Abstract
The discovery of somatic mutations in epidermal growth factor receptor (EGFR) and development of EGFR tyrosine kinase inhibitors (TKIs) have revolutionized treatment for lung cancer. However, resistance to TKIs emerges in almost all patients and currently no effective treatment is available. Here we show that β-catenin is essential for development of EGFR mutated lung cancers. β-catenin was upregulated and activated in EGFR mutated cells. Mutant EGFR preferentially bound to and tyrosine-phosphorylated β-catenin, leading to increase in β-catenin-mediated transactivation, particularly in cells harboring the gefitinib/erlotinib-resistant gatekeeper EGFR-T790M mutation. Pharmacological inhibition of β-catenin suppressed EGFR-L858R-T790M mutated lung tumor growth and genetic deletion of the β-catenin gene dramatically reduced lung tumor formation in EGFR-L858R-T790M transgenic mice. These data suggest that β-catenin plays an essential role in lung tumorigenesis and that targeting the β-catenin pathway may provide novel strategies to prevent lung cancer development or overcome resistance to EGFR TKIs.
Keywords: non-small cell lung cancer, epidermal growth factor receptor, β-catenin, T790M, phosphorylation
Introduction
Lung cancer is the leading cause of cancer death worldwide (1). The conventional treatment of advanced lung cancer is unsatisfactory; however, the discovery of epidermal growth factor receptor (EGFR) somatic kinase mutations provided the first glimpse of aberrant tyrosine kinase oncogene function in non-small cell lung cancer (NSCLC). Activating EGFR mutations (i.e., exon 19 deletions or the exon 21 L858R) are present in 10–40% of NSCLCs depending on race, gender, and smoking status (2). These mutations trigger the EGFR signaling pathway and promote EGFR-mediated pro-survival and anti-apoptotic signals (2). Most NSCLCs with activating EGFR mutations respond dramatically to gefitinib and erlotinib, which are reversible EGFR tyrosine kinase inhibitors (TKIs). However, almost all patients demonstrate progressive tumors within two years of continued drug exposure. The secondary EGFR-T790M mutation accounts for more than 50% of acquired resistance to gefitinib and erlotinib (2). Irreversible and second-generation EGFR inhibitors can partially overcome NSCLCs with T790M in vitro (3). This knowledge has spawned clinical trials of neratinib (4) and afatinib (5) in this patient population. However, these “second generation” inhibitors alone have not led to significant control of tumors with acquired resistance to gefitinib/erlotinib (4, 5). Thus, novel strategies to overcome the acquired resistance caused by T790M either by developing inhibitors specific to EGFR-T790M (6) or by developing inhibitors of T790M downstream targets are sorely needed.
β-catenin plays two major roles in normal cell homeostasis: it is a component of cell-cell adhesion structures and also a key player in the Wnt/β-catenin signaling pathway. As a cell adhesion molecule, membranous β-catenin links E-cadherin to α-catenin. In the nucleus, β-catenin acts as a transcriptional activator in conjunction with the TCF/LEF DNA binding proteins and regulates its target genes that are responsible for cellular proliferation and differentiation (7). In the absences of Wnt/β-catenin signaling, β-catenin is degraded by the destruction complex consisting of adenomatous polyposis coli (APC), axin, and glycogen synthase kinase 3β (GSK-3β). However, aberrant activation of the Wnt/β-catenin pathway can trigger tumorigenesis in several organs. In patients with familial adenomatous polyposis, mutations in APC lead to loss of APC function, stabilization of β-catenin, and constitutive activation of the Wnt/β-catenin pathway (8). In lung cancer, there is accumulating evidence that Wnt/β-catenin may be activated; however mutations in APC are uncommon (9). Recently, it has been shown that this pathway plays an important role in lung adenocarcinoma metastasis (10) but, the role of β-catenin signaling in NSCLC initiation/progression is not well understood.
In addition to serine/threonine phosphorylation, β-catenin can be tyrosine-phosphorylated by several tyrosine kinases including Src (11, 12) and EGFR (13). In physiological conditions, tyrosine-phosphorylation of β-catenin leads to dissociation of β-catenin from β-catenin/α-catenin/E-cadherin complexes, its stabilization with subsequent nuclear translocation, and finally increased transactivation of β-catenin (14). Moreover, it has been shown that β-catenin can also be tyrosine-phosphorylated by oncogenic Bcr-Abl or FLT3 mutant proteins, leading to its increased protein stability and transcriptional signaling activity (15, 16). These data strongly suggest that constitutively activated oncogenic kinases may contribute to cancer initiation and/or progression by increasing β-catenin activity.
In this study, we show that β-catenin is stabilized and activated through tyrosine-phosphorylation by mutant EGFRs. Inhibition of β-catenin was shown to suppress lung tumor growth in vitro and in vivo. Conditional deletion of the β-catenin gene (Ctnnb1) inhibited lung tumor formation induced by EGFR-L858R-T790M in transgenic mice. These data indicate that β-catenin plays an essential role on mutant EGFR-induced lung tumorigenesis.
Materials and Methods
Cell culture
A427, A549, NCI-H125, NCI-H1395, NCI-H1299, and NCI-H460 were purchased from the American Type Culture Collection. NCI-H1650, NCI-H3255, NCI-H1975, HCC827, PC9, and BEAS-2B cells were provided by Drs. Pasi Jänne and Geoffrey Shapiro (Dana-Farber Cancer Institute). 293T cells and COS7 cells were provided by Dr. Daniel G. Tenen (Beth Israel Deaconess Medical Center). These cells were regenotyped on a regular basis to confirm the presence of known EGFR mutations by standard Sanger sequencing.
Mice
The studies were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center. EGFR-L858R-T790M (EGFRTL)/CCSP-rtTA bi-transgenic mice and teto-Cre transgenic mice were previously reported (17, 18). Ctnnb1 floxed mice (19) and TOPGAL reporter mice (20) were purchased from the Jackson Laboratory. To induce EGFRTL expression and excise Ctnnb1, mice were fed with a doxycycline diet (Harlan Laboratories).
Plasmid constructs
FLAG-tagged and non-tagged pcDNA3-β-catenin plasmids were provided by Drs. Daniel G.Tenen and Akinobu Matsumoto (Beth Israel Deaconess Medical Center), respectively. pCG-LEF1 was provided by Rudolf Grosschedl (Max Planck Institute). Tyrosine to phenylalanine point mutations of β-catenin were generated using the QuikChange site-directed mutagenesis kit (Agilent technologies). pcDNA3.1 or MigR1 EGFR-mutant constructs were generated as described previously (21, 22).
Stable cell lines
Stable BEAS-2B cells expressing wild type EGFR and EGFR mutants were generated by retroviral infection using MigR1-EGFR constructs as described previously (22). β-catenin knockdown NCI-H1975 cells were generated by retroviral infection using pGIPZ constructs and GFP positive cells were sorted. shRNA sequences were 5’-GCTTCTAACACCGGAGGTCTT-3’ (control), 5’-TAAGGCTGCAGTTATGGTCCA-3’ (CTNNB1 #1), 5’-TTACCACTCAGAGAAGGAG-3’ (CTNNB1 #2), and 5’-TACTGTCCATCAATATCAG-3’ (CTNNB1 #3).
Real-time PCR assay
The mRNA levels of genes were measured as previously described (23). Primers for AXIN2 were 5’-TGTCCAGCAAAACTCTGAGG-3’ (forward) and 5’-GTGCAAAGACATAGCCAGAAC-3’ (reverse). Primers for GAPDH were 5’-CCACATCGCTCAGACACCAT-3’ (forward) and 5’-CCAGGCGCCCAATACG-3’ (reverse).
Luciferase reporter gene assay
293T cells (1×105 cells) in 24-well plates were transfected by Lipofectamine 2000 (Invitrogen) with 200 ng of TCF-reporter constructs (pTOPFLASH/pFOPFLASH) (24), and 100 ng of pcDNA3.1-EGFR, 100 ng of pCG-LEF1, and 500 ng of non-tagged pcDNA3-β-catenin. Cells were incubated overnight and cultured for another 24–28 hours in the presence of 100 ng/ml EGF. Luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega).
Wester blotting and immunoprecipitation
Cells lysates were prepared as previously described (23, 25). Nuclei and cytoplasmic protein were isolated as previously described (26). Protein lysates were subjected in SDS polyacrylamide and blotted on to PVDF membranes (Millipore).
