Abstract
Acinus has been reported to function in apoptosis, RNA processing and regulation of gene transcription including RA-dependent transcription. There are three different isoforms of Acinus termed Acinus-L, Acinus-S′ and Acinus-S. The isoforms of Acinus differ in their N-terminus while the C-terminus is consistent in all isoforms. The sub-nuclear localization of Acinus-L and Acinus-S′ was determined using fluorescence microscopy. Acinus-S′ colocalizes with SC35 in nuclear speckles while Acinus-L localizes diffusely throughout the nucleoplasm. RA treatment has little effect on the sub-nuclear localization of Acinus-L and Acinus-S′. The domains/regions necessary for the distinct sub-nuclear localization of Acinus-L and Acinus-S′ were identified. The speckled sub-nuclear localization of Acinus-S′ is dependent on its C-terminal RS- and RD/E-rich region but is independent of the phosphorylation status of Ser-453 and Ser-604 within this region. The unique N-terminal SAP-motif of Acinus-L is responsible for its diffuse localization in the nucleus. Moreover, the sub-nuclear localization of Acinus isoforms is affected by each other, which is determined by the combinatorial effect of the more potent SAP motif of Acinus-L and the C-terminal RS- and RD/E-rich region in all Acinus isoforms. The C-terminal RS- and RD/E-rich region of Acinus mediates the colocalization of Acinus isoforms as well as with its interacting protein RNPS1. In conclusion, the SAP motif is responsible for the difference in the nuclear localization between Acinus-L and Acinus-S′. This difference in the nuclear localization of Acinus-S′ and Acinus-L may suggest that these two isoforms have different functional roles.
Keywords: Acinus, nuclear speckles, SR/SR-related proteins, SAP motif, RS domain
Apoptotic Chromatin Condensation Inducer in the Nuclear (Acinus) was first identified in 1999 as a caspase-3-activated protein required for chromatin condensation during apoptosis (Sahara et al., 1999). Since that time Acinus has been implicated in a variety of other physiological processes. Cleavage of Acinus by caspase-3 was also suggested to be associated with non-apoptotic functions of caspase-3 including terminal erythroid cell differentiation and differentiation of monocytes into macrophages (Sordet et al., 2002; Zermati et al., 2001; Schwerk and Schulze-Osthoff, 2003). In addition, Acinus has also been shown to play a role in the regulation of gene expression. Our laboratory has previously reported that Acinus represses retinoic acid (RA)-responsive gene expression (Vucetic et al. 2008) and observed the upregulation of cyclin A1 expression by Acinus (Jang et al. 2008). Finally, Acinus has been implicated to play a role in pre-mRNA processing. Acinus has been shown to be a component of human spliceosomes (Rappsilber et al., 2002; Zhou et al., 2002), nuclear speckles, the storage site of pre-mRNA splicing factors (Saitoh et al., 2004; Spector and Lamond, 2011), and the exon junction complex (EJC) (Tange et al., 2005). It has also been shown to form a complex with two additional proteins, RNA-binding protein S1 (RNPS1) and Sin-3-associated protein of 18 kDa (SAP-18), termed ASAP (apoptosis- and splicing-associated protein) which inhibits splicing mediated by ASF-SF2 (SRSF1), SC35 (SRSF2) or RNPS1 in vitro (Schwerk et al., 2003).
Acinus is a SR-related protein (Boucher et al., 2001). Three isoforms, termed Acinus-L, Acinus-S and Acinus-S′, have been described which are most likely generated by alternative splicing and/or alternative promoter usage (Figure 1A) (Sahara et al, 1999). The protein sequence of Acinus displays high homology between human and mouse (>90% for Acinus-S/S′ and >80% for Acinus-L). The three isoforms share a common C-terminus which contains a RNA-recognition motif (RRM) and two regions rich in RS (arginine/serine) dipeptides. These are two typical structural features of SR/SR-related proteins. Interesting, between the two RS dipeptide repeat regions is a region containing RD/E dipeptide repeats (arginine/aspartic acid and arginine/glutamic acid). The three Acinus isoforms differ only in their N-termini: human Acinus-L has an unique 766 amino acid N-terminal region containing a SAP (after SAF-A/B, Acinus and PIAS) motif (Aravind and Koonin, 2000) and an additional RS domain, while human Acinus-S and Acinus-S′ have short unique N-termini which are 8 and 39 amino acids in length, respectively, which contain no identified conserved domains. The SAP motif is capable of binding to AT-rich chromosomal regions known as scaffold- or matrix-attachment regions (SARS/MARS) (Gohring et al., 1997; Kipp et al., 2000; Renz and Fackelmayer, 1996; Tan et al., 2002). To date, no distinct function(s) for any of these isoforms has been identified.
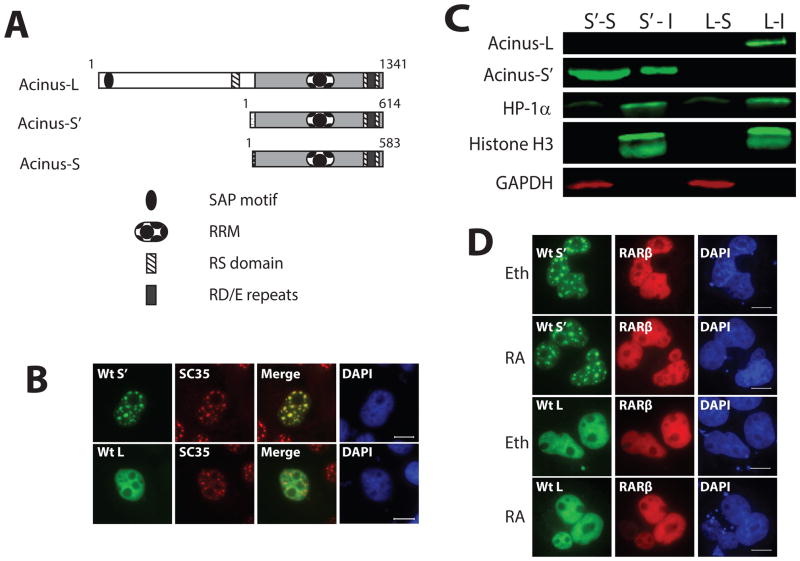
Figure 1. Acinus-L and Acinus-S′ have different sub-nuclear localization patterns.
A. Functional domains of the three human Acinus isoforms. B. The sub-nuclear localization of Acinus-L and Acinus-S′. C. Western blot analysis of Acinus-L and Acinus-S′ in soluble and insoluble fractions. D. RA does not affect the sub-nuclear localization of Acinus-L and Acinus-S′. In Panels B and D, COS7 cells were transfected with the indicated GFP-tagged wild type or mutant Acinus isoforms (B and D) and RFP-RARβ (D) expression vector DNAs. Twenty-four hr after transfection cells were fixed and proteins visualized using fluorescent microscopy (B) or treated with 10−6 M RA or Eth for an additional 5 hrs before fixation and visualization using fluorescent microscopy (D). Endogenous SC35 was labeled with anti-SC35 primary antibody and rhodamine conjugated secondary antibody (B). DAPI was used as a nuclear stain (B and D). Scale bar equals 10 μm (B and D). In Panel C, COS7 cells were transfected with wild type V5-Acinus-S′ (S′) or wild type V5-Acinus L (L) expression vector DNAs. Twenty-four hr after transfection soluble (S) and insoluble (I) fractions were prepared and analyzed by Western blot. Acinus-L and Acinus-S′ were detected using anti-V5 antibody. Histone H3 and HP1α were used as markers of the insoluble fraction and GAPDH was used as a marker of the soluble fraction.
