity of NIMA exposure determines whether tolerance or priming will occur.69 In this setting, tolerance may develop as a result of prolonged exposure to NIMA, both during pregnancy and thereafter during prolonged periods of breastfeeding. In support of this concept, oral exposure of mice to NIMA by breastfeeding led to improved outcome of offspring-to-mother bone marrow transplantation later in life.77 Furthermore, whereas chronic exposure to low doses of antigen often leads to development of Tregs78 or to cytotoxic T cell non-responsiveness, small doses induce specific cytotoxic T cell priming.79,80
Detection of functionally different types of T cells directed against fetal IPA in parous females
As extensively studied in murine pregnancy models, maternal Treg imprinting is essential for normal pregnancy outcome after both syngeneic and allogeneic pregnancy.81 In the human setting, fetal-specific CD8+ T lymphocytes have been observed in half of all pregnancies analyzed.82 These T cells typically become detectable after the first trimester, display an effector memory phenotype, and persist in the postnatal period. To study the functional features of such pregnancy-induced T cells, several studies analyzed the prevalence and functional properties of T cells specific for Y chromosome-encoded minor histocompatibility antigens. These so called HY antigens are small peptides derived from intracellular proteins, which are presented by HLA class I or class II alleles at the surface of male cells. To date, many different HY peptides have been identified which cause potent alloimmune responses in gender mismatched transplantation settings. Using a HY peptide/multimer-based sorting procedure, HY-specific T cells with classical cytolytic function have been isolated from blood samples collected either during82 or after66,83,84 male pregnancies. Using the trans-vivo delayed-type hypersensitivity (tvDTH) assay,85 also the presence of HY-specific Tregs in women with a documented pregnancy history has been reported.66 This assay detects the presence of Tregs, which are able to suppress footpad swelling induced by chemokines and cytokines released by co-activated recall antigen-specific memory T cells. In approximately half of the parous women analyzed, HY-specific Tregs could be detected up to several decades after the delivery of a son. Some women show profound HY-specific regulation, but others show only marginal suppression of recall-antigen-induced foot pad swelling responses in the presence of HY peptide(s).86 It is therefore tempting to speculate that either HY-specific regulatory T cells or cytolytic T cells dominate in some women with sons, but that these functionally different types of T cells co-exist in others.
The alloimmune status of individuals, who bear microchimeric cells
As discussed above, fetal and maternal microchimeric cells are found regularly among blood-borne cells and tissue-bound cells. By studying the offspring from specifically selected mouse breeding models, it has become clear that the levels of maternal microchimerism in blood and various organs such as the heart is directly correlated with the presence of CD4+ Tregs directed against NIMA antigens that have been encountered during fetal or neonatal life.67 Also in humans the putatively lifelong presence of maternal microchimeric cells is extensively documented. Likewise, the presence of male microchimerism in adult women has been widely reported. Despite the relatively high frequencies of chimerism detected in many women, there does not seem to be a correlation between the level or subtype(s) of circulating IPA-expressing chimeric cells and the presence of IPA-specific Treg.86 Interestingly, male chimerism is not necessarily restricted to women with male offspring, given that trans-maternal flow of male cells derived from a former pregnancy or from a twin sibling who died in utero can be transferred during a next pregnancy.47,55,87 Consequently, female cord blood units may comprise functional T cells directed against alloantigens, which are not expressed by the mother but which have been encountered through trans-maternal cell flow of chimeric cells.55 Likewise, the alloimmune T cell repertoire of multiparous women may contain considerable precursor frequencies of cells directed against the various mismatched HLA alleles expressed by either the maternal grandmother or by her offspring, or against mismatched minor histocompatibility antigens presented by shared HLA antigens.
Potential Clinical Benefits of Pre-Transplant Existing Microchimerism
Pre-transplant induced alloimmunization to mismatched alloantigens expressed by microchimeric cells may affect transplant outcome when such individuals enroll in a transplantation program. In case of durable maternal-specific tolerance in the child, this may have clinical consequences in case the child undergoes a transplant procedure later in life. Indeed, this has been found to be the case in the kidney transplant setting: graft survival was significantly higher in recipients of kidneys from siblings expressing NIMA than in recipients of kidneys from siblings expressing non-inherited paternal antigens (NIPA).88 The tolerogenic effect of having encountered NIMA prior to transplantion may be employed in the setting of unrelated donor and cord blood HSCT,89–91 for instance by including the NIMA in matching criteria.
HSCT using adult donors
Allogeneic HSCT is the curative treatment option for many hematological malignancies as well as for several life-threatening hematological diseases. The success of this treatment is largely determined by selection of the optimal stem cell donor, according to HLA typing of both the stem cell recipient and donor. One of the major causes of morbidity and mortality following adult or pediatric HSCT is GvHD, which may even occur in the case of full HLA compatibility between donor and recipient. Mismatched minor histocompatibility antigens expressed by the transplant recipient, but not the donor, are evoking the sometimes devastating immune responses in sibling transplantation. The involvement of alloreactive T cells in this process is illustrated by the detection of minor histocompatibility antigen-specific T lymphocytes in blood92 and tissues93 obtained from patients who developed GvHD after infusion of a Tcell-containing graft. GvHD can be even more severe in case of HLA mismatching, i.e., after transplantation with HLA-haploidentical donors (donors that share one haplotype with the recipient, e.g., parents to children or vice versa or transplants between haploidentical siblings). The so called graft-vs.-leukemia (GvL) reactions represent the other aspect of immunologic reactions mediated by the donor-host interactions. GvL reactions may contribute to regression of leukemia or prevention of a relapse after allogeneic transplantation. Lastly, rejection of a donor graft can be mediated by immunologic reactions against mismatched minor or major histocompatibility antigens.
Donor parity is a risk factor for the development of GvHD in both male and female recipients of a transplant from a HLA-identical donor.94 Interestingly, donor or recipient parity is also frequently associated with GvHD seen after HSCT between monozygotic twins, a setting in which neither HLA nor minor histocompatibility antigen disparities are involved.95 The donor parity effect has been attributed to the presence of fetal microchimeric cells in female donors. Such “foreign” microchimeric cells likely act as triggers of alloimmune responses by donor T cells, but the exact immunological mechanism by which fetal microchimerism could increase the GvHD risk in the recipient has yet to be determined.
Superior transplant outcome in recipients of maternal grafts
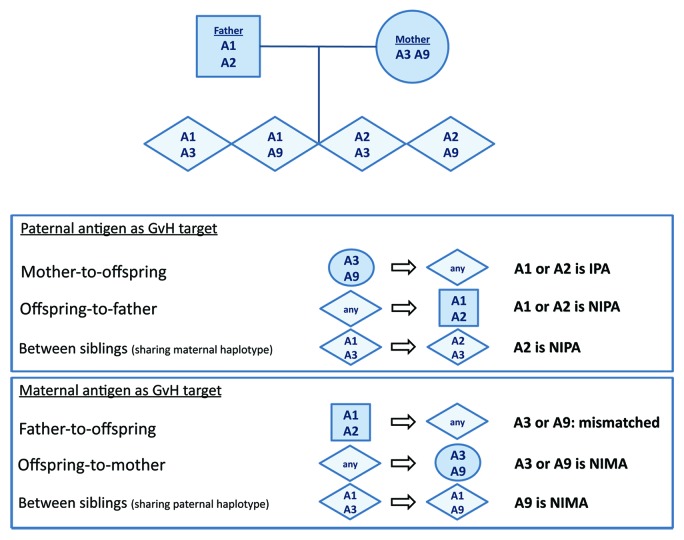
The persistence of bidirectional microchimerism between mother and child may have implications in family HSCT transplantation settings. Three studies have shown that in haploidentical transplantation, the parental origin of the mismatched HLA haplotype determines outcome. In the first study, Van Rood and colleagues analyzed a subset of haploidentical transplants (father-to-offspring, mother-to-offspring, or among HLA haploidentical siblings; see Fig. 1 for exemplary scheme).96 When the transplantation was performed among siblings who shared a paternal haplotype, the incidence of acute GvHD was significantly lower compared with all other donor-recipient combinations. This was attributed to the fact that the target for GvHD reactions was the maternal NIMA, to which the donor had been rendered tolerant during fetal life. All other situations (father-to-offspring, mother-to-offspring or transplants between siblings mismatched for the maternal NIPA) were associated with a higher incidence of acute and chronic GvHD. Notably, in the latter cases there had been no intrauterine exposure of the donor cells to GvHD targets present in the recipient. The incidence of graft rejection was lower when using maternal grafts compared with paternal grafts.
Figure 1. Exemplary scheme of shared and non-shared HLA antigens in the mother, the father, and the offspring. In this example the father has HLA-A1 and -A2 and the mother has HLA-A3 and -A9. In hematopoietic stem cell transplantation, the inherited paternal antigen (IPA), non-inherited paternal antigen (NIPA), and non-inherited maternal antigen (NIMA) have been indicated in case of a mother-to-offspring, offspring-to-mother, father-to-offspring, offspring-to-father or offspring-to-offspring combination.
In a Japanese study of T-cell-replete transplants, donors were selected to have detectable microchimerism (by PCR) for antigens present in the recipient.97 The donors were either siblings that shared paternal antigens (GvHD target was NIMA), mothers donating to offspring, or children who were used as donors for their mother. Fathers were not used as donors, nor were siblings who shared a maternal haplotype. Among 35 donor/recipient pairs, 15 were transplants from mother-to-offspring and the GvH reactivity was against the paternal HLA of the recipient (the IPA). In 20 cases, a transplant from offspring was infused in mother or in NIMA-incompatible offspring. The latter 20 cases had a significantly reduced risk of grade III-IV acute GvHD, presumably because exposure in utero to the NIMA had led to the development of NIMA-specific immune regulatory cells in the donor. Vice versa, it was proposed that the maternal grafts had developed persisting life-long immunity against IPA due to previous exposure of the maternal immune system to fetal antigens during pregnancy. Consequently, the presence of pregnancy-induced alloimmune effector T cells in the grafts of maternal donors may have caused more GvHD.
The third study by an Italian group emphasized the same principle in the setting of T-cell-depleted HSCT, which significantly reduces the GvHD risk. Pharmacological post-transplant immune suppression is not used in this transplant procedure. Comparing transplant outcomes from maternal and paternal grafts, they found that maternal grafts were associated with reduced rates of disease recurrence.98 This was seen in both female and male recipients. The improved overall survival mainly resulted from a lower incidence of relapse of malignant hematologic disease, as the incidence of acute GvHD was not significantly different between maternal and paternal grafts. It was proposed98 that in this case the primed maternal grafts mainly induce a beneficial GvL effect. GvHD is avoided because of profound T cell depletion. The authors speculate that the few T cells transferred with the graft (in this study, a median of 104/kg, i.e., a total of 0.5–1 million cells per graft), may have undergone unopposed proliferation after infusion in the recipient by virtue of the absence of pharmacologic GvHD prophylaxis. Two other retrospective studies confirm the superior survival of blood or marrow stem cell recipients given maternal grafts over recipients given paternal grafts.97,99 Whether pregnancy-induced memory T cells specific for shared paternal major or minor histocompatibility antigens83,100 expressed by the transplant recipient’s leukemic cells are among the expanding female donor-derived T cells remains to be studied.
