Many fungi can form biofilms, which may cause infection when present on implanted devices (e.g., mechanical heart valves). The genetic tractability of Candida albicans is helping to drive biofilm studies in fungi.
Abstract
A biofilm is a surface-associated microbial community. Diverse fungi are capable of biofilm growth. The significance of this growth form for infection biology is that biofilm formation on implanted devices is a major cause of recurrent infection. Biofilms also have limited drug susceptibility, making device-associated infection extremely difficult to treat. Biofilm-like growth can occur during many kinds of infection, even when an implanted device is not present. Here we summarize the current understanding of fungal biofilm formation, its genetic control, and the basis for biofilm drug resistance.
Much of the global microbial community grows on surfaces, including rock, sediment, bark, skin, and mucosal tissues (Kolter and Greenberg 2006). The growth form of microorganisms that is associated with a surface is called a biofilm. That definition is generally refined to include the specification that a true biofilm is one in which the organisms grow as a community, rather than separate surface-adherent cells. In addition, the cells of a true biofilm produce their own extracellular matrix material and manifest phenotypes that are distinct from the phenotypes of cells growing in suspension (called planktonic cells). It has long been realized that biofilm formation ability is widespread among bacteria, and although there are species-specific features of biofilm growth, there are also common themes that have emerged. In this review, we highlight the fact that biofilm formation ability is widespread among fungi as well, and discuss fungal biofilm structure and regulation.
The first question about fungal biofilm formation is “Why study it”? There are many answers of course, but the central significance of biofilms in the context of infection biology comes from the relationship of biofilms to device-associated infection. Implanted devices such as central venous catheters, indwelling bladder catheters, joint prostheses, and mechanical heart valves are in common use for the management of diverse medical conditions (Raad 1998; Darouiche 2001). The presence of such devices is a major risk factor for infection (Raad 1998; Costerton et al. 1999; Donlan 2002). These infections arise from microbial attachment to device surfaces and their growth into biofilms (Costerton et al. 1999; Hall-Stoodley et al. 2004; Kuhn and Ghannoum 2004). These biofilms are invariably recalcitrant to drug treatment for a variety of reasons, and thus can cause recurrent infections. Examination of devices isolated from infected patients has revealed that many bacterial and fungal species form biofilms that seed recurrent infections (Lynch and Robertson 2008). Among fungi, different Candida species are the most common colonizers of implanted medical devices (Kumamoto and Vinces 2005; Ramage et al. 2005). Fusarium species and Malessezia pachydermatis also form drug-resistant biofilms on implanted medical devices (Martinez and Fries 2010; Ramage et al. 2011). Table 1 describes biofilm characteristics for several fungi commonly associated with device-related biofilm infections.
Table 1.
Fungal biofilms and their relevance in human health
| Species | Devices associated with biofilm-related infection | Biofilm characteristics | ||
|---|---|---|---|---|
| Matrix components | Biofilm morphology | |||
| Candida species | Catheters (central and peripheral venous catheters, hemo- and peritoneal-dialysis catheters, urinary catheters), endotracheal tubes, cardiac devices (prosthetic valves, pacemakers), breast implants, voice prosthetic devices, and joint prostheses (Raad 1998; Donlan and Costerton 2002; Kojic and Darouiche 2004; Ramage et al. 2006) | Candida albicans | Glucose, DNA, small amounts of hexosamine, small amounts of protein, phosphorous, and uronic acid (Al-Fattani and Douglas 2006) | Complex morphology with blastospores and hyphae with ECM (Hawser and Douglas 1994; Chandra et al. 2001; Desai et al. 2013) |
| Candida glabrata | Higher concentration of carbohydrate and protein than Candida parapsilosis biofilms (Silva et al. 2009) | Layered clusters of blastospores with ECM (Silva et al. 2009; Kucharikova et al. 2011) | ||
| C. parapsilosis | Prominent presence of carbohydrates with less protein (Silva et al. 2009) | Strain-dependent variation in biofilm formation (Silva et al. 2009, 2011; Pannanusorn et al. 2013b) and clustered blastospores with ECM (Lattif et al. 2010) | ||
| Candida dubiliensis | - | Complex morphology with blastospores, pseudohyphae, and hyphae (Ramage et al. 2001) | ||
| Candida tropicalis | Hexosamine, small amounts of protein, phosphorous, and more uronic acid than C. albicans (Al-Fattani and Douglas 2006) | Clustered blastospores (Pannanusorn et al. 2013a) | ||
| Cryptococcus neoformans | Ventriculoatrial shunt (Walsh et al. 1986), prosthetic valve (Banerjee et al. 1997), and prosthetic dialysis fistula (Braun et al. 1994) | C. neoformans | Glucurunoxylomannan and sugars such as xylose, mannose, glucose, and galactoxylomannan (Martinez and Casadevall 2007) | Organized structure containing yeast cells surrounded by matrix material (Martinez and Casadevall 2007) |
| Trichosporon species | Dialysis graft (Krzossok et al. 2004) and breast implants (Reddy et al. 2002) | Trichosporon asahii | - | Biofilm consisting of yeast and hyphal cells embedded in ECM (Di Bonaventura et al. 2006) |
| Aspergillus fumigatus | Dialysis catheters (Bonfante et al. 2005; Jeloka et al. 2011) | A. fumigatus | Galactomannan, α-1,3-glucans, monosaccharides, polyols, melanins, and proteins such as hydrophobins (Ramage et al. 2011) and DNA (Rajendran et al. 2013) | Hyphal cells embedded in ECM (Loussert et al. 2010; Rajendran et al. 2013) |
ECM, extracellular matrix.
There are also situations in which the fungal growth form associated with an infection has biofilm-like properties, although there may not be a rigorous demonstration that the growth form has all of the defining features of a biofilm (Cushion et al. 2009; Mowat et al. 2009; Fanning and Mitchell 2012). An excellent illustration comes from the mold Aspergillus fumigatus, a common cause of nosocomial infection in immunocompromised individuals (Mowat et al. 2009). A. fumigatus is occasionally found to colonize implanted medical devices. However, most infections begin when inhaled conidia germinate in the upper airway and in lung tissue. This growth state may lack a clear substrate, but resembles a biofilm nonetheless because hyphae are enmeshed in an extracellular matrix (Loussert et al. 2010). Therefore, biofilm models have been developed for A. fumigatus that might mimic the growth state that occurs during infection (Loussert et al. 2010). Indeed, transcriptional profiling of this biofilm model found considerable similarity to the profile of A. fumigatus germlings in a lung infection model (McDonagh et al. 2008; Gibbons et al. 2012), thus validating the concept. Another interesting example comes from the study of Pneumocystis species, which cause pneumonia in immunocompromised patients. These species are challenging to study because they cannot be cultured outside of the host. Cushion and colleagues postulated that attachment of the organisms to tissue might generate a biofilm-like physiology that promotes growth and division (Cushion et al. 2009). Indeed, their in vitro model for biofilm development by Pneumocystis carinii and Pneumocystis murina enabled prolonged growth of the organisms in vitro compared with planktonic culture conditions (Cushion et al. 2009). Finally, mucosal infections, such as oropharyngeal candidiasis or vaginitis, are caused by true biofilms in which tissue, rather than a device, serves as the substrate for biofilm growth (Dongari-Bagtzoglou 2008; Ganguly and Mitchell 2011). In these situations, the biofilm-associated phenotype of drug resistance has not been shown. Nonetheless, the concept that a mucosal infection is a biofilm has proven useful by prompting the accurate prediction that known regulators of Candida albicans biofilm formation would also be required for mucosal infection (Ganguly and Mitchell 2011). These examples illustrate the principle that biofilm growth can provide a useful model for infection even if a canonical abiotic-surface biofilm is not clearly associated with infection.
GENETIC CONTROL OF FUNGAL BIOFILM DEVELOPMENT
What are the genes that govern biofilm formation? One can imagine a broad spectrum of gene functions that may be involved, affecting such biological properties as adherence, morphogenesis, quorum sensing, matrix production, cell wall biosynthesis, and metabolism. Diverse rationales have been used to choose candidate genes that may be required for normal biofilm formation. For example, investigators have chosen to analyze potential biofilm defects in mutants of random genes, mutants of genes with a particular class of product, known functions related to hyphal morphogenesis or azole drug resistance, and those differentially regulated during biofilm formation (Garcia-Sanchez et al. 2004; Murillo et al. 2005; Nobile et al. 2006, 2012; Bonhomme et al. 2011; Desai et al. 2013). These approaches have been most extensively applied to C. albicans, the focus of the discussion below, and have been implemented more recently with non-albicans Candida species as genetic tools have become available (Ramage et al. 2005; Finkel and Mitchell 2011).
