Abstract
Background:
Genes with recurrent codon-specific somatic mutations are likely drivers of tumorigenesis and potential therapeutic targets. Hypermutable cancers may represent a sensitive system for generation and selection of oncogenic mutations.
Methods:
We utilised exome-sequencing data on 25 sporadic microsatellite-instable (MSI) colorectal cancers (CRCs) and searched for base-specific somatic mutation hotspots.
Results:
We identified novel mutation hotspots in 33 genes. Fourteen genes displayed mutations in the validation set of 254 MSI CRCs: ANTXR1, MORC2, CEP135, CRYBB1, GALNT9, KRT82, PI15, SLC36A1, CNTF, GLDC, MBTPS1, OR9Q2, R3HDM1 and TTPAL. A database search found examples of the hotspot mutations in multiple cancer types.
Conclusions:
This work reveals a variety of new recurrent candidate oncogene mutations to be further scrutinised as potential therapeutic targets.
Keywords: oncogene, mutation hotspot, microsatellite instability, colorectal cancer, exome sequencing
The functional effect of somatic mutations can sometimes be predicted from mutation patterns. Inactivating mutations often truncate the protein product, and frequent occurrence of such changes in a gene is a sign of a possible tumour suppressor function, as exemplified by the APC gene in colorectal cancer (CRC; Fearon, 2011). The most pathognomonic feature of mutations activating a gene product is recurrent occurrence in one or few codons. Such a signal is a strong indicator of selective value. Importantly, these types of changes also provide possibilities for therapeutic approaches through tackling the abnormal activity of the specific mutant protein product. Examples of successful utilisation of such a strategy include, but are not limited to, inhibitors of BRAF Val600Glu and activated EGFR (Lynch et al, 2004; Bollag et al, 2010). This success is in many cases marred by the fact that targeted monotherapies often fail to cure a patient because of pre-existing resistant subclones. Whereas the most common mutation hotspots may at present be largely known, any additional hotspots would provide a valuable platform for cancer biology in pinpointing relevant genes and domains to study, as well as for development of potentially curative combination therapies for stratified cancer care.
Colorectal cancer with microsatellite instability (MSI) defines a subgroup of CRCs with distinct clinical characteristics. These hypermutable CRCs may represent a sensitive system for generation, selection and detection of mutation hotspots. Indeed, in MSI CRC multiple well-established oncogenes, in particular BRAF, KRAS, PIK3CA and CTNNB1, are known to display very specific mutation hotspots (Fearon, 2011).
We have previously mined exome-sequencing data of 25 MSI CRCs for hotspot point mutations (Gylfe et al, 2013). That study did not consider identical base-specific mutations in the discovery set. The rationale for exclusion of this important mutation type was that such mutation signals, according to our early experience with exome data, typically arose from uncalled polymorphic germline variants. Expansion of control data as well as developments in analysis pipeline currently enable much more robust calling of somatic mutations. Thus, we have here taken the study forward to search for novel recurrent somatic cancer mutations hitting exactly the same base position.
Materials and methods
Sample material
The discovery set (Supplementary Table S1) was composed of normal and fresh-frozen tumour tissue DNA samples of 25 sporadic MSI patients gathered from a population-based series of 1042 colorectal adenocarcinoma patients described previously (Aaltonen et al, 1998, Salovaara et al, 2000). The validation set consisted of 167 Finnish samples (42 Lynch syndrome and 125 sporadic tumours, Supplementary Table S2) obtained from the above-mentioned cohort or from a subsequently collected series of additional 472 Finnish CRCs (unpublished data), and 87 Danish MSI CRCs (Supplementary Table S3). In addition, DNA from 10 CRC cell lines (VACO5, CCL231, GP5D, HCA7, HCT-116, LOVO, LS174T, RKO, SNUC2B and DLD1) was available. The study has been reviewed and approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa, Finland. Either a signed informed consent or authorisation from the National Supervisory for Welfare and Health was obtained for all the samples.