Umbilical cord blood transplants
UCB transplantation is increasingly used in transplantation of children and adults, who lack a matched unrelated donor or extended family donor. Compared with transplants from adult donors with similar degrees of HLA disparity, UCB transplantation is associated with a reduced rate of disease recurrence and a decreased incidence of GvHD.101 The relapse rate may even be lower after double UCB transplantation than after a single UCB transplantation, although the latter hypothesis was not confirmed in a recent randomized study.102 The reason for the reduced rate of disease recurrence after UCB transplant has remained elusive, but may be indirectly related to maternal microchimerism. Emerging evidence indicates that the fetal immune system, and thus relevant for UCB grafts, is geared toward peripheral tolerance maintained by Tregs. This situation is thought to result in lifelong tolerance to NIMA encountered during fetal/neonatal life. Fetal tolerance to NIMA may be an important mechanism by which the GvHD risk is reduced after UCB transplantation. Van Rood and colleagues analyzed a group of UCB transplant recipients, of whom the complete HLA typing of the infant (donor of the UCB unit) and of the infant’s mother were available.103 Reexposure of cord blood to NIMA expressed by the recipient of the UCB graft improves transplant outcome in hematological malignancies. Most of the UCB recipients were HLA-mismatched with their donors at one or several antigens. But in some cases the HLA mismatch was identical to an HLA antigen present in the mother of the infant donor (NIMA match). In other cases, the mismatched HLA antigen was not present in the donor’s mother (no-NIMA match). Treatment-related mortality was significantly lower in the recipients of a NIMA-matched transplant, in part because of faster engraftment, but also because of a lower relapse rate. The authors speculated that cord blood, thought to be generally tolerant toward NIMA, might sometimes be sensitized, as was described for minor histocompatibility-antigen HA-1H specific T cells.104 In a more recent case-control study, the outcome of 48 NIMA-matched UCB transplants was compared with that of 116 non-NIMA-matched UCB transplants.90 The groups were matched for important covariates. Transplantation-related mortality was lower and overall survival significantly higher after NIMA-matched UCB transplantation compared with NIMA-mismatched UCB transplants. The effect of NIMA matching on relapse rate was not analyzed in this study.
Antileukemia effect and anti-IPA immunity exerted by microchimeric cells present in cord blood grafts
Van Rood and colleagues retrospectively analyzed in a large number of UCB transplants the HLA type of both mother and UCB, and deduced the IPA of the UCB.105 Transplants with IPA-targeting UCB were identified, in which the maternal lymphocytes would be primed against the IPA of the UCB and also against cells of the recipient who expresses the same IPA. Such was the case in most donor-recipient pairs. They compared the outcome of this group with the outcome of a limited number of cases where there was no IPA reactivity. They found an increased rate of disease recurrence in the latter group. Importantly, the decreased leukemia recurrence rate after IPA-targeted UCB transplantation was not associated with increased GvHD, possibly resulting from the effectiveness of co-transferred NIMA-specific fetal Tregs. Similar to possible mechanisms explaining the beneficial outcome of patients with a maternal graft, it was speculated in this particular paper105 and in related editorials,106,107 that maternal cells—earlier being primed against the IPA of the child and present in the UCB graft—confer a potent GvL effect. The results from these consecutive studies provide the first epidemiological evidence that anti-IPA immunity by maternal immune cells may play a role in the control of relapse in acute myeloid leukemia and acute lymphatic leukemia after unrelated cord blood transplantation.
The major challenge to date is how to address the actual role of microchimeric cells in UCB, a question that can only be addressed after isolation of these cells from the host background. One of the first questions arising from the proposed concept of antileukemia effects by maternal cells presumably present in cord blood grafts is which type of maternal cells is doing the job. These could be both CD4+ and CD8+ T cells, the importance of which has been described for GvL reactivity in mice.108,109 Furthermore, NK cells may contribute to the anti-leukemia effect (reviewed in ref. 110). Since anti-leukemic effects of NK cells are probably less prominent than those of T cells, the GvL effect by NK cells may come to the foreground after haploidentical transplantation in combination with rigorous T cell depletion. The same may hold true for UCB transplantation, given the significant delay in T cell recovery in this setting.111 Second, do microchimeric maternal cells require HLA specificity for targeting leukemic cells in the recipient after transplantation? Van Rood and colleagues showed that HLA antigens, i.e., those that the recipient shared with the cord blood, represent one target of relapse-reducing immunity. It has been proposed that eradication of leukemic cells results from Fas- or perforin-mediated mechanisms that are typically displayed by lymphocytes.110,112 Such mechanisms may even be more prevalent in situations of HLA-related, maternal-derived immunity. It is also possible that the GvL effect is established due to the mere presence of sensitized maternal cells within the graft, which are more immunogenic than the cord blood cells.
Detection and Enrichment of Microchimeric Targets
In the animal and human solid organ transplantation setting, chimerism could often not be detected by the serologic assays that were available at the time. Only after the introduction of the polymerase chain reaction (PCR), which led to a greatly enhanced sensitivity of chimerism measurement, it became possible to detect rare donor cells in the circulation and organs of the transplant recipient. An essential step in investigating the clinical consequences of microchimerism is the availability of sensitive tools to detect microchimerism. Furthermore, isolation of viable microchimeric cells from the host is essential for further characterization and functional analysis of such cells. In the next paragraphs an overview of the different methodologies used is given. We have summarized these methodologies in Table 2, along with the sensitivity, advantages, and disadvantages.
Table 2. Techniques for detecting of microchimerism1.
| Technique | Sensitivity2 | Advantages | Disadvantages |
|---|---|---|---|
| FISH | 10−2-10−6 (depending on target and method) | Identification in context of tissue morphology | Takes lot of time for analysis (up to 2 wk per sample) in case of high sensitivity |
| Nested PCR | 10−4 or lower | Higher sensitivity than direct PCR | High chance of false positivity by contamination; not quantitative |
| Real-time qPCR | 5x10−4-10−5 (depending on target) | Quantitative | Detection at low percentages requires running of multiplicates |
| Clamp PCR | 10−4, down to 10−5 or lower | Reduction of background levels | Cumbersome to find optimal probes; LNA modifications costly; novel methodology |
| WGA+qPCR | similar as qPCR | Allows more analyses per sample | No enhancement of sensitivity; requires additional manipulation of the sample |
| Single cell analysis | - | Information per cell | Cells need pre-enrichment; limited # of cells in analysis; may be most suitable for RNA |
| Digital PCR | 5 x 10−5-5 x 10−6 | Reliable and reproducible; background copies reduced | Conventional qPCR assays need to be optimized for digital PCR |
| HLA FACS sorting | ~10−4 | Generates viable chimeric cells for functional studies | Limited sensitivity; requires access to high-quality, allele-specific HLA mAbs |
1 FISH, fluorescence in situ hybridization; WGA, whole genome amplification; PCR, polymerase chain reaction; qPCR, quantitative PCR; LNA, locked nucleic acid; FACS, fluorescence-activated cell sorting; mAb, monoclonal antibody. 210−4 = 1 in 10,000 cells (0.01%); 10−5 = 1 in 100,000 cells (0.001%); 10−6 = 1 in 1,000,000 cells (0.0001%).
In situ detection of tissue-resident microchimeric cells
Fluorescence in situ hybridization (FISH) is a popular technique to identify microchimeric targets: chimeric cells can be identified within the context of tissue morphology and localization. Traditionally, sex chromosome-based XY-FISH has been used for chimerism analysis, whereby male cells are identified in a female genetic background. FISH can give false-positive results in rare cell conditions. The use of two different Y probes113 and the application of rehybridization with X and Y chromosomes with reverse colors114 may enhance reliability. Within the context of autoimmune disease, the group of Deforce has applied XY-FISH to count the number of Y-positive, fetal cells in women.115 With this approach they were able to distinguish 8–29 fetal cells/106 maternal cells in women with autoimmune disease from 0–2 fetal/106 maternal cells in healthy, pregnant women, either before delivery or several months postpartum.
PCR-based methodologies
Both classical and real-time quantitative PCR (qPCR) techniques have been used for detection of (micro)chimerism.116-119 Increased sensitivity may be achieved when the preliminary PCR is followed by second-round PCR with sequence-specific primers that target the first amplicon.120-123 However, as we (M.M., B.M.K.-M., J.J.M.D., M.E.; unpublished observations) and others124,125 have found, nested PCR is prone to decreased specificity and to contamination by nonspecific PCR products, which results in false-positive results.
Real-time qPCR offers an accurate and rapid tool compared with conventional PCR analysis of DNA tandem repeats, which is classically used to assess macrochimerism levels after HSCT.126 Multicopy genes localized on the Y-chromosome represent suitable targets for reproducible and sensitive analyses by qPCR.29,57,117,127,128 Its application is, however, restricted to sex-mismatched combinations, whereby male DNA targets are targeted in a female host. DYZ1 primers target a Y-chromosome selective sequence that is found 8 times in total in the DAZ2, DAZ3, and DAZ4 genes. The benefit of this assay is that it offers a relatively high sensitivity of 0.001% (1 in 100 000 cells detected; 10−5). This sensitivity is approximately 10-fold higher than that of single-copy genomic targets such as mismatched HLA genes and insertion/deletion regions.
One cell contains an estimated 7 pg DNA. A typical input of 0.5 μg DNA per conventional qPCR assay is expected to contain approximately 70 000 amplifiable targets. With 1 in 100 000 microchimeric targets, this would mean that on average 0.7 copies of DNA are available in the sample for amplification. According to expected stochastic results predicted by the Poisson equation, one copy is present in 82% of the time.138 For conventional qPCR, this means that in practice a single sample should be analyzed in multiplicate to detect microchimerism with a frequency ≤ 0.001%.
