Abstract
For more than a decade, protein-replacement therapy has been employed successfully for the treatment of Gaucher disease. Recently, a comparable therapy has become available for the related lipid-storage disorder Fabry disease. Two differently produced recombinant α-galactosidase A (α-gal A) preparations are used independently for this purpose. Agalsidase α is obtained from human fibroblasts that have been modified by gene activation; agalsidase β is obtained from Chinese hamster ovary cells that are transduced with human α-gal A cDNA. It has previously been claimed that α-gal A mRNA undergoes editing, which may result in coproduction of an edited protein (Phe 396 Tyr) that might have a relevant physiological function. We therefore analyzed the occurrence of α-gal A editing, as well as the precise nature, in this respect, of the therapeutic enzymes. No indications were obtained for the existence of editing at the protein or RNA level. Both recombinant enzymes used in therapy are unedited and are capable of functionally correcting cultured fibroblasts from Fabry patients in their excessive globotriaosylceramide accumulation. Although RNA editing is apparently not relevant in the case of α-gal A, a thorough analysis of the potential occurrence of editing of transcripts is nevertheless advisable in connection with newly developed protein-replacement therapies.
Introduction
Fabry disease (MIM 301500) is an X-linked lysosomal storage disorder caused by a deficiency of the lysosomal enzyme α-galactosidase A (α-gal A). This defect results in the progressive accumulation of glycosphingolipids in lysosomes of endothelial, perithelial, and smooth muscle cells of the vascular system, as well as renal epithelial cells, myocardial cells, and cells of the autonomic nervous system (Desnick et al. 1996). The accumulating glycosphingolipids contain terminal α-galactosyl moieties, such as globotriaosylceramide (Gb3); galabiosylceramide (Gb2); and, to a lesser extent, blood group B, B1, and P1 antigens (Martensson 1966; Wherrett and Hakomori 1973; Desnick et al. 1996) Fabry disease patients suffer from angiokeratomas, acroparesthesias, corneal dystrophy, renal failure, and myocardial and cerebral infarction, resulting in premature mortality. Since the α-gal A gene is located on the X chromosome (locus Xq22), all hemizygous males suffer from Fabry disease, although the majority of female heterozygous carriers also display attenuated forms of the disease. Gaucher disease, a related glycolipid storage disorder caused by a deficiency of the lysosomal β-glucocerebrosidase, can be treated by chronic intravenous administration of the enzyme (Barton et al. 1991). This so-called enzyme-replacement therapy is found to be remarkably effective for the non-neuronopathic variant of Gaucher disease, resulting in reversal of the major clinical symptoms. A comparable approach has recently been applied to the treatment of Fabry disease. Two differently produced enzyme preparations have independently been examined in clinical investigations: enzyme produced by Chinese hamster ovary (CHO) cells with classic recombinant technology (agalasidase β, Fabrazyme) and enzyme produced by cultured human skin fibroblasts with an activated promoter of the α-gal A gene (agalasidase α, Replagal). In both studies, promising lipid substrate reductions in tissue biopsies have been observed (Eng et al. 2001; Pastores and Thadhani 2001; Schiffmann et al. 2001) Very recently, both recombinant products have been registered by the European Medical Evaluation Agency (EMEA) in Europe, and their registration in the United States is pending.
The α-gal A protein is composed of two identical subunits of ∼49 kDa and is encoded by a mature transcript of 1,350 nucleotides (Calhoun et al. 1985; Bishop et al. 1986). Interestingly, the human α-gal A mRNA previously has been reported to undergo RNA editing: a uridylate residue at position 1187 of the mature transcript is converted to an adenylate residue (Novo et al. 1995). The result of this nucleotide conversion would be an amino acid substitution (Phe 396 Tyr) in the C-terminal part of the encoded protein, which theoretically could lead to two α-gal A proteins with different functions or modes of regulation in the human body. Examples of such a “single gene, multiple proteins” scenario have been described in detail for several human genes (for comprehensive reviews, see Chang et al. [1998] and Rueter and Emeson [1998]). For example, the human ApoB mRNA, L-glutamate receptor subunit mRNAs and serotonin receptor subunit mRNAs have all been shown to undergo different forms of RNA editing, resulting in the translation of different protein isoforms with different functions or physical properties in the human body (Chen et al. 1987; Powell et al. 1987; Burns et al. 1997). The report claiming the existence of editing of the human α-gal A transcript has so far apparently not raised concerns, despite the fact that it may have important implications for the products used in enzyme-replacement therapy for Fabry disease. The mRNA produced by gene activation in human fibroblasts should undergo editing. In the case of the classic recombinant technology using transduction with cDNA, the occurrence of RNA editing can not be predicted. We therefore studied the natural occurrence of “edited” α-gal A mRNA to gain insight into the biological significance of RNA editing of α-gal A in humans. Moreover, the editing status of the two therapeutic enzyme preparations was investigated.
