Abstract
Primary microcephaly (MIM 251200) is an autosomal recessive neurodevelopmental condition in which there is a global reduction in cerebral cortex volume, to a size comparable with that of early hominids. We previously mapped the MCPH1 locus, for primary microcephaly, to chromosome 8p23, and here we report that a gene within this interval, encoding a BRCA1 C-terminal domain–containing protein, is mutated in MCPH1 families sharing an ancestral 8p23 haplotype. This gene, microcephalin, is expressed in the developing cerebral cortex of the fetal brain. Further study of this and related genes may provide important new insights into neocortical development and evolution.
Introduction
Microcephaly is defined as a head circumference >3 standard deviations (SD) below the age-related mean. This reduction in head size is the consequence of a small brain (Ross and Frias 1977). Primary microcephaly (MIM 251200) is further defined by the absence of other syndromic features or significant neurological deficits (other than mild-to-moderate mental retardation). This entity is inherited as an autosomal recessive trait and is also known as “true microcephaly” or “microcephaly vera.”
Brain weight in primary microcephaly is markedly reduced (typically 430 g, compared with 1,450 g in unaffected adult males), and the cerebral cortex is disproportionately small (McCreary et al. 1996). Despite this marked reduction in size, the gyral pattern is relatively well preserved, with no major abnormality in cortical architecture (McCreary et al. 1996; Mochida and Walsh 2001). Furthermore, the only significant neurological deficit is a reduction in cognitive abilities (Bundey 1992).
Brain volume in primary microcephaly is comparable with that of early hominids (Wood and Collard 1999). This evolutionary parallel has been of longstanding interest, leading to the proposal that primary microcephaly could be an atavistic disorder (Komai et al. 1955; Mochida and Walsh 2001). Considerable expansion of the cerebral cortex has occurred during mammalian evolution, involving alterations in the timing, number, and pattern of cell divisions during neurogenesis (Kornack 2000). Therefore, mutation of genes controlling such processes could well underlie the pathogenesis of primary microcephaly.
Primary microcephaly is genetically heterogeneous, with five loci currently mapped (Jackson et al. 1998; Jamieson et al. 1999, 2000; Roberts et al. 1999; Moynihan et al. 2000; Pattison et al. 2000). We defined the first locus, MCPH1, on 8p23, by homozygosity mapping of consanguineous Pakistani families (Jackson et al. 1998). In the present report, to identify the MCPH1 gene, we searched for a founder effect in linked families and then sequenced positional candidates to identify the causative gene.
Patients and Methods
Subjects
The two families studied have five and two affected individuals, respectively (Jackson et al. 1998). Affected individuals in these families (aged 6–28 years) had head circumferences of 5–10 SD below the mean, mild-to-moderate mental retardation, and no focal neurological deficits or epilepsy. Linkage analysis had previously generated maximum multipoint LOD scores of 5.7 and 2.5, respectively. The study was approved by the Leeds Health Authority/United Leeds Teaching Hospitals National Health Service Trust Research Ethics Committee.
Contig Building
YAC clones were obtained from the Medical Research Council (MRC) Human Genome Mapping Project Centre. DNA was extracted using standard techniques and STS content determined by PCR. An in silico BAC contig was assembled using information from the Human Genome Project Working Draft at University of California Santa Cruz and BLAST sequence homology searches against the genome-survey sequence, nonredundant, and high-throughput genome-sequencing subdivisions of the National Center for Biotechnology Information (NCBI) sequence databases.
Genotyping
Genomic DNA was obtained from peripheral white blood cells by standard procedures. Primer sequences for known microsatellite markers were obtained from the Genome Database. Novel microsatellite markers were identified from human genome sequence in the public databases. Novel marker names, as depicted in figure 1, begin “ACxxx,” denoting the accession number of the sequenced BAC they are derived from, and end “CAyy,” indicating the sequence of the repeat and repeat number. Fluorescently labeled primers were used to amplify markers by PCR, which were electrophoresed on an ABI Prism 377 and were analyzed using Genescan and Genotyper (version 1.1.1) software (Applied Biosystems).
