Abstract
Background:
Pancreatic cancer has a poor prognosis because of its high refractoriness to chemotherapy and tumour recurrence, and these properties have been attributed to cancer stem cells (CSCs). MicroRNA (miRNA) regulates various molecular mechanisms of cancer progression associated with CSCs. This study aimed to identify the candidate miRNA and to characterise the clinical significance.
Methods:
We established gemcitabine-resistant Panc1 cells, and induced CSC-like properties through sphere formation. Candidate miRNAs were selected through microarray analysis. The overexpression and knockdown experiments were performed by evaluating the in vitro cell growth and in vivo tumourigenicity. The expression was studied in 24 pancreatic cancer samples after laser captured microdissection and by immunohistochemical staining.
Results:
The in vitro drug sensitivity of pancreatic cancer cells was altered according to the miR-1246 expression via CCNG2. In vivo, we found that miR-1246 could increase tumour-initiating potential and induced drug resistance. A high expression level of miR-1246 was correlated with a worse prognosis and CCNG2 expression was significantly lower in those patients.
Conclusions:
miR-1246 expression was associated with chemoresistance and CSC-like properties via CCNG2, and could predict worse prognosis in pancreatic cancer patients.
Keywords: microRNA, miR-1246, CCNG2, chemoresistance, cancer stem cell, pancreatic cancer
Pancreatic cancer has the worst prognosis of any major malignancy and is the fourth most common cause of cancer death each year in the United States. Even if it is diagnosed at an operable stage and curative resection is achieved, there is still a high incidence of recurrence (Hoyert et al, 2006). Although gemcitabine (GEM)-based chemotherapy has formed the core of multimodal therapy for pancreatic cancer (Oettle et al, 2007), it is rarely curative and only modestly effective against tumour recurrence. Cancer stem cells (CSCs) have been implicated in the clinical refractoriness, metastasis, and tumour recurrence of various types of cancer, including pancreatic cancer (Hermann et al, 2007; Li et al, 2007). The molecular mechanisms that link chemoresistance with CSCs remain unclear. Clarifying these mechanisms could improve prognosis of patients with pancreatic cancer.
The miRNAs are endogenous, 18–25-bp, single-stranded, non-coding modulators of the post-transcriptional process (Bartel, 2009). In the present study, we focused on miRNAs of GEM-resistant pancreatic cancer cell lines and CSC-like cell populations using sphere formation assays (Pastrana et al, 2011); pancreatic CSCs could be enriched as spheroid cells (Hermann et al, 2007). We performed comprehensive expression profiling of miRNAs in order to indentify the candidate miRNA and determine its clinical significance.
Materials and methods
Vector constructs and lentiviral production
The lentiviral pLenti-III-miR-1246 vector and its empty pLenti-III vector (Applied Biological Materials Inc., Richmond, Canada) were co-transfected with pCAG-HIVgp and pCMV-VSV-G-RSV-Rev into 293Ta cells (Gene Copoeia Inc., Rockville, MD, USA). Panc1 cells were infected with lentivirus and selected using 1 μg ml−1 puromycin for 4 weeks, to establish stable miR-1246 (Panc1-P-l-OE) and control (Panc1-P-l-C) transfectants. The CCNG2 lacking miR1246 target sites in 3′UTR were transfected into the Panc1-P-l-OE cells using FuGENE 6 Reagent (Promega, Madison, WI, USA).
Ethics statement on animals
All animal work was performed according to Animal Experiments Committee, Osaka University (the approval number, 24-122-011).
Animal experiments
Four different numbers of Panc1 cells, 1 × 102, 1 × 103, 1 × 104, and 1 × 105, were injected subcutaneously in 4- to 6-week-old female non-obese mice with diabetes/severe combined immunodeficiency (CLEA Japan, Tokyo, Japan). The mice were administered GEM intraperitoneally three times on days 35, 42, and 49 using the once-a-week protocol as to closely replicate clinical use. Mice were given 125 mg kg−1 GEM, or phosphate-buffered saline as control (Lonardo et al, 2011). The tumour volume was calculated as: (longest diameter) × (shortest diameter)2 × 0.5. The therapy was initiated when the tumour volumes were 60–100 mm3. The tumours were resected on day 53 after the cell injection. Total RNA was extracted from tumours. Immunohistochemistry was performed using polyclonal goat anti-human cyclinG2 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA).
Ethics statement on clinical samples
The use of resected samples was approved by the Human Ethics Review Committee of the Graduate School of Medicine, Osaka University (approval number 08226). Written informed consent was obtained from all patients included in the study.
Study on primary tumour samples
Between March 2007 and December 2010, 24 consecutive patients with pancreatic cancer were treated by histologically curative resection (R0) at Osaka University Hospital, and no preoperative therapy was given (Supplementary Table 3). Among 24 patients, 12 patients received adjuvant GEM chemotherapy. Immunohistochemical staining protocol is shown in the Supplementary Materials and Methods. To separate epithelial and mesenchymal parts, laser captured microdissection (LCM; LMD7000, Leica Microsystems GmbH, Wetzlar, Germany) was performed as 8-μm-thick sections from formalin-fixed paraffin-embedded (FFPE) samples. Total RNA was extracted using RNeasy FFPE Kit (Qiagen, Tokyo, Japan). The relative quantification of miRNA was studied by qRT–PCR using comparative CT method (2−ΔCT). Data were normalised using endogenous RNU48 control.
Statistical analysis
The clinicopathological parameters were compared using the Fisher's exact test, and the continuous variables were compared using the Student's t-test. The survival curves were computed using the Kaplan–Meier method. Statistical analysis was performed using JMP software version 10.0.2 (SAS Institute Inc., Cary, NC, USA).