Exome and whole-genome sequencing
Exomic regions from 25 MSI N/T pairs were captured with the Agilent SureSelect all exon kit v1 (Agilent, Santa Clara, CA, USA). Paired-end short-read sequencing was carried out with Illumina Genome Analyzer II machines (Illumina, San Diego, CA, USA) at the Karolinska Institutet, or the Finnish Institute for Molecular Medicine (FIMM) Genome and Technology Center, Finland. Details on read mapping and variant calling are described in the previous publication (Gylfe et al, 2013). Whole-genome sequencing (WGS) on 91 MSS CRCs and the matched normal samples was performed on the Illumina HiSeq 2000 platform with paired-end reads 100 bp in length (median coverage of at least × 40). Sequencing data analysis is described in detail in Pitkanen et al (2014).
Identification and validation of the mutation hotspots
An in-house comparative analysis tool, RikuRator (Katainen, 2013, http://hdl.handle.net/10138/39539), was used to visualise and analyse the exome data. The criterion for the mutation hotspot was that the same base had to be mutated in at least two tumours. All the hotspot mutations identified in the discovery set were validated by Sanger sequencing (N/T pairs). The confirmed somatic hotspot mutations were further sequenced in an additional set of 254 tumours and somatic origin of the detected mutations was confirmed by sequencing the respective normal DNA samples. The novel hotspot mutations that became validated (14 genes) were then screened in 10 CRC cell lines. See Supplementary Data for details.
Computational methods
Ensembl Variant Effect Predictor, including SIFT (http://sift.jcvi.org/) and PolyPhen (http://genetics.bwh.harvard.edu/pph2/), was used to predict the functional consequences of the variants (http://www.ensembl.org). Multispecies protein sequence alignments were performed with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Results
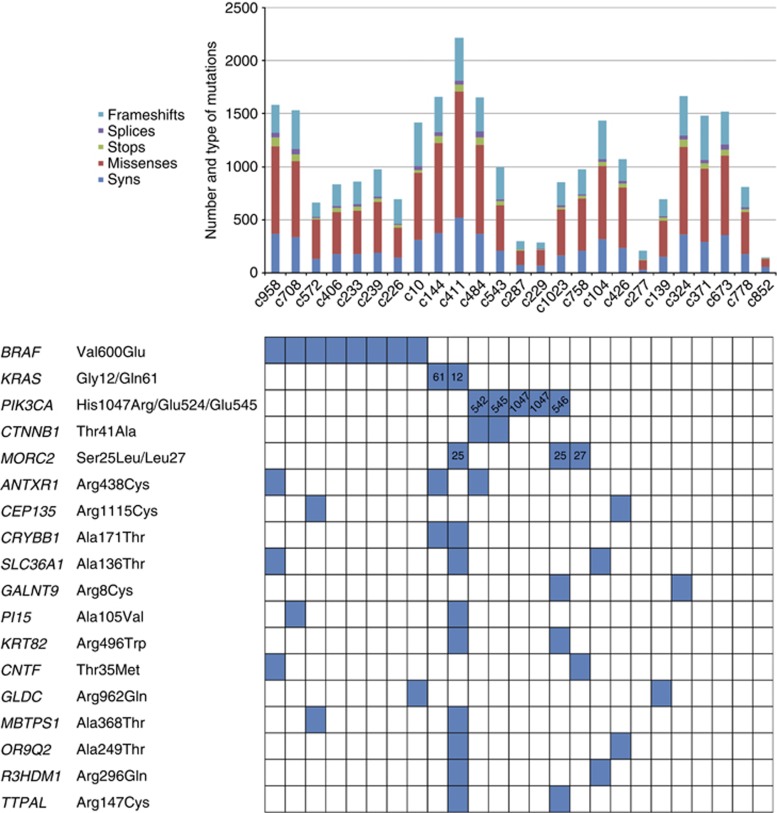
Exome-sequencing data of 25 sporadic MSI CRCs (Supplementary Table S1) were used as a discovery set to identify recurrent somatic missense mutations. Detailed information about the quality of the data can be found in our previous publication (Gylfe et al, 2013). Across all samples, 27 879 somatic coding mutations were identified, of which 21 167 were nonsynonymous and 5734 were synonymous (Figure 1). The most frequent single-nucleotide mutation types were C:G>T:A (48%), C:G>A:T (32%) and T:A>C:G (11%) substitutions, with the rest of the substitutions C:G>G:C (3.2%), T:A>A:T (2.9%) and T:A>G:C (2.4%) comprising only less than 9% of all mutations. This is a typical mutation spectrum in MSI CRCs (Alexandrov et al, 2013). We next identified somatic mutations hitting exactly the same base in two or more cancers. This produced a list of 42 genes with possible heterozygous somatic hotspot mutations of which mutations in 36 genes were confirmed to be real as well as somatic. Among these were previously known hotspot mutations in BRAF, PIK3CA and CTNNB1 (Table 1). Novel somatic mutation hotspots were found in 33 genes (Supplementary Table S4). The list of validated hotspot mutations was dominated by transition changes at CpG dinucleotides (32/33, 97%). When compared with the exome data, C:G>T:A mutation signature was overrepresented in the hotspots (97% vs 48%, P=5.33e-6, Fisher's test). The distribution of the mutations in the discovery set tumours is presented in Figure 1.
