Abstract
Mitotic bookmarking is an epigenetic control mechanism that sustains gene expression in progeny cells; it is often found in genes related to the maintenance of cellular phenotype and growth control. RUNX transcription factors regulate a broad spectrum of RNA Polymerase (Pol II) transcribed genes important for lineage commitment but also regulate RNA Polymerase I (Pol I) driven ribosomal gene expression, thus coordinating control of cellular identity and proliferation. In this study, using fluorescence microscopy and biochemical approaches we show that the principal RUNX co-factor, CBFβ, associates with nucleolar organizing regions (NORs) during mitosis to negatively regulate RUNX-dependent ribosomal gene expression. Of clinical relevance, we establish for the first time that the leukemogenic fusion protein CBFβ-SMMHC (smooth muscle myosin heavy chain) also associates with ribosomal genes in interphase chromatin and mitotic chromosomes to promote and epigenetically sustain regulation of ribosomal genes through RUNX factor interactions. Our results demonstrate that CBFβ contributes to the transcriptional regulation of ribosomal gene expression and provide further understanding of the epigenetic role of CBFβ-SMMHC in proliferation and maintenance of the leukemic phenotype.
Background
Runt-related transcription factors (RUNX) bookmark genes important for phenotype, but the mitotic behavior of RUNX cofactor, Core Binding Factor β (CBFβ) is unknown.
Results
CBFβ and leukemogenic fusion protein CBFβ-SMMHC associate with chromosomes during mitosis and regulate ribosomal genes.
Conclusion
CBFβ and CBFβ-SMMHC contribute to epigenetic control of ribosomal genes.
Significance
CBFβ-SMMHC alters regulation linking phenotypic control with cell growth, thereby promoting cancer.
Keywords: Leukemia, Epigenetics, Ribosomal RNA (rRNA), Molecular cell biology, Transcription coactivators, Core binding factor β, Mitotic bookmarking, Ribosomal gene expression
INTRODUCTION
Core Binding Factor β (CBFβ) heterodimerizes with the runt domain of RUNX transcription factors to enhance their affinity for DNA (1). Upon binding of CBFβ, the runt domain undergoes a conformational change that stabilizes DNA binding through hydrogen bonds and hydrophobic interactions (2). Biological functions of CBFβ can be inferred from the defective fetal liver hematopoiesis and central nervous system bleeding seen in Cbfβ knockout mice (3,4), and bone-development defects (5) in Cbfβ knock-in mice. CBFβ has also been implicated in several types of cancer (6-8). For example, acute myeloid leukemia (AML) of the subtype M4 with Eosinophilia (M4Eo) is characterized by a chromosomal translocation [Inv(16)] that fuses CBFβ with the MYH11 (Myosin Heavy Chain 11) gene to encode the CBFβ-SMMHC fusion protein (9), which sequesters RUNX1 in the cytoplasm (10,11) and acts as a transcriptional repressor (12). Thus, CBFβ plays a pivotal role in regulating RUNX-related transcriptional programs to ensure fidelity of tissue-specific development, the maintenance of cellular phenotype, as well as the onset and progression of cancer (13-15).
Mitotic bookmarking of genes by phenotype-specific transcription factors, a novel parameter of epigenetic control, establishes coordination between proliferation and cellular identity (16-19). RUNX transcription factors associate with (or bookmark) mitotic chromosomes to support post-mitotic gene expression via Pol II transcription and to regulate cell growth by modulating ribosomal gene transcription driven by Pol I (20-23). Transcription factors, such as RUNX1 and 2, and non-DNA binding protein complexes, such as Filamin A, have been implicated in the regulation of ribosomal gene expression through structural associations (24-28). The role of CBFβ, however, in mitotic bookmarking of genes has not yet been explored. Because CBFβ can be localized in nuclear and cytoskeletal structures (29,30) and because the nuclear envelope breaks down during compaction and partitioning of chromosomes (31), it is important to understand the dynamic distribution of CBFβ during mitosis. Here, we show that CBFβ associates with NORs during mitosis to negatively regulate RUNX-dependent ribosomal gene expression. Conversely, we demonstrate for the first time that the leukemogenic fusion protein, CBFβ-SMMHC, associates with mitotic chromosomes and promotes ribosomal gene transcription. Our results show that mitotic retention of CBFβ is important for sustaining competency of gene expression related to cell growth and phenotype in progeny cells. Our data also provide a disease perspective, supporting the view that CBFβ-SMMHC conveys information related to maintenance of the leukemic phenotype via mitotic bookmarking. .
EXPERIMENTAL PROCEDURES
Cell Culture and Synchronization
SaOS-2, HeLa and K562 cells were grown following cell culture conditions recommended by ATCC. ME-1 cells were grown in RPMI medium supplemented with 20% FBS, 2mM HEPES, and 1% Pen/Strep. Cells were blocked in mitosis by incubating with 100 ng/mL of colcemid (KaryoMax, Life Technologies, Carlsbad CA) for 16 h followed by shake-off to detach mitotic cells. Cell number, cell size and viability were calculated by trypan blue staining and using the Countess Automated Cell Counter (Life Technologies).
Primary and Secondary Antibodies
The following antibodies were used in this study: CBFβ rabbit polyclonal (ab33516, Abcam, Cambridge, MA), dilutions were 1:500 (IF, WB), 1:500 and 5 μg per IP (ChIP); CBFβ rabbit polyclonal (A-303-547A, Bethyl Labs, Montgomery, TX), dilutions were 1:1000 (WB) and 5 μg per IP; Runx1 (Cell Signaling Technology, Boston, MA), dilutions were 1:50 (IF) and 1:1000 (WB ); RUNX2 mouse monoclonal (8G5, MBL International, Woburn, MA), dilutions were 1:600 (IF) and 1:1000 (WB); UBF (F-9, Santa Cruz Biotechnology, Dallas, TX), dilutions were 1:500 (IF, WB); HDAC1 (H-51, Santa Cruz Biotechnology), 1:1000 dilution used for WB; beta-tubulin mouse monoclonal (T-4026, Sigma Aldrich, St. Louis, MO), 1:1000 dilution used for WB; lamin B1 (ab16048, Abcam) 1:1000 dilution used for WB; Normal IgG control (source) was used at 5 μg for IP. Secondary antibodies conjugated with HRP (Santa Cruz Biotechnology), were used at a dilution of 1:5000 for WB. Secondary antibodies conjugated with Alexa Fluor 488 or 594 (Life Technologies) were used at dilution of 1:500 for IF.
