Abstract
Oculocutaneous albinism (OCA) is a genetically heterogeneous disorder. There are four known types of OCA: OCA1–OCA4. The clinical manifestations of all types of OCA include skin and hair hypopigmentation and visual impairment. Although there are a few documented observations of high frequency of albinism among Native Americans, including the Hopi, Zuni, Kuna, Jemez, Laguna, San Juan, and Navajo, no causative molecular defect has been previously reported. In the present study, we show that albinism in one Native American population, the Navajo, is caused by a LINE-mediated 122.5-kilobase deletion of the P gene, thus demonstrating that albinism in this population is OCA2. This deletion appears to be Navajo specific, because this allele was not detected in 34 other individuals with albinism who listed other Native American origins, nor has it been reported in any other ethnic group. The molecular characterization of this deletion allele allowed us to design a three-primer polymerase chain reaction system to estimate the carrier frequency in the Navajo population by screening 134 unrelated normally pigmented Navajos. The carrier frequency was found to be ∼4.5%. The estimated prevalence of OCA2 in Navajos is between ∼1 per 1,500 and 1 per 2,000. We further estimate that this mutation originated 400–1,000 years ago from a single founder.
Introduction
Oculocutaneous albinism (OCA) is caused by a deficiency of melanin pigment in the skin, hair, and eye, which causes hypopigmentation in these tissues. Mutations in four genes have been reported in different types of human OCA. OCA1 (MIM #203100) is caused by mutations in the tyrosinase gene (Tomita et al. 1989), and people lacking a functional tyrosinase gene do not make melanin pigment. OCA2 (MIM #203200), OCA3 (MIM #203290), and OCA4 (MIM #606574) are due to mutations in the P gene (Gardner et al. 1992; Rinchick et al. 1993), the TYRP1 gene (Boissy et al. 1996), and the MATP gene (Newton et al. 2001), respectively. People with the last three types of OCA are hypopigmented but are still able to produce some melanin. In all forms of albinism, a deficit of melanin pigment in the developing eye leads to foveal hypoplasia and abnormal routing of the optic nerves, resulting in nystagmus, strabismus, and reduced visual acuity (King 1998).
The most common type of OCA is OCA2. In the United States, OCA2 frequencies in African American and white individuals are estimated to be 1:10,000 and 1:36,000, respectively (Witkop et al. 1989). The gene responsible for OCA2 encodes the P protein that is predicted to have 12 transmembrane domains, and, although its exact function is unknown, it may play a role in pH regulation of melanosomes (Puri et al. 2000).
Studies based on census data indicate high frequencies of albinism among several Native American populations. The frequencies of albinism in Native American tribes were reported to vary from 1:140 in the Jemez to 1:3,750 in the Navajo (Woolf 1965; Woolf and Dukepoo 1969). To date, no genetic defect has been reported for the albinism in Native Americans.
We have observed a high occurrence of albinism in the Navajo, who primarily live in northeastern Arizona. The phenotype of Navajos with albinism overlaps those described for OCA2 and the recently documented OCA4 (King 1998; Newton et al. 2001). To identify the causative genetic defect, we screened for mutations in the P and MATP genes, associated with OCA2 and OCA4, respectively. Although no mutations in the MATP gene were found, all Navajos with albinism in the present study were found to have a homozygous deletion of 122.5 kb of genomic DNA, including exons 10–20 of the P gene. We did not find this deletion allele in 34 other individuals with albinism who list various other Native American origins. After characterizing the breakpoints of the deletion, we designed a three-primer system to screen 134 unrelated normally pigmented Navajos and 42 Apaches by PCR and estimated the carrier frequency in the Navajo to be 4.5%. In contrast, we did not find any carrier of the deletion allele in the Apache, a tribe closely related to the Navajo. In addition, we tested the hypothesis that the deletion came from a founder mutation. The estimated age of the deletion is 400–1,000 years.
Subjects and Methods
Subjects and DNA Extraction
Navajo individuals N1, N4, and N5 all display hypopigmentation of the skin and have yellowish hair and visual problems consistent with OCA2 (or OCA4) phenotypes. N2 and N3, the parents of N4, were also included in the study. A total of 142 normally pigmented Navajo volunteers, among whom 134 are apparently unrelated, were chosen from different areas of the reservation, to obtain a representative and random sample of the Navajo tribe. A total of 42 normally pigmented Apache samples were also included in this study. In addition, 34 individuals with albinism who listed full or partial Native American origins (non-Navajo) were included in the present study. Control samples used in Southern blot analysis and PCR are from white individuals without deletions of the P gene. Written informed consent (approved by the institutional review boards of all participating centers) was obtained from all subjects. N5 and the white control DNAs were extracted from transformed lymphocyte cell lines, as described elsewhere (Durham-Pierre et al. 1994). The DNAs from all other individuals were extracted from buccal cells or from whole blood. Buccal cell DNA samples were isolated according to the procedure of Richards et al. (1993), whereas whole blood DNA was extracted using the procedure of Lahiri and Nurnberger (1991).
PCR Screening of the P and MATP Genes
Screening for mutations in the P and MATP genes that include 24 and 7 exons, respectively, was performed using procedures described elsewhere (Lee et al. 1995; Newton et al. 2001).
Southern Blot Analysis
N5 and control genomic DNAs (10 μg) were digested in parallel with different restriction enzymes, including BamHI, XbaI, and BglI. The digestion products were separated on 1% agarose gels and were transferred by the alkaline method to a nylon membrane (Hybond N+; Amersham). The membranes were hybridized with three distinct cDNA probes covering P gene exons 8–11, 11–15, and 15–20. The probes were obtained by PCR amplification from P gene cDNA. Probes were labeled with [32P]dCTP, using Random Primer labeling kit (Stratagene). The hybridization was performed in phosphate-buffered 7% SDS hybridization solution (Durham-Pierre et al. 1994). Blots were washed with 0.2× standard sodium citrate (1% SDS) at 60°C, prior to exposure to x-ray film (Kodak) at −80°C overnight.