For immunoprecipitation studies, 1 mg of total cell lysates was mixed with the first antibodies or normal mouse IgG (Santa Cruz Biotechnology) and then incubated with protein G Sepharose (GE Health Care Life Sciences). The beads were precipitated by centrifugation, washed three times with PBS, then suspended in sample buffer.
The antibodies used were: α-tubulin (Millipore); β-actin (Sigma-Aldrich); axin2 (Abcam); β-catenin (Santa Cruz Biotechnology); EGFR (Cell Signaling Technology); FLAG (Sigma-Aldrich); total-GSK3 (Cell Signaling Technology); phospho-Ser-9-GSK3β (Cell Signaling Technology); lamin B (Abcam); and phospho-tyrosine (Cell Signaling Technology).
Immunohistochemical analysis and X-gal Staining
The studies were approved by the Institutional Review Boards at Beth Israel Deaconess Medical Center, Keio University School of Medicine, and National University Health System in Singapore. A tissue microarray was constructed from tumor tissues of 148 patients with NSCLC obtained at the National University of Singapore as previously described (27). For conventional IHC analyses, four EGFR-mutated NSCLC patients (L858R and exon 19 deletion) with acquired gefitinib resistance were analyzed. They underwent computed tomography-guided pre- and post-treatment biopsies of their tumors at Keio University Hospital and Beth Israel Deaconess Medical Center. All tissues were embedded in paraffin, sectioned, and stained with H&E. Immunohistochemical staining was performed on formalin-fixed paraffin sections. All cases for quantitation of nuclear β-catenin staining were reviewed in a blinded fashion. For murine studies, mice were sacrificed and lungs were fixed in 10% neutral buffered formalin. The antibodies used were: β-catenin (Human: R&D systems or DAKO, Mouse: BD Transduction Laboratories); axin2 (Abcam); and TTF1 (Abcam). X-gal staining was performed as described (28).
Immunofluorescence analysis
Cells were plated in 6 well plates overnight and transfected with 1.5 µg pcDNA3.1-EGFR and 1.5 µg FLAG-tagged pcDNA3-β-catenin. Cells were incubated overnight and trypsinized the following day. Cells were then plated into the 2 well-chamber slides, incubated overnight, and stained with the following antibodies: FLAG (Sigma-Aldrich); β-catenin (BD Transduction Laboratories); Alexa 594-mouse and 488-rabbit (Invitrogen).
In vitro and in vivo drug treatment
For cell culture studies, both afatinib (LC Laboratory) and ICG-001 (Selleck Chemicals) were reconstituted in DMSO and used at the indicated concentrations. For in vivo studies, ICG-001 was suspended in 0.5% (w/v) methylcellulose (Sigma-Aldrich) and administered i.p. at 150 mg/kg/day. The stock solution was reconstituted every week and stored at 4°C. The same mice were imaged by MRI to determine the reduction in tumor volume after 2–3 weeks during the respective treatments.
Cell proliferation Assay
Growth inhibition was assessed by CellTiter 96 AQueous One solution proliferation kit (Promega) according to manufacturer’s instruction.
Colony Formation Assay
Anchorage-independent growth was assayed as previously described (29). A total of 5,000 cells were plated and colonies were counted after 14 days.
Magnetic Resonance Image scanning
All MRI was performed with a 4.7 Tesla Biospec 47/40 spectrometer using a birdcage radio-frequency coil with an inner-diameter of 30 mm (Bruker Instruments, Billerica, MA). Detailed procedure of MRI scanning was described previously (17). On each MRI image, areas indicating the pulmonary tumor were manually segmented and measured to calculate tumor volumes using NIH ImageJ software (version 1.46; http://rsb.info.nih.gov/ij/).
Statistical analysis
Differences between the experimental groups were tested with Fisher's exact test, Student’s t-test, or Wilcoxon signed-rank test. P-values of less than 0.05 were considered statistically significant. The Kaplan-Meier method was used to estimate overall survival. We performed our statistical analyses with STATA version 12.1 (STATA Corp).
Results
β-catenin is overexpressed in cells with EGFR activating mutations
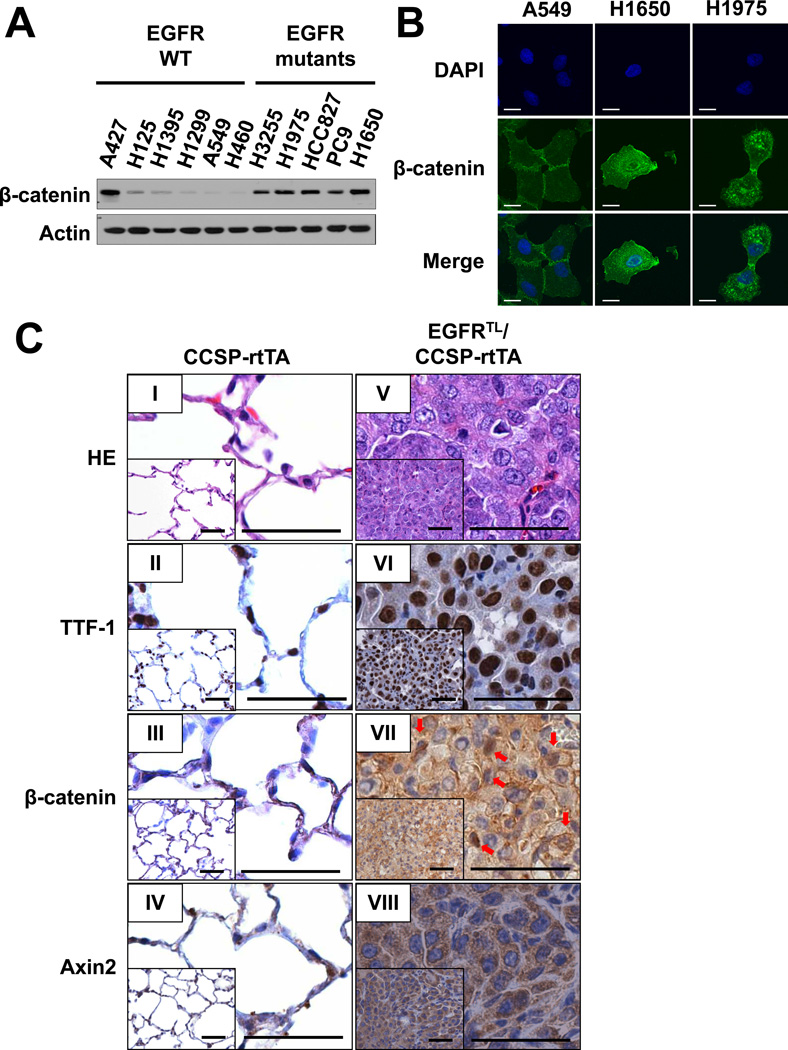
We previously identified cyclin D1 as one of the major effectors downstream of EGFR with activating mutations (23). As cyclin D1 is one of the target genes of the Wnt/β-catenin signaling pathway (7), we asked whether the pathway is activated in EGFR mutant cells. As shown in Fig. 1A, expression of β-catenin was increased in all EGFR mutant cells at similar levels to A427 cells carrying a β-catenin stabilizing mutation (30), compared to cells harboring wild type EGFR. In addition, immunofluorescence (IF) studies using confocal laser scanning microscopy showed much brighter signals in cells with EGFR mutations (NCI-H1650 and NCI-H1975) (Fig. 1B) as compared to A549 cells, which carry wild type EGFR. Interestingly, β-catenin was detected not only on the cell membrane but also in the cytoplasm and nucleus in cells with EGFR mutants. Conversely, it was localized on the cell surface and sites of cell-cell contact in A549 cells (Fig. 1B). These results prompted us to determine expression and localization of β-catenin in mutant EGFR-driven tumors in vivo. We have previously established the lung-specific conditional EGFR-L858R-T790M (EGFRTL) transgenic mice containing seven direct repeats of the tetracycline (tet)-operator sequence. These mice were crossed with CCSP-rtTA mice, which specifically express the reverse tetracycline transactivator protein (rtTA) in CCSP expressing cells (17). Consistent with our previous report (17), lung tumors were detected in EGFRTL/CCSP-rtTA mice treated with doxycycline but not in CCSP-rtTA mice (Fig. 1C: I and V). These tumors were positive for thyroid transcription factor-1 (TTF-1) (Fig. 1C: VI). More than 70% of tumor cells were positive for cytoplasmic β-catenin, whereas nuclear β-catenin was positive in approximately 20% of tumor cells (Fig. 1C: VII; arrows). Interestingly, cytoplasmic Axin2, which is a direct target gene of the Wnt/β-catenin pathway (31), was detected in almost all tumor cells (Fig. 1C: VIII), which indicates that β-catenin is indeed activated in EGFR mutant tumor cells.
