Abstract
Nitrogen mustard (NM), a structural analog of chemical warfare agent sulfur mustard (SM), forms adducts and crosslinks with DNA, RNA and proteins. Here we studied the mechanism of NM-induced skin toxicity in response to double strand breaks (DSBs) resulting in cell cycle arrest to facilitate DNA repair, as a model for developing countermeasures against vesicant-induced skin injuries. NM exposure of mouse epidermal JB6 cells decreased cell growth and caused S-phase arrest. Consistent with these biological outcomes, NM exposure also increased comet tail extent moment and the levels of DNA DSB repair molecules phospho H2A.X Ser139 and p53 Ser15 indicating NM-induced DNA DSBs. Since DNA DSB repair occurs via non homologous end joining pathway (NHEJ) or homologous recombination repair (HRR) pathways, next we studied these two pathways and noted their activation as defined by an increase in phospho- and total DNA-PK levels, and the formation of Rad51 foci, respectively. To further analyze the role of these pathways in the cellular response to NM-induced cytotoxicity, NHEJ and HRR were inhibited by DNA-PK inhibitor NU7026 and Rad51 inhibitor BO2, respectively. Inhibition of NHEJ did not sensitize cells to NM-induced decrease in cell growth and cell cycle arrest. However, inhibition of the HRR pathway caused a significant increase in cell death, and prolonged G2M arrest following NM exposure. Together, our findings, indicating that HRR is the key pathway involved in the repair of NM-induced DNA DSBs, could be useful in developing new therapeutic strategies against vesicant-induced skin injury.
Keywords: DNA double strand break (DSB) repair, Nitrogen mustard, NHEJ, HRR, NU7026, BO2, rad51 inhibitor
1. Introduction
Sulfur mustard (bis(2-chloroethyl)sulfide; SM) and its structural analog nitrogen mustard (bis(2-chloroethyl) methylamine; NM) are blister-causing chemical warfare agents, whose exposure can cause extensive damage to various tissues and organs including skin, eyes and lungs [1–3]. Though never used in the battlefield, NM was developed as a chemical warfare agent in 1940s by Germany and the United States [4], and poses a similar threat as SM for use as a warfare or terrorist agent. Like SM, cutaneous exposure to NM is also reported to cause toxicity to the continuously dividing skin basal epidermal cells [5]. This leads to the basal epidermal cell death and delayed vesication as well as other cutaneous injuries [5]. The insight into the mechanisms involved in these NM and SM-induced skin injuries is important for the development of effective therapies against the skin injuries by vesicant exposure.
Since DNA damage is the major consequence of vesicating agent-exposure which contributes to its genotoxicity [6–8], efforts have been directed to understand the signaling pathways involved in vesicant-induced DNA damage. SM/NM-induced cytotoxicity is mainly attributed to its alkylating properties. In aqueous solution, SM/NM can spontaneously lose a chloride ion and undergo nucleophilic substitution to form a cyclic sulfonium/aziridinium ion [9, 10]. This reactive intermediate can form a second sulfonium/aziridinium ion that can react with the solvent, or with nearby nucleophilic sites resulting in the formation of adducts or crosslinks. Potential targets include most cellular macromolecules including DNA, RNA and proteins [6, 11]. Cytotoxicity resulting from SM/NM exposure is attributed especially to its ability to induce DNA modifications. Interstrand crosslinks (ICLs) of DNA contribute significantly to SM/NM-induced cytotoxicity, and can result in the induction of cell cycle arrest, and cause inhibition of DNA synthesis and cell replication [6, 12].
There are two stages involved in ICL repair including recognition and incision of DNA ICLs, followed by the action of double strand break repair (DSB) pathways [13]. The first step in ICL repair involves the recognition and the incision of the DNA near the cross-link by nucleases, thus forming DNA DSBs [14, 15]. These DNA DSBs are mainly repaired by one of the two repair pathways, namely non homologous end joining (NHEJ) and homologous recombination repair (HRR) [16]. The NHEJ pathway, as the name indicates, involves a homology-independent DSB repair wherein broken DNA ends are ligated without the need for a homologous template. NHEJ repair starts with limited end-processing by the MRN (Mre11, Rad50, NBS1) complex. These DNA ends are then bound by ku70/ku80 heterodimer which recruits the DNA-PK catalytic subunit (DNA-PKcs) forming DNA-PK holoenzyme. DNA-PK, when bound to the broken ends, becomes activated, undergoes auto-phosphorylation and recruits DNA ligase IV along with its binding partners XRCC4 and XLF. This completes the repair process by resealing the broken ends [17]. By contrast, the HRR pathway requires that a homologous sequence in the genome be used as a template to complete repair of the DNA DSBs. The repair process begins with extensive end-processing of the broken ends from 5' to 3', which is regulated by MRN complex and exonucleases. The resulting 3' ssDNA tails are bound by RPA (replication protein A) which prevents the broken 3' ssDNA tails from binding to themselves [18, 19]. BRCA2 interacts with Rad51 and coordinates the binding of Rad51 filament to 3' ssDNA tails [20–22]. The Rad51 filament then searches and invades a homologous sequence with the help of Rad54, which stabilizes Rad51 filament and induces negative DNA supercoiling, resulting in a transient DNA strand separation. After synthesis of the repair branch, strand annealing is catalyzed by BRCA2 and Rad51, completing the DNA DSB repair process [18, 23]. In addition to its role in DNA DSB repair, rad51 has also be en shown to play an active role in the regulated processing of the ICLs [23, 24].
Apart from these repair pathways, vesicant-induced DNA damage has been reported to trigger the activation of other cell signaling pathways which in turn can initiate cell cycle arrest to facilitate repair, regulate transcription of repair molecules, or, in case of inadequate repair or excessive damage, activate the apoptotic pathway to remove damaged cells [25, 26]. These pathways are reported to include the activation of phosphatidylinositol-3 kinase-related kinase (PIKK) family proteins including Ataxia telangiectasia mutated (ATM) and Ataxia telangiectasia-Rad3-related (ATR) [27, 28]. Upon activation, these kinases phosphorylate and activate other molecules like p53, chk2, etc., which play important roles in regulating cell cycle arrest and apoptosis [26, 29, 30].
Although vesicant-induced DNA damage and signaling have been studied, the role DNA-damage repair mechanisms involved in NM/SM-induced DNA damage is not well examined in relevant skin injury models. A previous study in Chinese hamster ovary cells using SM and 2-chloroethyl ethyl sulfide (CEES), a monofunctional analog of SM, has reported the involvement of nucleotide excision repair (NER) in CEES-induced DNA monoadducts [31]. Studies in lymphoblastoid cells and Chinese hamster ovary mutant cell lines has shown the role of both NER and base excision repair (BER) in CEES-induced DNA damage [32] and role of HRR in response to bi-functional alkylating agents [33, 34]. However, these studies have not been carried out in skin epidermal cells, which can be employed as efficient skin injury models for screening therapeutics, and are reported as major targets of vesicating agents. Furthermore, these studies do not compare the activation of DNA repair pathways in a single cell line and also does not address skin toxicity by these vesicants [32].
Accordingly, the aim of the present study was to determine the activation and role of DNA DSB repair pathways after NM exposure in an epidermal keratinocyte cell line. Since NM is a bi-functional alkylating agent similar to SM, with comparable toxic effects and can be used in a laboratory setting, we have employed NM in this study. The toxic and DNA damaging effects of NM were studied employing JB6 mouse epidermal keratinocytes. The extent of involvement of DNA DSB repair pathways, NHEJ and HRR was determined using specific inhibitors of DNA-PK and Rad51 (key molecules involved in these pathways), NU7026 and BO2, respectively. Our studies using these inhibitors in JB6 cells have established that HRR plays the predominant role in response to NM-induced DNA damage relative to the NHEJ pathway.