CBFβ Inhibitor Treatment
CBFβ inhibitor (Calbiochem/EMD Millipore, Catalog # 219505) was dissolved in DMSO to a stock concentration of 85 mM. Working concentrations used in this study were 25, 50, 75 and 100 μM. Control cells were incubated with DMSO only. Final DMSO concentrations in cell cultures ranged from 0.025 to 0.1 %. In experiments with SaOS-2 cells, CBFβ inhibitor treatment was 3 days. ME-1 cells were treated with CBFβ inhibitor for 7 days.
Flow Cytometry
Cells were fixed and stained for flow cytometry using BD Cytofix fixation buffer (554655, BD Biosciences, Franklin Lakes, New Jersey) and BD Pharmingen™ PI/RNase staining buffer (550825, BD Biosciences), following the manufacturer’s instructions. Side versus forward scatter plots obtained on a BD Biosciences LSRII flow cytometer are shown.
rDNA Promoter Construct and transfection
The wildtype MR170-BH rDNA-promoter reporter plasmid (32) and RUNX binding site mutant rDNA-promoter reporter plasmid described previously (20) were transfected and detected using the experimental conditions developed in the Stein-Lian laboratory (20).
DNA Plasmids and Transfection Methods
CBFβ-GFP and CBFβ-MYH11-GFP were kindly provided by Dr. Paul Liu (10). Plasmids were transfected using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s recommendations.
Mitotic Chromosome Preparation
Mitotic cells were washed twice with 1X PBS, fixed with 4% paraformaldehyde (PFA), and attached to charged slides by using the Cytospin 4 (Thermo Shandon) for 5 min at 500 rpm. Mitotic-spread quality was monitored using phase-contrast microscopy and DAPI immunofluorescence. Spreads were maintained in 1X PBS for subsequent use.
Immunofluorescence Microscopy
Cells grown on coverslips and mitotic chromosomes were fixed in 4% PFA in PBS- (15 min, room temperature (RT)), permeabilized in 0.25% PBS-Triton X-100 for 15 min at RT, and blocked in 1% BSA in PBS for 1 h at RT. Primary antibodies were diluted in 1% BSA in PBS and incubated with samples for 1 h RT or overnight at 4°C. After successive washes in 1X PBS, secondary antibodies coupled to Alexa Fluor 488 and/or 594 were added for 1 h at RT in the dark. Samples were washed in PBS with a last wash in distilled water and mounted in Prolong-DAPI mounting reagent (Life Technologies). Cells were analyzed using an epifluorescence Zeiss microscope coupled with a Hammamatsu CCD camera. Images were acquired using the Zen 2011 imaging software (Zeiss) or ImageJ Software (Macphotonics).
Co-Immunoprecipitation and Western Blot Analysis
Cells were washed twice with ice-cold PBS and harvested in cold sonication buffer [150 mM NaCl, 50 mM Tris (pH 8), 1% NP-40, 0.5% deoxycholate, 25 mM MG132, and 2X protease inhibitor mixture (Roche, cOmplete EDTA-free)]. Cells were sonicated (QSonica Sonicator system fitted with a 1.6-mm tip) and centrifuged at 9391 RCF for 5 min at 4°C. Lysates were incubated overnight at 4°C with specific antibodies or normal IgG (Millipore, Billerica, MA). Lysates were then incubated with protein A/G beads (Santa Cruz Biotechnology) for 3 h, followed by 4 washes with ice cold wash buffer [50 mM NaCl, 20 mM Tris (pH 8.3), 0.5% Sodium deoxycholate, 0.5% Nonidet P-40, 2 mM EDTA, 25 μM MG132, and 2X protease inhibitor mixture]. Immunoprecipitated proteins were resolved by SDS/PAGE and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore). Blots were incubated with different primary antibodies followed by incubation with HRP-conjugated secondary antibodies. For CBFβ IP, Clean-Blot IP Detection Reagent was used (Cat. #21230, Thermo Scientific, Rockford, IL). Chemiluminescence (Perkin-Elmer Life Sciences, Boston, MA) was visualized using the BioRad ChemiDoc SRS device.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed using the EZ-ChIP kit (Millipore 17-371), following the manufacturer’s recommendations. DNA was sheared with a Covaris S-series S220 instrument (Woburn, MA) using the following conditions: Duty Cycle: 10%, Intensity: 3, Cycles per Burst: 200 to an average DNA fragment size of 350 base pairs. PCR primer sets used in qPCR to quantify ribosomal gene promoter regions in purified DNA fragments after ChIP were as described, as were negative control qPCR reactions (23).
RNA Isolation and cDNA synthesis
Total cellular RNA was isolated from cells using the RNAeasy mini kit (QIAGEN). cDNA was generated from RNA using the SuperScript First Strand kit (Life Technologies).
Real-time PCR
cDNA and DNA quantities were evaluated using real-time PCR (qPCR) with SYBR-green detection (Life Technologies) on the ViiA 7 Real Time PCR system (Life Technologies). RNA transcript levels were normalized using β-actin-encoding mRNA as the internal control. ChIP enrichment was determined as a percentage of input.
Isolation of Nuclear Fraction
Nuclei were purified using the protocol described in the Nuclei EZ Prep Nuclei Isolation Kit (Sigma Aldrich, St. Louis, MO). Additionally, cells were disrupted by several passes through a Dounce homogenizer and nuclei were centrifuged through a dense sucrose cushion (33) to obtain highest purity of nuclei. The quality of nuclear extraction was monitored by trypan blue staining and microscopic visualization at different time-points during the protocol. Nuclear extracts were subjected to SDS-PAGE and western blot.
