Abstract
In this review, we focus on two important steps in the formation of the embryonic heart: (i) the progressive addition of late differentiating progenitor cells from the second heart field that drives heart tube extension during looping morphogenesis, and (ii) the emergence of patterned proliferation within the embryonic myocardium that generates distinct cardiac chambers. During the transition between these steps, the major site of proliferation switches from progenitor cells outside the early heart to proliferation within the embryonic myocardium. The second heart field and ballooning morphogenesis concepts have major repercussions on our understanding of human heart development and disease. In particular, they provide a framework to dissect the origin of congenital heart defects and the regulation of myocardial proliferation and differentiation of relevance for cardiac repair.
Two major stages of embryonic heart development include: (1) the extension of the heart tube by the addition of second heart field progenitor cells from subpharyngeal mesoderm, and (2) the onset of chamber development by ballooning morphogenesis.
In this review, we consider the origin of cardiac progenitor cells in the early embryo and show how progressive specification, differentiation, and morphogenetic events lead to formation of the embryonic heart. We will focus on two conceptually important steps: (i) the regulation of late differentiating progenitor cells (the second heart field) from pharyngeal mesoderm that drives progressive heart tube extension during looping morphogenesis, and (ii) the emergence of patterned proliferation within the embryonic myocardium that generates distinct cardiac chambers. During the transition between these steps, there is a switch from proliferation of progenitor cells outside the early heart as the heart tube elongates to myocardial proliferation within the heart to promote atrial and ventricular chamber morphogenesis. Dissection of the genetic and cellular regulation and lineage relationships implicit in the second heart field and ballooning morphogenesis models are a major focus of ongoing research. Although emphasis will be placed on heart development in the early mouse embryo, with additional insights from avian and fish models, the second heart field and ballooning morphogenesis concepts have major biomedical repercussions on our understanding of human heart development and disease. We illustrate how they provide a framework to dissect the etiology of congenital heart defects, in addition to insights into the regulation of myocardial proliferation and differentiation of relevance for cellular and paracrine approaches to cardiac repair.
CARDIAC SPECIFICATION AND EARLY HEART TUBE DEVELOPMENT
Cells that give rise to the early heart tube are specified and differentiate in lateral anterior splanchnic mesoderm as a result of combinatorial signals from surrounding tissues. Cranial mesoderm is derived from progenitor cells that activate the bHLH transcription factor MESP1 in the primitive streak, under control of the T-box factor Eomesodermin (Saga et al. 2000; Costello et al. 2011). The pattern of inductive signals from adjacent endoderm and overlying ectoderm together with inhibitory signals from the embryonic midline and posterior region of the embryo refine the sites in which the cardiomyogenic transcriptional program is first activated (Marvin et al. 2001; Harvey 2002; Lopez-Sanchez and Garcia-Martinez 2011). These signals, including bone morphogenetic protein (BMP), fibroblast growth factor (FGF), and WNT signals, in addition to short range signaling including fibronectin mediated cascades, result in the activation of key upstream transcriptional regulators of the cardiac phenotype including genes encoding the transcription factors NKX2-5, GATA4, and TBX5, and chromatin remodeling protein SMARCD3 (BAF60c) (Lopez-Sanchez and Garcia-Martinez 2011; Cheng et al. 2013). Combinatorial transcription factor activity in turn activates the panoply of genes encoding sarcomeric components and the enzymatic machinery that define the differentiated myocardial phenotype (Bruneau 2002). Ectopic activation of SMARCD3, GATA4, and TBX5 has been shown to be sufficient to drive cardiomyogenesis in noncardiogenic regions of the embryo (Takeuchi and Bruneau 2009). Convergence of left and right precardiac regions in the embryonic anterior ventral midline results in the formation of the cardiac crescent and linear heart tube (Fig. 1). These early events in heart morphogenesis must be considered in the context of broad embryonic morphogenetic events including embryonic coelom formation and ventral folding of the embryo associated with foregut closure.
Figure 1.
Early events in heart tube formation. (A) The forming human heart in a ventral view and transverse section at stage 9 showing the heart tube open dorsally to foregut endoderm. (B) Quantitative 3D reconstruction of Ki67-positive cells at stage 10 showing the paucity of proliferating cells in the myocardium compared with the dorsal pericardial wall. Note the local increase in proliferation (arrowheads) representing the initiation of ballooning morphogenesis. (From Sizarov et al. 2011; reproduced, with permission.)