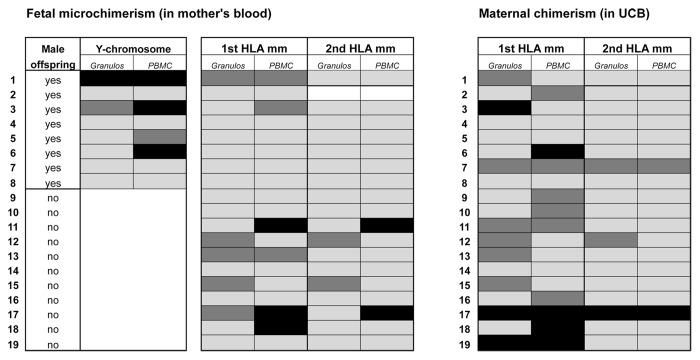
HLA represents a broad applicable target for microchimerism detection,116,117,129,130 as they are highly polymorphic131 and thus frequently mismatched between two individuals, irrespective whether there is a gender mismatch or not. Over the past years, several research groups, including those from Lee Nelson and Nathalie Lambert (Seattle WA) and from Diana Bianchi (Boston MA), have been developing HLA-allele specific qPCR assays. Using both DYZ1 and in-house developed HLA-directed primer sets, we have investigated presence of microchimerism in blood lymphocytes of 19 mother-child combinations in the PBMC fraction (Fig. 2). With only DYZ1, male microchimerism was detected in 4 out of 8 (50%) mothers who gave birth to a son. In the other 11 cases, DYZ1 was regarded as being not informative, since the women gave birth to a daughter. In line with other reports,132,133 we observed in a more recent analysis of a larger group of women with no sons, i.e., nulliparous women or females who had given birth to a daughter, a fairly high percentage of women displaying male microchimerism.86 With HLA-directed primers, 6 out of 19 (32%) showed weak or clear positivity of fetal microchimerism (Fig. 2). Two of these (#11 and #17; 11%) could be verified by a qPCR assay that targets a second fetus-specific HLA allele. Maternal microchimerism in the PBMC fractions of the 19 corresponding UCB samples was seen more frequently, namely in 10 cases (53%). Two of these (UCB7 and UCB 17) could be verified using a second HLA target.
Figure 2. Fetal and maternal microchimerism in mother-child combinations. Presence of microchimerism was investigated using Y-chromosome-specific and HLA-directed primer sets on DNA from umbilical cord blood (UCB) and peripheral blood of 19 mother-child combinations. Mean gravidity of the women was 2.8 ± 1.5, and mean parity was 1.4 ± 0.8. In most cases, blood was collected at a time point between 1 d before delivery and 1 d after delivery. Blood cells present in the Ficoll interface (indicated as ‘PBMC’) and in the Ficoll pellet (indicated as “granulocytes”) were separately processed for DNA extraction and microchimerism analysis by qPCR. Black boxes: positive signal (microchimerism detected). Dark gray boxes: weakly positive signal. Light gray boxes: negative signal (no microchimerism detected). Empty boxes: no informative marker available.
Insertion-deletion (indel) polymorphisms118,119,134,135 localized throughout the genome may represent complimentary targets for PCR, which in contrast to the HLA complex may enable clearer distinction of the specific allele from the off-target allele. Other targets that can be used to discriminate the chimeric population from a host are short-tandem repeats, genes encoding the Rhesus system, minor histocompatibility genes, and the killer-cell immunoglobulin-like receptor (KIR) genes. Mitochondrial DNA (mtDNA) may be a useful target for detecting microchimerism. This type of DNA is contained in the mitochondria on an extra-chromosomal and circular genome, and holds highly polymorphic regions in the displacement loop.136 Since every cell contains hundreds of mitochondria, each having many circular chromosomes, mtDNA represents a naturally enriched source of target DNA for microchimerism detection in the transplantation setting. Mitochondrial DNA is inherited solely from the mother, and thus its measurement cannot be applied for addressing pregnancy-related maternal and fetal microchimerism.
Enhanced PCR assays by probe modification
Novel PCR-based technology and modification of primer/probe sequences may further enhance sensitivity and specificity of PCR assays aimed at detecting microchimerism. Much of this technology has been pioneered in the field of cancer biology for detecting low-level cancer-related mutations.
Inclusion of one or more locked nucleic acid (LNA) modifications within primer sequences has resulted in improved results. LNAs enable high affinity binding of the probe to the complementary sequence, leading to enhanced sensitivity and specificity of the PCR assay. A relatively novel strategy is detection of the specific (microchimeric) allele in combination with blocking of amplification of the off-target (host) allele. CastPCR (Life Technologies) is aimed at detection of somatic mutations in cancer genes. Allele-specific qPCR primers that detect the mutant allele are combined with allele-specific Minor Groove Binder (MGB) blocker oligonucleotides, which suppress non-specific amplification of the off-target (wild type) allele. It is claimed that the assays can detect as few as 1 to 5 mutant copies in up to one million of background copies. PCR clamping with peptide nucleic acid (PNA)- or LNA-modified oligomers allows sensitive detection of targets present at a low frequency, and at the same time inhibits amplification of non-target (background) DNA.137 The combination of allele-specific priming, competitive probe blocking, and melt curve analysis has allowed single copy detection of variant alleles down to a sensitivity of at least 0.001% (10−5).138 The methodological approaches outlined above may be customized to detect appropriate target alleles for application in the microchimerism field.
Nucleic acid pre-amplification and single cell analysis
To multiply the amount of starting material before allele-specific qPCR analyses, whole-genome amplification with multiple displacement amplification (MDA) technology can be performed. Application of this technique gives up to 10 000-fold amplification of the whole genome.139,140 Due to the unbiased amplification of the genomic DNA, more tests can be performed by qPCR on one sample compared with situations without such pre-amplication. This may be a benefit, especially in the light of the need for running multiplicates of a certain microchimeric target allele in the same sample.
Since a cell contains multiple copies of mRNA transcripts, produced from any gene sequence encoded within the DNA, RNA lysates represent a naturally enriched source for the detection of microchimerism. On top of that, starting quantities down to the nanogram or even picogram range of RNA can be amplified in an unbiased manner. Availability of RNA amplication methodology has opened the possibility of expression profiling in single cells. Fluidigm has introduced the C1 Single-Cell AutoPrep System. Cells need to be enriched first, for example by FACS sorting, and 750–1000 cells (in 5 μL) are then entered into the microfluidic system. The C1 technology allows capturing of single cell in each chamber, the possibility of visual verification under the microscope, and profiling of each of the individual cells. The capillary system, 10 to 25 μm in size, captures single cells into 96 separate chambers, where cDNA is transcribed and pre-amplified. Subsequently, gene expression profiling and even whole transcriptome sequencing can be performed on each individual eluate. AmpliGrid technology (Advalytix) makes use of a similar approach: single cells are deposited into 48 different reaction sites by FACS sorting, which then undergo reverse transcription and real-time qPCR.
Digital PCR: a possible novel tool for microchimerism detection
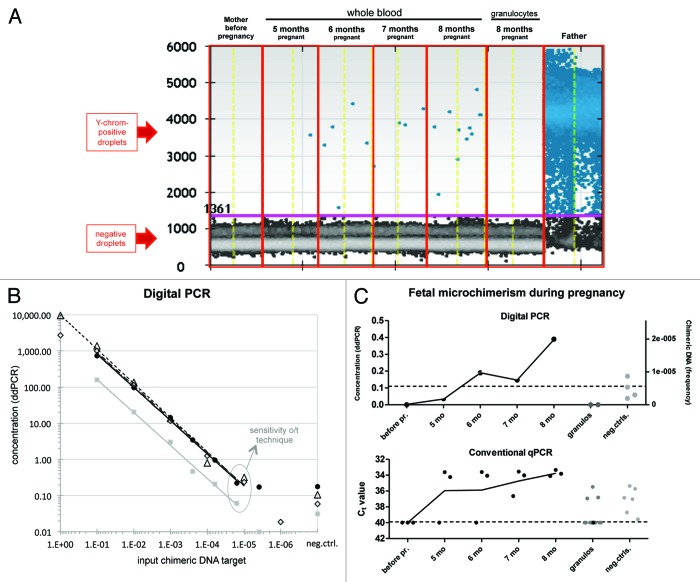
Digital PCR represents the latest generation in PCR technology: each sample is diluted into partitions (separate reaction chambers), and the number of partitions in which a reaction occurs are counted up.141 The benefit of ddPCR over conventional qPCR is enhanced accuracy and reproducibility. Fluidigm and Life Technologies provide a system of reaction chambers on chips (microscope slides) or plates. In droplet digital (dd)PCR (Bio-Rad and RainDance) the nucleic acids in the lysate are dispersed into thousands to millions droplets at a concentration of less than one genome equivalent per droplet. The droplets are subsequently transferred to tubes, and subjected to PCR to analyze per droplet whether a reaction has occurred (Fig. 3A). A Poisson algorithm is then applied for determination of absolute copy numbers.
Figure 3. Droplet digital PCR (Bio-Rad, QX100) results for microchimeric targets. (A) A pregnant woman was monthly monitored for fetal microchimerism by PCR for the Y chromosome in whole blood and in the granulocyte fraction. Each dot in the picture represents one droplet. The software distinguishes between positive droplets (the upper ones) and negative droplets (the lower ones) for the DNA target of interest. Each condition was analyzed in duplicate, and results were merged in the software. The total input of DNA per reaction was 270 ng. (B) DNA from one donor was diluted in DNA from another donor, in different ratios (10−1 to 5 × 10−6). Linear correlation between input and observed percentage was obtained at chimeric frequencies down to 2 × 10−5 (1 in 50 000 cells) for both Y-chromosome- (black circles, triangles, and diamonds) and indel-region (S09b)-targeting PCR assays (gray squares). The standard for the Y target was performed three times, each consisting of two to five replicates. (C) Comparison between ddPCR- and conventional qPCR results for the Y-chromosome PCR in pregnancy samples and in negative controls.
Digital PCR has been used to analyze rare DNA targets in samples, e.g., for detecting fetal DNA in maternal plasma142 and viral marker genes in single bacterial cells.143 This “needle-in-a-haystack” strategy makes the system the perfect tool to detect low numbers of microchimeric targets. Detection of chimeric DNA targets with a frequency of 1 in 10 000 background copies (0.01%; 10−4) with 95% reliability requires screening of at least 30 000 copies (~200 ng DNA) in total. When aiming to detect lower microchimeric frequencies, the amount of DNA as input needs to be increased to 0.5–1 μg and distributed over two to four wells. The number of positive and negative partitions from different wells can be added up and merged. Researchers have even been able to detect one mutant allele in a background of 200 000 wild type alleles (0.0005%; 5 × 10−6).141,144
Using ddPCR for the detection of Y chromosomes, we monthly monitored fetal microchimerism in peripheral blood samples obtained from a pregnant colleague known to carry a male fetus. Y chromosome-specific DNA was first detected in whole blood after 6 mo of pregnancy, and the extent of male DNA further increased over time (Fig. 3A). We obtained a sensitivity around 2 × 10−5 for PCRs targeting DYZ1 (Fig. 3B, black symbols) or an indel region (S09b; Fig. 3B, gray symbols). The multicopy benefit of the Y chromosome targeted by conventional qPCR does not apply for ddPCR. The extent of false positivity in negative controls is reduced in ddPCR compared with conventional qPCR (see Fig. 3C), since amplification of genomic targets in separately dispersed partitions increases PCR specificity by isolating the target signal from competing background.