Characterization of the Breakpoints of the Deletion
Primer pairs were designed at 2-kb intervals within introns 9 and 20, on the basis of the sequence data from the Celera database (Venter et al. 2001). The exact breakpoints were obtained by direct sequencing of the TA-cloned PCR fragment (Invitrogen) spanning the juncture of the breakpoints. We used primer pair MHB1334 and MHB1359 to amplify across the breakpoints. The sequences of the primers are as follows: MHB1334 (5′-CCATTTCTGCTGTTCTTCATCCC-3′) and MHB1359 (5′-GCCCAACCTTGAAAGTCTTGC-3′). The PCR was performed using ExTaq enzyme, as recommended by the supplier (Takara), in 50-μl reactions containing 100 ng genomic DNA, 0.5 μM of each primer, initially denatured for 2 min at 94°C, followed by 34 cycles of 1 min at 94°C, 1 min at 56°C, and 90 s at 72°C, and ending with a 5-min extension.
Estimation of the Deletion Frequency in the Navajo
A three-primer system was designed to distinguish the wild-type and the deletion allele in one PCR, using primers MHB1333, MHB1334, and MHB1370: MHB1333 (5′-CTGGAGGCAAGTAGATAGTGGAGC-3′), MHB1370 (5′-ATCCAAACCCTTCCCTGACCAC-3′), and the MHB1334 primer sequence shown above. Some of the PCR amplifications were performed in 50-μl reactions, as described above, with the exception of the annealing temperature (62°C). PCR products were separated on a 2% agarose gel (1% SeaKem GTG/1% NuSieve [BioWhittaker Molecular Applications]). Because of limited amounts of DNA, other amplifications were scaled down to 10-μl reactions containing 5 ng of genomic DNA.
Haplotype Analysis
On the basis of the Celera sequence data (Venter et al. 2001), three microsatellite markers and an SNP (T/C) were found in close proximity to the deletion breakpoints. One microsatellite marker, MHB001, is ∼8.6 kb upstream of the intron 9 breakpoint (IVS 9 +[5047–5093]), with 23 CA repeats in the Celera database. The other marker, MHB002, is ∼20 kb downstream of the intron 20 breakpoint (IVS 21 +[15106–15140]) and includes 17 CA repeats in the Celera database. Fluorescence-labeled primers were designed to amplify MHB001 and MHB002, and the fragments were run on an ABI 3100 sequencer. The data were analyzed by Genotyper (ABI). The sequences of the microsatellite primers are as follows: microsatellite MHB001 (MHB1387: 5′-[6FAM] CTGAGACTTCCACTTTGACCCATAG-3′), MHB1384 (5′-CATCTGTAATCCCAGCACTTTGG-3′), microsatellite MHB002 (MHB1388: 5′-[5HEX] GACATTGCTGAGTCTTGCTTGTTG-3′), and (MHB1386 (5′-GACCTTGTCCTCCAAAAACTGTAGG-3′).
Sequencing the PCR product amplified by MHB1334 and MHB1359 (that only amplify the deletion allele), allowed us to genotype the SNP T/C (IVS 9 +12,934), which is located 703 bp upstream of the intron 9 breakpoint. In addition, this product contained the (CA)10(TA)1(CA)4 repeat (MHB003) within the same PCR fragment that is 118 bp upstream of the intron 9 breakpoint. These data were obtained for all individuals with albinism, as well as for carriers.
Linkage Disequilibrium Analysis and Coalescence Estimates for the Founder Deletion
To choose the appropriate marker to date the deletion, it is important to use a marker with a high degree of variability to ensure generation of nonancestral genotypes. Also, the marker should not be too far from the deletion to equilibrate allele frequencies by recombination within the time frame of interest (Goldstein et al. 1999). Of the markers we have, MHB001 best meets the above criteria. MHB001 contains the largest number of repeats (23), and it is therefore likely to be the most variable, because long loci mutate more often than short loci (Ellegren 2000). In addition, it is only 8.6 kb from the deletion. Linkage disequilibrium (LD) analysis of the specific 466-bp allele of the marker MHB001 with the deletion was performed by determining the allele frequency in 102 Navajo control chromosomes and 12 unrelated Navajo deletion-bearing chromosomes. Fisher’s exact test was used to test the significance of the association of the 466-bp allele with the deletion. LD was quantified by the calculation of the parameter δ (Bengtsson and Thomson 1981). The δ value was calculated using the formula
 |
where p is the frequency of the ancestral allele (466-bp allele) in the deletion-bearing chromosomes and r is the frequency of the same allele in the control chromosomes. To estimate the coalescence time for the founder deletion, we used the formula
 |
where G is the generation number, r is the frequency of mutation and recombination events, and p is the proportion of the ancestral haplotype (Goldstein et al. 1999; Reich and Goldstein 1999). To estimate p for dating the deletion, we used all 15 deletion-bearing chromosomes, regardless of whether or not they were related. This is because the age estimation is independent of the topology of the population tree, as long as mutations at the marker locus have no selective effect; the correlations in the tree amount to a process of pseudoreplication of lineages (Reich and Goldstein 1999). However, when estimating the variance of p to obtain the variance of the date of the deletion, the use of correlated or uncorrelated deletion-bearing chromosomes in the analysis will result in a different estimation, because the variance of p is dependent on the demographic history of the population. For a tree typical of a constant-sized population with highly correlated lineages, the variance tends to be large. In contrast, in a tree typical of an expanding population with uncorrelated lineages, the variance tends to be smaller (Reich and Goldstein 1999). We used only 12 unrelated deletion-bearing chromosomes to obtain the minimum interval for the date of the deletion.