Results
Establishment of GEM-resistant pancreatic cancer cell clones
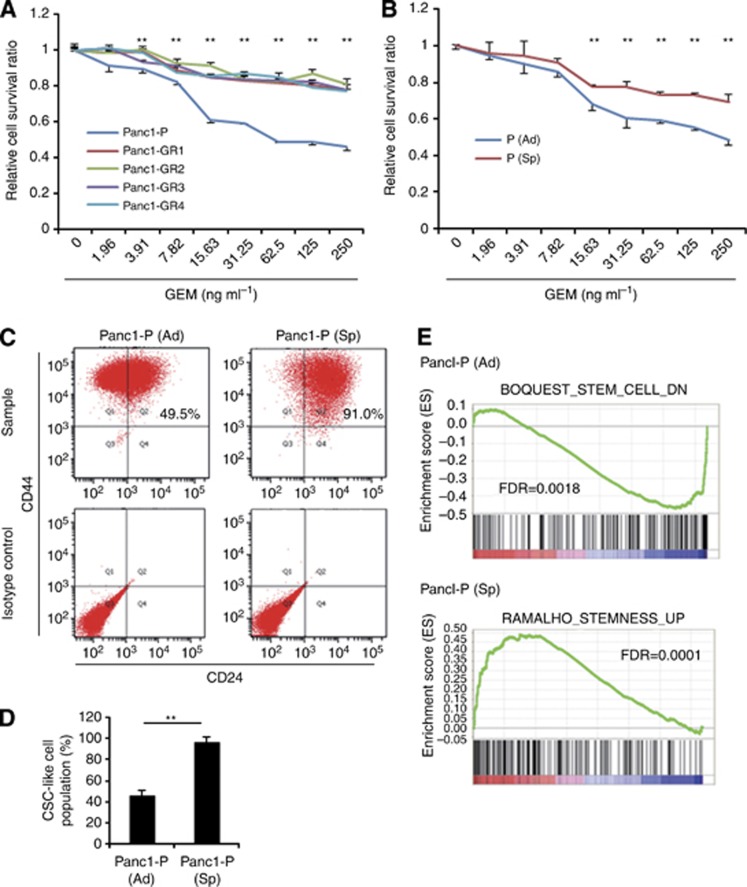
We established four independent GEM-resistant clones of Panc1 cells. Compared with the Panc1-P cells, the Panc1-GR cells were significantly more resistant to GEM (Figure 1A) and showed significantly more drug resistance to 5-FU at three densities (Supplementary Figure 1A). Several studies have recently reported that representative surface markers, such as CD44+CD24+, were useful to enrich the CSC-like cells from others in pancreatic cancer cell lines (Hermann et al, 2007; Li et al, 2007). Flow cytometry analysis of the CSC population in Panc1-GRs revealed that the CD44+CD24+ population was slightly increased (Figure 1B and Supplementary Figure 1B).
Figure 1.
Features of gemcitabine (GEM)-resistant pancreatic cancer cells (Panc1-GRs) and Panc1 parental CSC-like spheroid cells (Panc1-P (Sp)). (A) Growth-inhibitory effects of GEM on Panc1 parental (Panc1-P) cells and Panc1-GR (GR1, GR2, GR3, and GR4) cells were assessed by MTT assay. (B) Growth-inhibitory effects of GEM on normal adherent condition Panc1-P (Ad) and Panc1-P (Sp) cells were assessed by MTT assay. (C) The representative data of flow cytometry showed the CSC-like cell population of Panc1-P in Panc1-P (Ad) cells (left panel) and Panc1-P (Sp) cells (right panel). (D) The percentages of the CSC-like cell populations were shown in Pan1-P (Ad) and -P (Sp) cells. This assay was performed three times. (E) Gene set enrichment analysis (GSEA) of Panc1-P (Sp) cells compared with Panc1-P (Ad) cells was performed. GSEA-extracted representative gene sets enriched in those cells are shown. Data represent mean±s.d. of more than three experiments; **P<0.01.
Establishment of pancreatic CSC-like chemoresistant cells
The sphere formation and growth assays revealed that Panc1-P (Sp) was significantly resistant to GEM and 5-FU compared with Panc1-P (Figure 1B and Supplementary Figure 1D). Note that Panc1-P (Sp) cells were chemoresistant to GEM and 5-FU, although not exposed to agents. Flow cytometry analysis revealed that the CD24+CD44+ population nearly doubled due to sphere formation (Figure 1C and D).
Gene sets enriched in spheroid cells and adherent cells
Gene set enrichment analysis (GSEA) was performed between the coding gene expression profiles of the spheroid parental Panc1 (Panc1-P (Sp)) cells and those of the adherent parental cells (Panc1-P). The GSEA in spheroid cells revealed upregulation of stem cell pathway and downregulation of adherent pathway (Figure 1E and Supplementary Figure 2A and B).