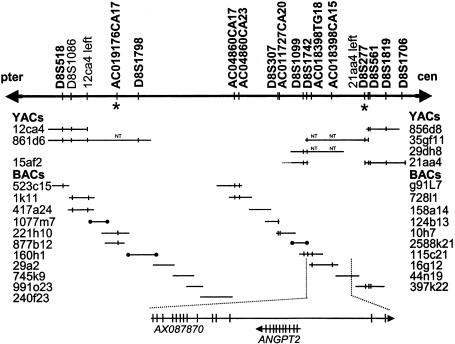
Figure 1.
Schematic of the 2.1-mb MCPH1 critical region. Microsatellite markers are in boldface type. An asterisk (*) denotes flanking markers for the ancestral haplotype. Sequenced BAC clones are shown as horizontal lines, and end-sequenced BAC clones as horizontal lines with blackened circles at the ends. The prefix “AC” denotes a novel microsatellite marker. Marker content determined by PCR (or electronically by BLAST) is indicated by vertical lines. NT = STS not tested against clone. The two genes in the critical region are shown, with their directions of transcription indicated by arrows. Vertical lines indicate the positions of individual exons.
Mutation Detection
Specific primers were designed using the publicly available genomic sequence to amplify individual exons of microcephalin. Primer sequences are available on request. Specifically, for exon 2 the following forward and reverse primers were used: dCTATTGGGCAGGGGATGCTG and dCCAATCAGAAGACTGTCATATGAATC. PCR amplification was performed using standard procedures, and the products were screened for mutations by sequencing using BigDye Terminator Cycle Sequencing kits (Applied Biosystems). Sequencing was performed using the same primers as for PCR amplification, and also with internal primers in the case of exon 8.
Restriction Digestion
The 74C→G mutation was confirmed using a TaqI restriction enzyme digestion. Primers TAQ1F (dGGTCATCCAATGGAACAGAAAATTATC) and TAQ1R (dGAATCAACAGCAAAATAAGTGTCTTACC) were used to amplify part of exon 2 from genomic DNA. A mismatch at the 3′ end of the TAQ1F primer creates a TaqI restriction site in the PCR product amplified from individuals with the mutation. PCR products were incubated at 65°C for a minimum of 3 h with TaqI and then were separated, by electrophoresis, on a 3% agarose gel (GibcoBRL).
RT-PCR of Human Fetal Tissues
The Clontech human fetal multiple tissue cDNA panel (MTC) was used according to the manufacturer's instructions. A 571-bp fragment of the microcephalin gene was amplified from the cDNAs by use of primers cA-F (dGATCCCGCCGTCTGTCATGGC) and c3-R (dCTCCTTCATCTCTTGTAATCTCTTC). These primers are separated by >32 kb in genomic DNA. GAPDH control primers used were (forward) dCGACCACTTTGTCAAGCTC and (reverse) dCAAGGGTCTACATGGCAAC, cDNA product size was 229 bp, and genomic product size was ∼330 bp.
Cloning of the Mouse Microcephalin Gene
Fragments of the mouse gene were identified by translated BLAST homology searches of publicly available mouse shotgun sequence. Primers designed from these sequence fragments were used in RT-PCR and “leapfrog” PCR (Gibbons et al. 1991) experiments, to clone and confirm the entire coding sequence of the mouse microcephalin transcript, AY070216.
In Situ Hybridization of RNA
Antisense and sense riboprobes from mouse cDNA clones, mISH1-s and mISH1-as, containing part of the mouse microcephalin cDNA (267–954 nt; 688 bp) in the pCR2.1 vector (Invitrogen) with opposite orientations were labeled in in vitro transcription reactions containing 35S-labeled CTP (1,250 Ci/mmol) (NEN) and T7 RNA polymerase according to the manufacturer’s protocol (Promega). Mouse embryo–section preparation and in situ hybridization are derived from methods described elsewhere (Cox et al. 1984).
