Abstract
Subtelomeric duplications of an obscure tubulin “genic” segment located near the telomere of human chromosome 4q35 have occurred at different evolutionary time points within the last 25 million years of the catarrhine (i.e., hominoid and Old World monkey) evolution. The analyses of these segments reported here indicate an exceptional level of evolutionary instability. Substantial intra- and interspecific differences in copy number and distribution are observed among cercopithecoid (Old World monkey) and hominoid genomes. Characterization of the hominoid duplicated segments reveals a strong positional bias within pericentromeric and subtelomeric regions of the genome. On the basis of phylogenetic analysis from predicted proteins and comparisons of nucleotide-substitution rates, we present evidence of a conserved b-tubulin gene among the duplications. Remarkably, the evolutionary conservation has occurred in a nonorthologous fashion, such that the functional copy has shifted its positional context between hominoids and cercopithecoids. We propose that, in a chimpanzee-human common ancestor, one of the paralogous copies assumed the original function, whereas the ancestral copy acquired mutations and eventually became silenced. Our analysis emphasizes the dynamic nature of duplication-mediated genome evolution and the delicate balance between gene acquisition and silencing.
Acquiring genetic diversity through gene duplication and divergent mutations is an important mechanism for evolution of the genome of a complex organism. Gene families are abundantly present and, in many cases, are part of a transcriptionally tightly regulated gene cluster at a particular genomic location. Tandem gene duplications and genomewide teraploidization events are believed to be largely responsible for shaping the present-day homeobox, hemoglobin, and immunoglobulin gene clusters in higher vertebrates (Shen et al. 1981; Ohno 1999; Zerucha and Ekker 2000). Other gene families have members scattered throughout the genome, and these gene duplications are mechanistically distinct from whole-genome or tandem duplication. Genes located in pericentromeric or subtelomeric domains are particularly prone to acquire genetic diversity, since frequent exchange of sequences occur between these dynamic (non-) homologous chromosome regions (Eichler et al. 1996; Pryde et al. 1997; Eichler 1998). For example, the large olfactory-receptor gene family has members for the most part distributed on human chromosome ends (Rouquier et al. 1998; Trask et al. 1998a, 1998b). Here, we present further insight into gene-duplication events on subtelomeric and pericentromeric regions, by analyses of gene-order conservation, phylogenetics, and fluorescent in situ hybridization (FISH). The data provides striking evidence for dynamic gene acquisition and silencing within evolutionary duplicated DNA segments.
Analysis of human chromosomal 4q35 segments, localized close to (<300 kb from) the telomere, has indicated a substantial amount of duplication throughout the genome (Grewal et al. 1999; van Geel et al. 1999). A tubulin-like element (TUBB4Q) residing in the region has at least seven homologous members in the genome (van Geel et al. 2000). High allelic sequence variability and lack of expression has suggested that this copy contains a pseudogene derivative (van Geel et al. 2000). The sequence dispersal and evolutionary history of 4qter was further analyzed by focusing in detail on this tubulin segment. The complete sequence of the human TUBB4Q members reported elsewhere (van Geel et al. 2000) was determined, and two additional members were identified, making the total number in humans at least 10. In addition, 12 members could be identified in both chimpanzees and baboons, and 2 members could be identified in squirrel monkeys, suggesting that duplications of this particular segment have occurred extensively in all these species. The structural ortholog of the HSA4q35 TUBB4Q sequence (for nomenclature, see table 1) was identified in baboon (PHA443J9 equals PHA IVq) and chimpanzee (PTR179F17 equals PTR IVq) by means of gene-order conservation with the flanking FRG1 gene (fig. 1). Baboon BAC-ends sequence (RPCI-41-209E1, RPCI-41-269I4, and RPCI-41-443J9) comparison with the human genomic chromosome 4q35 sequence (GenBank accession numbers AF250324, AF146191, U85056, U74497, and U74496) determined the baboon structural ortholog of HSA4q35 (homology >80%), since baboon has three tubulin loci flanked by FRG1 (see BACPAC Resources Home Page). Sequence orthology was also evident between the human HSA1q43-44, the chimpanzee PTR-Iq, and the baboon PHA Iq TUBB4Q members, which are the only members containing a 5′ truncated L1PA2 repeat within intron 3 (table 1). Moreover, this orthology was confirmed by phylogenetic and FISH analysis (see below). Consensus sequences of each genomic TUBB4Q member were coaligned by use of ClustalX software, version 1.8 (Thompson et al. 1994, 1997) and, in some instances, were manually edited. TUBB4Q comparative sequence alignments of all 36 operational taxonomic units (OTUs; sequences represented at the tips of the topological tree) denote genomic nucleotide-sequence identity of 78%–98% (GenBank accession numbers AF355105 to AF355140).
