Abstract
Hirschsprung disease (HSCR) is a common congenital disorder characterized by aganglionosis of the gut. The seemingly unrelated multiple endocrine neoplasia type 2 (MEN 2) is an autosomal dominant disorder characterized by medullary thyroid carcinoma (MTC), pheochromocytoma, and hyperparathyroidism. Yet, germline mutations in the RET proto-oncogene are associated with both MEN 2 and HSCR. In the former, gain-of-function mutations in a limited set of codons is found, whereas, in the latter, loss-of-function mutations are found. However, germline RET mutation is associated with only 3% of a population-based series of isolated HSCR, and little is known about susceptibility to sporadic MTC. We have found previously that specific haplotypes comprising RET coding single-nucleotide polymorphisms (SNPs) comprising exon 2 SNP A45A were strongly associated with HSCR, whereas haplotypes associated with exon 14 SNP S836S were associated with MTC. In this study, we describe three novel intron 1 SNPs, and, together with the coding SNP haplotypes, the data suggest the presence of distinct ancestral haplotypes for HSCR and sporadic MTC in linkage disequilibrium with a putative founding susceptibility locus/loci. The data are consistent with the presence of a very ancient, low-penetrance founder locus ∼20–30 kb upstream of SNP A45A, but the failure of the SNPs to span the locus presents challenges in modeling mode of transmission or ancestry. We postulate that this founding locus is germane to both isolated HSCR and MTC but also that different mutations in this locus would predispose to one or the other.
Introduction
Germline gain-of-function mutations of the RET proto-oncogene, encoding a receptor tyrosine kinase on 10q11.2, cause multiple endocrine neoplasia type 2 (MEN 2 [MIM 164761]) and loss-of-function mutations are associated with a small subset of Hirschsprung disease (HSCR [OMIM 142623]). Germline mutations in six different exons of the RET proto-oncogene (exons 10, 11, 13, 14, 15, and 16) account for at least 92% of cases of MEN 2, which is characterized by the triad of medullary thyroid carcinoma (MTC), pheochromocytoma, and hyperparathryoidism (Eng et al. 1996a). MEN 2, the heritable form of MTC, is believed to account for 25% of all MTC presentations. Thus, 75% are believed to be sporadic.
The etiology of sporadic MTC is not well understood (for review, see Eng 1999). Limited somatic genetic alterations—such as loss of heterozygosity of markers and somatic mutations in RET, principally M918T—have been described in sporadic MTC (Mulligan et al. 1993; Eng et al. 1994, 1995, 1996b; Hofstra et al. 1994) (reviewed in Eng 1999). Recently, we reported an over-representation of a germline RET sequence variant in exon 14, S836S (c.2439 C→T), among isolated patients from Germany affected with MTC, compared with a control population with the same geographic origin and ethnic and genetic backgrounds (Gimm et al. 1999). The association of S836S with MTC was confirmed, independently, in another series of European patients with MTC (Ruiz et al. 2001). These observations suggest that the phenomenon of association of S836S with MTC could be universal and that there is an ancestral low-penetrance susceptibility marker for MTC within the RET proto-oncogene (Ruiz et al. 2001). To date, S836S is the only germline susceptibility factor that has been identified for sporadic medullary thyroid cancer. Although various hypotheses have been suggested for its molecular mechanism, this remains unknown at this time (Gimm et al. 1999; Ruiz et al. 2001).
HSCR or aganglionic megacolon is a common disorder, occurring in 1 in 5,000 live births characterized by the absence of the intramural ganglia of Meissner and Auerbach in the hindgut and resulting in functional intestinal obstruction. HSCR most commonly presents in isolated cases, although it can be familial and may be inherited as autosomal dominant or autosomal recessive, with reduced penetrance and male predominance (Badner et al. 1990; Passarge 1967). The RET proto-oncogene is considered a major susceptibility gene for HSCR (Lyonnet et al. 1993; Edery et al. 1994; Romeo et al. 1994; Chakravarti 1996). Depending on the referral series, up to 50% of familial HSCR cases and anywhere between 10% and 35% of sporadic cases were reported to be accounted for by loss-of-function germline RET mutations (Angrist et al. 1995; Attié et al. 1995; Eng 1996). The only population-based series, however, estimates the frequency of germline RET mutation in 69 unselected patients with HSCR to be 7%, and only 3% of isolated patients with HSCR in this population-based series had traditional germline RET mutations (Svensson et al. 1998). Several other putative HSCR susceptibility genes have been proposed for syndromic and nonsyndromic HSCR (Puffenberger et al. 1995; Angrist et al. 1996; Amiel et al. 1996, 2001; Auricchio et al. 1996; Edery et al. 1996; Bidaud et al. 1997; Doray et al. 1998; Pingault et al. 1998; Southard-Smith et al. 1998; Wakamatsu et al. 2001).
The molecular mechanism responsible for the majority of isolated patients with HSCR remains unknown. It has been suggested that the sporadic cases of HSCR might be accounted for by a model of polygenic inheritance (Bolk et al. 2000). We have found a significant over-representation of the RET A45A (nt c135G→A, exon 2) and L769L (nt c2307T→G, exon 13) intragenic sequence variants in our cohort of patients with sporadic HSCR compared to the control group (P<.0006) (Borrego et al. 1999). These findings were independently confirmed in a HSCR cohort of German origin (Fitze et al. 1999). It was proposed that the A45A and L769L polymorphisms could act as low-penetrance alleles and/or as factors modifying the phenotypic expression or even be in linkage disequilibrium with an unknown functional variant (Borrego et al. 1998, 1999, 2000; Fitze et al. 1999). We subsequently studied the haplotypes on the basis of the combination of the seven polymorphic variants in the RET coding region. The over-represented haplotypes in HSCR were the so-called haplotype B (which includes only the A45A polymorphism) and haplotype C (which includes A45A and L769L). Analysis of phase-known genotypes (i.e., paired haplotypes) indicated an over-representation of genotypes BB, BC, and CC in the HSCR series versus the control group. On the basis of these data, it was proposed that the association of these genotypes with HSCR susceptibility occurs in an autosomal recessive manner in an additive dose-dependent manner (Borrego et al. 2000).
The existing data for HSCR suggest one of two hypotheses. Either the sequence variation representing each of the “at risk” haplotypes is itself functional and leads to low-penetrance loss of function, or the “at risk” haplotypes represent linkage disequilibrium with the actual low-penetrance susceptibility locus. There also appear to be consistent haplotypes containing S836S in patients with MTC, and the same two hypotheses hold as with HSCR: either the variant is functional, or these haplotypes are in linkage disequilibrium with a low-penetrance susceptibility locus. Because the haplotypes containing S836S in sporadic MTC never coincide with the HSCR “risk” haplotypes, we might postulate the existence of a single modifying/regulating locus of RET function where different specific alterations lead to opposing functional effects, conferring specific susceptibility to either MTC or HSCR. To begin testing these hypotheses, we sought to identify and characterize novel SNPs upstream of the earlier examined haplotypes and to use all of the data in haplotype analyses.
