Abstract
Columnar epithelia (e.g., kidney, intestine) and hepatocytes embody the two major organizational phenotypes of non-stratified epithelial cells. Columnar epithelia establish their apical and basal domains at opposing poles and organize in monolayered cysts and tubules, in which their apical surfaces form a single continuous lumen whereas hepatocytes establish their apical domains in the midst of their basolateral domains and organize a highly branched capillary luminal network, the bile canaliculi, in which a single hepatocyte can engage in lumen formation with multiple neighbors. To maintain their distinct tissue architectures, columnar epithelial cells bisect their luminal domains during symmetric cell divisions, while the cleavage furrow in dividing hepatocytes avoids bisecting the bile canalicular domains. We discuss recently discovered molecular mechanisms that underlie the different cell division phenotypes in columnar and hepatocytic model cell lines. The serine/threonine kinase Par1b determines both the epithelial lumen polarity and cell division phenotype via cell adhesion signaling that converges on the small GTPase RhoA.
Keywords: hepatocyte polarity, epithelial cells, mitotic spindle orientation, columnar and hepatocyte polarity, Par1b/MARK2, LGN/NuMA, RhoA
Epithelial cells constitute the biggest cell pool in the mammalian organism, with ~60% of mammalian cell types being of epithelial or epithelial-derived origin.1 Epithelial cells are found in a wide variety of tissues, such as the skin, lung, intestine, kidney, and liver, where they are situated at the interface between the organisms exterior and interior environment. Their major function is to protect the organism’s interior milieu (i.e., the blood) by physically separating it from the exterior environment, and also to regulate the transport of molecules (e.g., nutrients) between the environments.1,2 Epithelial cells are polarized cells in the sense that they have specific plasma membrane domains (also referred to as surfaces) that face either the exterior or the interior environment, or neighboring cells. The apical membrane faces the external environment or ‘lumen’ of the organism, such as the interior of the gut or lung. The basal membrane faces the interior milieu of the organism and is typically in contact with the extracellular matrix (ECM) and, ultimately, underlying blood vessels. The lateral plasma membrane domains contact neighboring cells via cell-adhesion protein complexes such as adherens junctions, desmosomes, and gap junctions.3 The basal and lateral membrane domains are often commonly referred to as the basolateral membrane, and the apical and basolateral plasma membrane domains are separated, and its protein and lipid composition maintained, by tight junctions.3-5 Most epithelial organs are created by epithelial cells of the columnar (they are typically taller than they are wide) polarity type and are aligned in tight single-cell monolayers that wrap around a central cavity or lumen, a hallmark of columnar polarity.1 Architecturally, columnar epithelial cells create hollow tubes that ultimately develop into interconnected tubular networks (Fig. 1, “Columnar”).
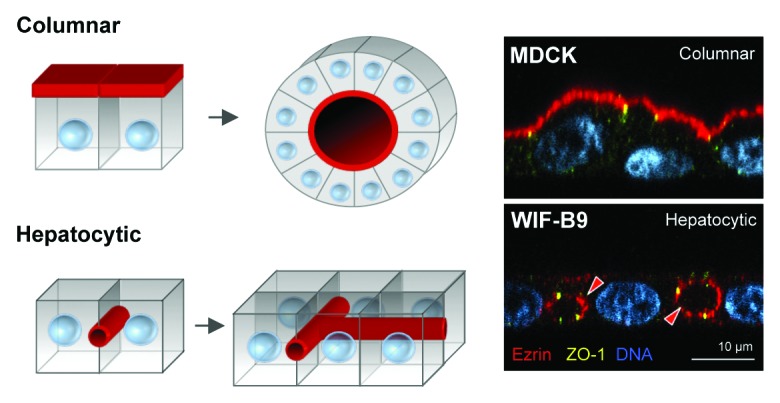
Figure 1. Columnar vs. hepatocytic polarity. Columnar epithelial cells form monolayers where multiple cells surround a central lumen (i.e., columnar polarity), whereas hepatocytes organize around tubular networks were the luminal domain is shared by no more than two cells (i.e., hepatocytic polarity), and each cell can have multiple luminal domains. Red arrowheads indicate the luminal domains marked by Ezrin. MDCK and WIF-B9 cells are kidney- and hepatocyte-derived culture models, respectively.