2. Materials and methods
2.1. Cell culture and treatments
JB6 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA) and cultured in minimal essential medium (MEM; Gibco BRL, Grand Island, NY) containing 5% heat-inactivated fetal bovine serum (FBS) and 25 µg/mL gentamycin. Cells were grown under standard culture conditions at 37 °C in a humidified 5% CO2 incubator. NM (mechlorethamine hydrochloride; 98%) was obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO) and the stock solution of NM was prepared fresh in dimethyl sulfoxide (DMSO) for the exposures. For the inhibitor studies, NU7026 and BO2 were obtained from Sigma and EMD Millipore (Billerica, MA), respectively, and the stocks were prepared in DMSO. Cells were treated with either 10 µM NU7026 1 h before NM exposure, or with 12.5 µM BO2 along with NM exposure and desired assays were carried out 4 to 48 h following these treatments. Unless stated otherwise, the final concentration of DMSO in the culture medium during treatments did not exceed 0.1% (v/v). All NM preparations were carried out in a continuously operated chemical and biological safety hood, and exposures were initiated under a safety laminar hood using all required and approved personal protective equipment.
2.2. Cell growth and cell cycle analysis
Cell viability was determined by Trypan blue exclusion assay. Cells were exposed to NM and harvested at desired time points by brief trypsinization, and washed and resuspended in cold PBS. Trypan blue (10 µL) was added to 90 µL of cell suspension and cells were counted for total and Trypan blue-positive (nonviable) cells using a hemocytometer. For cell cycle analysis, after desired exposures, cells were collected by brief trypsinization, washed twice with PBS and incubated in 0.5 mL of saponin/PI solution (0.3% saponin, 25 µg/mL PI, 0.1 mM EDTA, and 10 µg/mL RNase in PBS) at 4°C for 24 h in the dark. Cell cycle distribution was then analyzed using flow cytometry at the University of Colorado Cancer Center FACS analysis core laboratory.
2.3. Western blot analysis
Following exposures, whole cell lysates were prepared and protein estimation was done by Lowry method (Bio-Rad Laboratories, Richmond, CA). Immunoblot analysis using 80–150 µg of protein per sample was carried out on 6 – 12% Tris-glycine gels via western blotting as reported earlier [27, 30]. Membranes were blocked using Odyssey blocking buffer for 1 h at room temperature and probed using primary antibodies for H2A.X Ser139, p53 Ser15, total p53 (Cell Signaling; Beverly, MA), DNA-PK (Novus biological; Littleton, CO), DNA-PK Ser2056 (Biorbyt; Cambridge, UnitedKingdom) and rad51 (Santa Cruz Biotechnology; Dallas, TX) overnight at 4°C, followed by the incubation with IRDye® 800CW Conjugated, Goat Anti- Rabbit or Anti-Mouse IgG Polyclonal secondary antibody for 1 h at room temperature. The membranes were then visualized using Odyssey™ Infrared Imager (LI-COR Biosciences Lincoln, NE). To ensure equal protein loading, each membrane was stripped and reprobed with β-actin antibody (Sigma-Aldrich, St. Louis, MO). Quantitative analysis of the immunoblots was performed employing NIH ImageJ software and the values were normalized to the loading control.
2.4. Immunofluorescence confocal microscopy
Cells were plated overnight on cover slips placed in 12-well plates and the desired exposures were carried out. Following exposures, cells were washed with PBS and fixed in 3% formaldehyde for 30 min. Cells were next washed twice with PBS, incubated with ice cold methanol at −20°C for 10 min and then washed with PBS. Cells were incubated in Cas-block blocking solution for 1 h followed by overnight incubation with H2A.X Ser139 or rad51 (Abcam; Cambridge, MA) antibody. Cells were then washed thrice with PBS and finally incubated with Texas red-tagged goat anti-rabbit or Alexa fluor goat anti-mouse secondary antibody along with 4′,6′-diamidino-2-phenylindole (DAPI) for 45 min. The coverslips were mounted on microscope slides, cell images were captured at 400X magnification on a Nikon D Eclipse C1 confocal microscope (Nikon, Instruments Inc., Melville, NY), and images were processed by EZ-C1 Freeviewer software.
2.5. Comet assay
Single cell gel electrophoresis (SCGE), also known as alkaline comet assay (pH ≥ 13), was used to measure DNA damage following exposures and treatments [30, 35, 36]. Briefly, 150 µl of cell suspension was mixed with 500 µl of 1% low-melting point agarose and added to a 1% normal-melting point agarose-precoated slide. Thereafter, slides were left at 4 °C in the dark overnight in lysis solution (2.5 M NaCl, 100 mM EDTA, 10 mM Tris–Base pH 10.5 to which 1% Triton X-100, and 1% DMSO were freshly added). After lysis, slides were washed with ddH2O for 1 h, transferred to an electrophoresis unit, covered with fresh electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH ≥13), left for unwinding of DNA for 30 min and electrophoresed for 20 min at 22 V and 200 mA for 20 min. Slides were then neutralized with neutralization buffer (500 mM Tris-HCl, pH 8.0), washed with ddH2O and stained with 3 µg/ml of propidium iodide (PI). Slides were then dried overnight in dark and scored for comets using fluorescence microscopy under a Nikon inverted microscope (Nikon Eclipse TE300) at 200X magnification. Images were captured using CoolSNAPES CCD camera. Fifty cells per slide were analyzed and scored in triplicate using Komet 5.5 software (ANDOR Technology, South Windsor, CT). DNA damage is represented as tail extent moment (TEM), which is the product of tail length and percent tail DNA.