Table 1.
Localization of Human and Orthologous Primate TUBB4Q Members[Note]
| Human PAC Clone | HumanCytogeneticLocation | Orthologous ChimpanzeeCopy and Location | Orthologous Baboon Copy and Location | Paralogous Chimpanzee Copies | Paralogous Baboon Copies |
| RPCI-1-68H18 | HSAYq11 | PTR90F1, PTRYq | PHA90O17a | PTR1G20 | PHA1F5 |
| RPCI-1-120A17 | HSA12cen-q11 | PTR112D9, PTR XIIcen | PTR15E12 | PHA38E15 | |
| RPCI-1-122D15 | HSA9q34 | PTR44L22 | PHA122M7 | ||
| RPCI-1-191O24 | HSA16q24 | PTR51H24 | PHA140N17 | ||
| RPCI-1-226K22 | HSA4q35 | PTR179F17, PTR IVq | PHA443J9, PHA IVq | PTR58N10 | PHA209E1 |
| RPCI-1-250K1 | HSA1q42.3 | PTR62G8, PTR Iqb | PHA63K1, PHA Icen | PTR91D7 | PHA256F15 |
| RPCI-1-261A3 | HSA10P15 | PTR6H21, PTR Xp | PHA268O18 | ||
| RPCI-3-466K14 | HSA1q43-44 | PTR135K19, PTR Iq | PHA38L1, PHA Iq | PHA269I4 | |
| RPCI-3-492O24 | HSA18p11 | ||||
| RPCI-3-510E24 | HSA12cen-p11 | ?c |
Note.— The nomenclature of the TUBB4Q members is defined by their species acronym (HSA, Homo sapiens; PTR, Pan troglodytes; PHA, Papio hamadryas; SSC, Saimiri sciureus), followed by the genomic location in humans or the clone identification number in chimpanzees and baboons. Roman numerals behind the chimpanzee (PTR) and baboon (PHA) acronyms refer to the human syntenic chromosome positions, for clarity.
Location not determined.
Second copy on Iq.
Copy expected but not found.
Figure 1.
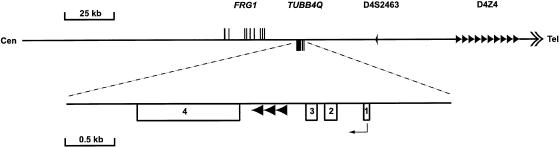
Organization of the human chromosome 4q35 region. Exons of the FRG1 and TUBB4Q genes are displayed on the forward and reverse strand, respectively. Specific family members of the tubulin located on chromosome 4q35 (TUBB4Q) were isolated by means of a specific probe situated proximally to the TUBB4Q sequence for hybridization on human (RPCI-1, 3; 6× genomic redundancy), chimpanzee (RPCI-43; 3.5× genomic redundancy) and baboon (RPCI-41; 10.4× genomic redundancy) genomic PAC and BAC libraries (for details, see the BACPAC Resources Home Page). Positive individual clones (RPCI-1/3; 61 positives, RPCI-43; 57 positives and RPCI-41; 158 positives) were subsequently rehybridized, with TUBB4Q (exon 4) and FRG1 (human cDNA) probes, for clone verification and genomic gene-order conservation, respectively. The D4Z4 repeat locus is represented by the arrow array, and the telomere is represented by a double arrow. The TUBB4Q genomic exon structure is enlarged underneath, with the putative direction of transcription and the arrowheads indicating the 110-bp repeats.