However, not all epithelial tissues develop columnar type of epithelial polarity and tubular architecture. The liver is an important metabolic organ and is responsible for the generation of bile salts, cholesterol homeostasis, plasma protein production, detoxification of the blood, and hormone and cytokine production. The epithelial cell of the liver, the hepatocyte, constitutes ~78–85% of the liver cell mass6,7 and provides most liver functions. In the adult healthy liver, hepatocytes are aligned in one or two-cell thick cords and are highly polarized. Similar to columnar epithelial cells, the basal membranes of hepatocytes are in contact with the ECM and blood via endothelial-lined sinusoids (also known as the space of Disse), and their lateral membranes are used to contact neighboring hepatocytes. During liver development, hepatocytes form small apical domains enclosed by tight junctions at the lateral membrane of two hepatocytes, that can later in development merge together and form canalicular structures that circumvent entire hepatocytes, a hallmark of hepatocyte polarity.8-12 Hepatocytes use the canaliculi to secrete and drain bile and it is commonly referred to as the bile canalicular network (Fig. 1, “Hepatocytic”).
It is important to realize that while adult hepatocytes show polarized plasma membrane domains like other epithelial cells, their polarity phenotype and the 3-dimensional tubular architecture that they create is different from a columnar epithelium. As described above, columnar epithelial cells form monolayers of multiple cells surrounding a central lumen (i.e., columnar polarity), but hepatocytes do not. In fact, from the apical domain point-of-view, the apical domain of hepatocytes is only shared by not more than two hepatocytes (i.e., hepatocytic polarity). It is because of this specific apical domain organization that bile canaliculi are able to completely circumvent entire hepatocytes.
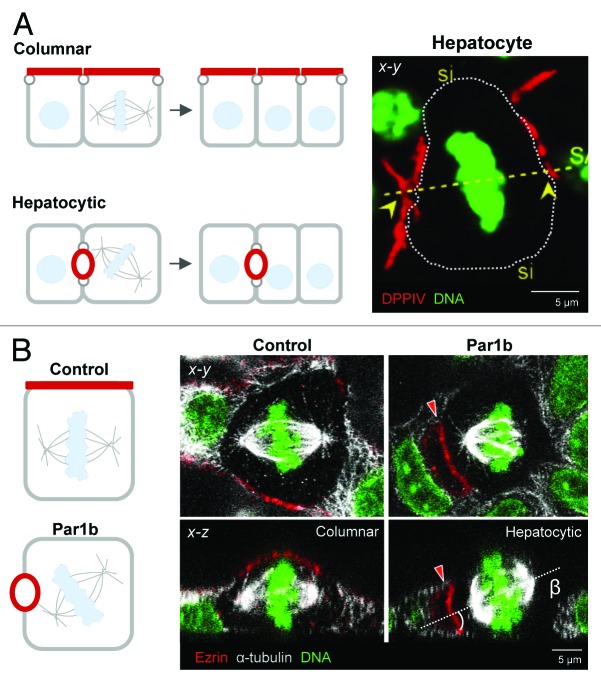
How tubular networks are formed is a subject of intense research. Recently, the orientation of cell division was found to be an important design principle for generating and maintaining columnar epithelial tissue (i.e., tubular) architecture, and failure to properly orient cell divisions correlates with tumorigenesis.1,13-17 The orientation of the mitotic spindle during mitosis dictates the position of the cleavage furrow, which is established perpendicular to the spindle pole axis. Thereby, mitotic spindle orientation guides both the positioning of the daughter cells within the epithelial tissue and the equal or asymmetric inheritance of cellular domains and cytoplasm by the two daughters. Columnar epithelial cells arranged in monolayers predominantly align their mitotic spindle with the substratum (e.g., ECM) and divide symmetrically, while preserving their membrane domain identities, as shown for kidney-derived MDCK cells.18 That is to say, epithelial cells symmetrically ‘segregate’ their apical and basolateral membrane domains to both daughter cells during cell division, and the daughter cells are then positioned in the plane of the monolayer, preserving tubular architecture (Fig. 2A “Columnar”; Fig. 2B, Control).