2.6. Statistical analysis
Data were analyzed by one-way analysis of variance (one-way ANOVA) using SigmaStat 3.5 software (Jandel scientific, San Rafael, CA) to determine the statistical significance of differences between different groups, followed by Tukey test for multiple comparisons. Differences were considered significant if the P-value was ≤ 0.05.
3. Results
3.1. NM exposure caused decreased cell growth and induced arrest in S-phase of cell cycle
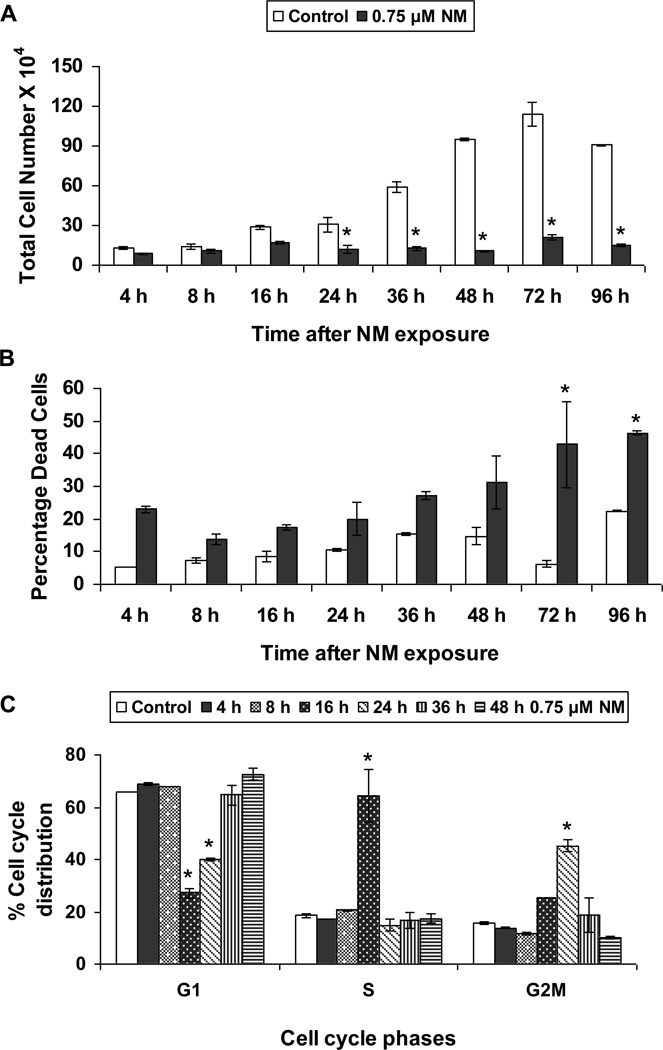
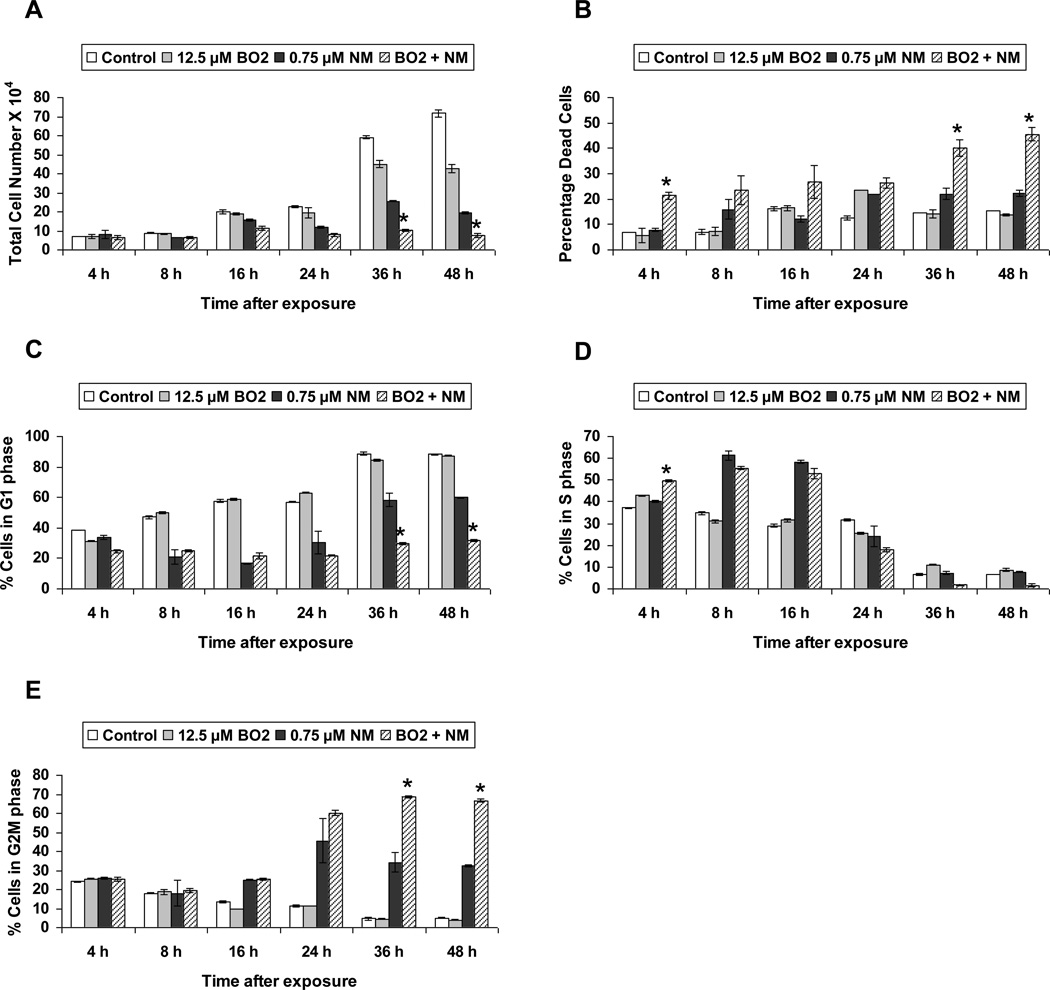
The aim of our study was to analyze DNA repair mechanisms activated after NM exposure, so we started our studies by determining a concentration of NM that does not cause more than 50% reduction in cell viability after 24 h of exposure (data not shown). Based on results from these experiments, we chose a level of 0.75 µM NM and conducted a time course (4 to 72 h) study using trypan blue exclusion assay. As shown in Figure 1 a and b, NM exposure caused a huge decrease in total cell numbers that was significant starting at 24 h after exposure, with a concomitant increase in percent dead cells when compared to controls. Nevertheless, the percent decrease in total cell numbers did not translate into an identical increase in percent dead cells, which led us to consider that NM exposure induced growth arrest.
Fig. 1. Effect of NM exposure on cell proliferation and cell cycle progression in JB6 cells.
JB6 cells plated overnight in 60 mm plates were exposed to either DMSO or 0.75 µM NM for 4 to 96 h. At the end of the exposure times, all cells, including the floating dead cells were collected, subjected to Trypan blue exclusion assay using a hemocytometer for counting the cells. The total number of cells (a) and the percentage dead cells (b) were calculated as described under material and methods. Following similar exposures, FACS analysis was carried out to determine the cell cycle progression following NM exposure (c). Cells collected after 4 to 48 h of exposures were incubated overnight with saponin/PI at 4°C and subjected to FACS analysis as described in material and methods. Data are presented as mean ± SEM, n = 3. * P< 0.05 as compared to control.
Based on the above results that suggested that NM exposure could induce growth arrest, we next analyzed the effect of NM on cell cycle progression using FACS analysis. In a time-kinetic study design, after 16 h of NM exposure, 64% of cells were arrested in the S-phase of the cell cycle. These cells had entered G2M phase at 24 h, and by 36 h and 48 h after NM exposure the cells moved into G1 phase (Fig 1c).
3.2. NM-induced DNA damage indicated by activation of DNA damage response molecules and an increase in the comet tail extent moment
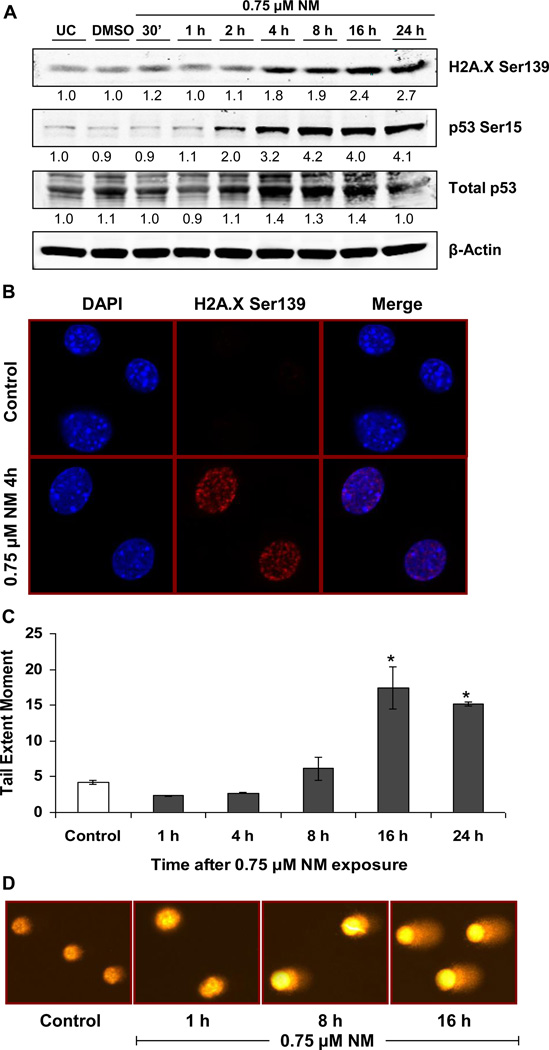
Next, we investigated the DNA damaging properties of NM that could lead to an arrest in S-phase of cell cycle progression and cause cell death. First we studied activation of known DNA damage markers, which are phosphorylated H2A.X at Ser139, p53 at Ser15 and the total p53 levels [37, 38]. Accordingly, an increase in the phosphorylation of H2A.X and p53, as well as an increase in total p53 levels, was observed within 4 h after NM exposure, which persisted for 24 h (Fig 2a). This activation of DNA damage response was further confirmed by immunofluorescence detection of H2A.X Ser139. Exposure to NM, induced nuclear H2A.X Ser139 foci formation which was observed as increased red fluorescence when compared to control cells (Fig 2b). We next confirmed the ICL forming ability of NM in JB6 cells through indirect comet assay, where the ability of NM to inhibit the comet formation by hydrogen peroxide is related to its ICL forming capacity (data not shown). Based on the fact that DNA ICLs are processed into DNA DSBs and that our results show an increase in DNA damage markers with NM exposure, we further quantified DNA damage employing comet assay, which has been previously used to quantify DNA damage following SM analog exposure [30]. A time-kinetic study from 1 h to 24 h using the concentration of 0.75 µM NM further confirmed the DNA damage pattern measured as TEM in comet assay, which increased between 4 and 8 h after NM exposure, with a substantial increase of 4-fold by 16 h that persisted at 24 h after NM exposure (Fig 2c and d). These results, along with those showing arrest of cells in S-phase from 8 h and an increase in DNA DSB response molecules from 4 h after initiation of NM exposure, suggested that most NM-induced DNA ICLs are processed within 4 and 8 h after exposure resulting in the formation of DNA DSBs.