To solve the evolutionary relationship of this complex series of duplication events, phylogeny among the different TUBB4Q members in the catarrhine clade (hominoids and Old World monkeys) were determined with the platyrrhine clade (squirrel monkey) as an outgroup. Although we used the neighbor-joining method for phylogenetic analysis, topological-tree construction is quite solid, since the maximum-parsimony and maximum-likelihood methods resulted in similar trees. The topology of the tree indicates three catarrhine branch clusters—that is, clades (monophyletic groups)—in which all three species are represented (fig. 2). On the basis of this phylogenetic reconstruction, the human TUBB4Q members on HSA1q42.3, HSA1q43-44, and HSAYq11 have orthologous members in both chimpanzees and baboons (fig. 2; table 1). Thus, these members predate the catarrhine radiation (>25 million years). In addition, the phylogenetic analysis specifies that only some human members (HSA10p15 and HSA12cen-q11) are clustering with chimpanzee members, suggesting orthology in chimpanzees but not in baboons (fig. 2; table 1). All other members show only intraspecies clustering. Surprisingly, the TUBB4Q members that are supposed to be orthologous with HSA4q35 by means of gene-order conservation (described above), do not group together in the phylogenetic analysis. In addition, the two human members on HSA1 and the two on HSA12 have a unilateral close relationship, which implies they occurred through separate intrachromosomal evolutionary duplication events. Timing of the duplication events was estimated by sequence-divergence calculations among human-chimpanzee orthologous members. The substitution rate (r) was estimated from ancient TUBB4Q copies that probably were not under selective pressure during their evolutionary existence (pseudogenes, since they lack a functional open reading frame [ORF]). The human and Old World monkey (cercopithecidae) lineages divergence was set at 25 million years ago (Goodman 1999) to estimate the r value in our tree. From the HSA1q42.3, HSA1q43-44, and HSAYq11 phylogenetic catarrhine tree branches, the HSA1q43-44 catarrhine branch was discarded, since the substitution rate was much slower (2× slower), compared with the other two branch clusters. The average K value of the HSA1q42.3 and HSAYq11 catarrhine clusters was used to calculate r by the neighbor-joining method (PAUP): r=K/(2T)=0.156/(2×25×106)=3.12×10-9 bp/site/year. This rate provided a calibrated value for the evolutionary genetic distance between the different sequence taxa. We estimate the hominoid duplication events to have occurred after the divergence of the human and baboon lineages some 17.3, 15.1, and 7.2 million years ago, respectively, for the HSA12cen-q11, HSA12cen-p11, and HSA10p15 clusters.
Figure 2.
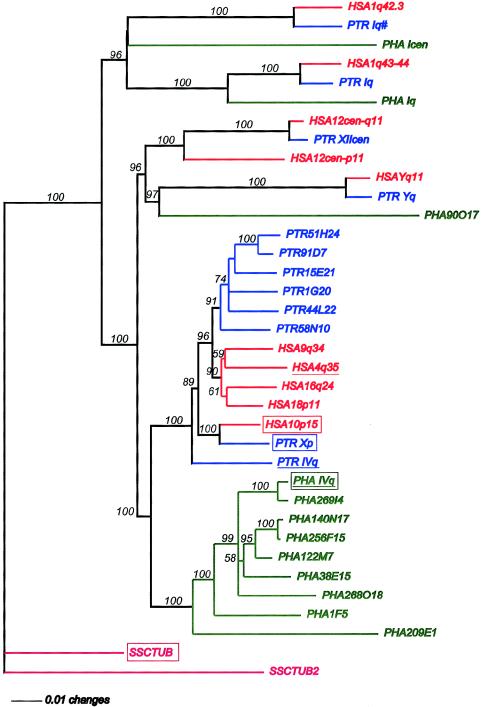
Phylogenetic analysis of TUBB4Q genomic sequence members from human (“HSA”; red), chimpanzee (“PTR”; blue), baboon (“PHA”; green), and squirrel monkey (“SSC”; pink), using the distance-matrix method (the heuristic computations under default parameter settings, with the maximum-parsimony and maximum-likelihood methods resulting in similar tree topologies). Phylogenetic trees and bootstrap analysis were generated with PAUP version 4.0b4a (Sinauer Associates). The tree’s branches are scaled and thus are proportional to the number of changes. The sequences of squirrel monkeys were used as an outgroup, because the New World monkeys (platyrrhine) are regarded as more diverged, relative to the catarrhine clade. The orthologs of the HSA4q35 tubulin in the chimpanzee and baboon are underlined. The boxed tubulins are highly conserved and are potential functional genes. With the bootstrap technique (heuristic search and 1,000 bootstrap replicates) the statistical confidence level of the phylogenetic analysis is estimated (percentages at branches, with a 50% cutoff value). For roman numerals, see the note to table 1. All TUBB4Q-related sequences from each individual PAC or BAC clone were isolated by PCR amplification, with initially different combinations of HSA4q35-specific TUBB4Q primers (van Geel et al. 2000). Sequences were analyzed by the phred, phrap, and consed software tools (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998). Within each species, segments of different duplicated TUBB4Q copies were distinguished by paralogous sequence variants. Individual BAC/PAC clones were selected to retrieve the complete sequence of each TUBB4Q member (three single clones of each copy, if available), by use of HSA4q35-specific primers or newly synthesized copy-specific primers for PCR and sequencing. In a few instances, fewer than five sequence differences were identified among the copies (>2.7 kb of sequence). Such copies were treated as allelic polymorphism, as a conservative estimate. The TUBB4Q members from squirrel monkeys were isolated by primer design on particularly highly conserved regions of the HSA10p15, PTR6H21, and PHA443J9 members (5′-CATGAGGGAGATCGTGCTCAC-3′, 5′-GCCACCTCCTCCTCAGCATA-3′) and by PCR on total genomic DNA isolated from a specific cell-line (CCL-194, ATCC). The single PCR product amplified from squirrel monkeys was cloned by use of the TA-Cloning kit (Invitrogen). Sequence analysis was performed on miniprep DNA of three clones with vector (PCR2.1)-specific primers (SP6, T7). Two distinct nonallelic members (SSCTUB and SSCTUB2) could be distinguished by sequence alignment.
