Abstract
A number of common contiguous gene syndromes have been shown to result from nonallelic homologous recombination (NAHR) within region-specific low-copy repeats (LCRs). The reciprocal duplications are predicted to occur at the same frequency; however, probably because of ascertainment bias and milder phenotypes, reciprocal events have been identified in only a few cases to date. We previously described seven patients with dup(17)(p11.2p11.2), the reciprocal of the Smith-Magenis syndrome (SMS) deletion, del(17)(p11.2p11.2). In >90% of patients with SMS, identical ∼3.7-Mb deletions in 17p11.2 have been identified. These deletions are flanked by large (∼200 kb), highly homologous, directly oriented LCRs (i.e., proximal and distal SMS repeats [SMS-REPs]). The third (middle) SMS-REP is inverted with respect to them and maps inside the commonly deleted genomic region. To investigate the parental origin and to determine whether the common deletion and duplication arise by unequal crossovers mediated through NAHR between the proximal and distal SMS-REPs, we analyzed the haplotypes of 14 families with SMS and six families with dup(17)(p11.2p11.2), using microsatellite markers directly flanking the SMS common deletion breakpoints. Our data indicate that reciprocal deletion and duplication of 17p11.2 result from unequal meiotic crossovers. These rearrangements occur via both interchromosomal and intrachromosomal exchange events between the proximal and distal SMS-REPs, and there appears to be no parental-origin bias associated with common SMS deletions and the reciprocal duplications.
Introduction
Smith-Magenis syndrome (SMS [MIM 182290]) is characterized by multiple congenital anomalies and mental retardation and is associated with an interstitial deletion of chromosome 17p11.2 (Smith et al. 1986; Stratton et al. 1986; Greenberg et al. 1991, 1996; Chen et al. 1996). Features of patients with SMS include neurobehavioral abnormalities, such as aggressive and self-injurious behaviors, sleep disturbances, delayed speech and motor development, midface hypoplasia, short stature, and brachydactyly. The majority (>90%) of patients with SMS have a common ∼3.7-Mb deletion, as defined by a unique de novo junction fragment identified by pulsed-field gel electrophoresis (PFGE) (Greenberg et al. 1991; Guzzetta et al. 1992; Juyal et al. 1996; Chen et al. 1997; Bi et al. 2002). Physical mapping studies have shown that the SMS common deletion region is flanked by large (∼200 kb), highly homologous, low copy repeats (LCRs) (i.e., proximal and distal SMS repeats [SMS-REPs]) (Chen et al. 1997; Park et al. 2002). A third copy of inverted orientation, middle SMS-REP, has been identified within the SMS common deletion region (Chen et al. 1997; Park et al. 2002). SMS-REPs share ∼160 kb of >98% sequence identity (Park et al. 2002). Given the direct orientation and extent of homology between proximal and distal SMS-REPs, we have proposed that SMS-REPs act as substrates for nonallelic homologous recombination (NAHR), or unequal crossing over, resulting in deletions and duplications of the intervening chromosomal region (Chen et al. 1997).
LCRs flanking deletion breakpoints have been identified in other contiguous-gene syndromes, such as Williams-Beuren syndrome (WBS) (Pérez Jurado et al. 1996, 1998; Osborne et al. 1997), Prader-Willi/Angelman syndromes (PWS/AS) (Amos-Landgraf et al. 1999; Christian et al. 1999), DiGeorge/velocardiofacial syndromes (DGS/VCFS) (Edelmann et al. 1999a, 1999b), and neurofibromatosis type 1 (NF1) (Dorschner et al. 2000). In each case, the microdeletion is proposed to result from NAHR between the flanking LCRs (Urbán et al. 1996; Baumer et al. 1998; Amos-Landgraf et al. 1999; López Correa et al. 2000; Trost et al. 2000; reviewed by Inoue and Lupski [2002] and by Stankiewicz and Lupski [2002a and 2002b]).
Although it is anticipated that the reciprocal duplications may occur at the same frequency as deletions, only a few such reciprocal deletion/duplication syndromes have been reported. The best-characterized example is that of hereditary neuropathy with liability to pressure palsies (HNPP) and Charcot-Marie-Tooth disease type 1A (CMT1A). CMT1A is associated with a 1.4-Mb duplication that results from NAHR between highly homologous 24-kb LCRs in 17p12, termed “CMT1A-REPs,” whereas HNPP results from the reciprocal deletion (Pentao et al. 1992; Chance et al. 1994; Reiter et al. 1996; Inoue et al. 2001). PWS/AS commonly result from a 4-Mb deletion, with breakpoints mapping within LCRs on chromosome 15 (Christian et al. 1995; Amos-Landgraf et al. 1999). An apparent reciprocal duplication of 15q11-q13, between the common PWS/AS deletion breakpoints, has been identified in at least 13 patients to date (Browne et al. 1997; Repetto et al. 1998; Thomas et al. 1999; Roberts et al. 2002), although, as is the case with WBS, DGS/VCFS, or NF1, physical evidence of a predicted specific and recurrent junction fragment for a reciprocal duplication remains to be demonstrated.