Figure 2. Columnar and hepatocytic division phenotypes are regulated by Par1b. (A) Columnar epithelial cells orient their metaphase plates perpendicular to the lumen. The resulting cleavage furrow bisects their luminal surface (red domains in the schematics, marked by dipeptidyl peptidase-IV (DPPIV)). Hepatocytes attach their astral microtubules adjacent to their luminal domain(s), thereby avoiding the bisection of their lumina during cell divisions. In cultured hepatocytic cells with a single luminal surface, as depicted in the schematic, the luminal domain is distributed to only one of the daughters. In multipolar hepatocytes in vivo, as shown in the fluorescent image on the right, one of the two DPPIV-positive luminal surfaces (yellow arrowheads) will segregate to each daughter. Si = sinusoids. (B) Par1b overexpression in MDCK cells promotes polarization with lateral rather than apical lumina (see x-z views, the apical domains (red) are marked by Ezrin) and mitotic spindles that are oriented toward the lateral lumen, instead of aligning with the basal surface. The β angle represents the angle between the spindle axis (dashed line) and the substratum.
However, symmetric inheritance of plasma membrane domains poses a problem for hepatocytes and hepatic polarity development, as this would induce and enforce the generation of columnar tubular structures with columnar polarity, and would therefore be incompatible with the formation of a canalicular network that is so unique for the liver architecture. Thus, hepatocytes must use a different orientation of the cell division mechanism to maintain hepatocyte polarity, and ultimately allow for the formation of the bile canalicular network. Indeed, earlier work in fixed rat liver tissue slices revealed that after partial hepatectomy proliferating hepatocytes asymmetrically segregate apical plasma membrane domains during cell division,19,20 though the 3-dimensional mechanics and molecular mechanisms have, until now, remained unknown. Using HepG2 and WIFB9 cell lines as in vitro cell models for hepatocyte polarity21-23 and studying cell division in regenerating rodent livers after partial hepatectomy, we found that, in compliance with earlier work, hepatocytes display a different mode of cell division orientation compared with columnar epithelial cells.24,25 Specifically, hepatocytes orient their mitotic spindle poles toward an area near the apical plasma membrane domain, which we call the apicolateral membrane domain, and during cytokinesis asymmetrically segregate their apical plasma membrane domain to daughter cells (Fig. 2A, “Hepatocytic”). By doing this, hepatocytes maintain their hepatic polarized state, and would ultimately be allowed to form and maintain the bile canalicular network to serve the unique liver architecture.