Material and Methods
Isolation and Culturing of Human Macrophages and Fibroblasts
Human peripheral blood monocytes were isolated and cultured as described elsewhere (Hollak et al. 1994). Spontaneous differentiation of monocytes into activated macrophages occurs on prolonged cell culture (Hollak et al. 1994). Human skin fibroblasts were cultured as described elsewhere (Wanders et al. 1992).
RNA and DNA Isolation
Total RNA from macrophages was isolated by use of the RNAzol B RNA isolation kit (Tel-Test), as described elsewhere (Boot et al. 1998). Total RNA from fibroblasts was isolated as described in (IJlst et al. 1994). DNA was isolated according to standard protocols (Sambrook et al. 1989).
Amplification of DNA and RNA Fragments (RT-PCR)
For PCR amplification of DNA fragments, the following protocol was used: 1 ng of recombinant DNA or 500 ng of total human DNA was incubated with 20 pmol oligonucleotides for 3 min at 94°C, after which it was amplified in 35 cycles of 30 s at 94°C, 30 s at 48°C, and 1 min at 72°C. The exact annealing temperature depended on the length and GC content of the oligonucleotides used in a particular experiment: 4°C per G or C, plus 2°C per A or T, minus 5°C. For these reactions, 1 U of Taq polymerase and buffers were used according to the manufacturer's instructions (Promega).
For amplification of RNA segments (RT-PCR), the PCR protocol was preceded by cDNA synthesis: 1 μg oligo (dT)12–18 was added to 10 μg of human total RNA, was denatured for 4 min at 74°C, and then was immediately put on ice and added to the RT mix containing 1× first strand buffer, 10 mM DTT, 500 μM each dNTP, 20 U of RNAsin (Promega), and 200 U Superscript II (Gibco BRL) in a total volume of 30 μl. The mixture was incubated for 2 h at 45°C, followed by 5 min at 95°C; 1 μl of the mixture was used for PCR, essentially as described above.
Oligonucleotides used were: DB001S, GAATTCACCATGCAGCTGAGGAACCCAGA; DB002S, GAATTCCTGGACAATGGATTGGCAAGG; DB003AS, GAATTCTTAAAGTAAGTCTTTTAATGACAT; DB005S, GCAGGCTTCCCTGGGAGT; DB006AS, TAAAGGAGCAGCCATGATAGC; DB007S, CCTCTGCATTGATAACTGTTGGATGGCTCCCC; DB008AS, GGGGAGCCATCCAACAGTTATCAATGCAGAGG; DB009S, GGAAGCTAGGGTACTATGAATGGACTTCAAGG; DB010AS, CCTTGAAGTCCATTCATAGTACCCTAGCTTCC; and DB011AS, TTGCGGCCGCTTAAAGTAAGTCTTTTAATGACAT. Underlined nucleotides indicate mismatches introduced for site-directed mutagenesis (see below).
Cloning of the Human α-gal A cDNA
The complete ORF encoding human α-gal A (including the signal peptide sequence; see Genbank accession number gi:3808177) was amplified in an RT-PCR procedure, making use of the oligonucleotides DB001S and DB011AS (see above). The amplified fragment was cloned into the pGEM-T vector (Promega), and positive clones were digested with EcoRI and NotI. The resulting 1,299-bp EcoRI-NotI fragment was then cloned into pcDNA3.1(+) (Invitrogen), which had been digested with EcoRI and NotI, resulting in plasmid pDB007.
Site-Directed Mutagenesis and Sequencing of Clones
Site-directed mutagenesis was performed by use of the QuikChange Site-Directed Mutagenesis Kit (Stratagene). In short, this procedure made use of plasmid pDB007, which contains the correct full-length α-gal A ORF. In this clone, the edited sequence was introduced artificially by use of the oligonucleotides DB009S and DB010AS (see above; mutated nucleotides underlined). The resulting clone was named pDB009. The catalytically inactive α-gal A clone was generated according to the same procedure, making use of the oligonucleotides DB007S and DB008AS (see above; mutated nucleotides underlined). The resulting clone was named “pDB008” and encodes an α-gal A protein that contains an asparagine residue instead of an aspartate residue at position 93.
Nucleotide sequences of both strands of positive clones were verified by use of universal M13 primers and the oligonucleotides listed above, on an Applied Biosystems 377A automated DNA sequencer, following ABI protocols.
Expression of Recombinant Proteins in COS-1 Cells
Transient transfection of COS-1 cells with different α-gal A constructs was performed by the DEAE-Dextran method, essentially as described elsewhere (Lopata et al. 1984).
Therapeutic Enzymes
Agalsidase α (Replagal, TKT) and agalsidase β (Fabrazyme, Genzyme) were obtained from the local hospital pharmacy.