In the present work, we investigated the nuclear localization of Acinus-L and Acinus-S′ to gain insight into the function of individual Acinus isoforms. We demonstrate the distinct sub-nuclear localization of Acinus-L and Acinus-S′. Acinus-S′ colocalizes with SC35 in nuclear speckles while Acinus-L localizes diffusely throughout the nucleoplasm. The speckled sub-nuclear localization of Acinus-S′ is dependent on its C-terminal RS- and RD/E rich regions. Interestingly, the diffuse nucleoplasmic localization of Acinus-L is dependent on the unique N-terminal SAP motif. Finally, the sub-nuclear localization of Acinus isoforms and potentially other SR/SR-related proteins is determined by the combinatorial effect of the more potent SAP motif of Acinus-L and the C-terminal RS- and RD/E rich regions in all Acinus isoforms.
MATERIALS AND METHODS
Cell culture and transient transfection
Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 μg/ml penicillin, and 100 units/ml streptomycin. The cells were maintained in a humidified 5% CO2 incubator at 37°C. All transfections were performed using GenJet™ DNA In Vitro Transfection Reagent (SignaGen, Rockville, MD) according to the manufacturer’s recommendations. After 6 hr of transfection, the medium was changed to vitamin A-free Dulbecco’s modified Eagle’s medium supplemented with 10% charcoal/dextran-treated fetal bovine serum. After 24 hr of transfection, in some experiments cells were treated with 10−6 M RA (generous gift from Hoffman-LaRoche, Nutley, NJ) or ethanol carrier and cultured for up to 24 hr.
Plasmids for fluorescence microscopy
pcDNA-DEST53-Acinus-L (GFP-Acinus-L) and pcDNA-DEST53-Acinus-L (ΔN) [GFP-Acinus-L (ΔN)] were constructed using Invitrogen Gateway® cloning technology. The coding sequences of full-length and the N-terminal 131 amino acid truncated human Acinus-L were amplified using the plasmid pCAGGS-Acinus-L-FLAG (a kind gift from Dr. Christian Schwerk, Heidelberg University, Germany) as the template. pcDNA-DEST53-Acinus-L (Δ14 aa) [GFP-Acinus-L (Δ14 aa)] and pcDNA-DEST53-Acinus-L (Δ35 aa) [GFP-Acinus-L (Δ35 aa)] were constructed using QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Inc. Wilmington, DE) to delete 14 amino acids (amino acids 85-98) or 35 amino acids (amino acids 72-106) of human Acinus-L, respectively. pEGFP-C3-Acinus-S′ (EGFP-Acinus-S′) was constructed by inserting the full length coding sequence of human Acinus-S′ into pEGFP-C3 (Clontech BD Biosciences, Mountain View, CA). pEGFP-C3-SAP-Acinus-S′ (EGFP-SAP-Acinus-S′) was constructed by inserting N-terminal 1-393 nucleotides (amino acids 1-131) of the coding sequence of human Acinus-L into pEGFP-C3-Acinus-S′. pEGFP-C3-Acinus-S′ (ΔC) [EGFP-Acinus-S′(ΔC)] and pEGFP-C3-SAP-Acinus-S′ (ΔC) [EGFP-SAP-Acinus-S′ (ΔC)] were constructed using QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Inc. Wilmington, DE) for introducing a premature stop codon to generate a C-terminal 205 amino acid truncated human Acinus-S′ and a C-terminal 205 amino acid truncated SAP-Acinus-S′ using the plasmids pEGFP-C3-Acinus-S′ and pEGFP-C3-SAP-Acinus-S′, respectively, as the templates. pmKate2-Acinus-S′ (mKate2-Acinus-S′) was constructed by inserting full-length coding sequence of human Acinus-S′ into pmKate2-C plasmid vector (Evrogen, Moscow, Russia). pRFP-RARβ was constructed by inserting the full length coding sequences of mouse RARβ into pDsRed-Monomer-C1 (Clontech BD Biosciences). pCMV-3XFLAG-RNPS1 (3XFLAG-RNPS1) was a kind gift from Dr. Akila Mayeda (Fujita Health University, Japan) (Sakashita et al, 2004).
Fluorescence and Immunofluorescence
COS-7 and 293A cells were seeded on glass coverslips. At the end of the transfection and treatment period, cells expressing GFP-tagged, EGFP-tagged or mKate-2 tagged proteins were fixed by immersion of the coverslips in 3.7% formaldehyde at room temperature for 30 min followed by three washes with PBS and one wash with ddH2O. To detect FLAG-tagged proteins, cells were fixed by immersion of the coverslips in 3.7% formaldehyde at room temperature for 30 min, followed by poration by immersion in 0.18% Triton X100 in PBS for 5 min and then washed 3 times with PBS. Blocking was performed by incubating cells with 1% BSA in PBS for 30 min at room temperature. Mouse monoclonal anti-FLAG M2 (F1804, Sigma Aldrich, St. Louis, MO) diluted with blocking buffer (1:500) was then added to the cover slips and incubated for 1 hr at room temperature. After incubation with the primary antibody, cells were washed 3 times with PBS followed by the addition of rhodamine conjugated bovine anti-mouse IgG (sc-2368, Santa Cruz, Santa Cruz, CA) diluted with blocking buffer (1:400) and incubated in the dark for 45 min at room temperature. After secondary antibody incubation, cells were washed 3 times with PBS. To detect endogenous SC35, the cells were fixed by immersion of the coverslips in a solution containing 2% formaldehyde and 0.2% Triton X-100 in PBS, and then incubated in acetone for 5 min at −20°C. Cells were processed as described for detection of FLAG-tagged proteins except that the primary antibody was mouse monoclonal anti-SC35 (ab11826, Abcam, Cambridge, MA) diluted in PBS (1:1000) for 1 hr at room temperature. In all cases, final processed coverslips were mounted with ProLong Gold Antifade Reagent with DAPI (Life Technologies, Carlsbad, CA) on microscope slides. Imaging was performed using a fluorescence microscope (OLYMPUS BX41) with filters for blue (DAPI), green (FITC) and red (TRITC) fluorescence. An Olympus Digital Camera Spot-Xplorer and the Spot Advanced Software (Oympus) was used to capture and merge images.
Soluble and Insoluble Chromatin Fractionation
Soluble and insoluble chromatin fractions were prepared as described by Masaki et al., 2007. Briefly, COS-7 cells were washed with PBS and extracted with lysis buffer containing 20 mM Tris-HCl, pH 7.5, 250 mM NaCl, 1mM EDTA, 1% NP-40, 1mM PMSF, 0.5 μg/ml leupeptin and 0.5 μg/ml pepstatin A. Following incubation on ice for 20 min, the soluble fraction was separated from the pellet by centrifugation at 22,500g for 20 min. The pellet (insoluble fraction) was washing with the lysis buffer and centrifuged at 22,500g for an additional 20 min. Proteins in the soluble and insoluble fractions were analyzed by Western blot.