To understand mechanistically how the two epithelial cell division phenotypes are linked to the two distinct polarity phenotypes, it helps to break down mitotic spindle orientation into spindle position in the x-y plane (birds-eye or planar view) and in the x-z plane (side view). In fact, most studies consider only one of these dimensions. In the x-z view, columnar, symmetrically dividing cells align their metaphase spindle with the basement membrane. This horizontal spindle orientation depends on the local positioning of two cortical cues that capture the two sets of astral microtubules at opposite lateral cell membranes in equal-distance from the basal domain. The attachment cues are an evolutionary conserved module that consists of the Gαi subunit of a trimeric G-protein that is anchored to the cortex via a myristoyl group and its binding partners, the proteins LGN (leucine-glycine-asparagine repeat protein) and NuMA (nuclear and mitotic apparatus) (reviewed in ref. 26). The latter is a nuclear protein that only becomes available for cortical complex formation after the nuclear envelope breaks down at the onset of prophase. NuMA mediates the interaction of astral MTs with the cortex via Dynein, a minus-end directed microtubules (MTs) motor. When anchored to the membrane by NuMA and walking along astral MTs toward the spindle poles, Dynein can exert pulling forces on the MTs that bring the spindle into place. When either Gαi, LGN, NuMA, or Dynein are depleted, spindles no longer align with the basal surface but become “tilted.” A similar phenotype is also observed when the spindle attachment module is present along the entire cell cortex. In HeLa cells, the restriction of NuMA to a tight patch on the lateral domain is due to β1-integrin mediated signaling processes that involve PI3K and Abl kinase activity as well as myosin X.27,28 Curiously, none of these ECM-signaling events appeared to operate in polarized MDCK cells when they were tested side-by-side with HeLa cells, which lack cell-cell adhesion junctions. Instead, independent work in Drosophila neuroepithelia and in MDCK cells established cell-cell adhesion proteins as the domineering cues in polarized epithelial cells, specifically the Adenomatosis Polyposis coli protein (APC), which has MT-tip binding abilities and could therefore directly capture astral MTs.29 In MDCK cells, APC-depletion or depletion of E-cadherin, which is instrumental in recruiting APC to adherens junctions (AJ), did not prevent cortical LGN/NuMA, but nevertheless caused tilted spindles.30 These findings led to the concept that ECM-signaling governs x-z spindle position via LGN/NuMA in non-adherent cells while cell-cell adhesion proteins serve as spindle attachment cues in polarized epithelia. Several findings, however, didn’t fit this simple model: β1-integrin depletion in Drosophila follicle epithelia caused tilted spindles and integrin signaling determined spindle positioning in mammalian basal keratinocytes; thus ECM-signaling does have a dominant role in epithelial spindle orientation in vivo.31,32 Furthermore, the LGN/NuMA module, which in mitosis colocalizes with adhesion markers at the lateral domain, overrides any cell-cell adhesion-mediated cues when it is ectopically activated in MDCK cells.33 We have now demonstrated that function-blocking β1-integrin antibodies indeed abolish spindle alignment with the substratum in MDCK cells, and further determined that the recruitment of LGN/NuMA to the metaphase cortex is dependent on collagen-IV mediated ECM-signaling in MDCK and HepG2 cells,24 although laminin-1 might also play a role (Slim, van IJzendoorn, unpublished data). In both cell lines, the position of a NuMA patch always correlated with a spindle pole facing NuMA. How does ECM/integrin signaling at the basal domain translate into discrete LGN/NuMA recruitment at the lateral cell cortex in epithelial cells? When cells enter mitosis they disassemble their focal adhesions leading to cell rounding and their cell cortex becomes stiff. Both these changes, one at the basal, the other at the lateral surface, are known to require RhoA activity.34 These observations made us wonder whether RhoA signaling could link basal ECM-signaling to lateral membrane organization. Indeed, we found, utilizing a FRET-based biosensor, that the presence of NuMA at the cortex always coincided with high RhoA activity, while RhoA was less active at the NuMA-negative cortex. Furthermore, depletion of RhoA or pharmacological inhibition of the RhoA effector Rho-kinase abolished LGN and NuMA from the metaphase cortex and resulted in tilted spindles, and HepG2 cell multilayering.35 Thus, ECM-signaling appears to drive NuMA positioning by activating RhoA at discrete cortical sites. What are those sites? In MDCK and HepG2 cells NuMA localizes where cell-cell adhesion junctions are present. They are connected to a circumferential actin belt that is under tension and likely requires RhoA to sustain high myosin II activity. Although we have not tested this hypothesis directly, we observed that non-polarized mitotic HepG2 cells lacked patches of high RhoA activity and were deficient in the recruitment of NuMA. Therefore, adherens junctions are good candidates to serve as sites of high RhoA activity required for LGN/NuMA recruitment and might function synergistically with the ECM signals to position the spindle parallel to the substratum in MDCK cells.