Figure 1. β-catenin is highly expressed in cells with EGFR activating mutations.
(A) Immunoblots of extracts from cancer cell lines harboring wild type or mutant EGFR. (B) Confocal images of A549, NCI-H1650, NCI-H1975 cells stained with anti β-catenin antibody and visualized by Alexa Fluor 488. Scale Bar = 10 µm. (C) Immunohistochemical analysis of lungs isolated from a CCSP-rtTA mouse (I, II, III, IV) and lung tumors from an EGFR-L858R (LR)-T790M (TM) (EGFR-LR-TM)/CCSP-rtTA double transgenic mouse (V, VI, VII, VIII). Both animals were sacrificed after 20 weeks of doxycycline administration. Scale bar = 50 µm.
EGFR mutants increase expression and activity of β-catenin
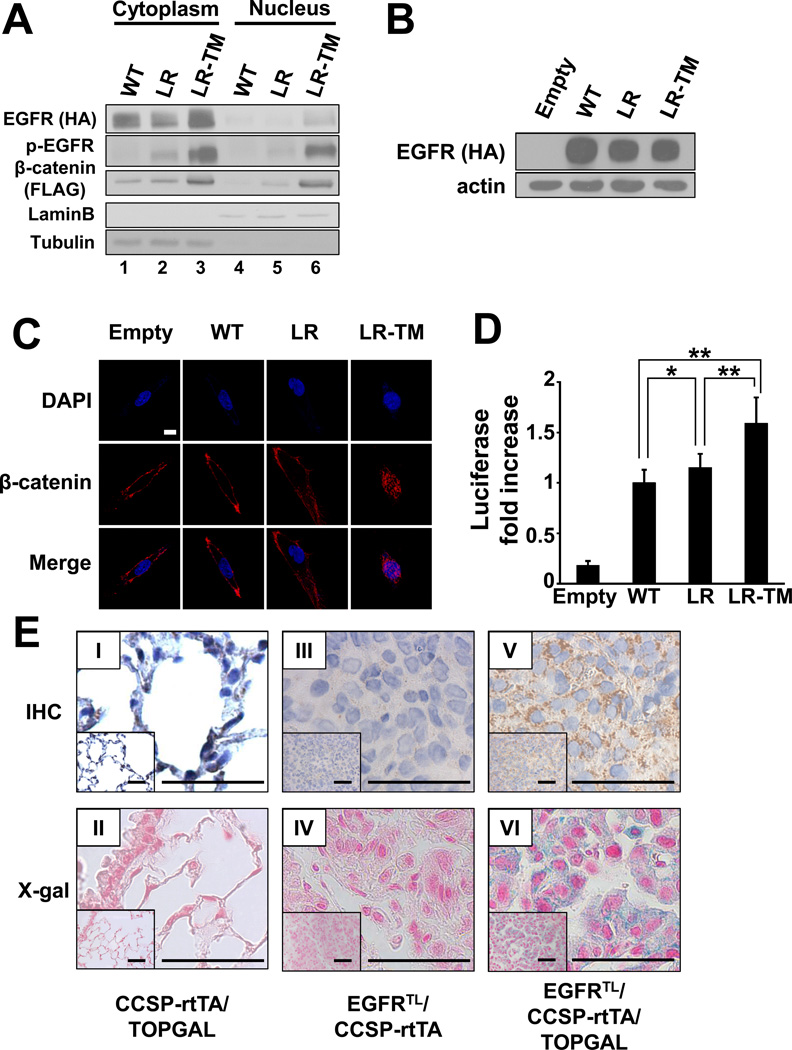
Next, we investigated the effects of mutant EGFR on stability and localization of β-catenin. When 293T cells were co-transfected with constructs containing β-catenin and either wild type EGFR, EGFR-L858R, or EGFR-L858R-T790M, expression of cytosolic and nuclear β-catenin was highest in cells transfected with EGFR-L858R-T790M (Fig. 2A). Lower expression of β-catenin was detected in cells transfected with EGFR-L858R, but the expression was still at higher level compared to cells transfected with wild type EGFR (Fig. 2A). Next, to examine the effect of EGFR mutants on β-catenin localization, we generated stable cell lines expressing either wild type EGFR, EGFR-L858R, or EGFR-L858R-T790M (Fig. 2B) using human immortalized bronchial epithelial cells (BEAS-2B) (32). Confocal microscopy analysis demonstrated that expression of nuclear β-catenin was highest in BEAS-2B cells expressing EGFR-L858R-T790M. Although cytoplasmic β-catenin was increased in BEAS-2B cells expressing EGFR-L858R, expression of nuclear β-catenin was modest compared to BEAS-2B cells expressing wild type EGFR (Fig. 2C). As nuclear translocation of β-catenin is required for its transcriptional activity, we attempted to determine whether activated EGFR mutants enhance transactivation using the TCF optimal promoter - luciferase reporter (pTOPFLASH) (24). The luciferase reporter containing the mutant motif (pFOPFLASH) was used to measure background activity. As shown in Fig. 2D, the TCF/LEF transactivation was highest in cells transiently transfected with EGFR-L858R-T790M, whereas cells transfected with EGFR-L858R showed slightly, but significantly, enhanced transactivation compared to cells transfected with wild type EGFR.
Figure 2. β-catenin is stabilized and activated by EGFR mutants.
(A) Immunoblots of fractionated extracts from 293T cells co-transfected with constructs containing FLAG-tagged β-catenin and either HA-tagged wild type EGFR (WT), EGFR-L858R (LR), or EGFR-L858R-T790M (LR-TM). (B) Immunoblots of whole extracts from BEAS-2B cells stably expressing HA-tagged wild type EGFR (WT), EGFR-L858R (LR), or EGFR-L858R-T790M (LR-TM). BEAS-2B cells infected with MigR1 empty vector were used as a control. (C) Confocal images of stable BEAS-2B cells described in (B). Cells were incubated with anti- β-catenin antibody and visualized by Alexa Fluor 488. Scale bar = 10 µm. (D) Transactivation of β-catenin measured by luciferase activity. Data represents mean ± standard deviation from eight independent experiments. * indicates p ≤ 0.05. ** indicates p ≤ 0.01. (E) Immunohistochemical analysis of β-galactosidase (I, III, V) and X-gal staining (II, IV, VI). Lungs isolated from CCSP-rtTA/TOPGAL (I, II), EGFRTL/CCSP-rtTA (III, IV), and EGFRTL/CCSP-rtTA/TOPGAL (V, VI) are shown. Scale bar = 50 µm.
Furthermore, to detect transactivation of β-catenin directly in lung tumor, we crossed EGFRTL/CCSP-rtTA mice to TOP galactosidase (TOPGAL) reporter mice (20). In this system, expression of β-galactosidase is mediated by TCF/LEF transactivation. Immunohistochemical (IHC) analysis showed that β-galactosidase was strongly positive in lung tumors isolated from EGFRTL/CCSP-rtTA/TOPGAL mice (Fig. 2E: V), whereas no staining was detected in EGFRTL/CCSP-rtTA mice (Fig. 2E: III). Consistent with this data, X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside) staining demonstrated β-galactosidase activity in lung tumors isolated from EGFRTL/CCSP-rtTA/TOPGAL mice (Fig. 2E: VI), but not in EGFRTL/CCSP-rtTA mice (Fig. 2E: IV). Alveolar cells were weakly positive for both β-galactosidase and X-gal staining in CCSP-rtTA/TOPGAL mice (Fig. 2E: I and II, respectively). These results indicate that β-catenin is indeed activated in EGFR mutated lung tumors and imply that EGFR mutants, particularly EGFR-L858R-T790M, increase expression and activity of β-catenin.
