Classical genetic techniques have been used to elucidate the basis of many monogenic skin diseases (e.g., keratin mutations in epidermolysis bullosa simplex). Advanced approaches are now being used to dissect complex diseases.
Abstract
The skin is composed of a variety of cell types expressing specific molecules and possessing different properties that facilitate the complex interactions and intercellular communication essential for maintaining the structural integrity of the skin. Importantly, a single mutation in one of these molecules can disrupt the entire organization and function of these essential networks, leading to cell separation, blistering, and other striking phenotypes observed in inherited skin diseases. Over the past several decades, the genetic basis of many monogenic skin diseases has been elucidated using classical genetic techniques. Importantly, the findings from these studies has shed light onto the many classes of molecules and essential genetic as well as molecular interactions that lend the skin its rigid, yet flexible properties. With the advent of the human genome project, next-generation sequencing techniques, as well as several other recently developed methods, tremendous progress has been made in dissecting the genetic architecture of complex, non-Mendelian skin diseases.
The skin is the largest organ in the body, and serves as a protective barrier against injury, possessing both flexible and rigid properties governed by the types of molecules expressed in each cell that permit proper association and communication between individual cells. However, these properties are severely compromised in many inherited skin diseases, or genodermatoses—a broad class of dermatological abnormalities attributed to genetic mutations.
Several different cell types of both mesenchymal and epithelial origin make up the skin: the ectodermally derived epidermis is composed of keratinocytes, and the underlying dermis consists of a heterogeneous population of mesenchymal cells that includes fibroblasts that secrete the connective tissue matrix. In all stratified squamous epithelia, such as the epidermis, esophagus, and cornea, there are undifferentiated and differentiated keratinocytes expressing specific type I (acidic) and type II (basic) keratin intermediate filaments, as well as structural proteins required for intercellular adhesion.
Undifferentiated keratinocytes in the basal layer are in direct contact with the basement membrane that separates the epidermis from the underlying dermis, and they express important proteins required for this contact as well as the overall structural integrity of the membrane. Mutations have been identified in many of the genes encoding these structural proteins, underlying the pathologies of several genodermatoses, many of which have been recapitulated in mouse models harboring mutations in these same genes. Notably, the phenotypes in human genetic skin diseases as well as those observed in the corresponding mouse models have taught us that many of these molecules play important roles in other organs as well.
Throughout the past several decades, substantial progress has been made in elucidating the genes that underlie a variety of skin diseases and a wide range of phenotypes. Several classes of molecules have been directly implicated in the pathogenesis of these diseases, and the majority of these findings have been achieved through the use of classical genetic techniques, which will be covered extensively in this chapter. These techniques have been instrumental in defining the genetic basis of monogenic (single-gene) skin disorders. More recently, the genetic bases of more complex, non-Mendelian skin diseases have begun to emerge and several loci have been implicated in their pathogeneses.
With the advent of the Human Genome Project, genome-wide association studies (GWAS), and more recently, next-generation sequencing (NGS) techniques, geneticists have been able to more readily identify candidate loci and take a more integrative approach utilizing expression data and functional genomics to infer the consequence and function of many of the molecules perturbed in these complex diseases. Moreover, cutting-edge techniques in large-scale profiling of DNA methylation have been instrumental in revealing both global and more localized aberrant methylation patterns in many autoimmune-related skin diseases. Although most (if not all) of the mutations identified in monogenic skin diseases reside in protein-coding genes, recent evidence from large-scale expression (microarrays, RNA sequencing, etc.) studies have uncovered an important role for a class of small noncoding RNAs, microRNAs, in the pathogenesis of several complex skin diseases.
This chapter will cover the wide spectrum and discovery of genetic perturbances as well as unusual mechanisms underlying both monogenic and complex human skin diseases.
THE DISCOVERY OF GENETIC MUTATIONS UNDERLYING SEVERAL HUMAN MONOGENIC SKIN DISEASES
One overarching goal in the study of inherited skin diseases is to find the causative gene(s) responsible for the phenotype manifested in a given genodermatosis and define its role in maintaining the integrity of the skin. Several more recent methods have been developed over the past decade to accomplish this, but the classical genetic approaches used to map, sequence, and characterize mutations and their effects at the mRNA and protein levels have proven to be the most fruitful, identifying hundreds of genetic mutations in dozens of monogenic Mendelian skin diseases. In the downstream analysis, the biochemical and structural nature of the molecules, domains of expression, and functions have been determined using well-developed in situ, in vitro, and in vivo methods, including mouse models that recapitulated the disease phentoypes.
The Classical Genetic Approach Used to Identify Pathogenic Mutations
Two major approaches have been used to isolate the gene responsible for a given phenotype: functional and positional cloning. The suitability of each approach for a given disease depended on what prior biochemical information was known about the defective gene product, as well as whether the inherited disease was sufficiently prevalent to study multiple affected individuals within a kindred (Yamanishi 1996). Although functional cloning exploits the known role of the candidate gene based on biochemical and genetic functional assays to isolate (clone) the gene, followed by mapping it to a locus, positional cloning relies only on the inheritance of the disease among related individuals (i.e., using linkage or haplotype analysis) to find the common region (mapping the gene), followed by functional studies.
Functional Cloning of Genes Underlying Several Inherited Skin Diseases
Over the course of the past several decades, histological, ultrastructural, and immunological analyses have provided molecular clues into the defects underlying the pathologies of several inherited skin diseases. This information had greatly aided researchers in isolating the gene by knowing its function, using a “reverse-genetics” approach to first study it through the use of transgenic mice, and then mapping it to a locus. The functional cloning approach has thus been most suitable for skin diseases in which information on the defective gene product (i.e., a keratin intermediate filament) and its general biological function was available.
Keratins in epidermolysis bullosa simplex (EBS) and epidermolytic hyperkeratosis (EH)
A classic example of the functional cloning and reverse genetic approach lies in the discovery of mutations in keratins expressed in the basal and suprabasal layers of the epidermis that underlie the autosomal dominant blistering skin diseases, epidermolysis bullosa simplex (EBS), and epidermolytic hyperkeratosis (EH), respectively (Coulombe et al. 1991; Vassar et al. 1991; Fuchs et al. 1992). EBS is characterized by keratinocyte fragility within the basal layer that ultimately leads to blistering of the skin. Ultrastructural analyses of EBS patient skin had previously revealed abnormal subcellular localization and aggregation of keratin intermediate filaments, suggesting a role for keratins in the pathology of EBS (Pearson and Spargo 1961; Anton-Lamprecht and Schnyder 1982; Anton-Lamprecht 1983; Buchbinder et al. 1986).
Vassar et al. (1991) explored the relationship between keratin intermediate filaments and genetic skin diseases by generating a transgenic mouse, in which the human promoter for KRT14 (type I keratin, basally expressed) drives a truncated murine K14 sequence, eliminating the last 138 amino acids of the protein. The mutation behaved in a dominant negative fashion, because heterodimerization between type I and II keratins is essential for their proper function, and the intraepithelial blistering and cytolysis phenotypes of the newborn mutant mice accurately recapitulated those observed in EBS patients. Moreover, amplification and sequencing analysis of KRT14 transcripts from EBS patients revealed point mutations in a critical region of the protein, and functional in vitro studies showed perturbation of the keratin network and filament assembly in keratinocytes transfected with the mutated human cDNA sequence (Coulombe et al. 1991).