Spindle orientation in the x-y dimension also depends on ECM-signaling mechanisms.36 When mitotic cells round up, their sole connections to the substratum are thin retraction fibers that correspond to the former cell adhesion points. The position of these retraction fibers serves as guideposts for the placement of the spindle. It is the tension in these fibers, which pin the cell down like the guylines of a tent, that convey a signal for x-y spindle positioning. It is tempting to speculate that RhoA activity is highest where retraction fibers are most abundant and attracts the Gαi/LGN/NuMA module to these x-y positions (Fig. 3, HeLa). Polarized epithelial cells have few focal adhesions and consequently feature few retraction fibers in mitosis. It is conceivable, as discussed above, that the cell-cell adhesion belt provides the RhoA cue in this case. However, the adhesion belt in monopolar columnar epithelial cells spans the entire cell circumference, suggesting that x-y spindle orientation in columnar epithelial cells is either random or that symmetry is broken by upstream signals that are likely non-cell autonomous. The latter would dictate the direction into which an epithelium expands. For columnar polarized epithelial cells growing as monolayers in culture, x-y spindle orientation is indeed unimportant. That LGN/NuMA are nevertheless restricted to two crescents rather than forming a continuous belt (Fig. 3, MDCK), can be explained by removal of LGN/NuMA from the areas of the x-y cortex that are not initially involved in MT-anchoring: chromatin that aligns at the metaphase plate emits a gradient of active Ran GTPase that antagonizes cortical LGN/NuMA at sites where it comes closest to the cortex, which is perpendicular to the spindle poles and the anchored astral MTs.37 In contrast to columnar epithelial cells, the lumen architecture of HepG2 cells results in a sub-luminal LGN/NuMA belt that is too narrow to anchor both astral MT fans. Even if the spindle would manage to curl around the luminal domain, the supposed Chromatin-Ran-gradient would likely remove the entire LGN/NuMA population. Instead, the subluminal NuMA patch anchors only one astral MT fan with the other facing the opposite basolateral surface. LGN/NuMA thereby serves as cue for the x-y spindle orientation. Indeed, depletion of LGN abolished the alignment of the spindle with the luminal domain in HepG2 cells.25 In multipolar hepatocytes, a second sub-luminal domain provides the anchor site for the other set of astral MTs (see Figure 2A). Thus, in MDCK cells, where spindle orientation in the x-y position is less important than in the x-z position for the maintenance of cell polarity and tissue architecture, Gαi/LGN/NuMA ensure spindle alignment with the basal domain but do not define its x-y-orientation. In HepG2 cells where x-y spindle orientation is equally important, the NuMA module also ensures that one of the spindle poles faces the luminal region in the x-y dimension.
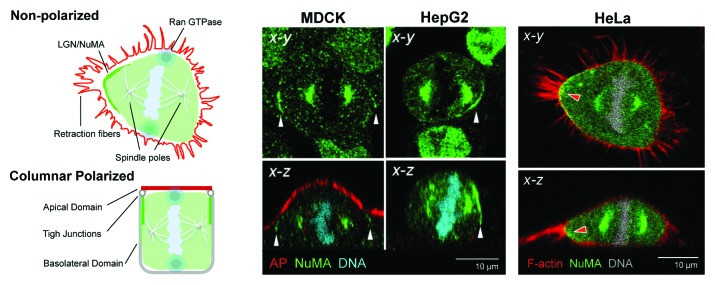
Figure 3. NuMA localization in polarized MDCK and HepG2 and in non-polarized HeLa cells. In polarized epithelial cells, such as MDCK and HepG2 cells, cortical NuMA in metaphase localizes below or adjacent to the luminal domain, coinciding with cell-cell adhesion sites (white arrowheads). The schematic illustrates the example of columnar epithelia (“Columnar Polarized”), In transformed epithelial cells such as HeLa cells, which lack adherens junctions, cortical NuMA coincides with the strongest retraction fibers (schematic “Non-Polarized”). In both instances, these are cortical areas under tension that likely feature high RhoA activity. Metaphase chromatin emits a Ran GTPase gradient that antagonizes cortical NuMA where the chromosomes come closest to the cortex, (blue circles in the schematics), resulting in two NuMA crescents. F-actin and an apical marker (AP) are in red.