β-catenin is upregulated in human lung tumors with EGFR mutations
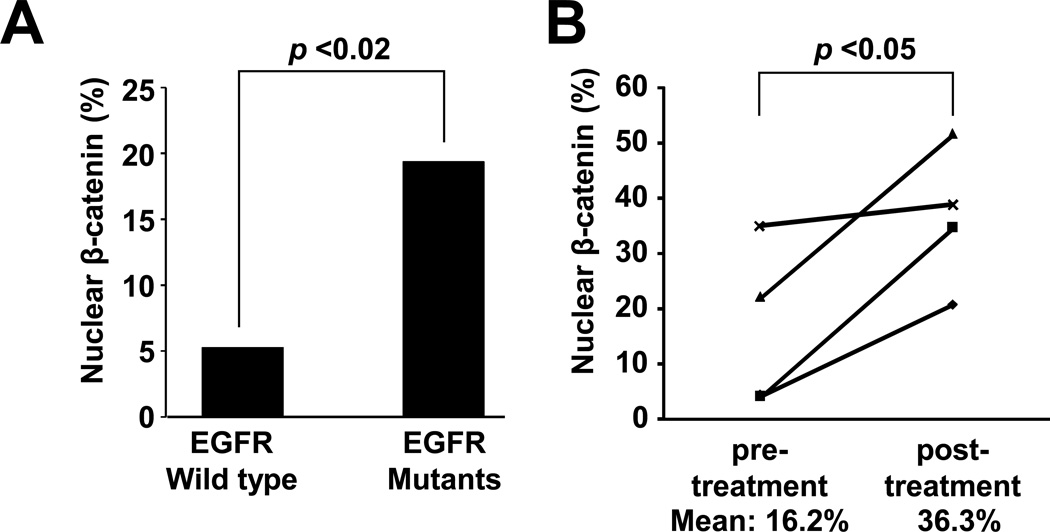
Our in vitro results prompted us to examine expression of β-catenin in primary human lung cancer specimens. IHC analysis of tissue microarray composed of 148 human NSCLC samples (Fig. 3A and Supplementary Fig. S1) demonstrated that expression of nuclear β-catenin was significantly higher in lung tumor specimens with EGFR-L858R or exon 19 deletions than tumors expressing wild type EGFR (19% [6/31] versus 6.0% [7/117]; p < 0.05, Fisher's exact test). However, this dataset did not include tumors harboring EGFR-T790M. As β-catenin expression (Fig. 2A and 2C) and activation (Fig. 2D) were higher in cells transfected with EGFR-L858R-T790M than EGFR-L858R, we next asked whether expression and/or localization of β-catenin are altered by emergence of T790M in human NSCLC. We obtained pre-and post-gefitinib biopsies from four patients who initially responded, but became resistant to TKIs due to development of T790M (Supplementary Table). While cytoplasmic and nuclear β-catenin (Supplementary Fig. S2: red arrows) were detected in both pre-treatment (Supplementary Fig. S2: I, III, V, VII) and post-treatment tissues (Supplementary Fig. S2: II, IV, VI, VIII), the cytoplasmic staining pattern was more intense and there were significantly more nuclear staining-positive cells in post-treatment tumors than pre-treated tumors (Fig. 3B). These results indicate that β-catenin is upregulated and activated in human NSCLC specimens and suggest that emergence of T790M may increase β-catenin activity.
Figure 3. β-catenin is activated in human NSCLC tumors harboring EGFR mutants.
(A) Frequency distribution of nuclear β-catenin based on immunohistochemistry on human tissue microarray. P-value was calculated using Fisher's exact test. (B) Frequency of nuclear β-catenin in 4 pairs of pre- and post-TKIs NSCLC tumors. Each symbol represents an individual patient. The detailed information can be found in Supplementary Table. P-value was calculated using Student’s t-test.
Tyrosine phosphorylation of β-catenin is mediated by EGFR mutants and enhances its activity
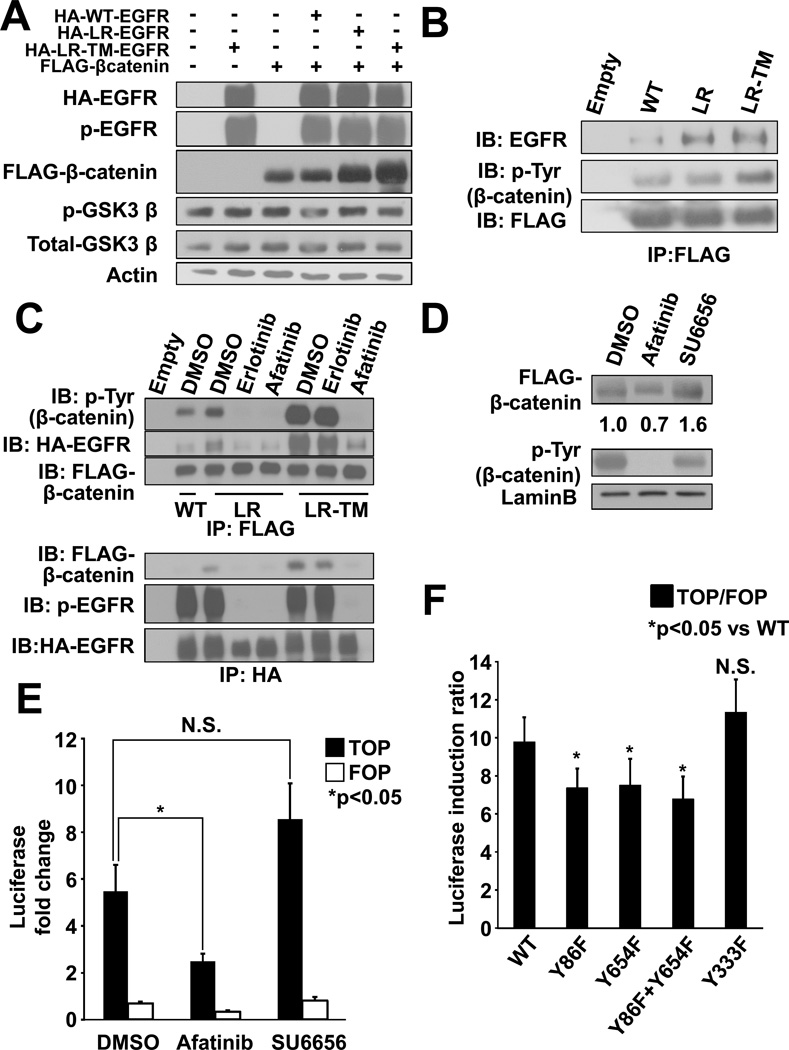
To investigate the mechanism of β-catenin stabilization, we first examined whether EGFR mutants enhance GSK3β phosphorylation. Although levels of β-catenin were higher in cells transfected with mutant EGFR than in cells with wild type EGFR, phosphorylation of GSK3β (serine 9) was not increased (Fig. 4A). These results suggest that GSK3β is not solely responsible for stabilization of β-catenin.
Figure 4. EGFR mutants tyrosine-phosphorylate β-catenin and increase expression and activity.
(A) Immunoblots of 293T cell extracts transiently transfected with constructs containing HA-tagged EGFR (wild type [WT], L858R [LR], or L858R-T790M [LR-TM]) and FLAG-tagged β-catenin. (B) Immunoprecipitation (IP) and immunoblot (IB) analyses showing binding of β-catenin and mutant EGFR. (C) IP and IB analyses showing tyrosine-phosphorylation of β-catenin by mutant EGFR. 293T cells were transiently transfected with constructs containing HA-tagged EGFR-L858R-T790M and FLAG-tagged β-catenin and treated with either 0.1% DMSO, 1 µM erlotinib, or 1 µM afatinib. (D) Immunoblots of nuclear extracts from 293T transiently transfected with constructs containing HA-tagged EGFR-L858R-T790M and FLAG-tagged β-catenin. Cells were treated with either 0.1% DMSO, 1 µM afatinib, or 1 µM SU6656. (E) Transactivation of β-catenin measured by luciferase activity. Cells were treated with either 0.1% DMSO, 1 µM afatinib, or 1 µM SU6656. Data represents mean ± standard deviation from three independent experiments. * indicates p ≤ 0.05. N.S.= not significant. (F) Transactivation of β-catenin measured by luciferase activity. A plasmid encoding for WT β-catenin and its tyrosine-to-phenylalanine (Y-to-F) mutants Y86F, Y654F, Y86F-Y654F, and Y333F were transiently transfected with EGFR-L858R-T790M together with either pTOPFLASH or pFOPFLASH plasmids. Data represents mean ± standard deviation from four independent experiments. * indicates p ≤ 0.05. N.S. indicates not significant.