Enrichment of microchimeric cells from bulk populations
With percentages of microchimerism of 0.01 or lower it would not be practically and economically feasible to analyze ≥10 000 single cells in search of that one microchimeric target. Instead, enrichment of chimeric targets may be reached by laser pressure catapulting of individual cells in a cytospin. Researchers from Austria have used laser microdissection to catapult rare, single candidate target cells from a total cytospin onto AmpliGrid slides.145-147 To verify the genomic identity of the target cell, it was subsequently subjected to whole genome amplification146 and DNA fingerprint analysis. DNA profiling on single cells could be used in 86% of the times to verify the genomic background of the cell.145 Full DNA profiles could be obtained with fewer than 10 cells (~70 pg) as input after on-chip DNA amplification.148,149
Once processed for qPCR or in situ analysis, functional studies of chimeric cells are precluded. One method for pre-enriching chimeric cell populations may be to separate cell subpopulations from the total lysate using magnetic cell sorting (MACS). This strategy especially holds true when clinical importance of the chimerism is expected to be related to a particular cell lineage where it resides. In such case, lineage-specific analysis of chimerism may be more informative.150,151 In most samples, purity of >90% was reached after MACS for the T cell (CD3+), monocyte (CD14+), and granulocyte (CD15+) subsets.152 In the majority of samples the subpopulations provided at least 0.5 ng/μL of DNA, which was a prerequisite for reliable qPCR-based chimerism analysis.152
Viable microchimeric cells can also be targeted and separated from the host environment by FACS sorting. Flow cytometry with anti-HLA mAbs is already used as a monitoring tool for chimerism after HLA-mismatched HSCT.153 In the late seventies, Herzenberg and colleagues described a FACS enrichment procedure of chimeric cells: anti-HLA-A2 antibodies were used to target fetal cells in a background of maternal peripheral blood lymphocytes.154 We have developed a set of 120 human HLA-specific monoclonal antibodies in our laboratory. Single HLA mAb labeling of cell populations that are present in a frequency of 0.4% or lower leads to suboptimal separation from the host cells. Double HLA antibody labeling was found to be essential for optimal separation of low-frequency microchimeric cells.155,156 Purity of the sorted samples was verified by qPCR analysis for HLA class I and class II alleles.156 We further applied HLA-targeted FACS sorting on UCB samples to separate microchimeric maternal cells. The methodology is applicable in case the HLA type of the minor cell population is known, for example for microchimerism enrichment in pregnancy and transplantation, and in case the frequencies of microchimeric cells are relatively high, as is often seen in UCB samples. For detection of fetal HLA targets in maternal blood before delivery, one could target HLA antigens from both paternal haplotypes, provided that the antibodies used for labeling the chimeric cells do not cross react as is often the case with HLA-specific antibodies.
Summary and Objectives for Future Research
We aimed to compose a comprehensive overview of the literature on naturally-acquired and transplant-associated microchimerism, and describe its effect on clinical transplant outcome. The results from literature reports on these complex topics are often contradictory. Furthermore, there is a requirement for systematic and in depth studies on the (patho)physiologic mechanisms of microchimerism and corresponding cell types in order to understand the interesting but as yet poorly understood epidemiological data obtained from clinical transplant settings.
Adult and cord blood HSCT may benefit from HLA matching criteria whereby the IPA and NIMA are taken into account. The NIMA effect in solid organ transplantation observed in studies that appeared in the previous century seems to have become less prominent nowadays, probably as a result of the change in the immunosuppressive medication regime over the years. To establish whether persistence of the NIMA effect later in life is enforced by prolongation of microchimeric conditions in the host would be subject of investigation. It remains elusive whether increased frequencies of maternal chimerism in cord blood samples are related to NIMA-specific Tregs. Likewise, several questions remain to be solved around the concept of antileukemia effects by maternal cells with anti-IPA immunity present in cord blood grafts. Do microchimeric maternal cells require HLA specificity for targeting leukemic cells in the recipient after transplantation? And why do the microchimeric anti-IPA-directed maternal cells cause no harm to the cord blood cells present in the same storage bag? Are maternal effector cells actively suppressed by (maternal) regulatory cells, and if so, which circumstances in the recipient could stimulate the anti-leukemic effect of the microchimeric maternal cells after infusion into the recipient?
We expect that recent developments in molecular biological tools for microchimerism detection, along with possibilities of isolating microchimeric cells, will propel future research into clarifying functional mechanisms (in)directly associated with the pre-transplant presence of chimeric cells. Viably isolated cells may eventually be cloned and non-specifically increased in numbers using conventional culture conditions. The methodology of HLA-targeted cell sorting may represent an additional tool for phenotypic characterization of microchimeric cells allowing molecular and functional studies of these cells in relation to the host environment. Such studies will step by step unravel the immunobiology behind the epidemiological findings as discussed in this review.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to the Macropa Foundation for financially supporting microchimerism research performed at the Leiden University Medical Center. We thank Carin van der Keur (Dept of IHB) for processing blood samples from mothers and their offspring, as shown in Figure 2. We are grateful to our colleague Monica Mirabile and her partner George van Zanten for participating in the monitoring study, and to Eveline Vreeswijk for collecting the corresponding blood samples. We thank Eddy van Collenburg (Bio-Rad) and Berit Kemps (Dept of IHB, LUMC) for technical support in generating the results obtained by the QX100 digital droplet PCR. Astrid van Halteren is a scholar of the National Blood Foundation/American Association of Blood Banks.
References
- 1.Owen RD. Immunogenetic consequences of vascular anastomoses between bovine twins. Science. 1945;102:400–1. doi: 10.1126/science.102.2651.400. [DOI] [PubMed] [Google Scholar]
- 2.Crow JF. A golden anniversary: cattle twins and immune tolerance. Genetics. 1996;144:855–9. doi: 10.1093/genetics/144.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunsford I, Bowley CC, Hutchison AM, Thompson JS, Sanger R, Race RR. A human blood-group chimera. Br Med J. 1953;2:81. doi: 10.1136/bmj.2.4827.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth PB, Plaut G, James JD, Ikin EW, Moores P, Sanger R, Race RR. Blood chimerism in a pair of twins. Br Med J. 1957;1:1456–8. doi: 10.1136/bmj.1.5033.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholas JW, Jenkins WJ, Marsh WL. Human blood chimeras a study of surviving twins. Br Med J. 1957;1:1458–60. doi: 10.1136/bmj.1.5033.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billingham RE, Brent L, Medawar PB. Actively acquired tolerance of foreign cells. Nature. 1953;172:603–6. doi: 10.1038/172603a0. [DOI] [PubMed] [Google Scholar]
- 7.Hasek M. Tolerance phenomena in birds. Proc R Soc Lond B Biol Sci. 1956;146:67–77. doi: 10.1098/rspb.1956.0072. [DOI] [PubMed] [Google Scholar]
- 8.Hasek M, Hraba T, Hort J. Acquired immunological tolerance of heterografts. Nature. 1959;183:1199–200. doi: 10.1038/1831199a0. [DOI] [PubMed] [Google Scholar]
- 9.Owen RD, Wood HR, Foord AG, Sturgeon P, Baldwin LG. Evidence for actively acquired tolerance to Rh antigens. Proc Natl Acad Sci USA. 1954;40:420–4. doi: 10.1073/pnas.40.6.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunker PA. Chimerism in transfusion medicine: the grandmother effect revisited. Chimerism. 2013;4:119–25. doi: 10.4161/chim.26912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sachs DH. Mixed chimerism as an approach to transplantation tolerance. Clin Immunol. 2000;95:S63–8. doi: 10.1006/clim.1999.4814. [DOI] [PubMed] [Google Scholar]
- 12.Sykes M. Mixed chimerism and transplant tolerance. Immunity. 2001;14:417–24. doi: 10.1016/S1074-7613(01)00122-4. [DOI] [PubMed] [Google Scholar]
- 13.Scandling JD, Busque S, Dejbakhsh-Jones S, Benike C, Millan MT, Shizuru JA, Hoppe RT, Lowsky R, Engleman EG, Strober S. Tolerance and chimerism after renal and hematopoietic-cell transplantation. N Engl J Med. 2008;358:362–8. doi: 10.1056/NEJMoa074191. [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, Cosimi AB, Spitzer TR, Tolkoff-Rubin N, Suthanthiran M, Saidman SL, Shaffer J, Preffer FI, Ding R, Sharma V, et al. HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med. 2008;358:353–61. doi: 10.1056/NEJMoa071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sachs DH, Sykes M, Kawai T, Cosimi AB. Immuno-intervention for the induction of transplantation tolerance through mixed chimerism. Semin Immunol. 2011;23:165–73. doi: 10.1016/j.smim.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai T, Sachs DH, Sykes M, Cosimi AB, Immune Tolerance Network HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med. 2013;368:1850–2. doi: 10.1056/NEJMc1213779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard RJ, Sutherland DE, Lum CT, Lewis WI, Kim TH, Slavin S, Najarian JS. Kidney allograft survival in dogs treated with total lymphoid irradiation. Ann Surg. 1981;193:196–200. doi: 10.1097/00000658-198102000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storb R, Yu C, Sandmaier BM, McSweeney PA, Georges G, Nash RA, Woolfrey A. Mixed hematopoietic chimerism after marrow allografts. Transplantation in the ambulatory care setting. Ann N Y Acad Sci. 1999;872:372–5, discussion 375-6. doi: 10.1111/j.1749-6632.1999.tb08481.x. [DOI] [PubMed] [Google Scholar]