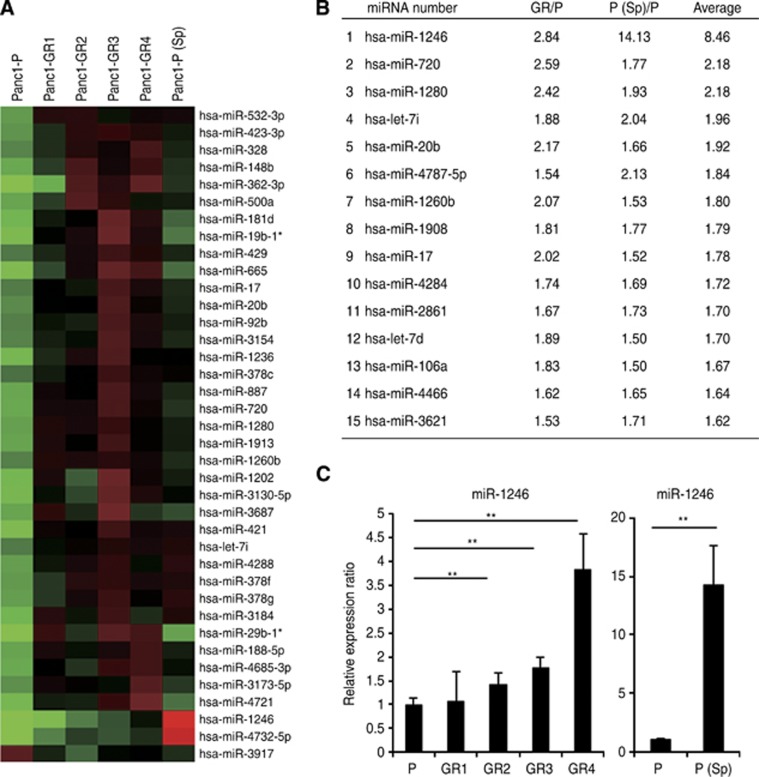
Outstanding expression of miR-1246 revealed by miRNA microarray analysis
To identify the candidate miRNAs related to chemoresistance and cancer stemness, we performed miRNA microarray studies. The data showed that, among the 1719 miRNAs, the miRNA expression levels of 92 in the Panc1-GR cells (Supplementary Table 1) and 216 in the Panc1-P (Sp) cells (Supplementary Table 2) were altered by an average of >1.5-fold relative to the parental levels. The heat map showed 38 common miRNAs, which included 37 miRNAs (>1.5-fold) and 1 miRNA (<0.66-fold) of the parental levels (Figure 2A). By excluding 23 miRNAs that showed undetectable or extremely low expression and non-functional miRNAs, we found 15 candidate miRNAs were expressed strongly in both Panc1-GRs and Panc1-P (Sp) cells (Figure 2B). miR-1246 showed the highest alteration (8.46 average fold change: 14.13-fold increase in the Panc1-P (Sp) and 2.84-fold increase in the Panc1-GR). The fold change of the miR-1246 expression level was outstanding in Panc1-P (Sp) cells. The real-time qRT–PCR confirmed the miR-1246 upregulation in Panc1-GRs and -P (Sp) cells (Figure 2C). We then assessed miR-1246 for further analysis.
Figure 2.
miRNA microarray analysis and the ranking of candidate miRNAs. (A) The heat map revealed the miRNAs whose expression levels were altered >1.5-fold relative to Panc1-P both in Panc1-GRs and -P (Sp) cells. *non-functional miRNA. (B) The list showed the ranking of candidate miRNAs according to the ratio of the change in expression level. From the above 38 miRNAs (A), 15 miRNAs were selected. (C) Real-time qRT–PCR showed the expression of miR-1246 in Panc1-P, -GRs, and -P (Sp) cells. Data represent mean±s.d. of more than three experiments; **P<0.01.
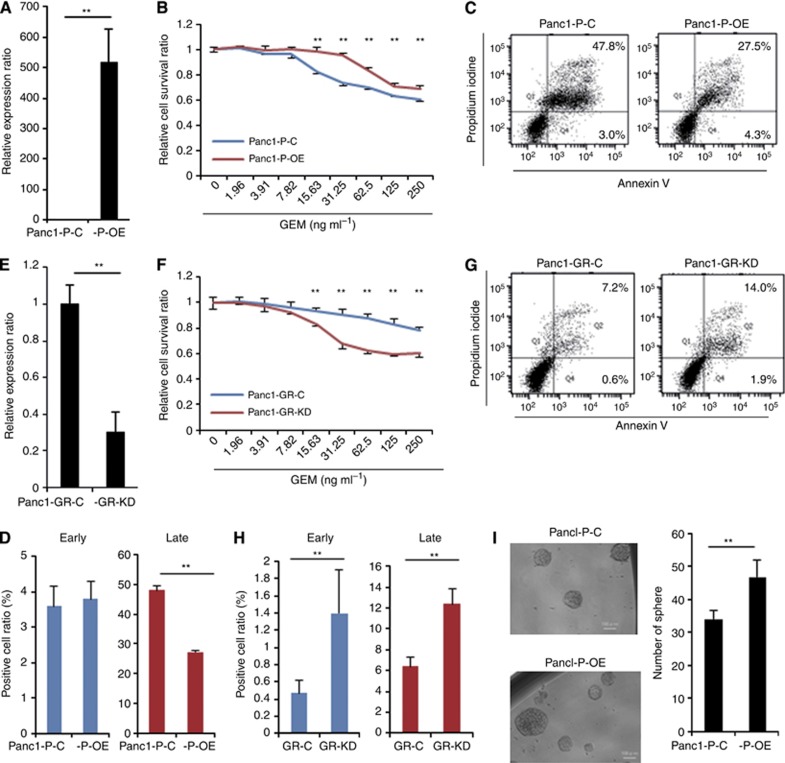
Association of miR-1246 expression with the resistance to GEM and sphere-forming ability
To evaluate the effect of miR-1246 on the response to GEM in Panc1 cells, pre- or anti-miR-1246 was introduced into Panc1-P/-GR4. Real-time qRT–PCR confirmed that the transfection with pre-miR-1246 resulted in the marked overexpression of mature miR-1246 (Panc1-P-OE; Figure 3A). The proliferation rate of the Panc1-P-OE cells was slightly increased compared with control Panc1-P-C. The MTT assay demonstrated that transfection of pre-miR-1246 into Panc1-P resulted in resistance to GEM (Figure 3B). In contrast, anti-miR-1246 transfection into Panc1-GR4, whose endogenous levels of miR-1246 was 3.5-fold higher than that of Panc1-P (Figure 2C), resulted in sufficient inhibition of miR-1246 by qRT–PCR (Panc1-GR-KD; Figure 3E). MTT assay demonstrated a significant reduction of chemoresistance to GEM in the Panc1-GR-KD cells (Figure 3F). The proliferation rate of the Panc1-GR-KD cells was not changed compared with control Panc1-GR-C. These results indicated that miR-1246 induced GEM-resistance in Panc1 cells. We assessed the sphere-forming ability. The proliferation ratio of spheres in Panc1-P-OE was significantly higher than that in Panc1-P-C (Supplementary Figure 1C). Panc1-P-OE spheres increased compared to Panc1-P-C (Figure 3I). We confirm the results with another pancreatic cancer cell line, MiaPaCa2 with endogenous miR-1246 decreased (40% of Panc1; Supplementary Figure 4A). The pre-miR-1246 transfection resulted in replenishment of miR-1246 (Supplementary Figure 4C). The MTT assay demonstrated that miR-1246 anti-sensitised MiaPaCa2 to GEM treatment (Supplementary Figure 4E). Although CSC-like cell population was relatively rare (Supplementary Figure 4B), the miR-1246 OE (Sp) enriched by sphere assay revealed that the proliferation rate was higher than control cells (Supplementary Figure 4D). The miR-1246 OE sphere increased significantly compared to control (Supplementary Figure 4F).