Only a few patients with dup(17)(p11.2p11.2) have been ascertained. It is predicted that the incidence of dup(17)(p11.2p11.2) is equal to that of SMS (1:20,000), but duplications remain underdetected. This may result from an ascertainment bias, because patients with duplications usually exhibit a significantly milder phenotype than do patients with deletions, and G-banded duplications can be more difficult to identify than deletions. Alternatively, duplication gametes may be at a selective disadvantage. This discrepancy in ascertainment for predicted reciprocal duplications may be common to other contiguous gene deletion syndromes with breakpoints flanked by LCRs. Individuals with reciprocal duplications may not be ascertained, because they exhibit a relatively milder phenotype or are unaffected.
Elsewhere, we reported seven individuals with the duplication dup(17)(p11.2p11.2), the predicted reciprocal homologous recombination product of the common SMS deletion (Potocki et al. 2000). A unique junction fragment of the same apparent size was identified in all seven patients by use of PFGE, indicating a precise and recurrent recombination and further suggesting that the seven duplications were of the same size. Microsatellite analysis of one pedigree with dup(17)(p11.2p11.2) revealed that the SMS reciprocal duplication was due to unequal meiotic crossing over between the proximal and distal SMS-REPs. This led us to analyze additional pedigrees with dup(17)(p11.2p11.2) and to further investigate, by segregation of genetic markers, the hypothesis that unequal meiotic crossing over between proximal and distal SMS-REPs results in both dup(17)(p11.2p11.2) and the SMS microdeletion.
Subjects and Methods
Subjects
For our DNA analysis we collected 14 families of patients with SMS and six families of patients with dup(17)(p11.2p11.2). All patients with SMS met diagnostic criteria for SMS (Chen et al. 1996). Peripheral blood samples from patients and family members were obtained after informed consent. The presence of a deletion in patients with SMS was confirmed by FISH analysis, using probes specific for FLII (the human ortholog of Drosophila melanogaster flightless–I [fliI]) (Chen et al. 1995) and ZNF179 (Zhao et al. 1998)—both mapping within the SMS common deletion region—and using the peripheral myelin protein 22 gene, PMP22, mapping within the commonly duplicated CMT1A region (Patel et al. 1992), as a control.
PFGE was performed on the patient samples, as described elsewhere (Chen et al. 1997; Potocki et al. 2000), to determine whether their deletions and duplications represent the repeat-mediated common rearrangements. A deletion or duplication is considered common if the breakpoints map within the proximal and distal SMS-REPs. Coincidentally, common deletions and duplications are each distinguished by a unique de novo ∼1.1-Mb band corresponding to the SMS rearrangement-specific common junction fragment.
Two of the patients with dup(17)(p11.2p11.2) reported here (patients 1006 and 1364) were reported elsewhere (Potocki et al. 2000). However, at the time of that report, siblings were not available for allele phasing for either patient. In addition, the father of patient 1364 is deceased, and microsatellite analysis was previously uninformative.
Genotyping
We determined both the parental origin of the rearranged chromosomes and the recombination mechanism resulting in the deletion or duplication by microsatellite haplotype reconstruction and the segregation of marker genotypes, using genomic DNA purified from peripheral blood. Nine microsatellite markers were used to reconstruct haplotypes, including three within the common SMS region (D17S2256, D17S2257, and D17S805), three centromeric to the proximal SMS-REP (D17S842, D17S841, and D17S1871) and three telomeric to the distal SMS-REP (D17S955, D17S122, and D17S1857). Two novel microsatellite markers (D17S2256 and D17S2257) were developed on the basis of BAC genomic sequence available from the National Center for Biotechnology Information. D17S2256 represents a (TG)20 direct repeat within RP11-1084K4; D17S2257 represents a (CA)29 direct repeat within RP11-189D22. Oligonucleotide primer sequences flanking each microsatellite were designed using Primer3 (Whitehead Institute; sequences available at the Lupski Lab Web site) or obtained from the Genome Database. The 5′ ends of forward primers were end-labeled with fluorescent dyes of 6-FAM, TET, or HEX (Applied Biosystems). PCR was performed in a final volume of 20 μl containing genomic DNA (100 ng), 10× buffer (Qiagen), dNTPs (2.5 mM each; Invitrogen), primers (15 pmol each) and HotStarTaq (0.75 U; Qiagen). Initial denaturation was at 95°C for 15 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 55°–67°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were visualized by 2% agarose gel electrophoresis and were diluted 2–15 times, according to the band intensity. We mixed diluted PCR products (1.5 μl) with 3.5 μl formamide loading dye and TAMRA 500 standard (Applied Biosystems), and we performed electrophoresis on 5% denaturing polyacrylamide gel in the 377-96 DNA sequencer (Applied Biosystems). Sizes and relative intensities of the peaks were calculated by use of GENESCAN (v. 2.1) and GENOTYPER (v. 2.5) software (Applied Biosystems). Genotypes were analyzed according to the Manual of Linkage Mapping Set (Applied Biosystems). Phases of parental haplotypes were defined on the basis of the most parsimonious explanation for observed genotypes in the siblings and under the assumption of no recombination.