MYOCARDIAL PROGENITOR CELLS IN SUBPHARYNGEAL MESODERM: THE SECOND HEART FIELD
The early myocardium, in which cardiac contractions commence, is initially a trough open dorsally onto the ventral foregut endoderm and contiguous along its entire length with more medial splanchnic mesoderm that forms the dorsal coelomic wall of the pericardial cavity (Fig. 1A). Myocardial specification and differentiation are ongoing processes that continue in splanchnic mesoderm for a 3 d period in the mouse, throughout the time of heart tube elongation and cardiac looping. Thus initial growth of the heart tube is driven by cells that are added along its entire length. Subsequently, the dorsal mesocardium, by which the cardiac trough is suspended in the pericardial cavity, breaks down dorsally, isolating the ventral heart tube from initially contiguous splanchnic mesodermal cells in the dorsal pericardial wall (Kelly and Buckingham 2002). These cells maintain continuity with the early heart tube at the venous and arterial poles of the heart, positioned posteriorly and anteriorly, respectively. Fluorescent dye labeling and proliferation studies have revealed that these cells divide rapidly (Fig. 1B) and are displaced toward the poles of the heart tube where they progressively differentiate as the linear dimensions of the heart increase rapidly during looping morphogenesis (Tirosh-Finkel et al. 2006; Abu-Issa and Kirby 2008; van den Berg et al. 2009). Addition of progenitor cells to the heart may in fact be a major driver of rightward looping, the regulation of which is as yet poorly understood at the molecular and cellular levels. The rate of displacement of these cells toward the poles of the heart tube has been estimated at 70 μm/h in avian embryos (van den Berg et al. 2009). These subpharyngeal cells are termed the second heart field and have been shown by genetic tracing and retrospective lineage analysis in the mouse to contribute to a large part of the definitive heart including atrial and inflow myocardium at the venous pole and right ventricular and outflow tract myocardium at the arterial pole (Buckingham et al. 2005; Kelly 2012). In contrast, early differentiating cardiac cells forming the cardiac crescent are termed the first heart field and give rise to part of the left ventricle. The embryonic heart is thus comprised of cardiomyocytes derived from the cardiac crescent and linear heart as well as those derived from second heart field progenitor cells in pharyngeal mesoderm (Fig. 2). Retrospective lineage analysis and genetic tracing using Cre recombinase support a two lineage model of heart development corresponding to the contributions of the first and second heart fields (Cai et al. 2003; Meilhac et al. 2004). Furthermore, a population of late differentiating cardiomyocytes has been found to add to the poles of the frog and fish heart suggesting that this mechanism for heart tube elongation is evolutionarily conserved across vertebrate species (de Pater et al. 2009; Gessert and Kuhl 2009; Hami et al. 2011; Lazic and Scott 2011; Zhou et al. 2011). The potential of the zebrafish model for forward genetic screens suggests that this will be an informative system to probe the genetic mechanisms underlying heart tube elongation.
Figure 2.
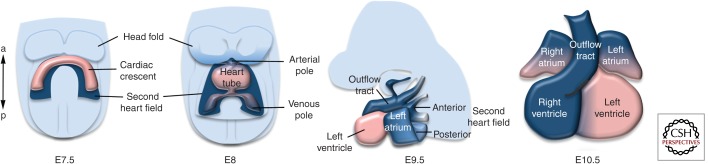
The contribution of the second heart field to heart tube extension. Cartoon showing the progressive addition of second heart field progenitor cells (dark blue) to the elongating heart tube between 7.5 and 9.5 d of mouse development. In the midgestation heart (right), second heart field–derived parts of the heart are indicated in blue. (From Kelly 2012; reproduced, with permission.)