One useful approach to define biofilm gene functions has been to identify transcription factors that are required for biofilm formation. The rationale behind the approach came from the suspicion that many gene products—such as the large families of ALS (agglutinin-like sequence) proteins, CFEM (common in fungal extracellular membranes) proteins, and proteins resembling Hyr1—might have compensatory or overlapping functions in biofilm formation (Nobile and Mitchell 2005). The idea was that a single transcription factor is often required for expression of many functionally related genes, and thus a defect in a transcription factor mutant may have a more pronounced phenotypic result than a defect in a single target gene. (The approach was based on the analysis of meiotic gene regulation in Saccharomyces cerevisiae [Mitchell 1994], in which many meiotic regulatory mutations had more dramatic phenotypes than target gene mutants.) Moreover, the relevant target genes could be identified through expression profiling (Nobile and Mitchell 2005) or, in subsequent studies, chromatin-immunoprecipitation analysis (Nobile et al. 2012). The approach of screening a panel of transcription factor mutants for biofilm defects was productive in yielding the first biofilm regulator, Bcr1 (Nobile and Mitchell 2005), which was not required for hyphal morphogenesis in the biofilm (see below). A more recent implementation of this approach has led to the finding that six key biofilm regulators all govern each other’s expression (Nobile et al. 2012), and a large spectrum of regulators of adherence converge on the regulation of a limited number of cell surface proteins (Finkel et al. 2012). We think that this approach remains an excellent strategy for dissection of biological processes.
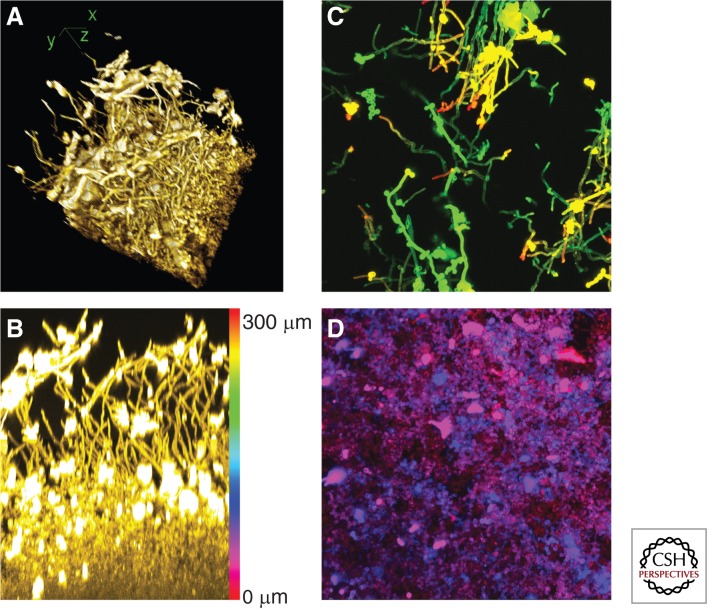
Among the most central conclusions from studies of C. albicans biofilm-defective mutants is the idea that an intact hyphal morphogenesis pathway is critical for accumulation of full biofilm biomass. This conclusion was first suggested by analysis of an uncharacterized hyphae-defective mutant and the hyphae-defective efg1Δ/Δ mutant (Lewis et al. 2002; Ramage et al. 2002b), and has been reinforced by random mutant screens, screens of transcription factor mutants and protein kinase mutants, and a recent comprehensive transcription factor mutant screen (Finkel and Mitchell 2011; Nobile et al. 2012; Bonhomme and d’Enfert 2013). The requirement for hyphal morphogenesis regulators remains under diverse in vitro culture conditions, as well as in animal biofilm infection models (Lewis et al. 2002; Garcia-Sanchez et al. 2004; Nobile et al. 2012). In some ways, this conclusion was expected because hyphae are a prominent feature of biofilms formed under most biofilm culture conditions (Fig. 1A). Under in vitro culture conditions, biofilm hyphae do not seem to be highly aggregated, but instead appear to be organized in a parallel array, much like tree trunks in a forest (Fig. 1B).
Figure 1.
Four views of a C. albicans biofilm. These images portray a single biofilm grown in yeast-peptone-dextrose medium at 37°C. It was embedded and stained with Alexafluor 594-conjugated Concanavalin A. Each image processes a portion of a confocal laser scanning microscope dataset with National Institutes of Health ImageJ software. (A) Three-dimensional reconstruction of the biofilm sample. Hyphae at the top of the biofilm are readily visible. x-, y-, and z-axes are diagrammed. (B) Side projection view of the x–z plane. Approximately 25% of the x-axis from the sample was used, permitting visualization of hyphae in the upper portion of the biofilm. Brightly stained aggregates are extracellular matrix material. The color scale bar represents the depth of the biofilm, 300 μm, and indicates the pseudocolor scale used for apical projections in C and D. (C) Apical projection of upper (substrate-distal) 100-μm region. This image shows the x–y plane. Hyphae extend from 200 μm from the substrate (green) to 300 μm from the substrate (red). Some yeast cells and extracellular material are apparent. (D) Apical projection of basal (substrate-proximal) 100-μm region. This image once again shows the x–y plane. Yeast cells are evident from the substrate level (red) to 100 μm above the substrate (blue), although light penetration limits the resolution of the image. Amorphous extracellular material is also evident.
We think that there are two critical roles for hyphae in C. albicans biofilm formation: adherence and metabolite exchange. With respect to adherence, hyphae of C. albicans express numerous adhesins, and their ability to adhere to other cells and surfaces is vital for biofilm stability. Consistent with this conclusion, some biofilm-defective mutants, such as bcr1Δ/Δ or rhr2Δ/Δ, are able to form hyphae under biofilm conditions, but fail to express hyphal adhesin genes (Nobile et al. 2006; Finkel et al. 2012; Desai et al. 2013). For these mutants, overexpression of adhesin genes ALS1, ALS3, or HWP1 restores the ability to form biofilms (Nobile et al. 2006; Desai et al. 2013). In fact, overexpression of the transcription factor gene BCR1 improves biofilm formation ability by the hyphae-defective, biofilm-defective tec1Δ/Δ mutant (Nobile et al. 2006), and overexpression of an ALS3-AGα1 hybrid adhesin gene improves biofilm formation ability by the hyphae-defective, biofilm-defective efg1Δ/Δ mutant (Zhao et al. 2006). In the overexpression-rescue experiments with tec1Δ/Δ or efg1Δ/Δ strains, the improvement in biofilm formation is not accompanied by restoration of ability to produce hyphae. These experiments indicate that the key role of hyphal adhesins in biofilm formation can be separated from a contribution of hyphal cell morphology. Recent comprehensive identification of transcription factors that govern adherence of yeast-form cells (also called blastospores) to silicone suggests that there may be additional biofilm adherence determinants, beyond the well-characterized hyphal adhesins (Finkel et al. 2012). This inference is consistent with the fact that yeast-form cells occupy the basal biofilm layers in most in vitro–grown biofilms (Fig. 1D).
In addition, hyphal cells may have a functional role in biofilm formation that is independent of their adhesins. Specifically, the parallel hyphae of in vitro biofilms provide good solvent access to the lower layer of the biofilm (Fig. 1B). Bacterial biofilm growth depends critically on solvent channels that presumably facilitate removal of metabolic wastes and entry of nutrients (Davies et al. 1998; Wilking et al. 2013). For example, Pseudomonas aeruginosa biofilms are a collection of mushroom-like microcolonies whose spacing creates solvent channels (Davies et al. 1998). Perhaps filamentous hyphae and pseudohyphae in fungal biofilms function similarly to improve metabolite exchange among biofilm cells.
The possibility that there is a distinctive physiology of biofilm cells was first indicated by transcriptomic comparisons of biofilm and planktonic cells. Among these studies we note, in particular, the foundational work of the d’Enfert laboratory, in which a core set of biofilm-induced genes was defined through multiple biofilm-planktonic comparisons (Garcia-Sanchez et al. 2004). One large subset of these genes was associated with hyphal development, as might be expected from the discussion above. In addition, a substantial proportion of these core up-regulated genes were associated with physiological functions such as protein synthesis, amino acid synthesis, and glycolytic flux. Analysis of suitable mutant strains (e.g., gcn4Δ/Δ, tye7Δ/Δ) has confirmed the functional requirement for these physiological processes in optimal biofilm growth (Garcia-Sanchez et al. 2004; Bonhomme et al. 2011).