RESULTS
CBFβ Associates with RUNX2 on Mitotic Chromosomes
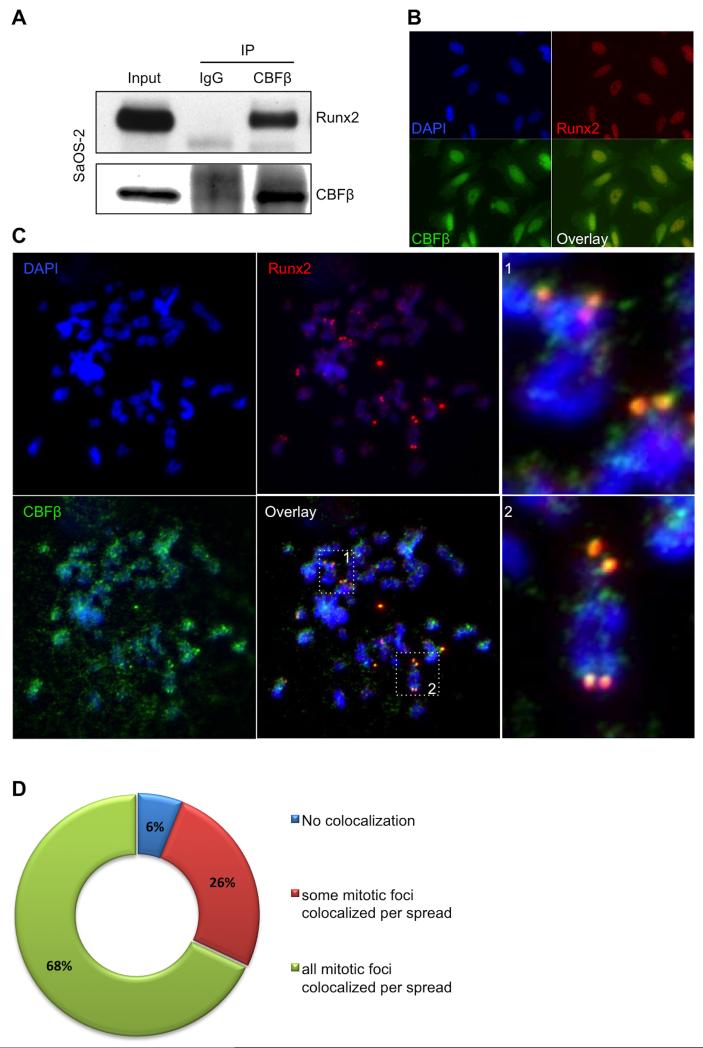
During mitosis, RUNX2 has been shown to localize in the NORs to regulate ribosomal gene expression (20). To determine the extent to which the RUNX transcription complex is epigenetically maintained, we explored whether CBFβ, the heterodimerization partner of RUNX proteins, also associates with NORs during mitosis. Initially, we used the osteosarcoma cell line, SaOS-2, that abundantly expresses both RUNX2 and CBFβ proteins, to verify that we could detect Runx2 in immunoprecipitate obtained using an anti-CBFβ antibody—indicating that the proteins interact and can be detected in our hands (Fig. 1A). Next, we examined the subcellular localization of CBFβ relative to RUNX2 during interphase in SaOS-2 cells using immunofluorescence (IF) analysis. Both CBFβ and RUNX2 were found throughout the nucleus of cells, and CBFβ also localized to the cytoplasm (Fig. 1B) as described previously (30). CBFβ antibody specificity was verified by including a blocking peptide that prevented detection of a CBFβ signal by IF analysis (data not shown). To address the localization of CBFβ in the NORs during mitosis, SaOS-2 cells were blocked with colcemid, mitotic spreads were prepared, and IF analysis for CBFβ and RUNX2 was performed (Fig. 1C and D). As previously shown, these data suggest that RUNX2 is present at specific loci in several mitotic chromosomes, including but not confined to the acrocentric chromosomes where the NORs reside, often symmetrically localized on sister chromatids (20). CBFβ immunofluorescence colocalized with these intense RUNX2 foci, but also was present at additional chromosomal regions (Fig. 1C, boxes 1 and 2). These results suggest that CBFβ associates with RUNX2 at NORs during mitosis, which raises the question of whether CBFβ is required for RUNX2 regulation of ribosomal gene expression.
FIGURE 1. Analysis of CBFβ/RUNX2 colocalization during mitosis.
A, Immunoprecipitation of CBFβ was performed from asynchronous SaOS-2 whole cell lysates. A 22 KDa band corresponding to CBFβ (lower panel) was detected in the input (5%) and the CBFβ-IP samples, but not in those immunoprecipitated with the control, normal IgG. RUNX2 (60 KDa) was also seen only in the CBFβ-IP samples (Upper panel), indicating that is was co-immunoprecipitated with CBFβ and suggesting that RUNX2 and CBFβ remain as a complex. B, IF microscopy images of interphase SaOS-2 cells using antibodies for RUNX2 and CBFβ, and DAPI stain. Fluorescence data indicate that CBFβ is detected in both nuclei and cytoplasm, and RUNX2 is mainly located in the nucleus. In agreement with the IP results, co-localization of CBFβ with RUNX2 is observed (overlay image); C, IF of chromosomal spreads from SaOS-2 cells previously blocked in mitosis indicates that CBFβ is frequently located in chromosomes and co-localizes with RUNX2 in NORs (dotted squares 1 and 2 in overlay, enlarged on the right); D, Pie chart representing the distribution of co-localization of CBFβ with RUNX2 in mitotic spreads.