It has been shown that tyrosine-phosphorylation of β-catenin enhances its stabilization, nuclear translocation, and/or transcriptional activity (11–13). As EGFR-L858R and L858R-T790M show high levels of constitutive tyrosine kinase activity (3, 33), we first asked whether EGFR mutants tyrosine-phosphorylate β-catenin. Immunoprecipitation experiments demonstrated that EGFR mutants preferentially bound to and proportionally tyrosine-phosphorylated β-catenin (Fig. 4B). Phosphorylation and binding to EGFR were inhibited by both erlotinib and afatinib in cells transfected with EGFR-L858R, but only by afatinib in cells with EGFR-L858R-T790M (Fig. 4C). Given that irreversible inhibitors such as afatinib can inhibit phosphorylation of both EGFR-L858R and EGFR-L858R-T790M in vitro (3), these data indicate that EGFR mutants bind to and tyrosine-phosphorylate β-catenin. Furthermore, inhibition of EGFR-mediated tyrosine-phosphorylation of β-catenin by afatinib resulted in less accumulation of β-catenin in the nucleus (Fig. 4D and Supplementary Fig. S3), suggesting that tyrosine-phosphorylation of β-catenin by EGFR mutants, at least in part, contributes to its nuclear translocation.
Next, we investigated whether tyrosine-phosphorylation of β-catenin leads to an increase in its activity. We focused on EGFR-L858R-T790M, as β-catenin was more efficiently phosphorylated and translocated into the nucleus in this model. Transactivation of β-catenin was inhibited by afatinib, which inhibits EGFR-L858R-T790M kinase activity (Fig. 4E). β-catenin is phosphorylated at Y86 by Src and oncogenic Bcr-Abl (15, 34) and at Y654 by activated wild type EGFR (35) and oncogenic FLT3-ITD (16). Based on the structural similarity in the tyrosine kinase domain of various oncogenes (36), we hypothesized that Y86 and Y654 residues of β-catenin would be phosphorylated by EGFR mutants and affect activation of β-catenin. β-catenin Y333, which has been shown to be phosphorylated by Src and bound to PKM resulting in nuclear translocation of β-catenin (12), was also tested. We created single β-catenin Y-to-F mutants (Y86F, Y654F and Y333F), plus the double-mutant Y86F+Y654F. All β-catenin Y-to-F mutants showed lower levels of tyrosine-phosphorylation (Supplementary Fig. S4) and decreased transactivation (Fig. 4F) compared to wild-type β-catenin, again suggesting that tyrosine-phosphorylation of β-catenin by EGFR mutants plays an important part for transactivation. Interestingly, Y333F had little impact on tyrosine-phosphorylation and transactivation of β-catenin (Fig. 4F). These results are consistent with the transactivation assay, which showed that SU6656, a Src inhibitor, failed to inhibit nuclear translocation (Fig. 4D) and transactivation of β-catenin (Fig. 4E). Taken together, these results suggest that tyrosine-phosphorylation of β-catenin by EGFR mutants, but not Src, contributes in part to its stabilization, translocation, and transactivation.
Inhibition of β-catenin suppresses tumor growth
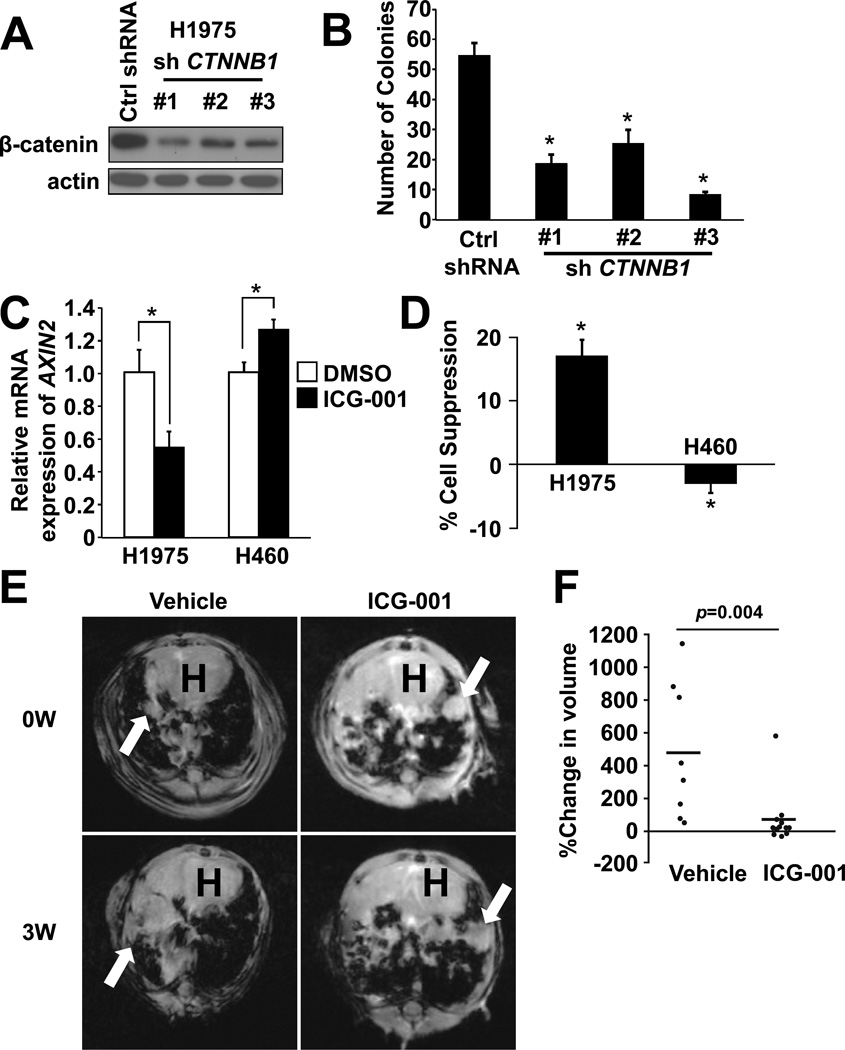
Based on our observations that β-catenin is constitutively activated downstream of mutant EGFR-L858R-T790M, we hypothesized that inhibition of β-catenin would lead to suppression of tumor growth. To test this hypothesis, we generated β-catenin knockdown cells using NCI-H1975 cells harboring EGFR-L858R-T790M (Fig. 5A). Soft agar colony formation assay demonstrated that the number of colonies was significantly suppressed in β-catenin knockdown NCI-H1975 cells compared to control cells (Fig. 5B). Furthermore, we used ICG-001, a small molecule that specifically blocks the CBP-β-catenin interaction (37). Treatment with ICG-001 led to suppression of AXIN2 (Fig. 5C) and suppressed cell growth in NCI-H1975 cells. ICG-001 failed to suppress NCI-H460 cell growth harboring wild type EGFR (Fig. 5D). Interestingly, AXIN2 was not suppressed at this concentration in NCI-H460 cells (Fig. 5C). These in vitro results prompted us to model the effects of ICG-001 in lung tumors in the EGFRTL/CCSP-rtTA mice described above. Although the body weight changes showed no significant difference between two groups, there was a trend towards kept body weight in ICG-001 treatment group, which may support the notion that ICG-001 treatment was effective against cachexia by suppressing tumors (Supplementary Fig. S5). Magnetic resonance imaging (MRI) studies demonstrated that ICG-001 significantly suppressed tumor growth when compared to the control treatment (Fig. 5E and 5F).
Figure 5. Pharmacological inhibition of β-catenin suppresses tumor growth.
(A) Immunoblots of β-catenin knockdown NCI-H1975. (B) Colony formation assay of β-catenin knockdown NCI-H1975 as described in (A). Colony numbers were counted after 14 days. Data represents mean ± standard deviation from three independent experiments. * indicates p ≤ 0.05. (C) Relative AXIN2 mRNA levels in NCI-H1975 and NCI-H460 cells treated with either 0.1% DMSO or 0.3125 µM ICG-001 were determined by real-time PCR. Data represents mean ± standard deviation from three independent experiments. * indicates p ≤ 0.05. (D) Cell proliferation assay of NCI-H1975 and NCI-H460 cells treated as in (C). Data represents mean ± standard deviation from three independent experiments. * indicates p ≤ 0.05 vs cells treated with 0.1% DMSO. (E) Representative MRI photographs of mice. Mice were treated with doxycycline for 11–15 weeks and treated with either vehicle or ICG-001. Arrows indicate lung tumors. Note that tumor size remained unchanged in mice treated with ICG-001, whereas tumors grew in control mice. “H” indicates location of the heart. (F) Changes in tumor volume were compared to baseline in mice treated with either vehicle (n=8) or ICG-001(n=12). P-value was calculated using Wilcoxon signed-rank test.