Material and Methods
Patients and Control Subjects
In the present work, we have included 103 patients affected with clinically sporadic HSCR, their unaffected parents, 51 patients affected with sporadic MTC, and 100 normal control subjects. Traditional germline MEN 2–defining RET mutations are absent in the 51 patients with sporadic MTC. HSCR and MTC cohorts have been described in our previous publications (Borrego et al. 1999, 2000; Ruiz et al. 2001). Normal control subjects were unselected, unrelated race-, age-, and gender-matched individuals from Spain. Informed consent was obtained in accordance with the approved protocols of the respective institutional review boards for the protection of human subjects.
Sequence Analysis of Intron 1 of RET
Genomic DNA was obtained from peripheral blood leukocytes using standard protocols (Dracopoli et al. 1994). To meticulously search for new SNPs within intron 1 of the RET proto-oncogene, we performed enzymatic mutation detection (EMD), beginning from 3′ of the intron, on overlapping fragments of 300 bp to 1 kb in length, according to the manufacturer's recommendations (del Tito et al. 1998) (fig. 1A and table 1). When any putative DNA variant was detected, we proceeded to direct sequencing of the fragments, by use of conditions published elsewhere (Marcos et al. 2000), to characterize the changes observed.
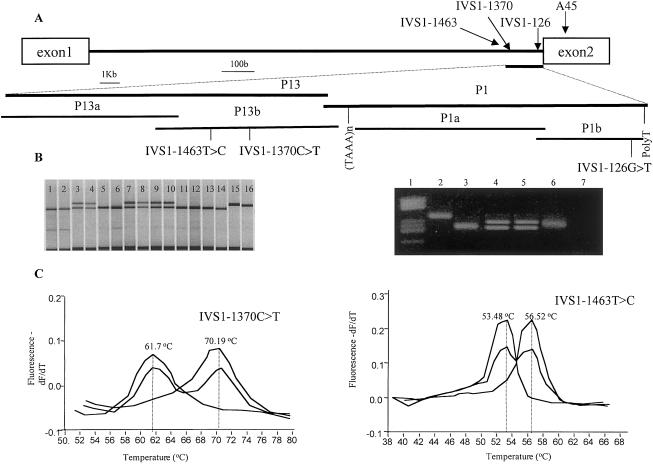
Figure 1.
Characterization of novel SNPs in RET intron 1. A, Schematic representation on 3′ end of intron 1 of the RET proto-oncogene. Codon 45 and the 3 IVS1 SNPs are indicated. B, left panel, genotyping of IVS1-126G→T by fluorescent SSCP. Lanes 1, 2, 5, 6, and 11–14, genotype −/−. Lanes 3, 4, and 7–10, genotype +/−. Lanes 15 and 16, genotype +/+. Right panel, genotyping of IVS1-126G→T by differential restriction with NlaIII. Lane 2, a nondigested sample. Lane 3, genotype −/−. Lanes 4 and 5, genotype +/−. Lane 6, genotype +/+. C, FRET analysis of IVS1-1370C→T and IVS1-1463T→C. In each case, amplicons containing the polymorphisms have a melting point (MP) which is >9°C lower (in the case of IVS1-1370C→T) and 3°C higher (in the case of IVS1-1463T→C) than the amplicons with the wild-type sequence. Genotype −/− , +/−, and +/+ are represented in red, blue, and green, respectively.
Table 1.
PCR Conditions of Individual Fragments within Intron 1 of RET Proto-Oncogene[Note]
|
Primers(5′→3′) |
|||||
| Fragment | Forward | Reverse | Size(bp) | AnnealingTemperature(°C ) | Cycles |
| P1 | M13F*-GTGGAAGTTGTGGTGAGCCAAG | M13R*-GTGGGAAGTAGGGAAGTGAGTGAG | 1,057 | 67 | 35 |
| P13 | M13F-TGGCACAATCTCGGCTCACTAC | M13R-CTTGGCTCACCACAACTTCCAC | 1,051 | 68 | 35 |
| P1b | M13F-ATGACTTTCCTGTAAAGTGC | M13R-GGAGTTTTTCATCTCTGTTC | 338 | 54 | 35 |
| P1a | M13F-AGATAAGATGCACGGACCTTAG | M13R-GCAACAGTTGCCAAAAAATG | 596 | 60 | 35 |
| P13b | M13F-CCAGAAGTGGGATTGGTAG | M13R-TCACCACAACTTCCACCTC | 597 | 59 | 35 |
| P13a | M13F-GTGCAATGGCACAATCTC | M13R-GAAAAACAAGAAGGTAGTTCCC | 552 | 58 | 35 |
Note.— Primers were designed using the PRIME command of GCG-Wisconsin software, and all of them had universal M13 primers attached at the 5′ ends. M13F*: CGCCAGGGTTTTCCCAGTCACGAC; M13R*: TTTCACACAGGAAACAGCTATGAC. Fragments P1 and P13 were each divided into two smaller fragments (a and b), to obtain better results in the performance of EMD technology. Fragments were overlapping, except for P1a and P13b, which were separated by the STR (TAAAn).
Genotyping of SNPs
The variant IVS1-126G→T was genotyped using fluorescent SSCP analysis, after digestion of the PCR product with MspI (Roche), under conditions described elsewhere (Marcos et al. 2000). In addition, the results were confirmed by differential restriction digestion with NlaIII (New England Biolabs), since this enzyme loses a restriction site in the presence of the variant (fig. 1B).
To detect IVS1-1370C→T and IVS1-1463T→C, we have developed an automated method using fluorescence resonance energy transfer (FRET) technology. Internal probes were designed according to the sequences of interest and were purchased from TIB Molbiol. The whole procedure, including real-time PCR and a melting curve, was performed in a LightCycler machine (Roche) (fig. 1C and table 2).
Table 2.
FRET Conditions for Detection of the Polymorphisms within Intron 1 of RET Proto-Oncogene[Note]
|
Melting Conditions after Amplification |
||||
| PolymorphismGenotyped | Probesa | TemperatureTransition(°C) | TransitionRate(°C/s) | Gains (F1:F2:F3) |
| IVS1-1370C→T | Anchor: 5′- YgCTCACTCAgCCACAgCCgAggC pSensor: 5′- TgCACCgTgCCCCTgCTTg X | From 50 to 95 | .40 | |
| IVS1-1463T→C | Anchor: 5′- TTTTTgggAACTACCTTCTTgTTTTTCATSensor: 5′- Y-ACTgTATATTATTTTCCACTCACCgA p | From 40 to 95 | .20 | 1:45:30 |
Note.— We used 50 cycles of PCR at an annealing temperature of 66°C.
Boldface italics denote the nucleotides that are sensitive for the wild-type or the polymorphic allele. “X” represents fluorescein, and “Y” represents LC-Red 640. Some probes have modifications (represented by “p”) in their 3′ that disable extension of Taq polymerase.
Phase of haplotypes was determined by typing of the corresponding parents of patients with HSCR and control subjects where available (86% of patients with HSCR and 65% of control subjects had parents available for typing [data not shown]). Haplotypes (i.e., phase) were similar in patients and control subjects, and, thus, haplotypes from patients with MTC were inferred from the haplotypes available in patients with HSCR and control subjects. The median age of sporadic MTC diagnosis is 55–60 years, and, hence, the parents of patients with MTC were, in general, deceased.