Fig. 2. Effect of NM exposure on the activation of DNA damage response molecules and formation of DNA DSBs in JB6 cells.
JB6 cells were exposed to either DMSO or 0.75 µM NM for 30 min to 24 h. Following exposures, whole cell lysates were prepared and subjected to SDS PAGE followed by western blot analysis for H2A.X Ser139 and p53 Ser15 using 80 µg protein as detailed under Materials and Methods (a). Total p53 protein levels and β-Actin levels were determined by stripping and reprobing the membranes with total p53 and β-Actin antibodies (a); densitometry values represented as numbers under the bands were calculated employing NIH ImageJ software. Following similar exposures, cells were subjected to immunofluorescence for assessing the accumulation of H2A.X Ser139 at the DNA damage sites, seen in the form of foci in the DAPI stained nucleus (b). Cell images were captured at 400× magnification using a Nikon D Eclipse C1 confocal microscope as described under Materials and Methods. Cells exposed to DMSO or 0.75 µM NM for 1 to 24 h were subjected to comet assay as described under Materials and Methods (c) and the representative pictures of NM exposed cells at various time points were captured using a fluorescence microscope at 200× original magnification (d). Data are presented as mean ± SEM, n =3. * P< 0.05 as compared to control.
3.3. NM induced activation of DNA DSB repair pathways NHEJ and HRR
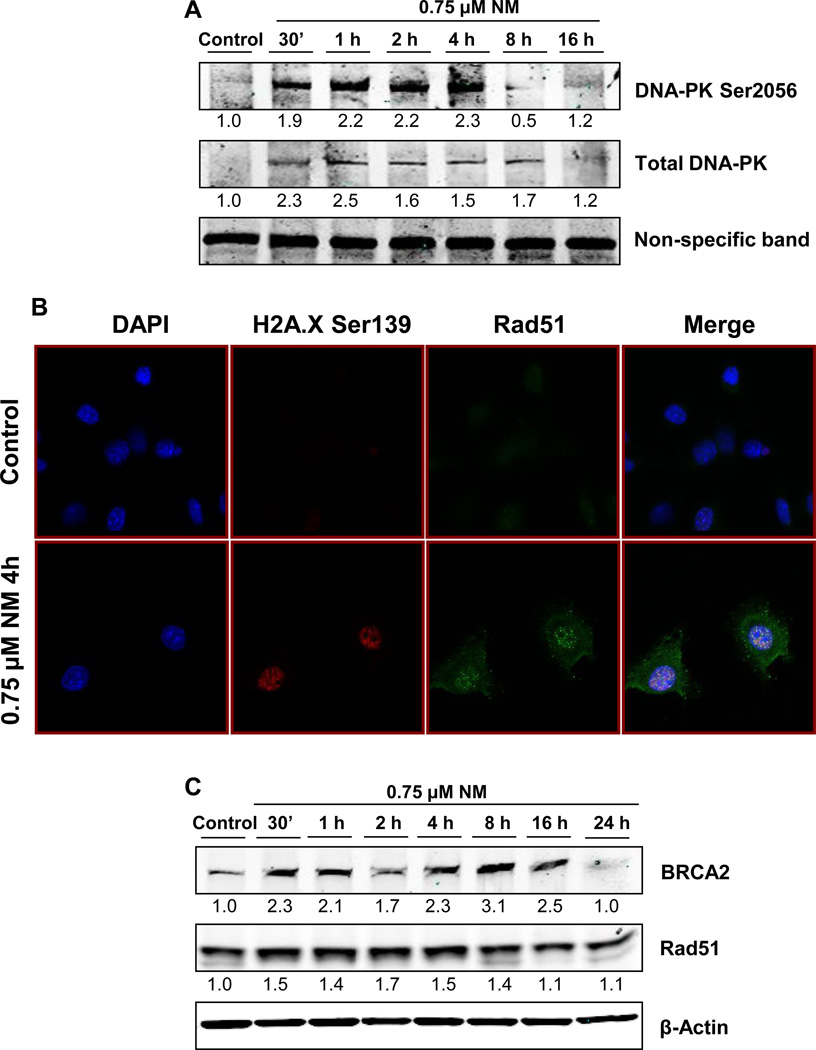
DNA ICLs are first processed by exonucleases to form DNA DSBs. Upon formation of DSBs, the two DNA DSB repair pathways, NHEJ and HRR, are activated, which help in repairing the damage. Our next objective was to determine if both the repair pathways are activated by NM-induced DNA damage in epidermal keratinocytes. Accordingly, we assessed activation of the NHEJ pathway by analyzing activation of DNA-PK, the key molecule involved in repair through this pathway in mammalian cells [39, 40]. DNA-PK is known to localize at DNA DSBs, then becoming activated by auto-phosphorylation at Ser2056 and Thr2609. DNA-PK, apart from its role in DNA repair, also plays a role in the phosphorylation of DNA damage-response molecules, namely p53 and H2A.X [41, 42]. Exposure of JB6 cells to NM caused an increase in the levels of total DNA-PK between 30 min and 4 h post-exposure. An increase in the phosphorylation of DNA-PK at Ser2056 was also observed, where a maximal effect was seen at 4 h after NM exposure, indicating activation of the NHEJ repair pathway (Fig 3a). Next, we assessed activation of the HRR pathway through Rad51 foci formation and through increased levels of BRCA2 and rad51 [43]. The process of HRR repair includes formation of rad51 foci at DNA DSB sites. This was confirmed through immunofluorescence assay, where NM exposure induced a significant increase in nuclear rad51 foci (Fig 3b). A time-kinetic immunoblotting analysis of NM-exposed cells further supported the activation of this pathway through increased BRCA2 and rad51 levels (Fig 3c). These studies demonstrated that NM exposure in JB6 cells caused activation of both DSB repair pathways, NHEJ and HRR.
Fig. 3. Effect of NM exposure on the activation of DNA DSB repair pathways NHEJ and HRR.
To determine the activation of NHEJ pathway, whole cell lysates prepared from JB6 cells exposed to either DMSO or 0.75 µM NM for 30 min to 16 h, were subjected to SDS PAGE followed by western blot analysis for DNA-PK Ser2056 and total DNA-PK using 150 µg protein as detailed under Materials and Methods and the densitometry values represented as numbers under the bands were calculated employing NIH ImageJ software (a). Similarly, cells were subjected to immunofluorescence after 4 h of exposures to determine the activation of HRR pathway seen as rad51 foci formation in the nucleus (b). Cell images were captured at 400× magnification using a Nikon D Eclipse C1 confocal microscope as described under Materials and Methods. The activation of HRR was further confirmed through immunoblot analysis for BRCA2 and rad51 as described under material and methods (c).