Deletion of Ctnnb1 significantly inhibits lung tumor formation
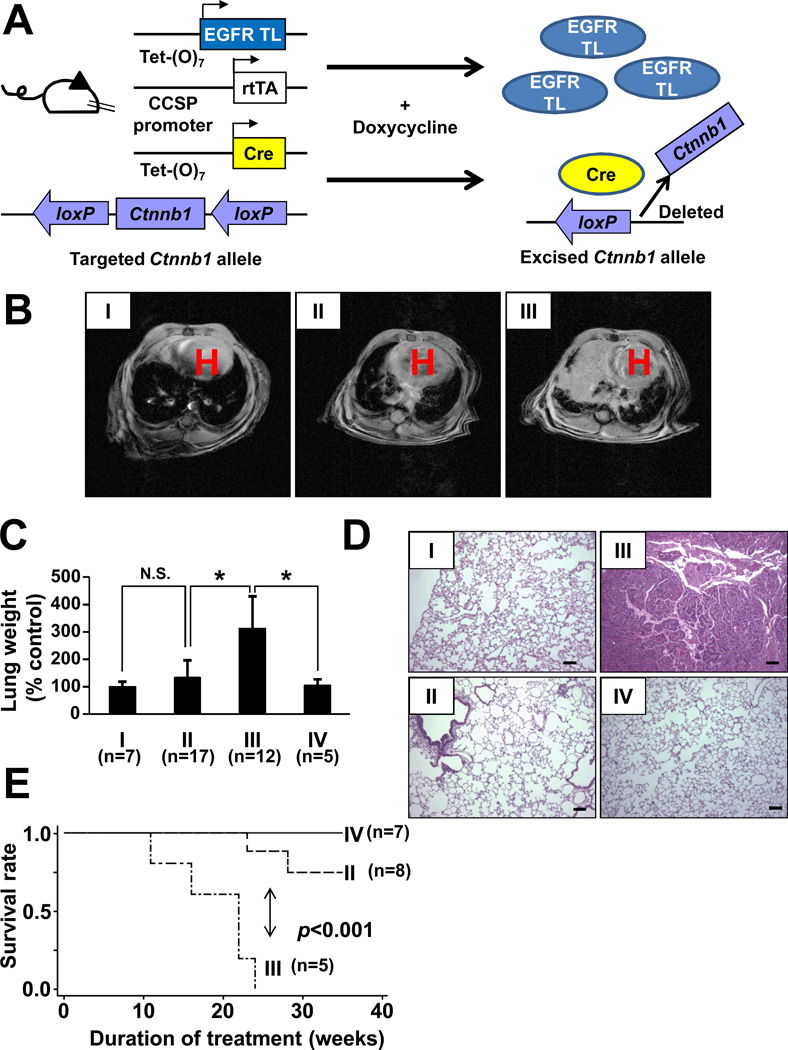
Next, we sought to determine whether β-catenin plays an important role in lung cancer development induced by EGFR mutants. To test this hypothesis, we generated a lung cancer mouse model in which Ctnnb1 can be conditionally deleted. We crossed EGFRTL/CCSP-rtTA, teto-Cre (38), and conditional β-catenin knockout mice (Ctnnb1 F/F) (19). In the resultant mice, EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/F mice (Fig. 6A; genotype is shown in Supplementary Fig. S6A), cre recombination and induction of mutated proteins (EGFR-L858R-T790M) occurred selectively in pulmonary epithelial cells with doxycycline treatment (39). MRI studies demonstrated that no or significantly smaller tumors were detected in EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/F mice compared to EGFRTL/CCSP-rtTA/teto-Cre/ Ctnnb1 F/+ mice (Fig. 6B) regardless of EGFR-L858R-T790M induction in both groups (Supplementary Fig. S6B). Lungs from EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/+ mice were more than threefold heavier than those from control mice, whereas lungs from EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/F mice showed no significant difference in weight compared to control CCSP-rtTA/Ctnnb1 F/+ mice (Fig. 6D). Hematoxylin and eosin (H&E) stain demonstrated lung tumors consistent with adenocarcinoma in EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/+(Fig. 6D, III). In contrast, deletion of Ctnnb1 in both alleles (EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/F) significantly inhibited tumor formation (Fig. 6D, II). Complete deletion of Ctnnb1 showed no impact on normal lung structure (Fig. 6D, IV) with morphology indistinguishable from control mice (Fig. 6D, I) even after 41 weeks of doxycycline treatment. Interestingly, we observed histologically detectable small tumor-clusters in 77% (17/22) of EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/F mice when they were treated with doxycycline for more than 25 weeks (Supplementary Fig. S7A). Interestingly, when we isolated these histologically detectable tumors by laser capture microdissection in EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/F mice, we found that β-catenin was not excised in most of these tumors (67% [8/12]). (Supplementary Fig. S7B). Regardless of these small tumors, EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/F mice showed significantly longer survival (p < 0.001) than EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/+ mice (Fig. 6E). Taken together, these observations suggest that β-catenin is required for lung tumor initiation and/or development induced by EGFR-L858R-T790M.
Figure 6. Deletion of Ctnnb1 impairs tumor formation in vivo.
(A) Scheme of EGFR-TL, Cre, and Ctnnb1construct for conditional transgenic mice. (B–E) Generation of lung specific conditional Ctnnb1knockout / EGFR-L858R-T790M transgenic mice. I: CCSP-rtTA/Ctnnb1 F/+ ; II: EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/F; III: EGFRTL/CCSP-rtTA/teto-Cre/Ctnnb1 F/+; IV: CCSP-rtTA/teto-Cre/Ctnnb1 F/F. (B) MRI images of transgenic mice treated with doxycycline for 10 weeks. Arrows indicate lung tumors. “H” indicates location of the heart. (C) Lung weight isolated from mice treated with doxycycline for 8–28 weeks. Data represents mean ± standard deviation. * indicates p ≤ 0.01. (D) H&E staining of lungs isolated from mice. Scale bar = 100 µm. (E) Survival curves after administration of doxycycline. P-value was determined by the log-rank test.
Discussion
EGFR TKIs have improved outcomes for patients with EGFR mutated NSCLC, however acquired resistance to TKIs is an inevitable consequence of tumor selection over continuous drug pressure. The EGFR-T790M mutation has emerged as the major mechanism of resistance to erlotinib and gefitinib. Although irreversible EGFR inhibitors are effective in lung cancer cells with EGFR-T790M mutation in vitro (3), clinical trials of these same compounds, such as afatinib or neratinib, have been disappointing (4, 5). Therefore, in addition to ongoing efforts to develop EGFR-T790M selective inhibitors, elucidating the precise mechanisms by which oncogenic EGFR mutations initiate and induce progressive lung tumors should be part of the research strategy to overcome and prevent resistance to clinically-available EGFR TKIs. In this study, we demonstrated that β-catenin is required for tumor development and progression induced by TKI-sensitive or resistant EGFR mutants. Our results give strong evidence for crosstalk between the oncogenic EGFR signaling and Wnt/β-catenin signaling for lung tumorigenesis.
Our data show that β-catenin is upregulated and translocated into nucleus in cells harboring EGFR mutants, particularly EGFR-T790M. Nuclear translocation of β-catenin is a hallmark of activation of β-catenin signaling, and indeed, β-catenin is activated in EGFR mutant cells in vitro (Fig. 2D) and in vivo (Fig. 2E). Our observations are supported by previous observations that β-catenin-TCF/LEF target genes such as cyclin D1 and c-Myc are upregulated in NSCLC harboring EGFR-T790M (23, 40). Inhibition of serine/threonine phosphorylation by GSK3β is a well-recognized mechanism of β-catenin stabilization (8). However, our data demonstrate that this may not be a sole mechanism of β-catenin upregulation in mutant EGFR transfected cells (Fig. 4A). It has been shown that tyrosine-phosphorylation contributes to activation of β-catenin (14) and our data suggest that EGFR mutants preferentially bound to and tyrosine-phosphorylate β-catenin, and modulate its activity. However, it is possible that other mechanism(s) is involved in β-catenin activation. For example, it has been reported that methylation is detected in the APC promoter region in 96% of primary lung cancer tissues and hypermethylation is associated with poor survival (41, 42). Thus, methylation of the APC promoter can occur in EGFR mutated tumors and affect β-catenin activity in EGFR mutated tumors. Future studies are required to uncover the precise mechanisms of β-catenin activation.