Statistical Analysis
Allelic frequencies of the three new RET polymorphic loci were determined, and haplotypes were constructed (tables 3 and 4). The frequencies of each haplotype were compared between the patients with MTC and control subjects, as well as between patients with HSCR and control subjects and between patients with MTC and patients with HSCR. Comparisons were performed using either χ2 analysis with Yates's correction or, when appropriate, Fisher's two-tailed exact test. Nominal statistical significance was considered when P<.05.
Table 3.
Comparative Studies between the Groups, using χ2 Analysis with Yates's Correction
|
Results of Comparison |
||||||
| HSCR vs. Control |
sMTC vs. Control |
sMTC vs. HSCR |
||||
| HSCR Alleles | Control Alleles | sMTC Alleles | Control Alleles | sMTC Alleles | HSCR Alleles | |
| IVS1-126G→T: | ||||||
| No. of polymorphic T alleles | 44 | 70 | 49 | 70 | 49 | 44 |
| No. of wild-type G alleles | 162 | 130 | 53 | 130 | 53 | 162 |
| χ2 with Yates's correction (P value) | 8.69 (P=.0032053) | .28 (P=.0385776) | 21.79 (P=.000003) | |||
| IVS1-1370C→T: | ||||||
| No. of polymorphic T alleles | 39 | 94 | 40 | 94 | 40 | 39 |
| No. of wild-type C alleles | 167 | 106 | 62 | 106 | 62 | 167 |
| χ2 with Yates's correction (P value) | 35.03 (P<.000001) | 1.36 (P=.2438972) | 13.67 (P=.0002175) | |||
| IVS1-1463T→C: | ||||||
| No. of polymorphic C alleles | 84 | 163 | 89 | 163 | 89 | 84 |
| No. of wild-type T alleles | 122 | 37 | 13 | 37 | 13 | 122 |
| χ2 with Yates's correction (P value) | 68.94 (P<.000001) | 1.23 (P=.2674883) | 57.99 (P<.000001) | |||
Table 4.
Polymorphic Frequencies in Patients with sMTC, Control Subjects, and Patients with HSCR
|
Frequency in |
|||
| Allele | PatientswithsMTC | ControlSubjects | PatientswithHSCR |
| IVS1-126G→T: | |||
| Polymorphic T | .48 | .35 | .21 |
| Wild-type G | .52 | .65 | .79 |
| IVS1-1370C→T: | |||
| Polymorphic T | .39 | .47 | .21 |
| Wild-type C | .61 | .53 | .79 |
| IVS1-1463T→C: | |||
| Polymorphic C | .9 | .82 | .41 |
| Wild-type T | .1 | .18 | .59 |
Haplotype analyses were performed by reconstructing and comparing the transmitted versus nontransmitted haplotypes to patients with HSCR, with the understanding that meiotic recombination is negligible for a single generation across such a short region. This approach is consistent with the haplotype relative risk approach of Falk and Rubinstein (1987) and is largely robust to population stratification (described by Ott [1999], p. 293).
Full multipoint model-based analyses of a founding locus were performed using DMLE+ (Reeve and Rannala 2002), which uses Monte Carlo integration to approximate Bayesian posteriors for locus position and age. Calculations were performed on a Pentium 4 workstation, using recommended burn-in and sampling intervals and a variety of modeling assumptions and parameter ranges, including population growth rates. The composite likelihood ratio of DISMULT (Terwilliger 1995) was also computed, using a Unix workstation. Numerical difficulties in evaluating the likelihood over such short intervals was resolved by artificially inflating the map distances, while interpreting the mutation age as increased by the same proportion.
In addition to these approaches, simple haplotype methods were used to test against the null hypothesis of equal transmission probabilities. These included specification of putative ancestral haplotypes and permutation testing by drawing 10,000 random permutations of the transmission status of all haplotypes. For each permutation, the maximum nominal χ2 statistic was recorded in the association of each haplotype (vs. all others combined) with transmission status. A somewhat more structured approach is described below.
The degree of linkage disequilibrium between disease and a marker locus is often described as pexcess, which measures the proportion of alleles at a locus caused by the putative founding mutation (de la Chapelle and Wright 1998). If an allele is associated with disease (e.g., the polymorphic allele A at codon 45 (Borrego et al. 1999, 2000), then we can compare the proportion of transmitted chromosomes which have the allele (paffected) with the proportion in nontransmitted chromosomes (pnormal). From these, we can compute pexcess=(paffected-pnormal)/(1-pnormal).
If there is a predominant ancestral haplotype, then the value of pexcess will tend to reach a maximum at markers very near the mutation and to descend as one moves away from the mutation. Moreover, the maximum value of pexcess (attained at the mutation locus) represents the proportion of transmitted chromosomes that contain the ancestral mutation. For a rare recessive disease, this can be, at most, 1, and for a rare dominant disease, it can be, at most, 0.5. The estimate of pexcess at A45A is ∼0.45 (derived from the frequency estimates of Borrego et al. [1999]), which suggests that, if the mutation exists, it is responsible for a large portion of sporadic HSCR in this population. Often, pexcess will not achieve the theoretical maximum value because of the presence of additional founding mutations that account for some of the sporadic cases.
To generalize this approach to multiple markers, we used a method (Gao and Wright 1999) that examines haplotypes in varying window widths of successive SNPs to find the window in which haplotypes (all treated separately) are most strikingly associated with disease, as judged by a χ2 statistic. The null distribution of this maximal statistic can be generated by recomputing it over permutation samples of transmission status. Also, we recorded, for each SNP, the single haplotype (over all window widths) most associated with transmission, which was then treated as if it were a single allele in a pexcess plot. The result is a plot of so-called hexcess (similar to pexcess, but with less error variation) for the excess of the haplotype among transmitted chromosomes as a function of physical position, and it is similar to the multiple-SNP haplotype-based disequilibrium measure described by Daly et al. (2001). Under the assumption that the ancestral haplotype has been correctly identified, hexcess at the mutation also represents the proportion of transmitted chromosomes containing the mutation.
Finally, noting that expected linkage disequilibrium decays exponentially with increasing recombination fraction (see, e.g, Hastbacka et al. 1992), simple linear regression of log(hexcess) on genomic position was performed, under the assumption that all transmitted chromosomes contain the ancestral mutation. This simple moment-based approach is based on our method for testing against a null hypothesis, but with no specification of the complicated dependencies arising under the alternative. Thus, it does not naturally give rise to CIs or precise inferential statements about the mutation age or location.
Results
Identification of Three Novel RET Intron 1 SNPs
Using a series of bioinformatics manipulations within GenBank and the National Center for Biotechnology Information (NCBI), we uncovered a 505-kb genomic contig on 10q23 that contained the entire RET sequence (GenBank accession number AC010864 [subfile of NT_033987]). Using the Blast 2 sequences tool at the NCBI Web site, we aligned the AC010864 sequence with those of exons 1 and 2 of the RET proto-oncogene described earlier. The result was a segment of contig AC010864 that contained the complete sequence of intron 1 of RET (23,127 bp). We were able to identify three novel SNPs within intron 1: IVS1-126 G→T, IVS1-1370 C→T, and IVS1-1463 T→C (fig. 1).
