Mutations in numerous genes cause congenital heart disease (CHD). Studies focused on specific CHD genes (e.g., NKX2-5 and GATA4) have expanded our understanding of heart development and the molecular pathogenesis of CHD.
Abstract
Study of monogenic congenital heart disease (CHD) has provided entry points to gain new understanding of heart development and the molecular pathogenesis of CHD. In this review, we discuss monogenic CHD caused by mutations of the cardiac transcription factor genes NKX2-5 and GATA4. Detailed investigation of these genes in mice and humans has expanded our understanding of heart development, shedding light on the complex genetic and environmental factors that influence expression and penetrance of CHD gene mutations.
THE IMPACT OF CONGENITAL HEART DISEASE
With a prevalence of approximately 0.8 per 1000 births, congenital heart defects are the most common major malformations seen at birth, accounting for nearly one-third of all major congenital anomalies (Reller et al. 2008; Dolk et al. 2011). An estimated 32,000 infants with congenital heart disease (CHD) are born each year in the United States, and ∼25% of them require invasive treatment in the first year of life (Roger et al. 2012). Twenty-four percent of infants who die of a birth defect have a heart defect, making it the most common congenital defect contributing to death in that first year. In the past four decades, extraordinary advances in management have transformed the severe CHD prognosis, so that the large majority of patients survive into adulthood (Khairy et al. 2010). More than half of all survivors of severe CHD are now adults, and most individuals with even complex CHD are now expected to reach reproductive age (Friedberg et al. 2009; Harris 2011). With burgeoning survival and rapid advances in genetic technologies, there has never been a more compelling time to unravel the complex mechanisms underpinning congenital cardiac malformations.
MONOGENIC CHD
The majority of CHD occurs sporadically in nonsyndromic patients. However, syndromic and familial CHD provide opportunities to identify key regulators of heart development, monogenic mutations of which cause CHD. An overview of currently known genes that when mutated cause human CHD is provided in Table 1.
Table 1.
Genes implicated in isolated, nonsyndromic CHD
| Gene | Phenotype | OMIM |
|---|---|---|
| ACTC1 | ASD | 102540 |
| ACVR1 / ALK2 | AVSD | 102576 |
| ACVR2B | Heterotaxy | 602730 |
| ALDH1A2 | TOF | 603687 |
| ANKRD1 | TAPVR | 609599 |
| CCDC11 | Heterotaxy | 614759 |
| CFC1 | Heterotaxy, TGA, DORV | 605194 |
| CITED2 | ASD, VSD | 602937 |
| CRELD1 | AVSD, heterotaxy | 607170 |
| ELN | SVAS | 130160 |
| FLNA | Cardiac valvular dysplasia, X-linked | 300017 |
| FOG2 | TOF, DORV | 603693 |
| FOXF1 | Misalignment of pulmonary veins | 601089 |
| FOXH1 | VSD, TGA | 603621 |
| GATA4 | VSD, ASD, AVSD | 600576 |
| GATA6 | TOF, AVSD, ASD, PTA | 601656 |
| GDF1 | TOF, TGA | 602880 |
| GJA1 | HLHS, AVSD | 121014 |
| HAND2 | TOF | 602407 |
| IRX4 | VSD | 606199 |
| JAG1 | TOF | 601920 |
| LEFTY2 | HLHS, AVSD | 601877 |
| MED13L | TGA | 608771 |
| MYH6 | ASD | 160710 |
| NKX2-5 | TOF, HLHS, ASD, VSD, conotruncal heart defects | 600584 |
| NKX2-6 | PTA | 611770 |
| NODAL | Heterotaxy | 601265 |
| NOTCH1 | Aortic valve disease | 190198 |
| PDGFRA | TAPVR | 173490 |
| SMAD6 | Aortic valve disease | 602931 |
| TAB2 | Bicuspid AoV, LVOTO | 605101 |
| TBX1 | TOF | 602054 |
| TBX5 | ASD, VSD | 601620 |
| TBX20 | ASD | 606061 |
| TDGF1 | VSD, TOF | 187395 |
| TFAP2B | PDA | 601601 |
| TLL1 | ASD | 606742 |
| VEGFA | Bicuspid AoV, AS, coarctation, VSD, PDA | 192240 |
| ZFPM2 | TOF | 603693 |
| ZIC3 | Heterotaxy | 300265 |
| MYH11 | Aortic aneurysm | 160745 |
A list of known genes implicated in monogenic, nonsyndromic CHD was assembled by searching Online Mendelian Inheritance in Man (OMIM) and recent reviews (Wessels and Willems 2010; Fahed et al. 2013).
ASD, atrial septal defect; AVSD, atrioventricular septal defect; TOF, tetralogy of Fallot; TAPVR, total anomalous pulmonary venous drainage; TGA, transposition of the great arteries; DORV, double outlet right ventricle; VSD (ventricular septal defect); SVAS, supravalvar aortic stenosis; PTA, persistent truncus arteriosus; HLHS, hypoplastic left heart syndrome; AoV, aortic valve; LVOTO, left ventricular outflow tract obstruction; AS, aortic stenosis; PDA, patent ductus arteriosus.
The overall recurrence risk of nonsyndromic CHD is between 2% and 10%, depending on the defect and sex of the parent concerned (Wessels and Willems 2010). Among affected relatives, CHD may vary significantly in specific type and severity (Whittemore et al. 1994; Gill et al. 2003). The variable penetrance and phenotype intuitively suggest a multifactorial mode of inheritance for CHD, first proposed by Nora as early as 1968 (Nora 1968).
However, detailed studies of several monogenic CHD disease genes in mice and humans over the past decade have broadened our understanding of how the complex interaction of stochastic factors, the environment, and modifier genes influence phenotypic expression of CHD gene mutations. These studies indicate that variable expression and incomplete penetrance of many rare single gene mutations provide an alternative mechanism that accounts for epidemiological CHD recurrence risk (Bruneau et al. 2001; Rajagopal et al. 2007; Winston et al. 2010, 2012). In this work, we will review, in depth, the literature on monogenic mutation of two cardiac transcription factors, NKX2-5 and GATA4, and reflect on how the interplay of human genetics and murine modeling has informed our understanding of CHD pathogenesis and genetics. We also touch on monogenic mutation of T-box genes insofar as they reinforce or add to our understanding of variable CHD penetrance and expression.
NKX2-5
The NKX2-5 gene in humans encodes a cardiac-specific homeobox transcription factor. NKX2-5 transcripts are detected in murine cardiomyocytes at the onset of cardiomyocyte differentiation, with continued expression through embryonic, fetal, and adult life (Lints et al. 1993). In four families with familial, autosomal-dominant CHD, genome-wide linkage studies showed that mutations in NKX2-5 segregated with CHD (Schott et al. 1998). Two of the families shared an NKX2-5 missense mutation in the region encoding the DNA-binding domain, whereas the remaining two harbored mutations that prematurely terminated translation just carboxy terminal to the DNA-binding domain. Most affected family members had secundum atrial septal defects (ASDs) and progressive atrioventricular block (AVB), but others had tetralogy of Fallot, ventricular septal defects (VSDs), and left ventricular hypertrophy with or without ASD or AVB.