REGULATING PROLIFERATION AND DIFFERENTIATION IN SUBPHARYNGEAL CARDIAC PROGENITOR CELLS
Second heart field cells in subpharyngeal mesoderm are characterized by a number of core properties. First, the continued proliferation of these cells allows separation of the sites of proliferation and differentiation during early heart tube formation, potentially facilitating early cardiac function. Proliferation in subpharyngeal mesoderm is highest in the posterior region of the dorsal pericardial wall in avian embryos although high rates of proliferation are observed throughout the dorsal pericardial wall in the mouse embryo (van den Berg et al. 2009; de Boer et al. 2012). Progenitor cell proliferation in pharyngeal mesoderm is regulated by canonical WNT, FGF, and Hedgehog signaling pathways (Dyer and Kirby 2009; Rochais et al. 2009). Differentiation is tightly orchestrated and triggered as cells approach the ends of the heart tube. The intercellular signaling events associated with heart tube elongation involve complex exchange between pharyngeal mesoderm and surrounding cell types, in particular pharyngeal endoderm and neural crest cells. At the arterial pole of the heart tube BMP signaling antagonizes the pro-proliferative role of FGF signaling and promotes progressive myocardial differentiation (Hutson et al. 2010; Tirosh-Finkel et al. 2010). Neural crest cells play an important role in mediating the BMP inhibition of FGF signaling during second heart field differentiation. Absence of neural crest derived mesenchyme in the pharyngeal region results in increased progenitor cell proliferation and impacts negatively on heart tube elongation. This role of neural crest derived cells in the pharyngeal region precedes their requirement within the outflow tract for arterial pole septation. Notch and noncanonical WNT signaling also regulate differentiation during second heart field deployment (High et al. 2009; Rochais et al. 2009). At the venous pole of the heart, WNT, Hedgehog, and BMP signaling have been shown to play important roles in regulating progenitor cell deployment and atrial septation (Xie et al. 2012; Briggs et al. 2013).
Gene regulatory networks controlling proliferation and differentiation in the second heart field have been identified, including regulatory nodes controlled by key transcription factors such as ISL1, NKX2-5, or TBX1 (Cai et al. 2003; Liao et al. 2008; Chen et al. 2009). TBX1, for example, promotes differentiation delay through multiple mechanisms, including direct interference with intracellular components of the BMP signaling cascade and negative regulation of Mef2c transcript and SRF protein levels (Chen et al. 2009; Fulcoli et al. 2009; Pane et al. 2012). Myocardial differentiation at the arterial pole of the heart tube is reinforced by BMP driven microRNA repression of Isl1 and Tbx1 (Wang et al. 2010). These transcription factors thus intersect with signaling pathway activity to control second heart field development. Analysis of cis-regulatory elements has provided insights into the wiring of transcriptional networks operative in the second heart field. Examples include two enhancers regulating Mef2C expression in the second heart field, one activated by ISL1 and GATA4 (Dodou et al. 2004), and one by NKX2-5 and FOXH1 (von Both et al. 2004), and an Isl1 enhancer activated in the second heart field by FOXC2 and GATA4 (Kang et al. 2009; Kappen and Salbaum 2009). Recently, the enhancer responsible for Fgf10 expression in the SHF has been shown to be regulated by TBX1, ISL1, and NKX2-5, with competition between ISL1 and NKX2-5 for similar homeodeomain binding sites (Watanabe et al. 2012). This is functionally important because NKX2-5 can act as a repressor in the heart tube leading to down-regulation of both Fgf10 and Isl1 on differentiation (Prall et al. 2002). In the case of NKX2-5, transcriptome analysis of mutant versus normal cells revealed many candidate targets in the SHF where NKX2-5 mainly functions as an activator (Prall et al. 2002). Despite these advances, systematic dissection of SHF regulatory networks remains to be performed. There is also little information about transcriptional cofactors in the second heart field or how genomic context, through transcription factors bound to adjacent sites, modulates transcriptional output. Furthermore, heterogeneity in the progenitor cell population complicates the definition of gene regulatory networks that should ideally be analyzed at the single cell level, with the challenge of relating information based on dissociated cells back to localization in the second heart field.