Results
Molecular Evidence for the Common-Size Deletion/Duplication of 17p11.2
To confirm the presence of a deletion or duplication, FISH analysis, using probes specific to the SMS common-deletion region, was performed on patient samples. Both FLII and ZNF179 map within the SMS common-deletion region and are deleted in all 14 patients with SMS, while all patients are not deleted for PMP22, suggesting the common deletion. The duplication in the six patients with dup(17)(p11.2p11.2) was determined, using FISH, to be tandem in orientation, as described elsewhere (Potocki et al. 2000). The majority of SMS deletions and reciprocal duplications have common breakpoints, although deletions and duplications of different sizes have been identified. PFGE was performed on patient samples to determine whether the deletion/duplication breakpoints were common. The ∼1.1-Mb SMS rearrangement-specific common junction fragment was identified in all patients with SMS and dup(17)(p11.2p11.2), indicating that all deletions/duplications were the common or predominant rearrangements.
Further Genetic Confirmation That dup(17)(p11.2p11.2) Occurs via Unequal Crossovers
We have previously reported seven patients with dup(17)(p11.2p11.2), five of which were paternal in origin, suggesting a potential parent-of-origin bias with the duplication (Potocki et al. 2000). To further investigate a potential bias, we determined the parent of origin in one uninformative patient included in the group reported elsewhere (Potocki et al. 2000) and in four additional families of patients with dup17(p11.2p11.2). We have collected siblings of patients 1006 and 1364, who were reported elsewhere (Potocki et al. 2000), to provide additional evidence of unequal crossovers in those families. The parental origin of the duplication is made evident by duplication of one or both of one parent's alleles for loci D17S2256, D17S2257, and D17S805. Four of the six duplicated chromosomes were of paternal origin, and the remaining two duplications were maternally derived (figs. 1 and 2; table 1). To determine whether a parent-of-origin bias exists, the data from five families of patients with dup(17)(p11.2p11.2) reported elsewhere (Potocki et al. 2000) were combined with the present data. In total, 8 of the 11 duplications were paternally derived, and three were maternal in origin. The binomial distribution was applied to these data and the apparent parent-of-origin bias was not found to be significant (P=.227).
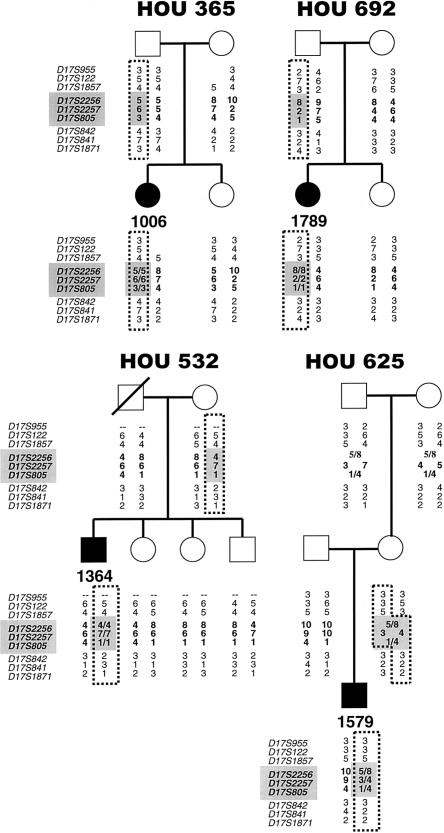
Figure 1.
Haplotypes of four patients with dup(17)(p11.2p11.2) and their families. Blackened symbols indicate affected individuals. To the left of each pedigree is a list of microsatellite markers used for genotyping; those within the SMS common-deletion region are shaded and in bold. The allele numbers are located under each family member. The genotypes of markers within the SMS common-deletion region are in bold and are shaded in each patient and the parent of origin. The dotted lines outline alleles inherited by the patient from the parent of origin. Loci D17S955 and D17S122 are deleted in the mother and patient in family HOU 365. D17S955 was not informative for family HOU 532. (The allele numbers for HOU 365 and HOU 532 that were reported elsewhere [Potocki et al. 2000] were changed to remain consistent with those in the present report.)
Figure 2.