Similar to EBS is the rare ichthyosis, epidermolytic hyperkeratosis (EH; previously termed bullous congenital ichthyosiform erythroderma), characterized by cytolysis in the suprabasal layers of the epidermis and hyperproliferation of the basal keratinocytes. It had been known at the time that undifferentiated keratinocytes expressing KRT5 and KRT14 turn off these genes to then express KRT1 and KRT10, markers of terminal differentiation (Fuchs and Green 1980; Moll et al. 1982; Eichner et al. 1984; Roop et al. 1987; Kopan and Fuchs 1989). Given the similarities between EBS and EH pathology and ultrastructural findings, Fuchs et al. (1992) investigated whether the genetic basis of EH was a defect involving terminal differentiation of keratinocytes by generating dominant-negative Keratin 10 transgenic mice. The newborn mutant mice showed dry, blistering skin and cytolysis in the suprabasal layers, accurately phenocopying the defects observed in the EH pathology.
LAMA5 in junctional epidermolysis bullosa (JEB)
Another classic example of functional cloning used to identify and isolate skin disease genes includes the LAMA5 (LAMA3/LAMB3/LAMC2) genes in the junctional form of epidermolysis bullosa. In the junctional form of EB, characterized by blistering and erosions of the skin and mucous membranes, ultrastructural findings revealed abnormalities in the hemidesmosomes (structures connecting the basal keratinocytes to the basement membrane) (Tidman and Eady 1986), and immunostaining using antibodies against the basement membrane marker, Laminin-332 revealed an absence of immunoreactivity in the skin of junctional EB patients (Verrando et al. 1991; Meneguzzi et al. 1992; Vailly et al. 1994), making LAMA5 an excellent candidate gene for the disease. Subsequently, mutations were identified in the α3, β3, and γ2 chains of LAMA5 in patients with the lethal form (Herlitz) of junctional EB (Pulkkinen et al. 1994a,b; McGrath et al. 1995a), and linkage analysis in pedigrees showed a strong association between the LAMA5 gene and the disease phenotype (Aberdam et al. 1994), providing convincing evidence for the involvement of this gene in the pathology of junctional EB.
COL7A1 in dystrophic epidermolysis bullosa (DEB)
In the dominant and recessive forms of dystrophic EB (DEB), a severe mutilating genodermatosis, previous ultrastructural analysis of patient skin had revealed a defect in the anchoring fibrils that reside within the papillary dermis beneath the basement membrane (Tidman and Eady 1985; Uitto and Christiano 1992; Uitto et al. 1992), where these fibrils were scarce in the dominant forms and almost completely absent in the recessive forms. Because type VII collagen (COL7A1) is the major component of the anchoring fibrils (Burgeson et al. 1990; Uitto et al. 1992), it was hypothesized that a defect in this protein product could lead to the separation of the dermis from the epidermis at the level of the anchoring fibrils observed in DEB (Uitto and Christiano 1992). COL7A1 was then mapped to chromosome 3p21 (Parente et al. 1991) and intragenic as well as flanking polymorphic markers were identified (Christiano et al. 1994b), which provided the means for establishing strong linkage between the disease phenotype and the COL7A1 gene in several families (Ryynanen et al. 1991; Hovnanian et al. 1992). Subsequently, a large number of mutations were then identified in both the recessive and dominant forms of DEB, providing convincing evidence for COL7A1 as the gene underlying the pathology (Dang and Murrell 2008).
Positional Cloning of Genes Underlying Several Inherited Skin Diseases
For skin diseases in which the molecular defect was not known, but a clear inheritance pattern was observed among related individuals, positional cloning, or “forward genetics” was used (Fig. 1). This strategy entails the identification of the common region that completely cosegregates with the disease phenotype (mapping), isolation of the gene (cloning), followed by characterization of its function. One of the most widely used methods to accomplish this is linkage analysis, a tool that enables the mapping of a gene or locus by analyzing its segregation with genetic markers [i.e., microsatellite markers, SNPs (single nucleotide polymorphisms)] of known chromosomal position. The power of linkage is based on the genetic principles of segregation and independent assortment, such that two traits located on separate chromosomes (or very far apart on the same chromosome) segregate independently, and thus have a recombination fraction of 50%. The closer the traits are, the less frequent recombinants are found; a recombination fraction (RF) of 0 implies the traits are tightly linked. In pedigree analysis, LOD (logarithm of the odds) scores are generated to calculate the likelihood that two loci are linked (Morton 1955); positive scores indicate linkage (≥ 3.0) and negative scores indicate that linkage is less likely (≤ −2.0).
Figure 1.
The classical genetic approach of functional and positional cloning used to elucidate the genetic basis of many inherited skin diseases. In positional cloning, or “forward genetics,” inheritance of a given trait is prevalent enough in related individuals to identify the shared genetic region. The trajectory for this approach is mapping, identification of mutations, cloning, followed by functional characterization of the protein product. In functional cloning, or “reverse genetics,” information regarding the functional properties of the protein product is exploited to then isolate the gene and map it to a locus. The trajectory for this approach is functional characterization of the protein product, cloning, identification of mutations, followed by mapping the trait among related individuals.
Many disease genes underlying several human skin diseases have been identified using this method. Interestingly, many of these discoveries were made in a very short period of time, such that several reports on a given skin disease as well as the mouse model were published in the same year, as in the case of KRT14 underlying the pathology of EBS (Bonifas et al. 1991; Vassar et al. 1991).
KRT9 in epidermolytic palmoplantar keratoderma (EPPK)
In the case of epidemolytic palmoplantar keratoderma (EPPK), an autosomal dominant skin disease marked by hyperkeratosis on the palms and soles, the etiology had not previously determined through the use of ultrastructural or histological methods. Reis et al. (1992) mapped the gene to chr17q11-23 using linkage with microsatellite markers in a kindred of seven generations, obtaining a LOD score of Z = 6.66 at a recombination fraction (θ) of 0. Interestingly, the type I (acidic) keratin cluster of genes map to this region and the KRT9 gene was deduced to be the candidate gene because it mapped to this region and is the only keratin to be specifically expressed in the terminally differentiated epidermis of palms and soles—the sites of pathology for the EPPK phenotype (Reis et al. 1992). Importantly, subsequent investigation led to the identification of several mutations within the rod domain of the KRT9 gene in unrelated kindreds (Reis et al. 1994).
Calcium pumps in Hailey–Hailey and Darier’s disease
Position cloning was also useful in elucidating the etiology of another skin disease, Hailey–Hailey disease, in which there is a loss of cellular adhesion between epidermal cells (acathyolysis) resulting in crusted plaques, erosions, and blistering (Ikeda et al. 1994). Although ultrastructural analysis of the lesional areas revealed subcellular abnormalities with respect to the localization of keratin intermediate filaments, the gene encoding a protein involved in keratinocyte adhesion had not been identified. Linkage using microsatellite markers initially localized the gene to chromosome 3q with a maximum combined LOD score of 14.6 (θ = 0) in four families (Ikeda et al. 1994), and several groups in the following years (Peluso et al. 1995; Richard et al. 1995; Ikeda et al. 2001) narrowed the region down to chromosome 3q21-24, where the gene was eventually found to be ATP2C1, a calcium pump encoding the calcium/magnesium ATPase of the Golgi apparatus that is required for the integrity of the normal epidermal calcium gradient (Hu et al. 2000; Sudbrak et al. 2000).