Western Blot
Western blot analysis was performed essentially as previously described (Vucetic et al., 2008; Zhao et al., 2009). Primary antibodies used were goat anti-GAPDH (sc-48176, Santa Cruz), mouse anti-V5 (R960-25, Life Technologies), mouse anti-FLAG (F1804, Sigma-Aldrich,), mouse anti-H3 histone (10799, Abcam) and rabbit anti-HP1α (2616, Cell Signaling, Beverly, MA). Secondary antibodies used were donkey anti-mouse IRDye 800CW, donkey anti-rabbit IRDYE 800CW, and donkey anti-goat IRDye 680CW purchased from LI-COR, Lincoln, NE. Images were captured and quantitated using the LI-COR Odyssey instrument and software. GAPDH levels were used as the loading control.
RT-PCR analysis
Total RNA was isolated from P19, HeLa and 293A cells using RNA-Bee™ reagent (Tel-Test Inc. Friendswood, TX) following the manufacturer’s protocol. cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Foster City, CA) using random primers and PCR was performed using Go Taq Flexi DNA Polymerase (Promega, Madison, WI) following the respective manufacturer’s protocol. PCR primers were as follows: All Human Acinus Isoforms - Forward: 5′-CAAGAGGAAGATCTCCGTTGTCT-3′ and Reverse: 5′-AGGGGTTTGATGTCGGGGA-3′; Human Acinus-L - Forward: 5′-ATGTGGAGACGGAAACATC-3′ and Reverse: 5′-CTTCGGGCATCTTCGGTAATTT-3′; Human Acinus-S/S′ - Forward: 5′-TGCCGCTCCGCCAATACAAT-3′ and Reverse: 5′-GGCA-AACCCCGAGTTCGGCT-3′; All Mouse Acinus Isoforms - Forward: 5′-CAAGAGGAAAA-TCTCCGTTGTCT-3′ and Reverse: 5′-AGGGGTTTGATGTCGGGGA-3′; Mouse Acinus-L - Forward: 5′-ATGTGGGGACGGAAACGAC-3′ and Reverse: 5′-CTTCGGGCATCTTCGG-TAATTT-3′; Mouse Acinus-S/S′ - Forward: 5′-GAGTCTGCTTACTACCAGCAACCTG-3′ and Reverse: 5′-GCAAACCCCGAGTTCGGCTG-3′. The standard PCR parameters were 94°C for 2 min to completely denature the template DNA, followed by various cycle numbers (20–35) of the following 3 steps: denaturation at 94°C for 15 sec, annealing at 55–65°C depending on the melting temperature of the primer pair for 30 sec, elongation at 68°C for 40 sec, a final extension at 60 °C for 10 min, and hold at 4°C. The PCR products were resolved by polyacrylamide gel electrophoresis and visualized by ethidium bromide staining.
RESULTS
The Sub-Nuclear Localization of Acinus-L Is Distinct from Acinus-S′
Acinus (without an isoform designation) has been identified by mass spectrometry to be a component of interchromatin granule clusters which are dynamic nuclear structures enriched in pre-mRNA splicing factors (Saitoh et al., 2004). Acinus-S has been shown to colocalize with SC35 in nuclear speckles (Hu et al., 2005; Jang et al., 2008), however the sub-nuclear localization of Acinus-L and Acinus-S′ has not been reported. In an attempt to further characterize each of the Acinus isoforms, GFP- tagged human Acinus-L (GFP-Acinus-L) and GFP-tagged human Acinus-S′ (GFP-Acinus-S′) were expressed individually in COS-7 cells and their localization patterns were examined by fluorescence microscopy. Both Acinus-S′ and Acinus-L localize exclusively in the nucleus. As expected, GFP-Acinus-S′ co-localizes with SC35 (nuclear speckle marker) in nuclear speckles (Figure 1B). Unexpectedly, GFP-Acinus-L diffusely localizes throughout the nucleus (Figure 1B). The same distribution pattern for GFP-Acinus-L and GFP-Acinus-S′ was also observed in the nucleus of 293A cells (Data Not Shown).
Acinus-L but not Acinus-S′ interacts with chromatin
To determine if nuclear Acinus interacts with chromatin, we extracted soluble and insoluble chromatin fractions from COS-7 cells transfected with either V5-tagged Acinus-L or V5-tagged Acinus-S′ using 1% NP-40 and 250 mM NaCl. Figure 1C shows that ~70% of Acinus-S′ associates with the soluble fraction and the remaining ~30% associates with the insoluble fraction. On the other hand, Acinus-L associates only with the insoluble fraction containing histone H3 and HP1a. These results suggest that Acinus-L but not Acinus-S′ interacts with chromatin.
RA Treatment Has Little Effect on the Sub-Nuclear Localization of Acinus
Since Acinus-S′ interacts with RARs and regulates RA-responsive gene expression (Vucetic et al., 2008), the effect of RA on the sub-nuclear localization of Acinus-S′ and Acinus-L was examined. GFP-Acinus-L or GFP-Acinus-S′ were expressed with RFP-tagged RARβ (RFP-RARβ) in COS-7 cells. Cells were then treated with ethanol (Eth, as a control) or RA for 5 hr. Analysis of the localization of these proteins by fluorescence microscopy demonstrated that all three proteins localize in the nucleus: RARβ and Acinus-L diffusely localize throughout the nucleus, and Acinus-S′ localizes mainly in nuclear speckles. Furthermore, RA treatment (for 1 hr, 5 hr or 24 hr) had no obvious effect on the sub-nuclear localization of either Acinus-L or Acinus-S′ (Figure 1D and Date Not Shown).
The C-terminal RS- and RD/E-rich Region of Acinus-S′ Is Responsible for Its Nuclear Speckled Localization
All three isoforms of Acinus have a degenerate RS domain in their C-termini, in which the RS repeat is continuous with RD/E dipeptides. The RS domain has been shown to target some SR pre-mRNA splicing factors to nuclear speckles (Hedley et al., 1995; Li and Bingham, 1991; Misteli et al., 1998). In order to determine if the C-terminal RS and RD/E-rich region is also required for the nuclear speckle localization of Acinus-S′, GFP-tagged C-terminal truncated Acinus-S′ [GFP-Acinus-S′ (ΔC)], which terminates at amino acid 409 and lacks the region containing RS and RD/E dipeptide repeats, was expressed in COS-7 cells. GFP-Acinus-S′ (ΔC) diffusely localizes throughout the nucleus (Figure 2A), indicating that the C-terminal RS and RD/E-rich region is necessary for Acinus-S′ to localize in nuclear speckles.