Subsequently, targeted sequencing of NKX2-5 in cohorts of patients with different forms of nonfamilial CHD revealed that NKX2-5 mutations contribute to nonsyndromic, ostensibly sporadic CHD that affects diverse chambers and structures, with or without conduction system disease (Fig. 1). Review of NKX2-5 variants and associated CHD shows a lack of a discernable relationship between mutation location and phenotype. Indeed, the same mutation yields diverse phenotypes. Some ostensibly sporadic CHD-associated NKX2-5 mutations arose de novo, whereas others were inherited from a parent without clinically detectable disease, indicative of incomplete penetrance.
Figure 1.
Summary of germline nonsynonymous NKX2-5 mutations associated with cardiac malformations. a.a., amino acid; AVB, atrioventricular block; ASD, atrial septal defect; TOF, tetralogy of Fallot; VSD, ventricular septal defect; PS, pulmonary stenosis; PTA, persistent truncus arteriosus; IAA, interrupted aortic arch; HLHS, hypoplastic left heart syndrome; PA, pulmonary atresia; coarct, coarctation of the aorta; pulm, pulmonary; L-TGA, levo-transposition of the great arteries; PDA, persistent ductus arteriosus; LVH, left ventricular hypertrophy; LVNC, left ventricular noncompaction; SVAS, supravalvar aortic stenosis; Anom sys veins, anomalous drainage of the systemic veins; LV, left ventricular; MVP, mitral valve prolapse, DORV, double outlet right ventricle.
Nkx2-5 regulation of heart development has been studied extensively in mouse models. Embryos engineered to lack expression of Nkx2-5 died at midgestation with severe heart defects (Lyons et al. 1995). Hearts formed, but development arrested at the looping heart tube stage, yielding unlooped hearts with one atrial and one ventricular chamber. Subsequently, studies revealed that Nkx2-5 regulates expression of a number of other cardiac genes and transcription factors, including Hand1 (Tanaka et al. 1999), a gene essential for left ventricular development (Srivastava et al. 1997). Nkx2-5 also regulates cardiac progenitor expansion and differentiation and cardiac outflow tract morphology by controlling expression of the morphogen BMP2 (Prall et al. 2007). Conditional inactivation of Nkx2-5 at later stages of heart development revealed that it controls cardiac trabeculation through another cardiac growth factor, BMP10 (Pashmforoush et al. 2004). Nkx2-5 is also a critical regulator of the formation of the central conduction system, as Nkx2-5-deficient mice suffer central conduction system hypoplasia (Jay et al. 2004a,b; Pashmforoush et al. 2004).
Affected human patients carry heterozygous mutations that either reduce the amount of gene product (haploinsufficiency) or produce a mutant gene product with dominant negative activity. Mice bearing heterozygous mutations for engineered mutant alleles often better model this situation than homozygous mutants, although reduced penetrance and severity of disease in heterozygotes complicates their analysis. Heterozygous Nkx2-5 mice were initially described as phenotypically normal, but additional scrutiny after the discovery of human NKX2-5 mutations showed that 40% of Nkx2-5 heterozygous mice in the inbred C57BL/6 strain background have ASD and/or VSD, and ∼10% perish in the newborn period (Biben et al. 2000; Tanaka et al. 2002; Winston et al. 2010, 2012).
Survival and the incidence of heart defects are affected by the genetic background: unlike highly inbred Nkx2-5+/− C57BL/6 mice, first-generation (F1) progeny of Nkx2-5+/− C57BL/6 mice crossed different inbred strains (FVB/N or A/J) had very low incidence of heart defects, and death from severe heart defects was not detected. The dependence of survival and heart defects on strain background suggested the hypothesis that inbred strains carry alleles of modifier genes that influence the risk of CHD. The alleles do not cause defects per se because the wild-type F1 pups are normal. Phenotypic analysis of the Nkx2-5+/− second-generation (F2) progeny of F1 intercrosses or F1 backcrosses to their parental strains supported this hypothesis (Winston et al. 2012). Defects recurred in all the F2 crosses, and the incidence of specific defects, such as common atrioventricular canal, varied between the crosses.
To map the modifier genes, genetic linkage analyses were performed on 597 hearts (233 membranous VSD, 80 muscular VSD, and 284 unaffected) selected from more than 3100 Nkx2-5+/− pups in the C57BL/6 × FVB/N F2 intercross. Loci on chromosomes 6, 8, and 10 clearly influenced susceptibility to membranous VSD. The chromosome 6 locus might also affect muscular VSD susceptibility, but the chromosome 8 and 10 loci do not (Winston et al. 2012). Thus, inbred strains carry polymorphisms in modifier genes that influence the susceptibility of specific developmental pathways to Nkx2-5 mutation. Maximal genetic heterogeneity, as seen in the F1, confers the greatest protection from heart defects.
In summary, heterozygous mutations of NKX2-5 cause human CHD with highly variable penetrance and expression. Studies in mouse models show that Nkx2-5 is a critical regulator of heart development, and robust, error-free heart development requires a full dose of Nkx2-5. Genetic modifiers clearly determine the penetrance and expression of CHD caused by Nkx2-5 mutation.
GATA4
GATA4, a zinc-finger transcription factor, has been shown to play critical roles in cardiac development (Pikkarainen et al. 2004; Zhou et al. 2012). GATA4 haploinsufficiency was first linked to CHD by the observation of microdeletion of 8p23.1, the locus that contains GATA4, in patients with CHD (Pehlivan et al. 1999). Garg et al. (2003) showed that GATA4 missense mutations segregate with CHD in two large pedigrees with septal defects. All family members with GATA4 mutation in both pedigrees had ASDs; other cardiac malformations that were observed in some, but not all, patients were VSD, pulmonary stenosis, and atrioventricular septal defect (AVSD). Of note, the cardiac conduction system was unaffected in these families.