Figure 1.
Pattern of mutations in the discovery set of 25 MSI CRCs.
Per-sample mutation rates and types are presented on the top. Distribution of the hotspots in the discovery set tumours is depicted with blue boxes for both the previously known and 14 novel candidate oncogenes (bottom).
Table 1. Genes with validated somatic hotspot mutations.
| |
|
|
|
Discovery set |
Validation set |
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Genome position | cDNA change | Protein change | n out of 25 | % | Finnish n=167 (%) | Danish n=87 (%) | Total n=254 | % | COSMIC |
|
BRAF |
chr7:140453136 |
c.1799T>A |
Val600Glu |
8 |
32 |
68/167 (41) |
36/82 (44) |
104/249 |
42 |
COSM476 |
|
KRAS |
chr12:25398285-4,25398282 |
c.34G–5G, c.37G>T |
Gly12/Gly13Asp |
1 |
4 |
25/165 (15) |
13/82 (16) |
38/247 |
15 |
See COSMIC database |
|
PIK3CA ex20 |
chr3:178952085 |
c.3140A>G |
His1047Arg |
2 |
8 |
15/139 (11) |
7/68 (10) |
22/207 |
11 |
COSM775 |
|
PIK3CA ex9 |
chr3:178936082-3/178936091-2/178936094-6 |
c.1624G–5A/c.1633G–4A/c.1636C–8G |
Glu542/Glu545/Gln546 |
3 |
12 |
8/137 (6) |
4/80 (5) |
12/217 |
5.5 |
See COSMIC database |
|
CTNNB1 |
chr3:41266124 |
c.121A>G |
Thr41Ala |
2 |
8 |
11/140 (7.9) |
4/76 (5.3) |
15a/216 |
6.9 |
COSM5664 |
|
ANTXR1 |
chr2:69409751 |
c.1312C>T |
Arg438Cys |
3 |
12 |
4/165 (2.4) |
3/86 (3.5) |
7/251 |
2.8 |
COSM1641964 |
|
CEP135 |
chr4:56890689 |
c.3343C>T |
Arg1115Cys |
2 |
8 |
2/165 (1.2) |
1/80 (1.3) |
3/245 |
1.2 |
|
|
CRYBB1 |
chr22:26997907 |
c.511G>A |
Ala171Thr |
2 |
8 |
1/163 (0.6) |
2/85 (2.4) |
3/248 |
1.2 |
|
|
MORC2 |
chr22:31345795/31345789 |
c.74C>T/c.80A>T |
Ser25Leu/Lys27Met |
3b |
12 |
2c/163 (1.2) |
0/81 (0) |
2/244 |
0.8 |
COSM184853 (Lys27Arg) |
|
SLC36A1 |
chr5:150844717 |
c.406G>A |
Ala136Thr |
3 |
12 |
2/166 (1.2) |
0/87 (0) |
2/253 |
0.8 |
COSM420825 |
|
GALNT9 |
chr12:132688193 |
c.22C>T |
Arg8Cys |
2 |
8 |
2d/165 (1.2) |
0/73 (0) |
2/238 |
0.8 |
|
|
PI15 |
chr8:75756256 |
c.314C>T |
Ala105Val |
2 |
8 |
1e/161 (0.6) |
1/86 (1.2) |
2/247 |
0.8 |
|
|
KRT82 |