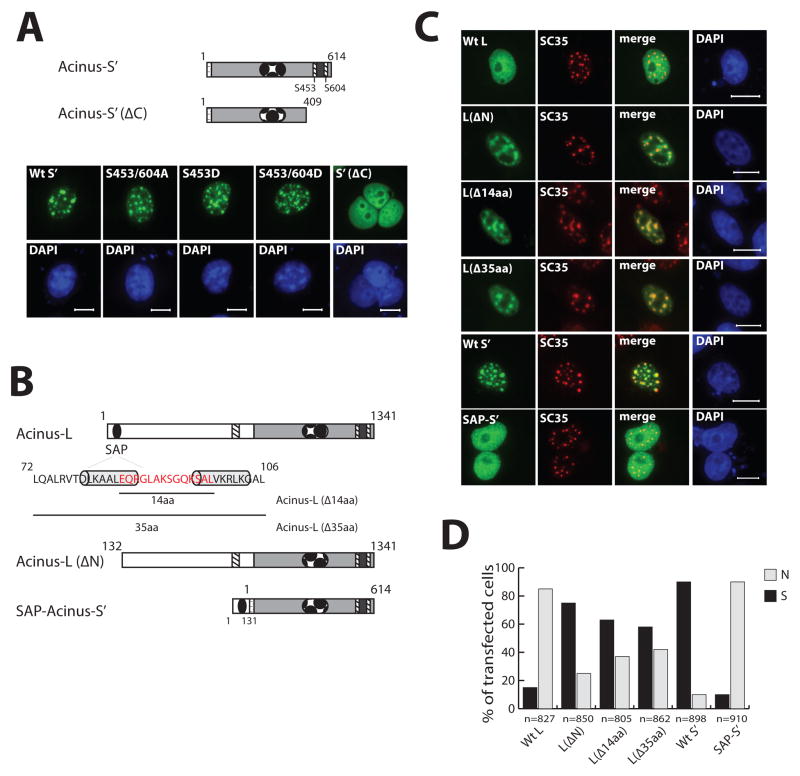
Figure 2. The role of the C-terminal RS- and RD/E-rich region and N-terminal SAP motif in the localization of Acinus-S′ and Acinus-L.
A. C-terminal RS- and RD/E-rich region is required for localization of Acinus-S′ in nuclear speckles. B. Schematic diagram of wild and mutant forms of Acinus-S and Acinus-L used in Panel C. C. SAP motif of Acinus-L is responsible for its diffuse nuclear localization. D. Quantitation of sub-nuclear localization of wild type and mutant Acinus isoforms. In Panels A and C, COS7 cells were transfected with the indicated GFP-tagged wild type or mutant Acinus isoform (A and C) expression vector DNAs. Twenty-four hr after transfection cells were fixed and proteins visualized using fluorescent microscopy (A and C. Endogenous SC35 was labeled with anti-SC35 primary antibody and rhodamine conjugated secondary antibody (C). DAPI was used as a nuclear stain (A and C). Scale bar equals 10 μm (A and C). Panel D shows quantitation of cellular localization data from Panel C, total number of observed transfected cells (n); diffuse nuclear localization (N); speckled nuclear localization (S).
Phosphorylation Status of Ser-453 and Ser-604 of Acinus-S′ Has No Effect on Its Sub-Nuclear Localization
Phosphorylation/dephosphorylation of the RS domain has been suggested to regulate the exchange rate of SR proteins between nuclear speckles and the nucleoplasm (Spector and Lamond, 2011). It has been reported that Ser-422 and Ser-573 located in the C-terminus of human Acinus-S can be phosphorylated by Akt, and Ser-422 can also be phosphorylated by SRPK2 (Hu et al., 2005; Jang et al., 2008). The S422D mutant of Acinus-S, mimicking phosphorylation at Ser-422, was previous reported to localize diffusely throughout the nucleus, while the S422A and the S422/573A mutants of Acinus-S, mimicking dephosphorylation at Ser-422 or at Ser-422 and Ser-573, were shown to localize in nuclear speckles (Jang et al., 2008). The difference between the amino acid sequence of Acinus-S and that of Acinus-S′ resides only in their short unique N-termini. Ser-422 and Ser-573 of Acinus-S correspond to Ser-453 and Ser-604 of Acinus-S′ (Figure 2B).
To determine if the phosphorylation status of Acinus-S′ at Ser-453 and Ser-604 affects the sub-nuclear localization of Acinus-S′, the S453/604A, the S453D and the S453/604D mutants of GFP-Acinus-S′ were created and expressed individually in COS-7 cells. As shown in Figure 2A, each of these mutant proteins localize in nuclear speckles comparable to that of wild type Acinus-S′. Also, the localization pattern of these mutant proteins in 293A cells is the same as that in COS-7 cells (data not shown). Taken together, these data demonstrate that the C-terminal RS and RD/E rich region of Acinus-S′ is required for its localization in nuclear speckles however the phosphorylation status of Ser-453 and Ser-604 alone does not affect its sub-nuclear localization pattern.
The SAP motif of Acinus-L Is Responsible for Its Diffuse Localization in the Nucleus
Both Acinus-L and Acinus-S′ have the same C-terminal region rich in RS and RD/E dipeptides, but they localize differently in the nucleus as shown above in Figure 1. In order to identify the region of Acinus-L responsible for its diffuse localization within the nucleus, the functional domains of Acinus-L and Acinus-S′ were compared. Acinus-L has a unique SAP motif in its N-terminus and an additional RS domain in its central region, both of which are absent in Acinus-S′ (Figures 1A and 2B). Since the RS domain targets proteins to nuclear speckles, it is unlikely that the unique RS domain in Acinus-L is responsible for its diffuse localization in the nucleus. Instead, the N-terminal SAP motif is likely to have the potential to target Acinus-L to diffusely localize within the nucleus because the SAP motif in other proteins, such as SAF-A/B and PIAS, has been shown to bind AT-rich chromosomal regions (SARs/MARs) (Gohring et al., 1997; Okubo et al., 2004; Renz and Fackelmayer, 1996).
To test this hypothesis, GFP-tagged N-terminal truncated Acinus-L lacking the first 131 amino acids which includes the SAP motif [GFP-Acinus-L ΔN)] and GFP-tagged Acinus-S′ fused with the N-terminal 131 amino acids from Acinus-L at its N-terminus (GFP-SAP-Acinus-S′) were constructed (Figure 2B). GFP-Acinus-L (ΔN) and GFP-SAP-Acinus-S′ were expressed in COS-7 cells individually. GFP-Acinus- L (ΔN) localizes in nuclear speckles like Acinus-S′ instead of diffusely throughout the nucleus in over 75% of the transfected cells (Figure 2C and D). On the other hand, GFP-SAP-Acinus-S′ was found in over 90% of the transfected cells to localize diffusely throughout the nucleus similar to Acinus-L (Figure 2C and D). To extend these findings, two mutants of the SAP motif in GFP-Acinus-L were prepared and their localization patterns were studied. These two mutants are: (1) a deletion of the 14-amino acid region critical to the putative DNA-binding ability of the SAP motif (Gohring et al., 1997), and (2) a 35-amino acid deletion that completely removes the SAP motif (Figure 2B). Analysis of the distribution pattern of both mutant proteins showed a speckled nuclear localization in about 60% of the transfected cells (Figure 2C and D). Taken together, these data demonstrate that the SAP motif is a critical factor responsible for the difference in the sub-nuclear localization between Acinus-L and Acinus-S′, probably due to its AT-rich chromosomal region-binding ability.