Pedigrees and genotype plots of two dup(17)(p11.2p11.2) families and one SMS family. A, An example of paternal interchromosomal recombination resulting in a duplication. Pedigree and genotype plots of family HOU 660, showing inheritance by patient 1618 of both alleles from his father and one allele from his mother for marker D17S2256. Peaks labeled 213, 215, 217 and 225 correspond to alleles 4, 5, 6 and 10, respectively, in the pedigree. B, dup(17)(p11.2p11.2) patient 1913 who inherited two copies of her father’s allele and one copy of her mother’s allele for marker D17S805, demonstrating a paternal intrachromosomal event leading to duplication. Peaks labeled 223, 231 and 233 correspond to alleles 1, 4 and 5, respectively, in the pedigree. C, Genotype plots and pedigree of family HOU 421 showing inheritance by patient 1144 of one allele from his mother and none from his father for marker D17S2256. Peaks and alleles are labeled as in panel A. There is a crossover between D17S1857 and D17S2256 on the patient’s maternal chromosome.
Table 1.
Origin and Mechanism of dup(17)(p11.2p11.2) Rearrangement[Note]
|
Recombination Mechanism |
|||
| ParentalOrigin | Interchromosomal | Intrachromosomal | Unknown |
| Maternal | 2 | 1 | 0 |
| Paternal | 4 | 4 | 0 |
Note.— Includes data from the article by Potocki et al. (2000).
Unequal crossovers between the proximal and distal SMS-REPs were apparent in one informative family with dup(17)(p11.2p11.2) reported by Potocki et al. (2000). To further confirm the duplication is due to unequal crossing over, we analyzed segregation of marker haplotypes in the current six families with dup17(p11.2p11.2). In total, two duplications occurred via interchromosomal recombination and four were a result of intrachromosomal exchange (figs. 1 and 2; table 1). Paternal interchromosomal recombination was demonstrated in one family (HOU 660) by the presence of two distinct paternal alleles and one maternal allele for markers D17S2256, D17S2257, and D17S805 (fig. 2). Paternal intrachromosomal recombination was indicated by the presence of a higher-intensity peak, corresponding to a double dose of one of the paternal alleles for markers D17S2256, D17S2257, and D17S805 in three families (HOU 365, HOU 692, and HOU 724) (figs. 1 and 2). One maternal duplication resulted from interchromosomal recombination (HOU 625), and one duplication arose from maternal intrachromosomal exchange (HOU 532) (fig. 1). Patient 1006 (HOU 365) has an apparent crossover on her maternal chromosome, between markers D17S842 and D17S1871, and loci D17S955 and D17S122 are deleted, as they are in her mother. The PMP22 gene was deleted in both individuals, and both display a phenotype consistent with HNPP (Potocki et al. 1999). Locus D17S955 was uninformative in family HOU 532. The haplotype of the deceased father is inferred from those of his children. A sister of patient 1364 has an apparent crossover on her paternal chromosome, between markers D17S1857 and D17S2256, and patient 1618 (family HOU 660; fig. 2) has a crossover on his maternal chromosome between loci D17S2257 and D17S842. Phasing of the patient, maternal, and grandparental chromosomes was not possible for markers D17S2256 and D17S805 in family HOU 625.
Genetic Evidence That Unequal Crossovers Generate del(17)(p11.2p11.2)
Because of the paucity of genetic markers between the proximal SMS-REP and the centromere, previous haplotype analysis of families with SMS has not been performed. Here we have determined the parent-of-origin and recombination mechanism of the SMS deletion in 14 families. All of the patients analyzed were hemizygous for markers D17S2256, D17S2257, and D17S805, located within the common SMS deletion region (figs. 2 and 3). Eight of the rearranged chromosomes were of paternal origin, as is demonstrated by the absence of maternal alleles for markers D17S2256, D17S2257, and D17S805, and six of the deletions were maternal in origin (figs. 2 and 3; table 2).
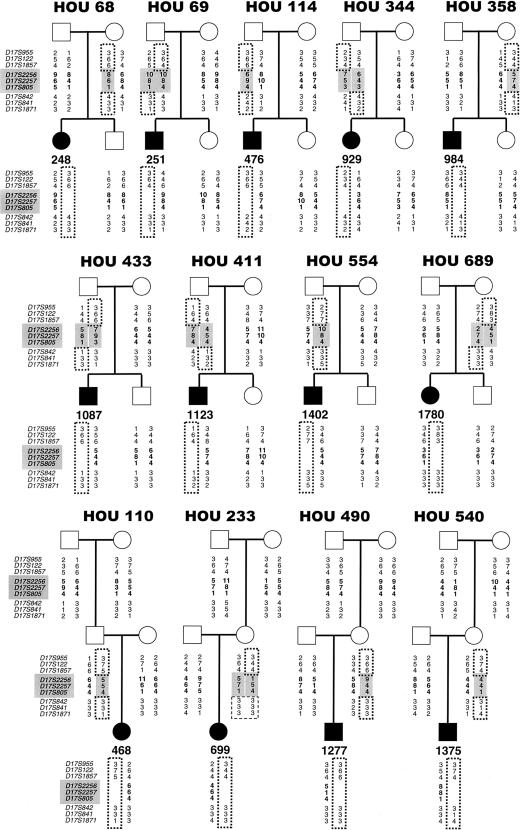
Figure 3.