Analysis of CBFβ Association with Pol-I Ribosomal Machinery
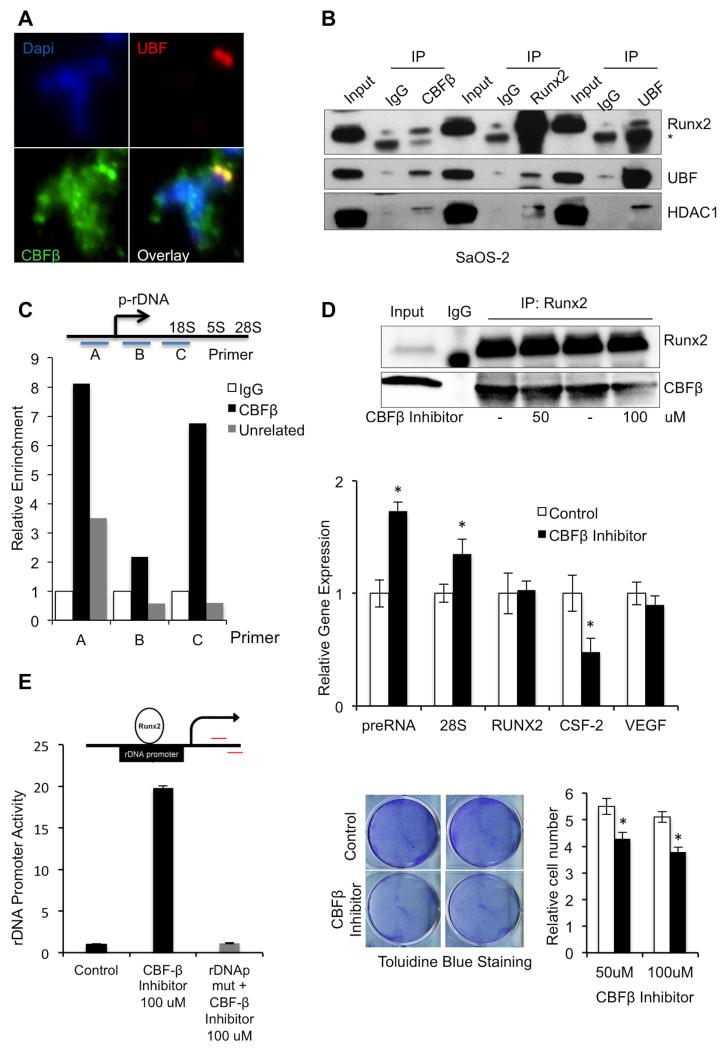
Previous studies have shown that RUNX2 localizes at NORs (20,23) and associates with both Upstream Binding Factor (UBF), an essential component of the ribosomal gene regulatory machinery and Histone deacetylase 1 (HDAC1), a RUNX2 cofactor that has been shown to be involved in rRNA gene expression (27). To investigate whether CBFβ is also involved in these associations, we immunolabeled SaOS-2 mitotic chromosomes; the IF data indicate that UBF and CBFβ colocalize at the NORs during mitosis (Fig. 2A). We then conducted co-immunoprecipitation experiments using CBFβ, RUNX2 and UBF antibodies with lysates from asynchronous SaOS-2 cells. The IP reaction products were analyzed by Western blotting, using RUNX2, UBF and HDAC1. The data indicate that each IP captured not only the cognate protein, but also the other two, supporting the hypothesis that these proteins reside in a complex (Fig. 2B). To confirm that CBFβ associates with the ribosomal gene transcription machinery, we performed ChIP using an antibody for CBFβ. We detected specific CBFβ enrichment in three different ribosomal gene promoter regions (Fig. 2C) using real-time PCR with primer sets described previously (27). As controls, relative enrichment was compared to that seen in ChIP samples immunoprecipitated using normal IgG and an unrelated antibody. Taken together, these results demonstrate that CBFβ associates with Pol-I regulatory complexes of ribosomal RNA genes.
FIGURE 2. CBFβ Association with Pol-I ribosomal machinery.
A, Immunofluorescence staining in mitotic spreads with UBF CBFβ primary antibodies indicates that, like UBF, CBFβ is present in NORs during mitosis; B, Whole cell lysates from asynchronous SaOS-2 cells were immunoprecipitated using CBFβ, RUNX2 or UBF antibodies; normal IgG was used as a nonspecific control. The IP products were evaluated by Western blot with RUNX2, UBF and HDAC1. Signal from IgG heavy chain [IgG] was detected in some samples, depending on the primary and secondary antibodies used. The results suggest that these proteins associate in cells; C, To confirm that CBFβ associates with the ribosomal gene transcription complex, ChIP was performed in SaOS-2 cells using a CBFβ, unrelated, or normal IgG antibody. The immunoprecipitated DNA fragments were amplified using real-time PCR with one of 3 primer sets that span the ribosomal gene promoter (top, (27)). Relative enrichment of all three ribosomal gene promoter regions was seen in samples that used CBFβ antibody for ChIP compared to that seen in control ChIP samples (bottom). D, To evaluate the effect of the CBFβ/RUNX2 complex on ribosomal gene expression, SaOS-2 cells were treated with CBFβ inhibitor (or DMSO-only as a control) for 3 days. Samples were then subjected to IP using RUNX2 antibodies and immunoprecipitated protein was evaluated by Western blot (upper panel). RT-qPCR analysis of ribosomal gene expression was conducted using methods and primers described previously (23,27). This analysis suggests that CBFβ inhibition results in increased levels of preRNA and 28S ribosomal gene transcripts and decreased CSF-2 expression levels; RUNX2 and VEGF levels were equivalent with and without CBFβ inhibitor treatment (middle panel). Data are presented as mean ± standard deviation (SD) (n=3). (*) indicates P <0.05 when compared to DMSO-treated control cells; t- test analysis was performed. The effects of CBFβ inhibitor (black bars), compared to DMSO-only treatment (white bars) on cell proliferation were evaluated using Toluidine Blue staining and relative cell count (n=3, mean ± SD (*) indicates P < 0.05; t-test). Both analyses suggest that CBFβ inhibition reduces cell proliferation (lower panels). E, Analysis of CBFβ inhibition on rDNA promoter activity via RUNX2. An rDNAp (top) was transfected into SaOS-2 cells and its activity was measured upon treatment with CBFβ inhibitor (bottom). The CBFβ inhibitor did not increase promoter activity of the rDNAp containing a mutated RUNX site (rDNApmut). Data are presented as mean ± SD (n=3).
CBFβ Regulates Ribosomal Gene Expression
In osteoblastic cells, RUNX2 plays an important role in negatively regulating the expression of ribosomal genes (20). To provide mechanistic insight into the role of its primary co-transcription factor, CBFβ, in regulating ribosomal gene expression, we pursued two approaches. First, we examined ribosomal gene expression levels in conditions that perturb the CBFβ/RUNX2 complex using a CBFβ inhibitor designated “17”. This inhibitor has been shown to bind to CBFβ and allosterically prevent formation of complexes with RUNX1 (34). The CBFβ inhibitor decreased CBFβ/RUNX2 interaction in SaOS-2 cells as detected by immunoprecipitation (Fig. 2D, top panel). Real-time PCR (RT-qPCR) analysis of ribosomal gene expression in treated cells showed that CBFβ inhibition resulted in increased levels of preRNA and 28S ribosomal gene transcripts, but did not affect RUNX2 mRNA levels (Fig. 2D, middle panel). In addition, CBFβ inhibition downregulated expression of the RUNX-related Colony Stimulating Factor 2 gene (CSF-2). Expression levels of RUNX2 and the related gene, VEGF, remained unchanged. As expected, CSF-2 inhibition correlated with a decrease in cell proliferation detected by toluidine blue staining and cell count in cells treated with CBFβ inhibitor (Fig. 2D, lower panel). These results suggest that CBFβ plays a role in modulating the expression of ribosomal genes. We next evaluated the effect of CBFβ inhibitor on the activity of a reporter construct containing either a normal or mutated ribosomal DNA promoter (rDNAp) sequence (Fig. 2E). CBFβ inhibitor dramatically increased ribosomal gene promoter activity in the wildtype construct, but had no effect on the rDNA reporter with a mutated RUNX-recognition sequence (rDNApmut). These findings suggest that CBFβ supresses ribosomal gene expression via RUNX transcription factor binding to the ribosomal promoter. Taken together, our results indicate that CBFβ interacts with the ribosomal gene transcription machinery via RUNX2 to regulate rDNA expression.