chr12:52788815 |
c.1486C>T |
Arg496Trp |
2 |
8 |
0/165 (0) |
1/87 (1.1) |
1/252 |
0.4 |
COSM468531 |
|
CNTF |
chr11:58390329 |
c.104C>T |
Thr35Met |
2 |
8 |
0/164 (0) |
1/87 (1.1) |
1/251 |
0.4 |
|
|
GLDC |
chr9:6556244 |
c.2885G>A |
Arg962Gln |
2 |
8 |
0/164 (0) |
1/85 (1.2) |
1/249 |
0.4 |
|
|
MBTPS1 |
chr16:84120995 |
c.1102G>A |
Ala368Thr |
2 |
8 |
1/166 (0.6) |
0/87 (0) |
1/253 |
0.4 |
|
|
OR9Q2 |
chr11:57958707 |
c.745G>A |
Ala249Thr |
2 |
8 |
1/165 (0.6) |
0/86 (0) |
1/251 |
0.4 |
COSM1218891 |
|
R3HDM1 |
chr2:136393737 |
c.887G>A |
Arg296Gln |
2 |
8 |
1/165 (0.6) |
0/86 (0) |
1/251 |
0.4 |
|
| TTPAL | chr20:43109078 | c.439C>T | Arg147Cys | 2 | 8 | 1/166 (0.6) | 0/86 (0) | 1/252 | 0.4 | COSM32271 |
7 × Ser45Pro/Ser45Phe.
2 × Ser25Leu, 1 × Lys27Met.
Lys27Arg, Ser25Leu.
1 × Arg8Ser, 1 × Arg8His.
Ala105Thr.
Next, the 33 recurrent somatic missense mutations, as well as known hotpots in BRAF, KRAS, PIK3CA and CTNNB1, were screened in a validation set of 254 MSI CRCs (Supplementary Tables S2 and S3). Fourteen of the novel genes displayed hotspot mutations also in the validation set samples: ANTXR1, CEP135, CRYBB1, MORC2, SLC36A1, GALNT9, PI15, KRT82, CNTF, GLDC, MBTPS1, OR9Q2, R3HDM1 and TTPAL (Table 1, Supplementary Table S4). Twenty-eight (11%) of the validation set tumours displayed at least one of these hotspot mutations (Supplementary Figure S1). BRAF, KRAS, CTNNB1 and PIK3CA were mutated at frequencies of 42%, 15%, 6.9% and 5.5–11%, respectively (Table 1). The most frequent novel hotspot mutation encountered in the validation set was ANTXR1 Arg438Cys (7 out of 251, 2.8% Figure 2A). The next frequent hotspots were CEP135 Arg1115Cys and CRYBB1 Ala171Thr (1.2%). Distribution of mutations in MORC2 differed somewhat from the other genes (3xSer25Leu, Lys27Met, Lys27Arg). Distribution of the mutations in the validation set tumours is presented in Supplementary Figure S1. Proposed functions of the 14 validated genes are summarised in Supplementary Table S5.
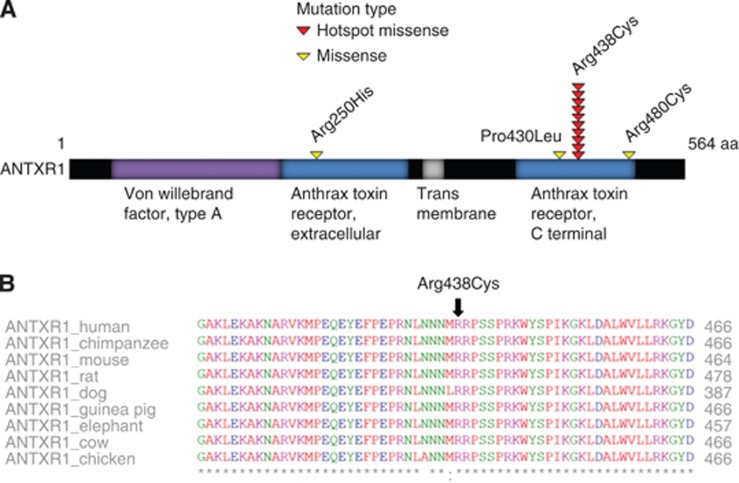
Figure 2.