Results
Characterization of the Deletion of the P Gene
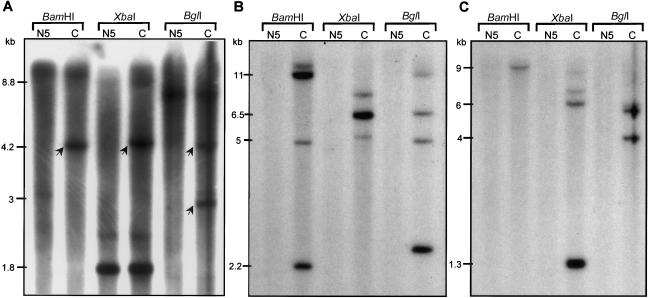
We initiated our molecular analysis by focusing on DNA from one of the subjects with OCA, N5. In our attempts to screen for mutations of the P gene by PCR amplification and sequencing, we found that exons 10–20 could not be amplified, even though all other exons could easily be amplified. This suggested that exons 10–20 were deleted. All other amplified exons were subjected to direct sequencing, and no mutations were found, with the exception of a few SNPs, all in homozygous states. In addition, no mutation was found when screening the MATP gene. To verify the existence of the P gene deletion, a series of Southern blot analyses were performed (fig. 1). We used three separate cDNA probes, corresponding to exons 8–11, 11–15, and 15–20. As shown in figure 1, hybridization with the exon 8–11 fragment revealed some missing fragments, whereas hybridization with the exon 11–15 or 15–20 fragments did not generate any signal for N5 DNA. This was in agreement with our prediction that exons 10–20 were deleted.
Figure 1.
Southern blot analysis of BamHI, XbaI, and BglI digests of genomic DNA from both a Navajo individual with OCA2 (N5) and a control subject (C). A, The hybridization was performed with a cDNA probe corresponding to exons 8–11 of the P gene. The arrows show the fragments missing in N5. B, The cDNA probe corresponds to exons 11–15 of the P gene. Note that no hybridizing fragments are detected in N5. C, The cDNA probe corresponds to exons 15–20 of the P gene. Note that no hybridizing fragments are detected in N5. Sizes of the hybridization fragments (calculated from molecular standards) are shown to the left of each panel.
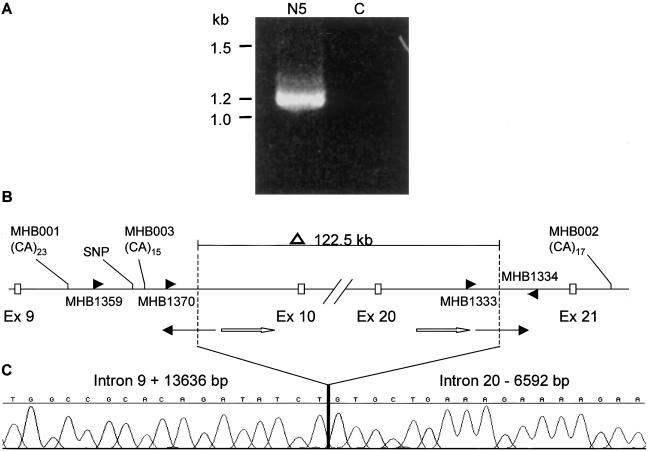
To pinpoint the breakpoints of the deletion, a series of primer pairs corresponding to the genomic sequences within intron 9 and intron 20 were designed on the basis of genomic sequences from the Celera human genome database and were used to amplify genomic DNA from both N5 and control individuals. In this way, we narrowed down the breakpoints at each end of the deletion to ∼2-kb regions. Subsequently, we used pairs of forward primers within intron 9 and reverse primers within intron 20 to try to amplify across the breakpoints by PCR. As shown in figure 2A, primer pair MHB1334 and MHB1359 successfully generated a distinct fragment of 1.2 kb in N5 DNA, whereas no fragment was amplified in the control DNA. The breakpoint-spanning PCR fragment was TA cloned and was then subjected to direct DNA sequencing. This resulted in the identification of breakpoints within intron 9 (IVS 9 +13,636 bp) and intron 20 (IVS20 −6,592 bp) (fig. 2B and 2C).
Figure 2.
A, Breakpoint-spanning fragment amplified by primers MHB1359 and MHB1334. In a Navajo individual with OCA2 (N5), a 1.2-kb fragment amplified, whereas in the control subject (C), no fragment was detected. B, Sequences flanking the breakpoints. The breakpoints are represented as vertical dashed lines, which are located at positions intron 9 + 13636 bp and intron 20 −6592 bp. As shown above, 122.5 kb of the P gene is deleted, including exons 10–20. The primers, MHB1359/1334, were used to amplify across the breakpoints (panel A) and the three primers, MHB1370/1333/1334, were used for detecting carriers. Microsatellite markers MHB001, MHB002, MHB003, and the SNP were used in haplotype analysis. MHB001, located 8.6 kb upstream of the left-hand breakpoint, consists of a perfect (CA)23 repeat (corresponding to the 466-bp allele). The SNP (T/C) is 703 bp upstream of the left-hand breakpoint. MHB003 is 118 bp upstream of the left-hand breakpoint and consists of compound repeat (CA)10(TA)1(CA)4. MHB002 is 20 kb downstream of the deletion and consists of a perfect (CA)17 repeat (corresponding to the 370-bp allele). At the breakpoint juncture, two LINE sequences (thin arrows) are at each end and are oriented in opposite directions. The two homologous LINE sequences (white arrows) are 3,032 bp downstream of the left-hand breakpoint and 1,481 bp upstream of right-hand breakpoint. The homologous LINE sequences share 85% identity in a 745-bp stretch and are oriented in the same direction. C, Chromatograph of the sequence flanking the breakpoints.