Revealing insight into biofilm cell physiology came in part from comparative transcriptomic studies (Rossignol et al. 2009). The Butler and d’Enfert laboratories focused on Candida parapsilosis, a highly significant cause of device-associated infection. C. parapsilosis, like C. albicans, is a member of the CTG clade, but it makes only pseudohyphae, not true hyphae. The study found a highly significant correlation between the gene-expression signature of biofilm cells and the gene-expression response to hypoxia, or low oxygen conditions. Functional analysis in C. albicans has extended this hypothesis with the finding that adaptation to hypoxia is critical for biofilm integrity (Bonhomme et al. 2011). Thus, the hypoxia response is a major driving force behind the gene expression and physiology of biofilm cells.
Among the most perplexing observations regarding C. albicans biofilm gene expression is that biofilm cells up-regulate genes for sulfur assimilation and the biosynthesis of methionine and cysteine (Garcia-Sanchez et al. 2004; Murillo et al. 2005). These genes are also up-regulated as part of the hypoxic response (Stichternoth and Ernst 2009). It seems possible that these two sulfur amino acids may be overrepresented in biofilm translation products, or perhaps there are greater demands for the methyl donor S-adenosyl-methionine in biofilm cells as compared with planktonic cells. It has also been suggested that the reducing environment created by cysteine may be necessary to detoxify reactive oxygen species in biofilm cells (Stichternoth and Ernst 2009). In view of the key role of hypoxic adaptation in biofilm formation, we propose an additional metabolic explanation for this regulatory response. C. albicans cannot ferment sugars, and thus it usually reduces oxygen through aerobic respiration to balance reducing equivalents from glycolysis. When oxygen is limiting, it may adapt by using other reduction reactions to achieve this balance. The sulfur assimilation pathway includes multiple reduction steps to convert sulfate to sulfide. We suggest that C. albicans uses sulfate as an alternative electron acceptor under hypoxic conditions to balance reducing equivalents that are generated from glycolysis.
Biofilms are not, simply, slightly hypoxic aggregates of yeast cells and hyphae; a biofilm community has complex phenotypes. For example, biofilm cells produce an extracellular matrix and are resistant to many antimicrobial agents, as discussed below. In addition, yeast cells are not restricted to the lower layers of the biofilm (Fig. 1C), and the yeast cells in the upper biofilm sites have distinct properties from planktonic yeast cells (see below) (Uppuluri et al. 2010). Many other biofilm phenotypes have been assayed in focused studies or more broad-ranging screens. Among the most remarkable findings in this context is the discovery that yeast-form cells released from C. albicans biofilms have properties that are distinct from planktonic yeast-form cells (Uppuluri et al. 2010). Most noteworthy is the fact that released yeast-form cells are hypervirulent in a mouse-disseminated infection model compared with a planktonic inoculum (Uppuluri et al. 2010). Biofilm cell release is governed by several hyphal regulators, as well as HSP90 (Robbins et al. 2011). This result has led to the proposal that HSP90 inhibitors could be used to reduce disseminated infection rates (Robbins et al. 2011). Therefore, it appears that C. albicans biofilms promote infection not only by serving as a pathogen reservoir, but also through physiological changes that augment the virulence of dispersed cells.
Gene-expression profiling of Candida biofilms has been useful not only for inferring key cellular processes, but also for defining specific gene functions that are required for normal biofilm formation (Bonhomme et al. 2011; Desai et al. 2013). A recent comparison of A. fumigatus biofilm and planktonic transcriptomes has the potential to open the door to functional analysis in that organism (Gibbons et al. 2012; Muszkieta et al. 2013). Biofilm up-regulated genes were enriched for such functions as translation, stress responses, transport, and cell wall/cell surface features. These are just the sorts of gene functions that may reflect common themes in fungal biofilm formation. Interestingly, there was also enrichment for secondary metabolism genes, which may represent a unique feature of A. fumigatus biofilm formation. Given the many differences between molds and yeasts, we anticipate that functional analysis of A. fumigatus biofilm formation will reveal many distinctive processes not evident in Candida biofilms.
The basidiomycete Cryptococcus neoformans is a distant relative of the ascomycetes such as Candida and Aspergillus, and several novel findings have emerged from the study of C. neoformans biofilms and cell–cell adherence. Cell–cell communication is critical for C. neoformans biofilm formation and is coupled to adherence in a novel way. Specifically, the surface/secreted protein Cfl1 functions both as an adhesin and a signaling molecule that induces hyphal development (Wang et al. 2012, 2013). CFL1 is under the control of the mating regulator Znf2 (Wang et al. 2012, 2013), and it has yet to be determined whether it functions only in mating rather than in overall biofilm formation. Nonetheless, a recent study revealed that conditioned medium indeed stimulates C. neoformans biofilm formation (Albuquerque et al. 2013), in keeping with the idea that secreted products promote, rather than inhibit, biofilm formation. In contrast, the most well-studied quorum-sensing molecule from C. albicans, farnesol (Nickerson et al. 2006; Madhani 2011), inhibits biofilm formation when supplied exogenously in pure form (Ramage et al. 2002a; Cao et al. 2005), although effects of farnesol from endogenous biofilms may be more difficult to predict (Ganguly et al. 2011). Clearly, our understanding of C. neoformans biofilm formation is in its infancy, and presents an opportunity for the discovery of novel mechanisms that support biofilm formation.
Although we usually think of biofilm formation as a single process, it has become clear that overall biofilm development depends greatly on the environment. For example, Soll and colleagues have shown that the structures of biofilms grown in different media have very different structures (Daniels et al. 2013). Thus, it is likely that the relative prominence of many regulatory relationships in biofilm formation may be environmentally contingent. This conclusion was anticipated by the diverse biofilm transcriptomic datasets of the d’Enfert laboratory (Garcia-Sanchez et al. 2004), and the finding that several mutant strains are biofilm defective under some conditions, but not others (Richard et al. 2005; Nobile et al. 2006). In addition, biofilms of mating-competent C. albicans strains, which are homozygous at the MTL locus, create biofilms with some genetic and environmental requirements that are distinct from those of the more frequently isolated MTLa/MTLα strains (Sahni et al. 2010; Park et al. 2013; Si et al. 2013). In fact, Fanning et al. (2012a) have shown that the biofilm regulator Bcr1 has distinct sets of responsive genes in the same strain during biofilm growth in vitro and on the oral mucosa in vivo. Epistasis analysis of mutations in BCR1 and the cAMP-dependent protein kinase gene TPK1 provide insight in this regard; loss of Tpk1 function permits biofilm formation by a bcr1Δ/Δ strain. However, loss of Tpk1 function does not restore expression of many major Bcr1-dependent genes, but instead activates alternative genes that can compensate for the defect in Bcr1 (Fanning et al. 2012a). Tpk1 and Bcr1 thus govern alternative target gene sets, and either one can promote biofilm formation (Fanning et al. 2012a). The ability of fungi to form biofilms in diverse environments may reflect reliance on a portfolio of gene functions, groups of which enable adaptation to a subset of environmental conditions.
MULTISPECIES BIOFILMS
One of the most important environmental factors that can modulate fungal biofilm formation and its consequences is the presence of bacteria. Clinical and experimental data underscore the significance of these interactions, as explained in several excellent recent reviews (Morales and Hogan 2010; Peleg et al. 2010, 2012a; Harriott and Noverr 2011). Some of the most dramatic results come from studies with C. albicans and Staphylococcus aureus: coinfection causes much greater mortality than single-species infection in a mouse model (Carlson 1982). In addition, C. albicans enables S. aureus biofilm formation in serum and decreases its susceptibility to vancomycin (Harriott and Noverr 2009). These simple experiments underscore the need to understand polymicrobial interactions to guide diagnosis and therapy.
The distinct properties of mixed-species biofilms arise from both direct cell–cell contact and the impact of secreted molecules. Among the best-characterized bacterial receptors on C. albicans is the surface adhesin Als3. Jenkinson and colleagues used heterologous expression to show that Als3 and Eap1 are sufficient to promote binding to the oral bacterium Streptococcus gordonii (Nobbs et al. 2010). Remarkably, Als3 is also responsible for C. albicans binding to S. aureus (Peters et al. 2012b). Given that ALS3 and EAP1 are both greatly induced during biofilm formation, a C. albicans biofilm would provide an environment that could maintain these bacteria as coaggregates. Jenkinson and colleagues underscore that S. gordonii is a pioneer colonizer of the oral cavity, and thus argue that coaggregate formation may increase levels of C. albicans carriage (Nobbs et al. 2010).