Schematic representation of functional domains and mutations of ANTXR1.
(A) Mutation hotspot in ANTXR1 is in the anthrax-binding domain. Other mutations, Arg250His (MSS), Pro430Leu (MSS) and Arg480Cys (MSI) are all located in the same functional domain. (B) The hotspot amino acid is depicted with the black arrow in the multispecies comparison (ClustalW2) and is highly conserved between species.
Multispecies protein alignments indicated that in 10 out of 14 genes (ANTXR1, MORC2, CEP135, GALNT9, PI15, KRT82, GLDC, MBTPS1, R3HDM1 and TTPAL) the targeted hotspot amino acids were highly conserved. Multispecies protein alignment for ANTXR1 is shown in Figure 2B. Functional predictions with PolyPhen2 and SIFT indicated damaging or deleterious effect for amino-acid substitutions in 11 genes (ANTXR1, MORC2, CEP135, GALNT9, KRT82, PI15, SLC36A1, GLDC, MBTPS1, R3HDM1 and TTPAL; Supplementary Table S4).
We next examined the presence of the hotspot mutations in CRC cell lines and MSS CRCs. Sequencing of the 14 validated hotspots in 10 CRC cell lines revealed MORC2 Ser25Ala in HCA7 and OR9Q2 Ala249Thr in DLD1. To study MSS CRCs, we utilised WGS data on 91 MSS CRCs (unpublished data). We could not find any of the 33 hotspot mutations in MSS CRCs. When examining the complete coding sequence, mutations were found in ANTXR1, GALNT9, KRT82, R3HDM1, TTPAL, MTMR12, NLRP10, DHX58, PTPRS and KCNB1 (Supplementary Table S4). As ANTXR1 was the strongest hit in our MSI CRC mutation screen, we examined the two MSS CRC mutations in this gene, Arg250His and Pro430Leu, by Sanger sequencing and confirmed their somatic nature also with this method.
To examine the presence of the 33 hotspot mutations across cancer types and cell lines, cBioPortal (Cerami et al, 2012), International Cancer Genome Consortium Data Portal (Hudson et al, 2010) and COSMIC database (Forbes et al, 2011) were utilised. In CRCs, identical hotspot mutations were found in ANTXR1, OR9Q2, CTTNBP2, HEXIM2 and PALD1 (Supplementary Table S6). In MORC2, mutations Lys27Arg and Lys27Thr were found. In addition, in KCNB1 and NLRP10 examples of hotspot codon mutations were found, although the exact base changes were different from those seen in our discovery set data. Mutations in ANTXR1, OR9Q2, CTTNBP2, HEXIM2, PALD1, NLRP10 and MORC2 were observed in MSI CRCs. In addition, one TCGA CRC sample displayed ANTXR1 Arg439Gln, hitting adjacent to our hotspot. MSI CRC cell lines HCT-15 and HCT-116 displayed hotspot mutations in OR9Q2 and HEXIM2, respectively.
In extracolonic cancers, hotspot mutations in ANTXR1, CRYBB1, PALD1, SLC36A1, KCNB1, SETD9, TMEM53, PTPRS, KRT82, PI15 and GALNT9 were encountered (Supplementary Table S6). We specifically reviewed the TCGA data on endometrial and stomach MSI cancers. Among the 127 MSI-H endometrial cancers, one example of hotspot mutation in TMEM53 was found. Among 64 MSI-H stomach cancers, hotspot mutations in ANTXR1, CRYBB1 and KCNB1 were encountered, each once. Thus, although the hotspot mutations were not common, they occurred in a wide spectrum of different tumour types.