Sequence analyses of the breakpoints and the flanking sequences revealed a few salient features (fig. 2B and 2C). First, both breakpoints were found to be exactly end-joined. Second, both breakpoints were found within long interspersed nucleotide elements (LINEs) (revealed by RepeatMasker software). However, the breakpoint LINEs are oppositely oriented, and no sequence homology between them was found at the breakpoint juncture. Further analysis of the sequences flanking both breakpoints, using RepeatMasker revealed significant percentages of repeat sequences in both breakpoints’ flanking regions. The BLAST2 analysis of the two flanking sequences revealed two homologous sequences of LINE/L1s in proximity to both breakpoints, oriented in the same direction, that share 85% identity in a stretch of 745 bp. One of them, which is part of the LIPA 3 family sequence of LINE/L1s, is 3,032 bp downstream of the intron 9 breakpoint. The other one, which is part of the LIPA10 family sequence of LINE/L1s, is 1,481 bp upstream of the intron 20 breakpoint.
The deletion removes 122,569 bp of the P gene, including exons 10–20, and leads to OCA2. Although the deletion does not disrupt the reading frame of the P gene, it removes 345 amino acids of the total 838 amino acids (41.1% of the P protein). Calculations performed with the use of TMHMM software predict that the topology of the encoded protein will be dramatically changed. The 12 transmembrane domains of the wild-type P protein are reduced to 5, and so the predicted orientation of some of the remaining transmembrane domains of the P protein is changed. Thus, we are confident that the deletion results in a null allele. Moreover, all of the Navajo individuals with OCA were homozygous for the deletion. In addition, pedigree analysis of proband N4 and his normally pigmented parents (N2/N3 carriers; fig. 3) is consistent with this deletion resulting in a null allele.
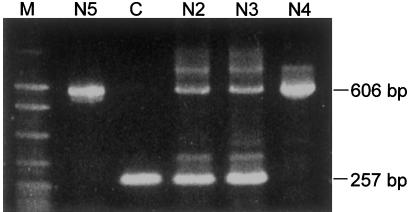
Figure 3.
PCR analysis of the deletion in affected individuals (N4 and N5), parents of N4 (N2 and N3, who are obligate carriers), and a control subject (C). Primers are shown in fig. 2B. In the control subject, amplification with primers MHB1333/1334 results in a 257-bp fragment, whereas, in carriers, primers MHB1370/1334 gave an additional 606-bp fragment. In Navajos with OCA2, only the 606-bp fragment is amplified. M = molecular ladder.
Estimation of Albinism Frequency in the Navajo Population
The identification of the breakpoints of the deletion allowed us to design a PCR system, using three primers to screen for carriers. As shown in figure 3, amplification of the deletion allele (N5 and N4) gave rise to a 606-bp fragment, and amplification of a nondeletion (control) allele gave a 257-bp fragment. DNA from obligate carriers (N2 and N3) gave both fragments after PCR amplification. Using this PCR system, we screened 142 normally pigmented Navajos (134 of whom were unrelated) and detected 7 carriers, among whom 6 were unrelated (see table 1). On the basis of this observation, the carrier frequency is estimated to be 4.5% (95% CI 1.66%–9.49%) (VassarStats). For a given allele frequency, the proportion of individuals homozygous for an allele is a function of the amount of inbreeding in the population. The OCA2 individual frequency can be estimated, according to Wright’s equilibrium law (Li 1955), as R=q2+αpq, where R is the frequency of the affected individuals, q is the frequency of the mutation allele, p is the frequency of wild type allele, and α is the inbreeding coefficient. Spuhler and Kluckhohn (1953) obtained an α value of 0.0080 for Ramah Navajo in New Mexico, a relatively low inbreeding coefficient. After adjustment for this coefficient, the calculated OCA2 frequency in the Navajo is 1:1,477. However, the Ramah Navajo are a relatively small and isolated community in comparison with the main Navajo reservation. Therefore, the α value for the majority of Navajos living on the main reservation could be smaller than that of the Ramah Navajo (but not less than zero). At an α value of zero, the estimation of the OCA2 frequency is ∼1:2,000. Thus, our estimation of the OCA2 frequency among the Navajo is ∼1:1,500–1:2,000, significantly higher than that seen in African Americans (1:10,000) and white Americans (1:36,000) (Witkop et al. 1989). The estimation from the present molecular analysis is consistent with the estimation, reported by Woolf (1965) and based on census data, that the frequency of albinism in Navajo is >1:3,750.
Table 1.
The Genotypes of the Markers in all Carriers and Affected Individuals[Note]
| Subject | MHB001(bp) | SNP T/C | MHB003 | MHB002(bp) | Notes |
| N1 | 466/466 | C | (CA)15 | 370/370 | Affected |
| N2 | 464/466 | C | (CA)15 | 370/370 | Carrier N4 parent |
| N3 | 464/466 | C | (CA)15 | 370/370 | Carrier N4 parent |
| N4 | 466/466 | C | (CA)15 | 370/370 | Affected |
| N5 | 466/466 | C | (CA)15 | 370/370 | Affected |
| JJDa | 464/466 | C | (CA)15 | 370/370 | Carrier |
| JJSa | 464/466 | C | (CA)15 | 370/370 | Carrier |
| 4C | 462/466 | C | (CA)15 | 370/370 | Carrier |
| Nav4 | 466/462 | C | (CA)15 | 370/370 | Carrier |
| NM9 | 466/458 | C | (CA)15 | 370/370 | Carrier |
| NN2 | 462/464 | C | (CA)15 | 370/370 | Carrier |
| NN13 | 462/466 | C | (CA)15 | 370/370 | Carrier |
Note.— The study includes a group of three homozygotes and nine carriers, among whom the three homozygotes are unrelated to one another and the six carriers are not related. (Two carriers [N2 and N3] were not included in the carrier screening, because their status as obligate carriers had been known from their affected child, N4.) The marker MHB003 and the SNP were only genotyped in deletion-bearing chromosomes.
JJD is the father of JJS.