Figure 3.
Association of miR-1246 expression with the resistance to GEM and sphere-forming ability. (A) Real-time qRT–PCR showed the expression level of miR-1246 in Panc1-P cells transfected with pre-miR-1246 (Panc1-P-OE) and transfected with the negative control (Panc1-P-C). (B) MTT assay demonstrated relative cell survival ratio of Panc1-P-C and Panc1-P-OE cells to GEM, respectively. (C) The representative data of Annexin V assay showed the distribution of the early and late apoptotic cells in the Panc1-P-C and Panc1-P-OE cells after GEM exposure for 72 h. (D) The percentages of early and late apoptotic cells in the Panc1-P-C and Panc1-P-OE cells after GEM exposure for 72 h. This assay was performed three times. (E) Real-time qRT–PCR showed the expression level of miR-1246 in Panc1-GR cells transfected with anti-miR-1246 (Panc1-GR-KD) and transfected with negative control (Panc1-GR-C). (F) MTT assay demonstrated relative cell survival ratio of Panc1-GR-C and Panc1-GR-KD to GEM. (G) The representative data of Annexin V assay showed the distribution of the early and late apoptotic cells in Panc1-GR-C and Panc1-GR-KD cells after GEM exposure for 72 h. (H) The percentages of early and late apoptotic cells in Panc1-GR-C and Panc1-GR-KD cells after GEM exposure for 72 h. This assay was performed three times. (I) Sphere formation assay was performed in Panc1-P-C and Panc1-P-OE cells. The representative image of spheres (left) and the number of spheres (right) were shown. Bar=100 μm. Data represent mean±s.d. of three experiments; **P<0.01.
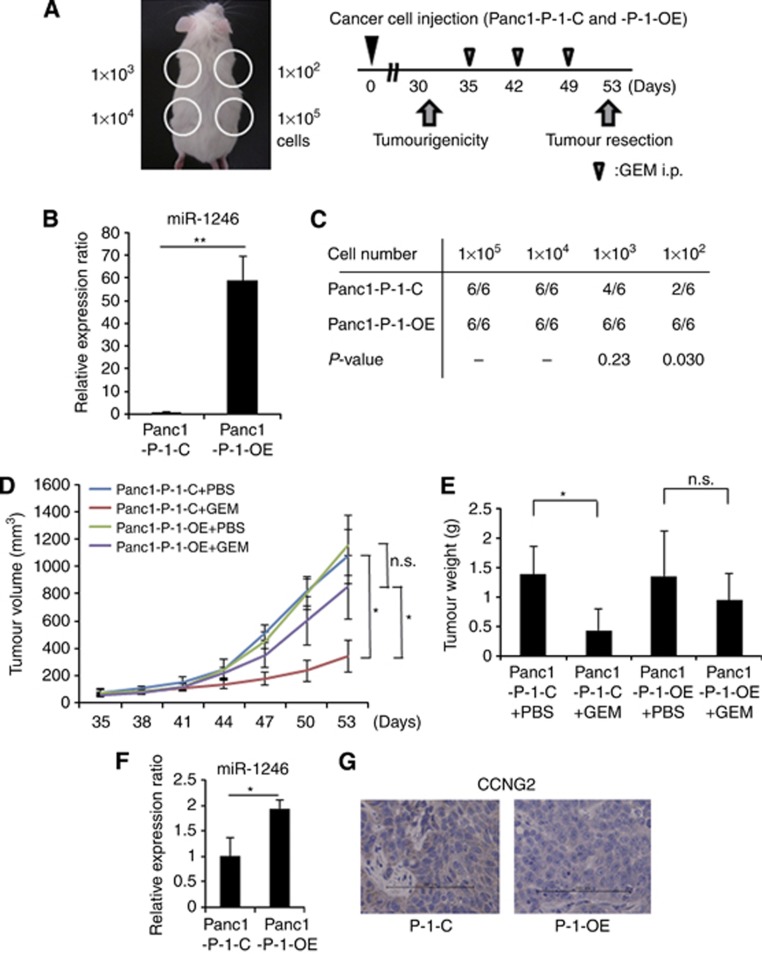
miR-1246 induced tumourigenicity and refractoriness to GEM
Although pre-miR-1246 expression continued for a week, it decreased gradually (Supplementary Figure 3A; the data of anti-miR-1246 in Supplementary Figure 3B). To assess miR-1246 functions relating to tumourigenicity and refractoriness to GEM in vivo (Figure 4A), we established Panc1 (Panc1-P-l-OE) cells that overexpressed miR-1246 continuously in high level (Supplementary Figure 3C). We confirmed that the expression level of miR-1246 in Panc1-P-l-OE was ∼50-fold higher than that in Panc1-l-C (Figure 4B). The tumourigenicity of Panc1-P-l-OE was significantly increased compared with Panc1-P-l-C (at 1 × 102 cell injection; Figure 4C). The tumour volume and weight of Panc1-P-l-C, but not Panc1-P-l-OE, was significantly inhibited by GEM treatment compared with controls at day 53 (Figure 4D and E). The miR-1246 expression in the Panc1-P-l-OE tumours was significantly two times higher than the expression in Panc1-P-l-C at day 53 (Figure 4F).