The Leukemogenic Fusion Protein CBFβ-SMMHC is Retained in Mitotic Chromosomes
Our laboratory has demonstrated that the leukemogenic chromosomal translocation fusion protein AML1/ETO (35) is retained in the NORs of human AML-derived Kasumi-1 cells in mitosis and epigenetically mediates cell growth through upregulation of ribosomal gene transcription (22,36). Whether other leukemogenic translocation fusion proteins are also involved in mitotic retention/bookmarking remains to be resolved. We therefore investigated the CBFβ fusion protein CBFβ- SMMHC that is associated with the M4Eo type of leukemia.
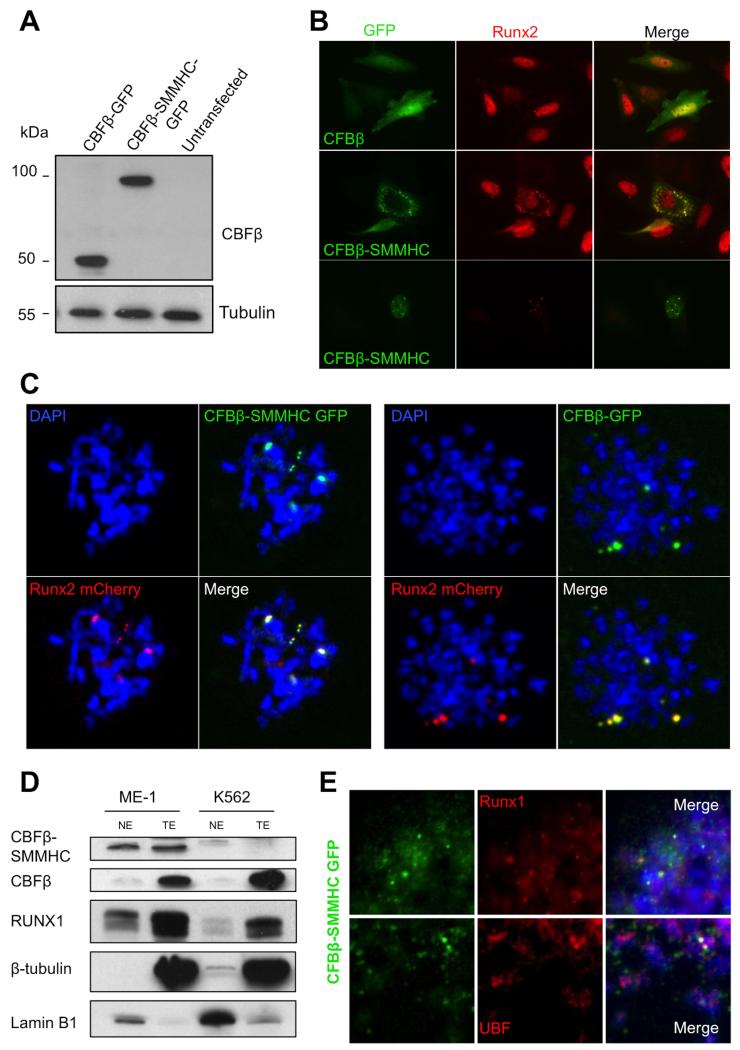
First, we expressed CBFβ-GFP or CBFβ-SMMHC-GFP proteins in HeLa and SaOS-2 cells to monitor their localization in mitotic chromosomes. The identities of both the wild type and fusion proteins were first confirmed in HeLa cells by Western blot analysis for CBFβ, using tubulin as a loading control (Fig. 3A).
FIGURE 3. Leukemogenic CBFβ-SMMHC associates with NORs in mitotic chromosomes.
A, HeLa cells were transfected with CBFβ-GFP or CBFβ-SMMHC-GFP expression plasmids and western blot analysis from total cell extracts was performed. Detection with anti-CBFβ antibody shows proteins of sizes that correspond to exogenous (expression-plasmid-encoded) proteins (~54Kda and ~96 KDa, respectively); β-tubulin was used as a loading control. B, SaOS-2 cells transfected with CBFβ-GFP or CBFβ-SMMHC-GFP were fixed and labeled using an anti-RUNX2 antibody; GFP was detected by fluorescence only. CBFβ-SMMHC-GFP but not CBFβ-GFP caused cytoplasmic sequestration of RUNX2 as well as atypical distribution of RUNX2 in the nucleus; C, IF of chromosomal spreads from HeLa cells transfected with RUNX2-mCherry and CBFβ-GFP or CBFβ-SMMHC-GFP indicate that both normal and leukemic proteins remained associated with NORs during mitosis. D, Western blot of nuclear and total extracts from ME-1 and K562 leukemia cell lines to detect protein levels of CBFβ, CBFβ-SMMHC, and RUNX1; lamin B1 (nuclear) and β-tubulin (cytoplasmic) were used as loading and cell fractionation controls. E, Mitotic spreads from ME-1 cells expressing CBFβ-SMMHC-GFP were labeled with DAPI and examined via IF with antibodies to detect RUNX1 (upper panel) or UBF (lower panel).
By fluorescence microscopy, CBFβ-GFP showed normal distribution throughout the nucleus and cytoplasm of SaOS-2 cells during interphase (Fig. 3B, upper panel; compare with Fig. 1B). It has been reported that CBFβ-SMMHC perturbs normal RUNX2 localization, causing it to localize in the cytoplasm, rather than in the nucleus (10,29). In a separate study, CBFβ-SMMHC was shown to localize to the nucleus and cytoplasmic membrane (37). In our studies, we observed both phenomena, with some cells showing punctate staining from CBFβ-SMMHC-GFP in the cytoplasm (Fig. 3B, middle) and others in the nucleus (Fig. 3B, lower panel).