The combinatorial effect of the SAP motif and the C-terminal RS- and RD/E-rich region determines the sub-nuclear localization of Acinus isoforms
It is very interesting that the N-terminal SAP motif of Acinus-L leads to the sub-nuclear localization of Acinus-L within the nucleoplasm rather than nuclear speckles even though it also contains a C-terminal RS domain (a nuclear speckle-targeting signal). This suggests that the SAP motif is more potent than the RS domain in determining the sub-nuclear localization of Acinus-L. To examine the relative role of the SAP motif and the RS and RD/E rich region in determining the nuclear localization of Acinus isoforms, mKate2-tagged wild type Acinus-S′ (mKate2-Acinus-S′) was coexpressed with GFP-SAP-Acinus-S′ in COS7 cells. The use of SAP-Acinus-S′ instead of Acinus-L allows the direct examination of the effect of the N-terminal 131 amino acids which includes the SAP motif without interference with other unique regions of Acinus-L. As shown in Figure 3A, when the plasmid DNA ratio of GFP-SAP-Acinus-S′ to mKate2-Acinus-S′ was 2 to 1, the sub-nuclear localization of mKate2-Acinus-S′ completely changes from nuclear speckles to the nucleoplasm where it colocalizes with GFP-SAP-Acinus-S′. On the other hand, partial movement of GFP-SAP-Acinus-S′ from the nucleoplasm to nuclear speckles to colocalize with mKate2-Acinus-S′ requires a plasmid DNA ratio of mKate2-Acinus-S′ to GFP-SAP-Acinus-S′ of at least 6:1 (Figure 3B). No colocalization of mKate2-Acinus-S′ with GFP-SAP-Acinus-S′ in nuclear speckles is observed when the plasmid DNA ratio of mKate2-Acinus-S′ to GFP-SAP-Acinus-S′ are 2:1 or 4:1 (data not shown). These data indicate that the sub-nuclear localization of Acinus isoforms is dependent on the level of expression (ratio of the two plasmid DNAs transfected) of the two proteins. The protein with the higher expression level maintains its sub-nuclear localization, while the protein with the lower expression level is prone to change its sub-nuclear localization and colocalize with the one with higher expression level. In addition, the SAP motif of Acinus-L likely competes with the RS- and RD/E- rich region to determine the sub-nuclear localization of Acinus isoforms with the SAP motif appearing to be more potent than the RS- and RD/E- rich region.
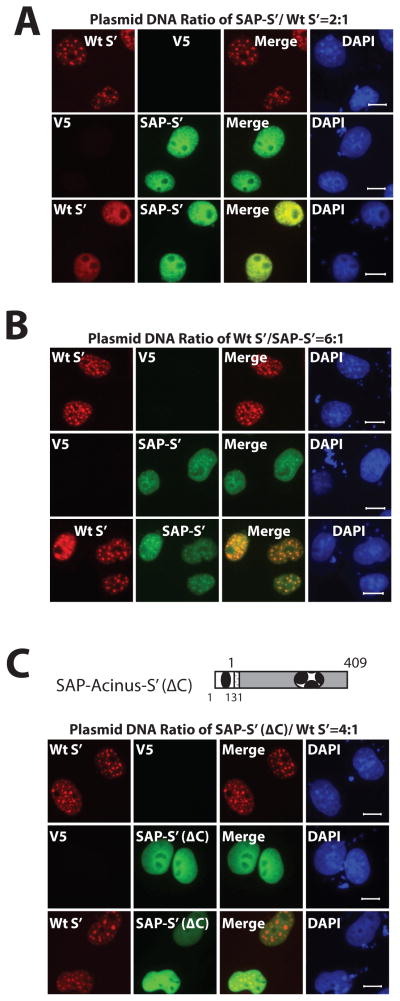
Figure 3. The combinatorial effect of the SAP motif and the C-terminal RS- and RD/E-rich region determines the sub-nuclear localization of Acinus isoforms.
A and B. SAP motif competes with the RS- and RD/E-rich region to determine the sub-nuclear localization of Acinus isoforms. C The C-terminal RS- and RD/E-rich region of Acinus mediates the co-localization of Acinus isoforms. COS7 cells were transfected with the indicated ratios of mKate-tagged wild type Acinus-S′ (A and B) and either GFP-tagged wild type SAP-Acinus-S′ (A and B) or GFP-tagged SAP-Acinus-S′ (ΔC) (C) expression plasmid DNAs. V5 empty vector DNA was used as the control. Twenty-four hr after transfection, cells were fixed and Acinus proteins were directly visualized using fluorescent microscopy. DAPI was used as a nuclear stain. Scale bar equals 10 μm.
Since the RS domain of several SR proteins has been reported to be structurally disordered and to mediate self-association and aggregation of these proteins (Ge et al., 1991; Krainer et al., 1991; Nikolakali et al., 2008; Haynes and Iakouchea, 2006), the localization of mKate2-Acinus-S′ and GFP-SAP-Acinus-S′ (ΔC) was examined. As shown in Figure 3C, mKate2-Acinus-S′ maintains its speckled sub-nuclear localization even when the plasmid DNA ratio of GFP-SAP-Acinus-S′ (ΔC) to mKate2-Acinus-S′ is increased to 4:1 (compared to 2:1 for GFP-SAP-Acinus-S′ and mKate2-Acinus-S′ in Figure 3A). These data indicate that without the C-terminal RS- and RD/E-rich region of GFP-SAP-Acinus-S′, the sub-nuclear localization of mKate2-Acinus-S′ is unaffected even though the relative level of expression of GFP-SAP-Acinus-S′ (ΔC) is high. This supports the conclusion that the RS- and RD/E-rich region is necessary for the colocalization of GFP-SAP-Acinus-S′ and mKate2-Acinus-S′ most likely by mediating self-association and aggregation of these proteins.
Sub-nuclear localization of other SR-related proteins is influenced by Acinus isoforms
To determine if this effect of the C-terminal region of Acinus extends to the localization of other interacting SR-related proteins, the sub-nuclear localization of RNPS1 was studied. RNPS1 is a SR-related protein that has been reported to bind to Acinus (all three isoforms) along with SAP18 to form a stable trimeric complex in vivo termed ASAP which is also part of the EJC (Schwerk et al., 2003; Tange et al., 2005). Based on this information, it is likely that RNPS1 colocalizes with Acinus in the nucleus. To examine if Acinus-L or Acinus-S′ affects the sub-nuclear localization of RNPS1 in the nucleus, FLAG-tagged RNPS1 (3xFLAG-RNPS1) and GFP-Acinus-S′ or GFP-Acinus-L were coexpressed in COS-7 cells. As shown in Figure 4A, when RNPS1 is expressed alone it localizes partly in nuclear speckles and partly in the nucleoplasm. However, RNPS1 becomes localized mainly in nuclear speckles where it colocalizes with Acinus-S′ when they are coexpressed in COS-7 cells. On the other hand, co-expression of RNPS1 with Acinus-L results in the sub-nuclear localization of RNPS1 diffusely throughout the nucleus where it colocalizes with Acinus-L (Figure 4A).