Similar to Hailey–Hailey disease is another autosomal dominant genodermatosis, Darier’s disease, characterized by acanthyolysis (cell separation) of the suprabasal epidermal cells resulting in warty papules and plaques as well as nail abnormalities. Although electron microscopy analysis revealed structural defects, including abnormal subcellular distribution of desmosomal proteins, the pathophysiology remained unclear. Two groups initially and simultaneously mapped Darier’s disease to chromosome 12q using linkage analysis in multiple large kindreds (Bashir et al. 1993; Craddock et al. 1993). Speculation arose as to whether the defect resided in a keratin because the KRT1 and KRT5 genes had already been mapped to chromosome 12q, and ultrastructural findings had indicated a defective desmosomal-keratin filament complex (Burge 1994). However, mutations were identified in a calcium pump ATP2A2, which encodes the sarco/endoplasmic reticulum calcium ATP-ase, defining a key role for this gene in maintaining cell-to-cell adhesion and keratinization (Sakuntabhai et al. 1999).
DSG4 in Localized Autosomal Recessive Hypotrichosis (LAH)
One of the most elegant examples of positional cloning lies in the discovery of a novel desmoglein, DSG4, as the underlying cause of localized autosomal recessive hypotrichosis (LAH) in humans (Kljuic et al. 2003). Mutations identified in individuals with LAH, in conjunction with comparative genomics studies revealing mutations in the lanceolate hair mouse, defined the critical role for DSG4, a suprabasally expressed desmoglein, in the skin and hair follicle. Importantly, this was one of the first reports of positional cloning that was accelerated by the Human Genome Project (2003) because DSG4 had not been identified by previous methods.
The genes underlying these and several other genodermatoses were identified and cloned using this classical genetic approach. In several of the studies previously mentioned in the Functional Cloning of Genes Underlying Several Inherited Skin Diseases section, linkage studies were subsequently (or simultaneously) used to test complete cosegregation of the candidate genes with the disease phenotype, providing strong evidence for the involvement of these genes in their associated skin diseases.
Methods for Characterizing and Validating the Mutations Identified
Upon the identification of the pathogenic gene segregating with a disease phenotype, several techniques were used to identify and characterize the nature of the mutation at the DNA, RNA, and protein levels. In most cases, this process began with the sequential amplification of all the exons in the gene using intronic primers and DNA isolated from peripheral blood (or tissue) from control and affected individuals. The widely used method, single-strand conformation polymorphism (SSCP), a type of heteroduplex analysis, was then used to screen for mutations based on the premise that even a single base change in a nucleic acid sequence would alter the conformation of the single-stranded DNA under denaturing conditions, altering the migration of the band compared to the control sequence (Ganguly et al. 1993). Depending on whether a shift in mobility was observed would determine subsequent direct sequencing or cloning of the amplified product to be sequenced. Another method used to analyze the consequence of the change in nucleic acid sequence is restriction enzyme analysis, exploiting the potential that the mutation either deleted or created a novel restriction site, which can be used as a diagnostic screening method for the mutation in affected and unaffected individuals.
To analyze the consequence of the mutation at the level of the transcript, particularly in the case of splice site mutations, amplification of the cDNA sequence flanking the mutation on both ends was performed, followed by gel electrophoresis (and even direct sequencing) to determine whether an exon had been spliced out. This was the case for a splice site mutation identified in the DSG1 gene in a case of striate palmoplantar keratoderma (SPPK), in which SSCP revealed abnormal migration of the intron 2-intron 3 amplicon, and amplification of the mutant transcript revealed that exon 3 was spliced out of the processed transcript (Rickman et al. 1999).
Importantly, these methods (Christiano et al. 1997) for direct mutation detection were key in the identification and characterization of hundreds of intragenic mutations across a wide range of genodermatoses, identifying well over 100 sequence variants in dystrophic epidermolysis bullosa alone (Dang and Murrell 2008).
Types of Mutations Identified and the Wide Spectrum of Their Phenotypic Consequences
For any given genodermatosis, there exists a broad range of mutations that have been identified with varying consequences. Different mutations in the same gene have been reported to cause both dominant and recessive forms of the same disease, as in the case of COL7A1 in dystrophic epidermolysis bullosa. Moreover, the proteins encoded by these genes can have either broad or very specific domains of expression in the skin and other stratified squamous epithelia, governing the phenotypic outcome on multiple sites of the body.
Classes of Alterations in the Nucleic Acid Sequence
For the most part, the intragenic mutations identified in monogenic skin diseases are small—point mutations (substitutions, splice site mutations), INDELS, etc.—but their phenotypic consequences are profound, especially considering the number of skin diseases that are inherited in an autosomal-dominant manner. In many cases, these mutations occur in critical domains of the protein and, depending on their nature, may lead to nonsense-mediated decay, decreasing the mRNA levels. This is especially the case for mutations that create a frameshift, followed by a premature termination codon, as has been reported in several cases of recessive dystrophic epidermolysis bullosa (Christiano et al. 1994a). On the other hand, in-frame mutations can also have a significant impact on the size of the protein, shown by splice-site mutations that can lead to the removal of a critical exon, as in the previously mentioned case of DSG1 in SPPK (Rickman et al. 1999). Similarly, single base substitutions (especially at the 5′ end of the gene) can result in a premature termination codon during translation that leads to the production of a truncated protein, as observed with several DSG1 mutations identified in SPPK cases (Hunt et al. 2001).
Whereas in these cases the dominant effect was caused by haploinsufficiency, mutations in intermediate filaments (i.e., keratins) lead to a dominant-negative effect, as the resulting heterotypic association between type I and II keratins renders the complex dysfunctional, phenocopying the loss-of-function mutation in the complementary keratin it pairs with (i.e., K5 and K14 mutations in EBS). Similarly, the mutations in COL7A1 that underlie dominant DEB (DDEB) act in a dominant-negative manner by interfering with the production of the normal allele, and because COL7A1 is a homotrimer of three α1 chains, very few functional anchoring fibrils are present in the papillary dermis in DDEB (Uitto et al. 1994).
Considering these genetic mutations, some additional interesting phenomena are observed in skin diseases. In rare instances, compound heterozygosity for two recessively inherited alleles of a gene can be observed in genodermatoses, creating an intermediate phenotype. McGrath and colleagues reported two unrelated patients with skin fragility and hypohidrotic ectodermal dysplasia that were heterozygous for two different mutations in the same gene, PKP1, an accessory protein of the desmosome (McGrath et al. 1997, 1999), resulting in an entirely new genodermatosis. Additional interesting phenomena observed in hereditary skin diseases include the association of particular nucleotide substitution with the generation of a premature termination codon and how the location of a mutation relative to the entire sequence influences the phenotype and inheritance. An example of the former is the classic C→T substitution observed in cases of junctional EB as well as dystrophic EB in the LAMA3/LAMB3/LAMC2 and COL7A1 genes, respectively (Aberdam et al. 1994; Baudoin et al. 1994; Christiano et al. 1994a; Hovnanian et al. 1994; Pulkkinen et al. 1994b), which is thought to occur as a result of spontaneous deamination of a 5-methylcytocine to a thymine, acting as a mechanism to generate premature termination codons from arginine codons in heritable diseases (Kivirikko et al. 1995). In the case of the latter, although recessive mutations in structural proteins are relatively rare, the COL7A1 and KRT14 genes have been reported to underlie both recessive and dominant forms of DEB and EBS, respectively (Coulombe et al. 1991; Christiano et al. 1993, 1994c; Hovnanian et al. 1993, 1994), showing how the precise location and nature of the mutation is critical in governing the phenotypic output.