Finally, we examined expression of the ANTXR1 Arg438Cys mutant allele in five tumours, as well as the Pro430Leu and Arg250His from MSS CRCs. The mutant allele was absent in all seven cDNAs. Four tumours with exon 1 variant rs28365986 (Arg7Lys) expressed both alleles.
Discussion
Comprehensive knowledge on mutations underlying human cancers is essential for improved cancer diagnostics, personalised care and novel therapeutic interventions. Most cancer genes that are mutated at high frequency have already been discovered; however, the comprehensive catalogue of mutations occurring at intermediate (2–20%) or low frequencies is far from complete (Lawrence et al, 2014). Oncogenes are often found to be recurrently mutated at preferred amino-acid-coding positions, and it has been suggested that the pattern rather than the frequency should be used as criteria for defining new driver genes (Vogelstein et al, 2013). Mutation hotspots are markers of selection in tumorigenesis, as well as attractive therapeutic targets. Here, we aimed at identifying new oncogenic drivers of cancer by examining MSI CRCs as a discovery set. These tumours might be ideal for detection of novel oncogenic mutations, by forming a sensitive model system generating frequent mutations for possible selection during tumour progression.
We identified 33 candidate oncogenes of which 14 displayed hotspot mutations in a validation set. ANTXR1 (also known as TEM8) stood out as the most frequent target for hotspot mutations (2.8%). ANTXR1 encodes a single-pass cell-surface protein that was originally identified based on its overexpression in the tumour-infiltrating vasculature in human CRC (St Croix et al, 2000). It has been shown to have an important role in tumour angiogenesis (Chaudhary et al, 2012) and has an ability to promote cell migration by linking the extracellular matrix to the actin cytoskeleton (Werner et al, 2006). We found no expression of the ANTXR1 mutant allele in the seven primary tumours examined. The combination of selection of specific mutations and subsequent loss of expression of the mutant allele, as observed in the resulting cancer, is unusual. Whereas lack of expression of the mutant allele in the mature tumours may make the change unattractive as a therapeutic target, the exact mechanisms of tumorigenesis involved should be a particularly intriguing subject of cancer biology research.
In addition to CRC, the newly discovered hotspot mutations appear to have a role in the genesis of a wide variety of cancer types. Hotspot mutations were encountered in many extracolonic cancers, albeit with low frequency. Whereas some of the common cancer hotspot mutations have been successfully utilised as targets for therapy, targeted monotherapy often fails to cure the patient due to pre-existing resistant subclones (Diaz et al, 2012). The catalogue of mutation hotspots reported in this study should serve as a valuable resource for focused and fruitful cancer biology work, to pave the way towards identification of multiple druggable targets—and combination treatment protocols aiming at cure—for an increasing proportion of tumours.
Acknowledgments