ENCODING DIVERSITY IN THE SECOND HEART FIELD
The progressive contribution of pharyngeal mesoderm to different regions of the elongating heart tube suggests that future cardiac regions are prepatterned in the progenitor cell population. For example, a specific myocardial region surrounding the base of the pulmonary trunk appears to derive from a Tbx1 dependent subpopulation of progenitor cells (Théveniau-Ruissy et al. 2008). How and when different regions of the definitive heart are prepatterned in the progenitor cell population is poorly understood. However, the importance of anterior-posterior patterning within the second heart field is illustrated by the expression profile of anterior Hox genes in progenitor cells in the posterior region of the second heart field that give rise to both the venous pole of the heart and the Tbx1-dependent subpulmonary myocardial domain at the arterial pole (Bertrand et al. 2011). Consistent with these observations, retrospective clonal analysis has revealed a clonal relationship between cardiomyocytes in the outflow tract and venous pole of the heart (Lescroart et al. 2012). Retinoic acid signaling has been shown to operate upstream of Hox gene expression in defining the posterior boundary of the second heart field as well as playing a later role in promoting distal outflow tract development (Ryckebusch et al. 2008; Sirbu et al. 2008). In addition to anterior–posterior patterning, intersection between second heart field regulators and the downstream laterality gene Pitx2 plays a role in conferring left identity to structures at the arterial and venous poles of the heart tube (Liu et al. 2002).
Analysis of populations of embryonic stem cell derived cells expressing Isl1 and Tbx1 has shown that these genes are expressed in multipotent cardiovascular progenitor cells that can give rise to endothelial and smooth muscle cells in addition to cardiac myocytes (Laugwitz et al. 2005; Chen et al. 2009). The time at which these different cell types diverge in vivo and how this multipotency is encoded in subpharyngeal mesoderm during heart tube extension is currently unclear, although clonal analysis has identified multipotent progenitor cells in the avian second heart field (Hutson et al. 2010). Multipotency in pharyngeal mesoderm extends to a skeletal muscle fate. Isl1, Tbx1, and Fgf10 are expressed in the progenitor cells of a subset of craniofacial skeletal muscles termed branchiomeric muscles involved in mastication, facial expression, and laryngeal and pharyngeal function (Kelly 2010). Branchiomeric skeletal muscles activate the MyoD family of myogenic determination genes in the core mesoderm of each pharyngeal or branchial arch. Retrospective clonal analysis has shown that branchiomeric skeletal muscles share a clonal relationship with second heart field derived parts of the heart, but not with the left ventricle, demonstrating that common cardiac and skeletal muscle progenitor cells exist in pharyngeal mesoderm after the split between the first and second cardiac lineages (Lescroart et al. 2010). Furthermore, clonally distinct populations of mesoderm exist at the level of different pharyngeal arches: First arch derived skeletal muscles involved in jaw closure share a lineage relationship with the right ventricle and second arch derived muscles involved in facial expression share a lineage relationship with outflow tract myocardium (Lescroart et al. 2010). These common lineages reflect the fact that the linear heart tube forms from anterior cranial mesoderm at the level of the future face and progressively moves in a posterior direction in the embryo as pharyngeal arch and arch artery morphogenesis proceeds. In the protochordate Ciona intestinalis, Isl1 expressing cells adjacent to cardiac progenitor cells have been shown to have a skeletal rather than cardiac muscle fate, suggesting an evolutionary origin for the vertebrate second heart field by which a population of Isl1 positive cells may have adopted a cardiac progenitor cell fate during vertebrate radiation (Stolfi et al. 2010). Recently, Wang and colleagues have shown that antagonism between the ascidian NKX2-5 and TBX1 homologs regulates cardiac versus skeletal muscle fate in the ascidian second heart field (Wang et al. 2013).
THE BIOMEDICAL IMPACT OF THE SECOND HEART FIELD MODEL
The outflow tract and venous pole of the heart are hotspots of congenital heart defects in human patients. Dissecting the mechanisms involved in the morphogenesis of these parts of the heart is thus an essential step toward understanding the etiology of human disease. Underdevelopment of the Tbx1-dependent myocardial domain at the base of the pulmonary trunk, for example, is considered to be the primary defect in hearts with tetralogy of Fallot, characterized by pulmonary atresia, a ventricular septal defect, overriding aorta, and right ventricular hypertrophy (van Praagh 2009). DiGeorge syndrome patients, haploinsufficient for a multigene deletion on chromosome 22 that includes TBX1, display a range of conotruncal congenital heart defects, including tetralogy of Fallot. The contribution of progenitor cells in the posterior region of the second heart field to the venous pole of the heart has also been associated with congenital heart defects including atrial and atrioventricular septal defects. These anomalies result from failure of development of a structure called the dorsal mesenchymal protrusion that plays a critical role in atrial and atrioventricular septation. The dorsal mesenchymal protrusion is derived from Isl1 and Tbx5 expressing progenitor cells in the posterior second heart field and requires Hedgehog and BMP signaling for its correct development (Snarr et al. 2007; Hoffmann et al. 2009; Briggs et al. 2013). It is important to note that perturbations of the final stages of heart tube extension have a greater likelihood of being encountered in human congenital heart defects than more severe earlier perturbations. Investigation of the mechanisms of the terminal stages of second heart field addition to the heart tube is thus likely to yield clinically relevant insights into the etiology of common human congenital heart defects. In addition to providing insights into normal and pathological heart development, identification of the signals and transcription factors regulating cardiac progenitor cell fate and triggering differentiation in the early embryo will contribute to cellular and paracrine approaches to myocardial repair.