Enzyme Activity Measurement
The enzymatic activity of α-gal A was measured using the fluorogenic substrate 4-methylumbelliferyl-α-D-galactopyranoside (4-MU-galactopyranoside, Sigma), essentially as described elsewhere (Ioannou et al. 1992). In brief, 25 μl of cleared medium was incubated with 100 μl reaction mixture containing the 4-MU substrate (final concentration 3.5 mM) in 100 mM citrate/200 mM phosphate buffer pH 4.6, at 37°C, for 1 h. Reactions contained 100 mM N-acetylgalactosamine to inhibit α-galactosidase B activity (Mayes et al. 1981). Reactions were terminated by the addition of 2 ml 300 mM Glycine/NaOH buffer, pH 10.6, and fluorescent 4-methyl-umbelliferone was measured with a fluorimeter (Perkin-Elmer) at 445 nm.
Specific activity of the commercial α-gal A preparations was determined using the protein measurement procedure by Lowry with human albumin as reference protein. Specific activity of α-gal A enzyme produced in COS-1 cells was determined following 10% SDS-PolyAcrylAmide gel electrophoresis and silverstaining with a commercial α-gal A preparation (Agalsidase β, Fabrazyme, Genzyme) as reference protein.
Globotriaosylceramide (Gb3) Measurement
The levels of Gb3 in fibroblasts were determined following lipid extraction of harvested cells using the thin layer chromatographic procedure described by Desnick and coworkers (1971).
Generation and Purification of Antibodies
280 μg of recombinant human α-gal A protein (Genzyme), either native or denatured, was used to immunize rabbits. Generated antibodies were purified by the use of a MAbTrap G II column (Pharmacia Biotech), according to the manufacturer’s instructions. This procedure made use of Protein G, which has affinity for rabbit IgGs, coupled to a Sepharose matrix.
Purification of Human α-gal A
Human α-gal A was comparably purified from various sources (culture media of transfected COS-1 cells; urine samples from a healthy male) by use of sequential concanavalin A sepharose column chromatography, immunoaffinity precipitation, and preparative isoelectric focusing. In brief, fractions containing enzyme activity were concentrated using the Amicon Diaflo system, with a 10-kDa cutoff membrane, and were washed with a 100-mM potassium-phosphate buffer, pH 6.5, containing 50 mM NaCl (buffer A). The concentrated fractions were next applied to a 30-ml ConA-Sepharose column that had been equilibrated with buffer A. Unbound proteins were removed by washing with 10-column volumes of buffer B (100 mM potassium phosphate pH 6.5; 500 mM NaCl). Bound proteins were eluted from the ConA column with buffer B containing 100 mM methylmannoside and were collected in 1-ml fractions. Fractions containing α-gal A activity were pooled and concentrated using Millipore Ultrafree-MC centrifugal filter units with a molecular cutoff of 10 kDa. This concentrated material was then subjected to immunoprecipitation using anti-galactosidase A antiserum covalently coupled to CNBr-activated Sepharose 4B according to the manufacturer’s instructions (Pharmacia Biotech). After extensive washing in phosphate buffered saline, the bound enzyme was released by incubation in 0.1 M citric acid (pH 3.5). Following neutralization with potassium phosphate buffer, the fraction was subjected to preparative isoelectric focusing, using Servalyt 4-9 T (Serva) as carrier ampholytes. After focusing, the gel was divided into 30 fractions, and proteins were extracted by the addition of 1 ml water followed by centrifugation. The supernatants were then tested for α-gal A activity.
Determination of Amino Acid Sequences of α-gal A
For matrix-assisted laser desorption ionization (MALDI) analysis, protein-containing gel slices were S-alkylated, were digested with trypsin (Boehringer-Mannheim, sequencing grade), and were extracted (Shevchenko et al. 1996). Only the peptides eluted with 20 mM NH4HCO3 were used in the MALDI analysis. After drying in a vacuum centrifuge, the peptides were dissolved in 10 ml 60% acetonitrile, 1% formic acid. 0.5 ml of this solution was mixed with 0.5 ml of a solution of 52 mM α-cyano-4-hydroxycinnamic acid in 49% ethanol/49% acetonitrile/2% TFA and 1 mM ammonium acetate. Prior to dissolving, the α-cyano-4-hydroxycinnamic acid (Sigma) was washed briefly with acetone. The mixture was spotted on a targetplate and allowed to dry at room temperature. Reflectron MALDI-TOF spectra were acquired on a Micromass Tof Spec 2EC (Micromass). The resulting peptide spectra were used to search a nonredundant protein sequence database (SWISS-PROT/TREMBL) using the Protein Probe program.