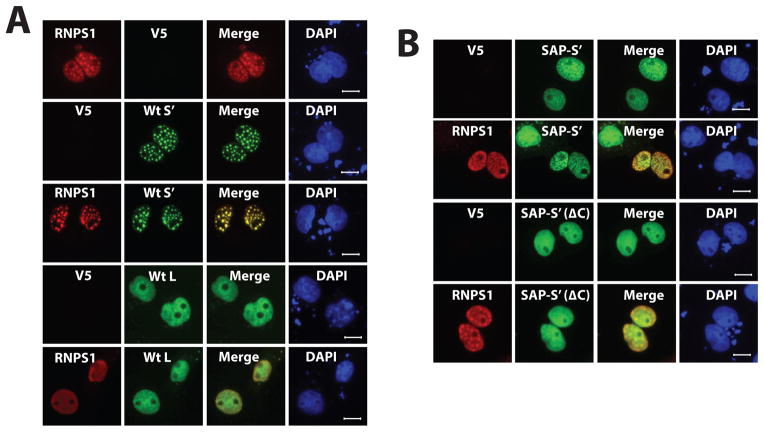
Figure 4. RNPS1 sub-nuclear localization is influenced by Acinus isoforms.
A. RNPS1 and Acinus isoforms co-localize in the nucleus. B. The C-terminal RS- and RD/E-rich region of Acinus is necessary for its co-localization with RNPS1. COS7 cells were transfected with one of the following GFP-tagged Acinus isoforms [wild type Acinus-S′ (A), wild type Acinus-L (A), SAP-Acinus-S′ (B) or SAP-Acinus-S (ΔC) (B)] and 3XFLAG-tagged RNPS1 (A and D) expression plasmid DNAs at a ratio of 6:1.. Twenty-four hr after transfection, cells were fixed and Acinus proteins were directly visualized using fluorescent microscopy. RNPS1 was detected by immunofluorescent microscopy following staining with anti-FLAG primary antibody and rhodamine conjugated second antibody. V5 empty vector DNA was used as the control. DAPI was used as a nuclear stain. Scale bar equals 10 μm.
The formation of the ternary ASAP has been reported to require the RRM domain of RNPS1, the UBL (ubiquitin-like) domain of SAP18 and a region of Acinus centered on (but not limited to) an uncharacterized conserved motif in the C- terminus (Murachelli et al., 2012). In order to determine if Acinus through its C-terminus affects the sub-nuclear localization of RNPS1, 3xFLAG-RNPS1 was coexpressed with GFP-SAP-Acinus-S′ or with GFP-SAP-Acinus-S′ (ΔC) in COS-7 cells. Figure 4B shows that the colocalization of RNPS1 with Acinus isoforms is dependent on the C-terminal RS- and RD/E-rich region since coexpression of GFP-SAP-Acinus-S′ but not GFP-SAP-Acinus-S′ (ΔC) results in their colocalization (Figure 4B). Taken together, these data demonstrate that Acinus associates with other Acinus isoforms, RNPS1 and potentially other SR/SR related proteins to determine their sub-nuclear localization via their common C-terminal RS- and RD/E-rich region with the SAP motif containing protein (diffusely localizing in the nucleoplasm) dominating over proteins with only the C-terminal RS- and RD/E-rich region (localizing in nuclear speckles).
The Distribution Pattern of Acinus-S′ in the Nucleus Correlates with the Relative Expression Level of Endogenous Acinus-L and Endogenous Acinus-S/S′
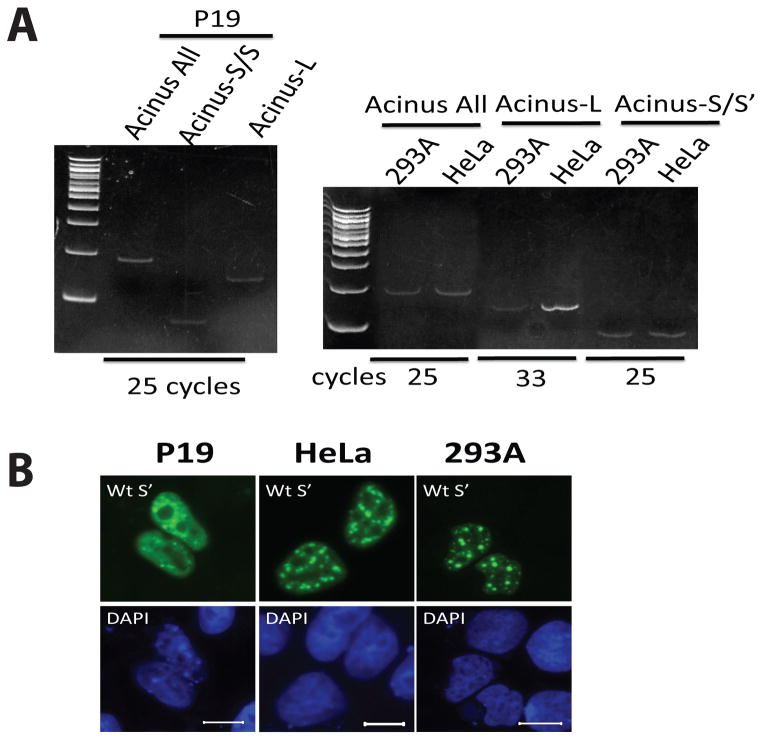
Data shown in Figure 3 suggests that the sub-nuclear localization of endogenous Acinus isoforms could be affected by the relative expression levels of Acinus-L and Acinus-S/S′ within different cell types. In order to compare the expression level of endogenous Acinus-L and Acinus-S/S′, the mRNA levels of endogenous Acinus-L and Acinus-S/S′ in three cell lines were examined by RT-PCR. Figure 5A shows that with 25 PCR cycles, both endogenous Acinus-L and Acinus-S/S′ mRNA in P19 cells can be detected by ethidium bromide staining and the density of the RT-PCR product bands of mRNAs for both endogenous Acinus isoforms are similar (Figure 5A Left Panel), suggesting that the expression levels of endogenous Acinus-L and Acinus-S/S′ in P19 cells are comparable. However, in 293A and HeLa cells, only endogenous Acinus-S/S′ mRNA can be detected with 25 PCR cycles by ethidium bromide staining; endogenous Acinus-L mRNA can be detected by ethidium bromide staining only when the PCR cycle number is increased to over 30 (Figure 5A Right Panel). These data indicate that the expression level of endogenous Acinus-L is much lower than that of endogenous Acinus-S/S′ in both 293A and HeLa cells, and the ratio of the expression level of Acinus-L to Acinus-S/S′ in HeLa cells is somewhat higher than that in 293A cells.
Figure 5. The nuclear distribution of Acinus-S′ in cell lines with different relative expression levels of endogenous Acinus-L.