Secreted small molecules, such as quorum-sensing molecules and metabolites, are also an important factor in fungal–bacterial interaction. For example, the Hogan group has shown that the C. albicans quorum-sensing molecule, farnesol, inhibits expression of P. aeruginosa virulence factors by affecting its quinolone signal production machinery (Cugini et al. 2007). Jabra-Rizk et al. (2006) have shown that farnesol inhibits S. aureus biofilm formation, thus this fungal quorum-sensing molecule impacts both Gram-negative and -positive bacteria. Bacteria are all too happy to return the favor; the P. aeruginosa quorum-sensing molecule, homoserinelactone, inhibits hyphal formation by C. albicans through inhibition of the Ras-adenylate cyclase pathway (Davis-Hanna et al. 2008; Hall et al. 2011). Given that yeast-form cells are most easily dispersed from C. albicans biofilms (Uppuluri et al. 2010), one would expect the presence of P. aeruginosa to promote C. albicans biofilm dispersal.
We have provided only a small sampling of the types of interactions that may confer unique properties of polymicrobial biofilms. Clearly, there is much to learn, and clinical observations serve as a useful guide to significant interactions (Morales and Hogan 2010; Peleg et al. 2010; Harriott and Noverr 2011; Peters et al. 2012a). It seems likely that mixed species interactions are among the driving forces behind the evolution of fungal biofilm gene functions.
BIOFILM COMPARATIVE GENOMICS
Can the detailed knowledge of C. albicans biofilm formation be applied to the biofilms of other fungal pathogens? The Butler laboratory has pioneered the use of comparative genomics to explore the functions of C. parapsilosis orthologs of C. albicans biofilm regulators. They have found that both Bcr1 and Efg1 are required for C. parapsilosis biofilm formation (Ding and Butler 2007; Ding et al. 2011; Connolly et al. 2013). Bcr1 has analogous functions, to some extent, in both species; the bcr1Δ/Δ mutant C. parapsilosis biofilms have reduced biomass, and there is no defect in morphogenesis of the filamentous cells—pseudohyphae—that this species produces (Ding and Butler 2007; Ding et al. 2011). However, there is limited overlap among Bcr1 target genes in the two species. The shared targets include cell surface CFEM protein genes, an expanded gene family in C. parapsilosis that is involved in heme-iron acquisition (Ding and Butler 2007; Almeida et al. 2009). However, orthologs of the C. albicans biofilm adhesin genes ALS1, ALS3, and HWP1 are not dependent on Bcr1 for expression in C. parapsilosis (Ding and Butler 2007; Ding et al. 2011). In fact, the Bcr1-dependent genes that govern C. parapsilosis biofilm formation are unknown at present; several CFEM protein mutants tested do not affect biofilm biomass or structure (Ding et al. 2011). The Efg1 orthologs of C. parapsilosis and C. albicans have greater overlap among their target genes (Connolly et al. 2013), which include several transcription factor genes that are members of the C. albicans biofilm regulatory network (Nobile et al. 2012). The emerging conclusion from these studies is that biofilm regulators are conserved, whereas their target genes are not. One caveat is that there may be many examples of transcription factors whose target genes are contingent on the environment; as noted by the Butler group, the number of Bcr1- and Efg1-dependent genes detected in different studies of C. albicans varied greatly (Ding et al. 2011; Fanning et al. 2012b; Nobile et al. 2012; Connolly et al. 2013).
BIOFILM EXTRACELLULAR MATRIX
Production of an extracellular matrix is one of the distinguishing characteristics of both eukaryotic and prokaryotic biofilms (Hawser et al. 1998; Donlan and Costerton 2002; O’Toole 2003). In many organisms, this material is critical for biofilm formation, providing the scaffold for surface adhesion and cellular aggregation to maintain the biofilm architecture (Flemming and Wingender 2010). Extracellular matrix has also been shown in some organisms to assist with retention of water and absorption of nutrients. In addition, the material may be degraded during nutrient-limiting conditions by microbially produced enzymes to provide a carbon or nitrogen source (Flemming and Wingender 2010). One of the most medically important attributes of the extracellular matrix is its ability to provide protection from environmental insults, including host defenses and antimicrobial therapies (Costerton et al. 1999; Donlan 2001).
Hawser et al. (1998) first described the presence of C. albicans biofilm matrix material and its variation with environmental conditions. The clinical relevance of this material is evident by its presence on the catheters of patients with Candida biofilm (Paulitsch et al. 2009). Animal models of device-associated infections also reveal abundant biofilm matrix material and provide an opportunity to study matrix function (Andes et al. 2004; Nett et al. 2010b; Johnson et al. 2012). In vivo, the matrix is postulated to contain both the microbial-derived components identified in vitro, as well as host proteins, such as plasma and salivary proteins that adsorb to the surface of medical devices (Nett and Andes 2006).
The Douglas group has performed several key investigations to examine the content of Candida biofilm matrix (Table 1) (Baillie and Douglas 2000). They then probed the functions of matrix components in adherence through enzymatic digestion and measurement of cellular detachment (Al-Fattani and Douglas 2006). This study pointed to roles for matrix protein, chitin, extracellular DNA (eDNA), and β-1,3 glucan in surface adhesion or cellular cohesion.
Matrix protein constituents have been identified by Lopez-Ribot and colleagues through 2D PAGE and mass spectrometry (Thomas et al. 2006). Surprisingly, the most abundant biofilm matrix proteins were very similar to those in planktonic culture supernatants. Thus, matrix protein function during in vitro growth reflects a change in the state of secreted proteins rather than a change in their identity. Matrix proteins included both predicted secretome components, as well as many predicted cytoplasmic proteins. It seems likely that cell lysis contributes to the matrix through release of proteins and DNA. Although matrix eDNA has a role in biofilm structure and drug susceptibility (discussed below), the roles of the individual matrix proteins in biofilm properties is not well understood.
Interestingly, the matrix content appears to vary significantly among Candida species (Al-Fattani and Douglas 2006; Silva et al. 2009). For example, compared with C. albicans, Candida tropicalis biofilms have higher hexosamine content (27%) and are not disrupted by deoxyribonuclease (DNase) treatment (Al-Fattani and Douglas 2006). Also, matrix composition is dependent on environmental and media conditions. For example, C. albicans biofilms formed in RPMI (Roswell Park Memorial Institute) medium have an ∼1000×-fold higher concentration of matrix eDNA when compared with yeast nitrogen base medium (Martins et al. 2010).
Like Candida, Aspergillus biofilms have been linked to infection of medical devices (Ramage et al. 2011). However, biofilm-like growth has also been associated with the most common Aspergillus infections, including invasive pulmonary aspergillosis and aspergilloma (Loussert et al. 2010). As a clinical niche, the lung varies greatly from the environments for most Candida biofilm infections. The ability of the most common Aspergillus species, A. fumigatus, to form a multicellular community with a surrounding extracellular matrix material has been shown both in vitro and in vivo (Beauvais et al. 2007; Loussert et al. 2010). Ultrastructure analysis shows that these biofilms are composed of hyphae coalescing into a three-dimensional structure covered in dense extracellular material with embedded air channels (Beauvais et al. 2007).
Beauvais et al. (2007) found that the extracellular matrix of in vitro A. fumigatus biofilms is composed of galactomannan, α-1,3 glucan, monosaccharides, polyols, melanin, and protein (Table 1). Immunolabeling showed that galactomannan was deposited throughout the cell wall and matrix. In contrast, the α-1,3 glucan label was the highest in the amorphous extracellular matrix material near the hyphal surface. The protein fraction of matrix was small (2%) and included several major secreted antigens, as well as a group of hydrophobins. These hydrophobic surface-active proteins, commonly associated with aerial growth, are presumed to play a role in cell–cell adhesion in the biofilm. Recent analysis also has shown that eDNA accumulates in the matrix of A. fumigatus biofilms in vitro (Rajendran et al. 2013; Shopova et al. 2013). This eDNA is identical to genomic DNA and is probably released through autolysis (Rajendran et al. 2013).
By studying the resected aspergillomas of several patients, Loussert et al. (2010) shed light on the in vivo composition of A. fumigatus biofilms. Similar to in vitro biofilms, galactomannan was identified throughout the fungal cell wall and extracellular matrix of in vivo biofilms on immunolabeling. Also, α-1,3 glucan was found in the matrix material located most proximal to the cell wall surface. Compared with in vitro biofilms, in vivo biofilms appeared to have a higher concentration of the recently identified cell wall polysaccharide galactosaminogalactan. Recent infection model studies indicate that galactosaminogalactan mediates adherence, but also masks β-1,3 glucan to limit host recognition (Gravelat et al. 2013). The presence of melanin in the biofilm matrix was confirmed in vivo, whereas antigenic proteins were not as apparent in vivo. When comparing the aspergillomas of patients with a murine model of invasive pulmonary aspergillosis, many similarities were noted, including the presence of both matrix galactomannan and galactosaminogalactan (Loussert et al. 2010). However, unlike in vitro biofilms and the aspergilloma biofilm, the extracellular matrix of the pulmonary aspergillosis biofilms did not appear to have a high concentration of α-1,3 glucan.