Results
Occurrence of Edited α-gal A Transcripts in Humans
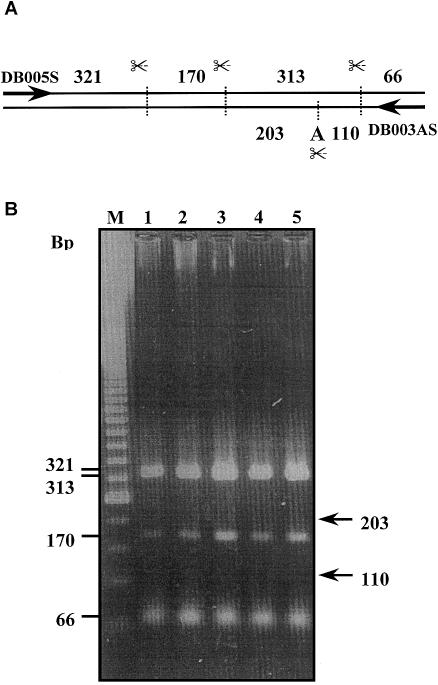
The reported editing of human α-gal A mRNA involves a single-nucleotide conversion in the 3′ end of the coding region (nucleotide 1187; see Novo et al. 1995). To investigate the frequency of the editing phenomenon, we performed RT-PCR analyses on mRNA isolated from macrophages and fibroblasts from different individuals. To be sure that transcripts encoded by a single gene were analyzed, only RNA isolated from males was analyzed. As outlined in figure 1, an 870-bp fragment of the α-gal A mRNA was amplified that subsequently was digested with RsaI and was examined on agarose gels. Since the described conversion of uridylate residue 1187 into an adenylate residue should give rise to the creation of an additional RsaI restriction site, amplified fragments containing the edited nucleotide sequence should be discriminated in this analysis (fig. 1A). However, no digestion fragments derived from edited α-gal A mRNA were detected in any of the macrophage RNA samples (fig. 1B, lanes 3–5; note the absence of DNA fragments of 203 and 110 bp). Since Novo et al. reported editing levels of 63% for α-gal A mRNA isolated from fibroblasts, our RT-PCR analyses also included two fibroblast RNA samples as positive controls (lanes 1 and 2). Also these RNA samples did not show the presence of digested PCR fragments derived from edited α-gal A mRNA (fig. 1B). The unexpected result could not be attributed to an inactive RsaI enzyme, since RsaI digestion fragments from unedited α-gal A mRNA were detectable. This finding therefore rather suggested that the investigated macrophages and fibroblasts seemed to be devoid of edited α-gal A mRNA (fig. 1B).
Figure 1.
Apparent absence of α-gal A mRNA editing. RT-PCR followed by RsaI digestion of macrophage- and fibroblast-derived RNA. A, Schematic overview of expected RsaI digestion fragment pattern. B, lanes 1 and 2, patterns for fibroblasts of two control subjects; lanes 3–5, patterns for macrophages of three control subjects.
The analysis of the editing status of fibroblast α-gal A mRNA was further extended. We isolated mRNA from fibroblasts of 10 different males and amplified part of the α-gal A mRNA containing the putative editing site. From each amplified mRNA fragment, the nucleotide sequence of at least 10 independent clones was determined. This extensive analysis made clear that none of the amplified α-gal A mRNA fragments contained an edited sequence, indicating that α-gal A mRNA is most likely not edited in the fibroblasts from the 10 donors tested (table 1).
Table 1.