Selected human PAC clones, each containing one individual member of the TUBB4Q family, were localized by FISH and PCR-based monochromosomal hybrid mapping (table 1). It is interesting that, although duplicated segments have been described for other genes that are exclusively dispersed over either centromeres or telomeres (Kermouni et al. 1995; Eichler et al. 1997; Trask et al. 1998a), we here provide evidence that duplications have also occurred between pericentromeric and subtelomeric regions of the primate genome.
Orthology of the different catarrhine TUBB4Q members was primarily defined from both the perspective of gene-order conservation and that of phylogenetic analysis (see above). To specifically verify this distinct orthology, positional conservation was tested, by FISH analysis, in specific orthologous members of the chimpanzee and baboon species. In concordance with the human chromosomal positions, specific signals were identified on chimpanzee and baboon syntenic chromosomal locations (fig. 3; table 1). However, the phylogenetically orthologous baboon copy from HSA1q42.3 was located near the centromere of PHA Iq, instead of near a telomeric position. This contradictory location could possibly be ascribed to an evolutionary intrachromosomal inversion event during the evolving baboon or hominoid genomes.
Figure 3.
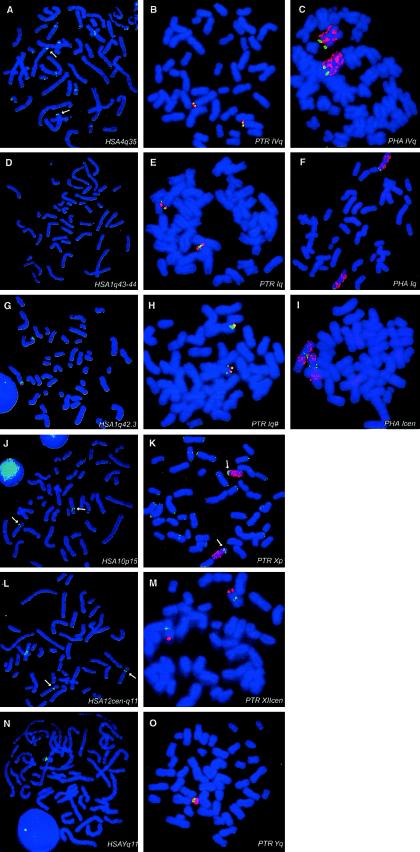
FISH analysis of orthologous TUBB4Q members (contained in PACs or BACs) on human (A, D, G, J, L, and N), chimpanzee (B, E, H, K, M, and O) and baboon (C, F, and I) chromosomal metaphase spreads. Primate chromosomes were cohybridized (Wirth et al. 1999; Shan et al. 2000) with chromosome-specific DNA libraries or telomeric YACs for specific chromosome identification. A, HSA4q35 with PAC RPCI-1-226K22. B, PTR179F17 with BAC RPCI-43-179F17 (green) and a human chromosome 4–specific telomeric YAC 908f5 (red; containing STS D4S2930). C, PHA443J9 with BAC RPCI-41-443J9 (green) and a human chromosome 4 paint (red). D, HSA1q43-44 with PAC RPCI-3-466K14. E, PTR135K19 with BAC RPCI-43-135K19 (green) and a human chromosome 1–specific telomeric YAC 848h10 (red; containing D1S423). F, PHA38L1 with BAC RPCI-41-38L1 (green) and a human chromosome 1 paint (red). G, HSA1q42.3 with PAC RPCI-1-250K1. H, PTR62G8 with BAC RPCI-43-62G8 (green) and a human chromosome 1–specific telomeric YAC 848h10 (red). I, PHA63K1 with BAC RPCI-41-63K1 (green) and a human chromosome 4 paint (red). J, HSA10P15 with PAC RPCI-1-261A3. K, PTR6H21 with BAC RPCI-43-6H21 (green) and a human chromosome 10 paint (red). L, HSA12cen-q11 with PAC RPCI-1-120A17. M, PTR122D9 with BAC RPCI-43-122D9 (green) and a human chromosome 12–specific telomeric YAC 751b8 (red; containing D12S1599). N, HSAYq11 with PAC RPCI-1-68H18. O, PTR90F1 with BAC RPCI-43-90F1 (green) and a human chromosome Y paint (red). Arrows indicate the specific hybridization signals. The PHA90O17 TUBB4Q copy, presumed to be located on the Y chromosome, could not be verified, since our metaphase speads were from a female baboon. Chromosomal localization of some of the TUBB4Q members was determined by use of copy-specific primers on a monochromosomal somatic hybrid panel (Coriell), under stringent PCR amplifications.