Classes of Molecules Perturbed and Their Multifunctional Roles in Other Organs
Components of the desmosome
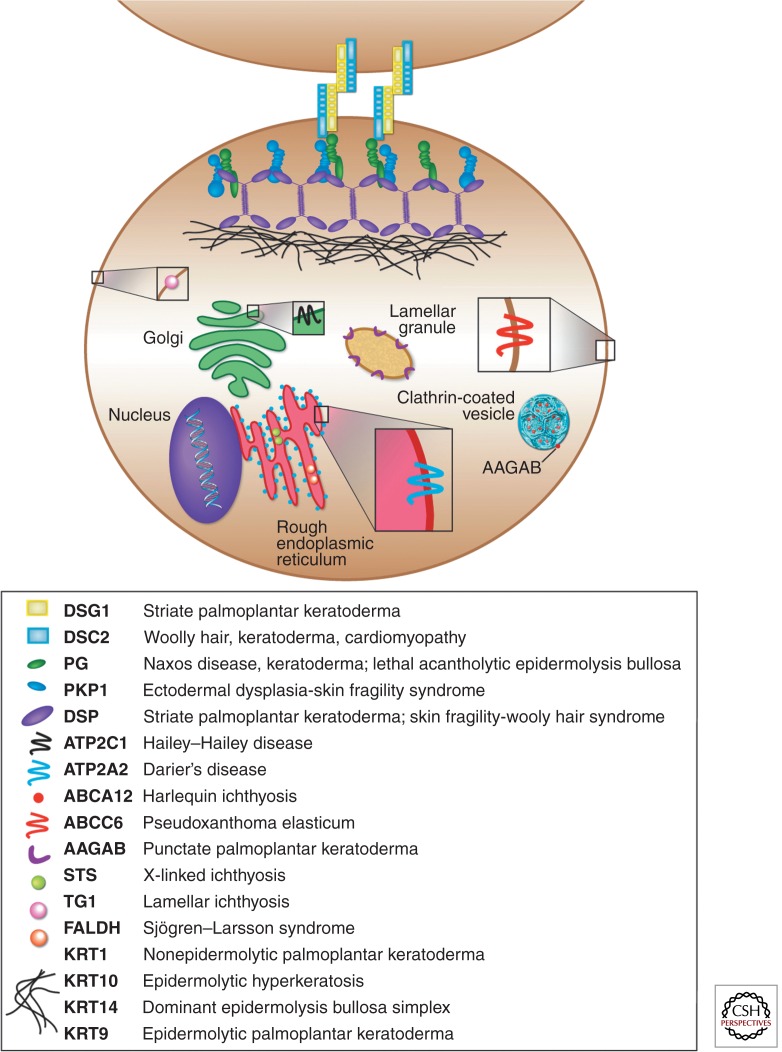
A great number of mutations have been identified in genes encoding proteins required for cellular adhesion between keratinocytes and the basement membrane, maintaining the overall structural integrity of the skin. The desmosome, a junction found on epithelia, cardiac muscle, brain meninges, and follicular dendritic cells, plays a key role in intracellular adhesion between keratinocytes in the epidermis and is composed of calcium-dependent adhesion molecules (desmogleins and desmocollins), as well as several accessory proteins that function to link these membrane proteins to intermediate filaments of the cytoskeleton (Fig. 2). Mutations in a plakophilin gene, PKP1, which associates with desmogleins and desmocollins, have been reported in skin fragility syndrome with hypohidrotic ectodermal dysplasia (McGrath et al. 1999). Similarly, dominant mutations in the amino terminus of the DSG1 gene have been reported in cases of striate PPK (Rickman et al. 1999; Hunt et al. 2001), characterized by hyperkeratotic bands of the palms and soles, and interestingly, the phenotype partly recapitulated what was observed in the Dsg3 transgenic mouse with an amino-terminal deletion (Allen et al. 1996). Furthermore, patients with the autoimmune diseases pemphigus vulgaris and pemphigus foliaceous, characterized by acanthyolysis and severe blistering, produce autoantibodies against DSG3/DSG4 and DSG1, respectively (Stanley 1993; Kljuic et al. 2003).
Figure 2.
Classes of molecules perturbed in skin diseases, illustrated at the subcellular level. Two epidermal keratinocytes are connected by the desmosome (desmoglein, desmocollin, plakoglobin, plakophilin, desmoplakin), which is connected to the keratin intermediate filament network. Other molecules, such as lipid and ion transporters (i.e., ATP2C1, ATP2A2, ABCC6, AAGAB, ABCA12), as well as enzymes in biosynthetic pathways (i.e., STS, TG1, FALDH) are depicted as residents of their respective organelles (i.e., endoplasmic reticulum, Golgi apparatus). All proteins are color coded to match the corresponding monogenic skin diseases they are associated with.
The intermediate filament (IF) network
As previously mentioned, several mutations in keratin genes have been identified in EBS, EH, as well as other skin diseases. Disruption of the intermediate filament network leads to acanthyolysis within the basal and suprabasal layers of the epidermis, ultimately resulting in blistering and/or hyperkeratinization of the skin. Because the IF network is intimately linked to the desmosome via desmosomal accessory proteins (i.e., desmoplakins) (Fig. 2), mutations in keratin genes phenocopy mutations in desmosomal proteins. This is exemplified in the nonepidermolytic form of PPK, in which a mutation has been reported in the nonhelical head domain of KRT1, which may interact with a desmoplakin protein (Kimonis et al. 1994; Rickman et al. 1999).
Calcium ion pumps and ABC/vesicle transporter proteins
Desmosomal cadherins, being calcium-dependent adhesion proteins, require proper regulation of intracellular and extracellular calcium levels to function. Along these lines, mutations have been reported in the calcium pump ATP2A2 and ATP2C1 genes in Darier’s and Hailey–Hailey diseases, respectively, consistent with previous electron microscopy studies that revealed loss of desmosomal attachments between the keratinocytes (Burge and Garrod 1991). Interestingly, mutations in ATP-binding cassette (ABC) transporters, MRP6 (ABCC6) and ABCA12, have been reported in the connective tissue disorder pseudoxanthoma elasticum (PXE) (Ringpfeil et al. 2000) and the very severe and lethal ichthyosis, Harlequin ichthyosis (Kelsell et al. 2005), respectively. Importantly, these transporters are crucial to mediating lipid transport and maintaining the overall integrity of the epidermal barrier (Fig. 2).
Mutations in the AAGAB gene, encoding the α- and γ-adaptin-binding protein p34 that binds clathrin adaptor complexes and is involved in vesicle transport (Fig. 2), have been identified in patients with a punctate form of palmoplantar keratoderma (Giehl et al. 2012; Pohler et al. 2012). The overall function of this gene is consistent with the ultrastructural findings in lesional plantar skin, which revealed vesicle defects in basal keratinocytes (Pohler et al. 2012). Importantly, the identification of mutations in various transporter proteins in skin diseases suggests an important role in maintaining the structural and functional integrity of the epidermis.