Results
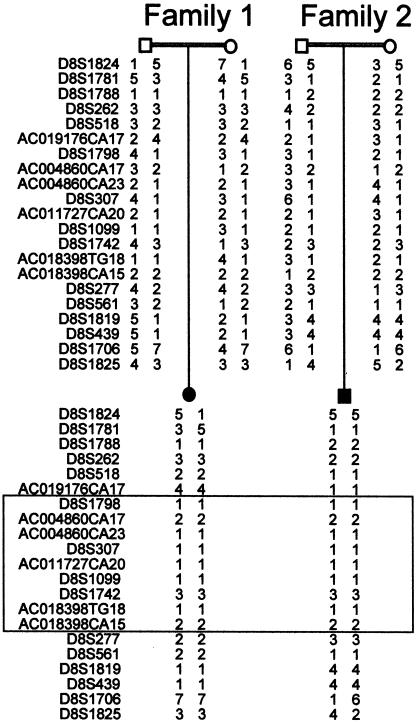
We previously reported localization of the MCPH1 primary microcephaly locus to a 13-cM interval on chromosome 8p23, by analysis of consanguineous families originating from the Mirpur region of northern Pakistan (Jackson et al. 1998). We have since constructed a partial yeast artificial chromosome (YAC) and an electronic BAC contig of the region (fig. 1). A large number of known and novel markers were mapped onto the contig and genotyped in families linked to the MCPH1 locus (Jackson et al. 1998). An ancestral haplotype of nine microsatellite markers was thus identified (fig. 2). This refined the critical region to a 2.1-mb interval between the microsatellite markers AC019176CA17 and D8S277. Two genes lie in this interval, angiopoietin-2 (ANGPT2) (Grosios et al. 1999) and an uncharacterized gene, AX087870.
Figure 2.
Genotyping results from one affected individual and parents from families 1 and 2, showing a region of identical alleles shared between both families (boxed), indicative of a founder effect.
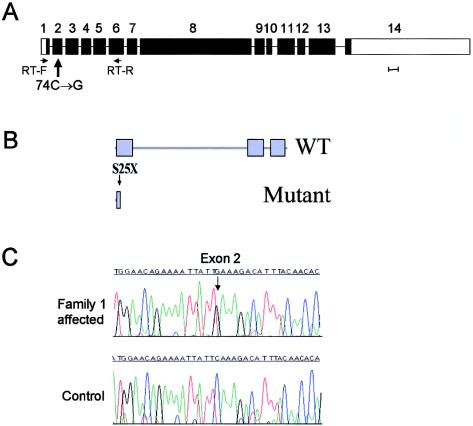
All exons of ANGPT2 were screened for mutations, but no changes were found. However, when AX087870 was screened, a C→G mutation at coding base pair 74 was found in exon 2, creating a premature stop codon (S25X). This mutation consistently segregated with the microcephaly phenotype in both the families sharing the ancestral haplotype. All seven affected individuals were homozygous for the mutation, and their eight parents, (obligate carriers), were heterozygous for this mutation. This sequence change was not present in 202 Pakistani control alleles. The mutation considerably truncates the 835–amino acid (aa) AX087870 protein, which we have termed “microcephalin” (fig. 3).
Figure 3.
Microcephalin structure and mutation. A, Gene organization. Scale bar represents 100 bp of exon sequence. B, Predicted domain structure of the wild-type (WT) and mutant protein. BRCT domains are indicated by blue boxes. The position of the S25X mutation is indicated by an arrow. C, Nonsense mutation in exon 2 in affected individual from family 1.
Microcephalin is predicted to contain three BRCA1 C-terminal (BRCT) domains, with the S25X mutation occurring in the first of these motifs. These domains interact to form homo/hetero BRCT multimers (Huyton et al. 2000). Interactions also occur with other proteins and directly with DNA at double-strand breaks. Sequence database searches identify several human proteins with homology, based primarily on the C-terminal paired BRCT domains. They include the BRCA1 protein itself, a DNA topoisomerase II–binding protein, and a cloned transcript (BAA13389) with homology to Schizosaccharomyces pombe rad4+/cut5+.
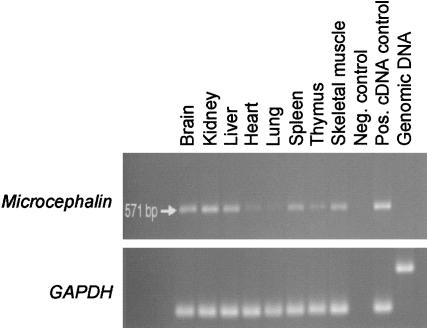
RT-PCR of human fetal tissues confirms that microcephalin is expressed in fetal brain (fig. 4). A similar level of expression is also present in fetal liver and kidney, and transcripts are detectable at low levels in a range of other fetal tissues (fig. 4), as well as in a number of adult tissues (data not shown).