Intuition suggests that the many molecular aspects that amount to the distinct polarity phenotype and tissue organization of monolayered epithelial cells and hepatocytes must result from many molecular signaling pathways and processes that are fundamentally distinct in these epithelia. It is remarkable therefore, that a ubiquitously expressed serine/threonine kinase, Par1b/MARK2 can single-handedly convert the two phenotypes into each other in vitro. Ever since one of our labs reported that Par1b overexpression in MDCK cells causes a switch to a hepatocyte lumen polarity phenotype as found in cultured hepatocytic HepG2 and WIFB cell lines,38,39 we wondered how far the phenotypic conversion goes. We have now demonstrated that MDCK-Par1b cells, like the hepatocytic lines, also feature tilted metaphase spindles that orient toward their lateral luminal domain and give rise to asymmetric divisions and asymmetrical inheritance of apical plasma membrane domains24,25 (Fig. 2B). Remarkably, Par1b not only promotes both these aspects of hepatocyte polarity in MDCK cells, but does so via common signaling mechanisms: Par1b inhibits the deposition of a basement membrane,40 and both Par1b-phenotypes are overcome when the basal lamina deposited by MDCK-Par1b cells is supplemented with exogenous collagen-IV.24 Defective ECM-signaling in turn reduces RhoA activity. Inhibition of RhoA indeed causes tilted spindles and lateral lumen polarity in MDCK cells suggesting that the two polarity aspects are intimately linked. Our recent evidence suggests that the converse also applies, namely that HepG2 cells adopt features of the columnar phenotype when Par1b levels are reduced. Par1b-depletion in columnar epithelia leads to a disorganized monolayer, and complete abrogation of all Par1 activity (there is at least one additional Par1 isoform present in most cells) likely interferes with cell-cell adhesion.41 Reduced Par1b levels in WIF-B9 cells resulted in areas of the monolayer that exhibited a chickenwire arrangement of tight junction and apical junctional markers and in the establishment of a luminal domain at the apex, although the phenotype reversal was not perfect and many cells simply lost polarity. The metaphase spindle in Par1b-depleted HepG2 cells always aligned with the substratum as observed in columnar epithelial cells, which resulted in symmetric divisions where the apical domain was divided between daughters. Even in HepG2 cells that maintained lateral lumen organization, the x-y spindle alignment mechanism was abolished, resulting in more divisions in which the luminal domain was divided between daughters. Altogether, these features resemble the domain and spindle organization of MDCK cells. The combined gain and loss-of function data thus implicate Par1b as a key determinant in the branching of the two epithelial phenotypes that are exemplified by kidney and hepatocyte epithelia.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
A.M. acknowledges funding by RO1 NIDDK R01KD064842–07.