PATTERNS OF INTRACARDIAC PROLIFERATION AND THE INITIATION OF CHAMBER MORPHOGENESIS
From midgestation in the mouse, or the fifth week of human gestation, addition of pharyngeal mesoderm to the poles of the heart is complete and there is a shift from proliferation in extracardiac progenitor cell populations to intracaradiac myocardial proliferation as the main driver of cardiac growth. The emergence of patterned gene expression domains in the embryonic heart drives chamber morphogenesis and initiates septation and conduction system differentiation. Ventricular and atrial working chamber myocardium develops on the outer curvature of the embryonic heart while cardiac cushions develop from endocardium underlying atrioventricular canal and outflow tract myocardium (Fig. 3A). BMP and Notch signaling play upstream roles in restricting the expression patterns of transcription factors of the T-box gene family to different regions of the developing heart. Overlapping expression patterns of T-box containing transcription factor encoding genes plays a central role in the emergence of cardiac form and early establishment of the cardiac conduction system (Fig. 3B) (Hoogaars et al. 2007; Greulich et al. 2011). The transcriptional repressors TBX2 and TBX3 play a role in induction of cardiac cushions and in repressing the working myocardial program in atrioventricular myocardium (Habets et al. 2002). The T-box transcriptional activators TBX20 and TBX5 restrict Tbx2 and Tbx3 expression to the atrioventricular canal region and compete with these factors for interaction with core myocardial transcription factors such as NKX2-5, respectively (Habets et al. 2002; Singh et al. 2009). An important outcome of these patterning events is the emergence of chamber-specific transcriptional programs and proliferative centers on the outer curvature of the embryonic heart, resulting in the development of right and left atrial and ventricular chambers through a process termed ballooning morphogenesis (Christoffels et al. 2000). In addition to patterned proliferation, cellular mechanisms contribute to ballooning morphogenesis. DiI labeling and Cre genetic tracing experiments have shown that descendants of Tbx2 expressing cells in the outflow tract and atrioventricular canal adopt a working myocardial phenotype and contribute to growth of right and left ventricular free walls (Rana et al. 2007; Aanhaanen et al. 2009).
Figure 3.
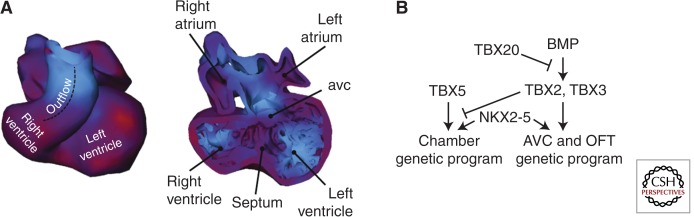
The emergence of patterned proliferation in the embryonic heart. (A) Reconstructions of the myocardium and lumen of a mouse heart at day 11 of development in ventral views showing elevated BrdU incorporation in forming camber myocardium at the outer curvature of the heart tube. (B) Patterned proliferation in the heart is regulated by transcription factors including T-box family members and NKX2-5 (right). (Panel A reproduced, with permission, from de Boer et al. 2012.)
Growth of the ventricular wall is accompanied by the development of distinct trabeculated and compact myocardial layers. Analysis of patterns of proliferation during formation of the four-chambered mouse heart has revealed that proliferative centers at the base of the trabecules contribute to growth of the compact myocardial layer (de Boer et al. 2012). Growth of ventricular myocardium occurs in response to Notch and Neuregulin signaling from endocardium at the base of the trabecular pits and FGF signals from the epicardium and endocardium (Grego-Bessa et al. 2007; Peshkovsky et al. 2011). The regulation of ventricular growth through signals from adjacent endocardium in response to hemodynamic and flow related forces highlights the interplay between form and function in cardiac morphogenesis. Dissecting how extrinsic functional parameters such as blood flow intersect with intrinsic genetic regulation to pattern growth of the ventricular myocardium is a major challenge.