Figure 4.
Expression pattern of microcephalin; RT-PCR analysis of human fetal tissues. Upper panel, a 570-bp microcephalin fragment amplified by use of primers from exons 1 and 6, to include the entire first BRCT domain. Lower panel, GAPDH control.
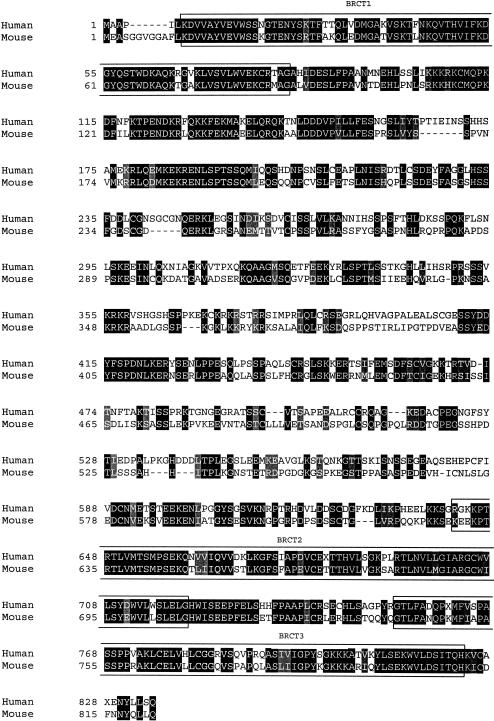
The mouse ortholog of microcephalin was next cloned using RT-PCR. This protein contains 822 amino acids and has only 57% identity to the human protein (fig. 5). The most conserved regions are the BRCT domains, where there is 80% identity. The mouse gene maps to chromosome 8A2, a region that also contains the Angpt2 gene and shows conserved synteny with human 8p23.
Figure 5.
ClustalX alignment of human and mouse microcephalin proteins. Identical amino acids shaded in black, conservative substitutions in gray. BRCT domains (as predicted by SMART) are boxed.
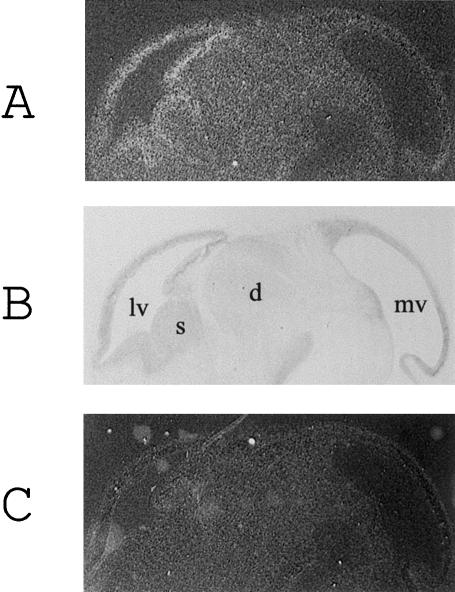
In situ hybridization experiments on fetal mouse sections confirm that microcephalin is expressed during neurogenesis. In fetal brain, high levels of gene expression localize to the developing forebrain (fig. 6) and, in particular, to the walls of the lateral ventricles. In this region, progenitor cells divide to produce neurons that migrate to form the cerebral cortex (Angevine and Sidman 1961). Hence, the expression pattern of microcephalin in the brain is consistent with a role in neurogenesis and in regulation of the size of the cerebral cortex.
Figure 6.
Expression of microcephalin mRNA in the fetal mouse brain. A, B, and C Expression of microcephalin mRNA in 7-μm E13.5 fetal mouse sagittal brain sections. Antisense microcephalin riboprobe (A), toluidine blue counterstained section (B), sense microcephalin riboprobe (C). lv = lateral ventricle; s = corpus striatum; d = diencephalon; mv = mesencephalic vesicle.