3.4. Inhibition of NHEJ caused increased NM-induced cell death but did not alter NM-induced cell cycle arrest
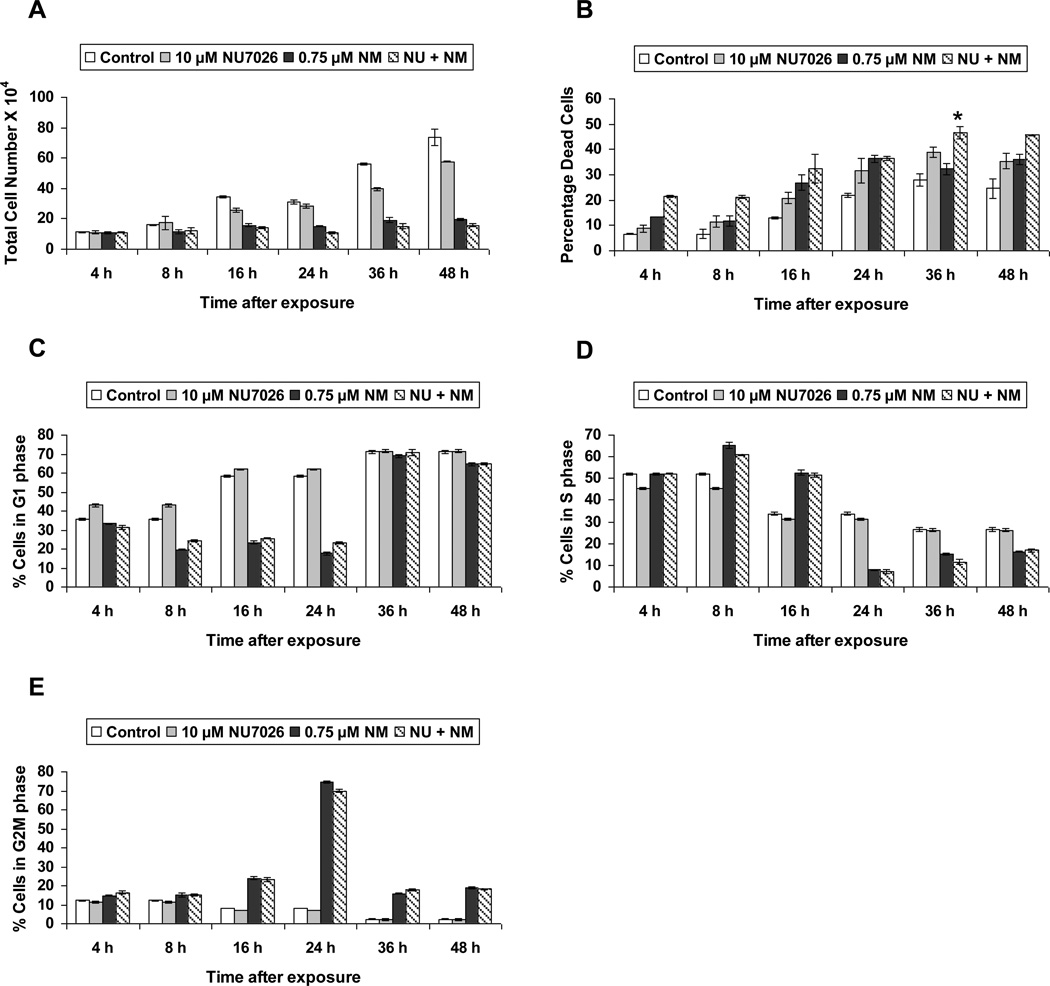
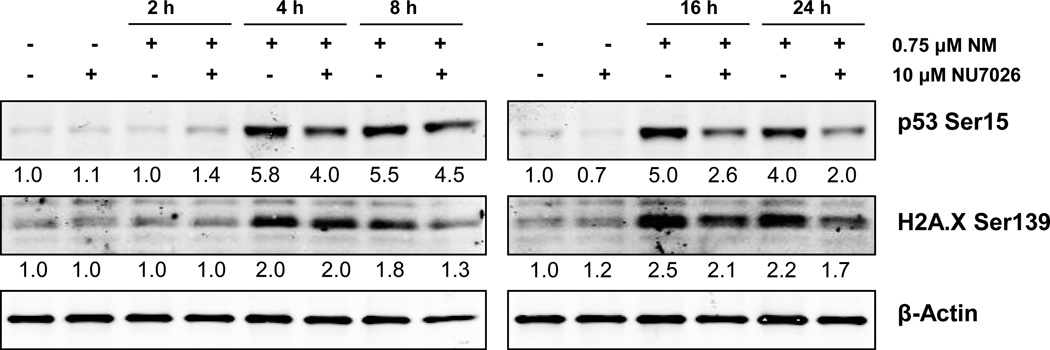
Next we analyzed the role of NHEJ pathway in the cellular response to NM-induced cytotoxicity. To accomplish this, we employed NU7026, a competitive inhibitor of DNA-PK. This compound acts by inhibiting auto-phosphorylation of DNA-PK at DNA strand breaks, thereby resulting in inhibition of the NHEJ repair pathway [44, 45]. We used a concentration of 10 µM NU7026 1 h prior to NM exposure. Our trypan blue exclusion assay showed that NU7026 treatment prior to NM exposure did not cause a significant decrease in total cell number when compared to NM exposure alone (Fig 4a). However, NU7026 pretreatment caused a significant increase (14% as compared to NM exposure) in the percentage dead cells at 36 h post-NM exposure (Fig 4b). We next examined the effect of NHEJ pathway inhibition on NM-induced cell cycle arrest, by following a time-kinetic FACS analysis of cells treated with 10 µM NU7026 1 h prior to NM exposure. Our results showed that inhibition of the NHEJ pathway did not have a significant effect on NM-induced cell cycle arrest in JB6 cells (Fig 4c, d and e). Furthermore, we assessed the effect of NHEJ inhibition on NM-induced activation of DNA damage markers. Our immunoblotting results showed that NU7026 pretreatment caused a decrease in phosphorylation of both H2A.X Ser139 and p53 Ser15 (Fig 5). This could be a result of inhibition of DNA-PK, which, apart from its DNA repair properties, can also induce phosphorylation of p53 and H2A.X [41, 42, 46]. These immunoblot results confirm inactivation of DNA-PK activity by NU7026.
Fig. 4. Effect of DNA-PK inhibition on NM-induced cytotoxicity.
JB6 cells were treated with 10 µM NU7026 1 h prior to 0.75 µM NM exposure for 4 to 48 h. At the end of the treatment times, all the cells were collected and subjected to trypan blue exclusion staining to determine total cell number (a) and percentage dead cells (b) as described earlier in material and methods. Following similar treatments, cells were collected and incubated with saponin/PI for 24 h at 4°C and analyzed for the G1 (c), S (d) and G2M (e) phases of cell cycle progression using FACS analysis as detailed under materials and methods. Data are presented as mean ± SEM, n =3. * P< 0.05 as compared to 0.75 µM NM.
Fig. 5. Effect of DNA-PK inhibition on NM-induced phosphorylation of H2A.X at Ser139 and p53 at Ser15 in JB6 cells.
Whole cell lysates were prepared from JB6 cells treated with 10 µM NU7026 1 h prior to 0.75 µM NM exposure for 4 to 24 h. 80 µg protein was subjected to SDS PAGE followed by western blot analysis for H2A.X Ser139 and p53 Ser15 as detailed under Materials and Methods and the densitometry values represented as numbers under the bands were calculated employing NIH ImageJ software.
3.5. Inhibition of HRR sensitized cells towards NM-induced cytotoxicity
Rad51 and BRCA2 are the key molecules involved in homologous recombination repair [21]. To study the effect of HRR inhibition on NM-induced cytotoxicity, we employed BO2, a new small molecule inhibitor of rad51 which acts by inhibiting the DNA strand exchange activity of rad51 [47, 48]. Our cell viability study employing trypan blue exclusion assay showed that 12.5 µM BO2, when administered throughout NM exposure, caused a significant decrease in total cell numbers at 36 and 48 h when compared to 0.75 µM NM exposure alone (Fig 6a). There was also a significant increase in percent cell death observed with BO2 treatment in NM-exposed cells at 4, 36 and 48 h of exposure (Fig 6b). Cell cycle analysis of JB6 cells exposed to BO2 along with NM showed that rad51 inhibition did not have a significant effect on NM-induced cell cycle arrest through 24 h of NM exposure, but by 36 and 48 h, rad51 inhibition caused accumulation of NM-exposed cells in G2M phase, preventing them from entering G1 phase (Fig 6c, d and e).
Fig. 6. Effect of rad51 inhibition on NM-induced cytotoxicity.
JB6 cells treated with 12.5 µM BO2 along with 0.75 µM NM exposure for 4 to 48 h, were collected and subjected to trypan blue exclusion staining to determine total cell number (a) and percentage dead cells (b) as described earlier in material and methods. Following similar treatments, cells were collected and incubated with saponin/PI for 24 h at 4°C and analyzed for the percentage cells in G1 phase (c), S phase (d) and G2M phase (e) of cell cycle progression using FACS analysis as detailed under materials and methods. Data are presented as mean ± SEM, n =3. * P< 0.05 as compared to 0.75 µM NM.