(A) The mRNA level of endogenous Acinus-L, Acinus-S/S′ and all isoforms of Acinus in P19, HeLa and 293A cell lines. (B) The nuclear distribution of GFP-Acinus-S′ in P19, HeLa and 293A cell lines. In Panel A, RNA from P19, HeLa and 293 cells was isolated and RT-PCR was performed using primer pairs specific for detecting endogenous levels of mRNA for Acinus-L, Acinus-S/S′ or all isoforms of Acinus. PCR cycle numbers are indicated in the figure. PCR product of each sample was loaded and resolved on polyacrylamide gel and stained by ethidium bromide. In Panel B, GFP-Acinus-S′ expression vector DNA was transfected into P19, HeLa or 293A cells. Twenty-four hr after transfection, cells were fixed and fluorescence microscopy was performed to detect exogenous Acinus-S′ sub-nuclear localization. DAPI was used as a nuclear stain. Scale bar equals 10 μm.
In order to examine if the relative level of expression of Acinus-L and Acinus-S/S′ correlates with the sub-nuclear distribution of Acinus-S′, the sub-nuclear localization of GFP-Acinus-S′ in P19 cells was examined by fluorescence microscopy and compared with that in HeLa and 293A cells. As shown in Figure 5B, the sub-nuclear localization of GFP-Acinus-S′ is substantially less speckled in P19 cells compared with that in HeLa and 293A cells. This interesting observation supports the idea that the distribution pattern of endogenous Acinus-S/S′ isoforms is affected by the relative expression level of Acinus-L.
DISCUSSION
These studies have determined the following new information about Acinus isoforms: (1) Acinus-L localizes diffusely throughout the nucleus while Acinus-S′ localizes in nuclear speckles. (2) The domains responsible for each of their distinct sub-nuclear localization patterns have been identified. The C-terminal RS- and RD/E-rich region targets Acinus-S′ to nuclear speckles, while the SAP motif of Acinus-L is responsible for its diffuse localization in the nucleus. (3) Furthermore, the sub-nuclear localization of Acinus isoforms and potentially other interacting SR/SR-related proteins is determined by the combinatorial effect of the SAP motif of Acinus-L and the C-terminal RS- and RD/E-rich region in all Acinus isoforms.
Acinus has three isoforms, termed Acinus-L, Acinus-S and Acinus-S′, which are most likely generated by alternative promoter usage and/or alternative splicing. Proteomic analysis has identified Acinus (without an isoform designation) as a component of nuclear speckles (Saitoh et al., 2004) and Acinus-S has been shown to localize in nuclear speckles using fluorescence microscopy (Hu et al., 2005; Jang et al., 2008). The current studies using fluorescence microscopy have extended these findings by demonstrating that Acinus-S′ also co-localizes with SC35 in nuclear speckles while unexpectedly, Acinus-L diffusely localizes throughout the nucleus. Since nuclear speckles are thought to be storage/reassembly sites for pre-mRNA slicing factors including snRNP, SR/SR-related proteins and some other splicing regulatory proteins, it is likely that at least Acinus-S′ and Acinus-S participate in regulating pre-mRNA splicing. In support of this, proteomic analysis has identified Acinus (without an isoform designation) in the human spliceosome (Rappsilber et al., 2002; Zhou et al., 2002).
All three isoforms of Acinus have a common C-terminal region that contains a RRM domain and a C-terminus rich in RS, RD and RE dipeptide repeats, which defines Acinus as a SR-related protein. The RS domain has been shown to target some SR splicing factors to nuclear speckles (Caceres et al., 1997; Hedley et al., 1995; Li and Bingham, 1991). Consistent with these findings, our studies demonstrate that the C-terminal RS- and RD/E rich region is necessary for Acinus-S′ to localize in nuclear speckles. However the phosphorylation status of Ser-453 and Ser-604 alone does not affect its sub-nuclear localization pattern and has no effect on its exchange rate between nuclear speckles and the nucleoplasm. It is unclear why the S453D mutant of Acinus-S′ remains localized in nuclear speckles (data presented here) while the homologous S422D mutant of Acinus-S is diffusely distributed throughout the nucleus (Jang et al., 2008). Possible reasons for this difference might be related to the unique N-termini of Acinus-S′ and Acinus-S and/or the post-translational modifications of other amino acid residues in addition to Ser-453 and Ser-604 within the RS domains of Acinus-S′ are important for its sub-nuclear localization.
Although Acinus-L also has the same C-terminus, the C-terminal-RS- and RD/E-rich region fails to localize Acinus-L to nuclear speckles. Data presented demonstrate that the unique N-terminal SAP motif of Acinus-L targets it to the nucleoplasm. Removal of the SAP motif results in the colocalization of Acinus-L with SC35 in nuclear speckles, while fusion of the SAP motif to the N-terminus of Acinus-S′ leads to the redistribution of Acinus-S′ from nuclear speckles to the nucleoplasm. Since the SAP motif of several proteins, such as SAF-A/B and PIAS, has been shown to bind to AT-rich chromosomal regions (SARs/MARs) (Gohring et al., 1997; Okubo et al., 2004; Renz and Fackelmayer, 1996), it is possible that Acinus-L also binds AT-rich chromosomal regions and contributes to chromosomal organization. In support of this, our data demonstrates that Acinus-L associates only with the insoluble fraction containing histone H3 and HP1a. Therefore, the localization of Acinus-L in nucleoplasm instead of nuclear speckles is likely due to its binding to AT-rich chromosomal region through its unique SAP-motif.
In addition to targeting proteins to nuclear speckles, the RS domain has been suggested to be a protein interaction domain (Graveley, 2000). Several in vitro experiments demonstrate that SR/SR-related proteins including Acinus produced in bacteria have a tendency to self-associate due to the intrinsic structural disorder of the unmodified RS domain (Ge et al., 1991; Krainer et al., 1991; Nikolakaki et al., 2008). Our results demonstrate that wild type Acinus-S′ colocalizes with SAP-Acinus-S′ either in the nucleoplasm or in nuclear speckles depending on their relative expression levels and that the C-terminal RS- and RD/E-rich region is required for this colocalization. This suggests that Acinus isoforms may self-associate in vivo through their C-terminal RS-and RD/E-rich region, and that the SAP motif competes with the C-terminal RS- and RD/E-rich region with the former being more potent than the latter in the determination of the sub-nuclear localization of Acinus isoforms and potentially other interacting SR/SR-related proteins. Interestingly, we found that the sub-nuclear distribution pattern of exogenous Acinus-S′ correlates with the relative expression levels of endogenous Acinus-L and endogenous Acinus-S/S′. In addition, the sub-nuclear localization of an interacting SR-related protein, RNPS1, is also affected by Acinus isoforms. Since nuclear speckles are storage sites for splicing factors and the nucleoplasm is where both transcription and splicing take place, it is possible that the relative expression levels of Acinus-L and Acinus-S′/S can regulate the accessibility of Acinus isoforms and their binding partners to where pre-mRNAs are transcribed and spliced by determining their sub-nuclear localization.