To examine the localization of CBFβ-SMMHC during mitosis, mitotic chromosomal spreads were prepared from HeLa cells transfected with both RUNX2-mCherry and CBFβ-MYH11-GFP or CBFβ-GFP expression plasmids. In our earlier experiments, CBFβ and RUNX2 co-localized on mitotic chromosomes (Fig. 1). We saw comparable results in HeLa cells, with both normal CBFβ and leukemia-related CBFβ-SMMHC retained in mitotic foci with RUNX2 (Fig. 3C). To confirm these observations for RUNX1, we used the human leukemic ME-1 cell line that endogenously expresses both RUNX1 and CBFβ-SMMHC and K562 cells that do not express the leukemic fusion protein (38). As expected, CBFβ-SMMHC was detected in nuclear and total cell extracts from ME-1 cells but not in the leukemic K562 cells (negative control) (Fig. 3D). Endogenous RUNX1 and CBFβ were detected in total cell extracts and to a lesser extent in nuclear extracts of both cell lines. Tubulin and lamin B1 were used to monitor cell fractionation. Immunofluorescence analysis of mitotic chromosome spreads from ME-1 cells expressing CBFβ-SMMHC-GFP showed co-localization of the fusion protein with endogenous RUNX1 (Fig 3E, upper panels) as well as with UBF (Fig. 3E, lower panels). Based on these results we conclude that the leukemogenic CBFβ-SMMHC fusion protein associates with NORs in mitotic chromosomes.
CBFβ-SMMHC Regulates Ribosomal Gene Expression
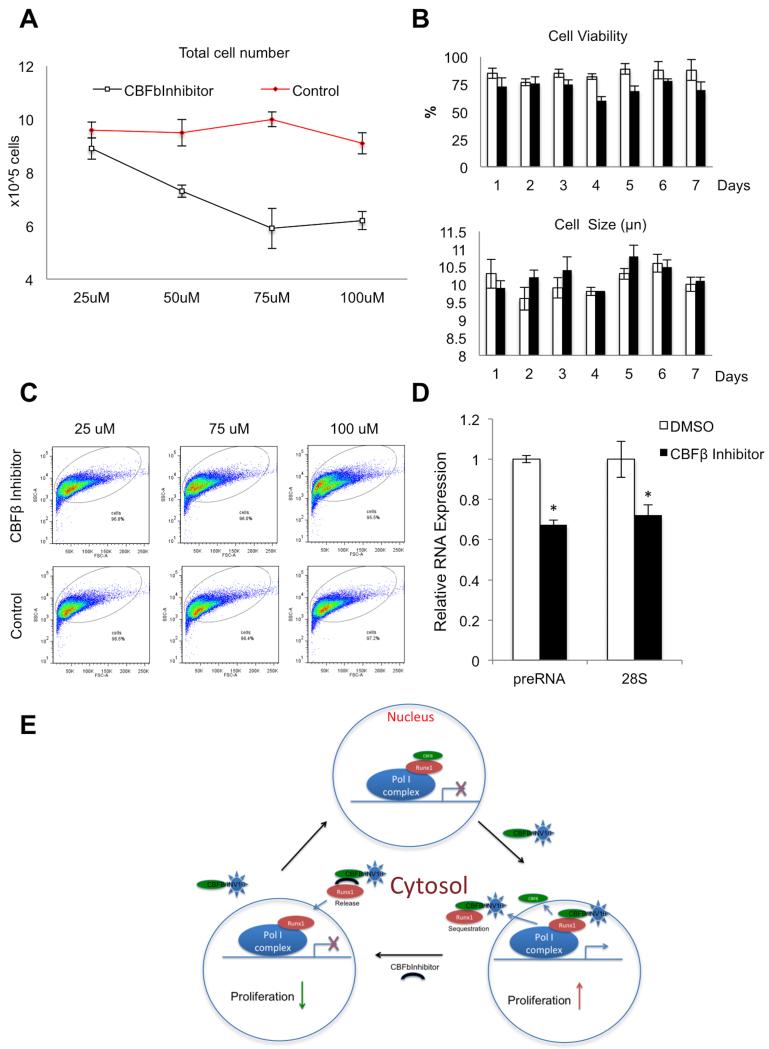
With strong evidence that CBFβ regulates ribosomal gene expression (Fig. 2) and CBFβ-SMMHC associates with mitotic chromosomes in NORs (Fig. 3), we investigated whether CBFβ-SMMHC likewise regulates expression of ribosomal genes (Fig. 4). First we analyzed ribosomal gene expression in ME-1 cells treated with CBFβ inhibitor compared to untreated controls, using cell proliferation as a read-out. As reported previously (34), our data indicate that CBFβ inhibitor decreased proliferation of ME-1 cells with increasing dose (Fig. 4A). CBFβ inhibitor did not significantly affect cell viability as detected by trypan blue staining, nor did it alter cell size as determined by quantitative analysis of cell diameter (Fig. 4B). However, it did induce changes in cellular morphology. Cells treated with increasing concentrations of CBFβ inhibitor or DMSO-only were analyzed by flow cytometry, and graphed as side versus forward scatter plots (Fig. 4B). At the higher levels of CBFβ inhibitor, we detected a cell population with increased side scatter, indicating changes in cell density and granularity. These observations are consistent with reported changes in cell shape, lower nuclear to cytoplasmic ratio and lobulation of nuclei after CBFβ inhibitor treatment (34). RT-qPCR analysis of ribosomal transcript levels in ME-1 cells treated with inhibitor showed that treated cells had decreased levels of preRNA and 28S RNA levels, in comparison with control cells (Fig. 4D). These results suggest that in cells expressing the leukemogenic CBFβ fusion protein, the CBFβ inhibitor modulates the dynamics of the CBFβ-SMMHC/RUNX1 complex to downregulate ribosomal gene expression. This is in contrast to the inhibitor’s effect on normal wildtype CBFβ, where it results in the de-repression (upregulation) of RNA expression. Taken together, our findings suggest a novel epigenetic regulatory role of CBFβ-SMMHC in acute myeloid leukemia of the subtype M4Eo.
FIGURE 4. Effects of CBFβ inhibition on ribosomal gene expression and proliferation in ME-1 cells.
ME-1 cells are derived from an AML patient; they express both CBFβ and CBFβ-SMMHC proteins. A, Dose-response of CBFβ inhibitor on ME-1 cell proliferation. Cells were incubated with or without CBFβ inhibitor at different concentrations (x-axis) and cell number was counted (y-axis) after 7 days of treatment. Data are presented as mean ± SD (n=3). B, ME-1 cell viability (top graph) and cell size (lower graph) were assayed, using trypan blue assay and Countess instrument respectively, after treatment with 100 μM CBFβ inhibitor (black bars) or DMSO-only (white bars). Data are presented as mean ± SD (n=3); C, Flow cytometry was performed to detect changes in morphology of ME-1 cells treated with or without different concentrations of CBFβ inhibitor. Side versus forward scatter plots are presented. Note a shift of the ME-1 population in cells treated with the inhibitor versus control cells; D, RT-qPCR analysis in ME-1 cells suggest that relative levels of preRNA and 28S ribosomal gene transcripts were lower in cells treated with CBFβ inhibitor compared to DMSO-treated control cells (n=3, mean ± SD (*) indicates P < 0.05 ; t-test).