Founder Effect of the Deletion
This deletion allele was found only in the Navajo population. We did not find this allele in 34 people with albinism who listed other Native American origins. This suggested that the allele is Navajo specific and that it probably came from a founder mutation. To define the haplotype(s) associated with the deletion, we genotyped the three microsatellite markers and one SNP that are in close proximity to the breakpoints in all deletion chromosomes. As shown in figure 2B, marker MHB001 is 8.6 kb upstream of the left-hand breakpoint, whereas MHB002 is 20.8 kb downstream of the right-hand breakpoint. These two markers were genotyped by PCR with fluorescence-labeled primers (as described in the “Subjects and Methods” section). To better define the phases of the other two markers, MHB003 and the SNP, we used the primer pair MHB1359 and MHB1334 for amplification. This primer pair amplifies only the deletion-bearing chromosomes. The PCR amplicons were then subjected to direct sequencing. The putative ancestral haplotype that is associated with the deletion was inferred from the genotypes of the markers. As shown in table 1, the haplotypes of markers MHB003 and the SNP, which are amplified only in deletion-bearing chromosomes, are (CA)15 and C, respectively, in all affected individuals and carriers. The putative ancestral haplotypes for the other two markers, MHB001 and MHB002, are inferred from the genotypes of the affected individuals to be 466 bp and 370 bp, respectively. Furthermore, all carriers but one (NN2) have the same haplotypes for these two loci as do the affected individuals. Therefore, the most likely ancestral haplotype that is associated with the deletion is (listed 5′ to 3′) MHB001 = 466 bp, SNP = C, MHB003 = (CA)15, and MHB002 = 370 bp, (see table 1; fig. 2B). Furthermore, our sequence analysis of the 15 deletion-bearing chromosomes (see table 1; fig 2C) revealed that 13 of the 15 shared the exact same sequence throughout the 1.2-kb fragment (including the breakpoint). In the two remaining chromosome samples, the breakpoint region could not be unambiguously determined because of the absence of sufficient DNA. Given these data, the deletion is almost certainly from a founder mutation.
Coalescence Estimates for the Founder Deletion
Founder mutations occur on a single chromosome and result in LD with the flanking polymorphic markers. With time, this disequilibrium will decay, because of recombination and new mutations of these polymorphic markers. Therefore, knowing the level of LD and the mutation and recombination rate of the markers, we can date the founder mutation by equation 2. Marker MHB001 was used to estimate the date of the founder mutation. To assess the variability of and the frequencies of different alleles of this marker, 15 deletion chromosomes and 102 nondeletion (Navajo) chromosomes were genotyped at this locus (see table 2). MHB001 has six alleles, and the expected heterozygosity (H) of this marker is 70.2%. Fisher’s exact test showed that there was a significantly higher frequency of the 466-bp allele in deletion chromosomes than in nondeletion ones (P=.0012), indicating that this allele is in pairwise LD with the deletion allele. A measure of LD, δ, was calculated, according to equation 1, to be 0.86 for the deletion and the 466-bp allele of MHB001. (Another marker, MHB002, was also genotyped in the Navajo background chromosomes; it has only two alleles in this population, with an H value of 0.053, which precludes its use for dating the deletion.) Thus, the variability of the MHB001 locus and the presence of LD between the 466-bp allele and the deletion make it useful for dating the deletion. In our samples, 14 of 15 deletion-bearing chromosomes have alleles of 466 bp at the MHB001 locus. In dating the deletion by use of equation 2, we did not take the relationship of all 15 deletion-bearing chromosomes into consideration, for the reasons described in the “Subjects and Methods” section. Thus, the p here is 14/15. The reported mutation rate of an autosomal CA repeat is 5.6×10-4/generation (Weber and Wong 1993). On the basis of the data presented by Collins et al. (1996) (Genetic Location Database), we calculated the local recombination rate of the 15q11-12 region (where the P gene is located) to be 3.13 cM/Mb. Therefore, the recombination rate within 8.6 kb is 2.7×10-4/generation. The sum of the mutation rate and the recombination rate (8.3×10-4/generation) is the value r in equation 2. Applying p and r to equation 2, the mutation was calculated to have arisen 83.1 generations ago. If we assume 22 years for each generation, it occurred 1,828 years ago.
Table 2.
Alleles of MHB001 in Navajo Control and Deletion Chromosomes[Note]
|
Frequency in |
||
| Allele Size(bp) | Control Chromosomes(n = 102) | Deletion Chromosomes(n = 12) |
| 458 | .02 | 0 |
| 460 | .04 | 0 |
| 462 | .127 | .083a |
| 464 | .323 | .083a |
| 466 | .412 | .917 |
| 468 | .078 | 0 |
Note.— The expected heterozygosity of MHB001 is calculated to be 70.2%. Fisher’s exact test of frequencies of 466-bp allele in deletion and control chromosomes showed a significant difference (P=.0012 in a two-tailed test).
The carrier NN2 has the genotype of 462/464, with the deletion chromosome having either 462-bp or 464-bp allele (see table 1).
However, both the variability of our estimation of the ancestral haplotype frequency (p) and the mutation rate of the marker could skew our mutation-age estimation. It is difficult to calculate a CI for the estimation of G, because the mutation-bearing chromosomes are not independent under the assumption of LD (Risch et al. 1995; Rannala and Slatkin 1998). The degree of nonindependence, the variance of p, and the CI are strongly influenced by the demographic history of the population, as described in the “Subjects and Methods” section. Here we provide a minimum CI for G by assuming the star genealogy typical of an expanding population in which all lineages are uncorrelated: p is the observed proportion of the ancestral haplotype (11 of the 12 independent chromosomes), and the 95% CI of the estimation of p will be 0.6462–0.9851 for a binomial distribution (VassarStats). When this is applied to equation 2, the range of the mutation age is calculated to be 396–11,574 years. This interval is appropriate only for a rapidly expanding population (like the Navajo population), and the true CI may be even larger. To further assess the age of this deletion, we analyzed 42 normally pigmented Apache (a closely related Native American tribe) samples for this deletion, and no deletion was detected. Moreover, there are no reports of Apaches with OCA (J. Broon [Indian Health Service {IHS} White River] and B. Berkovich [IHS San Carlos], personal communications). These observations confirm the previous observation of absence of albinism among the Apache (Woolf 1965). It is believed that the Navajo and the Apache became distinct groups ∼600–1,000 years ago (Acrey 1982). If the deletion arose before the division into two populations, the deletion allele may also exist in the Apache population. The absence of albinism and of this allele in the Apache population indicates that this deletion is unlikely to have occurred before the time when the Apache and the Navajo became distinct populations (unless the deletion allele was lost or is present at very low frequency in Apache). Thus, our best estimate of the age of the deletion is 400–1,000 years.