- 19.Sayegh MH, Fine NA, Smith JL, Rennke HG, Milford EL, Tilney NL. Immunologic tolerance to renal allografts after bone marrow transplants from the same donors. Ann Intern Med. 1991;114:954–5. doi: 10.7326/0003-4819-114-11-954. [DOI] [PubMed] [Google Scholar]
- 20.Helg C, Chapuis B, Bolle JF, Morel P, Salomon D, Roux E, Antonioli V, Jeannet M, Leski M. Renal transplantation without immunosuppression in a host with tolerance induced by allogeneic bone marrow transplantation. Transplantation. 1994;58:1420–2. [PubMed] [Google Scholar]
- 21.Sorof JM, Koerper MA, Portale AA, Potter D, DeSantes K, Cowan M. Renal transplantation without chronic immunosuppression after T cell-depleted, HLA-mismatched bone marrow transplantation. Transplantation. 1995;59:1633–5. [PubMed] [Google Scholar]
- 22.Starzl TE. Immunosuppressive therapy and tolerance of organ allografts. N Engl J Med. 2008;358:407–11. doi: 10.1056/NEJMe0707578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spitzer TR, Sykes M, Tolkoff-Rubin N, Kawai T, McAfee SL, Dey BR, Ballen K, Delmonico F, Saidman S, Sachs DH, et al. Long-term follow-up of recipients of combined human leukocyte antigen-matched bone marrow and kidney transplantation for multiple myeloma with end-stage renal disease. Transplantation. 2011;91:672–6. doi: 10.1097/TP.0b013e31820a3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leventhal J, Abecassis M, Miller J, Gallon L, Ravindra K, Tollerud DJ, King B, Elliott MJ, Herzig G, Herzig R, et al. Chimerism and tolerance without GVHD or engraftment syndrome in HLA-mismatched combined kidney and hematopoietic stem cell transplantation. Sci Transl Med. 2012;4:24ra28. doi: 10.1126/scitranslmed.3003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreani M, Nesci S, Lucarelli G, Tonucci P, Rapa S, Angelucci E, Persini B, Agostinelli F, Donati M, Manna M. Long-term survival of ex-thalassemic patients with persistent mixed chimerism after bone marrow transplantation. Bone Marrow Transplant. 2000;25:401–4. doi: 10.1038/sj.bmt.1702151. [DOI] [PubMed] [Google Scholar]
- 26.Andreani M, Manna M, Lucarelli G, Tonucci P, Agostinelli F, Ripalti M, Rapa S, Talevi N, Galimberti M, Nesci S. Persistence of mixed chimerism in patients transplanted for the treatment of thalassemia. Blood. 1996;87:3494–9. [PubMed] [Google Scholar]
- 27.Serafini G, Andreani M, Testi M, Battarra M, Bontadini A, Biral E, Fleischhauer K, Marktel S, Lucarelli G, Roncarolo MG, et al. Type 1 regulatory T cells are associated with persistent split erythroid/lymphoid chimerism after allogeneic hematopoietic stem cell transplantation for thalassemia. Haematologica. 2009;94:1415–26. doi: 10.3324/haematol.2008.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burlingham WJ. Chimerism after organ transplantation: is there any clinical significance? Clin Transplant. 1996;10:110–7. [PubMed] [Google Scholar]
- 29.Starzl TE, Demetris AJ, Murase N, Ildstad S, Ricordi C, Trucco M. Cell migration, chimerism, and graft acceptance. Lancet. 1992;339:1579–82. doi: 10.1016/0140-6736(92)91840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starzl TE, Demetris AJ, Trucco M, Murase N, Ricordi C, Ildstad S, Ramos H, Todo S, Tzakis A, Fung JJ, et al. Cell migration and chimerism after whole-organ transplantation: the basis of graft acceptance. Hepatology. 1993;17:1127–52. doi: 10.1002/hep.1840170629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bettens F, Tiercy JM, Campanile N, Giostra E, Majno P, Rubbia L, Roosnek E, Mentha G, Villard J. Microchimerism after liver transplantation: absence of rejection without abrogation of anti-donor cytotoxic T-lymphocyte-mediated alloreactivity. Liver Transpl. 2005;11:290–7. doi: 10.1002/lt.20360. [DOI] [PubMed] [Google Scholar]
- 32.Pujal JM, Grinyó JM, Gil-Vernet S, Caldes A, Hernández P, Mestre M, Encuentra M, Perez-Garcia A, Gallardo D. Early hematopoietic microchimerism predicts clinical outcome after kidney transplantation. Transplantation. 2007;84:1103–11. doi: 10.1097/01.tp.0000286172.57076.df. [DOI] [PubMed] [Google Scholar]
- 33.Bonilla WV, Geuking MB, Aichele P, Ludewig B, Hengartner H, Zinkernagel RM. Microchimerism maintains deletion of the donor cell-specific CD8+ T cell repertoire. J Clin Invest. 2006;116:156–62. doi: 10.1172/JCI26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander SI, Smith N, Hu M, Verran D, Shun A, Dorney S, Smith A, Webster B, Shaw PJ, Lammi A, et al. Chimerism and tolerance in a recipient of a deceased-donor liver transplant. N Engl J Med. 2008;358:369–74. doi: 10.1056/NEJMoa0707255. [DOI] [PubMed] [Google Scholar]
- 35.Suberbielle C, Caillat-Zucman S, Legendre C, Bodemer C, Noël LH, Kreis H, Bach JF. Peripheral microchimerism in long-term cadaveric-kidney allograft recipients. Lancet. 1994;343:1468–9. doi: 10.1016/S0140-6736(94)92583-6. [DOI] [PubMed] [Google Scholar]
- 36.Schlitt HJ, Hundrieser J, Hisanaga M, Uthoff K, Karck M, Wahlers T, Wonigeit K, Pichlmayr R. Patterns of donor-type microchimerism after heart transplantation. Lancet. 1994;343:1469–71. doi: 10.1016/S0140-6736(94)92584-4. [DOI] [PubMed] [Google Scholar]
- 37.Elwood ET, Larsen CP, Maurer DH, Routenberg KL, Neylan JF, Whelchel JD, O’Brien DP, Pearson TC. Microchimerism and rejection in clinical transplantation. Lancet. 1997;349:1358–60. doi: 10.1016/S0140-6736(96)09105-2. [DOI] [PubMed] [Google Scholar]
- 38.Schlitt HJ, Hundrieser J, Ringe B, Pichlmayr R. Donor-type microchimerism associated with graft rejection eight years after liver transplantation. N Engl J Med. 1994;330:646–7. doi: 10.1056/NEJM199403033300919. [DOI] [PubMed] [Google Scholar]
- 39.Sivasai KS, Alevy YG, Duffy BF, Brennan DC, Singer GG, Shenoy S, Lowell JA, Howard T, Mohanakumar T. Peripheral blood microchimerism in human liver and renal transplant recipients: rejection despite donor-specific chimerism. Transplantation. 1997;64:427–32. doi: 10.1097/00007890-199708150-00010. [DOI] [PubMed] [Google Scholar]
- 40.Lagaaij EL, Cramer-Knijnenburg GF, van Kemenade FJ, van Es LA, Bruijn JA, van Krieken JH. Endothelial cell chimerism after renal transplantation and vascular rejection. Lancet. 2001;357:33–7. doi: 10.1016/S0140-6736(00)03569-8. [DOI] [PubMed] [Google Scholar]
- 41.Mengel M, Jonigk D, Marwedel M, Kleeberger W, Bredt M, Bock O, Lehmann U, Gwinner W, Haller H, Kreipe H. Tubular chimerism occurs regularly in renal allografts and is not correlated to outcome. J Am Soc Nephrol. 2004;15:978–86. doi: 10.1097/01.ASN.0000120369.92378.54. [DOI] [PubMed] [Google Scholar]
- 42.Grimm PC, Nickerson P, Jeffery J, Savani RC, Gough J, McKenna RM, Stern E, Rush DN. Neointimal and tubulointerstitial infiltration by recipient mesenchymal cells in chronic renal-allograft rejection. N Engl J Med. 2001;345:93–7. doi: 10.1056/NEJM200107123450203. [DOI] [PubMed] [Google Scholar]
- 43.Quaini F, Urbanek K, Beltrami AP, Finato N, Beltrami CA, Nadal-Ginard B, Kajstura J, Leri A, Anversa P. Chimerism of the transplanted heart. N Engl J Med. 2002;346:5–15. doi: 10.1056/NEJMoa012081. [DOI] [PubMed] [Google Scholar]
- 44.Koopmans M, Kremer Hovinga IC, Baelde HJ, Fernandes RJ, de Heer E, Bruijn JA, Bajema IM. Chimerism in kidneys, livers and hearts of normal women: implications for transplantation studies. Am J Transplant. 2005;5:1495–502. doi: 10.1111/j.1600-6143.2005.00858.x. [DOI] [PubMed] [Google Scholar]
- 45.Maloney S, Smith A, Furst DE, Myerson D, Rupert K, Evans PC, Nelson JL. Microchimerism of maternal origin persists into adult life. J Clin Invest. 1999;104:41–7. doi: 10.1172/JCI6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gammill HS, Guthrie KA, Aydelotte TM, Adams Waldorf KM, Nelson JL. Effect of parity on fetal and maternal microchimerism: interaction of grafts within a host? Blood. 2010;116:2706–12. doi: 10.1182/blood-2010-02-270942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gammill HS, Nelson JL. Naturally acquired microchimerism. Int J Dev Biol. 2010;54:531–43. doi: 10.1387/ijdb.082767hg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Rood JJ, Eernisse JG, Van Leeuwen A. Leucocyte antibodies in sera from pregnant women. Nature. 1958;181:1735–6. doi: 10.1038/1811735a0. [DOI] [PubMed] [Google Scholar]
- 49.Payne R, Rolfs MR. Fetomaternal leukocyte incompatibility. J Clin Invest. 1958;37:1756–63. doi: 10.1172/JCI103768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hönger G, Fornaro I, Granado C, Tiercy JM, Hösli I, Schaub S. Frequency and determinants of pregnancy-induced child-specific sensitization. Am J Transplant. 2013;13:746–53. doi: 10.1111/ajt.12048. [DOI] [PubMed] [Google Scholar]
- 51.Cuddapah SC, Gadi VK, de Laval de Lacoste B, Guthrie KA, Nelson JL. Maternal and fetal microchimerism in granulocytes. Chimerism. 2010;1:11–14. doi: 10.4161/chim.1.1.13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams Waldorf KM, Gammill HS, Lucas J, Aydelotte TM, Leisenring WM, Lambert NC, Nelson JL. Dynamic changes in fetal microchimerism in maternal peripheral blood mononuclear cells, CD4+ and CD8+ cells in normal pregnancy. Placenta. 2010;31:589–94. doi: 10.1016/j.placenta.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loubière LS, Lambert NC, Flinn LJ, Erickson TD, Yan Z, Guthrie KA, Vickers KT, Nelson JL. Maternal microchimerism in healthy adults in lymphocytes, monocyte/macrophages and NK cells. Lab Invest. 2006;86:1185–92. doi: 10.1038/labinvest.3700471. [DOI] [PubMed] [Google Scholar]