It is noteworthy to highlight that expression and activity of β-catenin, as well as tyrosine-phosphorylation of β-catenin, is enhanced in EGFR mutants bearing T790M than EGFR mutants with the classical sensitizing L858R mutation alone. Although a significant difference between wild type EGFR tumors and EGFR sensitizing mutant tumors was found, only less than 20% of EGFR sensitizing mutant tumors were positive for nuclear β-catenin in our tissue microarray analysis (Fig.3A). Given that less nuclear β-catenin was detected in cells with EGFR-L858R than ones with EGFR-L858R-T790M (Fig. 2A and 2C), one explanation may be that the tissue microarray sample set did not include tumors harboring EGFR-T790M. Interestingly, when lung tumors became resistant to erlotinib/gefitinib due to emergence of EGFR-T790M, an increase in the number of nuclear β-catenin positive cells was observed in our small size analysis (16.2% in pre-treatment vs. 36.3% in post-treatment: Fig. 3B and supplementary Fig. S2). Large-scale analysis will be required to further elucidate the role of β-catenin pathway in emergence of EGFR-T790M and resistance to EGFR TKIs.
Based on the observations that β-catenin activity was upregulated by EGFR-T790M, one can speculate that increased activation of β-catenin alone can lead to, or at least contribute to, resistance to EGFR TKIs. Interestingly, somatic mutations of β-catenin with concomitant EGFR-T790M mutation have been detected in 5% of EGFR TKI-resistant tumors in one series (43) and in vitro overexpression of β-catenin (or expression of activated form of β-catenin) leads to resistant to reversible EGFR TKIs in EGFR mutant cell lines lacking the T790M mutation (44, 45). Furthermore, acquired resistance to TKIs in EGFR mutant cells that undergo epithelial-to-mesenchymal transition can be concurrently seen with β-catenin signaling activation (46). Therefore, β-catenin modulation can be a means to overcome or delay resistance to gefitinib, erlotinib or other EGFR TKIs. In this report, we demonstrate that ICG-001 suppressed lung tumor growth in EGFR-T790M bearing in vivo preclinical models (Fig. 5). However, we note that ICG-001 failed to completely eradicate tumors or achieve perfect downstream inhibition. The doses used in our study did not seem to be toxic (Supplementary Fig. S5) and it is certainly possible that serum concentration did not reach the levels that inhibit β-catenin-CBP binding in vivo. More efficient β-catenin inhibitors will be required for further preclinical studies and subsequent clinical development.
The model of cooperation between EGFR mutants and β-catenin in lung tumors enhances the knowledge that the β-catenin pathway is involved in oncogene kinase-driven tumors. Mouse models of Kras G12D have demonstrated that activation of Wnt/β-catenin accelerates tumor development by converting bronchial clara cells to more proliferative embryonic progenitor cells (47). In addition, activating mutations of Ctnnb1 and mutations of Fgfr3 cooperatively induce lung tumors (48). Gene expression analysis of clinical lung cancer samples has identified that activation of Wnt/β-catenin is critical for tumor metastasis (10). Our own conditional knockout model clearly suggests that β-catenin plays a significant role on lung tumorigenesis induced by EGFR-L858R-T790M, since both deletion of β-catenin gene and induction of EGFR-L858R-T790M occur in the same cells (Fig. 6). Given that expression of an activated β-catenin alone causes lung tumors in none (47) or only a small subset of mice (49), it is predicted that tumor initiation likely requires concomitant genetic alterations in addition to activation of Wnt/β-catenin signaling. One possible explanation is that activation of β-catenin by EGFR mutants may “prime” pulmonary cells to be susceptible to other downstream signaling aberrations and subsequent transformation into tumor cells. Indeed, it has been shown that expression of activated β-catenin leads to expansion of lung-specific progenitor cells (50).
In this study, we used the EGFR-L858R-T790M inducible mouse model to show that β-catenin plays an essential role in lung tumor development in vivo. While β-catenin was most stabilized (Fig. 4A) and activated (Fig. 2D) in cells harboring EGFR-L858R-T790M, EGFR-L858R was still be a more potent activator of β-catenin (Fig. 2D) than wild type EGFR. Therefore, the question remains as to whether activation of β-catenin is specific for EGFR-T790M or also important for tumor development by non-T790M mutants such as EGFR-L858R. Furthermore, it is still unclear whether β-catenin is activated in lung tumors driven by other oncogenic kinases. β-catenin can also be tyrosine-phosphorylated and activated by oncogenic Bcr-Abl or FLT3 mutant proteins in leukemia (15, 16). Therefore, it is possible that other oncogenic receptor tyrosine kinases such as EML4-ALK translocations may also activate β-catenin. Future studies are required to answer these questions.
In summary, we demonstrate that β-catenin is involved in and required for lung tumor formation induced by EGFR mutants. These results uncover the mechanisms by which EGFR mutants lead to lung tumorigenesis and suggest that modulation of the β-catenin activity could be a novel therapeutic approach not only to inhibit lung cancer development, but also to overcome resistance to EGFR TKIs.
Supplementary Material
Acknowledgments
We thank Rudolf Grosschedl (Max Planck Institute, Germany), Daniel Tenen, and Akinobu Matsumoto (Beth Israel Deaconess Medical Center, Boston) for providing DNA constructs; Hideo Watanabe (Dana Farber Cancer Institute, Boston) and Balazs Halmos (Columbia University, New York) for critical reading the manuscript and helpful discussion; and Bhavin Thakkar (National University of Singapore, Singapore), Michelle Farley and David Bennett (Beth Israel Deaconess Medical Center) for technical assistance.
Financial Support:
This work was supported by National Institution of Health grants (R00CA126026 and R01CA169259 to S.S.K., P50CA090578 to D.B.C. and K.K.W, and R01CA122794, CA140594, CA163896, CA166480, CA154303, and CA120964 to K.K.W); an Lung Cancer Foundation of America-International Association to Study Lung Cancer grant (D.B.C.); an American Cancer Society grant RSG 11–186 (D.B.C.); Bonnie J. Addario Lung Cancer Foundation grant (S.N.); and the Singapore National Research Foundation and the Ministry of Education under the Research Center of Excellence Programme (R.A.S).
Footnotes
Disclosure of Potential Conflicts of Interest:
D.L. is currently an employee of Pfizer. D.B.C has previously received consulting fees from AstraZeneca, Roche and Pfizer.