Analysis of α-gal A mRNA Editing in Independent Fibroblast and Macrophage Clones
| Male | RNASource | No. ofClonesSequenced | No. of Clonesin Unedited/EditedSequence |
| M | Fibroblast | 24 | 24/0 |
| 97-3138 | Fibroblast | 10 | 10/0 |
| 92-052 | Fibroblast | 10 | 10/0 |
| 98-335 | Fibroblast | 10 | 10/0 |
| 96-229 | Fibroblast | 10 | 10/0 |
| 99-3138 | Fibroblast | 10 | 10/0 |
| 92-137 | Fibroblast | 10 | 10/0 |
| 90-225 | Fibroblast | 10 | 10/0 |
| 94-285 | Fibroblast | 10 | 10/0 |
| 98-361 | Fibroblast | 10 | 10/0 |
| WDJ | Macrophage | 29 | 29/0 |
Analysis of α-gal A Protein by MALDI Mass Spectrometry
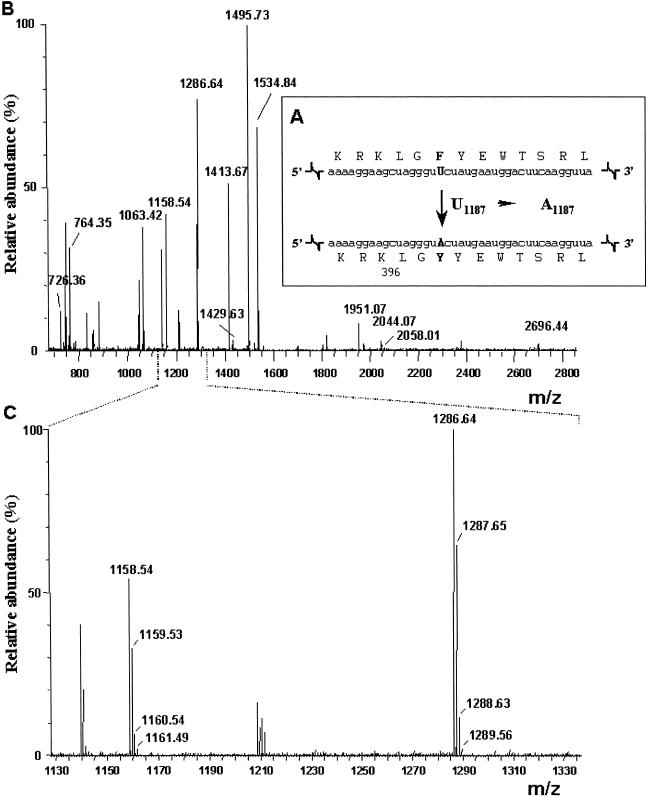
The two commercial α-gal A preparations were investigated by mass spectrometry as described in the “Material and Methods” section. Identical results were obtained for both preparations. As illustrated in figure 2B and table 2, a peptide mass fingerprinting analysis clearly identified 13 tryptic peptides derived from α-gal A (giving a coverage of 24%). The part of the protein derived from the putative editing domain (see fig. 2A and 2B) could easily be detected. It was observed as two peptide peaks at mass/charge (m/z) values of 1,158.54 and 1,286.64, caused by the presence of an additional lysine residue at the N-terminus of one of the two peptides (fig. 2C and table 2). This pattern is often found when trypsine is confronted with two successive basic residues (in this instance, RK). These two peptides were clearly derived from unedited α-gal A mRNA sequences (see table 1). The peptides derived from the “edited” protein sequence (fig. 2A) would be expected at m/z values of 1,174 and 1,302 (table 1). However, as can be seen in the enlarged mass spectrum (fig. 1C), these peptides could not be detected. Since the predicted peptides result from a very minor, F→Y change in the middle of the peptides, we would not expect this absence to be due to a difference in efficiency of ionization or recovery of the respective peptides. This seems especially true in the light of the fact that more profound differences—like the absence of the N-terminal lysine residue in one of the peptides—do not lead to clear differences in ionization or recovery of these peptides. These results show that both α-gal A enzyme preparations do not contain “edited” α-gal A protein.
Figure 2.
Recombinant α-galactosidase contains only “unedited” peptide sequence. A, Schematic representation of the editing site of human α-gal A mRNA and the resulting amino acid change. B, Full mass spectrum of agalsidase β. The X-axis indicates mass/charge values (m/z); the Y-axis indicates the relative intensity of detected ions (%). Identified peptides derived from α-gal A are indicated with their mass value. C, Enlargement of the 1,130–1,330 m/z range of panel B, with the isotopic distribution around the m/z 1,158.54 and 1,286.64 peaks. X-axis and Y-axis as in panel B.
Table 2.
Identification of Peptide Sequences of Recombinant Human α-gal A[Note]
| PredictedMolecularWeight | ObservedMolecularWeight | Sequence |
| 726.3973 | 726.36 | (R)FPHGI R(Q) |
| 764.3977 | 764.35 | (K)QGYQL R(Q) |
| 1,063.4454 | 1,063.42 | (R)QYCNH WR(N) + |
| 1,158.5505 | 1,158.54 | (K)LGFYE WTSR(L) |
| 1,286.6455 | 1,286.64 | (R)KLGFY EWTSR(L) * |
| 1,413.6659 | 1,413.67 | (R)TPTMG WLHWE R(F) |
| 1,429.6608 | 1,429.63 | (R)TPTMG WLHWE R(F) # |
| 1,495.7103 | 1,495.73 | (K)SILDW TSFNQ ER(I) |
| 1,534.8164 | 1,534.84 | (R)LQADP QRFPH GIR(Q) * |
| 1,951.0785 | 1,951.07 | (K)ALLQD KDVIA INQDP LGK(Q) * |
| 2,044.0758 | 2,044.07 | (K)GVACN PACFI TQLLP VKR(K) * + + |
| 2,058.1278 | 2,058.01 | (K)GVACN PACFI TQLLP VKRK(L) ** |
| 2,696.4657 | 2,996.44 | (K)ALLQD KDVIA INQDP LGKQG YQLR(Q) ** |
| 1,174.5499 | Not found | (K)LGYYE WTSR(L) @ |
| 1,302.6449 | Not found | (R)KLGYYE WTSR(L) @ |
Note.— Indicated are molecular weights of peptides predicted on the basis of the α-gal A cDNA sequence and the molecular weights obtained by mass spectrometry. Shown in the right column are the amino acid sequences that comply with the observed masses; parentheses indicate neighboring amino acids that are not part of the peptide. The last two predicted molecular weights are based on the putative edited sequence of the α-gal A mRNA sequence (see fig. 1a). Symbols: + = carboxyamidomethylcysteine(s); * = missed cleavage(s); # = methionine sulfoxide; @ = “Edited” peptide.