Discussion
We report here the first demonstration of a molecular defect underlying primary microcephaly. We have identified an ancestral haplotype shared by two MCPH1 families, and we show that affected individuals in both families are homozygous for a truncating mutation of a novel gene, microcephalin. This gene encodes a BRCA1 C-terminal (BRCT) domain–containing protein of as-yet-undefined function. In situ hybridization demonstrates that microcephalin is expressed in particular around the lateral ventricles of the developing forebrain, suggesting that the gene has a role in regulating the size of the cerebral cortex. The gene is also expressed in other regions of the brain, at lower levels, as well as in other tissues. Expression at sites other than those in which the phenotype manifests is frequently seen in genes causing developmental disorders (e.g., Dixon et al. 1997; Shahbazian et al. 2002). A variety of factors may account for restriction of phenotype to specific tissues in different conditions, and the way that mutation of microcephalin causes a reduction of brain size, in the apparent absence of other phenotypic effects, will require further investigation.
BRCT domains are known to be present in several key proteins controlling the cell cycle (Huyton et al. 2000). Therefore, mutation in microcephalin may cause primary microcephaly by perturbing normal cell-cycle regulation in neural progenitors. The length of the cell-cycle and duration of neurogenesis are crucial in determining total cortical cell number, and such parameters have significantly altered during evolution from rodents to primates (Kornack 2000). However, other factors may also be involved in pathogenesis of primary microcephaly. Neural cell number is also dependent on the number of founder cells at the start of neurogenesis, the relative proportions of asymmetric and symmetric cell division, and the extent of cell death (Kornack 2000). In particular, BRCT domains are found in many DNA-repair proteins (Huyton et al. 2000), and loss of function of DNA-repair genes can lead to excessive apoptotic cell death during neurogenesis (Mochida and Walsh 2001). In this respect, it is interesting to note that nibrin, the protein mutated in Nijmegen breakage syndrome, also contains a BRCT domain (Varon et al. 1998). Nijmegen breakage syndrome manifests as microcephaly, in combination with growth retardation, immunodeficiency, chromosome instability, and a marked predisposition to malignancy (International Nijmegen Breakage Syndrome Study Group 2000).
Identification of the microcephalin gene will facilitate the clinical discrimination of genetic from nongenetic disorders involving microcephaly and may aid the identification of the pathway(s) mutated in the other forms of autosomal recessive primary microcephaly, MCPH2-5 (Mochida and Walsh 2001). Identification of further mutations alongside functional characterization of microcephalin will be useful to confirm its role in the pathogenesis of microcephaly. Functional studies of microcephalin should provide insights into the regulation of cortical neural cell number, potentially aiding understanding of neural progenitor/stem cell regulation and evolution of the cerebral cortex.
Acknowledgments
We thank the families, for their participation in this study, and C. Inglehearn, T. J. Keen, and D. T. Bonthron, for discussions. A.P.J. and A.J.M. are MRC Clinician Scientists. C.G.W. is a Wellcome Trust Research Leave Senior Fellow. Research in the authors’ laboratories is also supported by the MRC, the Wellcome Trust, Yorkshire Cancer Research, and the West Riding Medical Research Trust.
Footnotes
Nucleotide sequence data reported herein are available in the DDBJ/EMBL/GenBank databases; for details, see the Electronic-Database Information section of this article.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Genome Database, http://www.gdb.org/
- Human Genome Project Working Draft at University of California Santa Cruz, http://genome.cse.ucsc.edu/
- MRC Human Genome Mapping Project Centre, http://www.hgmp.mrc.ac.uk/
- NCBI, http://www.ncbi.nlm.nih.gov/ (for microcephalin cDNA, AK022909, AX087870; genomic, AX087869; protein CAC34661; ANGPT2, NM_001147; DNA topoisomerase II binding protein, BAA34202. Mouse microcephalin cDNA, AY070216)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for primary microcephaly [MIM 251200])
- SMART, http://smart.embl-heidelberg.de/
References