Enzymes in biosynthetic pathways
Yet another group of molecules perturbed in skin diseases includes enzymes important for catalyzing key steps in biosynthetic and metabolic pathways in skin homeostasis (Fig. 2). The first metabolic enzyme described to be associated with a genodermatosis is the steroid sulfatase gene (arylsulfatase C) in X-linked ichthyosis (XLI), where fibroblasts cultured from these patients were found to be deficient in hydrolase 3β-hydroxysteroid sulfate sulfatase (Webster et al. 1978). The gene encoding this enzyme was then mapped to Xp22.3 (Tiepolo et al. 1980) and mutations were identified in several cases of XLI, in which 80%–90% of these mutations are complete deletions of the gene (Reed et al. 2005).
Another key example is the keratinocyte-specific transglutaminase (TGM1) enzyme required during terminal differentiation of keratinocytes for the cross-linking of proteins that make up the cell envelope in the stratum corneum (Fig. 2) (Epstein 1996), in which mutations in this gene cause lamellar ichthyosis (Hohl et al. 1993; Russell et al. 1994, 1995; Huber et al. 1995), a type of skin disease characterized by excessive thickening of the stratum corneum (Hohl et al. 1993). Similarly, Sjögren–Larsson syndrome, an inherited disorder characterized by ichthyosis, mental retardation, as well as spasticity, is associated with a deficiency of fatty acid dehydrogenase activity, as cultured fibroblasts and keratinocytes from patients possessed decreased levels and activity of this enzyme (Rizzo 1993). Importantly, mutations have been identified in the fatty aldehyde dehydrogenase (FALDH) gene in patients with Sjögren–Larsson syndrome (De Laurenzi et al. 1996), making this the first report of an ALDH gene underlying a cutaneous disorder and revealing an essential role for FALDH in the metabolism of lipids of the lamellar bodies residing in the stratum corneum (Fig. 2).
Localized versus generalized phenotypic consequences
Interestingly, the degree of phenotypic severity of genodermatoses can be highly variable depending on the expression domains of the molecules underlying the pathologies. Mutations in KRT9, a keratin specifically expressed in the epidermis of palms and soles, lead to epidermolytic PPK, which only affects those areas (Reis et al. 1994), whereas mutations in KRT14 can affect the epithelium of the entire body. Similarly, mutations in molecules required for the structural integrity of the skin often affect the ectodermal appendages, such as the hair follicle and sebaceous gland (i.e., PKP1 in skin fragility syndrome with ectodermal dysplasia), as well as the nail bed (i.e., K6A, K16, K17 in PPK associated with pachyonychia concenita, a nail disorder) (Bowden et al. 1995; McLean et al. 1995; McGrath et al. 1997, 1999).
Importantly, many of the molecules that underlie the pathologies of genodermatoses play key roles in other stratified squamous epithelia as well as other organs, such that their loss of function produces multiple and severe phenotypes affecting several parts of the body. This is the case for components of the desmosome, as mutations in murine plakoglobin 1 lead not only to skin blistering, but also to cardiac rupture caused by the loss of function of the desmosomes in the myocardium (Koch et al. 1997). Similarly, MRP6 (ABCC6) underlies the connective tissue disorder, PXE, which also manifests cardiovascular and ophthalmologic abnormalities (Neldner 1988; Ringpfeil et al. 2000). Thus, the discoveries of the genes and their functions in maintaining the structural integrity of the skin have also shed light on the similar properties between the skin and other organs, significantly impacting research in other fields.
POLYGENIC HUMAN SKIN DISEASES
The etiopathology of complex skin diseases is composed of a combination of genetic plus environmental factors. Although the classical genetic approach has been widely successful in identifying the pathogenic genes underlying skin diseases inherited in a Mendelian fashion, other approaches have been developed to define the genetic basis of the more complex, non-Mendelian human skin diseases, such as psoriasis, vitiligo, atopic dermatitis, alopecia areata, and androgenetic alopecia, in which several loci have been implicated. However, over the past decade, powerful tools have been developed that circumvent the limitations of preexisting methods and have aided in uncovering the genetic complexity of many of these diseases.
Strategy Used to Dissect the Genetic Architecture of Polygenic Skin Diseases
From Twin Studies to Parametric Linkage Analyses
A powerful method to assess the degree of inheritance of complex, polygenic traits is the analysis of its manifestation in monozygotic (identical) and dizygotic (fraternal) twins. Several studies have been performed in various geographic regions on twins who possess skin diseases that clearly show a genetic component but are not inherited in a Mendelian fashion. In the case of psoriasis, the concordance rate among monozygotic (MZ) twins (72% in Northern Europe, 35% in Australia) is higher than that reported in dizygotic (DZ) twins (15%–23% in Northern Europe, 12% in Australia), showing a clear genetic component to the disease (Farber et al. 1974; Brandrup et al. 1978; Pisani and Ruocco 1984; Larsen et al. 1986; Duffy et al. 1993; Schultz Larsen 1993). Similarly, a 55% concordance rate among MZ twins (0% rate among DZ twins) has been reported in the autoimmune hair loss disease, alopecia areata (Rodriguez et al. 2010), and in the case of atopic dermatitis (AD), a chronic inflammatory skin disease with impairment of the epidermal skin barrier, there is a high familial occurrence (MZ twins = 80%, DZ twins = 20%), strongly suggesting a genetic basis (Larsen et al. 1986; Schultz Larsen 1993). However, in the case of vitiligo, an autoimmune disease affecting the melanocytes that results in depigmentation of skin, hair, and mucous membranes, the largest twin study reported only a 23% concordance among monozygotic twins, strongly suggesting the presence of epigenetic and environmental components to the disease (Alkhateeb et al. 2003).
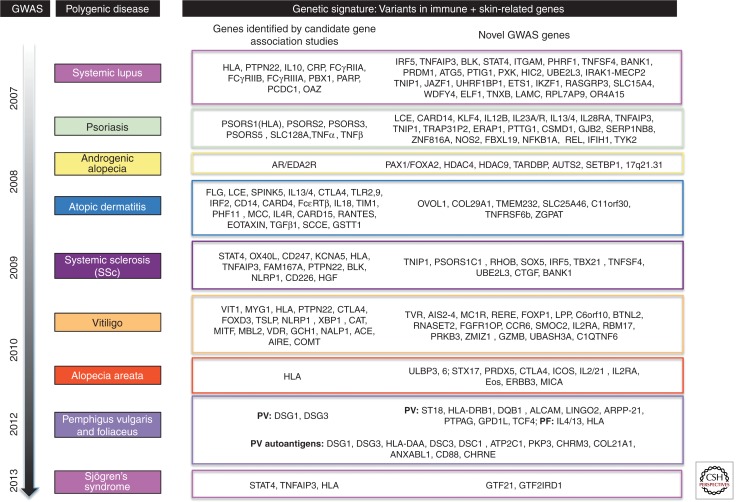
One of the earliest approaches used to determine the genetic basis of non-Mendelian skin diseases was parametric linkage analysis, in which a set of parameters is established for a given trait based on epidemiological evidence. In psoriasis, a chronic inflammatory disorder characterized by a thickened interfollicular epidermis as a result of excessive proliferation of keratinocytes, linkage and association studies were used to find the common region associated with the disease phenotype in multiple kindreds. The first study, performed in the early 1970s, revealed strong linkage to the HLA region (HLA-B13) on chromosome 6p (Russell et al. 1972)—and then subsequent genome-wide scans revealed strong associations of familial psoriasis with chromosomes 17q, 8q, 16q, 1q, 4q, and 20p (Tomfohrde et al. 1994; Matthews et al. 1996; Nair et al. 1997; Trembath et al. 1997; Capon et al. 1999). A more robust approach involving the analysis of 68 affected sibpairs from 41 multiplex families was undertaken by Trembath et al. (1997), identifying four regions of potential linkage on chromosomes 2, 8, 6, and 20, the strongest evidence for linkage disequilibrium being the MHC region at chromosome 6p21. To date, well over a dozen distinct loci have been reported to be associated with psoriasis, many of which contain immune-related genes (Fig. 3) (Bowcock and Krueger 2005; Tsoi et al. 2012).