We thank Inga-Lill Svedberg, Iina Vuoristo, Alison Ollikainen, Sini Nieminen, Marjo Rajalaakso, Sirpa Soisalo, Kirsi Pylvänäinen and Tuula Lehtinen for technical assistance. This work was supported by grants from the Academy of Finland (Finnish Center of Excellence Program 2012–2017), the Finnish Cancer Society, the Sigrid Juselius Foundation and SYSCOL (an EU FP7 Collaborative Project). Personal grants were received from the Academy of Finland (no. 260370 to PV) and from the University of Helsinki (postdoctoral grant for ST).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, De La Chapelle A. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, Van Buuren MM, Van 'T Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, Mcdermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D'andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, Mcarthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary A, Hilton MB, Seaman S, Haines DC, Stevenson S, Lemotte PK, Tschantz WR, Zhang XM, Saha S, Fleming T, St Croix B. TEM8/ANTXR1 blockade inhibits pathological angiogenesis and potentiates tumoricidal responses against multiple cancer types. Cancer Cell. 2012;21:212–226. doi: 10.1016/j.ccr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylfe AE, Kondelin J, Turunen M, Ristolainen H, Katainen R, Pitkanen E, Kaasinen E, Rantanen V, Tanskanen T, Varjosalo M, Lehtonen H, Palin K, Taipale M, Taipale J, Renkonen-Sinisalo L, Jarvinen H, Bohm J, Mecklin JP, Ristimaki A, Kilpivaara O, Tuupanen S, Karhu A, Vahteristo P, Aaltonen LA. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology. 2013;145:540–543. doi: 10.1053/j.gastro.2013.05.015. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, Guttmacher A, Guyer M, Hemsley FM, Jennings JL, Kerr D, Klatt P, Kolar P, Kusada J, Lane DP, Laplace F, Youyong L, Nettekoven G, Ozenberger B, Peterson J, Rao TS, Remacle J, Schafer AJ, Shibata T, Stratton MR, Vockley JG, Watanabe K, Yang H, Yuen MM, Knoppers BM, Bobrow M, Cambon-Thomsen A, Dressler LG, Dyke SO, Joly Y, Kato K, Kennedy KL, Nicolas P, Parker MJ, Rial-Sebbag E, Romeo-Casabona CM, Shaw KM, Wallace S, Wiesner GL, Zeps N, Lichter P, Biankin AV, Chabannon C, Chin L, Clement B, De Alava E, Degos F, Ferguson ML, Geary P, Hayes DN, Johns AL, Kasprzyk A, Nakagawa H, Penny R, Piris MA, Sarin R, Scarpa A, Van De Vijver M, Futreal PA, Aburatani H, Bayes M, Botwell DD, Campbell PJ, Estivill X, Grimmond SM, Gut I, Hirst M, Lopez-Otin C, Majumder P, Marra M, Mcpherson JD, Ning Z, Puente XS, Ruan Y, Stunnenberg HG, Swerdlow H, Velculescu VE, Wilson RK, Xue HH, Yang L, Spellman PT, Bader GD, Boutros PC, Flicek P, Getz G, Guigo R, Guo G, Haussler D, Heath S, Hubbard TJ, Jiang T, Jones SM, Li Q, López-Bigas N, Luo R, Muthuswamy L, Ouellette BF, Pearson JV, Puente XS, Quesada V, Raphael BJ, Sander C, Shibata T, Speed TP, Stein LD, Stuart JM, Teague JW, Totoki Y, Tsunoda T, Valencia A, Wheeler DA, Wu H, Zhao S, Zhou G, Stein LD, Guigó R, Hubbard TJ, Joly Y, Jones SM, Kasprzyk A, Lathrop M, López-Bigas N, Ouellette BF, Spellman PT, Teague JW, Thomas G, Valencia A, Yoshida T, Kennedy KL, Axton M, Dyke SO, Futreal PA, Gerhard DS, Gunter C, Guyer M, Hudson TJ, McPherson JD, Miller LJ, Ozenberger B, Shaw KM, Kasprzyk A, Stein LD, Zhang J, Haider SA, Wang J, Yung CK, Cros A, Liang Y, Gnaneshan S, Guberman J, Hsu J, Bobrow M, Chalmers DR, Hasel KW, Joly Y, Kaan TS, Kennedy KL, Knoppers BM, Lowrance WW, Masui T, Nicolás P, Rial-Sebbag E, Rodriguez LL, Vergely C, Yoshida T, Grimmond SM, Biankin AV, Bowtell DD, Cloonan N, deFazio A, Eshleman JR, Etemadmoghadam D, Gardiner BB, Kench JG, Scarpa A, Sutherland RL, Tempero MA, Waddell NJ, Wilson PJ, McPherson JD, Gallinger S, Tsao MS, Shaw PA, Petersen GM, Mukhopadhyay D, Chin L, DePinho