- 54.Cai J, Lee J, Jankowska-Gan E, Derks R, Pool J, Mutis T, Goulmy E, Burlingham WJ. Minor H antigen HA-1-specific regulator and effector CD8+ T cells, and HA-1 microchimerism, in allograft tolerance. J Exp Med. 2004;199:1017–23. doi: 10.1084/jem.20031012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dierselhuis MP, Blokland EC, Pool J, Schrama E, Scherjon SA, Goulmy E. Transmaternal cell flow leads to antigen-experienced cord blood. Blood. 2012;120:505–10. doi: 10.1182/blood-2012-02-410571. [DOI] [PubMed] [Google Scholar]
- 56.Stevens AM, Hermes HM, Kiefer MM, Rutledge JC, Nelson JL. Chimeric maternal cells with tissue-specific antigen expression and morphology are common in infant tissues. Pediatr Dev Pathol. 2009;12:337–46. doi: 10.2350/08-07-0499.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A. 1996;93:705–8. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams KM, Lambert NC, Heimfeld S, Tylee TS, Pang JM, Erickson TD, Nelson JL. Male DNA in female donor apheresis and CD34-enriched products. Blood. 2003;102:3845–7. doi: 10.1182/blood-2003-05-1570. [DOI] [PubMed] [Google Scholar]
- 59.Nelson JL. The otherness of self: microchimerism in health and disease. Trends Immunol. 2012;33:421–7. doi: 10.1016/j.it.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Donoghue K, Chan J, de la Fuente J, Kennea N, Sandison A, Anderson JR, Roberts IA, Fisk NM. Microchimerism in female bone marrow and bone decades after fetal mesenchymal stem-cell trafficking in pregnancy. Lancet. 2004;364:179–82. doi: 10.1016/S0140-6736(04)16631-2. [DOI] [PubMed] [Google Scholar]
- 61.Khosrotehrani K, Johnson KL, Cha DH, Salomon RN, Bianchi DW. Transfer of fetal cells with multilineage potential to maternal tissue. JAMA. 2004;292:75–80. doi: 10.1001/jama.292.1.75. [DOI] [PubMed] [Google Scholar]
- 62.Rossi G. Nature of stem cell involved in fetomaternal microchimerism. Lancet. 2004;364:1936. doi: 10.1016/S0140-6736(04)17469-2. [DOI] [PubMed] [Google Scholar]
- 63.Barker JN, Weisdorf DJ, DeFor TE, Blazar BR, McGlave PB, Miller JS, Verfaillie CM, Wagner JE. Transplantation of 2 partially HLA-matched umbilical cord blood units to enhance engraftment in adults with hematologic malignancy. Blood. 2005;105:1343–7. doi: 10.1182/blood-2004-07-2717. [DOI] [PubMed] [Google Scholar]
- 64.Kang HJ, Kho SH, Jang MK, Lee SH, Shin HY, Ahn HS. Early engraftment kinetics of two units cord blood transplantation. Bone Marrow Transplant. 2006;38:197–201. doi: 10.1038/sj.bmt.1705423. [DOI] [PubMed] [Google Scholar]
- 65.Mold JE, Michaëlsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, Lee TH, Nixon DF, McCune JM. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322:1562–5. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Halteren AG, Jankowska-Gan E, Joosten A, Blokland E, Pool J, Brand A, Burlingham WJ, Goulmy E. Naturally acquired tolerance and sensitization to minor histocompatibility antigens in healthy family members. Blood. 2009;114:2263–72. doi: 10.1182/blood-2009-01-200410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dutta P, Molitor-Dart M, Bobadilla JL, Roenneburg DA, Yan Z, Torrealba JR, Burlingham WJ. Microchimerism is strongly correlated with tolerance to noninherited maternal antigens in mice. Blood. 2009;114:3578–87. doi: 10.1182/blood-2009-03-213561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dutta P, Burlingham WJ. Stem cell microchimerism and tolerance to non-inherited maternal antigens. Chimerism. 2010;1:2–10. doi: 10.4161/chim.1.1.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Opiela SJ, Adkins B. The pendulum swings: Tolerance versus priming to NIMA. Chimerism. 2010;1:36–8. doi: 10.4161/chim.1.1.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andrassy J, Kusaka S, Jankowska-Gan E, Torrealba JR, Haynes LD, Marthaler BR, Tam RC, Illigens BM, Anosova N, Benichou G, et al. Tolerance to noninherited maternal MHC antigens in mice. J Immunol. 2003;171:5554–61. doi: 10.4049/jimmunol.171.10.5554. [DOI] [PubMed] [Google Scholar]
- 71.Molitor-Dart ML, Andrassy J, Kwun J, Kayaoglu HA, Roenneburg DA, Haynes LD, Torrealba JR, Bobadilla JL, Sollinger HW, Knechtle SJ, et al. Developmental exposure to noninherited maternal antigens induces CD4+ T regulatory cells: relevance to mechanism of heart allograft tolerance. J Immunol. 2007;179:6749–61. doi: 10.4049/jimmunol.179.10.6749. [DOI] [PubMed] [Google Scholar]
- 72.Ridge JP, Fuchs EJ, Matzinger P. Neonatal tolerance revisited: turning on newborn T cells with dendritic cells. Science. 1996;271:1723–6. doi: 10.1126/science.271.5256.1723. [DOI] [PubMed] [Google Scholar]
- 73.Anderson CC, Matzinger P. Immunity or tolerance: opposite outcomes of microchimerism from skin grafts. Nat Med. 2001;7:80–7. doi: 10.1038/83393. [DOI] [PubMed] [Google Scholar]
- 74.Chan WF, Razavy H, Luo B, Shapiro AM, Anderson CC. Development of either split tolerance or robust tolerance along with humoral tolerance to donor and third-party alloantigens in nonmyeloablative mixed chimeras. J Immunol. 2008;180:5177–86. doi: 10.4049/jimmunol.180.8.5177. [DOI] [PubMed] [Google Scholar]
- 75.Mold JE, Venkatasubrahmanyam S, Burt TD, Michaëlsson J, Rivera JM, Galkina SA, Weinberg K, Stoddart CA, McCune JM. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science. 2010;330:1695–9. doi: 10.1126/science.1196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Claas FH, Gijbels Y, van der Velden-de Munck J, van Rood JJ. Induction of B cell unresponsiveness to noninherited maternal HLA antigens during fetal life. Science. 1988;241:1815–7. doi: 10.1126/science.3051377. [DOI] [PubMed] [Google Scholar]
- 77.Aoyama K, Koyama M, Matsuoka K, Hashimoto D, Ichinohe T, Harada M, Akashi K, Tanimoto M, Teshima T. Improved outcome of allogeneic bone marrow transplantation due to breastfeeding-induced tolerance to maternal antigens. Blood. 2009;113:1829–33. doi: 10.1182/blood-2008-05-155283. [DOI] [PubMed] [Google Scholar]
- 78.Jonuleit H, Schmitt E, Schuler G, Knop J, Enk AH. Induction of interleukin 10-producing, nonproliferating CD4(+) T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J Exp Med. 2000;192:1213–22. doi: 10.1084/jem.192.9.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adkins B, Jones M, Bu Y, Levy RB. Neonatal tolerance revisited again: specific CTL priming in mouse neonates exposed to small numbers of semi- or fully allogeneic spleen cells. Eur J Immunol. 2004;34:1901–9. doi: 10.1002/eji.200324271. [DOI] [PubMed] [Google Scholar]
- 80.Opiela SJ, Levy RB, Adkins B. Murine neonates develop vigorous in vivo cytotoxic and Th1/Th2 responses upon exposure to low doses of NIMA-like alloantigens. Blood. 2008;112:1530–8. doi: 10.1182/blood-2007-08-106500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rowe JH, Ertelt JM, Xin L, Way SS. Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature. 2012;490:102–6. doi: 10.1038/nature11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lissauer D, Piper K, Goodyear O, Kilby MD, Moss PA. Fetal-specific CD8+ cytotoxic T cell responses develop during normal human pregnancy and exhibit broad functional capacity. J Immunol. 2012;189:1072–80. doi: 10.4049/jimmunol.1200544. [DOI] [PubMed] [Google Scholar]
- 83.Verdijk RM, Kloosterman A, Pool J, van de Keur M, Naipal AM, van Halteren AG, Brand A, Mutis T, Goulmy E. Pregnancy induces minor histocompatibility antigen-specific cytotoxic T cells: implications for stem cell transplantation and immunotherapy. Blood. 2004;103:1961–4. doi: 10.1182/blood-2003-05-1625. [DOI] [PubMed] [Google Scholar]
- 84.Piper KP, McLarnon A, Arrazi J, Horlock C, Ainsworth J, Kilby MD, Martin WL, Moss PA. Functional HY-specific CD8+ T cells are found in a high proportion of women following pregnancy with a male fetus. Biol Reprod. 2007;76:96–101. doi: 10.1095/biolreprod.106.055426. [DOI] [PubMed] [Google Scholar]
- 85.VanBuskirk AM, Burlingham WJ, Jankowska-Gan E, Chin T, Kusaka S, Geissler F, Pelletier RP, Orosz CG. Human allograft acceptance is associated with immune regulation. J Clin Invest. 2000;106:145–55. doi: 10.1172/JCI9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dierselhuis MP, Jankowska-Gan E, Blokland E, Pool J, Burlingham WJ, van Halteren AG, Goulmy E. HY Immune Tolerance Is Common in Women without Male Offspring. PLoS One. 2014;9:e91274. doi: 10.1371/journal.pone.0091274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Bellefon LM, Heiman P, Kanaan SB, Azzouz DF, Rak JM, Martin M, Roudier J, Roufosse F, Lambert NC. Cells from a vanished twin as a source of microchimerism 40 years later. Chimerism. 2010;1:56–60. doi: 10.4161/chim.1.2.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burlingham WJ, Grailer AP, Heisey DM, Claas FH, Norman D, Mohanakumar T, Brennan DC, de Fijter H, van Gelder T, Pirsch JD, et al. The effect of tolerance to noninherited maternal HLA antigens on the survival of renal transplants from sibling donors. N Engl J Med. 1998;339:1657–64. doi: 10.1056/NEJM199812033392302. [DOI] [PubMed] [Google Scholar]