Studies of Association Between the Three Intron 1 SNPs and HSCR or MTC
Patients with HSCR versus control subjects
The three newly identified intron 1 SNPs were found to be under-represented in the HSCR cohort compared to the control population (table 3). For example, of a total of 206 HSCR chromosomes, there were 44 chromosomes (21%) with the T variant at the IVS1-126G→T locus, and 162 (79%) the wild-type G allele. The T variant was significantly under-represented when compared to the control series (χ2 with Yates's correction 8.69, P=.0032). Similarly, the IVS1-1370C→T and IVS1-1463T→C polymorphic alleles (T and C, respectively) were also under-represented in the HSCR cohort compared to those observed in the control group (table 3; P≪.000001).
Patients with MTC versus control subjects
In contrast to the HSCR cohort, the IVS1-126G→T polymorphic allele was over-represented in patients with sporadic MTC. Of 102 MTC chromosomes studied, 49 (48%) carried the polymorphic T allele, and 53 (52%) carried the wild-type G (χ2=4.28; P=.038 with Yates's correction) (table 3). We did not find any statistically significant differences when comparing the allelic frequencies of the IVS1-1370C→T and IVS1-1463T→C loci between MTC and control subjects (table 3).
Patients with HSCR versus patients with MTC
One highlight of the results obtained in the analysis of allele frequencies of the newly identified loci was the inversion in the frequency of IVS1-126G→T when comparing the series of patients with MTC to the HSCR series. The frequency observed for this marker in the control population appeared in a range intermediate between those observed in patients of both pathologies (T: 70 [35%]; G: 130 [65%]). In comparison of the allele distribution of the SNPs between cohorts (HSCR vs. MTC), it was remarkable to note an inversion of the allele frequencies for each genotyped marker (tables 3 and 4).
Haplotype Analysis Using Intron 1 SNPs: Evidence of Association of Haplotype 0 and HSCR, and Haplotype 2 with MTC
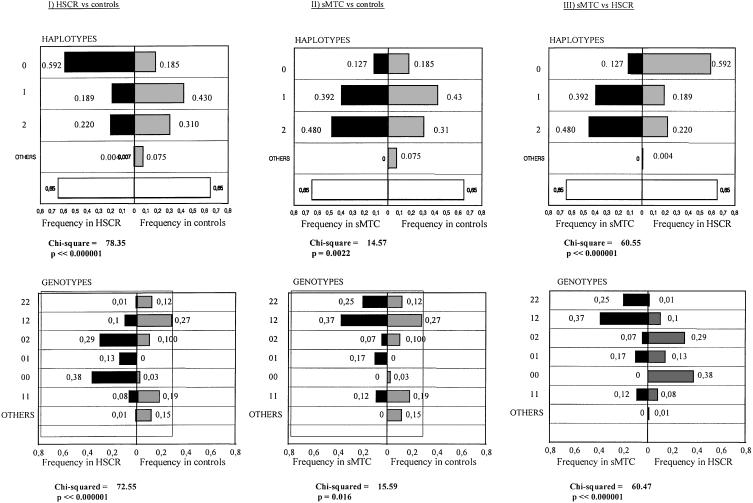
Using the information on the segregation of the SNPs identified in HSCR patients and their parents, we performed a construction of haplotypes comprising different combinations of the three SNPs at the 3′ end of RET intron 1 (table 5). The method and the group of patients used for the haplotype construction was similar to that described elsewhere (Borrego et al. 2000). We found a total of five haplotypes in the patients with HSCR and their parents (haplotypes 0–4). As the recombination events between the markers under study were predicted to be extremely rare (distance between SNPs <1,200 bp), we considered each haplotype as an individual allele within the same locus (3′ intron 1, RET proto-oncogene). Parents of MTC or control subjects were not available, so the haplotype construction in both groups was inferred from the haplotype of the HSCR patients. The haplotype distribution observed in the MTC, control, and HSCR groups was different. Thus, haplotype distribution of each cohort compared to the control group (MTC vs. control subjects or HSCR vs. control subjects), was found to be statistically significant (P<.0023) (fig. 3).
Table 5.
Haplotypes Based in the Combination of Three SNPs Located within Intron 1 of RET[Note]
|
Presence/Absence of |
|||
| Haplotype | IVS1-126G→T | IVS1-1370C→T | IVS1-1463T→C |
| 0 | − | − | − |
| 1 | − | + | + |
| 2 | + | − | + |
| 3 | − | − | + |
| 4 | + | + | + |
Note.— + = present; − = absent.
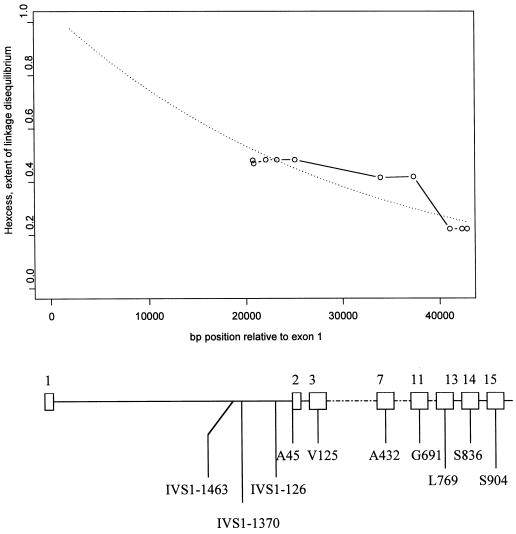
Figure 2.
Excess proportion of ancestral haplotypes among transmitted chromosomes to HSCR patients (see text). The fitted line of expected linkage disequilibrium suggests that the ancestral mutation lies in or near the 5′ region of RET—that is, within intron 1. A schematic of partial RET gene structure on which the positions of the 10 SNPs are placed lies below the plot. Numbers on top of the gene schematic are exon numbers, and the codes below represent the nature and position of the SNPs.
Figure 3.
Comparison of the frequencies and distribution of RET IVS1 haplotypes and genotypes between HSCR or patients with MTC and control subjects. χ2 (Yates's correction) and P values are denoted below each comparison.
We verified that haplotype 0 was the most common in the HSCR group (59%) (fig. 3). It is important to note that haplotype 0 is in complete linkage disequilibrium with all the risk haplotypes described so far (Borrego et al. 1999, 2000). That is, all the risk haplotypes share the same sequence of SNPs in this genomic segment of intron 1 (haplotype 0). This is consistent with our studies of linkage disequilibrium mapping and would place the locus of susceptibility to HSCR at ∼20 kb from the A45A marker (see below as well).
Haplotype 2 is the most frequent in patients with sporadic MTC (48%) (haplotype that contains IVS1-126G→T and IVS1-1463T→C in disequilibrium), whereas the frequency of haplotype 0 is notably low (12.7%). The MTC patients that carry S836S also carry intron 1 haplotype 2, which suggests that S836S and haplotype 2 are in linkage disequilibrium. These data have been confirmed in three patients with MTC who were homozygous for haplotype 2 and heterozygous for S836S.