Acinus-L localizes diffusely throughout the nucleus due to its unique N-terminal SAP motif which likely binds to AT-rich chromosomal regions (SARs/MARs) (Aravind and Koonin, 2000; Takata et al., 2009). SARs/MARs anchor chromatin loops comprising gene transcription and replication units to the nuclear scaffold/nuclear matrix. SARs/MARs are located near the boundaries of actively transcribed genes and have been implicated in the regulation of gene transcription. The SAP motif-containing proteins, such as SAF-A/B, PIAS (Gross et al., 2001; Nayler et al., 1998; Vizlin-Hodzic et al., 2011; Xiao et al., 2012) and Acinus-L (see Results) have been shown to regulate gene transcription and/or pre-mRNA splicing. The association of SARs/MARs with SAP motif-containing proteins brings these proteins close to their target genes and hence increases the opportunities for interaction between these SAP motif-containing proteins and other regulatory proteins. This can enhance their regulatory effect on transcription and splicing of target genes. In addition, the potential function of these SAP motif-containing proteins in chromosomal organization might also contribute to their regulation.
It is important to note that our results show that the distribution pattern of Acinus-S′ in the nucleus correlates with the relative expression level of endogenous Acinus-L compared to the expression level of endogenous Acinus-S/S′. In light of possible unique functions of each of the Acinus isoforms, it is logical to hypothesize that the relative levels of endogenous Acinus isoforms within various cell types may play a role in determining different patterns of gene expression and splicing in addition to different capacities to differentiate and to undergo apoptosis. The exact nature of these unique, isoform-specific functions of Acinus remain to be determined.
Acknowledgments
Contract Grant Sponsor: National Institutes of Health
Contract Grant Number: DK067558
We thank Mr. Zhenping Zhang and Ms. Dorret Lynch for their expert technical assistance. This work was supported by a grant to D.R.S. from the National Institutes of Health (DK067558).
References
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Boucher L, Ouzounis CA, Enright AJ, Blencowe BJ. A genome-wide survey of RS domain proteins. RNA. 2001;7:1693–1701. [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Gohring F, Schwab BL, Nicotera P, Leist M, Fackelmayer FO. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 1997;16:7361–7371. doi: 10.1093/emboj/16.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M, Liu B, Tan J, French FS, Carey M, Shuai K. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene. 2001;20:3880–3887. doi: 10.1038/sj.onc.1204489. [DOI] [PubMed] [Google Scholar]
- Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34:305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley ML, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci U S A. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yao J, Liu Z, Liu X, Fu H, Ye K. Akt phosphorylates acinus and inhibits its proteolytic cleavage, preventing chromatin condensation. EMBO J. 2005;24:3543–3554. doi: 10.1038/sj.emboj.7600823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SW, Yang SJ, Ehlen A, Dong S, Khoury H, Chen J, Persson JL, Ye K. Serine/arginine protein-specific kinase 2 promotes leukemia cell proliferation by phosphorylating acinus and regulating cyclin A1. Cancer Res. 2008;68:4559–4570. doi: 10.1158/0008-5472.CAN-08-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipp M, Gohring F, Ostendorp T, van Drunen CM, van Driel R, Przybylski M, Fackelmayer FO. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol Cell Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Li H, Bingham PM. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- Masaki H, Nishida T, Kitajima S, Asahla K, Teraoka H. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J Biol Chem. 2007;282:33034–33042. doi: 10.1074/jbc.M703245200. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murachelli AG, Ebert J, Basquin C, Le Hir H, Conti E. The structure of the ASAP core complex reveals the existence of a Pinin-containing PSAP complex. Nat Struct Mol Biol. 2012;19:378–386. doi: 10.1038/nsmb.2242. [DOI] [PubMed] [Google Scholar]
- Nayler O, Stratling W, Bourquin JP, Stagljar I, Lindemann L, Jasper H, Hartmann AM, Fackelmayer FO, Ullrich A, Stamm S. SAF-B protein cuples transcription and pre-mRNA splicing to SAR. MAR elements. Nucleic Acids Res. 1998;26:3542–3549. doi: 10.1093/nar/26.15.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki E, Drosou V, Sanidas I, Peidis P, Papamarcaki T, Iakoucheva LM, Giannakouros T. RNA association or phosphorylation of the RS domain prevents aggregation of RS domain-containing proteins. Biochim Biophys Acta. 2008;1780:214–225. doi: 10.1016/j.bbagen.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Okubo S, Hara F, Tsuchida Y, Shimotakahara S, Suzuki S, Hatanaka H, Yokoyama S, Tanaka H, Yasuda H, Shindo H. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J Biol Chem. 2004;279:31455–31461. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz A, Fackelmayer FO. Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Res. 1996;24:843–849. doi: 10.1093/nar/24.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara S, Aoto M, Eguchi Y, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401:168–173. doi: 10.1038/43678. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol Cell Biol. 2004;24:1174–1187. doi: 10.1128/MCB.24.3.1174-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Prasad J, Degenhardt K, Erdjument-Bromage H, White E, Tempst P, Kidd VJ, Manley JL, Lahti JM, Reinberg D. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol Cell Biol. 2003;23:2981–2990. doi: 10.1128/MCB.23.8.2981-2990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K. Non-apoptotic functions of caspases in cellular proliferation and differentiation. Biochem Pharmacol. 2003;66:1453–1458. doi: 10.1016/s0006-2952(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Sordet O, Rebe C, Plenchette S, Zermati Y, Hermine O, Vainchenker W, Garrido C, Solary E, Dubrez-Daloz L. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood. 2002;100:4446–4453. doi: 10.1182/blood-2002-06-1778. [DOI] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000646. pii: a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H, Nishijima H, Ogura S, Sakaguchi T, Bubulya PA, Mochizuki T, Shibahara K. Proteome analysis of human nuclear insoluble fractions. Genes Cells. 2009;14:975–990. doi: 10.1111/j.1365-2443.2009.01324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JA, Hall SH, Hamil KJ, Grossman G, Petrusz P, French FS. Protein inhibitors of activated STAT resemble scaffold attachment factors and function as interacting nuclear receptor coregulators. J Biol Chem. 2002;277:16993–17001. doi: 10.1074/jbc.M109217200. [DOI] [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizlin-Hodzic D, Runnverg R, Ryme J, Simonsson S, Simonsson T. SAF-A forms a complex with BRG1 and both components are required for RNA polymerase II mediated transcription. PLoS One. 2011;6:e28049. doi: 10.1371/journal.pone.0028049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic Z, Zhang Z, Zhao J, Wang F, Soprano KJ, Soprano DR. Acinus-S′ represses retinoic acid receptor (RAR)-regulated gene expression through interaction with the B domains of RARs. Mol Cell Biol. 2008;28:2549–2558. doi: 10.1128/MCB.01199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xioa R, Tang P, Yang B, Huang J, Zhou Y, Shao C, Li H, Sun H, Zhang Y, Fu XD. Nuclear matrix factor hnRNP U/SAT-A exerts a global control of alternative splicing regulating U2 snRNP maturation. Mol Cell. 2012;45:656–668. doi: 10.1016/j.molcel.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhang ZP, Vucetic Z, Soprano KJ, Soprano DR. HACE1: A novel repressor of RAR transcriptional activity. J Cell Biochem. 2009;107:482–493. doi: 10.1002/jcb.22146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zermati Y, Garrido C, Amsellem S, Fishelson S, Bouscary D, Valensi F, Varet B, Solary E, Hermine O. Caspase activation is required for terminal erythroid differentiation. J Exp Med. 2001;193:247–254. doi: 10.1084/jem.193.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider SP, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]