- 89.Hirayama M, Azuma E, Komada Y. Tolerogenic effect of non-inherited maternal antigens in hematopoietic stem cell transplantation. Front Immunol. 2012;3:135. doi: 10.3389/fimmu.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rocha V, Spellman S, Zhang MJ, Ruggeri A, Purtill D, Brady C, Baxter-Lowe LA, Baudoux E, Bergamaschi P, Chow R, et al. Eurocord-European Blood and Marrow Transplant Group and the Center for International Blood and Marrow Transplant Research Effect of HLA-matching recipients to donor noninherited maternal antigens on outcomes after mismatched umbilical cord blood transplantation for hematologic malignancy. Biol Blood Marrow Transplant. 2012;18:1890–6. doi: 10.1016/j.bbmt.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hirayama M, Azuma E, Ito T, Keida Y, Komada Y. A feasibility study on the prediction of acute graft-vs.-host disease before hematopoietic stem cell transplantation based on fetomaternal tolerance. Chimerism. 2013;4:84–6. doi: 10.4161/chim.24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mutis T, Gillespie G, Schrama E, Falkenburg JH, Moss P, Goulmy E. Tetrameric HLA class I-minor histocompatibility antigen peptide complexes demonstrate minor histocompatibility antigen-specific cytotoxic T lymphocytes in patients with graft-versus-host disease. Nat Med. 1999;5:839–42. doi: 10.1038/10563. [DOI] [PubMed] [Google Scholar]
- 93.Kim YH, Faaij CM, van Halteren AG, Schrama E, de Jong TA, Schøller J, Egeler RM, Pavel S, Vyth-Dreese FA, van Tol MJ, et al. In situ detection of HY-specific T cells in acute graft-versus-host disease-affected male skin after sex-mismatched stem cell transplantation. Biol Blood Marrow Transplant. 2012;18:381–7. doi: 10.1016/j.bbmt.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 94.Loren AW, Bunin GR, Boudreau C, Champlin RE, Cnaan A, Horowitz MM, Loberiza FR, Porter DL. Impact of donor and recipient sex and parity on outcomes of HLA-identical sibling allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2006;12:758–69. doi: 10.1016/j.bbmt.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 95.Adams KM, Holmberg LA, Leisenring W, Fefer A, Guthrie KA, Tylee TS, McDonald GB, Bensinger WI, Nelson JL. Risk factors for syngeneic graft-versus-host disease after adult hematopoietic cell transplantation. Blood. 2004;104:1894–7. doi: 10.1182/blood-2004-02-0508. [DOI] [PubMed] [Google Scholar]
- 96.van Rood JJ, Loberiza FR, Jr., Zhang MJ, Oudshoorn M, Claas F, Cairo MS, Champlin RE, Gale RP, Ringdén O, Hows JM, et al. Effect of tolerance to noninherited maternal antigens on the occurrence of graft-versus-host disease after bone marrow transplantation from a parent or an HLA-haploidentical sibling. Blood. 2002;99:1572–7. doi: 10.1182/blood.V99.5.1572. [DOI] [PubMed] [Google Scholar]
- 97.Tamaki S, Ichinohe T, Matsuo K, Hamajima N, Hirabayashi N, Dohy H, Japan Society of Hematopoietic Cell Transplantation Superior survival of blood and marrow stem cell recipients given maternal grafts over recipients given paternal grafts. Bone Marrow Transplant. 2001;28:375–80. doi: 10.1038/sj.bmt.1703146. [DOI] [PubMed] [Google Scholar]
- 98.Stern M, Ruggeri L, Mancusi A, Bernardo ME, de Angelis C, Bucher C, Locatelli F, Aversa F, Velardi A. Survival after T cell-depleted haploidentical stem cell transplantation is improved using the mother as donor. Blood. 2008;112:2990–5. doi: 10.1182/blood-2008-01-135285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kolb HJ, Bigalke I, Tischer J, et al. Risk factors of HLA-haploidentical transplantation using the combination of unmodified marrow and CD6-depleted G-CSF mobilized blood cells. Blood. 2005;106:2907. [abstract] [Google Scholar]
- 100.van Kampen CA, Versteeg-van der Voort Maarschalk MF, Langerak-Langerak J, van Beelen E, Roelen DL, Claas FH. Pregnancy can induce long-persisting primed CTLs specific for inherited paternal HLA antigens. Hum Immunol. 2001;62:201–7. doi: 10.1016/S0198-8859(01)00209-9. [DOI] [PubMed] [Google Scholar]
- 101.Eapen M, Rubinstein P, Zhang MJ, Stevens C, Kurtzberg J, Scaradavou A, Loberiza FR, Champlin RE, Klein JP, Horowitz MM, et al. Outcomes of transplantation of unrelated donor umbilical cord blood and bone marrow in children with acute leukaemia: a comparison study. Lancet. 2007;369:1947–54. doi: 10.1016/S0140-6736(07)60915-5. [DOI] [PubMed] [Google Scholar]
- 102.Wagner JE. No Survival Advantage After Double Umbilical Cord Blood (UCB) Compared to Single UCB Transplant in Children with Hematological Malignancy: Results of the Blood and Marrow Transplant Clinical Trials Network (BMT CTN 0501) [abstract]. ASH Annual Meeting 2012;120:359. [Google Scholar]
- 103.van Rood JJ, Stevens CE, Smits J, Carrier C, Carpenter C, Scaradavou A. Reexposure of cord blood to noninherited maternal HLA antigens improves transplant outcome in hematological malignancies. Proc Natl Acad Sci U S A. 2009;106:19952–7. doi: 10.1073/pnas.0910310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mommaas B, Stegehuis-Kamp JA, van Halteren AG, Kester M, Enczmann J, Wernet P, Kögler G, Mutis T, Brand A, Goulmy E. Cord blood comprises antigen-experienced T cells specific for maternal minor histocompatibility antigen HA-1. Blood. 2005;105:1823–7. doi: 10.1182/blood-2004-07-2832. [DOI] [PubMed] [Google Scholar]
- 105.van Rood JJ, Scaradavou A, Stevens CE. Indirect evidence that maternal microchimerism in cord blood mediates a graft-versus-leukemia effect in cord blood transplantation. Proc Natl Acad Sci U S A. 2012;109:2509–14. doi: 10.1073/pnas.1119541109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burlingham WJ, Nelson JL. Microchimerism in cord blood: mother as anticancer drug. Proc Natl Acad Sci U S A. 2012;109:2190–1. doi: 10.1073/pnas.1120857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Milano F, Lee Nelson J, Delaney C. Fetal maternal immunity and antileukemia activity in cord-blood transplant recipients. Bone Marrow Transplant. 2013;48:321–2. doi: 10.1038/bmt.2012.157. [DOI] [PubMed] [Google Scholar]
- 108.Truitt RL, Atasoylu AA. Contribution of CD4+ and CD8+ T cells to graft-versus-host disease and graft-versus-leukemia reactivity after transplantation of MHC-compatible bone marrow. Bone Marrow Transplant. 1991;8:51–8. [PubMed] [Google Scholar]
- 109.Chakraverty R, Eom HS, Sachs J, Buchli J, Cotter P, Hsu R, Zhao G, Sykes M. Host MHC class II+ antigen-presenting cells and CD4 cells are required for CD8-mediated graft-versus-leukemia responses following delayed donor leukocyte infusions. Blood. 2006;108:2106–13. doi: 10.1182/blood-2006-03-007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ringdén O, Karlsson H, Olsson R, Omazic B, Uhlin M. The allogeneic graft-versus-cancer effect. Br J Haematol. 2009;147:614–33. doi: 10.1111/j.1365-2141.2009.07886.x. [DOI] [PubMed] [Google Scholar]
- 111.Jain N, Liu H, Artz AS, Anastasi J, Odenike O, Godley LA, Joseph L, Marino S, Kline J, Nguyen V, et al. Immune reconstitution after combined haploidentical and umbilical cord blood transplant. Leuk Lymphoma. 2013;54:1242–9. doi: 10.3109/10428194.2012.739688. [DOI] [PubMed] [Google Scholar]
- 112.Hsieh MH, Patterson AE, Korngold R. T-cell subsets mediate graft-versus-myeloid leukemia responses via different cytotoxic mechanisms. Biol Blood Marrow Transplant. 2000;6:231–40. doi: 10.1016/S1083-8791(00)70005-X. [DOI] [PubMed] [Google Scholar]
- 113.Mergenthaler S, Babochkina T, Kiefer V, Lapaire O, Holzgreve W, Hahn S. FISH analysis of all fetal nucleated cells in maternal whole blood: improved specificity by the use of two Y-chromosome probes. J Histochem Cytochem. 2005;53:319–22. doi: 10.1369/jhc.4A6404.2005. [DOI] [PubMed] [Google Scholar]
- 114.Christensen B, Philip J, Kølvraa S, Lykke-Hansen L, Hromadnikova I, Gohel D, Lorch T, Plesch A, Bang J, Smidt-Jensen S, et al. Fetal cells in maternal blood: a comparison of methods for cell isolation and identification. Fetal Diagn Ther. 2005;20:106–12. doi: 10.1159/000082432. [DOI] [PubMed] [Google Scholar]
- 115.Lepez T, Vandewoestyne M, Hussain S, Van Nieuwerburgh F, Poppe K, Velkeniers B, Kaufman JM, Deforce D. Fetal microchimeric cells in blood of women with an autoimmune thyroid disease. PLoS One. 2011;6:e29646. doi: 10.1371/journal.pone.0029646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Garavito G, Klein D, Denis M, Pugliese A, Ricordi C, Pastori RL. Real-time sequence-specific primer polymerase chain reaction amplification of HLA class II alleles: a novel approach to analyze microchimerism. Transplantation. 2002;73:822–5. doi: 10.1097/00007890-200203150-00031. [DOI] [PubMed] [Google Scholar]
- 117.Pujal JM, Gallardo D. PCR-based methodology for molecular microchimerism detection and quantification. Exp Biol Med (Maywood) 2008;233:1161–70. doi: 10.3181/0802-RM-35. [DOI] [PubMed] [Google Scholar]
- 118.Gineikiene E, Stoskus M, Griskevicius L. Single nucleotide polymorphism-based system improves the applicability of quantitative PCR for chimerism monitoring. J Mol Diagn. 2009;11:66–74. doi: 10.2353/jmoldx.2009.080039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alizadeh M, Bernard M, Danic B, Dauriac C, Birebent B, Lapart C, Lamy T, Le Prisé PY, Beauplet A, Bories D, et al. Quantitative assessment of hematopoietic chimerism after bone marrow transplantation by real-time quantitative polymerase chain reaction. Blood. 2002;99:4618–25. doi: 10.1182/blood.V99.12.4618. [DOI] [PubMed] [Google Scholar]
- 120.Lo YM, Patel P, Sampietro M, Gillmer MD, Fleming KA, Wainscoat JS. Detection of single-copy fetal DNA sequence from maternal blood. Lancet. 1990;335:1463–4. doi: 10.1016/0140-6736(90)91491-R. [DOI] [PubMed] [Google Scholar]
- 121.Thomas MR, Williamson R, Craft I, Yazdani N, Rodeck CH. Y chromosome sequence DNA amplified from peripheral blood of women in early pregnancy. Lancet. 1994;343:413–4. doi: 10.1016/S0140-6736(94)91248-3. [DOI] [PubMed] [Google Scholar]
- 122.Murata H, Nakauchi H, Sumida T. Microchimerism in Japanese women patients with systemic sclerosis. Lancet. 1999;354:220. doi: 10.1016/S0140-6736(99)00164-6. [DOI] [PubMed] [Google Scholar]
- 123.Miyashita Y, Ono M, Ono M, Ueki H, Kurasawa K. Y chromosome microchimerism in rheumatic autoimmune disease. Ann Rheum Dis. 2000;59:655–6. doi: 10.1136/ard.59.8.654b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carter AS, Bunce M, Cerundolo L, Welsh KI, Morris PJ, Fuggle SV. Detection of microchimerism after allogeneic blood transfusion using nested polymerase chain reaction amplification with sequence-specific primers (PCR-SSP): a cautionary tale. Blood. 1998;92:683–9. [PubMed] [Google Scholar]