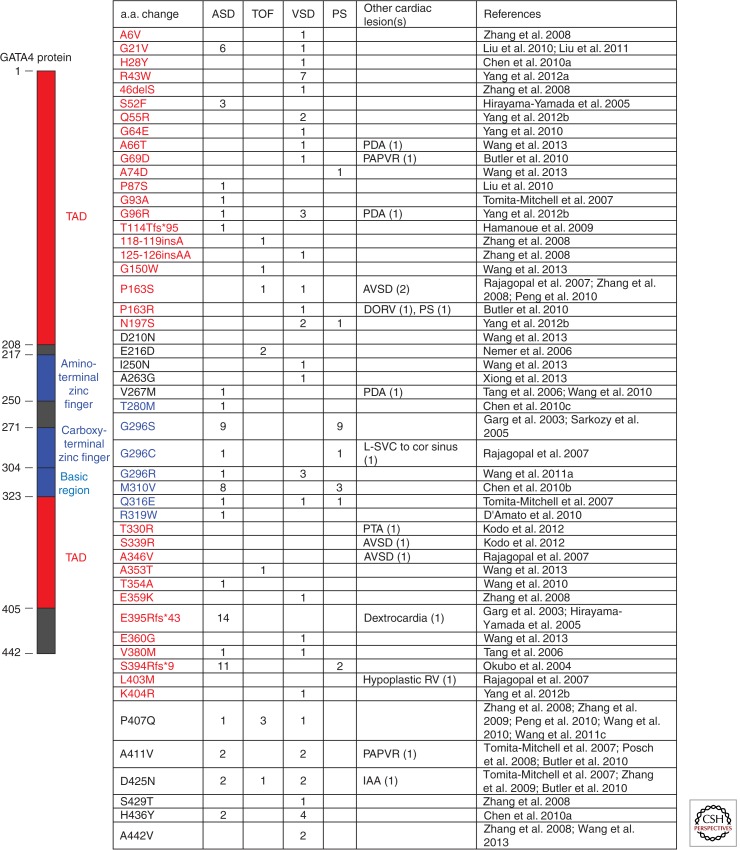
Subsequently, studies specifically investigating familial ASDs identified GATA4 mutation in four of 32 families with noted high penetrance of the phenotype in those families (Hirayama-Yamada et al. 2005; Sarkozy et al. 2005). GATA4 mutations have also been observed, albeit rarely, in cohorts with ostensibly sporadic CHD (Fig. 2). When identified, GATA4 mutations in sporadic heart disease (32 mutations identified in 2502 patients, or 1.3%) occurred in patients with VSDs (19), tetralogy of Fallot (6), ASD (3), AVSDs (3), and double inlet left ventricle (1). Mutation location does not appear predictive of phenotype (Fig. 2).
Figure 2.
Summary of germline nonsynonymous GATA4 mutations associated with human cardiac malformations. Abbreviations used: a.a., amino acid; ASD, atrial septal defect; TOF, tetralogy of Fallot; VSD, ventricular septal defect; PS, pulmonary stenosis; PDA, patent ductus arteriosus; PAPVR, partial anomalous pulmonary venous return; AVSD, atrioventricular septal defect; DORV, double outlet right ventricle; L-SVC to cor sinus, left superior vena cava draining to coronary sinus; PTA, persistent truncus arteriosus; RV, right ventricle; IAA, interrupted aortic arch; TAD, transcriptional activation domain.
The role of Gata4 in cardiac development has been studied in depth in mice lacking Gata4. Loss of Gata4 in all tissues (germline knockout) caused early embryonic lethality with cardiac bifida because of failure of normal embryonic folding (Kuo et al. 1997; Molkentin et al. 1997). Conditional Gata4-inactivation approaches revealed temporally and spatially restricted Gata4 function in heart development. In cardiomyocytes, loss of Gata4 impaired cardiomyocyte proliferation, resulting in myocardial hypoplasia and reduced cardiac trabeculation (Zeisberg et al. 2005). Cardiomyocyte Gata4 was also required for normal morphogenesis of the right ventricle, in part caused by Gata4 regulation of the gene Hand2, a key regulator of RV development (Zeisberg et al. 2005). Gata4 is also expressed at high levels in endocardium and endocardial cushions, and selective Gata4 ablation in these tissues revealed that Gata4 is required for formation of heart valve precursors, which are derived from endocardium of valve-forming regions (Rivera-Feliciano et al. 2006). Gata4 is also required in endocardium and endocardial cushion mesenchyme for later stages of heart valve development, as a point mutant Gata4 allele competent for valve precursor formation develops AVSDs (Rivera-Feliciano et al. 2006).
Rajagopal et al. (2007) used heterozygous Gata4 mice to define the phenotypic spectrum of heterozygous Gata4 mutation. In Gata4+/− mice on a highly inbred C57BL6 strain background, 76% of late gestation Gata4 heterozygous embryos had heart malformations including AVSD (59%), VSD (26%), and hypoplasia of the right ventricle (9%). These abnormalities were milder forms of abnormalities observed in conditional Gata4 knockout mice. Based on this phenotypic spectrum, Gata4 was sequenced in a human cohort that included septal defects, AVSD, and RV hypoplasia. Rare, nonsynonymous GATA4 sequence variants were found in patients with AVSD (2/43), ASD (1/8), and complex heart disease associated with right ventricular (RV) hypoplasia (1/9; the positive individual had double inlet left ventricle). Equally noteworthy was the absence of GATA4 mutations in those human cardiac phenotypes that were not present in the murine heterozygous mutant model. Specifically, there were no GATA4 mutations identified in any patient with a conotruncal anomaly (n = 34) or left-sided obstructive lesion (n = 81). This study illustrated that careful study of murine heterozygous mutant models can effectively direct patient selection and sequencing efforts.
In Gata4 heterozygous mutant mice, the frequency and type of CHD were strongly influenced by strain background: CHD occurred at 30% and 12% frequency in inbred FVB/N strain or mixed strain backgrounds, respectively, compared with 76% in the C57BL6 background. VSD frequency was similar between C57BL6 and FVB/N strains, but endocardial cushion defects were 14-fold less frequent (59% vs. 4%) in the FVB/N strain. A single-nucleotide-polymorphism-based whole genome scan for genetic modifiers did not identify strong modifier loci, although the study was relatively underpowered (25 affected, 13 unaffected; 80% likelihood of detecting linkage at logarithm of the odds (LOD) score 2.46 with relative risk of 23 or greater) compared with the Nkx2-5 modifier linkage scan. These results show that strain background strongly influences the expression and penetrance of CHD phenotypes in Gata4 heterozygous mice, but this strain effect is likely caused by multiple weaker modifiers.
Adult mice with heterozygous Gata4 mutation universally had left ventricular (LV) dysfunction, although the severity varied by strain background, with C57BL6 being more severely affected than FVB/N or mixed strains (Bisping et al. 2006; Rajagopal et al. 2007). Interestingly, LV dysfunction was not described in human pedigrees with Gata4 mutation, nor has it been reported in patients with sporadic Gata4 mutation. This discrepancy might reflect the studied murine Gata4 mutant allele, which expresses a truncated protein and therefore may have dominant negative activity, or may be a result of differences in dosage sensitivity between mouse and human. Heterozygous Gata4 mutation also sensitized mice to heart failure in a chronic pressure overload model, suggesting that patients with Gata4 mutation may also have a similar increased susceptibility to heart failure that may interact with volume or pressure loads associated with incompletely corrected structural heart disease.