Haplotypes of 13 patients with SMS and their families. The markers within the SMS common-deletion region are shaded and in bold, and their genotypes are shaded and in bold in the parents of origin and are in bold in the patients. The dotted lines outline alleles inherited by the patient from the parent of origin. The dashed lines in pedigree HOU 233 indicate that the deletion mechanism could not be determined for patient 699.
Table 2.
Origin and Mechanism of del(17)(p11.2p11.2) Rearrangement
|
Recombination Mechanism |
|||
| ParentalOrigin | Interchromosomal | Intrachromosomal | Unknown |
| Maternal | 1 | 4 | 1 |
| Paternal | 4 | 4 | 0 |
Unequal crossing over between the proximal and distal SMS-REPs was obvious in 13 of 14 patients, as evidenced by absence of a parental allele for each of three genetic markers (D17S2256, D17S2257, and D17S805) that map within the common-deletion region. Four of the paternal deletions were due to intrachromosomal exchange (HOU 110, HOU 114, HOU 421, and HOU 554), and four were interchromosomal (HOU 69, HOU 344, HOU 411, and HOU 433) (figs. 2 and 3; table 2). An interchromosomal recombination event was demonstrated by recombination between the markers directly flanking the SMS common-deletion region. Four of the maternal deletions were intrachromosomal (HOU 68, HOU 358, HOU 490, and HOU 540), and one was interchromosomal (HOU 689) (fig. 3 and table 2). The recombination mechanism (i.e., inter- vs. intrachromosomal) that resulted in the remaining maternal deletion (HOU 233), was not obvious. Patient 1402 has an apparent crossover on his maternal chromosome between markers D17S122 and D17S1857, and patient 1144 has an apparent crossover on his maternal chromosome between markers D17S1857 and D17S2256.
Discussion
One patient with dup(17)(p11.2p11.2) whom we have reported elsewhere (Potocki et al. 2000) demonstrated unequal crossing over between the proximal and distal SMS-REPs, resulting in a paternal interchromosomal duplication. Here, we provide further evidence supporting unequal crossing over in two families from that study (HOU 365 and HOU 532) and four additional families with dup(17)(p11.2p11.2). Of the 11 combined cases (7 from Potocki et al. [2000] and 4 from the present report), 6 resulted from interchromosomal recombination, and 5 were intrachromosomal, indicating that duplication occurs via both mechanisms at an approximately equal frequency (table 1). Likewise, we provide evidence showing that 5 of the 14 SMS deletions occurred via interchromosomal recombination and that 8 resulted from an intrachromosomal event (table 2). Since the SMS deletion and dup(17)(p11.2p11.2) represent reciprocal recombination events resulting from unequal crossing over between proximal and distal SMS-REPs, the data presented here for both can be combined. Of the 24 patients with informative SMS/dup(17)(p11.2p11.2), 11 rearrangements were interchromosomal, and 13 were intrachromosomal, further supporting the hypothesis that NAHR between proximal and distal SMS-REPs occurs as frequently between or within sister chromatids as between homologs.
Potocki et al. (2000) reported six patients with dup(17)(p11.2p11.2) in which the parental origin could be determined, and five of these were paternal in origin. This observation led to the hypothesis that there may be a parent-of-origin bias associated with this duplication. However, when these earlier data were pooled together with data reported here, 8 of 11 duplications were demonstrated to be paternal, and 3 were maternal (table 1). Thus, although a trend is evident (with eight paternal vs. three maternal duplications), these data do not support the significant association of a parent-of-origin bias with dup(17)(p11.2p11.2) (P=.227). Similarly, eight SMS deletions were paternal, and six were maternal in origin. The combined SMS and dup(17)(p11.2p11.2) parent-of-origin data show 16 paternal rearrangements and 9 of maternal origin, further indicating that unequal crossovers between proximal and distal SMS-REPs may occur as frequently on the maternal chromosome as the paternal chromosome (P=.230).