THE BIOMEDICAL IMPACT OF BALLOONING MORPHOGENESIS
Understanding the mechanisms underlying the shift to intracardiac proliferation at the onset of chamber morphogenesis and cardiac septation is of major biomedical importance. Indeed, failure of myocardial proliferation, frequently associated with altered flow patterns in the fetal heart, results in ventricular hypoplasia. Recent evidence has identified the regulation of myocardial proliferation as a key step in regenerative therapies for myocardial repair. In the zebrafish model, regeneration of damaged myocardium has been shown to primarily involve myocyte proliferation rather than de novo differentiation of resident stem cells (Kikuchi and Poss 2012). Similarly in the mouse, continued proliferation of myocytes in the first week after birth is associated with the ability of myocardial cells to repair cardiac damage by additional cycles of proliferation (Porrello et al. 2011, 2013). Dissecting the mechanisms regulating the onset of myocardial proliferation in the midgestation mouse heart will thus uncover mechanisms that may be used to trigger cell cycle reentry of postnatal myocytes.
CONCLUDING REMARKS
Understanding the origins of cardiac cells in the early embryo and the mechanisms regulating the orchestrated development of diverse lineages during cardiogenesis is an essential step toward the goals of identifying the etiology of common congenital heart defects and successfully regenerating myocardium after cardiac damage. Here we have discussed two major stages in the construction of the embryonic heart: heart tube extension by addition of progenitor cells from subpharyngeal mesoderm, and the onset of chamber development. The transition between these phases is marked by a shift in proliferation from progenitor cells outside the early heart tube to differentiated cardiomyocytes. Gaps in our current knowledge have been highlighted, including the mechanisms by which diversity is encoded in subpharyngeal mesoderm, and how functional parameters impact on patterned proliferation in the embryonic heart. Additional insights obtained in response to these and other questions from animal models including zebrafish, mouse, and chick, will undoubtedly contribute to identifying new mechanisms regulating heart development and pathology in man.
ACKNOWLEDGMENTS
Research in R.G.K.'s laboratory is supported by the Fondation pour la Recherche Médicale and EU FP7 contracts CardioGeNet and CardioNet. M.E.B. is supported by the Centre National de la Recherche Scientifique (CNRS) and Pasteur Institute.
Footnotes
Editors: Margaret Buckingham, Christine L. Mummery, and Kenneth R. Chien
Additional Perspectives on The Biology of Heart Disease available at www.perspectivesinmedicine.org
REFERENCES
- Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, et al. 2009. The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res 104: 1267–1274 [DOI] [PubMed] [Google Scholar]
- Abu-Issa R, Kirby ML. 2008. Patterning of the heart field in the chick. Dev Biol 319: 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Roux M, Ryckebusch L, Niederreither K, Dolle P, Moon A, Capecchi M, Zaffran S. 2011. Hox genes define distinct progenitor sub-domains within the second heart field. Dev Biol 353: 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs LE, Phelps AL, Brown E, Kakarla J, Anderson RH, van den Hoff MJ, Wessels A. 2013. Expression of the BMP receptor Alk3 in the second heart field is essential for development of the dorsal mesenchymal protrusion and atrioventricular septation. Circ Res 112: 1420–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. 2002. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res 90: 509–519 [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. 2005. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 6: 826–835 [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. 2003. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 5: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Fulcoli FG, Tang S, Baldini A. 2009. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res 105: 842–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Andersen P, Hassel D, Kaynak BL, Limphong P, Juergensen L, Kwon C, Srivastava D. 2013. Fibronectin mediates mesendodermal cell fate decisions. Development 140: 2587–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, et al. 2000. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol 223: 266–278 [DOI] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ. 2011. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol 13: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer BA, van den Berg G, de Boer PA, Moorman AF, Ruijter JM. 2012. Growth of the developing mouse heart: An interactive qualitative and quantitative 3D atlas. Dev Biol 368: 203–213 [DOI] [PubMed] [Google Scholar]