Tentative examination of potential ORFs of all identified tubulin members indicates four human members (HSA4q35, HSA10p15, HSA16q24, and HSA18p11), five members in both chimpanzee (PTR6H21 [PTR Xp], PTR51H24, PTR58N10, PTR91D7, and PTR179F17 [PTR IVq]) and baboon (PHA122M7, PHA140N17, PHA209E1, PHA269I4, and PHA443J9 [PHA IVq]), and one in squirrel monkey (SSCTUB), in which the ORF is not disrupted. The amino acid–sequence alignment (according to ClustalX; alignment available online) and subsequent phylogenetic analysis (fig. 4) reveals a strikingly closer relationship of the baboon PHA IVq TUBB4Q protein to the human HSA10p15 member (97.1% identity) than to its related HSA4q35 orthologous copy (88.5% identity). However, the chimpanzee orthologous members of HSA10P15 (PTR Xp) and HSA4q35 (PTR IVq) do cluster and indicate a monophyletic relationship (fig. 4). The protein sequence of the HSA10p15-PTR Xp clade evolved relatively slowly, compared with the other human and chimpanzee members. In addition, the only ORF-containing baboon member (PHA IVq) with orthologous counterparts in human (HSA4q35) and chimpanzee (PTR IVq) indicates minimal divergence from the HSA10p15 and PTR Xp members. Therefore, the sequences may be under evolutionary selective pressure because they represent genuine genes, even though we lack experimental evidence for transcriptional expression.
Figure 6 (Online Only) .
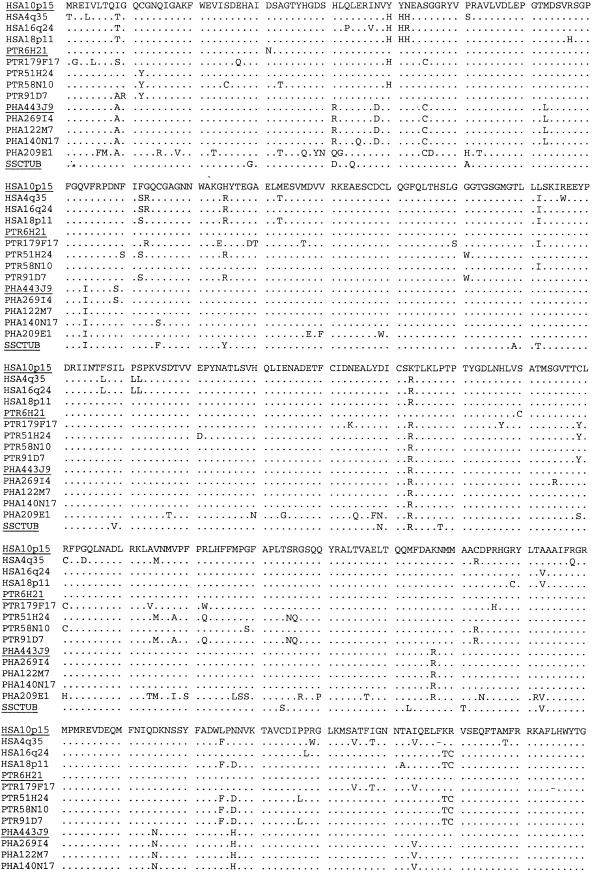
Pairwise alignment of 15 putative TUBB4Q protein sequences (452 sites). The member identification names are listed at the left, with potential functional genes underlined. Abbreviations: HSA (human), PTR (chimpanzee), PHA (baboon), and SSC (squirrel monkey). Identical amino acids are represented by an underscore and deletions by a dash.
Figure 4.
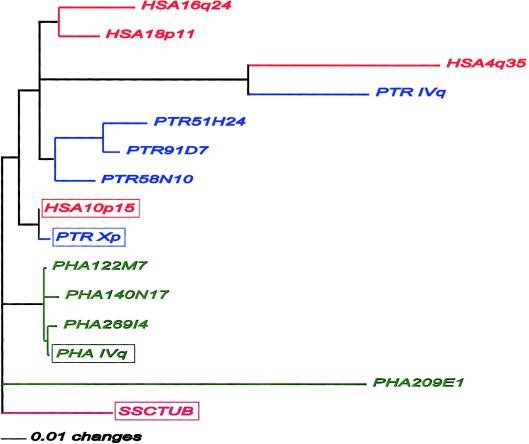
Phylogenic analysis of the protein sequences of TUBB4Q members of human (“HSA”; red), chimpanzee (“PTR”; blue), baboon (“PHA”; green), and squirrel monkey (“SSC”; pink) with an ORF, using the distance-matrix method. The squirrel-monkey sequence was used as an outgroup. The parsimony method resulted in a similar phylogenetic tree. The boxed tubulins are highly conserved and potentially functional genes. For roman numerals, see the note to table 1.