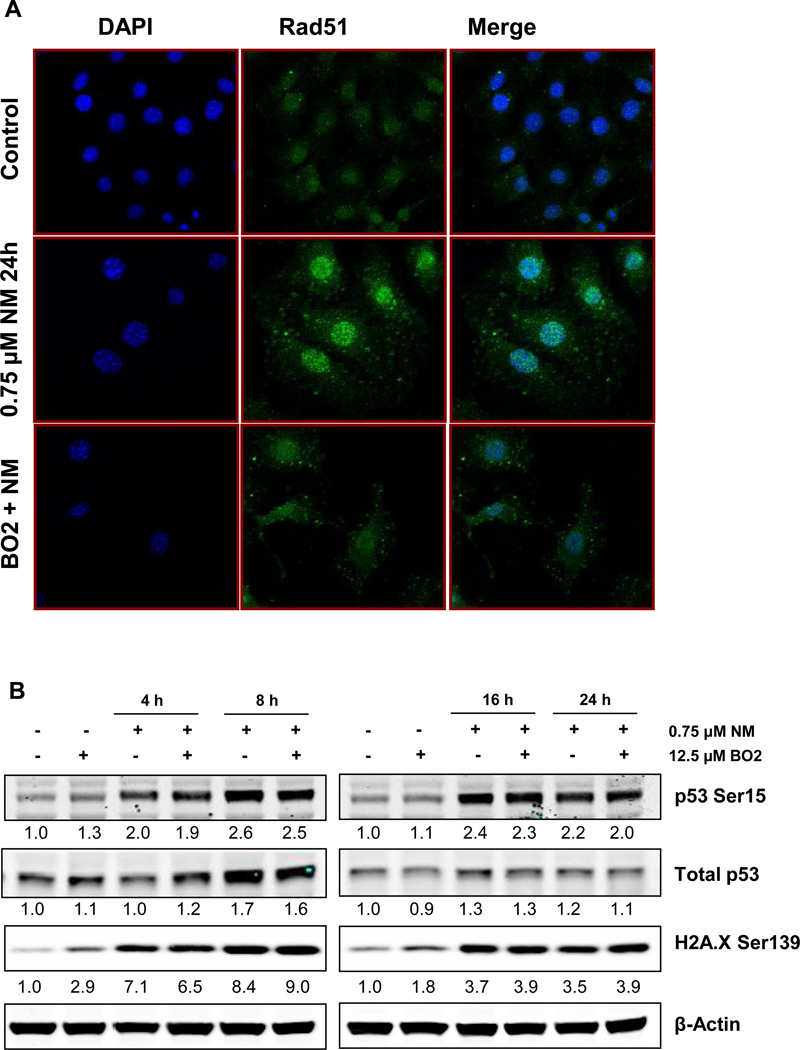
We also confirmed the rad51 inhibition efficacy of BO2 in JB6 cells through immunofluorescence, wherein 12.5 µM BO2 caused a significant decrease in NM-induced rad51 foci formation after 24 h of exposure (Fig 7a). In addition, an increase in cell size by 24h of NM exposure was also documented (Fig 7a). Our immunoblot analysis of cells exposed to BO2 along with NM showed that inhibition of HRR pathway caused persistence of DNA damage similar to that of NM exposure, as evidenced by increased phospho H2A.X Ser139 and p53 Ser15 levels through 24 h of exposure (Fig 7b).
Fig. 7. Effect of rad51 inhibition on NM-induced cytotoxicity.
JB6 cells were treated with 12.5 µM BO2 along with NM exposure for 24 h and were subjected to immunofluorescence to assess the accumulation of rad51 at the DNA damage sites, seen in the form of foci in the nucleus (a). Cell images were captured at 400× magnification using a Nikon D Eclipse C1 confocal microscope as described under Materials and Methods. Similarly, treated cells were collected between 4 h and 24 h of the treatments and whole cell lysates were prepared. 80 µg protein was subjected to SDS PAGE followed by western blot analysis for H2A.X Ser139 and p53 Ser15 as detailed under Materials and Methods and the densitometry values represented as numbers under the bands were calculated employing NIH ImageJ software (b).
4. Discussion
SM has been used as a chemical warfare agent for nearly a century, and despite its well-documented toxic effects, there is still a need for effective antidote(s) to counteract its damaging effects on various tissues, including the skin [49]. NM, which has a similar mechanism of action to SM, induces skin toxicity through its bifunctional alkylating effects, forming adducts and crosslinks with DNA, RNA and proteins [11]. Since keratinocytes are the major target of topically applied vesicating agents SM and NM, we used JB6 mouse skin keratinocytes to study the kinetics of NM-induced DNA damage and cytotoxicity. In addition, we explored the roles of DNA DSB repair mechanisms by employing specific DNA repair pathway inhibitors. Our studies demonstrated that HRR is the crucial DNA DSB repair pathway activated in response to NM. These findings, by clarifying the critical repair mechanism involved in repair of NM-induced DNA damage, the major event that leads to its cytotoxicity, could help define new rescue therapies for NM, and possibly SM, -induced skin injuries.
DNA damage is the major lesion following SM exposure [50]. Our previous studies using the monofunctional SM analog CEES have given a broader understanding of the role of oxidative stress and direct DNA damage in response to such exposure in both skin epidermal cells and dermal fibroblasts [30]. To better understand repair mechanisms activated in response to these alkylating agents, we employed NM. Since CEES is a monofunctional alkylating agent, it only forms adducts leading to DNA single strand breaks. By contrast, SM/NM cause ICLs and lead to the formation of DNA DSBs [11].
The damage to cells exposed to SM/NM, if not repaired, results in decreased cell viability as demonstrated in studies where SM caused dose- and time-dependent declines in cell viability of human epidermal keratinocytes [51, 52]. Accordingly, our time-kinetic studies employing 0.75 µM NM confirmed a decrease in cell viability starting as early as 16 h after NM exposure. However, this decrease in cell viability was not proportional to the increase in NM-induced cell death. This data directed us to the possibility that NM-induced decreased cell viability was due to decreased cell growth rather than an increased cell death. Previous studies indicated that exposure to SM causes cell cycle arrest via prolongation of the S phase of cell cycle progression [53, 54]. In fact, our cell cycle progression analysis also found that NM caused a prolonged S-phase arrest, after which these cells moved to G2M phase and later to G1 phase. Studies have shown that even the cells with unrepaired damaged DNA can initially continue through the cell cycle with prolonged S and G2M phases, but, may ultimately arrest in G1, where the fate of the cells is decided [55, 56]. In our study, cells moving from prolonged S and G2M phases may symbolize such phenomena, which might also be the reason that we see an increase in cell death starting 24 h in our cell viability studies, with the other possibility being that some cells are actually undergoing successful repair.
NM-induced DNA damage was indicated by an increase in the phosphorylation of H2A.X at Ser139 and p53 at Ser15. These markers have been established as indicators of DNA damage by our previous and various other studies [30, 32, 57, 58]. In the presence of DNA damage, MRN complex, H2A.X and various other factors localize to strand breaks aiding in recruitment of PIKK family protein kinases, which include ATM, ATR and DNA-PK. These kinases then become auto-phosphorylated in the presence of DNA damage, becoming activated, further phosphorylating and activating H2A.X, 53BP1, SMC1, p53, chk1, chk2 among other proteins, which are important in modulating cell cycle progression to facilitate the repair process, and, in the event of overwhelming damage, direct cells to apoptosis [6, 12]. The maximum increase of phospho H2A.X and p53 seen after NM exposure coincides with increases in tail extent moment measured via comet assay, which we noted to have begun between 4 and 8 h of exposure. This time interval (4 to 8 h after initiation of exposure) might be the time when maximal processing of NM-induced DNA ICLs is taking placing, leading to DNA DSB formation. Our study also shows the progression of events following NM exposure, where optimum DNA damage is followed by arrest of cells in S-phase, thus slowing cell cycle progression to facilitate repair.