RA, Thayer S, Muthuswamy L, Shazand K, Beck T, Sam M, Timms L, Ballin V, Lu Y, Ji J, Zhang X, Chen F, Hu X, Zhou G, Yang Q, Tian G, Zhang L, Xing X, Li X, Zhu Z, Yu Y, Yu J, Yang H, Lathrop M, Tost J, Brennan P, Holcatova I, Zaridze D, Brazma A, Egevard L, Prokhortchouk E, Banks RE, Uhlén M, Cambon-Thomsen A, Viksna J, Ponten F, Skryabin K, Stratton MR, Futreal PA, Birney E, Borg A, Børresen-Dale AL, Caldas C, Foekens JA, Martin S, Reis-Filho JS, Richardson AL, Sotiriou C, Stunnenberg HG, Thoms G, van de Vijver M, van't Veer L, Calvo F, Birnbaum D, Blanche H, Boucher P, Boyault S, Chabannon C, Gut I, Masson-Jacquemier JD, Lathrop M, Pauporté I, Pivot X, Vincent-Salomon A, Tabone E, Theillet C, Thomas G, Tost J, Treilleux I, Calvo F, Bioulac-Sage P, Clément B, Decaens T, Degos F, Franco D, Gut I, Gut M, Heath S, Lathrop M, Samuel D, Thomas G, Zucman-Rossi J, Lichter P, Eils R, Brors B, Korbel JO, Korshunov A, Landgraf P, Lehrach H, Pfister S, Radlwimmer B, Reifenberger G, Taylor MD, von Kalle C, Majumder PP, Sarin R, Rao TS, Bhan MK, Scarpa A, Pederzoli P, Lawlor RA, Delledonne M, Bardelli A, Biankin AV, Grimmond SM, Gress T, Klimstra D, Zamboni G, Shibata T, Nakamura Y, Nakagawa H, Kusada J, Tsunoda T, Miyano S, Aburatani H, Kato K, Fujimoto A, Yoshida T, Campo E, López-Otín C, Estivill X, Guigó R, de Sanjosé S, Piris MA, Montserrat E, González-Díaz M, Puente XS, Jares P, Valencia A, Himmelbauer H, Quesada V, Bea S, Stratton MR, Futreal PA, Campbell PJ, Vincent-Salomon A, Richardson AL, Reis-Filho JS, van de Vijver M, Thomas G, Masson-Jacquemier JD, Aparicio S, Borg A, Børresen-Dale AL, Caldas C, Foekens JA, Stunnenberg HG, van't Veer L, Easton DF, Spellman PT, Martin S, Barker AD, Chin L, Collins FS, Compton CC, Ferguson ML, Gerhard DS, Getz G, Gunter C, Guttmacher A, Guyer M, Hayes DN, Lander ES, Ozenberger B, Penny R, Peterson J, Sander C, Shaw KM, Speed TP, Spellman PT, Vockley JG, Wheeler DA, Wilson RK, Hudson TJ, Chin L, Knoppers BM, Lander ES, Lichter P, Stein LD, Stratton MR, Anderson W, Barker AD, Bell C, Bobrow M, Burke W, Collins FS, Compton CC, DePinho RA, Easton DF, Futreal PA, Gerhard DS, Green AR, Guyer M, Hamilton SR, Hubbard TJ, Kallioniemi OP, Kennedy KL, Ley TJ, Liu ET, Lu Y, Majumder P, Marra M, Ozenberger B, Peterson J, Schafer AJ, Spellman PT, Stunnenberg HG, Wainwright BJ, Wilson RK, Yang H. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katainen R.2013. Detection of pathogenic mutations from exome sequencing data. Master's thesis http://hdl.handle.net/10138/39539 .
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Pitkanen E, Cajuso T, Katainen R, Kaasinen E, Valimaki N, Palin K, Taipale J, Aaltonen LA, Kilpivaara O. Frequent L1 retrotranspositions originating from TTC28 in colorectal cancer. Oncotarget. 2014;5:853–859. doi: 10.18632/oncotarget.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salovaara R, Loukola A, Kristo P, Kaariainen H, Ahtola H, Eskelinen M, Harkonen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, Jarvinen H, Mecklin JP, Aaltonen LA, De La Chapelle A. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18:2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Werner E, Kowalczyk AP, Faundez V. Anthrax toxin receptor 1/tumor endothelium marker 8 mediates cell spreading by coupling extracellular ligands to the actin cytoskeleton. J Biol Chem. 2006;281:23227–23236. doi: 10.1074/jbc.M603676200. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.