Figure 3.
The genetic basis of complex, polygenic skin diseases. Several linkage and association studies have been performed over the course of the past few decades to elucidate the genes underlying the pathologies of several skin and autoinflammatory/autoimmune diseases, including psoriasis, systemic lupus erythematosus (SLE), androgenetic alopecia (AGA), atopic dermatitis (AD), systemic sclerosis (SSc), vitiligo (V), alopecia areata (AA), pemphigus vulgaris (PV), and foliaceus (PF). Several genes are common to these diseases, falling mostly into immune-related pathways, including inflammation, T-cell signaling, antigen presentation, and NF-κB signaling. In the past 5 years, numerous genome-wide association studies (GWAS) have been performed for these complex diseases and revealed additional candidate genes that play roles in these pathways. Autoantigens are listed for pemphigus vulgaris. All candidates are based on studies in the human conditions and do not include studies in mouse models. Genes with an asterisk were identified from association and linkage studies before the advent of the GWAS. For review on GWAS in polygenic skin diseases, see Zhang (2012). For additional review of the genetic association and linkage studies performed in polygenic skin diseases, see Cui et al. (2013) (SLE); Bussmann et al. (2011) (AD); Luo et al. (2013) (SSc); Sarig et al. (2012), Kalantari-Dehaghi et al. (2013), Yan et al. (2012) (PV); Toumi et al. (2013) (PF); Li et al. (2012) (AGA); Spritz (2011) (V); and Li et al. (2013) (SS).
Several linkage and association studies have been performed for atopic dermatitis, identifying linkage on 11 different chromosomes, where the only locus showing significant replication was 3p24 (Boguniewicz and Leung 2011). Within the association studies performed, only 13 out of the 81 genes examined were positively associated with other studies, with the filament aggregating protein, FLG, an epidermal differentiation complex (EDC) gene (also associated with ichthyosis vulgaris [Smith et al. 2006]) whose protein product makes up the cornified envelope of the stratum corneum, being the top candidate. Along these lines, mutations in the FLG gene, along with several other EDC genes (i.e., loricrin) and protease/antiprotease genes expressed in the stratum corneum, have been reported to be strongly associated with atopic dermatitis (Weidinger et al. 2006; Boguniewicz and Leung 2011).
Importantly, findings from twin studies, family-based linkage studies, as well as observed heritability among relatives provided several lines of evidence that suggest a genetic basis for several of these complex skin diseases.
Genome-Wide Association Studies (GWAS)
Nearly a decade ago, the GWAS became the gold standard technique for identifying common SNPs shared among unrelated individuals with a given complex trait. This method assays several thousand to more than a million SNPs in the genome, requiring a large sample size to produce a high resolution and power for detection of differences in allele frequencies between control and affected individuals.
Several GWAS studies have been performed on at least 11 polygenic dermatology-related diseases within the last 6 yr (Petukhova et al. 2010; Sarig et al. 2012; Zhang 2012; Luo et al. 2013), confirming previous findings from parametric linkage analysis and genome-wide scans as well as identifying new risk variants (Fig. 3). For some diseases, such as psoriasis, atopic dermatitis, and vitiligo, meta-analyses of GWAS studies have been performed, and integrating the data obtained from individual studies identified novel risk variants in these diseases (Jin et al. 2012; Paternoster et al. 2012; Tsoi et al. 2012). Interestingly, the combined findings across several dermatological diseases strongly suggests a common immune basis, as the SNPs with the highest level of significance are located within the HLA region and genes encoding MHC class I and II molecules, various cytokines, proteins involved in regulatory T-cell function/maintenance, as well as natural killer cell recognition (Petukhova et al. 2010; Zhang 2012). However, SNPs in several genes with defined roles in pathways specific to the sites of pathology for the phenotypes (i.e., epidermal proliferation and differentiation) have appeared in these lists, namely, the late cornified envelope (LCE) genes, encoding stratum corneum proteins in psoriasis (de Cid et al. 2009; Zhang et al. 2009), the TYR tyrosinase (melanosomal enzyme important for melanin biosynthesis) in vitiligo (Spritz 2008), as well as the androgen receptor (AR) gene in androgenetic alopecia (male-pattern baldness) (Fig. 3) (Ellis et al. 2001; Hillmer et al. 2005).
These studies also identified highly significant SNPs in novel genes that may play important roles in the effector cells and/or end organ, as in the case of the ULPB3 gene identified in the first GWAS performed in alopecia areata, where this encodes an NKG2D activating ligand not previously implicated in an autoimmune disease and whose expression is markedly up-regulated in the dermal sheath of patient hair follicles (Petukhova et al. 2010). Importantly, these common SNPs most likely serve as genetic susceptibility variants for these complex skin diseases. Moreover, in the case of the autoimmune diseases, the types of genes in which they are located suggest dysregulation at the both level of the effector cell and the end organ.
Large-Scale Expression Studies Using Microarrays and RNA-Seq
To gain insight into the transcriptional changes that ensue in polygenic skin diseases, researchers have embarked on large-scale expression studies, both array and nonarray based, for a subset of skin diseases. Both DNA microarrays and RNA sequencing studies have been performed in psoriasis (Bowcock et al. 2001; Zhou et al. 2003; Gudjonsson et al. 2009; Suarez-Farinas et al. 2010). One study in particular compared the transcriptomic profiles of psoriatic skin generated using both methods, and found 2629 and 569 differentially expressed genes (DEGs) using the RNA-seq and microarray approaches, respectively, showing that RNA-seq is better suited for identifying more DEGs and greater differences in expression of less-abundant transcripts in psoriatic skin (Jabbari et al. 2012). Transcriptional profiling has also been performed in vitiligo using DNA microarrays and qRT-polymerase chain reaction (PCR), identifying 30 DEGs (17 with decreased levels, mostly melanocyte specific; 13 with increased levels, mostly immune related) in lesional skin versus control, which strongly suggests aberrant innate immune function and activation in vitiligo skin (Yu et al. 2012). Similarly, transcriptional profiling in whole skin of alopecia areata patients primarily defined an immune-based signature, in addition to dysregulation of hair follicle and epidermal development-related genes (Subramanya et al. 2010). Importantly, these findings highlight the common collapse of immune function across the polygenic skin diseases.