Previous studies indicated that absence of HRR, NHEJ or NER pathways affects cells adversely in the event of SM exposure, and that absence of HRR has the most adverse effect following SM exposure [6, 32, 59]. However, the cell lines used to compare the role of the repair pathways in those studies were different for each of the pathways (human fibroblast cells, TK6 lymphoblastoid cells and Chinese hamster ovary cells). Therefore in our study we have aimed to use a single cell type to conduct a comparative study for the activation of the DNA DSB repair pathways following NM exposure. BER pathway was also shown to play a role in repair after exposure to half mustard (CEES) and NM [32, 60]. But studies have also shown that presence of active BER pathway further sensitized cells to these compounds and that the NER pathway assists in repair after SM/CEES exposure [61]. Our study, for the first time, has demonstrated the activation and role of the two DNA DSB repair pathways, NHEJ and HRR, in NM-induced DNA damage and cytotoxicity using a single epidermal keratinocyte cell line. As we have discussed earlier, DNA-PK, apart from its DNA damage repair functions, also acts as a protein kinase in response to DNA damage and phosphorylates DNA damage response molecules like H2A.X and p53. Accordingly, inhibition of DNA-PK, despite the presence of DNA damage, caused a decrease in p53 Ser15 and H2A.X Ser139, further confirming the inhibitory potential of NU7026. Furthermore, in spite of the slight increase in toxicity seen with NU7026 alone when compared to control, only a slight increase in NM-induced cell death following DNA-PK inhibition was observed, showing that the absence of the NHEJ pathway did not detectably affect cell cycle progression after NM exposure. By contrast, inhibition of HRR caused a significant decrease in total cell numbers and increased cell death, especially following 36 and 48 h of NM exposure. This coincided with the inability of these cells, unlike HRR-competent NM-exposed cells, to exit the G2M phase of the cell cycle. Our results, taken together, demonstrated activation of both NHEJ and HRR in response to NM-induced DNA damage, and that the activity of HRR was the major determinant of cell fate following NM exposure. These findings should be useful in rescue/repair of skin injury following NM, and potentially also SM, exposures. If an increase in HRR could be effected through the use of compounds such as rad51 stimulatory compound-1 (RS-1) [62], the increase in repair of damaged DNA could result in decreased cell death, preventing the release of pro-inflammatory molecules, and ultimately, excessive activation of inflammatory immune response [63]. At the same time, studies are also needed to explore potential adverse effects like, increases in mutations due to accelerated or faulty repair that might result when activating repair pathways through external stimuli. Finding a balance between advantageous and adverse effects of DNA repair activation could be a major new strategy in developing countermeasures against DNA damaging agents such as vesicants and associated damage to skin and other tissues and organs.
Research Highlights.
NM exposure caused growth arrest and increase in DNA double strand breaks in JB6 cells
NM exposure caused the activation of both DNA DSB repair pathways NHEJ and HRR
Unlike NHEJ, inactivation of HRR sensitized the cells towards NM-induced cytotoxicity
Acknowledgements
This work was supported by the Countermeasures Against Chemical Threats (CounterACT) Program, National Institutes of Health Office of the Director, and the National Institute of Environmental Health Sciences, [Grant U54ES-015678]. The study sponsors have no involvement in the study design; collection, analysis and interpretation of data; the writing of the manuscript; and the decision to submit the manuscript for publication.
List of Abbreviations
- ATM
Ataxia telangiectasia mutated
- ATR
Ataxia telangiectasia-Rad3-related
- BER
Base excision repair
- CEES
2-chloroethyl ethyl sulfide
- DAPI
4′,6′-diamidino-2-phenylindole
- DMSO
Dimethyl sulfoxide
- DSB
Double strand breaks
- FACS
Fluorescence activated cell sorting
- FBS
Fetal bovine serum
- HRR
Homologous recombination repair
- ICL
Interstrand crosslinks
- MEM
Minimal essential medium
- NER
Nucleotide excision repair
- NHEJ
Non homologous end joining
- NM
Nitrogen mustard
- PBS
Phosphate buffered saline
- PI
Propidium iodide
- SCGE
Single cell gel electrophoresis
- SM
Sulfur mustard
- TEM
Tail extent moment
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- 1.Ghanei M, Poursaleh Z, Harandi AA, Emadi SE, Emadi SN. Acute and chronic effects of sulfur mustard on the skin: a comprehensive review. Cutaneous and ocular toxicology. 2010;29:269–277. doi: 10.3109/15569527.2010.511367. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson WS. Delayed Mustard Gas Keratitis (Dichlorodiethyl Sulfide). A Report of Two Cases. Trans Am Ophthalmol Soc. 1947;45:81–92. [PMC free article] [PubMed] [Google Scholar]
- 3.Shohrati M, Peyman M, Peyman A, Davoudi M, Ghanei M. Cutaneous and ocular late complications of sulfur mustard in Iranian veterans. Cutaneous and ocular toxicology. 2007;26:73–81. doi: 10.1080/15569520701212399. [DOI] [PubMed] [Google Scholar]
- 4.Croddy, JJ EW. In: Weapons of mass destruction: an encyclopedia of worldwide policy, technology, and history. Croddy Eric JW, Larsen Jeffrey., editors. Volume 2. Santa Barbara, California: ABC-CLIO, Inc.; 2005. [Google Scholar]
- 5.Smith KJ, Smith WJ, Hamilton T, Skelton HG, Graham JS, Okerberg C, Moeller R, Hackley BE., Jr Histopathologic and immunohistochemical features in human skin after exposure to nitrogen and sulfur mustard. Am J Dermatopathol. 1998;20:22–28. doi: 10.1097/00000372-199802000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Hansson J, Lewensohn R, Ringborg U, Nilsson B. Formation and removal of DNA cross-links induced by melphalan and nitrogen mustard in relation to drug-induced cytotoxicity in human melanoma cells. Cancer Res. 1987;47:2631–2637. [PubMed] [Google Scholar]
- 7.Ludlum DB, Papirmeister B. DNA modification by sulfur mustards and nitrosoureas and repair of these lesions. Basic Life Sci. 1986;38:119–125. doi: 10.1007/978-1-4615-9462-8_12. [DOI] [PubMed] [Google Scholar]
- 8.Lakshmana Rao PV, Vijayaraghavan R, Bhaskar AS. Sulphur mustard induced DNA damage in mice after dermal and inhalation exposure. Toxicology. 1999;139:39–51. doi: 10.1016/s0300-483x(99)00097-9. [DOI] [PubMed] [Google Scholar]
- 9.Golumbic C, Fruton JS, Bergmann M. Chemical reactions of the nitrogen mustard gases; the transformations of methyl-bis(beta-chloroethyl)amine in water. J Org Chem. 1946;11:518–535. [PubMed] [Google Scholar]
- 10.Mann DJ. Aziridinium ion ring formation from nitrogen mustards: mechanistic insights from ab initio dynamics. J Phys Chem A. 2010;114:4486–4493. doi: 10.1021/jp9079553. [DOI] [PubMed] [Google Scholar]
- 11.Guainazzi A, Campbell AJ, Angelov T, Simmerling C, Scharer OD. Synthesis and molecular modeling of a nitrogen mustard DNA interstrand crosslink. Chemistry. 2010;16:12100–12103. doi: 10.1002/chem.201002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grillari J, Katinger H, Voglauer R. Contributions of DNA interstrand cross-links to aging of cells and organisms. Nucleic Acids Res. 2007;35:7566–7576. doi: 10.1093/nar/gkm1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 14.Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clingen PH, De Silva IU, McHugh PJ, Ghadessy FJ, Tilby MJ, Thurston DE, Hartley JA. The XPF-ERCC1 endonuclease and homologous recombination contribute to the repair of minor groove DNA interstrand crosslinks in mammalian cells produced by the pyrrolo[2,1-c][1,4]benzodiazepine dimer SJG-136. Nucleic Acids Res. 2005;33:3283–3291. doi: 10.1093/nar/gki639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 17.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Heyer WD. Homologous re combination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sleeth KM, Sorensen CS, Issaeva N, Dziegielewski J, Bartek J, Helleday T. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J Mol Biol. 2007;373:38–47. doi: 10.1016/j.jmb.2007.07.068. [DOI] [PubMed] [Google Scholar]