To summarize, GATA4 is a critical regulator of cardiac development and function, and humans with heterozygous GATA4 mutation develop ASDs and VSDs, pulmonary stenosis, endocardial cushion defects, and complex heart disease involving RV hypoplasia, such as double inlet left ventricle. This spectrum of heart defects is consistent with what is seen in mice with heterozygous Gata4 mutation, and the known roles of Gata4 in heart development. Expression and penetrance of Gata4 heterozygous mutation is strongly influenced by modifier genes, and this likely contributes to variable expression and penetrance observed in patients.
T-BOX TRANSCRIPTION FACTORS AND CHD
Here, we briefly discuss mutations of the T-box transcription factor genes TBX5 and TBX1, focusing on the issues of variable expression and penetrance raised by the analysis of NKX2-5 and GATA4 mutations. Readers are directed to recent reviews for more comprehensive enumeration of known CHD genes (Wessels and Willems 2010; Fahed et al. 2013).
Mutations of TBX5, encoding a member of the T-box family of transcription factors, cause Holt–Oram syndrome, characterized by heart and upper limb abnormalities (Mori and Bruneau 2004). An abnormal carpal bone is present in all cases, and some patients have more extensive upper limb involvement. Seventy-five percent have a congenital heart malformation and all patients, with or without CHD, are at risk of cardiac conduction disease. ASDs and VSDs are the most commonly described heart lesions. Occasionally, other forms of CHD have been reported, including hypoplastic left heart, persistence of the left superior vena cava, mitral valve prolapse, pulmonary stenosis, tetralogy of Fallot, truncus arteriosus, coarctation of the aorta, total anomalous pulmonary venous return, patent ductus arteriosus, tricuspid atresia, and AVSDs (Smith et al. 1979; Ruzic et al. 1981; Sahn et al. 1981; Glauser et al. 1989; Basson et al. 1994; Newbury-Ecob et al. 1996; Bruneau et al. 2001; Patel et al. 2012; Thal et al. 2012). Missense, insertion, deletion, and chromosomal translocation mutations have all been reported. Neither the type nor location of TBX5 mutation was predictive of heart or hand phenotypes (Brassington et al. 2003).
Homozygous knockout of Tbx5 in mouse embryos caused severe defects in the formation of the atria and left ventricle, consistent with expression of Tbx5 primarily in the first heart field, and embryos died by midgestation (Bruneau et al. 2001). As in patients, Tbx5 haploinsufficiency also causes highly penetrant heart and limb defects. Ninety percent of Tbx5del/+ mice died perinatally in an inbred 129/SvEv mouse strain background. Survival was exquisitely sensitive to Tbx5 gene dosage, as a mice heterozygous for a different, hypomorphic Tbx5 allele (Tbx5lox/+) that expressed 15% more Tbx5 suffered 27% perinatal lethality in the 129/SvEv background (Mori et al. 2006). Survival was also dependent on strain background, with 60% of Tbx5del/+ mice dying perinatally in the outbred Black Swiss strain background. All Tbx5lox/+ and Tbx5del/+ hearts had ASDs; other more severe heart defects were also identified in Tbx5del/+ mice (Bruneau et al. 2001), but their frequency and strain-dependence were not investigated in depth as they had been for Nkx2-5 and Gata4.
Another T-box transcription factor, TBX1, is the primary disease gene in DiGeorge syndrome, the second most common chromosomal cause of CHD after Down syndrome (DS) (Goodship et al. 1998). This syndrome, characterized by CHD, typical facies, and thymic and parathyroid hypoplasia is caused by chromosomal microdeletion of 22q11. Although there are 28 genes within this 3-Mb interval, TBX1 haploinsufficiency is thought to account for most of the disease manifestations, as rare, nondeleted DiGeorge patients have mutations localized to TBX1 (Yagi et al. 2003), and targeted mutation of Tbx1 recapitulates most aspects of the syndrome in mice (Jerome and Papaioannou 2001; Lindsay et al. 2001). Approximately 75% of patients with 22q11 microdeletion have CHD, most notably conotruncal and outflow tract abnormalities such as tetralogy of Fallot, interrupted aortic arch, and truncus arteriosus (Ryan et al. 1997). There is considerable phenotypic heterogeneity between patients with similar microdeletions, and even in monozygotic twins that share the same microdeletion (Yamagishi et al. 1998; Vincent et al. 1999).
Mouse models of both the 22q11 critical region microdeletion and targeted Tbx1 gene knockout have been generated. Interestingly, the relationship of gene dose to phenotype appears to differ between mice and humans. Whereas most patients with heterozygous 22q11 microdeletion show severe cardiac abnormalities, heterozygous mice did not develop these forms of CHD and display the same phenotypic heterogeneity (Lindsay 2001). Baldini et al. analyzed the phenotypes of mice with nine different Tbx1 genotypes that differed by the level of Tbx1 expression (Zhang and Baldini 2008). The cardiovascular phenotype of mice with 20% of normal Tbx1 expression closely mimicked humans with 22q11 microdeletion. Interestingly, the phenotypic response to Tbx1 dose was highly nonlinear and, furthermore, the aortic arch was significantly more sensitive to Tbx1 dose reduction than the cardiac outflow tract. Phenotypic variability was also dose sensitive, with the full spectrum of human cardiac phenotypes occurring at a specific level (18%) of Tbx1 expression, but not at higher or lower Tbx1 levels. Possibly, this critical level represents a precarious balance between normal and abnormal development that can be influenced by modifier genes, environmental factors, or stochastic events.
GENETIC MODIFERS AS INDEPENDENT CHD DISEASE GENES
By definition, genes that modify CHD risk in a sensitized background, such as NKX2-5 or GATA4 haploinsufficiency, regulate heart development. Thus, finding modifier genes in a sensitized genetic background is a potential strategy for candidate gene discovery. Although identification of specific genes that act as modifiers in Nkx2-5 or Gata4 heterozygous mice and their evaluation as disease genes in CHD patients will require further study, proof of principle has already been reported in DS (trisomy for human chromosome 21). DS is the leading risk factor for CHD, with nearly half of DS patients affected by some form of cardiac malformation, most classically AVSDs (Ferencz et al. 1989).
The incomplete penetrance and variable expression of trisomy 21 suggests that genetic modifiers interact with dosage-sensitive gene(s) on chromosome 21 to result in CHD. This hypothesis was tested by sequencing CRELD1, a cause of non-DS AVSD defect (Robinson et al. 2003), in DS patients with this form of CHD (Maslen et al. 2006; Li et al. 2012). Out of 135 patients sequenced, three individuals had two predicted damaging missense mutations, one of which had been previously identified in individuals with nonsyndromic AVSD. The genetic interaction of CRELD1 with dosage-sensitive loci that cause DS was studied by crossing the heterozygous Creld1 mice with a murine model of DS (Ts65Dn) (Li et al. 2012). Although Ts65Dn rarely (<5%) had septal defects and Creld1+/− mice were phenotypically normal, Ts65Dn::Creld1+/− mice had increased frequency of septal defects (33%). However, these septal defects were not AVSDs, but rather secundum ASDs and membranous VSDs. Overall, these data suggest that genetic modifiers alter the expression of DS, and genetic modifiers discovered in sensitized populations such as DS may also contribute to disease in nonsensitized individuals.