Potential functionality of the TUBB4Q cDNA sequences (theoretically extracted manually from the genomic sequences; alignment available on request) was tested statistically by comparison of the ratios of nonsynonymous (Ka) to synonymous (Ks) base substitution by use of MEGA version 2.0 (Kumar et al. 1994). Pseudogenes are not under functional constraints, and therefore nonsynonymous and synonymous base substitutions are expected to evolve at equal rates, unlike functional genes where nonsynonymous base substitutions are restricted (Meireles et al. 1999; Yang et al. 2000). The cDNAs with an intact ORF were jointly analyzed for the nucleotide-substitution pattern (table 2), whereas cDNAs with an interrupted ORF were already considered pseudogenes. The mean of the substitution ratio (Ka:Ks) of all functional genes in mammals is estimated to be ∼0.21 (Li 1997). Comparison of PHA IVq with the human HSA4q35 (Ka/Ks=0.199; Ka-Ks=-0.205±0.034; Z-test 6.062 with P<.0001) and chimpanzee PTR IVq (Ka/Ks=0.190; Ka-Ks=-0.183±0.031; Z-test 5.839 with P<.0001) ortholog TUBB4Q members indicates a higher Ka:Ks ratio (mean 0.195) than to the paralogous human HSA10p15 (Ka/Ks=0.065; Ka-Ks=-0.217±0.031; Z-test 6.912 with P<.0001) and chimp PTR Xp (Ka/Ks=0.075; Ka-Ks=-0.210±0.031; Z-test 6.780 with P<.0001) copies (mean 0.070). Similar ratios were calculated by comparison of the same catarrhine members to the squirrel monkey SSCTUB member (table 2), although we lack evidence for an orthologous relationship. The Ka:Ks ratios of HSA10p15 and PTR Xp to PHA IVq or SSCTUB, respectively, clearly indicate significance for selective constraint. In comparison, the Ka:Ks values of functional known b-tubulin genes among human and mouse vary between 0.022 (TUBB5) and 0.067 (TUBB2), which suggests higher selective pressure on these structural proteins than average. The Ka:Ks ratios of HSA4q35 and PTR IVq to the (orthologous) PHA IVq or SSCTUB are similar to the gene average, which may suggest that evolutionary selective pressure was long present but was recently lifted in humans and chimpanzees. This may explain the higher Ka:Ks ratios between HSA4q35 and PTR IVq (0.48; see table 3).
Table 2.
Pairwise Nonsynonymous (Upper Right Matrix) and Synonymous (Lower Left Matrix) Nucleotide Substitutions among 15 cDNAs from Different TUBB4Q Members Using the Distance Method from Nei-Gojobori (Jukes-Cantor)
|
Pairwise Ka and Ks Values for cDNA |
|||||||||||||||
| cDNA | HSA10p15 | HSA4q35 | HSA16q24 | HSA18p11 | PTR Xp | PTR IVq | PTR51H24 | PTR58N10 | PTR91D7 | PHA IVq | PHA269I4 | PHA122M7 | PHA140N17 | PHA209E1 | SSCTUB |
| HSA10p15 | .043 | .027 | .017 | .002 | .033 | .024 | .015 | .021 | .015 | .017 | .015 | .018 | .098 | .029 | |
| HSA4q35 | .108 | .038 | .041 | .045 | .051 | .056 | .044 | .051 | .051 | .051 | .049 | .052 | .126 | .067 | |
| HSA16q24 | .087 | .092 | .022 | .029 | .036 | .041 | .031 | .037 | .038 | .041 | .038 | .041 | .112 | .051 | |
| HSA18p11 | .055 | .084 | .037 | .019 | .044 | .027 | .018 | .022 | .027 | .029 | .027 | .030 | .109 | .040 | |
| PTR Xp | .065 | .111 | .113 | .080 | .035 | .026 | .017 | .023 | .017 | .019 | .017 | .020 | .100 | .031 | |
| PTR IVq | .083 | .106 | .098 | .062 | .087 | .051 | .038 | .048 | .043 | .043 | .041 | .044 | .119 | .060 | |
| PTR51H24 | .065 | .118 | .083 | .058 | .091 | .102 | .023 | .011 | .037 | .039 | .037 | .040 | .114 | .050 | |
| PTR58N10 | .084 | .138 | .094 | .076 | .095 | .114 | .065 | .020 | .027 | .029 | .027 | .030 | .105 | .039 | |
| PTR91D7 | .072 | .129 | .079 | .062 | .102 | .113 | .023 | .055 | .029 | .031 | .029 | .032 | .110 | .047 | |
| PHA IVq | .232 | .256 | .257 | .232 | .228 | .226 | .247 | .260 | .259 | .002 | .002 | .005 | .096 | .033 | |
| PHA269I4 | .236 | .267 | .259 | .237 | .241 | .240 | .252 | .264 | .264 | .010 | .002 | .005 | .099 | .035 | |
| PHA122M7 | .214 | .242 | .234 | .210 | .218 | .217 | .234 | .246 | .245 | .020 | .023 | .003 | .096 | .033 | |
| PHA140N17 | .223 | .255 | .247 | .223 | .227 | .228 | .245 | .257 | .256 | .026 | .030 | .020 | .097 | .036 | |
| PHA209E1 | .239 | .281 | .266 | .248 | .248 | .251 | .254 | .269 | .268 | .196 | .200 | .191 | .188 | .115 | |
| SSCTUB | .357 | .367 | .349 | .324 | .341 | .333 | .343 | .335 | .362 | .392 | .409 | .397 | .400 | .424 | |
Table 3.