- 20.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 23.Hinz JM. Role of homologous recombination in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- 24.Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehe K, Reisinger H, Szinicz L. Sulfur mustard induces apoptosis and necrosis in SCL II cells in vitro. J Appl Toxicol. 2000;20(Suppl 1):S81–S86. doi: 10.1002/1099-1263(200012)20:1+<::aid-jat684>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 26.Ruff AL, Dillman JF. Signaling molecules in sulfur mustard-induced cutaneous injury. Eplasty. 2007;8:e2. [PMC free article] [PubMed] [Google Scholar]
- 27.Tewari-Singh N, Gu M, Agarwal C, White CW, Agarwal R. Biological and molecular mechanisms of sulfur mustard analogue-induced toxicity in JB6 and HaCaT cells: possible role of ataxia telangiectasia-mutated/ataxia telangiectasia-Rad3-related cell cycle checkpoint pathway. Chem Res Toxicol. 2010;23:1034–1044. doi: 10.1021/tx100038b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jowsey PA, Williams FM, Blain PG. DNA damage, signalling and repair after exposure of cells to the sulphur mustard analogue 2-chloroethyl ethyl sulphide. Toxicology. 2009;257:105–112. doi: 10.1016/j.tox.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–942. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 30.Inturi S, Tewari-Singh N, Gu M, Shrotriya S, Gomez J, Agarwal C, White CW, Agarwal R. Mechanisms of sulfur mustard analog 2-chloroethyl ethyl sulfide-induced DNA damage in skin epidermal cells and fibroblasts. Free Radic Biol Med. 2011;51:2272–2280. doi: 10.1016/j.freeradbiomed.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matijasevic Z, Precopio ML, Snyder JE, Ludlum DB. Repair of sulfur mustard-induced DNA damage in mammalian cells measured by a host cell reactivation assay. Carcinogenesis. 2001;22:661–664. doi: 10.1093/carcin/22.4.661. [DOI] [PubMed] [Google Scholar]
- 32.Jowsey PA, Williams FM, Blain PG. DNA damage responses in cells exposed to sulphur mustard. Toxicol Lett. 2012;209:1–10. doi: 10.1016/j.toxlet.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Jowsey PA, Williams FM, Blain PG. The role of homologous recombination in the cellular response to sulphur mustard. Toxicol Lett. 2010;197:12–18. doi: 10.1016/j.toxlet.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Nikolova T, Ensminger M, Lobrich M, Kaina B. Homologous recombination protects mammalian cells from replication-associated DNA double-strand breaks arising in response to methyl methanesulfonate. DNA Repair (Amst) 2012;9:1050–1063. doi: 10.1016/j.dnarep.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 36.Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 37.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. The Journal of cell biology. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 39.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes & development. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yajima H, Lee KJ, Zhang S, Kobayashi J, Chen BP. DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J Mol Biol. 2009;385:800–810. doi: 10.1016/j.jmb.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An J, Huang YC, Xu QZ, Zhou LJ, Shang ZF, Huang B, Wang Y, Liu XD, Wu DC, Zhou PK. DNA-PKcs plays a dominant role in the regulation of H2AX phosphorylation in response to DNA damage and cell cycle progression. BMC Mol Biol. 11:18. doi: 10.1186/1471-2199-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao WT, Wang YT, Huang ZW, Fang J. BRCA2 affects the efficiency of DNA double-strand break repair in response to N-nitroso compounds with differing carcinogenic potentials. Oncol Lett. 2013;5:1948–1954. doi: 10.3892/ol.2013.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollick JJ, Golding BT, Hardcastle IR, Martin N, Richardson C, Rigoreau LJ, Smith GC, Griffin RJ. 2,6-disubstituted pyran-4-one and thiopyran-4-one inhibitors of DNA-Dependent protein kinase (DNA-PK) Bioorg Med Chem Lett. 2003;13:3083–3086. doi: 10.1016/s0960-894x(03)00652-8. [DOI] [PubMed] [Google Scholar]
- 45.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 46.An J, Huang YC, Xu QZ, Zhou LJ, Shang ZF, Huang B, Wang Y, Liu XD, Wu DC, Zhou PK. DNA-PKcs plays a dominant role in the regulation of H2AX phosphorylation in response to DNA damage and cell cycle progression. BMC Mol Biol. 2010;11:18. doi: 10.1186/1471-2199-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang F, Motlekar NA, Burgwin CM, Napper AD, Diamond SL, Mazin AV. Identification of specific inhibitors of human RAD51 recombinase using high-throughput screening. ACS Chem Biol. 6:628–635. doi: 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang F, Mazina OM, Zentner IJ, Cocklin S, Mazin AV. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. J Med Chem. 55:3011–3020. doi: 10.1021/jm201173g. [DOI] [PubMed] [Google Scholar]
- 49.Kehe K, Balszuweit F, Emmler J, Kreppel H, Jochum M, Thiermann H. Sulfur mustard research--strategies for the development of improved medical therapy. Eplasty. 2008;8:e32. [PMC free article] [PubMed] [Google Scholar]
- 50.Kehe K, Balszuweit F, Steinritz D, Thiermann H. Molecular toxicology of sulfur mustard-induced cutaneous inflammation and blistering. Toxicology. 2009;263:12–19. doi: 10.1016/j.tox.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 51.Mol MA, de Vries-van de Ruit AM. Concentration- and time-related effects of sulphur mustard on human epidermal keratinocyte function. Toxicol In Vitro. 1992;6:245–251. doi: 10.1016/0887-2333(92)90038-s. [DOI] [PubMed] [Google Scholar]
- 52.Ku WW, Bernstein IA. bis-(beta-chloroethyl)sulfide (BCES)-induced changes in epidermal cell homeostasis in vitro. Toxicol Appl Pharmacol. 1988;95:397–411. doi: 10.1016/0041-008x(88)90358-4. [DOI] [PubMed] [Google Scholar]
- 53.Savage JR, Breckon G. Differential effects of sulphur mustard on S-phase cells of primary fibroblast cultures from Syrian hamsters. Mutation research. 1981;84:375–387. doi: 10.1016/0027-5107(81)90205-0. [DOI] [PubMed] [Google Scholar]
- 54.Lin PP, Bernstein IA, Vaughan FL. Bis(2-chloroethyl)sulfide (BCES) disturbs the progression of rat keratinocytes through the cell cycle. Toxicol Lett. 1996;84:23–32. doi: 10.1016/0378-4274(95)03453-6. [DOI] [PubMed] [Google Scholar]
- 55.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13:738–747. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 56.Callegari AJ, Clark E, Pneuman A, Kelly TJ. Postreplication gaps at UV lesions are signals for checkpoint activation. Proc Natl Acad Sci U S A. 107:8219–8224. doi: 10.1073/pnas.1003449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tewari-Singh N, Jain AK, Inturi S, Agarwal C, White CW, Agarwal R. Silibinin attenuates sulfur mustard analog-induced skin injury by targeting multiple pathways connecting oxidative stress and inflammation. PloS one. 2012;7:e46149. doi: 10.1371/journal.pone.0046149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jain AK, Tewari-Singh N, Gu M, Inturi S, White CW, Agarwal R. Sulfur mustard analog 2-chloroethyl ethyl sulfide-induced skin injury involves DNA damage and induction of inflammatory mediators, in part via oxidative stress, in SKH-1 hairless mouse skin. Toxicology letters. 2011;205:293–301. doi: 10.1016/j.toxlet.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller C, Christodoulopoulos G, Salles B, Panasci L. DNA-Dependent protein kinase activity correlates with clinical and in vitro sensitivity of chronic lymphocytic leukemia lymphocytes to nitrogen mustards. Blood. 1998;92:2213–2219. [PubMed] [Google Scholar]
- 60.De Alencar TA, Leitao AC, Lage C. Nitrogen mustard- and half-mustard-induced damage in Escherichia coli requires different DNA repair pathways. Mutat Res. 2005;582:105–115. doi: 10.1016/j.mrgentox.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Matijasevic Z, Volkert MR. Base excision repair sensitizes cells to sulfur mustard and chloroethyl ethyl sulfide. DNA Repair (Amst) 2007;6:733–741. doi: 10.1016/j.dnarep.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 62.Jayathilaka K, Sheridan SD, Bold TD, Bochenska K, Logan HL, Weichselbaum RR, Bishop DK, Connell PP. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc Natl Acad Sci U S A. 2008;105:15848–15853. doi: 10.1073/pnas.0808046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi Y, Zheng W, Rock KL. Cell injury releases endogenous adjuvants that stimulate cytotoxic T cell responses. Proc Natl Acad Sci U S A. 2000;97:14590–14595. doi: 10.1073/pnas.260497597. [DOI] [PMC free article] [PubMed] [Google Scholar]