SOURCES OF PHENOTYPIC VARIABILITY AND THEIR POTENTIAL SIGNIFICANCE
A challenge in CHD genetics has been to understand variable penetrance and phenotypic expression of gene mutations. Careful study of the Nkx2-5+/− and Gata4+/− mouse models highlights the impact of genetic modifiers (Rajagopal et al. 2007; Winston et al. 2010, 2012). Nevertheless, mice with a well-characterized single-gene defect on defined genetic backgrounds and raised in controlled, uniform environments showed incomplete penetrance and variable expression. Why does CHD occur in some mice, but not others, even when genotype and environmental conditions are made as uniform as possible?
There might, of course, be unrecognized environmental factors. From the perspective of the embryo, at least three uncontrolled environmental variables existed in the Nkx2-5+/− F2 intercross described above: maternal age, paternal age, and litter size (Winston et al. 2012). Neither litter size nor paternal age had a significant effect on VSD risk, but maternal age was positively correlated with VSD risk. For example, pups born to old mothers were twice as likely to have membranous VSD as pups born to young mothers. Maternal age acted independently of identified genetic modifiers, and the effect of maternal age and genetic modifiers was additive. Each genetic or environmental modifier may have a small effect on risk in the experimental model, but their existence proves, in principle, that pathways can be manipulated to prevent CHD. A therapy that mimics the effect of a protective polymorphism or environmental modifier is arguably more plausible than repairing a mutant gene in the embryo.
Even after controlling for maternal age, apparently equivalent Nkx2-5 heterozygous mice have dichotomous outcomes: some developed CHD, but most did not. The apparent stochastic occurrence of CHD in the Nkx2-5 and Gata4 heterozygous murine models suggests that a normal gene dose is required to ensure that the complex process of heart development is robust. This can be understood using Waddington’s metaphor of “canalization,” which conceptualizes how the normal phenotype is buffered against genetic or environmental perturbation (Waddington 1942). Development is depicted on a topographical surface. Banks guide the course of development and buffer it against environmental or stochastic forces. Perturbations such as NKX2-5 or GATA4 mutation modify the topographical surface so that otherwise inconsequential environmental or stochastic forces have a chance to push development down alternative paths to an abnormal phenotype. Genetic modifiers cause more subtle alterations in the topographical surface, and may protect against a particular insult, but may increase susceptibility to another. A corollary of this hypothesis of developmental buffering is that canalization encourages the accumulation of cryptic genetic variation, enhancing the organism’s evolutionary fitness. Mutation reduces canalizing forces (e.g., by mutation of CHD disease genes such as NKX2-5 or GATA4, or chromosomal anomalies such as trisomy 21), exposing these cryptic variants as genetic modifiers and leading to altered susceptibility to perturbed development (Waddington 1942; Flatt 2005).
CONCLUDING REMARKS
Epidemiology of CHD shows that recurrence risk is sub-Mendelian and there is substantial variability in disease phenotype. These observations led Nora and others to propose that CHD is polygenic and multifactorial (Nora 1968). This model can now be refined in light of what we have learned from detailed studies on the expression and penetrance of monogenic mutations in inbred mice raised under tightly controlled conditions. Rare, moderate-effect gene mutations reduce “canalization” and increase susceptibility to a range of heart malformations. Environmental (e.g., maternal age) and genetic factors (e.g., modifier genes) modify the risk of developing CHD imposed by these disease gene mutations and influence the specific type of cardiac malformation. With decreased canalization caused by gene mutation, stochastic events become significant, so that mice with carefully controlled genotypes and environment develop divergent outcomes. Although the prevalence of mutations in any single gene in sporadic CHD appears low, current whole exome sequencing results suggest that mutations in a large number of genes cause CHD (Zaidi et al. 2013). Thus, CHD may be caused by a large number of moderate-effect, single-gene mutations that “decanalize” heart development, increase stochastic variation, and expose weak-effect modifier variants. Individually, each disease gene likely contributes to a small fraction of CHD, but, in aggregate, this disease model may account for a substantial portion of the CHD burden.
ACKNOWLEDGMENTS
W.T.P. is supported by the Boston Children’s Hospital Translational Research Program, an American Heart Association award, and funding from the National Heart, Lung, and Blood Institute (R01 HL095712). T.P. is supported by an award from the Irish Cardiac Society Brian McGovern Travelling Fellowship. P.Y.J. is supported by the American Heart Association, the Children’s Heart Foundation, the Washington University School of Medicine, and St. Louis Children’s Hospital Children’s Discovery Institute, and National Institutes of Health (R01 HL105857).