The haplotypes observed in the control series show a distribution intermediate to that observed in the MTC and HSCR cohorts. Haplotype 1 was the most common in the control subjects (43%). Furthermore, haplotype 4, which is not represented in any of the patient series being studied, appears with a certain frequency (4%). Haplotype 0 has a frequency of 18.5% in control subjects, compared with 59% in patients with HSCR and 12.7% in patients with MTC. The frequency of haplotype 2 in control subjects was 31%, compared with 21.3% in individuals with HSCR and 48% in individuals with MTC. These observations suggest that haplotype 0 could be a haplotype associated with HSCR, and haplotype 2, a haplotype associated with MTC (fig. 3; also see below).
Analysis of the IVS1 Genotypes in HSCR and MTC
We studied the genotype composition (pairs of haplotypes) in each study group (fig. 3). Genotypes 00 and 02 predominate in HSCR (70%), whereas, in MTC, genotypes 12 and 22 are more predominant (67%). In the control group, the most common genotype was 12, followed by 11 and 22. Analysis of the genotypes observed/expected in the study groups shows that the results are in Hardy-Weinberg equilibrium. If we compare the distribution of genotypes between both study groups (MTC and HSCR), the deviation is rather striking (fig. 3). As a whole, these findings corroborate the observations made in individual allele as well as haplotype frequency studies.
Haplotype Analysis Using All 10 RET SNPs: Evidence for a Novel Founding Locus for HSCR
Using the data from the same consecutive incident cases of isolated HSCR from the Western Andalusia region described in Borrego et al. (1999, 2000), we have analyzed the observed RET haplotypes and their association with HSCR for evidence of a mutation or variant contributing to sporadic cases of disease. This offers a potentially more parsimonious explanation for the linkage disequilibrium than the hypothesis that the polymorphisms in RET have subtle functional effects.
We analyzed the transmitted versus nontransmitted haplotypes using DMLE+ (see the “Material and Methods” section), which is a fully model-based approach for linkage disequilibrium mapping. The results were difficult to interpret, as the posterior for mutation location was extremely short (<1% of allowed range) and always placed at the extreme 5′ end of the allowed range. Mutation age estimates were very highly sensitive to starting values, despite long burn-in times for DMLE+ and ranged from 200 to 1,500 generations, under the assumption of an average correspondence of 1 cM = 1 Mb. We speculate that the difficulty in obtaining estimates stems from the failure of the SNPs to span the mutation, in contrast to classic examples used to evaluate the approach (e.g., diastrophic dysplasia [Reeve and Rannala 2002]). The program DISMULT achieved maximum-likelihood values at or just upstream of the most 5′ SNP IVS1-1463, with estimated mutation age (see the “Material and Methods” section) sharply changing from 4,100 to 8,000 generations in a short interval having high likelihood, and with estimated proportion of the mutation, among transmitted chromosomes, of ∼0.45.
Our haplotype-based linkage-disequilibrium analysis is in rough agreement with the DISMULT results, except that we believe the data do not preclude the possibility that a mutation may exist further upstream of the describe RET polymorphisms (5′ direction, centromeric).
The hexcess plot in figure 2 (see the “Materials and Methods” section) is maximized at ∼0.45, at the most 5′ SNPs, and reveals that the linkage disequilibrium descends toward the 3′ end of RET. The permutation-based empirical P value for the association of ancestral haplotype with transmission status was <.0001. The fact that the linkage disequilibrium levels off for a few markers at a time is expected—this reflects that historical crossovers occurred at only a few points across RET.
Most compelling is the fact that the “most significant” short haplotypes at each marker position (i.e., in χ2 tests of association with disease) are consistent with a single, longer ancestral haplotype. The ancestral haplotype for the 10 SNPs appears to be (5′→3′) WWWMWWWMWW, where M signifies mutant and W wild type. This is haplotype C from the Borrego et al. (2000) study, with the addition of the three new SNPs from intron 1. Haplotype MWMWMWWWWW is associated with disease (9 transmitted vs. 0 nontransmitted) but may be part of a different minor founding mutation.
The maximum value that the linkage-disequilibrium measure attains is not known and awaits further determination of SNPs in intron 1 and upstream of RET. An exponential fit to the decaying linkage-disequilibrium measure (which cannot exceed 1.0) in figure 2 (dashed line) suggests that the major mutation is likely to reside in intron 1 or just upstream of RET.
A recent refined genetic map indicates that the rate of recombination per unit of physical distance in the vicinity of RET is ∼0.8 cM/Mb (Kong et al. 2002). Combining this information with the fitted rate of decay results in an estimated time from the founding mutation of 3,800 generations. Such a surprisingly ancient estimate must be interpreted with caution, as the recombination rate and mutation age are confounded, and the resolution of such recombination rate estimates remains low. Moreover, although linkage disequilibrium is clearly present, the current data do not support the construction of plausible CIs. Nonetheless, the data are consistent with a founder mutation old enough to be widespread in European populations. This is further supported by the report from Fitze et al. (1999), in which genotypes of the same seven polymorphisms studied earlier by us (Borrego et al. 2000) display a very similar pattern of association with sporadic HSCR in a German population.
Discussion
Traditional germline mutations identified in the RET proto-oncogene have been associated with two different neurocristopathies (MEN 2 and HSCR). The phenotype observed in one is completely different from that in the other (for a review, see MIM 142623 and MIM 164761), and the molecular mechanism proposed for the two cases seem to be opposites, according to the available functional data. Mutations causing MEN 2 produce gain-of-function alterations in the cascade of RET signals, either because of constitutive activation or alteration of substrate specificity (Santoro et al. 1990; Songyang et al. 1995). In patients with HSCR, traditional RET mutations typically result in loss of function, including structural and functional haploinsufficiency and a deficit in the cascade of RET signals in the target tissues (Pasini et al. 1995; Pelet et al. 1998). It has recently been proposed that the mechanism of some HSCR mutations could consist of an activation of cryptic proapoptotic functional motifs in the altered RET protein that lead to premature apoptosis (Bordeaux et al. 2000). The observation that the RET receptor is involved both in tumorigenesis and in the development of the nervous system is reminiscent of that observed for the p75NTR and DCC receptors, in which the proapoptosis mechanism seems similar to that observed in both cases (Rabizdeh et al. 1993; Mehlen et al. 1998).
In contrast to the traditional mutations described for a subset of HSCR and the majority of MEN 2, the isolated forms of both pathologies—namely, MTC and HSCR—appear to be attributable to low-penetrance alleles, the downstream functional mechanism of which is unknown. These alleles have been identified using common polymorphisms throughout the RET proto-oncogene sequence and studying genetic association or disequilibrium of transmission in families affected with HSCR with incomplete penetrance (Borrego et al. 1998, 1999, 2000; Fitze et al. 1999; Gimm et al. 1999; Bolk et al. 2000; Griseri et al. 2000). Initially, direct action mechanisms of the linked coding RET coding SNPs, such as the activation of cryptic splicing sites or altered expression of the allele with the variant (producing over- or underexpression of the RET proto-oncogene) were proposed. As an alternative, the mechanism was also suggested to be the preferential use of specific tRNAs that reduced the efficiency of the translation of the allele that carried the variant, and the alteration of functional motifs of the RET sequence which constituted the targets for DNA- or RNA-binding proteins. Although some functional studies have been performed on these polymorphisms, no alteration has yet been described that explains the direct effect of any of the SNPs associated with both pathologies (Griseri et al. 2000). Besides the hypothesis of direct action of the markers linked to each pathology, a mechanism related to the existence of linkage disequilibrium of some functional allele in noncoding sequence with the markers studied has also been considered (Borrego et al. 1998, 1999, 2000; Fitze et al. 1999; Gimm et al. 1999; Bolk et al. 2000; Griseri et al. 2000).