Generation of “Edited” α-gal A Protein and Comparison with Other Enzyme Preparations
A cDNA construct was generated that contained the edited α-gal A nucleotide sequence (see the “Material and Methods” section for details). In parallel, COS-1 cells were transfected with either a construct containing the normal α-gal A nucleotide sequence (clone pDB007) or a construct containing an α-gal A nucleotide sequence with a point mutation in the putative catalytic site (clone pDB008 [Zeidner et al. 1993]) as positive and negative controls, respectively. Expression in COS-1 cells of the construct containing the putative catalytic-site mutation did not result in the synthesis of catalytically active α-gal A protein (pDB008). Expression of the construct containing the edited α-gal A nucleotide sequence (pDB009), however, led to the synthesis of normal amount of catalytically active α-gal A protein. Enzyme in the medium of the transfected COS-1 cells with wild-type and “edited” α-gal A cDNA was next purified by sequential concanavalin A chromatography, anti α-gal A immunoprecipitation, and isoelectric focusing. The specific activity of the wild-type and “edited” (Phe 396 Tyr) α-gal A produced in COS-1 cells was similar (4.1 ± 0.4 × 106 and 4.0 ± 0.2 × 106 nmol/mg/h (mean ± SD), respectively). The specific activity was comparable to that of the two therapeutic enzyme preparations agalsidase α (Replagal) and agalsidase β (Fabrazyme), being 4.7 ± 0.5 × 106 and 4.9 ± 0.5 × 106 nmol/mg.hour, respectively.
Functional Correction of Fabry Fibroblasts by α-gal A Preparations
Cultured fibroblasts from normal subjects and two patients with Fabry disease were incubated for 3 h, with either agalsidase α or agalsidase β preparation (0.1 mmol substrate hydrolysis per h), in the absence or presence of 5 mM mannose phosphate. Next, the medium was changed, and cells were harvested after extensive washing. The cellular α-gal A activity was determined. Table 3 shows the results of one such experiment mimicking enzyme-replacement therapy. Similar data were obtained in two other, independent experiments with cells derived from four different patients. As can be seen in table 3, the uptake of enzyme from both commercial preparations was highly similar and almost completely inhibited by the presence of mannose-6-phosphate in the culture medium. Impressive corrections in cellular α-gal A activity levels were observed in the Fabry fibroblasts even after a 3-d delay. In both enzyme preparations, cellular α-gal A activity was always at least 20%–30% of that immediately after the pulse labeling of the Fabry fibroblasts (n=4, in three independent experiments). Moreover, the enzyme uptake resulted in functional correction as illustrated by the marked reduction in Gb3 levels. In fibroblasts from one patient with Fabry disease, the lipid level was reduced by 86% and 82% following exposure to agalsidase α and β preparations (0.1 mmol/h for 3 h), respectively. In fibroblasts from another patient with Fabry disease, the reductions were 72% and 70%, respectively.
Table 3.
Correction of Cellular α-gal A Activity Following Exposure to Recombinant Enzyme Preparations[Note]
|
Enzyme Activity(nmol/h/mg protein) |
|||||
| Agalsidase α |
Agalsidase β |
||||
| Source ofFibroblasts | No Enzyme | −M6P | +M6P | −M6P | +M6P |
| Control subject 1 | 240 | 4,300 | 810 | 3,900 | 790 |
| Control subject 2 | 285 | 3,560 | 620 | 3,500 | 710 |
| Patient 1 | 25 | 4,640 | 955 | 3,360 | 680 |
| Patient 2 | 11 | 3,105 | 720 | 3,700 | 605 |
Note.— Cells were incubated for 3 h with 0.1 × 106 nmol/h α-gal A activity. Enzyme activity in cell extracts was determined as described in the “Material and Methods” section.