We show that rearrangements of maternal and paternal origin each occur via both inter- and intrachromosomal unequal crossing over; thus, our data provide no evidence for a significant association of sex-biased recombination mechanisms with SMS/dup(17)(p11.2p11.2). This finding contrasts with similar studies performed on the reciprocal deletion/duplication syndromes HNPP and CMT1A, in which all informative paternal CMT1A duplications (n=32) resulted from unequal interchromosomal crossing over between CMT1A-REPs, whereas the few informative maternal duplications (n=2) and HNPP deletions (n=2) resulted from intrachromosomal recombination (Lopes et al. 1997, 1998). The de novo CMT1A duplication event has been found to occur 10 times more frequently in male than in female patients (Palau et al. 1993; Lopes et al. 1997). Interestingly, genetic mapping in CEPH reference families reveals that male patients exhibit a lower recombination frequency in the CMT1A region than do female patients (.67cM/Mb vs. 5.5cM/Mb) (Inoue et al. 2001). This reduced male recombination frequency was hypothesized to result in an extended region of unsynapsed chromosome segments in meiosis, enabling the chromosomes to slip on each other, thus predisposing to unequal crossovers between misaligned CMT1A-REPs. Thus, reduced recombination has been proposed to potentially enable an increase of unequal crossovers and therefore may explain the high frequency of paternally derived duplications in CMT1A. On the basis of a comparison between the genetic and physical maps of 17p11.2, we reported elsewhere that male and female patients exhibit a reduced but equal rate of recombination in the SMS region (Bi et al. 2002). Therefore, the absence of a parent-of-origin bias for the reciprocal SMS del/dup(17)(p11.2p11.2) could still be consistent with a “reduced recombination/increased unequal crossover” hypothesis. Indeed, SMS deletions occur at a relatively equal frequency in male and female patients.
The molecular mechanisms resulting in several other chromosome deletions and duplications that cause contiguous gene syndromes have been reported elsewhere; DGS/VCFS, NF1, and PWS/AS deletions occur via both intra- and interchromosomal events (Carrozzo et al. 1997; Baumer et al. 1998; Robinson et al. 1998; Edelmann et al. 1999a; López Correa et al. 2000; Trost et al. 2000), whereas the majority of WBS deletions are due to interchromosomal recombination (Urbán et al. 1996). The parental origins of these rearrangements also have been investigated. The majority of NF1 deletions are maternal in origin (Lázaro et al. 1996), as are reciprocal duplications of the PWS/AS critical region (Browne et al. 1997; Repetto et al. 1998; Thomas et al. 1999; Roberts et al. 2002), but no consistent sex bias has been detected in WBS (Urbán et al. 1996) or DGS/VCFS (Trost et al. 2000).
Several chromosome deletions have been noted to be recurrent and to occur with a higher frequency in specific regions of the human genome. The breakpoints of such rearrangements, including all of those mentioned above, have been found to fall within recombination-prone LCR regions (Lopes et al. 1996; Chen et al. 1997; Lupski 1998; Pérez Jurado et al. 1998; Amos-Landgraf et al. 1999; Edelmann et al. 1999b; López Correa et al. 2000; Potocki et al. 2000). Since duplications are predicted to occur at the same frequency as deletion events resulting from LCR-mediated unequal crossing over, it remains a distinct possibility that most microdeletion syndromes have corresponding microduplication syndromes that represent the reciprocal recombination product.
In summary, our data provide genetic evidence that the SMS deletion and reciprocal duplication (17)(p11.2p11.2) occur by unequal crossing over, presumably mediated through NAHR between the proximal and distal SMS-REPs. These chromosomal rearrangements occur on paternal and maternal chromosomes and arise from both inter- and intrachromosomal exchange events. The present study provides further evidence for a model in which reciprocal deletion and duplication syndromes arise from unequal crossing over between LCRs.
Acknowledgments
We thank the patients with SMS and dup(17)(p11.2p11.2) and their families for their participation in this study. We also thank Dr. Lorraine Potocki, for procurement of samples, Dr. Pawel Stankiewicz, for helpful discussion, and Dr. E. O'Brian Smith, for assistance with statistical analyses. This work has been supported in part by National Institute of Child Health and Development grant P01 HD39420, National Institutes of Health (NIH) grant HD2406407 to the Baylor College of Medicine Mental Retardation Research Center, and NIH grant M01 RR00188 to the Texas Children's Hospital General Clinical Research Center.