Figure 4.
The function of miR-1246 in tumourigenicity and GEM-resistance in vivo. (A) The experimental design was shown. (B) Real-time qRT–PCR showed the expression level of miR-1246 in miR-1246 continuously expressing Panc1 (Panc1-P-l-OE) and control Panc1 (Panc1-P-l-C) cells. (C) The tumourigenic ability was evaluated on day 30 after Panc1 cell injection in the Panc1-P-l-C and Panc1-P-l-OE groups. (D) The therapy was initiated when the tumour volumes (1 × 105 cells injected) were between 60 and 100 mm3. (E) The weights of tumours were measured at day 53 after resection. (F) Real-time qRT–PCR showed the expression level of miR-1246 in the tumours of Panc1-P-l-OE and Panc1-P-l-C cell-injected groups. (G) Immunohistochemistry of the tumour specimens showed CCNG2 expression in the tumours of Panc1-P-l-OE and Panc1-P-l-C cell-injected groups. Bar=100 μm. Data represent mean±s.d. of three experiments; *P<0.05; **P<0.01.
miR-1246 inhibits response to GEM by targeting CCNG2
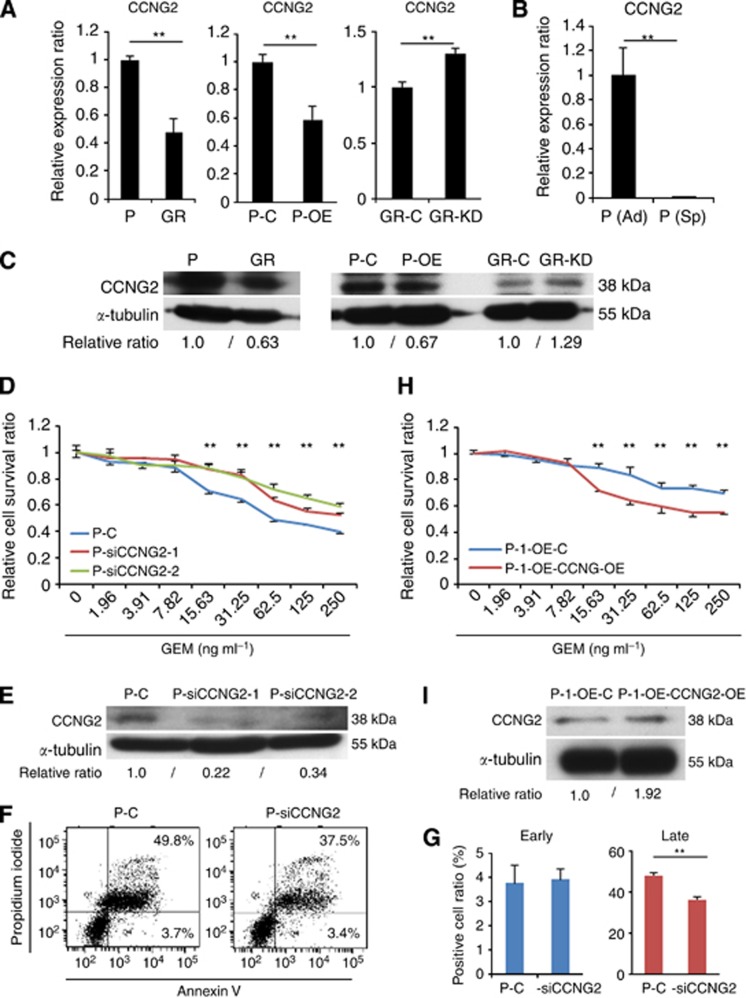
Few studies have reported miR-1246 expression in gastrointestinal cancers, and little is known about the function. As putative miR-1246 targets, the TargetScan (http://www.targetscan.org/) predicted 178 genes. Among them, CCNG2, a family of cyclins that is homologous to CCNG1 (Bates et al, 1996) and is also known as a tumour suppressor, was selected for further analysis. Western blotting and qRT–PCR showed that CCNG2 expression was lower in Panc1-GR cells than in Panc1-P cells (Figure 5A and C). Western blotting and qRT–PCR confirmed that pre-miR-1246 transfection decreased CCNG2 expression, whereas anti-miR-1246 increased it (Figure 5A and C), suggesting that CCNG2 was one of the target genes of miR-1246. CCNG2 expression was significantly lower in Panc1-P (Sp) than in control (Figure 5B). We are interested in a role of CCNG2 in the resistance to GEM. To this end, we performed knockdown of CCNG2 by siRNA. Western blotting confirmed the knockdown (Figure 5E). The MTT assay demonstrated that transfection of siCCNG2 elicited the resistance of Panc1-P to GEM (Figure 5D). Given reportedly CCNG2 regulated cell growth and induced apoptosis (Kim et al, 2004), we studied the involvement of apoptosis in present case. In Panc1-P, pre-miR-1246 significantly reduced the number of late apoptotic cells (Figure 3C and D). Similarly, pre-miR-1246 significantly reduced the early and late apoptosis in MiaPaCa2 cells (Supplementary Figure 4G and H). In contrast, the anti-miR-1246 increased the number of early and late apoptotic cells significantly in Panc1-GR (Figure 3G and H). Also, siCCNG2 significantly reduced late apoptosis (Figure 5F and G). These data suggested that miR-1246 regulated chemoresistance via CCNG2 expression that was involved in apoptosis. Furthermore, in order to study whether the ectopic CCNG2 expression can rescue the miR-1246-driven GEM-resistance, we transfected CCNG2 vector (lacking miR-1246 target sites) into Panc1-P-l-OE cells. The MTT assay demonstrated that ectopic CCNG2 significantly reduced chemoresistance to GEM in the Panc1-P-l-OE cells (Figure 5H); the elevation of CCNG2 expression was confirmed with western blotting (Figure 5I).