REFERENCES
- Basson CT, Cowley GS, Solomon SD, Weissman B, Poznanski AK, Traill TA, Seidman JG, Seidman CE. 1994. The clinical and genetic spectrum of the Holt–Oram syndrome (heart-hand syndrome). N Engl J Med 330: 885–891 [DOI] [PubMed] [Google Scholar]
- Beffagna G, Cecchetto A, Dal Bianco L, Lorenzon A, Angelini A, Padalino M, Vida V, Bhattacharya S, Stellin G, Rampazzo A, et al. 2012. R25C mutation in the NKX2.5 gene in Italian patients affected with non-syndromic and syndromic congenital heart disease. J Cardiovasc Med (Hagerstown) 14: 582–586 [DOI] [PubMed] [Google Scholar]
- Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, et al. 1999. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest 104: 1567–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, et al. 2000. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res 87: 888–895 [DOI] [PubMed] [Google Scholar]
- Bisping E, Ikeda S, Kong SW, Tarnavski O, Bodyak N, McMullen JR, Rajagopal S, Son JK, Ma Q, Springer Z, et al. 2006. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci 103: 14471–14476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassington AM, Sung SS, Toydemir RM, Le T, Roeder AD, Rutherford AE, Whitby FG, Jorde LB, Bamshad MJ. 2003. Expressivity of Holt–Oram syndrome is not predicted by TBX5 genotype. Am J Hum Genet 73: 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. 2001. A murine model of Holt–Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106: 709–721 [DOI] [PubMed] [Google Scholar]
- Chen MW, Pang YS, Guo Y, Pan JH, Liu BL, Shen J, Liu TW. 2010a. GATA4 mutations in Chinese patients with congenital cardiac septal defects. Pediatr Cardiol 31: 85–89 [DOI] [PubMed] [Google Scholar]
- Chen Y, Han ZQ, Yan WD, Tang CZ, Xie JY, Chen H, Hu DY. 2010b. A novel mutation in GATA4 gene associated with dominant inherited familial atrial septal defect. J Thorac Cardiovasc Surg 140: 684–687 [DOI] [PubMed] [Google Scholar]
- Chen Y, Mao J, Sun Y, Zhang Q, Cheng HB, Yan WH, Choy KW, Li H. 2010c. A novel mutation of GATA4 in a familial atrial septal defect. Clin Chim Acta 411: 1741–1745 [DOI] [PubMed] [Google Scholar]
- D’Amato E, Giacopelli F, Giannattasio A, D’Annunzio G, Bocciardi R, Musso M, Lorini R, Ravazzolo R. 2010. Genetic investigation in an Italian child with an unusual association of atrial septal defect, attributable to a new familial GATA4 gene mutation, and neonatal diabetes due to pancreatic agenesis. Diabet Med 27: 1195–1200 [DOI] [PubMed] [Google Scholar]
- Dolk H, Loane M, Garne E. 2011. Congenital heart defects in Europe: Prevalence and perinatal mortality, 2000 to 2005. Circulation 123: 841–849 [DOI] [PubMed] [Google Scholar]
- Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, et al. 2003. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: Associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol 41: 2072–2076 [DOI] [PubMed] [Google Scholar]
- Fahed AC, Gelb BD, Seidman JG, Seidman CE. 2013. Genetics of congenital heart disease: The glass half empty. Circ Res 112: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferencz C, Boughman JA, Neill CA, Brenner JI, Perry LW. 1989. Congenital cardiovascular malformations: Questions on inheritance. Baltimore-Washington Infant Study Group. J Am Coll Cardiol 14: 756–763 [DOI] [PubMed] [Google Scholar]
- Flatt T. 2005. The evolutionary genetics of canalization. Q Rev Biol 80: 287–316 [DOI] [PubMed] [Google Scholar]
- Friedberg MK, Silverman NH, Moon-Grady AJ, Tong E, Nourse J, Sorenson B, Lee J, Hornberger LK. 2009. Prenatal detection of congenital heart disease. J Pediatr 155: 26–31, 31.e1 [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. 2003. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424: 443–447 [DOI] [PubMed] [Google Scholar]
- Gill HK, Splitt M, Sharland GK, Simpson JM. 2003. Patterns of recurrence of congenital heart disease: An analysis of 6,640 consecutive pregnancies evaluated by detailed fetal echocardiography. J Am Coll Cardiol 42: 923–929 [DOI] [PubMed] [Google Scholar]
- Gioli-Pereira L, Pereira AC, Mesquita SM, Xavier-Neto J, Lopes AA, Krieger JE. 2010. NKX2.5 mutations in patients with non-syndromic congenital heart disease. Int J Cardiol 138: 261–265 [DOI] [PubMed] [Google Scholar]
- Glauser TA, Zackai E, Weinberg P, Clancy R. 1989. Holt–Oram syndrome associated with the hypoplastic left heart syndrome. Clin Genet 36: 69–72 [DOI] [PubMed] [Google Scholar]
- Goldmuntz E, Geiger E, Benson DW. 2001. NKX2.5 mutations in patients with tetralogy of fallot. Circulation 104: 2565–2568 [DOI] [PubMed] [Google Scholar]
- Goodship J, Cross I, LiLing J, Wren C. 1998. A population study of chromosome 22q11 deletions in infancy. Arch Dis Child 79: 348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Riveron JT, Pope M, Bu’lock FA, Thornborough C, Eason J, Setchfield K, Ketley A, Kirk EP, Fatkin D, Feneley MP, et al. 2012. Combined mutation screening of NKX2-5, GATA4, and TBX5 in congenital heart disease: Multiple heterozygosity and novel mutations. Congenit Heart Dis 7: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Roelens I, Sluysmans T, Gewillig M, Devriendt K, Vikkula M. 2002. Progressive AV-block and anomalous venous return among cardiac anomalies associated with two novel missense mutations in the CSX/NKX2-5 gene. Hum Mutat 20: 75–76 [DOI] [PubMed] [Google Scholar]
- Hamanoue H, Rahayuningsih SE, Hirahara Y, Itoh J, Yokoyama U, Mizuguchi T, Saitsu H, Miyake N, Hirahara F, Matsumoto N. 2009. Genetic screening of 104 patients with congenitally malformed hearts revealed a fresh mutation of GATA4 in those with atrial septal defects. Cardiol Young 19: 482–485 [DOI] [PubMed] [Google Scholar]
- Harris IS. 2011. Management of pregnancy in patients with congenital heart disease. Prog Cardiovasc Dis 53: 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama-Yamada K, Kamisago M, Akimoto K, Aotsuka H, Nakamura Y, Tomita H, Furutani M, Imamura S, Takao A, Nakazawa M, et al. 2005. Phenotypes with GATA4 or NKX2.5 mutations in familial atrial septal defect. Am J Med Genet A 135: 47–52 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Hiroi Y, Hosoda T, Utsunomiya T, Matsuo S, Ito T, Inoue J, Sumiyoshi T, Takano H, Nagai R, et al. 2002. Novel point mutation in the cardiac transcription factor CSX/NKX2.5 associated with congenital heart disease. Circ J 66: 561–563 [DOI] [PubMed] [Google Scholar]
- Jay PY, Harris BS, Buerger A, Rozhitskaya O, Maguire CT, Barbosky LA, McCusty E, Berul CI, O’brien TX, Gourdie RG, et al. 2004a. Function follows form: Cardiac conduction system defects in Nkx2-5 mutation. Anat Rec A Discov Mol Cell Evol Biol 280: 966–972 [DOI] [PubMed] [Google Scholar]
- Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, Kupershmidt S, Roden DM, Schultheiss TM, O’Brien TX, et al. 2004b. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest 113: 1130–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. 2001. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet 27: 286–291 [DOI] [PubMed] [Google Scholar]