- Angevine JB Jr, Sidman RL (1961) Autoradiographic study of cell migration during histiogenesis of cerebral cortex in the mouse. Nature 192:766–768 [DOI] [PubMed] [Google Scholar]
- Bundey S (1992) Microcephaly. In: Genetics and neurology: genetics in medicine and surgery. Churchill Livingstone, Edinburgh, pp 20–24 [Google Scholar]
- Cox KH, DeLeon DV, Angerer LM, Angerer RC (1984) Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol 101:485–502 [DOI] [PubMed] [Google Scholar]
- Dixon J, Hovanes K, Shiang R, Dixon MJ (1997) Sequence analysis, identification of evolutionary conserved motifs and expression analysis of murine tcof1 provide further evidence for a potential function for the gene and its human homologue, TCOF1. Hum Mol Genet 6:727–737 [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Asai DJ, Ching NS, Dolecki GJ, Mocz G, Phillipson CA, Ren H, Tang WJ, Gibbons BH (1991) A PCR procedure to determine the sequence of large polypeptides by rapid walking through a cDNA library. Proc Natl Acad Sci USA 88:8563–8567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosios K, Leek JP, Markham AF, Yancopoulos GD, Jones PF (1999) Assignment of ANGPT4, ANGPT1, and ANGPT2 encoding angiopoietins 4, 1 and 2 to human chromosome bands 20p13, 8q22.3→q23 and 8p23.1, respectively, by in situ hybridization and radiation hybrid mapping. Cytogenet Cell Genet 84:118–120 [DOI] [PubMed] [Google Scholar]
- Huyton T, Bates PA, Zhang X, Sternberg MJ, Freemont PS (2000) The BRCA1 C-terminal domain: structure and function. Mutat Res 460:319–332 [DOI] [PubMed] [Google Scholar]
- International Nijmegen Breakage Syndrome Study Group, (2000) Nijmegen breakage syndrome. Arch Dis Child 82:400–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, McHale DP, Campbell DA, Jafri H, Rashid Y, Mannan J, Karbani G, Corry P, Levene MI, Mueller RF, Markham AF, Lench NJ, Woods CG (1998) Primary autosomal recessive microcephaly (MCPH1) maps to chromosome 8p22-pter. Am J Hum Genet 63:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Fryns JP, Jacobs J, Matthijs G, Abramowicz MJ (2000) Primary autosomal recessive microcephaly: MCPH5 maps to 1q25-q32. Am J Hum Genet 67:1575–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Govaerts C, Abramowicz MJ (1999) Primary autosomal recessive microcephaly: homozygosity mapping of MCPH4 to chromosome 15. Am J Hum Genet 65:1465–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komai T, Kishimoto K, Ozaki Y (1955) Genetic study of microcephaly based on Japanese material. Am J Hum Genet 47:51–65 [PMC free article] [PubMed] [Google Scholar]
- Kornack DR (2000) Neurogenesis and the evolution of cortical diversity: mode, tempo, and partitioning during development and persistence in adulthood. Brain Behav Evol 55:336–344 [DOI] [PubMed] [Google Scholar]
- McCreary BD, Rossiter JP, Robertson DM (1996) Recessive (true) microcephaly: a case report with neuropathological observations. J Intellect Disabil Res 40:66–70 [DOI] [PubMed] [Google Scholar]
- Mochida GH, Walsh CA (2001) Molecular genetics of human microcephaly. Curr Opin Neurol 14:151–156 [DOI] [PubMed] [Google Scholar]
- Moynihan L, Jackson AP, Roberts E, Karbani G, Lewis I, Corry P, Turner G, Mueller RF, Lench NJ, Woods CG (2000) A third novel locus for primary autosomal recessive microcephaly maps to chromosome 9q34. Am J Hum Genet 66:724–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison L, Crow YJ, Deeble VJ, Jackson AP, Jafri H, Rashid Y, Roberts E, Woods CG (2000) A fifth locus for primary autosomal recessive microcephaly maps to chromosome 1q31. Am J Hum Genet 67:1578–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Jackson AP, Carradice AC, Deeble VJ, Mannan J, Rashid Y, Jafri H, McHale DP, Markham AF, Lench NJ, Woods CG (1999) The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1-13.2. Eur J Hum Genet 7:815–820 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Frias JL (1977) Microcephaly. In: Congenital malformations of the brain and skull Part 1. Vol. 30: Handbook of clinical neurology. Elsevier Holland Biomedical Press, Amsterdam, pp 507–524 [Google Scholar]
- Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY (2002) Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet 11:115–124 [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A (1998) Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93:467–476 [DOI] [PubMed] [Google Scholar]
- Wood B, Collard M (1999) The human genus. Science 284:65–71 [DOI] [PubMed] [Google Scholar]