Discussion
Fabry disease is an X-linked lysosomal storage disorder that affects ∼1/40,000 newborns (Desnick et al. 1996). Protein-replacement therapy using recombinant α-gal A is considered to allow correction of the harmful lysosomal accumulation of glycosphingolipids containing terminal α-galactosyl moieties in various cell types of patients with Fabry disease. Obviously, the quality of the enzyme product used in such replacement therapy is of key importance. In this connection, it is surprising that little attention has been paid to the report by Novo et al. (1995) that α-gal A mRNA undergoes RNA editing. On the basis of their findings, two distinct α-gal A proteins could, in theory, be encoded by a single α-gal A gene, with each of them having different physical properties and possibly different functions in the human body. At present, two different α-gal A preparations are employed that are produced by two entirely different techniques. Agalsidase α, produced by human fibroblasts following gene activation, could theoretically undergo editing. In contrast, agalsidase β, which is produced by transduction of Chinese hamster ovary cells with cDNA, can only undergo editing if this form of editing does not require the presence of intron sequences (compare Rueter and Emeson 1998). It is therefore of the utmost importance to know precisely which α-gal A protein forms are administered during replacement therapy and whether different α-gal A isoforms with different functions exist in vivo. In the present article, we report that both recombinantly produced α-galactosidases A used for therapy contain the unedited amino acid sequence only. Both recombinant enzyme preparations can functionally correct cultured fibroblasts from patients with Fabry disease, causing a correction in Gb3 lipid levels.
Our investigation also found no evidence for the existence of α-gal A editing in humans. This could be concluded from a detailed study of the editing status of human α-gal A transcripts. According to Novo et al. (1995), 40%–60% of the α-gal A transcripts isolated from fibroblast, muscle, and cerebellum contain the edited nucleotide sequence. In contrast, our RT-PCR analyses of macrophage and fibroblast RNA from different male individuals revealed no edited α-gal A transcripts. This was further substantiated by the α-gal A nucleotide-sequence determination of at least 10 independent clones obtained from fibroblast RNA from 10 different male individuals each (totalling >130 independent cDNA clones). These data—and the fact that α-gal A editing would involve the only known instance of U→A base conversion, for which no simple enzymatic pathway can be envisaged—lead us to conclude that α-gal A transcripts are not edited. Further support for this absence of editing of α-gal A transcripts comes from EST databases. Currently, the human EST database at the National Center for Biotechnology Information Web site contains over 60 α-gal A tags from different human tissues, of which 44 contain the putative editing domain. Of these 44 tags, however, none contains the edited sequence as reported by Novo et al. (1995). The reason for the discrepancy between our data and those of Novo and coworkers is hard to identify. One possible explanation would be the existence of polymorphic sequences in the α-gal A gene. Sequencing of the corresponding cDNA would then lead one to conclude (incorrectly) that the mRNA is edited, when compared to genomic database sequences. Novo and coworkers, however, compared their cDNA sequences with genomic sequences obtained from the same individual, making this explanation unlikely. A more plausible explanation would be that Novo et al. (1995) picked up a cDNA fragment containing a PCR mistake that contaminated subsequent PCR reactions. Such a contamination would be difficult to detect in cDNA PCR amplifications and cannot be detected in the genomic PCR reactions as carried out by Novo et al., since their approach makes use of primers that are specific for genomic sequences, not cDNA sequences.
In conclusion, our data show that patients with Fabry disease currently receive therapy with unedited α-gal A protein only and that edited α-gal A transcripts do not occur in humans in vivo. Although RNA editing is apparently not relevant in the case of α-gal A, it has to be realized that a thorough analysis of the potential occurrence of editing of transcripts is nevertheless advisable in connection with protein-replacement therapies.
Acknowledgments
We thank Jaap Willem Back for expert help with the mass spectrometry analyses. The Micromass Tof Spec 2EC was largely funded by a grant from the Council for Medical Sciences of the Netherlands Organisation for Scientific Research (NWO). We thank Thijs Hendriks for expert help in the generation of the anti–α-gal A antibodies. We thank Dr. R. J. A. Wanders and Mrs. E. M. Hogenhout for the donation of human fibroblast RNA. We thank Dr. R. Benne for stimulating discussions and editorial comments on the manuscript.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/ (for human EST database)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Fabry disease [MIM 301500])
References