Electronic-Database Information
Accession number and URLs for data presented herein are as follows:
- Genome Database, The, http://gdbwww.gdb.org/
- Lupski Lab Web site, http://imgen.bcm.tmc.edu/molgen/lupski/
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SMS [MIM 182290]) [PubMed]
- Whitehead Institute, http://www-genome.wi.mit.edu/
References
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer A, Dutly F, Balmer D, Riegel M, Tükel T, Krajewska-Walasek M, Schinzel AA (1998) High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum Mol Genet 7:887–894 [DOI] [PubMed] [Google Scholar]
- Bi W, Yan J, Stankiewicz P, Park S-S, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJM, Inoue K, Lupski JR (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne CE, Dennis NR, Maher E, Long FL, Nicholson JC, Sillibourne J, Barber JCK (1997) Inherited interstitial duplications of proximal 15q: genotype-phenotype correlations. Am J Hum Genet 61:1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozzo R, Rossi E, Christian SL, Kittikamron K, Livieri C, Corrias A, Pucci L, Fois A, Simi P, Bosio L, Beccaria L, Zuffardi O, Ledbetter DH (1997) Inter- and intrachromosomal rearrangements are both involved in the origin of 15q11-q13 deletions in Prader-Willi syndrome. Am J Hum Genet 61:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR (1994) Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3:223–228 [DOI] [PubMed] [Google Scholar]
- Chen K-S, Gunaratne PH, Hoheisel JD, Young IG, Gabor Miklos GL, Greenberg F, Shaffer LG, Campbell HD, Lupski JR (1995) The human homologue of the Drosophila melanogaster flightless-I gene (fliI) maps within the Smith-Magenis microdeletion critical region in 17p11.2. Am J Hum Genet 56:175–182 [PMC free article] [PubMed] [Google Scholar]
- Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Chen K-S, Potocki L, Lupski JR (1996) The Smith-Magenis syndrome [del(17)p11.2]: clinical review and molecular advances. Ment Retard Dev Disabil Res Rev 2:122–129 [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH (1999) Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 8:1025–1037 [DOI] [PubMed] [Google Scholar]
- Christian SL, Robinson WP, Huang B, Mutirangura A, Line MR, Nakao M, Surti U, Chakravarti A, Ledbetter DH (1995) Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. Am J Hum Genet 57:40–48 [PMC free article] [PubMed] [Google Scholar]
- Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K (2000) NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet 9:35–46 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE (1999a) Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet 64:1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RSK, Magenis E, Shprintzen RJ, Morrow BE (1999b) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 [DOI] [PubMed] [Google Scholar]
- Greenberg F, Guzzetta V, Montes de Oca-Luna R, Magenis RE, Smith ACM, Richter SF, Kondo I, Dobyns WB, Patel PI, Lupski JR (1991) Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(17)(p11.2). Am J Hum Genet 49:1207–1218 [PMC free article] [PubMed] [Google Scholar]
- Greenberg F, Lewis RA, Potocki L, Glaze D, Parke J, Killian J, Murphy MA, Williamson D, Brown F, Dutton R, McCluggage C, Friedman E, Sulek M, Lupski JR (1996) Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2). Am J Med Genet 62:247–254 [DOI] [PubMed] [Google Scholar]
- Guzzetta V, Franco B, Trask BJ, Zhang H, Saucedo-Cardenas O, Montes de Oca-Luna R, Greenberg F, Chinault AC, Lupski JR, Patel PI (1992) Somatic cell hybrids, sequence-tagged sites, simple repeat polymorphisms, and yeast artificial chromosomes for physical and genetic mapping of proximal 17p. Genomics 13:551–559 [DOI] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B (2001) The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res 11:1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR (2002) Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3:199–242 [DOI] [PubMed] [Google Scholar]
- Juyal RC, Figuera LE, Hauge X, Elsea SH, Lupski JR, Greenberg F, Baldini A, Patel PI (1996) Molecular analyses of 17p11.2 deletions in 62 Smith-Magenis syndrome patients. Am J Hum Genet 58:998–1007 [PMC free article] [PubMed] [Google Scholar]
- Lázaro C, Gaona A, Ainsworth P, Tenconi R, Vidaud D, Kruyer H, Ars E, Volpini V, Estivill X (1996) Sex differences in mutational rate and mutational mechanism in the NF1 gene in neurofibromatosis type 1 patients. Hum Genet 98:696–699 [DOI] [PubMed] [Google Scholar]
- Lopes J, LeGuern E, Gouider R, Tardieu S, Abbas N, Birouk N, Gugenheim M, Bouche P, Agid Y, Brice A (1996) Recombination hot spot in a 3.2-kb region of the Charcot-Marie-Tooth type 1A repeat sequences: new tools for molecular diagnosis of hereditary neuropathy with liability to pressure palsies and of Charcot-Marie-Tooth type 1A. French CMT Collaborative Research Group. Am J Hum Genet 58:1223–1230 [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Ravisé N, Vandenberghe A, Palau F, Ionasescu V, Mayer M, Lévy N, Wood N, Tachi N, Bouche P, Latour P, Ruberg M, Brice A, LeGuern E (1998) Fine mapping of de novo CMT1A and HNPP rearrangements within CMT1A-REPs evidences two distinct sex-dependent mechanisms and candidate sequences involved in recombination. Hum Mol Genet 7:141–148 [DOI] [PubMed] [Google Scholar]