- de Pater E, Clijsters L, Marques SR, Lin YF, Garavito-Aguilar ZV, Yelon D, Bakkers J. 2009. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 136: 1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. 2004. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 131: 3931–3942 [DOI] [PubMed] [Google Scholar]
- Dyer LA, Kirby ML. 2009. The role of secondary heart field in cardiac development. Dev Biol 336: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcoli FG, Huynh T, Scambler PJ, Baldini A. 2009. Tbx1 regulates the BMP-Smad1 pathway in a transcription independent manner. PLoS ONE 4: e6049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gessert S, Kuhl M. 2009. Comparative gene expression analysis and fate mapping studies suggest an early segregation of cardiogenic lineages in Xenopus laevis. Dev Biol 334: 395–408 [DOI] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, et al. 2007. Notch signaling is essential for ventricular chamber development. Dev Cell 12: 415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich F, Rudat C, Kispert A. 2011. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res 91: 212–222 [DOI] [PubMed] [Google Scholar]
- Habets PEMH, Moorman AFM, Clout DEW, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. 2002. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: Implications for cardiac chamber formation. Genes Dev 16: 1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hami D, Grimes AC, Tsai HJ, Kirby ML. 2011. Zebrafish cardiac development requires a conserved secondary heart field. Development 138: 2389–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RP. 2002. Patterning the vertebrate heart. Nat Rev Genet 3: 544–556 [DOI] [PubMed] [Google Scholar]
- High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, Kaestner KH, Pear WS, Epstein JA. 2009. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J Clin Invest 119: 1986–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AD, Peterson MA, Friedland-Little JM, Anderson SA, Moskowitz IP. 2009. Sonic hedgehog is required in pulmonary endoderm for atrial septation. Development 136: 1761–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WMH, Barnett P, Moorman AFM, Christoffels VM. 2007. T-box factors determine cardiac design. Cell Mol Life Sci 64: 646–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson MR, Zeng XL, Kim AJ, Antoon E, Harward S, Kirby ML. 2010. Arterial pole progenitors interpret opposing FGF/BMP signals to proliferate or differentiate. Development 137: 3001–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Nathan E, Xu SM, Tzahor E, Black BL. 2009. Isl1 is a direct transcriptional target of Forkhead transcription factors in second-heart-field-derived mesoderm. Dev Biol 334: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappen C, Salbaum JM. 2009. Identification of regulatory elements in the Isl1 gene locus. Int J Dev Biol 53: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RG. 2010. Core issues in craniofacial myogenesis. Exp Cell Res 316: 3034–3041 [DOI] [PubMed] [Google Scholar]
- Kelly RG. 2012. The second heart field. Curr Topics Dev Biol 100: 33–65 [DOI] [PubMed] [Google Scholar]
- Kelly RG, Buckingham ME. 2002. The anterior heart-forming field: Voyage to the arterial pole of the heart. Trends Genet 18: 210–216 [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Poss KD. 2012. Cardiac regenerative capacity and mechanisms. Annu Rev Cell Dev Biol 28: 719–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. 2005. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433: 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic S, Scott IC. 2011. Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev Biol 354: 123–133 [DOI] [PubMed] [Google Scholar]
- Lescroart F, Kelly RG, Le Garrec JF, Nicolas JF, Meilhac SM, Buckingham M. 2010. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development 137: 3269–3279 [DOI] [PubMed] [Google Scholar]
- Lescroart F, Mohun T, Meilhac SM, Bennett M, Buckingham M. 2012. Lineage tree for the venous pole of the heart: Clonal analysis clarifies controversial genealogy based on genetic tracing. Circ Res 111: 1313–1322 [DOI] [PubMed] [Google Scholar]
- Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. 2008. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol 316: 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liu W, Palie J, Lu MF, Brown NA, Martin JF. 2002. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development 129: 5081–5091 [DOI] [PubMed] [Google Scholar]
- Lopez-Sanchez C, Garcia-Martinez V. 2011. Molecular determinants of cardiac specification. Cardiovasc Res 91: 185–195 [DOI] [PubMed] [Google Scholar]
- Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. 2001. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev 15: 316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. 2004. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell 6: 685–698 [DOI] [PubMed] [Google Scholar]