Nonsynonymous versus Synonymous (Ka:Ks)
|
Ka:Ks for cDNA |
|||||
| cDNA | HSA10p15 | HSA4q35 | PTR Xp | PTR IVq | PHA IVq |
| HSA4q35 | .398 | ||||
| PTR Xp | .031 | .405 | |||
| PTR IVq | .398 | .481 | .402 | ||
| PHA IVq | .065 | .199 | .075 | .190 | |
| SSCTUB | .081 | .183 | .091 | .180 | .084 |
Four human members have structural orthologs in baboon and potentially are representative of the ancestral copy. Of these members only the baboon ortholog of HSA4q35 (PHA IVq) is likely a functional gene. The HSA1q42.3, HSA1q43-44, and HSAYq11 members are all considered pseudogenes in all species tested. It is likely that they were derived from their functional counterpart. Therefore, the progenitor of all derivative human TUBB4Q copies was almost certainly HSA4q35.
We hypothesize that the HSA4q35 tubulin was once a functional gene (fig. 5). Because of its proclivity to duplicate to subtelomeric locations, a novel tubulin member was transposed to 10p15, ∼7.3 million years ago. We propose that mutations have accumulated on the original copy since this time, while the HSA10p15 copy has been under selection pressure. Interestingly, no mutations have occurred to disrupt the ORF of the HSA4q35 TUBB4Q, making it a possible resource for the development of a future, highly specialized, evolved tubulin gene, expanding the present day b-tubulin gene family.
Figure 5.
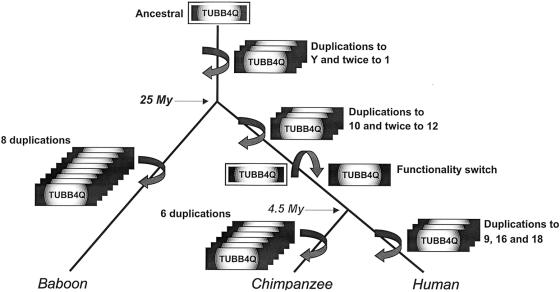
Schematic representation of the TUBB4Q duplication events during the catarrhine evolution. Within the catarrhine tree, duplication events are indicated by boxes and vertical circled arrows. The locations where the TUBB4Q members will finally end up in humans are indicated. The ancestral TUBB4Q member is highlighted, and the functionality switch is indicated, in the hominoid lineage, with a horizontal, circled arrow.
This duplicated segment has been part of the hominoid/cercopithecoid clades since early in their evolution and suggests an exceptional level of evolutionary instability for a genomic segment. Chromosomal location (pericentromeric or telomeric) is undoubtedly an important underlying aspect of genomic instability. Our study emphasizes the dynamic nature of duplication-mediated genome evolution and the delicate balance between gene acquisition and silencing.
Acknowledgments
This work was supported by the Muscular Dystrophy Association and by the German Research Foundation (DFG; research grant Ha1374/5-2). We would like to thank Barbara Swiatkiewicz and Joe Catanese, for their excellent assistance with the genomic libraries; Linda Haley and Sheila Sait, for the human FISH analyses; and Gert-Jan van Ommen, for critical reading of the manuscript.