References
- 1.Bryant DM, Mostov KE. From cells to organs: building polarized tissue. Nat Rev Mol Cell Biol. 2008;9:887–901. doi: 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Brien LE, Zegers MMP, Mostov KE. Opinion: Building epithelial architecture: insights from three-dimensional culture models. Nat Rev Mol Cell Biol. 2002;3:531–7. doi: 10.1038/nrm859. [DOI] [PubMed] [Google Scholar]
- 3.Giepmans BNG, van Ijzendoorn SCD. Epithelial cell-cell junctions and plasma membrane domains. Biochim Biophys Acta. 2009;1788:820–31. doi: 10.1016/j.bbamem.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Aijaz S, Balda MS, Matter K. Tight junctions: molecular architecture and function. Int Rev Cytol. 2006;248:261–98. doi: 10.1016/S0074-7696(06)48005-0. [DOI] [PubMed] [Google Scholar]
- 5.Shin K, Fogg VC, Margolis B. Tight junctions and cell polarity. Annu Rev Cell Dev Biol. 2006;22:207–35. doi: 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- 6.Blouin A, Bolender RP, Weibel ER. Distribution of organelles and membranes between hepatocytes and nonhepatocytes in the rat liver parenchyma. A stereological study. J Cell Biol. 1977;72:441–55. doi: 10.1083/jcb.72.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–89. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Feracci H, Connolly TP, Margolis RN, Hubbard AL. The establishment of hepatocyte cell surface polarity during fetal liver development. Dev Biol. 1987;123:73–84. doi: 10.1016/0012-1606(87)90429-5. [DOI] [PubMed] [Google Scholar]
- 9.Hubbard AL, Wall DA, Ma A. Isolation of rat hepatocyte plasma membranes. I. Presence of the three major domains. J Cell Biol. 1983;96:217–29. doi: 10.1083/jcb.96.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luzzatto AC. Hepatocyte differentiation during early fetal development in the rat. Cell Tissue Res. 1981;215:133–42. doi: 10.1007/BF00236254. [DOI] [PubMed] [Google Scholar]
- 11.Montesano R, Friend DS, Perrelet A, Orci L. In vivo assembly of tight junctions in fetal rat liver. J Cell Biol. 1975;67:310–9. doi: 10.1083/jcb.67.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood RL. An electron microscope study of developing bile canaliculi in the rat. Anat Rec. 1965;151:507–29. doi: 10.1002/ar.1091510403. [DOI] [PubMed] [Google Scholar]
- 13.Pease JC, Tirnauer JS. Mitotic spindle misorientation in cancer--out of alignment and into the fire. J Cell Sci. 2011;124:1007–16. doi: 10.1242/jcs.081406. [DOI] [PubMed] [Google Scholar]
- 14.Fischer E, Legue E, Doyen A, Nato F, Nicolas J-F, Torres V, Yaniv M, Pontoglio M. Defective planar cell polarity in polycystic kidney disease. Nat Genet. 2006;38:21–3. doi: 10.1038/ng1701. [DOI] [PubMed] [Google Scholar]
- 15.Quyn AJ, Appleton PL, Carey FA, Steele RJC, Barker N, Clevers H, Ridgway RA, Sansom OJ, Näthke IS. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6:175–81. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Noatynska A, Gotta M, Meraldi P. Mitotic spindle (DIS)orientation and DISease: cause or consequence? J Cell Biol. 2012;199:1025–35. doi: 10.1083/jcb.201209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–60. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinsch S, Karsenti E. Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarized MDCKII cells. J Cell Biol. 1994;126:1509–26. doi: 10.1083/jcb.126.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartles JR, Hubbard AL. Preservation of hepatocyte plasma membrane domains during cell division in situ in regenerating rat liver. Dev Biol. 1986;118:286–95. doi: 10.1016/0012-1606(86)90095-3. [DOI] [PubMed] [Google Scholar]
- 20.Stamatoglou SC, Enrich C, Manson MM, Hughes RC. Temporal changes in the expression and distribution of adhesion molecules during liver development and regeneration. J Cell Biol. 1992;116:1507–15. doi: 10.1083/jcb.116.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Decaens C, Durand M, Grosse B, Cassio D. Which in vitro models could be best used to study hepatocyte polarity? Biol Cell. 2008;100:387–98. doi: 10.1042/BC20070127. [DOI] [PubMed] [Google Scholar]