Figure 5.
The CCNG2 function as a target of miR-1246 in Panc1 cells. (A–C) Real-time qRT–PCR and western blotting demonstrated CCNG2 expression level in Panc1-P and Panc1-GR cells, Panc1-P-C and Panc1-P-OE cells, Panc1-GR-C and Panc1-GR-KD cells, and Panc1-P (Ad) and Panc1-P (Sp) cells. (D) MTT assay demonstrated the relative cell survival ratio of Panc1-P cells transfected with siCCNG2 (Panc1-P-siCCNG2-1, -2) and transfected with negative control (Panc1-P-C). (E) The protein level of CCNG2 in Panc1-P and GR cells, Panc1-P-C and Panc1-P-OE cells, and Panc1-GR-C and Panc1-GR-KD cells in western blotting. (F) The representative data of Annexin V assay showed the distribution of the early and late apoptotic cells in Panc1-P-C and Panc1-P-siCCNG2 after GEM exposure for 72 h. (G) The percentages of early and late apoptotic cells in Panc1-P-C and Panc1-P-siCCNG2 after GEM exposure for 72 h. This assay was performed three times. (H) MTT assay demonstrated relative cell survival ratio of Panc1-P-l-OE cells transfected with ectopic CCNG2 (Panc1-P-l-OE-CCNG2-OE) and transfected with control vector (Panc1-P-l-OE-C) to GEM. (I) Western blotting demonstrated CCNG2 expression level in Panc1-P-l-OE-CCNG2-OE and Panc1-P-l-OE-C. Data represent mean±s.d. of three experiments; **P<0.01.
As for the in vivo experiment mentioned above, qRT–PCR and immunohistochemistry demonstrated that the CCNG2 expression was decreased in both the mRNA (data not shown) and protein levels (Figure 4G) in the Panc1-P-l-OE tumours at day 53. This result supported the conclusion that CCNG2 was one of the target genes for miR-1246.
miR-1246 and CCNG2 expression in primary pancreatic cancer samples
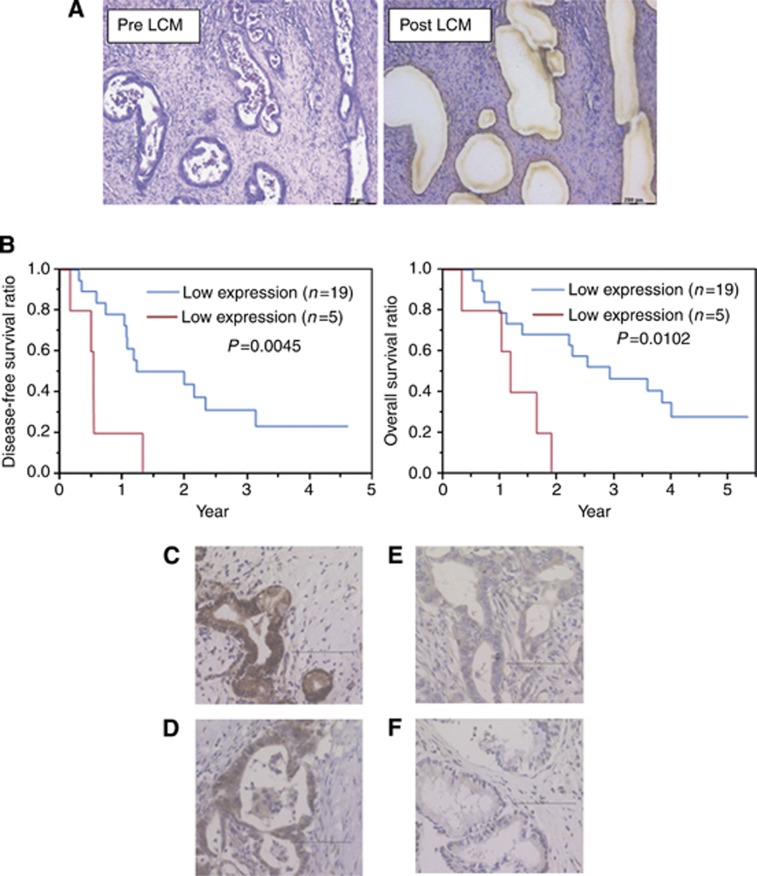
To study the clinical significance, we performed LCM to collect cancer sections from tumour tissues, qRT–PCR, and immunohistochemistry in 24 patients who underwent R0 resection (Figure 6A). The expression level of miR-1246 in each sample was shown in Supplementary Figure 5A. Of the 24 patients, the mean expression level of miR-1246 was 57.5 (/RNU48). We divided those patients into two groups by the mean value of miR-1246 expression (high or low). Disease-free survival ratio (P=0.0045) and overall survival ratios (P=0.0102) were significantly lower in the high miR-1246 expression group (Figure 6B).