- Pane LS, Zhang Z, Ferrentino R, Huynh T, Cutillo L, Baldini A. 2012. Tbx1 is a negative modulator of Mef2c. Hum Mol Genet 21: 2485–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkovsky C, Totong R, Yelon D. 2011. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev Dyn 240: 446–456 [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. 2011. Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. 2013. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci 110: 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prall OW, Elliott DA, Harvey RP. 2002. Developmental paradigms in heart disease: Insights from tinman. Ann Med 34: 148–156 [PubMed] [Google Scholar]
- Rana MS, Horsten NC, Tesink-Taekema S, Lamers WH, Moorman AF, van den Hoff MJ. 2007. Trabeculated right ventricular free wall in the chicken heart forms by ventricularization of the myocardium initially forming the outflow tract. Circ Res 100: 1000–1007 [DOI] [PubMed] [Google Scholar]
- Rochais F, Mesbah K, Kelly RG. 2009. Signaling pathways controlling second heart field development. Circ Res 104: 933–942 [DOI] [PubMed] [Google Scholar]
- Ryckebusch L, Wang Z, Bertrand N, Lin SC, Chi X, Schwartz R, Zaffran S, Niederreither K. 2008. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci 105: 2913–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Kitajima S, Miyagawa-Tomita S. 2000. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med 10: 345–352 [DOI] [PubMed] [Google Scholar]
- Singh R, Horsthuis T, Farin HF, Grieskamp T, Norden J, Petry M, Wakker V, Moorman AF, Christoffels VM, Kispert A. 2009. Tbx20 interacts with smads to confine Tbx2 expression to the atrioventricular canal. Circ Res 105: 442–452 [DOI] [PubMed] [Google Scholar]
- Sirbu IO, Zhao X, Duester G. 2008. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn 237: 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizarov A, Ya J, de Boer BA, Lamers WH, Christoffels VM, Moorman AF. 2011. Formation of the building plan of the human heart: Morphogenesis, growth, and differentiation. Circulation 123: 1125–1135 [DOI] [PubMed] [Google Scholar]
- Snarr BS, O’Neal JL, Chintalapudi MR, Wirrig EE, Phelps AL, Kubalak SW, Wessels A. 2007. Isl1 expression at the venous pole identifies a novel role for the second heart field in cardiac development. Circ Res 101: 971–974 [DOI] [PubMed] [Google Scholar]
- Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. 2010. Early chordate origins of the vertebrate second heart field. Science 329: 565–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. 2009. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459: 708–U112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveniau-Ruissy M, Dandonneau M, Mesbah K, Ghez O, Mattei MG, Miquerol L, Kelly RG. 2008. The del22q11.2 candidate gene Tbx1 controls regional outflow tract identity and coronary artery patterning. Circ Res 103: 142–148 [DOI] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Elhanany H, Rinon A, Tzahor E. 2006. Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development 133: 1943–1953 [DOI] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Zeisel A, Brodt-Ivenshitz M, Shamai A, Yao Z, Seger R, Domany E, Tzahor E. 2010. BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development 137: 2989–3000 [DOI] [PubMed] [Google Scholar]
- van den Berg G, Abu-Issa R, de Boer BA, Hutson MR, de Boer PA, Soufan AT, Ruijter JM, Kirby ML, van den Hoff MJ, Moorman AF. 2009. A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ Res 104: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praagh R. 2009. The first Stella van Praagh memorial lecture: The history and anatomy of tetralogy of Fallot. Semin Thorac Cardiovasc Surg Pediatr Card Surg Annu. 2009: 19–38 [DOI] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL. 2004. Foxh1 is essential for development of the anterior heart field. Dev Cell 7: 331–345 [DOI] [PubMed] [Google Scholar]
- Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. 2010. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a microRNA-mediated mechanism. Dev Cell 19: 903–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Razy-Krajka F, Siu E, Ketcham A, Christiaen L. 2013. NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLoS Biol 11: e1001725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Zaffran S, Kuroiwa A, Higuchi H, Ogura T, Harvey RP, Kelly RG, Buckingham M. 2012. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2-5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc Natl Acad Sci 109: 18273–18280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Hoffmann AD, Burnicka-Turek O, Friedland-Little JM, Zhang K, Moskowitz IP. 2012. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell 23: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, et al. 2011. Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature 474: 645–648 [DOI] [PMC free article] [PubMed] [Google Scholar]