Widespread Epigenetic Changes Associated with Complex Polygenic Skin Diseases
As previously mentioned, discordance between monozygotic twins for a given trait strongly suggests environmental factors and an epigenetic component that accounts for the variability, making twin studies an excellent model system for investigating the role of epigenetic factors in non-Mendelian skin diseases. Along these lines, several studies have been performed for diseases manifesting cutaneous symptoms, examining changes in genome-wide and promoter DNA methylation, as well as the expression of small noncoding RNAs that regulate gene expression levels.
Aberrant DNA Methylation in Autoimmune-Related Skin Diseases
Widespread changes in DNA methylation have been described extensively in cutaneous carcinomas, including melanoma and cutaneous lymphoma, as genomic instability is a key feature of cancer cells. However, global hypomethylation has also been observed in autoimmune-related skin diseases, such as systemic lupus erythematosous (SLE), in which DNMT1 transcription is reduced in T and B cells, resulting in global hypomethylation and hyperexpression of several methylation-sensitive autoimmune genes (Richardson et al. 1990; Balada et al. 2007; Zhou and Lu 2008). Similarly, global hypomethylation as a result of decreased DNMT1 transcription has been reported in atopic dermatitis PBMCs (Nakamura et al. 2006), which may lead to increased production of IgE and contribute to the pathogenesis of AD.
Localized aberrant DNA methylation has also been reported in various skin diseases, as in the case of systemic sclerosis (SSc), characterized by fibrosis as a result of excessive collagen deposition in skin as well as other tissues, in which methylation of the CpG islands of FL1 (a transcription factor that inhibits collagen production) is associated with reduced FL1 gene expression (Cross et al. 1994; Czuwara-Ladykowska et al. 2001; Wang et al. 2006). Similarly, hypomethylation of p16INK4A (antiapoptotic gene) has been reported in psoriasis, along with demethylation of one of the promoters of the SHP-1 gene, important for keratinocyte growth and proliferation (Ruchusatsawat et al. 2006; Zhang et al. 2007; Chen et al. 2008). Importantly, the findings from these studies provides additional evidence that the molecular defects underlying polygenic skin diseases transcend single-gene mutations, and may be characterized by more global changes at the chromatin level associated with a multitude of downstream effects on gene expression.
The Role of Small, Noncoding RNAs in the Pathogenesis of Polygenic Skin Diseases
In addition to modifications at the DNA and chromatin levels, a class of small noncoding RNAs, microRNAs (miRNAs) that regulate gene expression levels posttranscriptionally have been implicated in the pathogenesis of several autoimmune-related skin diseases. Deep sequencing and cloning of miRNAs, in conjunction with expression-based studies, have revealed a subset of these regulators that comprise the “T-cell signature,” and whose target genes possess defined roles in inflammation and/or epidermal specification/differentiation. Dysregulated expression of miR-146a, a negative regulator of the interferon-γ pathway, has been reported in SLE, where the target genes of miR-146a predominantly reside in the type I interferon pathway (Tang et al. 2009).
In psoriasis, aberrant expression of miR-146a, -125b, -203, and -21, among several others, has been reported to impact the transcript levels of the several immune-related target genes, including IRAK1/TRAF6 and TNF-α, leading to increased STAT3 activation and the subsequent formation of psoriatic plaques (Banerjee et al. 2011). Moreover, in a recent study, several known and novel lineage-specific miRNAs with roles in hematopoietic, angiogenic, and epithelial differentiation, including the novel keratinocyte-specific miR-135b, were identified as differentially expressed and/or processed in psoriatic skin compared to control (Joyce et al. 2011), suggesting an important role for these miRNAs in regulating transcript levels of both immune- and epidermal-related genes.
Perhaps one of the most well-characterized immune-related miRNAs is miR-155, implicated in several autoimmune skin diseases, including atopic dermatitis, in which its aberrant expression leads to dysregulated levels of the T-cell regulatory gene, CTLA4, and increased cellular proliferation (Sonkoly et al. 2010). Similarly, the miRNA-29 family (miR-29a, b, c) is known to regulate the transcript levels of type I and III collagens, and strong down-regulation of miR-29a expression is often found in the skin and fibroblasts from patients with systemic sclerosis, characterized by overproduction of dermal fibrillar collagens and modifying enzymes as well as ECM proteins (Asano 2010; Maurer et al. 2010).
UNUSUAL GENETIC MECHANISMS UNDERLYING MONOGENIC AND POLYGENIC SKIN DISEASES
Genetic Mosaicism
Mosaicism refers to the presence of two genetically distinct populations of cells in an individual that arise from a homogeneous zygote. In individuals with inherited skin diseases, mosaicism can be observed upon correction of the genetic mutation (revertant mosaicism) via mechanisms including gene conversion, intragenic recombination, back-mutation, and a second-site mutation that restores the protein product, creating a patchwork-like appearance when these clones proliferate in the skin. Similarly, mutations that arise very early on in development can lead to a mosaic appearance of a given dermatological condition (in the case of segmental disorders) that follows a clear, defined pattern in which the mutated, rather than healthy, tissue constitutes the mosaic patches of skin. A form of epigenetic mosaicism observed in dermatological diseases includes Lyonization, or X-chromosome inactivation, the biological process of dosage compensation that is considered to be functional X mosaicism.
These distinct mechanisms create striking phenotypes and have been observed in several monogenic and polygenic dermatological diseases. More importantly, mosaicism functions to protect against the severe consequences of a given mutation that may ordinarily result in lethality.
Revertant Mosaicism in Monogenic Skin Diseases
Several cases of revertant mosaicism, or clonal restoration of the mutant phenotype, have been reported, many of which are associated with the recessive, nonlethal (Herlitz) form of JEB. DNA sequencing of the COL17A1 and LAMB3 genes in the reverted skin of several EB patients revealed the occurrence of the four major mechanisms of reversion (gene conversion, intragenic recombination, back-mutation, and a second-site mutation) (Lai-Cheong et al. 2011). Mitotic gene conversion was observed in a patient with the non-Herlitz form of JEB, compound heterozygous for a frameshift and nonsense mutation, in which the gene conversion event led to restoration of the frameshift (Jonkman et al. 1997). Interestingly, this was actually the first report of a gene conversion event as a mechanism for gene correction.
Second-site and true back-mutations (correction of the original mutation) have also been reported in the reverted skin of patients with the non-Herlitz form of JEB, where DNA sequencing of reverted skin from different regions of the body in two JEB patients revealed multiple second-site mutations in the LAMB3 and COL17A1 genes that negate the effects of the original germline mutations (Pasmooij et al. 2005, 2007). Moreover, immunofluorescence staining on the reverted and nonreverted areas of the skin convincingly show these corrections and full restoration of protein levels. Revertant mosaicism was also reported in a case of RDEB, involving the COL7A1 gene in an individual compound heterozygous for a nonsense and frameshift mutation in which long-range PCR revealed the corrected paternally inherited frameshift mutation on the paternal allele in the reverted patches, suggesting that an intragenic crossover had restored the frameshift mutation, resulting in the production of functional protein product (Almaani et al. 2010).