- Khairy P, Ionescu-Ittu R, Mackie AS, Abrahamowicz M, Pilote L, Marelli AJ. 2010. Changing mortality in congenital heart disease. J Am Coll Cardiol 56: 1149–1157 [DOI] [PubMed] [Google Scholar]
- Kodo K, Nishizawa T, Furutani M, Arai S, Ishihara K, Oda M, Makino S, Fukuda K, Takahashi T, Matsuoka R, et al. 2012. Genetic analysis of essential cardiac transcription factors in 256 patients with non-syndromic congenital heart defects. Circ J 76: 1703–1711 [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev 11: 1048–1060 [DOI] [PubMed] [Google Scholar]
- Li H, Cherry S, Klinedinst D, DeLeon V, Redig J, Reshey B, Chin MT, Sherman SL, Maslen CL, Reeves RH. 2012. Genetic modifiers predisposing to congenital heart disease in the sensitized Down syndrome population. Circ Cardiovasc Genet 5: 301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA. 2001. Chromosomal microdeletions: Dissecting del22q11 syndrome. Nat Rev Genet 2: 858–868 [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, et al. 2001. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410: 97–101 [DOI] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. 1993. Nkx-2.5: A novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119: 969. [DOI] [PubMed] [Google Scholar]
- Liu XY, Yang YQ, Ma J, Lin XP, Zheng JH, Bai K, Chen YH. 2010. Novel GATA4 mutations identified in patients with congenital atrial septal defects. Zhonghua Xin Xue Guan Bing Za Zhi 38: 724–727 [PubMed] [Google Scholar]
- Liu XY, Wang J, Zheng JH, Bai K, Liu ZM, Wang XZ, Liu X, Fang WY, Yang YQ. 2011. Involvement of a novel GATA4 mutation in atrial septal defects. Int J Mol Med 28: 17–23 [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. 1995. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev 9: 1654–1666 [DOI] [PubMed] [Google Scholar]
- Maslen CL, Babcock D, Robinson SW, Bean LJ, Dooley KJ, Willour VL, Sherman SL. 2006. CRELD1 mutations contribute to the occurrence of cardiac atrioventricular septal defects in Down syndrome. Am J Med Genet A 140: 2501–2505 [DOI] [PubMed] [Google Scholar]
- McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. 2003. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol 42: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev 11: 1061–1072 [DOI] [PubMed] [Google Scholar]
- Mori AD, Bruneau BG. 2004. TBX5 mutations and congenital heart disease: Holt–Oram syndrome revealed. Curr Opin Cardiol 19: 211–215 [DOI] [PubMed] [Google Scholar]
- Mori AD, Zhu Y, Vahora I, Nieman B, Koshiba-Takeuchi K, Davidson L, Pizard A, Seidman JG, Seidman CE, Chen XJ, et al. 2006. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol 297: 566–586 [DOI] [PubMed] [Google Scholar]
- Nemer G, Fadlalah F, Usta J, Nemer M, Dbaibo G, Obeid M, Bitar F. 2006. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat 27: 293–294 [DOI] [PubMed] [Google Scholar]
- Newbury-Ecob RA, Leanage R, Raeburn JA, Young ID. 1996. Holt–Oram syndrome: A clinical genetic study. J Med Genet 33: 300–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora JJ. 1968. Multifactorial inheritance hypothesis for the etiology of congenital heart diseases. The genetic-environmental interaction. Circulation 38: 604–617 [DOI] [PubMed] [Google Scholar]
- Okubo A, Miyoshi O, Baba K, Takagi M, Tsukamoto K, Kinoshita A, Yoshiura K, Kishino T, Ohta T, Niikawa N, et al. 2004. A novel GATA4 mutation completely segregated with atrial septal defect in a large Japanese family. J Med Genet 41: e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang P, Saarel E, Bai Y, Luo C, Lv Q, Xu Y, Wang F, Fan C, Younoszai A, Chen Q, et al. 2011. A de novo mutation in NKX2.5 associated with atrial septal defects, ventricular noncompaction, syncope and sudden death. Clin Chim Acta 412: 170–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst S, Wollnik B, Rohmann E, Hintz Y, Glanzer K, Vetter H, Nickenig G, Grohe C. 2008. A novel stop mutation truncating critical regions of the cardiac transcription factor NKX2-5 in a large family with autosomal-dominant inherited congenital heart disease. Clin Res Cardiol 97: 39–42 [DOI] [PubMed] [Google Scholar]
- Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, et al. 2004. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117: 373–386 [DOI] [PubMed] [Google Scholar]
- Patel C, Silcock L, McMullan D, Brueton L, Cox H. 2012. TBX5 intragenic duplication: A family with an atypical Holt–Oram syndrome phenotype. Eur J Hum Genet 20: 863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, Wilson DB, Watson MS, Hing AV. 1999. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet 83: 201–206 [PubMed] [Google Scholar]
- Peng T, Wang L, Zhou SF, Li X. 2010. Mutations of the GATA4 and NKX2.5 genes in Chinese pediatric patients with non-familial congenital heart disease. Genetica 138: 1231–1240 [DOI] [PubMed] [Google Scholar]
- Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. 2004. GATA transcription factors in the developing and adult heart. Cardiovasc Res 63: 196–207 [DOI] [PubMed] [Google Scholar]
- Posch MG, Perrot A, Schmitt K, Mittelhaus S, Esenwein EM, Stiller B, Geier C, Dietz R, Gessner R, Ozcelik C, et al. 2008. Mutations in GATA4, NKX2.5, CRELD1, and BMP4 are infrequently found in patients with congenital cardiac septal defects. Am J Med Genet A 146A: 251–253 [DOI] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. 2007. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128: 947–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V, Srivastava D, et al. 2007. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol 43: 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch R, Hofbeck M, Zweier C, Koch A, Zink S, Trautmann U, Hoyer J, Kaulitz R, Singer H, Rauch A. 2010. Comprehensive genotype-phenotype analysis in 230 patients with tetralogy of Fallot. J Med Genet 47: 321–331 [DOI] [PubMed] [Google Scholar]
- Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. 2008. Prevalence of congenital heart defects in metropolitan Atlanta, 1998–2005. J Pediatr 153: 807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, Izumo S, Tabin CJ, Pu WT. 2006. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development 133: 3607–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SW, Morris CD, Goldmuntz E, Reller MD, Jones MA, Steiner RD, Maslen CL. 2003. Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am J Hum Genet 72: 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al. 2012. Executive summary: Heart disease and stroke statistics--2012 update: A report from the American Heart Association. Circulation 125: 188–197 [DOI] [PubMed] [Google Scholar]
- Ruzic B, Bosnar B, Beleznay O. 1981. An unusual type of congenital heart disease associated with the Holt–Oram-syndrome (author’s transl). Radiologe 21: 296–299 [PubMed] [Google Scholar]
- Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B, Prieur M, et al. 1997. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: A European collaborative study. J Med Genet 34: 798–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahn DJ, Goldberg SJ, Allen HD, Canale JM. 1981. Cross-sectional echocardiographic imaging of supracardiac total anomalous pulmonary venous drainage to a vertical vein in a patient with Holt–Oram syndrome. Chest 79: 113–115 [DOI] [PubMed] [Google Scholar]
- Sarkozy A, Conti E, Neri C, D’Agostino R, Digilio MC, Esposito G, Toscano A, Marino B, Pizzuti A, Dallapiccola B. 2005. Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J Med Genet 42: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. 1998. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281: 108–111 [DOI] [PubMed] [Google Scholar]
- Smith AT, Sack GH Jr, Taylor GJ. 1979. Holt–Oram syndrome. J Pediatr 95: 538–543 [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. 1997. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 16: 154–160 [DOI] [PubMed] [Google Scholar]
- Stallmeyer B, Fenge H, Nowak-Gottl U, Schulze-Bahr E. 2010. Mutational spectrum in the cardiac transcription factor gene NKX2.5 (CSX) associated with congenital heart disease. Clin Genet 78: 533–540 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. 1999. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 126: 1269–1280 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Berul CI, Ishii M, Jay PY, Wakimoto H, Douglas P, Yamasaki N, Kawamoto T, Gehrmann J, Maguire CT, et al. 2002. A mouse model of congenital heart disease: Cardiac arrhythmias and atrial septal defect caused by haploinsufficiency of the cardiac transcription factor Csx/Nkx2.5. Cold Spring Harb Symp Quant Biol 67: 317–325 [DOI] [PubMed] [Google Scholar]
- Tang ZH, Xia L, Chang W, Li H, Shen F, Liu JY, Wang Q, Liu MG. 2006. Two novel missense mutations of GATA4 gene in Chinese patients with sporadic congenital heart defects. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 23: 134–137 [PubMed] [Google Scholar]
- Thal S, Boyella R, Arsanjani R, Thai H, Juneman E, Movahed MR, Goldman S. 2012. Unusual combination of Holt-Oram syndrome and persistent left superior vena cava. Congenit Heart Dis 7: E46–E49 [DOI] [PubMed] [Google Scholar]
- Tomita-Mitchell A, Maslen CL, Morris CD, Garg V, Goldmuntz E. 2007. GATA4 sequence variants in patients with congenital heart disease. J Med Genet 44: 779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent MC, Heitz F, Tricoire J, Bourrouillou G, Kuhlein E, Rolland M, Calvas P. 1999. 22q11 deletion in DGS/VCFS monozygotic twins with discordant phenotypes. Genet Couns 10: 43–49 [PubMed] [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [DOI] [PubMed] [Google Scholar]
- Wang J, Li XM, Xin YF, Wang LJ, Xu WJ, Hu DY. 2010. Genetic screening for novel GATA4 mutations associated with congenital atrial septal defect. Zhonghua Xin Xue Guan Bing Za Zhi 38: 429–434 [PubMed] [Google Scholar]
- Wang J, Fang M, Liu XY, Xin YF, Liu ZM, Chen XZ, Wang XZ, Fang WY, Liu X, Yang YQ. 2011a. A novel GATA4 mutation responsible for congenital ventricular septal defects. Int J Mol Med 28: 557–564 [DOI] [PubMed] [Google Scholar]
- Wang J, Liu XY, Yang YQ. 2011b. Novel NKX2-5 mutations responsible for congenital heart disease. Genet Mol Res 10: 2905–2915 [DOI] [PubMed] [Google Scholar]
- Wang J, Lu Y, Chen H, Yin M, Yu T, Fu Q. 2011c. Investigation of somatic NKX2-5, GATA4 and HAND1 mutations in patients with tetralogy of Fallot. Pathology 43: 322–326 [DOI] [PubMed] [Google Scholar]
- Wang J, Xin YF, Liu XY, Liu ZM, Wang XZ, Yang YQ. 2011d. A novel NKX2-5 mutation in familial ventricular septal defect. Int J Mol Med 27: 369–375 [DOI] [PubMed] [Google Scholar]
- Wang E, Sun S, Qiao B, Duan W, Huang G, An Y, Xu S, Zheng Y, Su Z, Gu X, et al. 2013. Identification of functional mutations in GATA4 in patients with congenital heart disease. PLoS ONE 8: e62138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels MW, Willems PJ. 2010. Genetic factors in non-syndromic congenital heart malformations. Clin Genet 78: 103–123 [DOI] [PubMed] [Google Scholar]
- Whittemore R, Wells JA, Castellsague X. 1994. A second-generation study of 427 probands with congenital heart defects and their 837 children. J Am Coll Cardiol 23: 1459–1467 [DOI] [PubMed] [Google Scholar]
- Winston JB, Erlich JM, Green CA, Aluko A, Kaiser KA, Takematsu M, Barlow RS, Sureka AO, LaPage MJ, Janss LL, et al. 2010. Heterogeneity of genetic modifiers ensures normal cardiac development. Circulation 121: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JB, Schulkey CE, Chen IB, Regmi SD, Efimova M, Erlich JM, Green CA, Aluko A, Jay PY. 2012. Complex trait analysis of ventricular septal defects caused by Nkx2-5 mutation. Circ Cardiovasc Genet 5: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Li Q, Zhang C, Chen Y, Li P, Wei X, Zhou W, Li L, Shang X, Xu X. 2013. Analyses of GATA4, NKX2.5, and TFAP2B genes in subjects from southern China with sporadic congenital heart disease. Cardiovasc Pathol 22: 141–145 [DOI] [PubMed] [Google Scholar]
- Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, et al. 2003. Role of TBX1 in human del22q11.2 syndrome. Lancet 362: 1366–1373 [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Ishii C, Maeda J, Kojima Y, Matsuoka R, Kimura M, Takao A, Momma K, Matsuo N. 1998. Phenotypic discordance in monozygotic twins with 22q11.2 deletion. Am J Med Genet 78: 319–321 [PubMed] [Google Scholar]
- Yang YQ, Tang YQ, Liu XY, Lin XP, Chen YH. 2010. A novel GATA4 mutation leading to congenital ventricular septal defect. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 27: 512–516 [DOI] [PubMed] [Google Scholar]
- Yang YQ, Li L, Wang J, Liu XY, Chen XZ, Zhang W, Wang XZ, Jiang JQ, Liu X, Fang WY. 2012a. A novel GATA4 loss-of-function mutation associated with congenital ventricular septal defect. Pediatr Cardiol 33: 539–546 [DOI] [PubMed] [Google Scholar]
- Yang YQ, Wang J, Liu XY, Chen XZ, Zhang W, Wang XZ, Liu X, Fang WY. 2012b. Novel GATA4 mutations in patients with congenital ventricular septal defects. Med Sci Monit 18: CR344–CR350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. 2013. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498: 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. 2005. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest 115: 1522–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Baldini A. 2008. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum Mol Genet 17: 150–157 [DOI] [PubMed] [Google Scholar]
- Zhang W, Li X, Shen A, Jiao W, Guan X, Li Z. 2008. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet 51: 527–535 [DOI] [PubMed] [Google Scholar]
- Zhang WM, Li XF, Ma ZY, Zhang J, Zhou SH, Li T, Shi L, Li ZZ. 2009. GATA4 and NKX2.5 gene analysis in Chinese Uygur patients with congenital heart disease. Chin Med J (Engl) 122: 416–419 [PubMed] [Google Scholar]
- Zhou P, He A, Pu WT. 2012. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol 100: 143–169 [DOI] [PubMed] [Google Scholar]