Figure 6.
miR-1246 and CCNG2 expression in primary pancreatic cancer samples. (A) The representative image of pre/post (left/right) laser captured microdissection (LCM). Bar=200 μm. (B) Relationships between miR-1246 expression and disease-free survival, or overall survival. (C–F) Immunohistochemical staining of CCNG2 in 24 primary tumours. The CCNG2-positive cases show the diffuse (C) or spotted (D) nuclear patterns; CCNG2-negative ones depict cytoplasmic pattern (E; not stained in the nucleus) or the negative pattern (F; not stained in the nucleus or cytoplasm).
Subsequently, immunohistochemical staining for CCNG2 was performed in the corresponding 24 samples. The nuclei of normal pancreatic ductal cells were partially stained and the acinar cells were stained strongly in the cytoplasm and nuclei, which were used as positive controls (Supplementary Figure 5B and C). With cancer sections, although CCNG2 had been shown to appear not only in the nucleus but also in the cytoplasm, the functional CCNG2 protein may localise in the nucleus (Choi et al, 2009). We defined CCNG2-positive cases as those that showed diffuse or spotted nuclear patterns (Figure 6C and D), and CCNG2-negative cases as cytoplasmic pattern (not stained in the nucleus; Figure 6E) or an absent pattern (not stained in the nucleus or cytoplasm; Figure 6F) in pancreatic cancer lesions. Among the 24 patients examined, 15 (62.5%) showed positive staining, whereas 9 (37.5%) patients were negative for CCNG2.
Finally, we evaluated the patient backgrounds in low or high miR-1246 expression groups (Table 1 and Supplementary Table 3). The tumour location and CCNG2 expression in other clinicopathological factors showed significant difference between the two groups. The high miR-1246 expression group showed lower CCNG2 expression, whereas the low miR-1246 expression group showed high CCNG2 expression, with statistical significance (P=0.047). This result demonstrated that miR-1246 expression correlated inversely with CCNG2 expression in clinical samples, and an especially high expression level of miR-1246 predicted worse pancreatic cancer prognosis.
Table 1. The primary pancreatic cancer patients in the present study.
| |
miR-1246 expression |
|
|
|---|---|---|---|
| Low (n=19) | High (n=5) | P-value | |
| Age (<65:⩾65) | 7:12 | 0:5 | 0.14 |
| Sex (male:female) | 12:7 | 2:3 | 0.33 |
| Histopathological type (well or mod:poor) | 17:2 | 5:0 | 0.62 |
| Tumour size (mm) | 24.4±14.4 | 24.6±9.5 | 0.48 |
| Tumour location (head:body or tail) | 8:11 | 5:0 | 0.030 |
| Pathological depth of invasion depth pT (T1 or T2:T3) | 6:13 | 0:5 | 0.20 |
| Pathological lymph node metastasis pN (negative:positive) | 12:7 | 1:4 | 0.14 |
| Pathological stage (IA or IB or IIA:IIB or IV) | 12:7 | 1:4 | 0.14 |
| Adjuvant therapy (−/+) | 9:10 | 2:3 | 0.59 |
| CCNG2 expression (negative:positive) | 5:14 | 4:1 | 0.047 |
Abbreviations: mod=moderately differentiated; poor=poorly differentiated; well=well differentiated.
Discussion
Several miRNAs were reported in association with drug resistance against GEM: miR-15a (Zhang et al, 2010), miR-21 (Ali et al, 2010; Giovannetti et al, 2010), miR-200b and miR-200c (Ali et al, 2010; Li et al, 2009), miR-320c (Iwagami et al, 2013), and members of the let7 family (Li et al, 2009). Although these reports studied the bulk of tumours, we here identified chemoresistance- and cancer stemness-associated miRNAs. We focused on miR-1246, which was expressed in CSC-like spheroid cells. CSC-like spheroids showed chemoresistance for reagents, and the gene microarray analysis and GSEA revealed that the stemness-related pathways were increased, suggesting that the spheroids at least partially mimicked the CSC-like phenotype.
miR-1246 was reported as a diagnostic biomarker for oesophageal squamous cell carcinoma (Takeshita et al, 2013) and cervical cancer (Chen et al, 2014). However, few studies have reported the miR-1246 function in pancreatic cancer. In the present study, we demonstrated that miR-1246 induced chemoresistance and was related to cancer stemness in pancreatic cancer cell lines. Among miR-1246 targets, we focused on CCNG2 (Bates et al, 1996), a tumour suppressor gene. The CCNG2 expression was downregulated in the thyroid (Ito et al, 2003), oral (Kim et al, 2004), breast (Montagner et al, 2012), gastric (Sun et al, 2014a), oesophageal (Chen et al, 2013), prostate (Cui et al, 2014a), kidney (Cui et al, 2014b), and colorectal (Sun et al, 2014b) cancers. Antitumor agents induced CCNG2 expression and inhibited cancer (Kasukabe et al, 2008; Padua and Hansen, 2009; Zhao et al, 2011). We demonstrated that CCNG2 was decreased in pancreatic CSC-like spheroid cells and induced apoptosis, similarly to oral cancer (Kim et al, 2004). Given that CCNG2 deeply participated in cancer proliferation, invasion, chemoresistance, and differentiation, which characterise CSCs, CCNG2 may be involved at least partially in the maintenance of CSC-like spheroid cells. The present data showed CCNG2 expression was correlated inversely with miR-1246 expression, suggesting miR-1246 control CCNG2 function. We confirmed in vitro data by human primary tumours. Laser captured microdissection analysis and immunohistochemistry revealed the high miR-1246 expression and low CCNG2 expression in patients. The present study suggested that miR-1246-CCNG2 axis is critical for chemoresistance, and shows the candidacy as a bona fide useful predictive marker.