Discussion
The mouse and human P proteins are predicted to have 12 transmembrane domains and to share significant sequence homology with membrane transporters, some of which are involved in the transport of anions (Brilliant 2001). It has been suggested that the P protein might function to regulate the pH of the melanosome (Puri et al. 2000). Whatever its exact function is, the P protein is required for normal pigmentation. In the present study, we identified the causative genetic defect for the high prevalence of albinism in the Navajo to be a 122.5-kb deletion of the P gene. This deletion removes 41.1% of the encoded P protein, including seven of the transmembrane domains, thus leading to loss of function of the protein. The prevalence of albinism is reported to be high among other Native American tribes, including the Hopi, Zuni, Kuna, Jemez, Laguna, and San Juan (Woolf 1965). Thus, the question arises of whether this deletion is also responsible for the albinism that occurs in other Native American populations. To answer this question, we screened for the presence/absence of this deletion in 34 individuals with OCA who listed other Native American origins, and no deletion was found in any of these individuals. This suggests that more than one genetic defect is responsible for albinism in Native Americans. This is also in agreement with our conclusion that this deletion came from a Navajo founder ∼400–1,000 years ago.
Either genetic drift (e.g., resulting from a bottleneck effect) or heterozygote advantage can explain the high frequency of a recessive allele. The Navajo tribe is located in the southwestern part of the United States, covering parts of Arizona, New Mexico, and Utah. The Navajo may have arrived in the southwest as early as 1000 ad (Underhill 1956; Kluckohn and Leighton 1962). Many indigenous peoples suffered significant population losses after the arrival of Europeans in the region. During the late 1800s and early 1900s, a series of conflicts between the Navajo and U.S. Army led to forced relocation, during which many Navajos died and the total population dwindled to an estimated 8,000–10,000 people (Kluckohn and Leighton 1962; Williams 1992). The actual number of Navajos from whom the current population is derived would be much smaller than the total population, because a significant fraction of the population would have been of post-breeding age and because as many as half of the population might have been children of the same founders (Erickson 1999). The population has subsequently greatly expanded, and, as of the year 2000, >180,000 Navajos live in Arizona, New Mexico, and Utah, according to the U.S. Census Bureau. Therefore, the population history of the Navajo includes one or more known bottlenecks that could have led to the high frequency of the deletion allele from a single founder. In addition to the high frequency of OCA2 among the Navajo that is revealed by the present study, other uniquely frequent genetic disorders have been reported among the Southwestern Athabaskans (Navajos and Apaches) and are thought to result from a genetic bottleneck (Erickson 1999). Thus, it seems likely that the high prevalence of OCA2 among the Navajos could also be a consequence of a genetic bottleneck. Other explanations for the high frequency of albinism in the Hopi population have been proposed by Woolf and Dukepoo (1969), who believed that in addition to genetic drift, positive cultural selection, including a male-mating advantage for individuals with albinism, contributed to the high incidence of albinism among the Hopi. However, as was pointed out in their article, this explanation could hardly account for the high frequency of albinism in other tribes, because their cultures are not necessarily all favorable to individuals with albinism. Furthermore, Hedrick (in press) recently tested the cultural-selection hypothesis by population modeling and pointed out that any mating advantage of men with albinism was not likely to be a significant factor in the high incidence of albinism among the Hopi, given the previously assumed viability selection against individuals with albinism. Finally, heterozygote advantage has been used to explain the high frequencies of several recessive conditions, such as sickle cell anemia and cystic fibrosis (Gabriel et al. 1994; Aidoo 2002). However, we are not aware of any such advantage for OCA2 heterozygotes. The two possibilities of bottleneck effect and positive selection on heterozygotes could be ultimately distinguished by comparing the shape of the genealogy of the affected chromosomes with those of the genealogies of neutral genomic regions.