Candida MATRIX AND BIOFILM DRUG RESISTANCE
Several investigations have examined the contribution of the biofilm extracellular matrix to the drug resistance phenotype associated with the Candida biofilm lifestyle (Baillie and Douglas 2000; Al-Fattani and Douglas 2004, 2006; Samaranayake et al. 2005). These studies have used several experimental designs to test the hypothesis that matrix may prevent access of antifungals to the cells embedded in the biofilm, postulating that the material may either slow the rate of drug transport or specifically bind antifungals extracellularly. Together, these investigations found that an intact matrix is one of the most influential factors promoting the drug resistance of biofilms formed by different Candida species.
Early studies from the Douglas group implicated the C. albicans biofilm matrix in drug resistance (Baillie and Douglas 2000; Al-Fattani and Douglas 2004, 2006). To explore the possibility that matrix impedes drug access to biofilm cells, Samaranayake et al. (2005) measured drug penetration using filter-grown biofilms. C. albicans biofilms inhibited the penetration of all antifungals tested, including fluconazole, amphotericin B, and flucytosine. C. parapsilosis and Candida krusei biofilms also impeded amphotericin B, but flucytosine and fluconazole penetration was higher than for C. albicans biofilms. Hence, this investigation suggests that the matrix contributes biofilm antifungal resistance by limiting drug diffusion, although it is unclear that affecting the diffusion gradient would be sufficient to abrogate drug sensitivity entirely.
Several key observations led to the discovery of a drug sequestration activity for the Candida biofilm matrix. The Andes group added isolated Candida biofilm matrix to planktonic minimum growth inhibition concentration assays and found that this material was able to provide a degree of antifungal resistance to the nonbiofilm cells (Nett et al. 2007). To test the hypothesis that the matrix interacts with antifungals, preventing drug penetration into the cells, radiolabeled fluconazole was tracked through biofilms. The vast majority of the drug was found in association with matrix, implicating drug sequestration. To explore the contribution of individual matrix components, the biofilm matrix was exposed to enzymes targeting the constituents. Degradation of matrix β-1,3 glucan impaired the biofilm-associated resistance mechanism, indicating a role for this matrix component in protecting biofilms from antifungal drugs. The role for matrix β-1,3 glucan in biofilm drug resistance during clinical infection was supported by the synergistic action of glucanase and fluconazole on rat venous catheters infected with C. albicans (Nett et al. 2007).
Matrix β-1,3 glucan has been linked to resistance with other antifungals in distinct drug classes. Vediyappan et al. (2010) have provided evidence that matrix β-1,3 glucan binds and sequesters amphotericin B. A similar resistance mechanism has also been shown for flucytosine (Nett et al. 2010a). Furthermore, biofilms formed by other common Candida species, including Candida glabrata, C. parapsilosis, and C. tropicalis, also produce matrix β-1,3 glucan that sequesters antifungal and enhances drug resistance (Mitchell et al. 2013). Therefore, drug sequestration by matrix β-1,3 glucan is a common mechanism of biofilm-based resistance.
There are additional mechanisms that promote C. albicans biofilm drug resistance. For example, several efflux pump genes are up-regulated in biofilms (Mukherjee et al. 2003). The efflux pumps have a detectable role in fluconazole resistance in early biofilms, but their impact is more minor in mature biofilms with extensive matrix (Mukherjee et al. 2003). Finally, one of the most widespread biofilm resistance phenomena in bacteria, the presence of a small resistant subpopulation of “persister” cells (Lewis 2010), occurs in C. albicans biofilms as well (LaFleur et al. 2006). Interestingly, a study from the Thevissen group showed that cell surface superoxide dismutases are critical for the accumulation of persister cells (Bink et al. 2011). This study could be extended to other fungal biofilms readily through the use of superoxide dismutase inhibitors (Bink et al. 2011). Thus, biofilm drug resistance is clearly multifactorial, a “perfect storm” of matrix binding, efflux pump up-regulation, and persister cell accumulation.
Candida MATRIX GLUCAN PRODUCTION
What are the genes that govern matrix β-1,3 glucan production? In C. albicans, FKS1 encodes a glucan synthase responsible for manufacture of cell wall β-1,3 glucan during planktonic growth. Nett et al. (2010a,b) showed the role of FKS1 in matrix-mediated drug sequestration with two key observations: reduced expression of FKS1 expression causes reduced matrix β-1,3 glucan and increased sensitivity of biofilm cells to fluconazole, and increased expression of FKS1 increased matrix β-1,3 glucan and decreased sensitivity of biofilm cells to fluconazole. Fluconazole sensitivity was unaffected when the strains were grown under nonbiofilm conditions (Nett et al. 2010a,b). The phenotypes were shown both in vitro and in a rat venous catheter model of biofilm infection. Thus, Fks1 is required to generate matrix β-1,3 glucan and full levels of biofilm drug resistance.
To delineate the process by which glucan is modified and delivered to the extracellular biofilm matrix, Taff et al. (2012) used a candidate gene approach, examining a set of mutants based on transcriptional profiling of C. albicans in a rat venous catheter biofilm. They identified a role for two predicted glucan transferases (Bgl2 and Phr1) and an exoglucanase (Xog1) in both the production of biofilm matrix and development of the resistance to antifungals during biofilm growth. Mutants with disruption of one of these genes produced significantly less matrix β-1,3 glucan and displayed increased biofilm susceptibility to azole drugs compared with the parent strain both in vitro and in a rat venous catheter biofilm model. The isolated matrix of the mutant strains showed decreased capacity for antifungal sequestration. The gene products appear to have partially redundant functions, as the matrix sequestration defect was more extreme in double mutants than in single mutants. These gene products are proposed to modify matrix glucan generated by Fks1 for development of functional matrix capable of sequestering antifungal drugs and promoting biofilm antifungal drug resistance.
Several pathways regulate production of Candida biofilm matrix and drug resistance. Nobile et al. (2009) discovered a critical negative regulator of C. albicans biofilm matrix glucan production both in vitro and in vivo. In a screen of transcription factor mutants, the zap1–/– mutant was noted to have a glistening appearance that correlated with abundant matrix glucan. Through transcription profiling and chromatin immunoprecipitation, several groups of target genes were identified. First, Zap1 was found to repress transcription of two glucoamylase genes (GCA1 and GCA2) that may have a positive role in matrix production, likely by hydrolyzing insoluble carbohydrates. A second group of Zap1 targets includes three alcohol dehydrogenase genes (ADH5, CSH1, and LFD6) that may influence biofilm formation and matrix glucan production through quorum signal pathways. Interestingly, the expression of BGL2, PHR1, and XOG1 do not appear to be under Zap1 control (Taff et al. 2012). Thus, Zap1 may govern matrix production through mechanisms that feed into, or converge with, the FKS1 pathway defined above.
The Cowen group discovered heat shock protein, Hsp90, as novel regulator of C. albicans biofilm matrix production and drug resistance (Robbins et al. 2011). Disruption of this pathway increased the effectiveness of triazole drugs both in vitro and in a rat venous catheter model. Unlike planktonic conditions, this pathway was not modulated via calcineurin or Mkc1 pathways. The finding that inhibition of Hsp90p activity led to decreased β-1,3 glucan in the matrix of C. albicans biofilms suggests that Hsp90p contributes to the production of drug sequestering matrix. How Hsp90p specifically modulates this activity is unknown, but it is hypothesized to stabilize one or more of the proteins involved in production and regulation of matrix production, such as Fks1 or Zap1.
A search for additional regulators of biofilm matrix-associated drug resistance identified downstream components of the yeast protein kinase C (PKC) pathway (Nett et al. 2011). C. albicans mutants with disruption of either SMI1 or RLM1 were deficient in both matrix glucan production and susceptibility to azole drugs. These factors are thought to govern matrix-associated drug resistance through FKS1, as FKS1 overexpression restored the biofilm-associated drug-resistant phenotype. Surprisingly, upstream PKC pathway components do not appear to be involved in regulation of biofilm matrix production and drug resistance. This suggests that matrix-associated drug resistance and cell wall integrity are networked, but the biofilm drug resistance is triggered through a pathway distinct from the PKC pathway. The nature of the crosstalk that engages PKC pathway downstream components independently of PKC pathway upstream components is not understood at this time.