Together with our previous observations (Borrego et al. 1999, 2000), our current observations indicate that the A45A polymorphism, irrespective of the 3′ haplotype, is in linkage disequilibrium with a group of markers within intron 1 of the RET proto-oncogene (haplotype 0-A45A). In other words, the under-representation of the variant alleles at all three new loci within intron 1 described in the HSCR cohort can be explained by the polymorphic variant at codon 45 (A45A, nt c135G→A) to be in complete linkage disequilibrium with the wild-type allele (G) at IVS1-126G→T, the wild-type allele (C) at IVS1-1370C→T and the wild-type allele (T) at IVS1-1463T→C. More importantly, our statistical analysis of all 10 RET SNPs in HSCR (see the last subsection under “Results”) strongly suggest the existence of a low-penetrance locus of susceptibility for HSCR at a distance of <20 kb from RET codon 45, perhaps still within intron 1.
In contrast to isolated HSCR, individuals with sporadic MTC, originating in Germany and Spain, were shown to have an over-representation of S836S (Gimm et al. 1999; Ruiz et al. 2001). Similarly, the same marker has been attributed with a protector effect against the appearance of the HSCR phenotype (Griseri et al. 2000, 2002). Whether the sequence variation in and of itself conferred a subtle effect on protein expression, splicing, or function remains to be elucidated. Like the association of specific RET haplotypes with isolated HSCR, the existence of a functional locus in linkage disequilibrium with S836S has also been suggested as an alternative to the hypothesis of the direct effect of S836S (Gimm et al. 1999; Griseri et al. 2000, 2002; Ruiz et al. 2001). This latter hypothesis and those that invoke expressional variation are not mutually exclusive, as the function of the linked upstream putative susceptibility locus has yet to be elucidated. Thus, our current observations shed some light to differentiate amongst these hypotheses. We have noted that there is a MTC-specific risk haplotype within intron 1: haplotype 2, which is always associated with S836S, and both haplotype 2 and S836S are concurrently over-represented among our isolated MTC series. Our current data, therefore, strongly suggest that the association of S836S with MTC can be most plausibly explained by linkage disequilibrium between S836S and haplotype 2. Further, preliminary germline intron 1 haplotyping data in the MTC cohort of German origin show a high frequency of homozygosity for haplotype 2 (data not shown). This suggests the existence of a low-penetrance susceptibility locus for MTC in close proximity to that for isolated HSCR, within intron 1 of the RET proto-oncogene.
It is tempting to speculate about the nature of the susceptibility loci that could be located within the immense intron 1 of the RET proto-oncogene. Perhaps the most plausible hypothesis would be the existence of two independent mutations within the same functional motif—unknown at present—that would control the transcriptional activity of the RET proto-oncogene. Little is known about the transcriptional factors that bind to the putative RET promoter. For example, there is in vitro evidence that RAF-1, PAX3, SOX10, AP1, and AP2 could act as transcriptional factors that bind upstream of exon 1 (Carson et al. 1995; Capes-Davis et al. 1999; Lang et al. 2000). Using the MatInspector program (Quandt et al. 1995), bioinformatic analysis of the region harboring the three novel SNPs predicts four motifs that could be binding sites for transcription factors BRN2, NFAT, IRF1, and IRF2 at IVS1-1463T (wild type; HSCR-associated). When IVS1-1463 is altered to the polymorphic C (which is MTC-associated), the BRN2 site is obliterated and a new site for OCT1 appears. Furthermore, the two IRF sites are strengthened, whereas the NFAT site is weakened, in the presence of the variant. Similarly, this program also predicts that when the sequence is wild-type (G, HSCR-associated) at IVS1-126, there is the motif for a TCF1-binding site. When this sequence is variant, T (MTC-associated), an NFAT motif is created. It is interesting there is no difference at IVS1-1370 (both wild type) between haplotypes 0 (HSCR) and 2 (MTC), and no binding motifs are predicted at this site. Unfortunately, no in vitro or in vivo data exist regarding the plausibility of these binding motifs and actual binding sites within RET intron 1. To date, such bioinformatic mining for transcription binding sites has yet to yield functionally significant motifs for any gene in vivo. Therefore, further experimental work is required to prove the functional significance of these transcription factor binding sites predicted from bioinformatic analysis.
In summary, our data demonstrate that isolated MTC and HSCR are significantly associated with specific linked haplotypes (haplotypes 2 and 0, respectively) within RET intron 1. We believe that the data strongly suggest that each haplotype is likely to be in linkage disequilibrium with its respective putative low-penetrance susceptibility locus and that both loci may reside in a very small genomic segment (<30 kb). These loci might not only act in and of themselves as low-penetrance susceptibility loci for HSCR or MTC but would likely interact with other variants and with traditional germline HSCR-associated or MEN 2-associated mutations to modulate development of features, age at onset, and the like, as suggested by our prior, preliminary observations (Borrego et al. 1998).