- Lopes J, Vandenberghe A, Tardieu S, Ionasescu V, Lévy N, Wood N, Tachi N, Bouche P, Latour P, Brice A, LeGuern E (1997) Sex-dependent rearrangements resulting in CMT1A and HNPP. Nat Genet 17:136–137 [DOI] [PubMed] [Google Scholar]
- López Correa C, Brems H, Lázaro C, Marynen P, Legius E (2000) Unequal meiotic crossover: a frequent cause of NF1 microdeletions. Am J Hum Genet 66:1969–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR (1998) Charcot-Marie-Tooth disease: lessons in genetic mechanisms. Mol Med 4:3–11 [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Herbrick J-A, Greavette T, Heng HHQ, Tsui L-C, Scherer SW (1997) PMS2-related genes flank the rearrangement breakpoints associated with Williams syndrome and other diseases on human chromosome 7. Genomics 45:402–406 [DOI] [PubMed] [Google Scholar]
- Palau F, Löfgren A, De Jonghe P, Bort S, Nelis E, Sevilla T, Martin J-J, Vilchez J, Prieto F, Van Broeckhoven C (1993) Origin of the de novo duplication in Charcot-Marie-Tooth disease type 1A: unequal nonsister chromatid exchange during spermatogenesis. Hum Mol Genet 2:2031–2035 [DOI] [PubMed] [Google Scholar]
- Park S-S, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR (2002) Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res 12:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PI, Roa BB, Welcher AA, Schoener-Scott R, Trask BJ, Pentao L, Snipes GJ, Garcia CA, Francke U, Shooter EM, Lupski JR, Suter U (1992) The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat Genet 1:159–165 [DOI] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR (1992) Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet 2:292–300 [DOI] [PubMed] [Google Scholar]
- Pérez Jurado LA, Peoples R, Kaplan P, Hamel BCJ, Francke U (1996) Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am J Hum Genet 59:781–792 [PMC free article] [PubMed] [Google Scholar]
- Pérez Jurado LA, Wang Y-K, Peoples R, Coloma A, Cruces J, Francke U (1998) A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Hum Mol Genet 7:325–334 [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Koeuth T, Killian J, Iannaccone ST, Shapira SK, Kashork CD, Spikes AS, Shaffer LG, Lupski JR (1999) DNA rearrangements on both homologues of chromosome 17 in a mildly delayed individual with a family history of autosomal dominant carpal tunnel syndrome. Am J Hum Genet 64:471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Park S-S, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, Shaffer LG, Lupski JR (2000) Molecular mechanism for duplication 17p11.2: the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet 24:84–87 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 [DOI] [PubMed] [Google Scholar]
- Repetto GM, White LM, Bader PJ, Johnson D, Knoll JHM (1998) Interstitial duplications of chromosome region 15q11q13: clinical and molecular characterization. Am J Med Genet 79:82–89 [DOI] [PubMed] [Google Scholar]
- Roberts SE, Dennis NR, Browne CE, Willatt L, Woods CG, Cross I, Jacobs PA, Thomas NS (2002) Characterisation of interstitial duplications and triplications of chromosome 15q11-q13. Hum Genet 110:227–234 [DOI] [PubMed] [Google Scholar]
- Robinson WP, Dutly F, Nicholls RD, Bernasconi F, Peñaherrera M, Michaelis RC, Abeliovich D, Schinzel AA (1998) The mechanisms involved in formation of deletions and duplications of 15q11-q13. J Med Genet 35:130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ACM, McGavran L, Robinson J, Waldstein G, Macfarlane J, Zonona J, Reiss J, Lahr M, Allen L, Magenis E (1986) Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am J Med Genet 24:393–414 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002a) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 [DOI] [PubMed] [Google Scholar]
- ——— (2002b) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- Stratton RF, Dobyns WB, Greenberg F, DeSana JB, Moore C, Fidone G, Runge GH, Feldman P, Sekhon GS, Pauli RM, Ledbetter DH (1986) Interstitial deletion of (17)(p11.2p11.2): report of six additional patients with a new chromosome deletion syndrome. Am J Med Genet 24:421–432 [DOI] [PubMed] [Google Scholar]
- Thomas NS, Browne CE, Oley C, Healey S, Crolla JA (1999) Investigation of a cryptic interstitial duplication involving the Prader-Willi/Angelman syndrome critical region. Hum Genet 105:384–387 [DOI] [PubMed] [Google Scholar]
- Trost D, Wiebe W, Uhlhaas S, Schwindt P, Schwanitz G (2000) Investigation of meiotic rearrangements in DGS/VCFS patients with a microdeletion 22q11.2. J Med Genet 37:452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbán Z, Helms C, Fekete G, Csiszár K, Bonnet D, Munnich A, Donis-Keller H, Boyd CD (1996) 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. Am J Hum Genet 59:958–962 [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Chen K-S, Bejjani BA, Lupski JR (1998) Cloning, genomic structure, and expression of mouse ring finger protein gene Znf179. Genomics 49:394–400 [DOI] [PubMed] [Google Scholar]