Although we may never know the exact mechanism that led to the generation of the 122.5-kb deletion, the sequence itself contains some potential clues. Both breakpoints are within LINE sequences that lack any homology at the breakpoint juncture. In addition, the LINE sequences are oppositely oriented. These sequence features could not explain the occurrence of the deletion by homologous recombination (Ostertag et al. 2001). However, there are two homologous LINE sequences within the deletion that are oriented in the same direction. A LINE-mediated recombination between these homologous sequences would result in a smaller deletion than the one characterized in our study. However, the double-stranded breaks (DSBs) generated in the homologous recombination could be repaired by different mechanisms. Two major repair mechanisms for DSBs in mammals are homologous recombination repair (HRR) and nonhomologous end joining (NHEJ) (Karran 2000). Inappropriate repair of DSBs will cause chromosome rearrangements, such as inversions, large deletions, or translocations (Pastink et al. 2001). HRR requires extensive sequence homology between the two ends to join the breakpoints, whereas NHEJ simply ligates the two broken ends together without requiring sequence homology (Pfeiffer et al. 2000). In the present study, the lack of any homology between the two broken ends suggests that NHEJ was involved in the repair process. A salient feature of NHEJ is the further deletion of the junction sequences, enlarging the original deletion. Such deletions may result from DSB-flanking regions being degraded by exonucleolysis (Lehman 1994; Pfeiffer 1998). A plausible scenario for the origin of the deletion characterized in this study includes LINE-mediated homologous recombination between the two flanking homologous sequences (Burwinkel et al. 1998; Segal et al. 1999; Ostertag et al. 2001) generating DSBs. Subsequently, the homology-independent repair mechanism for DSB caused a larger deletion by exonucleolytic degradation of DSB-flanking regions (Lehman 1994; Pfeiffer 1998). Alternatively, there are reports of deletion in which only one breakpoint is within a LINE sequence and the other end lacks any apparent repetitive or homologous sequence (Henthorn et al. 1990; Drechsler et al. 1996). Mechanisms yet to be defined may have caused the deletion in this scenario. The deletion defined in this study is the second large deletion of the P gene reported so far. The first one is the 2.7-kb deletion that removes exon 7 of the P gene (Durham-Pierre et al. 1994). The P gene maps to a region very rich in repetitive sequences, in which chromosome rearrangements have been reported to cause disease. Examples include Prader-Willi syndrome and Angelman syndrome (Amos-Landgraf et al. 1999). On the other hand, chromosome rearrangements are rarely reported in the P gene. The identification of this deletion in the P gene suggests that more chromosome rearrangements involving the P gene may exist than are detected. In addition, we and others often identify only one of the two mutations in PCR screening of individuals with albinism. This points to the possibility of the existence of other mutations (including deletions) that are missed by the widely used PCR mutation screen strategy. Application of highly variable intragenic microsatellites and SNPs can help to identify heterozygous deletions (Ringpfeil et al. 2001; Huie et al. 2002).
In conclusion, we identified the causative genetic defect responsible for the high prevalence of albinism in the Navajo population (1:1,500–2,000) and showed for the first time, to our knowledge, that albinism in the Navajo is OCA2. The fact that we did not find the deletion allele in individuals with albinism of other Native American origins suggests the existence of other genetic defect(s) responsible for albinism in Native Americans.
Acknowledgments
We would like to thank all the Native Americans who participated in this study. We also would like to thank all our lab colleagues for helpful discussions and computer assistance. This work was supported by National Institutes of Health grant AR4596 (to M.H.B.). N.G. was supported by MARC program grant T34 GM08718.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST 2, http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html (for sequence homology search)
- Celera, http://www.celera.com/ (for OCA2 genomic DNA sequences data)
- Genetic location database, http://cedar.genetics.soton.ac.uk/public_html/ldb.html (for local recombination rate at 15q11-12)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for OCA1 [MIM #203100], OCA2 [MIM #203200], OCA3 [MIM #203290]), and OCA4 [MIM #606574])
- RepeatMasker, http://woody.embl-heidelberg.de/repeatmask/ (for detecting repetitive sequences)
- TMHMM (v. 2.0), http://www.cbs.dtu.dk/services/TMHMM-2.0/ (for membrane-topology predictions)
- U.S. Census Bureau, http://www.census.gov/ (for current Navajo population size)
- VassarStats, http://faculty.vassar.edu/lowry/prop1.html (for 95% CI of proportion for binomial distribution)
References
- Acrey BP (1982) Navajo history to 1846: the land and the people. Department of Curriculum Materials Development, Central Consolidated School District No. 22, Shiprock, New Mexico [Google Scholar]
- Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, Kariuki S, Nahlen BL, Lal AA, Udhayakumar V (2002) Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet 359:1311–1312 [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson BO, Thomson G (1981) Measuring the strength of associations between HLA antigens and diseases. Tissue Antigens 18:356–363 [DOI] [PubMed] [Google Scholar]
- Boissy RE, Zhao H, Oetting WS, Austin LM, Wildenberg SC, Boissy YL, Zhao Y, Sturm RA, Hearing VJ, King RA, Nordlund JJ (1996) Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with Brown oculocutaneous albinism: a new type of albinism classified as “OCA3.” Am J Hum Genet 58:1145–1156 [PMC free article] [PubMed] [Google Scholar]
- Brilliant MH (2001) The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment Cell Res 14:86–93 [DOI] [PubMed] [Google Scholar]
- Burwinkel B, Kilimann MW (1998) Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J Mol Biol 277:513–517 [DOI] [PubMed] [Google Scholar]
- Collins A, Frezal J, Teague J, Morton NE (1996) A metric map of humans: 23,500 loci in 850 bands. Proc Natl Acad Sci USA 93:14771–14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler M, Royer-Pokora B (1996) A LINE element is present at the site of a 300-kb deletion starting in intron 10 of the PAX6 gene in a case of familial aniridia. Hum Genet 98:297–303 [DOI] [PubMed] [Google Scholar]