- 22.Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, Schroer TA, Pagano RE, Hubbard AL. WIF-B cells: an in vitro model for studies of hepatocyte polarity. J Cell Biol. 1993;123:1761–75. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van IJzendoorn SC, Hoekstra D. (Glyco)sphingolipids are sorted in sub-apical compartments in HepG2 cells: a role for non-Golgi-related intracellular sites in the polarized distribution of (glyco)sphingolipids. J Cell Biol. 1998;142:683–96. doi: 10.1083/jcb.142.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lázaro-Diéguez F, Cohen D, Fernandez D, Hodgson L, van Ijzendoorn SCD, Müsch A. Par1b links lumen polarity with LGN-NuMA positioning for distinct epithelial cell division phenotypes. J Cell Biol. 2013;203:251–64. doi: 10.1083/jcb.201303013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slim CL, Lázaro-Diéguez F, Bijlard M, Toussaint MJM, de Bruin A, Du Q, Müsch A, van Ijzendoorn SCD. Par1b induces asymmetric inheritance of plasma membrane domains via LGN-dependent mitotic spindle orientation in proliferating hepatocytes. PLoS Biol. 2013;11:e1001739. doi: 10.1371/journal.pbio.1001739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werts AD, Roh-Johnson M, Goldstein B. Dynamic localization of C. elegans TPR-GoLoco proteins mediates mitotic spindle orientation by extrinsic signaling. Development. 2011;138:4411–22. doi: 10.1242/dev.070979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyoshima F, Nishida E. Spindle orientation in animal cell mitosis: roles of integrin in the control of spindle axis. J Cell Physiol. 2007;213:407–11. doi: 10.1002/jcp.21227. [DOI] [PubMed] [Google Scholar]
- 28.Matsumura S, Hamasaki M, Yamamoto T, Ebisuya M, Sato M, Nishida E, Toyoshima F. ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat Commun. 2012;3:626. doi: 10.1038/ncomms1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahmanyar S, Nelson WJ, Barth AIM. Role of APC and its binding partners in regulating microtubules in mitosis. Adv Exp Med Biol. 2009;656:65–74. doi: 10.1007/978-1-4419-1145-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.den Elzen N, Buttery CV, Maddugoda MP, Ren G, Yap AS. Cadherin adhesion receptors orient the mitotic spindle during symmetric cell division in mammalian epithelia. Mol Biol Cell. 2009;20:3740–50. doi: 10.1091/mbc.E09-01-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–80. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernández-Miñán A, Martín-Bermudo MD, González-Reyes A. Integrin signaling regulates spindle orientation in Drosophila to preserve the follicular-epithelium monolayer. Curr Biol. 2007;17:683–8. doi: 10.1016/j.cub.2007.02.052. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Z, Zhu H, Wan Q, Liu J, Xiao Z, Siderovski DP, Du Q. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J Cell Biol. 2010;189:275–88. doi: 10.1083/jcb.200910021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddox AS, Burridge K. RhoA is required for cortical retraction and rigidity during mitotic cell rounding. J Cell Biol. 2003;160:255–65. doi: 10.1083/jcb.200207130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrema H, Czajkowska D, Théard D, van der Wouden JM, Kalicharan D, Zolghadr B, Hoekstra D, van Ijzendoorn SCD. Rho kinase, myosin-II, and p42/44 MAPK control extracellular matrix-mediated apical bile canalicular lumen morphogenesis in HepG2 cells. Mol Biol Cell. 2006;17:3291–303. doi: 10.1091/mbc.E06-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Théry M, Racine V, Pépin A, Piel M, Chen Y, Sibarita J-B, Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–53. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 37.Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–7. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen D, Rodriguez-Boulan E, Müsch A. Par-1 promotes a hepatic mode of apical protein trafficking in MDCK cells. Proc Natl Acad Sci U S A. 2004;101:13792–7. doi: 10.1073/pnas.0403684101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen D, Brennwald PJ, Rodriguez-Boulan E, Müsch A. Mammalian PAR-1 determines epithelial lumen polarity by organizing the microtubule cytoskeleton. J Cell Biol. 2004;164:717–27. doi: 10.1083/jcb.200308104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen D, Fernandez D, Lázaro-Diéguez F, Müsch A. The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling. J Cell Biol. 2011;192:525–40. doi: 10.1083/jcb.201007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Böhm H, Brinkmann V, Drab M, Henske A, Kurzchalia TV. Mammalian homologues of C. elegans PAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Curr Biol. 1997;7:603–6. doi: 10.1016/S0960-9822(06)00260-0. [DOI] [PubMed] [Google Scholar]