Acknowledgments
Heather Dziema and Alexander Niess provided technical assistance. C.E. thanks Getachew Boru for helpful discussions. This study was partially funded by grants R01HD39058 from the National Institutes of Health (to S.B., F.A.W., G.A., and C.E.); P30CA16058 from the National Cancer Institute (to The Ohio State University Comprehensive Cancer Center); FIS 01/0551 from the Fondo de Investigación Sanitaria, Spain (to S.B. and G.A.); and CAA 116/00 and CAA 24/01 from the Consejeria de Salud de la Junta de Andalucia, Spain (to S.B. and G.A.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for RET sequence [accession number AC010864])
- NCBI, http://www.ncbi.nlm.nih.gov/ (for Blast 2 sequences tool)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HSCR [MIM 142623] and MEN 2 [MIM 164761])
References
- Amiel J, Attié T, Jan D, Pelet A, Edery P, Bidaud C, Lacombe D, Tam P, Simeoni J, Flori E, Nihoul-Fékété C, Munnich A, Lyonnet S (1996) Heterozygous endothelin receptor B (EDNRB) mutations in isolated Hirschsprung disease. Hum Mol Genet 5:355–357 [DOI] [PubMed] [Google Scholar]
- Amiel J, Espinosa-Parilla Y, Steffan J, Gosset P, Pelet A, Prieru M, Boute O, Choiset A, Lacombe D, Philip N, Le Merrer M, Tanaka H, Till M, Touraine R, Toutain A, Vekemans M, Munnich A, Lyonnet S (2001) Large-scale deletions and SMADIP1 truncating mutations in syndromic Hirschsprung disease with involvement of midline structures. Am J Hum Genet 69:1370–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrist M, Bolk S, Halushka M, Lapchak PA, Chakravarti A (1996) Germline mutations in glial cell line-derived neurotrophic factor (GDNF) and RET in a Hirschsprung disease patient. Nat Genet 14:341–344 [DOI] [PubMed] [Google Scholar]
- Angrist M, Bolk S, Thiel B, Puffenberger EG, Hofstra RM, Buys CHCM, Cass DT, Chakravarti A (1995) Mutation analysis of the RET receptor tyrosine kinase in Hirschsprung disease. Hum Mol Genet 4:821–830 [DOI] [PubMed] [Google Scholar]
- Attié T, Pelet A, Edery P, Eng C, Mulligan LM, Amiel J, Boutrand L, Beldjord C, Nihoul-Fékété C, Munnich A, Ponder BAJ, Lyonnet S (1995) Diversity of RET proto-oncogene mutations in familial and sporadic Hirschsprung disease. Hum Mol Genet 4:1381–1386 [DOI] [PubMed] [Google Scholar]
- Auricchio A, Casari G, Staiano A, Ballabio A (1996) Endothelin-B receptor mutations in patients with isolated Hirschsprung disease from a non-inbred population. Hum Mol Genet 5:351–354 [DOI] [PubMed] [Google Scholar]
- Badner JA, Sieber WK, Garver KL, Chakravarti A (1990) A genetic study of Hirschsprung disease. Am J Hum Genet 46:568–580 [PMC free article] [PubMed] [Google Scholar]
- Bidaud C, Salomon R, Vancamp G, Pelet A, Attié T, Eng C, Bonduelle M, Amiel J, Nihoul-Fékété C, Willems PJ, Munnich A, Lyonnet S (1997) Endothelin-3 gene mutations in isolated and syndromic Hirschsprung disease. Eur J Hum Genet 5:247–251 [PubMed] [Google Scholar]
- Bolk S, Pelet A, Hofstra RMW, Angrist M, Salomon R, Croaker D, Buys CHCM, Lyonnet S, Chakravarti A (2000) A human model for multigenic inheritance: phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc Natl Acad Sci USA 97:268–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeaux MC, Forcet C, Granger L, Corset V, Bidaud C, Billaud M, Bredesen DE, Edery P, Mehlen P (2000) The RET proto-oncogene induces apoptosis: a novel mechanism for Hirschsprung disease. EMBO J 19:4056–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego S, Eng C, Sánchez B, Sáez M-E, Navarro E, Antiñolo G (1998) Molecular analysis of RET and GDNF genes in a family with multiple endocrine neoplasia type 2A and Hirschsprung disease. J Clin Endocrinol Metab 83:3361–3364 [DOI] [PubMed] [Google Scholar]
- Borrego S, Saez ME, Ruiz A, Gimm O, Gao X, Lopez-Alonso M, Hernandez A, Wright FA, Antiñolo G, Eng C (2000) RET genotypes comprising specific haplotypes of polymorphic variants predispose to isolated Hirschsprung disease. J Med Genet 37:572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego S, Saez ME, Ruiz A, Gimm O, Lopez-Alonso M, Antiñolo G, Eng C (1999) Specific polymorphisms in the RET proto-oncogene are over-represented in individuals with Hirschsprung disease and may represent loci modifying phenotypic expression. J Med Genet 36:771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes-Davis A, Andrew SD, Hyland VJ, Twigg S, Learoyd DL, Dwight T, Marsh DJ, Robinson BG (1999) Glucocorticoids differentially inhibit expression of the RETproto-oncogene. Gene Expr 8:311–326 [PMC free article] [PubMed] [Google Scholar]
- Carson EB, McMahon M, Baylin SB, Nelkin BD (1995) Ret gene silencing is associated with Raf-1-induced medullary thyroid carcinoma cell line differentiation. Cancer Res 55:2048–2052 [PubMed] [Google Scholar]
- Chakravarti A (1996) Endothelin receptor-mediated signaling in Hirschsprung disease. Hum Mol Genet 5:303–307 [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Wright FA (1998) Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci USA 95:12416–12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Tito BJ, Poff HE, Novotny MA, Cartledge DM, Walker RI 2nd, Earl CD, Bailey AL (1998) Automated fluorescent analysis procedure for enzymatic mutation detection. Clin Chem 44:731–739 [PubMed] [Google Scholar]
- Doray B, Salomon R, Amiel J, Pelet A, Touraine R, Billaud M, Attié T, Bachy B, Munnich A, Lyonnet S (1998) Mutation of the RET ligands, neurturin, supports multigenic inheritance in Hirschsprung disease. Hum Mol Genet 7:1449–1452 [DOI] [PubMed] [Google Scholar]
- Dracopoli NH, Haines JL, Korf BR, Moir DT, Morton CC, Seidman CE, Seidman JG, Smith DR (1994) Current protocols in human genetics. Vol. 1. John Wiley and Sons, New York [Google Scholar]
- Edery P, Attié T, Amiel J, Pelet A, Eng C, Hofstra RMW, Martelli H, Bidaud C, Munnich A, Lyonnet S (1996) Mutation of the endothelin-3 gene in Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome). Nat Genet 12:442–444 [DOI] [PubMed] [Google Scholar]
- Edery P, Attié T, Mulligan LM, Pelet A, Eng C, Ponder BAJ, Munnich A, Lyonnet S (1994) A novel polymorphism in the coding sequence of the human RET proto-oncogene. Hum Genet 94:579–580 [DOI] [PubMed] [Google Scholar]
- Eng C (1996) The RET proto-oncogene in multiple endocrine neoplasia type 2 and Hirschsprung disease. N Engl J Med 335:943–951 [DOI] [PubMed] [Google Scholar]
- Eng C (1999) RET proto-oncogene in the development of human cancer. J Clin Oncol 17:380–393 [DOI] [PubMed] [Google Scholar]
- Eng C, Clayton D, Schuffenecker I, Lenoir G, Cote G, Gagel RF, Ploos van Amstel HK, et al (1996a) The relationship between specific RET proto-oncogene mutations and disease phenotype in multiple endocrine neoplasia type 2: International RET Mutation Consortium analysis. JAMA 276:1575–1579 [PubMed] [Google Scholar]
- Eng C, Mulligan LM, Healey CS, Houghton C, Frilling A, Raue F, Thomas GA, Ponder BAJ (1996b) Heterogeneous mutation of the RET proto-oncogene in subpopulations of medullary thyroid carcinoma. Cancer Res 56:2167–2170 [PubMed] [Google Scholar]