The Soll laboratory has also explored the genetic control of a panel of matrix-related phenotypes. They found that a mating-competent MTLa/MTLa C. albicans biofilm differs from a mating incompetent MTLa/MTLα biofilm in being more permeable to dyes and neutrophils and more sensitive to fluconazole (Srikantha et al. 2013). Their genetic studies indicated that in the MTLa/MTLa strain reduced activity of the Bcr1 transcription factor, a known positive regulator of biofilm formation, was the cause of the three phenotypes of dye and neutrophil permeability and fluconazole sensitivity (Yi et al. 2011). Accordingly, they developed an overexpression approach to identify several Bcr1 target genes that govern the three phenotypes (Srikantha et al. 2013). Notable among the gene products whose overexpression increases the level of dye and neutrophil permeability and fluconazole sensitivity were a set of cell surface proteins with the CFEM motif (Srikantha et al. 2013), which has been shown to mediate iron acquisition. The three phenotypes are all probably related to extracellular matrix accumulation or composition. Consistent with this idea, Noverr and colleagues have shown that Bcr1 is required for matrix accumulation in a vaginal colonization model (Harriott et al. 2010). It will be interesting to see whether the CFEM proteins have a structural role in the extracellular matrix, or if their effects are more indirect.
MATRIX eDNA AND DRUG RESISTANCE
The presence of eDNA in the Candida biofilm matrix is critical for biofilm integrity and maintenance (Martins et al. 2010). Degradation of matrix eDNA not only destroys biofilm architecture, but the addition of exogenous DNA promotes biofilm growth (Martins et al. 2010). Martins et al. (2012) explored the relationship between eDNA and C. albicans biofilm drug resistance through DNase treatment. Degradation of eDNA significantly improved the activity of amphotericin B against Candida biofilms, based on either metabolic activity measurements or cell viability assays. However, biofilms treated with DNase remained resistant to both fluconazole and caspofungin. This suggests that there may be a specific eDNA–amphotericin B interaction. An alternative possibility is that the matrix eDNA may interact with a variety of antifungals, but other resistance processes in play maintain biofilm resistance on eDNA degradation. How matrix eDNA contributes to Candida biofilm resistance remains unknown.
Aspergillus BIOFILM MATRIX AND DRUG RESISTANCE
Considerably less is known about the process of Aspergillus biofilm matrix production and how it impacts drug susceptibility during biofilm growth. The matrix of Aspergillus biofilms is thought to contribute to antimicrobial resistance, through either slowing drug transit or direct drug sequestration (Beauvais et al. 2007; Bugli et al. 2013; Rajendran et al. 2013). Extracellular matrix content increases as A. fumigatus biofilms mature, so matrix is thought to promote drug resistance during the later phases of biofilm growth (Beauvais et al. 2007).
One recently recognized matrix component shown to impact drug resistance in A. fumigatis is eDNA. Rajendran et al. (2013) showed that the content of genomic DNA in the matrix increased as in vitro A. fumigatus biofilms matured from 8–48 h. DNA release was linked to chitinase activity, suggesting that autolysis is the source of eDNA. Degradation of eDNA by DNase decreases the biomass of the biofilm and renders biofilms more susceptible both to amphotericin B and caspofungin, an echinocandin. Interestingly, exogenous DNA also promotes structural integrity of the biofilm and increases biofilm matrix carbohydrate production, suggesting a potential role for host DNA during in vivo Aspergillus biofilm formation (Rajendran et al. 2013; Shopova et al. 2013). Therefore, matrix eDNA appears to be important for both biofilm structural integrity and resistance to antifungals in A. fumigatus.
Much remains unknown about Aspergillus biofilm drug resistance and how the different components of the biofilm matrix influence this phenotype. An intriguing study by Bugli et al. (2013) found that administration of alginate lyase increased in vitro A. fumigatus biofilm susceptibility to amphotericin B. Alginate lyase degrades uronic-acid-containing carbohydrates and is thought to act on the extracellular matrix carbohydrates of Aspergillus biofilms. This study suggests that a previously unidentified matrix carbohydrate is contributing to antifungal drug resistance during biofilm growth. Further investigation is of interest to characterize this material and its specific role in resistance to antifungal treatment.
CONCLUDING REMARKS
The ability to form biofilms is widespread among pathogenic fungi. It is relevant to infection biology in the context of device-associated infection and as an in vivo growth form. Despite the broad significance of fungal biofilms, our understanding of the mechanisms that govern their formation, physiology, and drug resistance is limited. The genetic tractability of C. albicans has driven biofilm studies in that organism, and we now have candidate genes and processes that might be tested in other fungal pathogens. Molds and basidiomycetes may have distinct processes that mediate biofilm formation and our growing understanding of A. fumigatus and C. neoformans biofilms provides concepts to fuel testable hypotheses with related fungi.
There are many major questions on the horizon of fungal biofilm studies. For example, cell heterogeneity is an important attribute of all biofilms but, apart from recognizing yeast and hyphae, we have little understanding of how extensive cell heterogeneity may be in biofilms, how biofilm growth generates heterogeneity, and how heterogeneity may contribute to biofilm persistence or functional adaptation. Our understanding of mixed species interactions in biofilms remains in its infancy, and we suspect that computational analysis will be necessary to distill the bewildering spectrum of species and relative abundances of different organisms into meaningful scenarios for reductionist analysis. Relevant to both single-species and mixed-species biofilms, we have yet to define many of the extracellular small molecules that have a major impact on biofilm growth and integrity, yet small molecules are among the most promising targets for therapeutic intervention. Finally, the appreciation that biofilm growth is distinct from planktonic growth has driven new efforts to develop biofilm-based screens for antifungal activities (see Srinivasan et al. 2013, for example). Such efforts will be critical as the use of implanted devices and immunosuppressive therapies increases in the future.
ACKNOWLEDGMENTS
Biofilm research in our laboratories is supported by NIH Grants R01 AI073289 (D.R.A.), R01 AI067703 (A.P.M), and a Stupakoff fellowship (J.V.D.).
REFERENCES
- Albuquerque P, Nicola AM, Nieves E, Paes HC, Williamson PR, Silva-Pereira I, Casadevall A. 2013. Quorum sensing-mediated, cell density-dependent regulation of growth and virulence in Cryptococcus neoformans. MBio 5: e00986-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Fattani MA, Douglas LJ. 2004. Penetration of Candida biofilms by antifungal agents. Antimicrob Agents Chemother 48: 3291–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Fattani MA, Douglas LJ. 2006. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J Med Microbiol 55: 999–1008 [DOI] [PubMed] [Google Scholar]
- Almeida RS, Wilson D, Hube B. 2009. Candida albicans iron acquisition within the host. FEMS Yeast Res 9: 1000–1012 [DOI] [PubMed] [Google Scholar]
- Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun 72: 6023–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas LJ. 2000. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J Antimicrob Chemother 46: 397–403 [DOI] [PubMed] [Google Scholar]
- Banerjee U, Gupta K, Venugopal P. 1997. A case of prosthetic valve endocarditis caused by Cryptococcus neoformans var. neoformans. J Med Vet Mycol 35: 139–141 [PubMed] [Google Scholar]
- Beauvais A, Schmidt C, Guadagnini S, Roux P, Perret E, Henry C, Paris S, Mallet A, Prevost MC, Latge JP. 2007. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell Microbiol 9: 1588–1600 [DOI] [PubMed] [Google Scholar]
- Bink A, Vandenbosch D, Coenye T, Nelis H, Cammue BP, Thevissen K. 2011. Superoxide dismutases are involved in Candida albicans biofilm persistence against miconazole. Antimicrob Agents Chemother 55: 4033–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante L, Nalesso F, Cara M, Antonello A, Malagoli A, Pastori G, Guizzo M, D’Angelo A, Gambaro G. 2005. Aspergillus fumigatus peritonitis in ambulatory peritoneal dialysis: A case report and notes on the therapeutic approach. Nephrology (Carlton) 10: 270–273 [DOI] [PubMed] [Google Scholar]
- Bonhomme J, d’Enfert C. 2013. Candida albicans biofilms: Building a heterogeneous, drug-tolerant environment. Curr Opin Microbiol 16: 398–403 [DOI] [PubMed] [Google Scholar]
- Bonhomme J, Chauvel M, Goyard S, Roux P, Rossignol T, d’Enfert C. 2011. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol Microbiol 80: 995–1013 [DOI] [PubMed] [Google Scholar]
- Braun DK, Janssen DA, Marcus JR, Kauffman CA. 1994. Cryptococcal infection of a prosthetic dialysis fistula. Am J Kidney Dis 24: 864–867 [DOI] [PubMed] [Google Scholar]