DISCUSSION
Mitotic bookmarking of genes by RUNX proteins is essential for the maintenance of coordination between cell growth and cellular identity (19). In this study we show that CBFβ, the primary RUNX cofactor, associates with ribosomal genes on chromosomes during mitosis to epigenetically regulate RUNX-meditated ribosomal gene expression. In addition, we observed that the CBFβ association with gene loci on mitotic chromosomes is not confined to NORs but that this so-called mitotic bookmarking also occurs in other chromosomal regions, that are not associated with ribosomal genes. RUNX proteins have also been shown to associate with chromosomes in regions other than the NORs (20). Interestingly, CBFβ is rather broadly expressed, including in RUNX-negative cells such as HeLa and embryonic stem cells, indicating the strong possibility of a biological role apart from contributing to control by RUNX transcription factors. Further studies will be necessary to conclusively delineate other genes that are mitotically bookmarked by CBFβ.
Ribosomal gene expression is driven by the Pol-I regulatory machinery, of which UBF is an essential component. In our study, CBFβ, UBF and RUNX2 were immunoprecipitated together. We also found that perturbing CBFβ-RUNX interactions using CBFβ inhibitor resulted in an increase in rRNA gene expression, presumably by relieving RUNX suppression of rRNA genes. Our results suggest that CBFβ is a physiological mediator of rRNA transcription—it has been shown to interact with the cytoskeletal protein Filamin A in the cytoplasm, an interaction that prevents CBFβ from functioning as a regulatory partner of RUNX1 (30). Additionally, Filamin A can be localized to the nucleolus via a nucleolar localization signal (NoLS) to suppress ribosomal gene transcription (25). It will be informative to determine whether Filamin A/RUNX/CBFβ interactions modulate ribosomal gene transcription.
Many leukemias result from chromosomal translocations that alter RUNX biology; these include RUNX1-ETO (AML1-ETO) and CBFβ-MYH11. It has previously been shown that RUNX1-ETO functions to activate ribosomal gene transcription and upregulate protein synthesis (22). Of clinical relevance, we show that the leukemogenic fusion protein CBFβ-SMMHC is retained on mitotic chromosomes and regulates ribosomal gene transcription during interphase. Given the dynamics of CBFβ-SMMHC in the cytoskeleton and in the nuclear compartment, we propose the following mechanism for the regulation of ribosomal gene expression: In normal diploid cells, association of RUNX/CBFβ with ribosomal genes suppresses ribosomal gene transcription. When present, CBFβ-SMMHC competes with and displaces CBFβ from ribosomal gene promoters. As a consequence, CBFβ-SMMHC/RUNX dynamically relocates from the ribosomal gene promoter to the cytoskeletal compartment, thereby alleviating suppression of ribosomal gene transcription.
The CBFβ inhibitor that prevents RUNX proteins from complexing with CBFβ (34) may release cytoskeleton-sequestered RUNX from CBFβ-SMMHC to enter the nucleus, resulting in transcriptional repression of ribosomal genes (Fig. 4E). It is recognized that the CBFβ inhibitor used in this study targets both CBFβ and CBFβ-SMMHC. Availability of a selective inhibitor for CBFβ-SMMHC would be mechanistically informative for further investigations and could advance our understanding of the relationships between expression of the CBFβ-SMMHC fusion protein and control of cell growth. It also has the potential to provide a therapeutic option for treating patients carrying the CBFβ-MYH11 translocation by modulating the activity of ribosomal gene expression that is linked to proliferation and tumorigenesis.
Our findings reinforce the significance and expand mechanistic understanding of bookmarking genes on mitotic chromosomes as a novel regulatory mechanism that contributes to control of cell growth, phenotype identity and proliferation. Our results are consistent with functional involvement of CBFβ and CBFβ-SMMHC in mitotic bookmarking, expanding the cohort of regulatory molecules that are engaged in sustaining epigenetic control in normal and cancer cells.
Acknowledgements
We thank Paul Liu for the GFP plasmids used in this study, Sayyed K. Zaidi for helpful scientific recommendations, Lori Martin for editing assistance, and Shirwin Pockwinse at the University of Massachusetts Medical School for assistance with microscopy.
Footnotes
This work was supported by National Institutes of Health (NIH) grant P01 CA082834 (to GSS).
The abbreviations used are: CBFβ, Core Binding Factor β; SMMHC, Smooth Muscle Myosin Heavy Chain; RUNX, runt-related transcription factor; NORs, Nucleolar Organizing Regions; AML, Acute Myeloid Leukemia; M4Eo, M4 with Eosinophilia; MYH11, Myosin Heavy Chain 11; Pol I, RNA Polymerase I; Pol II, RNA Polymerase II; ChIP, Chromatin Immunoprecipitation; IF, Immunofluorescence; UBF, Upstream Binding Factor; CSF-2, Colony Stimulating Factor 2; rDNAp, ribosomal DNA promoter; rDNApmut, mutated RUNX recognition sequence; NoLS, Nucleolar Localization Signal; SD, standard deviation.