Authors’ Contributions
Conception and design: S. Nakayama, N. Sng, S. S. Kobayashi
Development of methodology: S. Nakayama, N. Sng, S. S. Kobayashi
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Nakayama, N. Sng, J. Carretero, R. Welner, Y. Hayashi, M. Yamamoto, A. J. Tan, N. Yamaguchi, H. Yasuda, D. Li, K. Soejima, R. A. Soo, D.B. Costa, S. S. Kobayashi
Analysis and interpretation of data (e.g., statistical analysis, biosta-tistics, computational analysis): S. Nakayama, N. Sng, J. Carretero, R. Welner, M. Hayashi, M, Yamamoto, A. J. Tan, N. Yamaguchi, H. Yasuda, D.B. Costa, K.-K. Wong, S. S. Kobayashi
Writing, review, and/or revision of the manuscript: S. Nakayama, N. Sng, J. Carretero, R. Welner, A. J. Tan, N. Yamaguchi, H. Yasuda, R. A. Soo, D.B. Costa, K.-K. Wong, S. S. Kobayashi
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S. Nakayama, N. Sng, S. S. Kobayashi
Study supervision: S.S. Kobayashi
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. Vol. 63. CA: a cancer journal for clinicians; 2013. pp. 11–30. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Stiegler AL, Boggon TJ, Kobayashi S, Halmos B. EGFR-mutated lung cancer: a paradigm of molecular oncology. Oncotarget. 2010;1:497–514. doi: 10.18632/oncotarget.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi S, Ji H, Yuza Y, Meyerson M, Wong KK, Tenen DG, et al. An alternative inhibitor overcomes resistance caused by a mutation of the epidermal growth factor receptor. Cancer research. 2005;65:7096–7101. doi: 10.1158/0008-5472.CAN-05-1346. [DOI] [PubMed] [Google Scholar]
- 4.Sequist LV, Besse B, Lynch TJ, Miller VA, Wong KK, Gitlitz B, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 5.Miller VA, Hirsh V, Cadranel J, Chen YM, Park K, Kim SW, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. The lancet oncology. 2012;13:528–538. doi: 10.1016/S1470-2045(12)70087-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression; Proceedings of the National Academy of Sciences of the United States of America; 2000. pp. 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 9.Van Scoyk M, Randall J, Sergew A, Williams LM, Tennis M, Winn RA. Wnt signaling pathway and lung disease. Transl Res. 2008;151:175–180. doi: 10.1016/j.trsl.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrens J, Vakaet L, Friis R, Winterhager E, Van Roy F, Mareel MM, et al. Loss of epithelial differentiation and gain of invasiveness correlates with tyrosine phosphorylation of the E-cadherin/beta-catenin complex in cells transformed with a temperature-sensitive v-SRC gene. J Cell Biol. 1993;120:757–766. doi: 10.1083/jcb.120.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoschuetzky H, Aberle H, Kemler R. Beta-catenin mediates the interaction of the cadherin-catenin complex with epidermal growth factor receptor. J Cell Biol. 1994;127:1375–1380. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilien J, Balsamo J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of beta-catenin. Curr Opin Cell Biol. 2005;17:459–465. doi: 10.1016/j.ceb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Coluccia AM, Vacca A, Dunach M, Mologni L, Redaelli S, Bustos VH, et al. Bcr-Abl stabilizes beta-catenin in chronic myeloid leukemia through its tyrosine phosphorylation. Embo J. 2007;26:1456–1466. doi: 10.1038/sj.emboj.7601485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kajiguchi T, Katsumi A, Tanizaki R, Kiyoi H, Naoe T. Y654 of beta-catenin is essential for FLT3/ITD-related tyrosine phosphorylation and nuclear localization of beta-catenin. Eur J Haematol. 2012;88:314–320. doi: 10.1111/j.1600-0609.2011.01738.x. [DOI] [PubMed] [Google Scholar]
- 17.Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Huettner CS, Zhang P, Van Etten RA, Tenen DG. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nature genetics. 2000;24:57–60. doi: 10.1038/71691. [DOI] [PubMed] [Google Scholar]
- 19.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 20.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 22.Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Science translational medicine. 2013;5 doi: 10.1126/scitranslmed.3007205. 216ra177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi S, Shimamura T, Monti S, Steidl U, Hetherington CJ, Lowell AM, et al. Transcriptional profiling identifies cyclin D1 as a critical downstream effector of mutant epidermal growth factor receptor signaling. Cancer research. 2006;66:11389–11398. doi: 10.1158/0008-5472.CAN-06-2318. [DOI] [PubMed] [Google Scholar]
- 24.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 25.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. discussion 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. The Journal of biological chemistry. 2011;286:20558–20568. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 28.Al Alam D, Green M, Tabatabai Irani R, Parsa S, Danopoulos S, Sala FG, et al. Contrasting expression of canonical Wnt signaling reporters TOPGAL, BATGAL and Axin2(LacZ) during murine lung development and repair. PloS one. 2011;6:e23139. doi: 10.1371/journal.pone.0023139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoemaker RH, Wolpert-DeFilippes MK, Kern DH, Lieber MM, Makuch RW, Melnick NR, et al. Application of a human tumor colony-forming assay to new drug screening. Cancer research. 1985;45:2145–2153. [PubMed] [Google Scholar]
- 30.Sunaga N, Kohno T, Kolligs FT, Fearon ER, Saito R, Yokota J. Constitutive activation of the Wnt signaling pathway by CTNNB1 (beta-catenin) mutations in a subset of human lung adenocarcinoma. Genes Chromosomes Cancer. 2001;30:316–321. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1097>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 31.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Ke Y, Reddel RR, Gerwin BI, Miyashita M, McMenamin M, Lechner JF, et al. Human bronchial epithelial cells with integrated SV40 virus T antigen genes retain the ability to undergo squamous differentiation. Differentiation. 1988;38:60–66. doi: 10.1111/j.1432-0436.1988.tb00592.x. [DOI] [PubMed] [Google Scholar]
- 33.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP; Proceedings of the National Academy of Sciences of the United States of America; 2008. pp. 2070–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piedra J, Martinez D, Castano J, Miravet S, Dunach M, de Herreros AG. Regulation of beta-catenin structure and activity by tyrosine phosphorylation. The Journal of biological chemistry. 2001;276:20436–20443. doi: 10.1074/jbc.M100194200. [DOI] [PubMed] [Google Scholar]
- 35.Miravet S, Piedra J, Castano J, Raurell I, Franci C, Dunach M, et al. Tyrosine phosphorylation of plakoglobin causes contrary effects on its association with desmosomes and adherens junction components and modulates beta-catenin-mediated transcription. Mol Cell Biol. 2003;23:7391–7402. doi: 10.1128/MCB.23.20.7391-7402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 37.Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005;24:3619–3631. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 38.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung; Proceedings of the National Academy of Sciences of the United States of America; 2002. pp. 10482–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perl AK, Tichelaar JW, Whitsett JA. Conditional gene expression in the respiratory epithelium of the mouse. Transgenic Res. 2002;11:21–29. doi: 10.1023/a:1013986627504. [DOI] [PubMed] [Google Scholar]
- 40.Weaver Z, Difilippantonio S, Carretero J, Martin PL, El Meskini R, Iacovelli AJ, et al. Temporal molecular and biological assessment of an erlotinib-resistant lung adenocarcinoma model reveals markers of tumor progression and treatment response. Cancer research. 2012;72:5921–5933. doi: 10.1158/0008-5472.CAN-12-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brabender J, Usadel H, Danenberg KD, Metzger R, Schneider PM, Lord RV, et al. Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 2001;20:3528–3532. doi: 10.1038/sj.onc.1204455. [DOI] [PubMed] [Google Scholar]
- 42.Usadel H, Brabender J, Danenberg KD, Jeronimo C, Harden S, Engles J, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer research. 2002;62:371–375. [PubMed] [Google Scholar]
- 43.Sequist L, Waltman B, Dias-Santagata D, Digumarthy S, Turke A, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011:3. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang X, Gu P, Zhou C, Liang A, Ren S, Liu F, et al. β-Catenin overexpression is associated with gefitinib resistance in non-small cell lung cancer cells. Pulmonary pharmacology & therapeutics. 2013 doi: 10.1016/j.pupt.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Casas-Selves M, Kim J, Zhang Z, Helfrich BA, Gao D, Porter CC, et al. Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer research. 2012;72:4154–4164. doi: 10.1158/0008-5472.CAN-11-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conacci-Sorrell M, Zhurinsky J, Ben-Ze'ev A. The cadherin-catenin adhesion system in signaling and cancer. J Clin Invest. 2002;109:987–991. doi: 10.1172/JCI15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pacheco-Pinedo EC, Durham AC, Stewart KM, Goss AM, Lu MM, Demayo FJ, et al. Wnt/beta-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. J Clin Invest. 2011;121:1935–1945. doi: 10.1172/JCI44871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmad I, Singh LB, Foth M, Morris CA, Taketo MM, Wu XR, et al. K-Ras and beta-catenin mutations cooperate with Fgfr3 mutations in mice to promote tumorigenesis in the skin and lung, but not in the bladder. Dis Model Mech. 2011;4:548–555. doi: 10.1242/dmm.006874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mucenski ML, Nation JM, Thitoff AR, Besnard V, Xu Y, Wert SE, et al. Beta-catenin regulates differentiation of respiratory epithelial cells in vivo. Am J Physiol Lung Cell Mol Physiol. 2005;289:L971–L979. doi: 10.1152/ajplung.00172.2005. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Goss AM, Cohen ED, Kadzik R, Lepore JJ, Muthukumaraswamy K, et al. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nature genetics. 2008;40:862–870. doi: 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.