Electronic-Database Information
The URL for data in this article is as follows:
- BACPAC Resources Home Page, http://www.chori.org/bacpac/ (for BAC-end sequencing method)
References
- Eichler EE (1998) Masquerading repeats: paralogous pitfalls of the human genome. Genome Res 8:758–762 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser HW (1997) Interchromosomal duplications of the adrenoleukodystrophy locus: a phenomenon of pericentromeric plasticity. Hum Mol Genet 6:991–1002 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL (1996) Duplication of a gene-rich cluster between 16p11.1 and Xq28: a novel pericentromeric-directed mechanism for paralogous genome evolution. Hum Mol Genet 5:899–912 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- Goodman M (1999) The genomic record of humankind's evolutionary roots. Am J Hum Genet 64:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202 [DOI] [PubMed] [Google Scholar]
- Grewal PK, van Geel M, Frants RR, de Jong P, Hewitt JE (1999) Recent amplification of the human FRG1 gene during primate evolution. Gene 227:79–88 [DOI] [PubMed] [Google Scholar]
- Kermouni A, Van Roost E, Arden KC, Vermeesch JR, Weiss S, Godelaine D, Flint J, Lurquin C, Szikora JP, Higgs DR, Marynen P, Renauld JC (1995) The IL-9 receptor gene (IL9R): genomic structure, chromosomal localization in the pseudoautosomal region of the long arm of the sex chromosomes, and identification of IL9R pseudogenes at 9qter, 10pter, 16pter, and 18pter. Genomics 29:371–382 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (1994) MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci 10:189–191 [DOI] [PubMed] [Google Scholar]
- Li W-H (1997) Molecular evolution. Sinauer Associates, Sunderland, MA, pp 1–487 [Google Scholar]
- Meireles CM, Czelusniak J, Goodman M (1999) The Tarsius gamma-globin gene: pseudogene or active gene? Mol Phylogenet Evol 13:434–439 [DOI] [PubMed] [Google Scholar]
- Ohno S (1999) The one-to-four rule and paralogues of sex-determining genes. Cell Mol Life Sci 55:824–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Gorham HC, Louis EJ (1997) Chromosome ends: all the same under their caps. Curr Opin Genet Dev 7:822–828 [DOI] [PubMed] [Google Scholar]
- Rouquier S, Taviaux S, Trask BJ, Brand-Arpon V, van den Engh G, Demaille J, Giorgi D (1998) Distribution of olfactory receptor genes in the human genome. Nat Genet 18:243–250 (erratum in Nat Genet 19:102) [DOI] [PubMed] [Google Scholar]
- Shan Z, Zabel B, Trautmann U, Hillig U, Ottolenghi C, Wan Y, Haaf T (2000) FISH mapping of the sex-reversal region on human chromosome 9p in two XY females and in primates. Eur J Hum Genet 8:167–173 [DOI] [PubMed] [Google Scholar]
- Shen SH, Slightom JL, Smithies O (1981) A history of the human fetal globin gene duplication. Cell 26:191–203 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask BJ, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, Kuo WL, Massa H, Morrish T, Naylor S, Nguyen OT, Rouquier S, Smith T, Wong DJ, Youngblom J, van den Engh G (1998a) Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet 7:13–26 [DOI] [PubMed] [Google Scholar]
- Trask BJ, Massa H, Brand-Arpon V, Chan K, Friedman C, Nguyen OT, Eichler E, van den Engh G, Rouquier S, Shizuya H, Giorgi D (1998b) Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum Mol Genet 7:2007–2020 [DOI] [PubMed] [Google Scholar]
- van Geel M, Heather LJ, Lyle R, Hewitt JE, Frants RR, de Jong PJ (1999) The FSHD region on human chromosome 4q35 contains potential coding regions among pseudogenes and a high density of repeat elements. Genomics 61:55–65 [DOI] [PubMed] [Google Scholar]
- van Geel M, van Deutekom JC, van Staalduinen A, Lemmers RJ, Dickson MC, Hofker MH, Padberg GW, Hewitt JE, de Jong PJ, Frants RR (2000) Identification of a novel beta-tubulin subfamily with one member (TUBB4Q) located near the telomere of chromosome region 4q35. Cytogenet Cell Genet 88:316–321 [DOI] [PubMed] [Google Scholar]
- Wirth J, Nothwang HG, van der Maarel S, Menzel C, Borck G, Lopez-Pajares I, Brondum-Nielsen K, Tommerup N, Bugge M, Ropers HH, Haaf T (1999) Systematic characterisation of disease associated balanced chromosome rearrangements by FISH: cytogenetically and genetically anchored YACs identify microdeletions and candidate regions for mental retardation genes. J Med Genet 36:271–278 [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerucha T, Ekker M (2000) Distal-less-related homeobox genes of vertebrates: evolution, function, and regulation. Biochem Cell Biol 78:593–601 [PubMed] [Google Scholar]