REFERENCES
- 1.Bravo J, Li Z, Speck NA, Warren AJ. The leukemia-associated AML1 (Runx1)--CBF beta complex functions as a DNA-induced molecular clamp. Nature structural biology. 2001;8:371–378. doi: 10.1038/86264. [DOI] [PubMed] [Google Scholar]
- 2.Nagata T, Gupta V, Sorce D, Kim WY, Sali A, Chait BT, Shigesada K, Ito Y, Werner MH. Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nature structural biology. 1999;6:615–619. doi: 10.1038/10658. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, Muenke M, Lian JB, Yang Y, Nuckolls GH, Stein GS, Liu PP. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nature genetics. 2002;32:639–644. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- 6.Davis JN, Rogers D, Adams L, Yong T, Jung JS, Cheng B, Fennell K, Borazanci E, Moustafa YW, Sun A, Shi R, Glass J, Mathis JM, Williams BJ, Meyers S. Association of core-binding factor beta with the malignant phenotype of prostate and ovarian cancer cells. Journal of cellular physiology. 2010;225:875–887. doi: 10.1002/jcp.22298. [DOI] [PubMed] [Google Scholar]
- 7.Mendoza-Villanueva D, Deng W, Lopez-Camacho C, Shore P. The Runx transcriptional co-activator, CBFbeta, is essential for invasion of breast cancer cells. Molecular cancer. 2010;9:171. doi: 10.1186/1476-4598-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Beau MM, Larson RA, Bitter MA, Vardiman JW, Golomb HM, Rowley JD. Association of an inversion of chromosome 16 with abnormal marrow eosinophils in acute myelomonocytic leukemia. A unique cytogenetic-clinicopathological association. The New England journal of medicine. 1983;309:630–636. doi: 10.1056/NEJM198309153091103. [DOI] [PubMed] [Google Scholar]
- 9.Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, Collins FS. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 10.Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Molecular and cellular biology. 1998;18:7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanno Y, Kanno T, Sakakura C, Bae SC, Ito Y. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor alpha (CBFalpha) subunit by the leukemia-related PEBP2/CBFbeta-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Molecular and cellular biology. 1998;18:4252–4261. doi: 10.1128/mcb.18.7.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutterbach B, Hou Y, Durst KL, Hiebert SW. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12822–12827. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo YH, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood. 2009;113:3323–3332. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo YH, Gerstein RM, Castilla LH. Cbfbeta-SMMHC impairs differentiation of common lymphoid progenitors and reveals an essential role for RUNX in early B-cell development. Blood. 2008;111:1543–1551. doi: 10.1182/blood-2007-07-104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo YH, Landrette SF, Heilman SA, Perrat PN, Garrett L, Liu PP, Le Beau MM, Kogan SC, Castilla LH. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer cell. 2006;9:57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Tai PW, Zaidi SK, Wu H, Grandy RA, Montecino M, van Wijnen AJ, Lian JB, Stein GS, Stein JL. The dynamic architectural and epigenetic nuclear landscape: developing the genomic almanac of biology and disease. J Cell Physiol. 2014;229:711–727. doi: 10.1002/jcp.24508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaidi SK, Van Wijnen AJ, Lian JB, Stein JL, Stein GS. Targeting deregulated epigenetic control in cancer. J Cell Physiol. 2013;228:2103–2108. doi: 10.1002/jcp.24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein GS, Zaidi SK, Stein JL, Lian JB, van Wijnen AJ, Montecino M, Young DW, Javed A, Pratap J, Choi JY, Ali SA, Pande S, Hassan MQ. Genetic and epigenetic regulation in nuclear microenvironments for biological control in cancer. Journal of cellular biochemistry. 2008;104:2016–2026. doi: 10.1002/jcb.21813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nature reviews. Genetics. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 21.Pande S, Ali SA, Dowdy C, Zaidi SK, Ito K, Ito Y, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. Journal of cellular physiology. 2009;218:473–479. doi: 10.1002/jcp.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakshi R, Zaidi SK, Pande S, Hassan MQ, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. Journal of cell science. 2008;121:3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, Shakoori AR, Montecino MA, Lian JB, van Wijnen AJ, Imbalzano AN, Stein GS, Stein JL. Phenotypic transcription factors epigenetically mediate cell growth control. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annual review of pharmacology and toxicology. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 25.Deng W, Lopez-Camacho C, Tang JY, Mendoza-Villanueva D, Maya-Mendoza A, Jackson DA, Shore P. Cytoskeletal protein filamin A is a nucleolar protein that suppresses ribosomal RNA gene transcription. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1524–1529. doi: 10.1073/pnas.1107879109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics : official journal of the DNA Methylation Society. 2009;4:374–382. doi: 10.4161/epi.4.6.9449. [DOI] [PubMed] [Google Scholar]
- 27.Ali SA, Dobson JR, Lian JB, Stein JL, van Wijnen AJ, Zaidi SK, Stein GS. A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation. Journal of cell science. 2012;125:2732–2739. doi: 10.1242/jcs.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali SA, Zaidi SK, Dobson JR, Shakoori AR, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4165–4169. doi: 10.1073/pnas.1000620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka Y, Watanabe T, Chiba N, Niki M, Kuroiwa Y, Nishihira T, Satomi S, Ito Y, Satake M. The protooncogene product, PEBP2beta/CBFbeta, is mainly located in the cytoplasm and has an affinity with cytoskeletal structures. Oncogene. 1997;15:677–683. doi: 10.1038/sj.onc.1201235. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida N, Ogata T, Tanabe K, Li S, Nakazato M, Kohu K, Takafuta T, Shapiro S, Ohta Y, Satake M, Watanabe T. Filamin A-bound PEBP2beta/CBFbeta is retained in the cytoplasm and prevented from functioning as a partner of the Runx1 transcription factor. Molecular and cellular biology. 2005;25:1003–1012. doi: 10.1128/MCB.25.3.1003-1012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nature reviews. Molecular cell biology. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 32.Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg ME, Bender TP. Identification of newly transcribed RNA. Current protocols in molecular biology / edited by Frederick M. Ausubel … [et al.] 2007 doi: 10.1002/0471142727.mb0410s78. Chapter 4, Unit 4 10. [DOI] [PubMed] [Google Scholar]
- 34.Gorczynski MJ, Grembecka J, Zhou Y, Kong Y, Roudaia L, Douvas MG, Newman M, Bielnicka I, Baber G, Corpora T, Shi J, Sridharan M, Lilien R, Donald BR, Speck NA, Brown ML, Bushweller JH. Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBFbeta. Chemistry & biology. 2007;14:1186–1197. doi: 10.1016/j.chembiol.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 36.Zaidi SK, Trombly DJ, Dowdy CR, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Epigenetic mechanisms in leukemia. Advances in biological regulation. 2012;52:369–376. doi: 10.1016/j.jbior.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Kanto S, Chiba N, Tanaka Y, Fujita S, Endo M, Kamada N, Yoshikawa K, Fukuzaki A, Orikasa S, Watanabe T, Satake M. The PEBP2beta/CBF beta-SMMHC chimeric protein is localized both in the cell membrane and nuclear subfractions of leukemic cells carrying chromosomal inversion 16. Leukemia. 2000;14:1253–1259. doi: 10.1038/sj.leu.2401821. [DOI] [PubMed] [Google Scholar]
- 38.Yanagisawa K, Horiuchi T, Fujita S. Establishment and characterization of a new human leukemia cell line derived from M4E0. Blood. 1991;78:451–457. [PubMed] [Google Scholar]