- Barton NW, Brady RO, Dambrosia JM, Di Bisceglie AM, Doppelt SH, Hill SC, Mankin HJ, Murray GJ, Parker RI, Argoff CE (1991) Replacement therapy for inherited enzyme deficiency–macrophage-targeted glucocerebrosidase for Gaucher's disease. N Engl J Med 324:1464–1470 [DOI] [PubMed] [Google Scholar]
- Bishop DF, Calhoun DH, Bernstein HS, Hantzopoulos P, Quinn M, Desnick RJ (1986) Human alpha-galactosidase A: nucleotide sequence of a cDNA clone encoding the mature enzyme. Proc Natl Acad Sci USA 83:4859–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot RG, Renkema GH, Verhoek M, Strijland A, Bliek J, de Meulemeester TM, Mannens MM, Aerts JM (1998) The human chitotriosidase gene: nature of inherited enzyme deficiency. J Biol Chem 273:25680–25685 [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387:303–308 [DOI] [PubMed] [Google Scholar]
- Calhoun DH, Bishop DF, Bernstein HS, Quinn M, Hantzopoulos P, Desnick RJ (1985) Fabry disease: isolation of a cDNA clone encoding human α-galactosidase A. Proc Natl Acad Sci USA 82:7364–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BHJ, Lau PP, Chan L (1998) Apoliprotein B mRNA editing. In: Grosjean H, Benne R (eds) Modification and editing of RNA. American Society for Microbiology, Washington, DC, pp 325–342 [Google Scholar]
- Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, Silberman SR, Cai SJ, Deslypere JP, Rosseneu M (1987) Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science 238:363–366 [DOI] [PubMed] [Google Scholar]
- Desnick RJ, Dawson G, Desnick SJ, Sweeley CC, Krivit W (1971) Diagnosis of glycosphingolipidoses by urinary-sediment analysis. N Engl J Med 284:739–744 [DOI] [PubMed] [Google Scholar]
- Desnick RJ, Ioannou YA, Eng ME (1996) α-galactosidase A deficiency: Fabry disease. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 6th ed. McGraw-Hill, New York, pp 2741–2784 [Google Scholar]
- Eng CM, Guffon N, Wilcox WR, Germain DP, Lee P, Waldek S, Caplan L, Linthorst GE, Desnick RJ (2001) Safety and efficacy of recombinant human α-galactosidase A replacement therapy in Fabry's disease. N Engl J Med 345:9–16 [DOI] [PubMed] [Google Scholar]
- Hollak CE, van Weely S, van Oers MH, Aerts JM (1994) Marked elevation of plasma chitotriosidase activity: a novel hallmark of Gaucher disease. J Clin Invest 93:1288–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJlst L, Wanders RJ, Ushikubo S, Kamijo T, Hashimoto T (1994) Molecular basis of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency: identification of the major disease-causing mutation in the alpha-subunit of the mitochondrial trifunctional protein. Biochim Biophys Acta 1215:347–350 [DOI] [PubMed] [Google Scholar]
- Ioannou YA, Bishop DF, Desnick RJ (1992) Overexpression of human alpha-galactosidase A results in its intracellular aggregation, crystallization in lysosomes, and selective secretion. J Cell Biol 119:1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata MA, Cleveland DW, Sollner-Webb B (1984) High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res 12:5707–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martensson E (1966) Neutral glycolipids of human kidney isolation, identification, and fatty acid composition. Biochim Biophys Acta 116:296–308 [DOI] [PubMed] [Google Scholar]
- Mayes JS, Scheerer JB, Sifers RN, Donaldson ML (1981) Differential assay for lysosomal α-galactosidases in human tissues and its application to Fabry's disease. Clin Chim Acta 112:247–251 [DOI] [PubMed] [Google Scholar]
- Novo FJ, Kruszewski A, MacDermot KD, Goldspink G, Gorecki DC (1995) Editing of human α-galactosidase RNA resulting in a pyrimidine to purine conversion. Nucleic Acids Res 23:2636–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastores GM, Thadhani R (2001) Enzyme-replacement therapy for Anderson-Fabry disease. Lancet 358:601–603 [DOI] [PubMed] [Google Scholar]
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J (1987) A novel form of tissue-specific RNA processing produces apolipoprotein- B48 in intestine. Cell 50:831–840 [DOI] [PubMed] [Google Scholar]
- Rueter SM, Emeson RB (1998) Adenoside-to-inosine conversion in mRNA. In: Grosjean H, Benne R (eds) Modification and editing of RNA. American Society for Microbiology, Washington, DC, pp 343–361 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schiffmann R, Kopp JB, Austin HA, III, Sabnis S, Moore DF, Weibel T, Balow JE, Brady RO (2001) Enzyme replacement therapy in Fabry disease: a randomized controlled trial. JAMA 285:2743–2749 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858 [DOI] [PubMed] [Google Scholar]
- Wanders RJ, IJlst L, Poggi F, Bonnefont JP, Munnich A, Brivet M, Rabier D, Saudubray JM (1992) Human trifunctional protein deficiency: a new disorder of mitochondrial fatty acid beta-oxidation. Biochem Biophys Res Commun 188:1139–1145 [DOI] [PubMed] [Google Scholar]
- Wherrett JR, Hakomori SI (1973) Characterization of a blood group B glycolipid, accumulating in the pancreas of a patient with Fabry's disease. J Biol Chem 248:3046–3051 [PubMed] [Google Scholar]
- Zeidner KM, Ioannou YA, Desnick RJ (1993) Human α-gal A active site residue: determination by homology, mutagenesis, and transient expression. Am J Hum Genet Suppl 53:1682 [Google Scholar]