- Bugli F, Posteraro B, Papi M, Torelli R, Maiorana A, Paroni Sterbini F, Posteraro P, Sanguinetti M, De Spirito M. 2013. In vitro interaction between alginate lyase and amphotericin B against Aspergillus fumigatus biofilm determined by different methods. Antimicrob Agents Chemother 57: 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YY, Cao YB, Xu Z, Ying K, Li Y, Xie Y, Zhu ZY, Chen WS, Jiang YY. 2005. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob Agents Chemother 49: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E. 1982. Synergistic effect of Candida albicans and Staphylococcus aureus on mouse mortality. Infect Immun 38: 921–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. J Bacteriol 183: 5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly LA, Riccombeni A, Grozer Z, Holland LM, Lynch DB, Andes DR, Gacser A, Butler G. 2013. The APSES transcription factor Efg1 is a global regulator that controls morphogenesis and biofilm formation in Candida parapsilosis. Mol Microbiol 90: 36–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: A common cause of persistent infections. Science 284: 1318–1322 [DOI] [PubMed] [Google Scholar]
- Cugini C, Calfee MW, Farrow JM III, Morales DK, Pesci EC, Hogan DA. 2007. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol 65: 896–906 [DOI] [PubMed] [Google Scholar]
- Cushion MT, Collins MS, Linke MJ. 2009. Biofilm formation by Pneumocystis spp. Eukaryot Cell 8: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KJ, Park YN, Srikantha T, Pujol C, Soll DR. 2013. Impact of environmental conditions on the form and function of Candida albicans biofilms. Eukaryot Cell 12: 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darouiche RO. 2001. Device-associated infections: A macroproblem that starts with microadherence. Clin Infect Dis 33: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298 [DOI] [PubMed] [Google Scholar]
- Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. 2008. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol 67: 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai JV, Bruno VM, Ganguly S, Stamper RJ, Mitchell KF, Solis N, Hill EM, Xu W, Filler SG, Andes DR, et al. 2013. Regulatory role of glycerol in Candida albicans biofilm formation. MBio 4: e00637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bonaventura G, Pompilio A, Picciani C, Iezzi M, D’Antonio D, Piccolomini R. 2006. Biofilm formation by the emerging fungal pathogen Trichosporon asahii: Development, architecture, and antifungal resistance. Antimicrob Agents Chemother 50: 3269–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Butler G. 2007. Development of a gene knockout system in Candida parapsilosis reveals a conserved role for BCR1 in biofilm formation. Eukaryot Cell 6: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Vidanes GM, Maguire SL, Guida A, Synnott JM, Andes DR, Butler G. 2011. Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS ONE 6: e28151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A. 2008. Pathogenesis of mucosal biofilm infections: Challenges and progress. Expert Rev Anti Infect Ther 6: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM. 2001. Biofilms and device-associated infections. Emerg Infect Dis 7: 277–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM. 2002. Biofilms: Microbial life on surfaces. Emerg Infect Dis 8: 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. 2002. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15: 167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Mitchell AP. 2012. Fungal biofilms. PLoS Pathog 8: e1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Xu W, Beaurepaire C, Suhan JP, Nantel A, Mitchell AP. 2012a. Functional control of the Candida albicans cell wall by catalytic protein kinase A subunit Tpk1. Mol Microbiol 86: 284–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. 2012b. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell 11: 896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Mitchell AP. 2011. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, et al. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog 8: e1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8: 623–633 [DOI] [PubMed] [Google Scholar]
- Ganguly S, Mitchell AP. 2011. Mucosal biofilms of Candida albicans. Curr Opin Microbiol 14: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S, Bishop AC, Xu W, Ghosh S, Nickerson KW, Lanni F, Patton-Vogt J, Mitchell AP. 2011. Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot Cell 10: 1448–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d’Enfert C. 2004. Candida albicans biofilms: A developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 3: 536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons JG, Beauvais A, Beau R, McGary KL, Latge JP, Rokas A. 2012. Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot Cell 11: 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat FN, Beauvais A, Liu H, Lee MJ, Snarr BD, Chen D, Xu W, Kravtsov I, Hoareau CM, Vanier G, et al. 2013. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β-glucan from the immune system. PLoS Pathog 9: e1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, Turner KJ, Chaloupka J, Cottier F, De Sordi L, Sanglard D, Levin LR, Buck J, Muhlschlegel FA. 2011. The quorum-sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans. Eukaryot Cell 10: 1034–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108 [DOI] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC. 2009. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob Agents Chemother 53: 3914–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC. 2011. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol 19: 557–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Lilly EA, Rodriguez TE, Fidel PL Jr, Noverr MC. 2010. Candida albicans forms biofilms on the vaginal mucosa. Microbiology 156: 3635–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser SP, Douglas LJ. 1994. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun 62: 915–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser SP, Baillie GS, Douglas LJ. 1998. Production of extracellular matrix by Candida albicans biofilms. J Med Microbiol 47: 253–256 [DOI] [PubMed] [Google Scholar]
- Jabra-Rizk MA, Meiller TF, James CE, Shirtliff ME. 2006. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother 50: 1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeloka TK, Shrividya S, Wagholikar G. 2011. Catheter outflow obstruction due to an aspergilloma. Perit Dial Int 31: 211–212 [DOI] [PubMed] [Google Scholar]
- Johnson CC, Yu A, Lee H, Fidel PL Jr, Noverr MC. 2012. Development of a contemporary animal model of Candida albicans-associated denture stomatitis using a novel intraoral denture system. Infect Immun 80: 1736–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic EM, Darouiche RO. 2004. Candida infections of medical devices. Clin Microbiol Rev 17: 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP. 2006. Microbial sciences: The superficial life of microbes. Nature 441: 300–302 [DOI] [PubMed] [Google Scholar]
- Krzossok S, Birck R, Henke S, Hof H, van der Woude FJ, Braun C. 2004. Trichosporon asahii infection of a dialysis PTFE arteriovenous graft. Clin Nephrol 62: 66–68 [DOI] [PubMed] [Google Scholar]
- Kucharikova S, Tournu H, Lagrou K, Van Dijck P, Bujdakova H. 2011. Detailed comparison of Candida albicans and Candida glabrata biofilms under different conditions and their susceptibility to caspofungin and anidulafungin. J Med Microbiol 60: 1261–1269 [DOI] [PubMed] [Google Scholar]
- Kuhn DM, Ghannoum MA. 2004. Candida biofilms: Antifungal resistance and emerging therapeutic options. Curr Opin Investig Drugs 5: 186–197 [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. 2005. Alternative Candida albicans lifestyles: Growth on surfaces. Annu Rev Microbiol 59: 113–133 [DOI] [PubMed] [Google Scholar]
- LaFleur MD, Kumamoto CA, Lewis K. 2006. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother 50: 3839–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattif AA, Mukherjee PK, Chandra J, Swindell K, Lockhart SR, Diekema DJ, Pfaller MA, Ghannoum MA. 2010. Characterization of biofilms formed by Candida parapsilosis, C. metapsilosis, and C. orthopsilosis. Int J Med Microbiol 300: 265–270 [DOI] [PubMed] [Google Scholar]
- Lewis K. 2010. Persister cells. Annu Rev Microbiol 64: 357–372 [DOI] [PubMed] [Google Scholar]
- Lewis RE, Lo HJ, Raad II, Kontoyiannis DP. 2002. Lack of catheter infection by the efg1/efg1 cph1/cph1 double-null mutant, a Candida albicans strain that is defective in filamentous growth. Antimicrob Agents Chemother 46: 1153–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loussert C, Schmitt C, Prevost MC, Balloy V, Fadel E, Philippe B, Kauffmann-Lacroix C, Latge JP, Beauvais A. 2010. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol 12: 405–410 [DOI] [PubMed] [Google Scholar]
- Lynch AS, Robertson GT. 2008. Bacterial and fungal biofilm infections. Annu Rev Med 59: 415–428 [DOI] [PubMed] [Google Scholar]
- Madhani HD. 2011. Quorum sensing in fungi: Q&A. PLoS Pathog 7: e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez LR, Casadevall A. 2007. Cryptococcus neoformans biofilm formation depends on surface support and carbon source and reduces fungal cell susceptibility to heat, cold, and UV light. Appl Environ Microbiol 73: 4592–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez LR, Fries BC. 2010. Fungal biofilms: Relevance in the setting of human disease. Curr Fungal Infect Rep 4: 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M, Uppuluri P, Thomas DP, Cleary IA, Henriques M, Lopez-Ribot JL, Oliveira R. 2010. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 169: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M, Henriques M, Lopez-Ribot JL, Oliveira R. 2012. Addition of DNase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses 55: 80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, Loss O, Cairns T, Goldman G, Armstrong-James D, et al. 2008. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog 4: e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AP. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev 58: 56–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KF, Taff HT, Cuevas MA, Reinicke EL, Sanchez H, Andes DR. 2013. Role of matrix β-1,3 glucan in antifungal resistance of non-albicans Candida biofilms. Antimicrob Agents Chemother 57: 1918–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales DK, Hogan DA. 2010. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog 6: e1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat E, Williams C, Jones B, McChlery S, Ramage G. 2009. The characteristics of Aspergillus fumigatus mycetoma development: Is this a biofilm? Med Mycol 47 (Suppl 1): S120–S126 [DOI] [PubMed] [Google Scholar]
- Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. 2003. Mechanism of fluconazole resistance in Candida albicans biofilms: Phase-specific role of efflux pumps and membrane sterols. Infect Immun 71: 4333–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo LA, Newport G, Lan CY, Habelitz S, Dungan J, Agabian NM. 2005. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot Cell 4: 1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszkieta L, Beauvais A, Pahtz V, Gibbons JG, Anton Leberre V, Beau R, Shibuya K, Rokas A, Francois JM, Kniemeyer O, et al. 2013. Investigation of Aspergillus fumigatus biofilm formation by various “omics” approaches. Front Microbiol 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett J, Andes D. 2006. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol 9: 340–345 [DOI] [PubMed] [Google Scholar]
- Nett J, Lincoln L, Marchillo K, Massey R, Holoyda K, Hoff B, VanHandel M, Andes D. 2007. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother 51: 510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Crawford K, Marchillo K, Andes DR. 2010a. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob Agents Chemother 54: 3505–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Sanchez H, Cain MT, Andes DR. 2010b. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J Infect Dis 202: 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. 2011. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell 10: 1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson KW, Atkin AL, Hornby JM. 2006. Quorum sensing in dimorphic fungi: Farnesol and beyond. Appl Environ Microbiol 72: 3805–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Vickerman MM, Jenkinson HF. 2010. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell 9: 1622–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 15: 1150–1155 [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog 2: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol 7: e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148: 126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA. 2003. To build a biofilm. J Bacteriol 185: 2687–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannanusorn S, Fernandez V, Romling U. 2013a. Prevalence of biofilm formation in clinical isolates of Candida species causing bloodstream infection. Mycoses 56: 264–272 [DOI] [PubMed] [Google Scholar]
- Pannanusorn S, Ramirez-Zavala B, Lunsdorf H, Agerberth B, Morschhauser J, Romling U. 2013b. Characterization of biofilm formation and the role of BCR1 in clinical isolates of Candida parapsilosis. Eukaryot Cell 13: 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YN, Daniels KJ, Pujol C, Srikantha T, Soll DR. 2013. Candida albicans forms a specialized “sexual” as well as “pathogenic” biofilm. Eukaryot Cell 12: 1120–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulitsch AH, Willinger B, Zsalatz B, Stabentheiner E, Marth E, Buzina W. 2009. In vivo Candida biofilms in scanning electron microscopy. Med Mycol 47: 690–696 [DOI] [PubMed] [Google Scholar]
- Peleg AY, Hogan DA, Mylonakis E. 2010. Medically important bacterial–fungal interactions. Nat Rev Microbiol 8: 340–349 [DOI] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. 2012a. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin Microbiol Rev 25: 193–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Ovchinnikova ES, Krom BP, Schlecht LM, Zhou H, Hoyer LL, Busscher HJ, van der Mei HC, Jabra-Rizk MA, Shirtliff ME. 2012b. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 158: 2975–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raad I. 1998. Intravascular-catheter-related infections. Lancet 351: 893–898 [DOI] [PubMed] [Google Scholar]
- Rajendran R, Williams C, Lappin DF, Millington O, Martins M, Ramage G. 2013. Extracellular DNA release acts as an antifungal resistance mechanism in mature Aspergillus fumigatus biofilms. Eukaryot Cell 12: 420–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. 2001. Biofilm formation by Candida dubliniensis. J Clin Microbiol 39: 3234–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL. 2002a. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol 68: 5459–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. 2002b. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett 214: 95–100 [DOI] [PubMed] [Google Scholar]
- Ramage G, Saville SP, Thomas DP, Lopez-Ribot JL. 2005. Candida biofilms: An update. Eukaryot Cell 4: 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Martinez JP, Lopez-Ribot JL. 2006. Candida biofilms on implanted biomaterials: A clinically significant problem. FEMS Yeast Res 6: 979–986 [DOI] [PubMed] [Google Scholar]
- Ramage G, Rajendran R, Gutierrez-Correa M, Jones B, Williams C. 2011. Aspergillus biofilms: Clinical and industrial significance. FEMS Microbiol Lett 324: 89–97 [DOI] [PubMed] [Google Scholar]
- Reddy BT, Torres HA, Kontoyiannis DP. 2002. Breast implant infection caused by Trichosporon beigelii. Scand J Infect Dis 34: 143–144 [DOI] [PubMed] [Google Scholar]
- Richard ML, Nobile CJ, Bruno VM, Mitchell AP. 2005. Candida albicans biofilm-defective mutants. Eukaryot Cell 4: 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins N, Uppuluri P, Nett J, Rajendran R, Ramage G, Lopez-Ribot JL, Andes D, Cowen LE. 2011. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog 7: e1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol T, Ding C, Guida A, d’Enfert C, Higgins DG, Butler G. 2009. Correlation between biofilm formation and the hypoxic response in Candida parapsilosis. Eukaryot Cell 8: 550–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, Soll DR. 2010. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: Insights into the evolution of new signal transduction pathways. PLoS Biol 8: e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaranayake YH, Ye J, Yau JY, Cheung BP, Samaranayake LP. 2005. In vitro method to study antifungal perfusion in Candida biofilms. J Clin Microbiol 43: 818–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopova I, Bruns S, Thywissen A, Kniemeyer O, Brakhage AA, Hillmann F. 2013. Extrinsic extracellular DNA leads to biofilm formation and colocalizes with matrix polysaccharides in the human pathogenic fungus Aspergillus fumigatus. Front Microbiol 4: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si H, Hernday AD, Hirakawa MP, Johnson AD, Bennett RJ. 2013. Candida albicans white and opaque cells undergo distinct programs of filamentous growth. PLoS Pathog 9: e1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Henriques M, Martins A, Oliveira R, Williams D, Azeredo J. 2009. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med Mycol 47: 681–689 [DOI] [PubMed] [Google Scholar]
- Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. 2011. Adherence and biofilm formation of non-Candida albicans Candida species. Trends Microbiol 19: 241–247 [DOI] [PubMed] [Google Scholar]
- Srikantha T, Daniels KJ, Pujol C, Kim E, Soll DR. 2013. Identification of genes upregulated by the transcription factor Bcr1 that are involved in impermeability, impenetrability, and drug resistance of Candida albicans a/α biofilms. Eukaryot Cell 12: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Leung KP, Lopez-Ribot JL, Ramasubramanian AK. 2013. High-throughput nano-biofilm microarray for antifungal drug discovery. MBio 4: pii: e00331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stichternoth C, Ernst JF. 2009. Hypoxic adaptation by Efg1 regulates biofilm formation by Candida albicans. Appl Environ Microbiol 75: 3663–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: Implications for drug resistance. PLoS Pathog 8: e1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DP, Bachmann SP, Lopez-Ribot JL. 2006. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics 6: 5795–5804 [DOI] [PubMed] [Google Scholar]
- Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Kohler JR, Kadosh D, Lopez-Ribot JL. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog 6: e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vediyappan G, Rossignol T, d’Enfert C. 2010. Interaction of Candida albicans biofilms with antifungals: Transcriptional response and binding of antifungals to β-glucans. Antimicrob Agents Chemother 54: 2096–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TJ, Schlegel R, Moody MM, Costerton JW, Salcman M. 1986. Ventriculoatrial shunt infection due to Cryptococcus neoformans: An ultrastructural and quantitative microbiological study. Neurosurgery 18: 373–375 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhai B, Lin X. 2012. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog 8: e1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tian X, Gyawali R, Lin X. 2013. Fungal adhesion protein guides community behaviors and autoinduction in a paracrine manner. Proc Natl Acad Sci 110: 11571–11576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA. 2013. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci 110: 848–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Sahni N, Daniels KJ, Lu KL, Srikantha T, Huang G, Garnaas AM, Soll DR. 2011. Alternative mating type configurations (a/α versus a/a or α/α) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol 9: e1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Daniels KJ, Oh SH, Green CB, Yeater KM, Soll DR, Hoyer LL. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152: 2287–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]