Acknowledgments
We thank the members of our laboratories for their fruitful discussion. This work was supported in part by a Grant-in-Aid for Scientific Research and a grant from the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology; a Grant-in-Aid from the Third Comprehensive 10-year Strategy for Cancer Control, Ministry of Health, Labor, and Welfare; a grant from the Kobayashi Cancer Research Foundation; a grant from the Princess Takamatsu Cancer Research Fund, Japan; a grant from the National Institute of Biomedical Innovation, Japan.
HI and MK received partial support from Chugai Co., Ltd., Yakult Honsha Co., Ltd., and Merck Co., Ltd. through institutional endowments.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang Z, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Rowan S, Vousden KH. Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene. 1996;13:1103–1109. [PubMed] [Google Scholar]
- Chen J, Yao D, Zhao S, He C, Ding N, Li L, Long F.2014MiR-1246 promotes SiHa cervical cancer cell proliferation, invasion, and migration through suppression of its target gene thrombospondin 2 Arch Gynecol Obstetin press. [DOI] [PubMed]
- Chen JQ, Liu CJ, Wen HX, Shi CL, Zhang HS, Li M, Sun GG. Changes in the expression of cyclin G2 in esophageal cancer cell and its significance. Tumour Biol. 2013;35:3355–3362. doi: 10.1007/s13277-013-1442-3. [DOI] [PubMed] [Google Scholar]
- Choi MG, Noh JH, An JY, Hong SK, Park SB, Baik YH, Kim KM, Sohn TS, Kim S. Expression levels of cyclin G2, but not cyclin E, correlate with gastric cancer progression. J Surg Res. 2009;157:168–174. doi: 10.1016/j.jss.2008.06.020. [DOI] [PubMed] [Google Scholar]
- Cui DW, Cheng YJ, Jing SW, Sun GG. Effect of cyclin G2 on proliferative ability of prostate cancer PC-3 cell. Tumour Biol. 2014;35:3017–3024. doi: 10.1007/s13277-013-1389-4. [DOI] [PubMed] [Google Scholar]
- Cui DW, Sun GG, Cheng YJ. Change in expression of cyclin G2 in kidney cancer cell and its significance. Tumour Biol. 2014;35:3177–3183. doi: 10.1007/s13277-013-1415-6. [DOI] [PubMed] [Google Scholar]
- Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, Danesi R, Campani D, Verheul HM, Boggi U. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Hoyert DL, Heron MP, Murphy SL, Kung HC. Deaths: final data for 2003. Natl Vital Stat Rep. 2006;54:1–120. [PubMed] [Google Scholar]
- Ito Y, Yoshida H, Uruno T, Nakano K, Takamura Y, Miya A, Kobayashi K, Yokozawa T, Matsuzuka F, Kuma K, Miyauchi A. Decreased expression of cyclin G2 is significantly linked to the malignant transformation of papillary carcinoma of the thyroid. Anticancer Res. 2003;23:2335–2338. [PubMed] [Google Scholar]
- Iwagami Y, Eguchi H, Nagano H, Akita H, Hama N, Wada H, Kawamoto K, Kobayashi S, Tomokuni A, Tomimaru Y, Mori M, Doki Y. miR-320c regulates gemcitabine-resistance in pancreatic cancer via SMARCC1. Br J Cancer. 2013;109:502–511. doi: 10.1038/bjc.2013.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukabe T, Okabe-Kado J, Honma Y. Cotylenin A, a new differentiation inducer, and rapamycin cooperatively inhibit growth of cancer cells through induction of cyclin G2. Cancer Sci. 2008;99:1693–1698. doi: 10.1111/j.1349-7006.2008.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shintani S, Kohno Y, Zhang R, Wong DT. Cyclin G2 dysregulation in human oral cancer. Cancer Res. 2004;64:8980–8986. doi: 10.1158/0008-5472.CAN-04-1926. [DOI] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardo E, Hermann PC, Mueller MT, Huber S, Balic A, Miranda-Lorenzo I, Zagorac S, Alcala S, Rodriguez-Arabaolaza I, Ramirez JC, Torres-Ruiz R, Garcia E, Hidalgo M, Cebrian DA, Heuchel R, Lohr M, Berger F, Bartenstein P, Aicher A, Heeschen C. Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9:433–446. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487:380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- Oettle H, Post S, Neuhaus P, Gellert K, Langrehr J, Ridwelski K, Schramm H, Fahlke J, Zuelke C, Burkart C, Gutberlet K, Kettner E, Schmalenberg H, Weigang-Koehler K, Bechstein WO, Niedergethmann M, Schmidt-Wolf I, Roll L, Doerken B, Riess H. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- Padua MB, Hansen PJ. Changes in expression of cell-cycle-related genes in PC-3 prostate cancer cells caused by ovine uterine serpin. J Cell Biochem. 2009;107:1182–1188. doi: 10.1002/jcb.22222. [DOI] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun GG, Hu WN, Cui DW, Zhang J. Decreased expression of CCNG2 is significantly linked to the malignant transformation of gastric carcinoma. Tumour Biol. 2014;35:2631–2639. doi: 10.1007/s13277-013-1346-2. [DOI] [PubMed] [Google Scholar]
- Sun GG, Zhang J, Hu WN. CCNG2 expression is downregulated in colorectal carcinoma and its clinical significance. Tumour Biol. 2014;35:3339–3346. doi: 10.1007/s13277-013-1440-5. [DOI] [PubMed] [Google Scholar]
- Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Komatsu A, Jitsukawa M, Matsubara H. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Liu Y, He H, Chen X, Chen J, Lu YC. Candidate genes influencing sensitivity and resistance of human glioblastoma to Semustine. Brain Res Bull. 2011;86:189–194. doi: 10.1016/j.brainresbull.2011.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.