- Eng C, Mulligan LM, Smith DP, Healey CS, Frilling A, Raue F, Neumann HPH, Pfragner R, Behmel A, Lorenzo MJ, Stonehouse TJ, Ponder MA, Ponder BAJ (1995) Mutation in the RET proto-oncogene in sporadic medullary thyroid carcinoma. Genes Chrom Cancer 12:209–212 [DOI] [PubMed] [Google Scholar]
- Eng C, Smith DP, Mulligan LM, Nagai MA, Healey CS, Ponder MA, Gardner E, Scheumann GFW, Jackson CE, Tunnacliffe A, Ponder BAJ (1994) Point mutation within the tyrosine kinase domain of the RET proto-oncogene in multiple endocrine neoplasia type 2B and related sporadic tumours. Hum Mol Genet 3:237–241 [DOI] [PubMed] [Google Scholar]
- Falk CT, Rubinstein P (1987) Haplotype relative risks: an easy reliable way to construct a proper control sample for risk calculations. Ann Hum Genet 51:227–233 [DOI] [PubMed] [Google Scholar]
- Fitze G, Schreiber M, Kuhlisch E, Schackert HK, Roesner D (1999) Association of the RET proto-oncogene codon 45 polymorphism with Hirschsprung disease. Am J Hum Genet 65:1469–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wright FA (1999) Nonparametric disequilibrium mapping when haplotypes are available. Am J Hum Genet Suppl 65:A250 [Google Scholar]
- Gimm O, Neuberg DS, Marsh DJ, Dahia PLM, Hoang-Vu C, Raue F, Hinze R, Dralle H, Eng C (1999) Over-representation of a germline RET sequence variant in patients with sporadic medullary thyroid carcinoma and somatic RET codon 918 mutation. Oncogene 18:1369–1370 [DOI] [PubMed] [Google Scholar]
- Griseri P, Pesce B, Patrone G, Osinga J, Puppa F, Sancarndi M, Hofstra R, Romeo G, Ravazzolo R, Devoto M, Ceccherini I (2002) A rare haplotype of the RET proto-oncogene is a risk-modifying allele in Hirschsprung disease. Am J Hum Genet 71:969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griseri P, Sancandi M, Patrone G, Bocciardi R, Hofstra R, Ravazzolo R, Devoto M, Romeo G, Ceccherini I (2000) A single-nucleotide polymorphic variant of the RET proto-oncogene is underrepresented in sporadic Hirschsprung disease. Eur J Hum Genet 8:721–724 [DOI] [PubMed] [Google Scholar]
- Hastbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E (1992) Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat Genet 2:204–211 [DOI] [PubMed] [Google Scholar]
- Hofstra RMW, Landsvater RM, Ceccherini I, Stulp RP, Stelwagen T, Luo Y, Pasini B, Höppener JWM, Ploos van Amstel HK, Romeo G, Lips CJM, Buys CHCM (1994) A mutation in the RET proto-oncogene associated with multiple endocrine neoplasia type 2B and sporadic medullary thyroid carcinoma. Nature 367:375–376 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Lang D, Chen F, Milewski R, Li J, Lu MM, Epstein JA (2000) Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J Clin Invest 106:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyonnet S, Bolino A, Pelet A, Abel L, Nihoul-Fekété C, Briard ML, Mok-Siu V, Kaariainen H, Martucciello G, Lerone M, Puliti A, Luo Y, Weissenbach J, Devoto M, Munnich A, Romeo G (1993) A gene for Hirschsprung disease maps to the proximal long arm of chromosome 10. Nat Genet 4:346–350 [DOI] [PubMed] [Google Scholar]
- Marcos I, Ruiz A, Blaschak CJ, Borrego S, Cutting GR, Antinolo G (2000) Mutation analysis of GABRR1 and GABRR2 in autosomal recessive retinitis pigmentosa. J Med Genet 37:E5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen P, Rabizadeh S, Snipas SJ, Assa-Munt N, Salvesen GS, Bredesen DE (1998) The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature 395:801–804 [DOI] [PubMed] [Google Scholar]
- Mulligan LM, Gardner E, Smith BA, Mathew CGP, Ponder BAJ (1993) Genetic events in tumor initiation and progression in multiple endocrine neoplasia. Genes Chrom Cancer 6:166–177 [DOI] [PubMed] [Google Scholar]
- Ott J (1999) Analysis of human genetic linkage, 3rd edition. Johns Hopkins Press, Baltimore [Google Scholar]
- Pasini B, Borrello MG, Greco A, Bongarzone I, Luo Y, Mondellini P, Alberti C, Miranda C, Arighi E, Bocciardi R, Seri M, Barone V, Radice MT, Romeo G, Pierotti MA (1995) Loss of function effect of RET mutations causing Hirschsprung disease. Nat Genet 10:35–40 [DOI] [PubMed] [Google Scholar]
- Passarge E (1967) The genetics of Hirschsprung's disease. N Engl J Med 276:138–141 [DOI] [PubMed] [Google Scholar]
- Pelet A, Geneste O, Edery P, Pasini A, Chappuis S, Attié T, Munnich A, Lenoir G, Lyonnet S, Billaud M (1998) Various mechanisms cause RET-mediated signaling defects in Hirschsprung's disease. J Clin Invest 101:1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault V, Bondurand N, Kuhlbroadt K, Goerich DE, Préhu M-O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Clayton Smith J, Read AP, Wegner M, Goossens M (1998) SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet 18:171–173 [DOI] [PubMed] [Google Scholar]
- Puffenberger EG, Hosoda K, Washington SS, Nakao K, de Wit D, Yanagisawa M, Chakravarti A (1994) A missense mutation of the endothelin B receptor gene in multigenic Hirschsprung's disease. Cell 79:1257–1266 [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T (1995) MatInd and MatInspector—new fast and versatible tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23:4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabizdeh S, Oh J, Zhong LT, Yang J, Bitler CM, Butcher LL, Bredesen DE (1993) Induction of apoptosis by the low-affinity NGF receptor. Science 261:345–348 [DOI] [PubMed] [Google Scholar]
- Reeve JP, Rannala B (2002) DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics 18:894–895 [DOI] [PubMed] [Google Scholar]
- Romeo G, Ronchetto P, Luo Y, Barone V, Seri M, Ceccherini I, Pasini B, Bocciardi R, Lerone M, Kääriäinen H, Martucciello G (1994) Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung's disease. Nature 367:377–378 [DOI] [PubMed] [Google Scholar]
- Ruiz A, Antiñolo G, Fernandez RM, Eng C, Marcos I, Borrego S (2001) Germline sequence variant S836S in the RET proto-oncogene is associated with low level predisposition to sporadic medullary thyroid carcinoma in the Spanish population. Clin Endocrinol 55:399–402 [DOI] [PubMed] [Google Scholar]
- Santoro M, Rosato R, Grieco M, Berlingieri MT, Luca-Colucci D'Amato G, de Franciscis V, Fusco A (1990) The ret proto-oncogene is consistently expressed in human pheochromocytomas and thyroid medullary carcinomas. Oncogene 5:1595–1598 [PubMed] [Google Scholar]
- Songyang Z, Carraway III KL, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BAJ, Mayer BJ, Cantley LC (1995) Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature 373:536–539 [DOI] [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ (1998) Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet 18:60–64 [DOI] [PubMed] [Google Scholar]
- Svensson P-J, Molander J-L, Eng C, Anvret M, Nordenskjöld A (1998) Low frequency of RET mutations in Hirschsprung disease in Sweden. Clin Genet 54:39–44 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, Taniguchi H, Kitoh H, Mutoh N, Yamanaka T, Mushiake K, Kato K, Sonata S, Nagaya M (2001) Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat Genet 27:369–370 [DOI] [PubMed] [Google Scholar]