- 125.Carter AS, Cerundolo L, Bunce M, Koo DD, Welsh KI, Morris PJ, Fuggle SV. Nested polymerase chain reaction with sequence-specific primers typing for HLA-A, -B, and -C alleles: detection of microchimerism in DR-matched individuals. Blood. 1999;94:1471–7. [PubMed] [Google Scholar]
- 126.Kletzel M, Huang W, Olszewski M, Khan S. Validation of chimerism in pediatric recipients of allogeneic hematopoietic stem cell transplantation (HSCT) a comparison between two methods: Real-time PCR (qPCR) vs. variable number tandem repeats PCR (VNTR PCR) Chimerism. 2013;4:1–8. doi: 10.4161/chim.23158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nelson JL, Furst DE, Maloney S, Gooley T, Evans PC, Smith A, Bean MA, Ober C, Bianchi DW. Microchimerism and HLA-compatible relationships of pregnancy in scleroderma. Lancet. 1998;351:559–62. doi: 10.1016/S0140-6736(97)08357-8. [DOI] [PubMed] [Google Scholar]
- 128.Lambert NC, Lo YM, Erickson TD, Tylee TS, Guthrie KA, Furst DE, Nelson JL. Male microchimerism in healthy women and women with scleroderma: cells or circulating DNA? A quantitative answer. Blood. 2002;100:2845–51. doi: 10.1182/blood-2002-01-0295. [DOI] [PubMed] [Google Scholar]
- 129.Nelson JL, Gillespie KM, Lambert NC, Stevens AM, Loubiere LS, Rutledge JC, Leisenring WM, Erickson TD, Yan Z, Mullarkey ME, et al. Maternal microchimerism in peripheral blood in type 1 diabetes and pancreatic islet beta cell microchimerism. Proc Natl Acad Sci U S A. 2007;104:1637–42. doi: 10.1073/pnas.0606169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lambert NC, Erickson TD, Yan Z, Pang JM, Guthrie KA, Furst DE, Nelson JL. Quantification of maternal microchimerism by HLA-specific real-time polymerase chain reaction: studies of healthy women and women with scleroderma. Arthritis Rheum. 2004;50:906–14. doi: 10.1002/art.20200. [DOI] [PubMed] [Google Scholar]
- 131.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, et al. Nomenclature for Factors of the HLA System, 2004. Hum Immunol. 2005;66:571–636. doi: 10.1016/j.humimm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 132.Yan Z, Lambert NC, Guthrie KA, Porter AJ, Loubiere LS, Madeleine MM, Stevens AM, Hermes HM, Nelson JL. Male microchimerism in women without sons: quantitative assessment and correlation with pregnancy history. Am J Med. 2005;118:899–906. doi: 10.1016/j.amjmed.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 133.Lambert NC, Pang JM, Yan Z, Erickson TD, Stevens AM, Furst DE, Nelson JL. Male microchimerism in women with systemic sclerosis and healthy women who have never given birth to a son. Ann Rheum Dis. 2005;64:845–8. doi: 10.1136/ard.2004.029314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee TH, Chafets DM, Reed W, Wen L, Yang Y, Chen J, Utter GH, Owings JT, Busch MP. Enhanced ascertainment of microchimerism with real-time quantitative polymerase chain reaction amplification of insertion-deletion polymorphisms. Transfusion. 2006;46:1870–8. doi: 10.1111/j.1537-2995.2006.00992.x. [DOI] [PubMed] [Google Scholar]
- 135.Wu D, Vu Q, Nguyen A, Stone JR, Stubbs H, Kuhlmann G, Sholl LM, Iafrate AJ. In situ genetic analysis of cellular chimerism. Nat Med. 2009;15:215–9. doi: 10.1038/nm.1862. [DOI] [PubMed] [Google Scholar]
- 136.Warner JB, Bruin EJ, Hannig H, Hellenkamp F, Hörning A, Mittmann K, van der Steege G, de Leij LF, Garritsen HS. Use of sequence variation in three highly variable regions of the mitochondrial DNA for the discrimination of allogeneic platelets. Transfusion. 2006;46:554–61. doi: 10.1111/j.1537-2995.2006.00775.x. [DOI] [PubMed] [Google Scholar]
- 137.Terahara T, Chow S, Kurogi H, Lee SH, Tsukamoto K, Mochioka N, Tanaka H, Takeyama H. Efficiency of peptide nucleic acid-directed PCR clamping and its application in the investigation of natural diets of the Japanese eel leptocephali. PLoS One. 2011;6:e25715. doi: 10.1371/journal.pone.0025715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou L, Wang Y, Wittwer CT. Rare allele enrichment and detection by allele-specific PCR, competitive probe blocking, and melting analysis. Biotechniques. 2011;50:311–8. [Google Scholar]
- 139.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A. 2002;99:5261–6. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–64. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baker M. Digital PCR hits its stride. Nat Methods. 2012;9:541–4. doi: 10.1038/nmeth.2027. [DOI] [Google Scholar]
- 142.Lo YM, Lun FM, Chan KC, Tsui NB, Chong KC, Lau TK, Leung TY, Zee BC, Cantor CR, Chiu RW. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci U S A. 2007;104:13116–21. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science. 2011;333:58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pekin D, Skhiri Y, Baret JC, Le Corre D, Mazutis L, Salem CB, Millot F, El Harrak A, Hutchison JB, Larson JW, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11:2156–66. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- 145.Kroneis T, Gutstein-Abo L, Kofler K, Hartmann M, Hartmann P, Alunni-Fabbroni M, Walcher W, Dohr G, Petek E, Guetta E, et al. Automatic retrieval of single microchimeric cells and verification of identity by on-chip multiplex PCR. J Cell Mol Med. 2010;14:954–69. doi: 10.1111/j.1582-4934.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kroneis T, Geigl JB, El-Heliebi A, Auer M, Ulz P, Schwarzbraun T, Dohr G, Sedlmayr P. Combined molecular genetic and cytogenetic analysis from single cells after isothermal whole-genome amplification. Clin Chem. 2011;57:1032–41. doi: 10.1373/clinchem.2011.162131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sedlmayr P, Kroneis T. Verification of the genomic identity of candidate microchimeric cells. Chimerism. 2011;2:63–4. doi: 10.4161/chim.17741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schmidt U, Lutz-Bonengel S, Weisser HJ, Sänger T, Pollak S, Schön U, Zacher T, Mann W. Low-volume amplification on chemically structured chips using the PowerPlex16 DNA amplification kit. Int J Legal Med. 2006;120:42–8. doi: 10.1007/s00414-005-0041-2. [DOI] [PubMed] [Google Scholar]
- 149.Vandewoestyne M, Van Nieuwerburgh F, Van Hoofstat D, Deforce D. Evaluation of three DNA extraction protocols for forensic STR typing after laser capture microdissection. Forensic Sci Int Genet. 2012;6:258–62. doi: 10.1016/j.fsigen.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 150.Miura Y, Tanaka J, Toubai T, Tsutsumi Y, Kato N, Hirate D, Kaji M, Sugita J, Shigematsu A, Iwao N, et al. Analysis of donor-type chimerism in lineage-specific cell populations after allogeneic myeloablative and non-myeloablative stem cell transplantation. Bone Marrow Transplant. 2006;37:837–43. doi: 10.1038/sj.bmt.1705352. [DOI] [PubMed] [Google Scholar]
- 151.Lim ZY, Pearce L, Ingram W, Ho AY, Mufti GJ, Pagliuca A. Chimerism does not predict for outcome after alemtuzumab-based conditioning: lineage-specific analysis of chimerism of specific diseases may be more informative. Bone Marrow Transplant. 2008;41:587–8. doi: 10.1038/sj.bmt.1705937. [DOI] [PubMed] [Google Scholar]
- 152.Willasch A, Eing S, Weber G, Kuçi S, Schneider G, Soerensen J, Jarisch A, Rettinger E, Koehl U, Klingebiel T, et al. Enrichment of cell subpopulations applying automated MACS technique: purity, recovery and applicability for PCR-based chimerism analysis. Bone Marrow Transplant. 2010;45:181–9. doi: 10.1038/bmt.2009.89. [DOI] [PubMed] [Google Scholar]
- 153.Schumm M, Feuchtinger T, Pfeiffer M, Hoelle W, Bethge W, Ebinger M, Kuci S, Handgretinger R, Lang P. Flow cytometry with anti HLA-antibodies: a simple but highly sensitive method for monitoring chimerism and minimal residual disease after HLA-mismatched stem cell transplantation. Bone Marrow Transplant. 2007;39:767–73. doi: 10.1038/sj.bmt.1705676. [DOI] [PubMed] [Google Scholar]
- 154.Herzenberg LA, Bianchi DW, Schröder J, Cann HM, Iverson GM. Fetal cells in the blood of pregnant women: detection and enrichment by fluorescence-activated cell sorting. Proc Natl Acad Sci U S A. 1979;76:1453–5. doi: 10.1073/pnas.76.3.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pei R, Chen T, Orpilla J, Lee JH. A simultaneous negative and positive selection method that can detect chimerism at a frequency of 1 per 10,000 by flow cytometry. Tissue Antigens. 1997;50:197–201. doi: 10.1111/j.1399-0039.1997.tb02859.x. [DOI] [PubMed] [Google Scholar]
- 156.Drabbels JJ, van de Keur C, Kemps BM, Mulder A, Scherjon SA, Claas FH, Eikmans M. HLA-targeted flow cytometric sorting of blood cells allows separation of pure and viable microchimeric cell populations. Blood. 2011;118:e149–55. doi: 10.1182/blood-2011-06-362053. [DOI] [PubMed] [Google Scholar]
- 157.Dutta P, Molitor-Dart M, Bobadilla JL, Roenneburg DA, Yan Z, Torrealba JR, Burlingham WJ. Microchimerism is strongly correlated with tolerance to noninherited maternal antigens in mice. Blood. 2009;114:3578–87. doi: 10.1182/blood-2009-03-213561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stevens AM, Hermes HM, Rutledge JC, Buyon JP, Nelson JL. Myocardial-tissue-specific phenotype of maternal microchimerism in neonatal lupus congenital heart block. Lancet. 2003;362:1617–23. doi: 10.1016/S0140-6736(03)14795-2. [DOI] [PubMed] [Google Scholar]
- 159.Nelson JL, Gillespie KM, Lambert NC, Stevens AM, Loubiere LS, Rutledge JC, Leisenring WM, Erickson TD, Yan Z, Mullarkey ME, et al. Maternal microchimerism in peripheral blood in type 1 diabetes and pancreatic islet beta cell microchimerism. Proc Natl Acad Sci U S A. 2007;104:1637–42. doi: 10.1073/pnas.0606169104. [DOI] [PMC free article] [PubMed] [Google Scholar]