- Durham-Pierre D, Gardner JM, Nakatsu Y, King RA, Francke U, Ching A, Aquaron R, del Marmol V, Brilliant MH (1994) African origin of an intragenic deletion of the human P gene in tyrosinase positive oculocutaneous albinism. Nat Genet 7:176–179 [DOI] [PubMed] [Google Scholar]
- Ellegren H (2000) Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet 16:551–558 [DOI] [PubMed] [Google Scholar]
- Erickson RP (1999) Southwestern Athabaskan (Navajo and Apache) genetic diseases. Genet Med 1:151–157 [DOI] [PubMed] [Google Scholar]
- Gabriel SE, Brigman KN, Koller BH, Boucher RC, Stutts MJ (1994) Cystic fibrosis heterozygote resistance to cholera toxin in the cystic fibrosis mouse model. Science 266:107–109 [DOI] [PubMed] [Google Scholar]
- Gardner JM, Nakatsu Y, Gondo Y, Lee S, Lyon MF, King RA, Brilliant MH (1992) The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science 257:1121–1124 [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Reich DE, Bradman N, Usher S, Seligsohn U, Peretz H (1999) Age estimates of two common mutations causing factor XI deficiency: recent genetic drift is not necessary for elevated disease incidence among Ashkenazi Jews. Am J Hum Genet 64:1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. Hopi indians, “cultural” selection, and albinism. Am J Phys Anthropol (in press) [DOI] [PubMed] [Google Scholar]
- Henthorn PS, Smithies O, Mager DL (1990) Molecular analysis of deletions in the human-globin gene cluster: deletion junctions and locations of breakpoints. Genomics 6:226–237 [DOI] [PubMed] [Google Scholar]
- Huie ML, Anyane-Yeboa K, Guzman E, Hirschhorn R (2002) Homozygosity for multiple contiguous single-nucleotide polymorphisms as an indicator of large heterozygous deletions: identification of a novel heterozygous 8-kb intragenic deletion (IVS7–19 to IVS15-17) in a patient with glycogen storage disease type II. Am J Hum Genet 70:1054–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P (2000) DNA double strand break repair in mammalian cells. Curr Opin Genet Dev 10:144–150 [DOI] [PubMed] [Google Scholar]
- King RA (1998) Genetic hypomelanoses: disorders characterized by generalized hypomelanoses. In: Nordlund JJ, Boissy RE, Hearing VJ, King RA, Ortonne J-P (eds) The pigmentary system: physiology and pathophysiology. Oxford University Press, New York, pp 553–575 [Google Scholar]
- Kluckhohn C, Leighton D (eds) (1962) The Navaho. Doubleday, New York [Google Scholar]
- Lahiri DK, Nurnberger JI (1991) A rapid nonenzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res 19:5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Nicholls RD, Jong MTC, Fukai K, Spritz RA (1995) Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics 26:354–363 [DOI] [PubMed] [Google Scholar]
- Lehman CW, Trautman JK, Carroll D (1994) Illegitimate recombination in Xenopus: characterization of end-joined junctions. Nucleic Acids Res 22:434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C (ed) (1955) Population genetics. University of Chicago Press, Chicago [Google Scholar]
- Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, Brilliant MH (2001) Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet 69:981–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr (2001) Biology of mammalian L1 retrotransposons. Annu Rev Genet 35:501–538 [DOI] [PubMed] [Google Scholar]
- Pastink A, Eeken JC, Lohman PH (2001) Genomic integrity and the repair of double-strand DNA breaks. Mutat Res 480–481:37–50 [DOI] [PubMed] [Google Scholar]
- Pfeiffer P (1998) The mutagenic potential of DNA double-stranded break repair. Toxicol Lett 96–97:119–129 [DOI] [PubMed] [Google Scholar]
- Pfeiffer P, Goedecke W, Obe G (2000) Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 15:289–302 [DOI] [PubMed] [Google Scholar]
- Puri N, Gardner JM, Brilliant MH (2000) Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J Invest Dermatol 115:607–613 [DOI] [PubMed] [Google Scholar]
- Rannala B, Slatkin M (1998) Likelihood analysis of disequilibrium mapping and related problems. Am J Hum Genet 62:459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB (1999) Estimating the age of mutations using variation at linked markers. In: Goldstein DB, Schlotterer C (eds) Microsatellites: evolution and applications. Oxford University Press, Oxford, pp 129–138 [Google Scholar]
- Richards B, Skoletsky J, Shuber AP, Balfour R, Stern RC, Dorkin HL, Parad RB, Witt D, Klinger KW (1993) Multiplex PCR amplification from the CFTR gene using DNA prepared from buccal brushes/swabs. Hum Mol Genet 2:159–163 [DOI] [PubMed] [Google Scholar]
- Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, Spritz RA, Avidano KM, Jong MTC, Nicholls RD (1993) A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 361:72–76 [DOI] [PubMed] [Google Scholar]
- Ringpfeil F, Nakano A, Uitto J, Pulkkinen L (2001) Compound heterozygosity for a recurrent 16.5-kb Alu-mediated deletion mutation and single-base-pair substitutions in the ABCC6 gene results in pseudoxanthoma elasticum. Am J Hum Genet 68:642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, de Leon D, Ozelius L, Kramer P, Almasy L, Singer B, Fahn S, Breakefield X, Bressman S (1995) Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet 9:152–159 [DOI] [PubMed] [Google Scholar]
- Segal Y, Peissel B, Renieri A, de Marchi M, Ballabio A (1999) LINE-1 elements at the sites of molecular rearrangements in Alport syndrome-diffuse leiomyomatosis. Am J Hum Genet 64:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuhler JN, Kluckhohn C (1953) Inbreeding coefficients of the Ramah Navaho population. Hum Biol 25:295–317 [PubMed] [Google Scholar]
- Tomita Y, Takeda A, Okinaga S, Tagami H, Shibahara S (1989) Human oculocutaneous albinism caused by single base insertion in the tyrosinase gene. Biochem Biophys Res Commun 164:990–996 [DOI] [PubMed] [Google Scholar]
- Underhill RM (1956) The Navajos. University of Oklahoma Press, Norman [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al. (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- Weber JL, Wong C (1993) Mutation of human short tandem repeats. Hum Mol Genet 2:1123–1128 [DOI] [PubMed] [Google Scholar]
- Williams J (1992) Trails of tears: American indians driven from their lands. Hendrick-Long, Dallas, pp 101–146 [Google Scholar]
- Witkop CJ Jr, Quevedo WC Jr, Frizpatrick TB, King RA (1989) Albinism. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic basis of inherited disease, 6th ed, Vol 2. McGraw-Hill, New York, pp 2905–2947 [Google Scholar]
- Woolf CM (1965) Albinism among indians in Arizona and New Mexico. Am J Hum Genet 17:23–35 [PMC free article] [PubMed] [Google Scholar]
- Woolf CM, Dukepoo FC (1969) Hopi indians, inbreeding, and albinism. Science 164:30–37 [DOI] [PubMed] [Google Scholar]