Although most cases of revertant mosaicism have been reported in individuals with the recessive forms of EB, this phenomenon has also been observed in a case of the dominantly inherited Dowling–Meara subtype of EBS, involving the KRT14 gene (Smith et al. 2004). Similarly, revertant mosaicism has been reported in several cases of another autosomal dominant skin disease, ichthyosis with confetti (IWC), involving loss of heterozygosity (LOH) of a mutation in the KRT10 gene and characterized by a high incidence of spontaneous reversions that correct the mutation, leading to many small patches of unaffected skin (hence, the name “confetti”) (Brusasco et al. 1994).
Segmental Manifestations of Monogenic and Polygenic Skin Diseases
Mosaicism for postzygotic mutations during development can lead to segmental manifestations of a given skin disease, in which the mutation is propagated through a particular cell lineage(s) causing the phenotype to appear linear, or otherwise, segmental. Often times, the patterns observed in segmental skin diseases are consistent with the lines of Blaschko, which delineate the visible patterns of cellular lineages in the skin concordant with the migration of these cells during the formation of the primitive streak early in development (Happle 1985).
For monogenic skin diseases, there are two types of segmental manifestations, termed type I and type II, in which type I manifestations arise from autosomal dominant postzygotic mutations in an otherwise healthy embryo, and type II manifestations occur as a result of LOH for the same autosomal dominant mutation but in a heterozygous background, creating segments of affected tissue that are superimposed onto the classical disease phenotype. Several examples of each class have been reported for Darier’s disease, epidermolysis hyperkeratosis of Broqc, and Hailey–Hailey, to name a few (Happle 1999; Sakuntabhai et al. 2000; Hwang et al. 2003; Wada et al. 2003). The segmental forms of the disease can most often be distinguished from the uniform phenotypes by eye, and in the case of type II segmental manifestations, DNA sequencing in the patches or segments of affected tissue superimposed onto the uniform disease can reveal the LOH that lead to homo- or hemizygosity of the given mutation.
Unlike monogenic diseases, in which the genetic basis is clear and a genetically “healthy” embryo can be distinguished from a genetically “affected” embryo, non-Mendelian polygenic skin diseases are not classified as “type I” and “type II,” but rather more broadly as “isolated” versus “superimposed.” Interestingly, psoriasis was the first polygenic skin disease for which the concept of LOH was proposed as a mechanism to explain the observed segmental manifestation (Happle 1991), and in the case of other diseases, including atopic dermatitis, SLE, and vitiligo, segmental manifestations of the disease phenotype have also been reported (Happle 2007).
Functional X-Chromosome Mosaicism
Another mechanism that gives rise to mosaicism is X-chromosome inactivation, the phenomenon of dosage compensation observed in females. Several X-linked skin diseases have been described, where the skin of males is completely affected and that of hemizygous female carriers possess patterns that often follow the lines of Blasckho. In the case of the X-linked dominant disorder, X-linked hypohidrotic ectodermal dysplasia, a skin disease that causes males to lose their ability to sweat, hemizygous female carriers show mosaicism for functional sweat pores, determined by a classical sweat test using starch and iodide in which the observed pattern follows the lines of Blaschko and is consistent with functional X inactivation (Traupe 1999). Mutations in the ectodysplasin-A (EDA) gene are responsible for the human disease phenotype (Monreal et al. 1998), and mutations in the mouse homolog cause the tabby phenotype of hypoplastic hair, teeth, and eccrine sweat glands (Srivastava et al. 1997; Grzeschik et al. 2007).
The genotypic and phenotypic consequences of some X-linked dominant skin disorders can be quite severe, leading to lethality in affected males. This is the case in patients with focal dermal hypoplasia (FDH), a disease characterized by multiple ectoderm and mesoderm defects, including dermal lesions of hypoplastic tissue, adipose tissue herniation, and linear pigmentary defects. The PORCN gene, a regulator of Wnt signaling, has been identified as the culprit underlying this disease based on linkage studies and mutations in PORCN that have been identified in FDH patients (Grzeschik et al. 2007).
Interestingly, in the case of incontinentia pigmenti, a rare X-linked dominant disease caused by mutations in the gene encoding the NF-κB essential modulator (NEMO), this disease is lethal in the majority of affected males; however, males who survive are mosaic for the mutation (Makris et al. 2000; Smahi et al. 2000; Berlin et al. 2002). Although some of the reported cases are XXY males (Klinefelter syndrome), in which case one of the X chromosomes would become inactivated, the other cases with normal karyotypes may be attributed to somatic mosaicism of a very early postzygotic or half-chromatid mutation, or even hypomorphic mutations with less deleterious effects.
Isodisomy
Uniparental disomy (UPD) is the inheritance of two chromosomes of one pair from one parent, where meiotic errors can lead to either heterodisomy (inheritance of two nonidentical homologs, meiosis I error) or isodisomy (inheritance of two identical homologs, meiosis II error). Importantly, uniparental isodisomy can lead to reduction to homozygosity for a given mutation. These events are quite rare, but nonetheless have been reported in several patients with severe skin diseases. Several genetic mechanisms can lead to isodisomy and are detectable using various cytogenetic and molecular techniques, such as karyotyping and microsatellite marker analysis.
Cases of isodisomy have been most frequently reported in the Herlitz form of JEB, but also in JEB with pyloric atresia and RDEB, where mutations in the LAMB3, LAM2C, ITGB4, and COL7A1 genes were present on one allele in the gametes from one parent, and the gametes from the other parent were unaffected (McGrath et al. 1995b; Fassihi et al. 2005, 2006; Natsuga et al. 2010). One mechanism to explain the isodisomy is nondisjunction followed by monosomy rescue, when upon the fusion of a gamete nullasomic for these respective chromosomes with one containing a duplicated chromosome possessing the mutant allele (monosomy rescue), isodisomy occurs and leads to reduction to homozygosity.
Although karyotyping analysis following chorionic villus sampling would not reveal any defects in chromosome number for these cases, analysis of microsatellites spanning the entire chromosome has revealed whether two chromosome homologs were inherited from the same parent. Interestingly, a case of isodisomy has been reported in Harlequin ichthyosis, where the mechanism for reduction to homozygosity of an ABCA12 mutation was proposed to be a trisomic rescue because nonmosaic trisomy was observed for the ABCA12-bearing chromosome, resulting in a uniparental isodisomy that was detected through karyotyping analysis (Castiglia et al. 2009).
It is often difficult to definitely prove the genetic mechanism by which UPD arises, and these rare events as the result of meiotic errors often produce normal karyotypes that make it difficult to identify UPD events during early embryonic development. A more robust prenatal diagnostic screening approach integrating changes in copy number with gene expression and perhaps even microsatellite marker analysis would be required to more effectively identify UPD and facilitate gene therapy treatments for these patients.
Position Effects
A position effect is defined as a change in gene expression as a result of altering the location of the gene relative to its chromosomal surroundings in which several genetic mechanisms, including chromosomal rearrangements, insertions, deletions, and copy number variants, can lead to the separation of the gene from a tissue- or temporal-specific modifier element (Kleinjan and van Heyningen 1998). This definition of position effect is not to be confused with the classical phenomenon of position effect variegation, first reported in Drosophila at the white+ locus, which describes the heritably stable inhibition of gene expression as a result of juxtaposition with heterochromatin (Weiler and Wakimoto 1995).
Position effects on single genes as a result of large chromosomal rearrangements or copy number variants have been reported in several human genetic diseases, including ectodermal dysplasias, which are characterized by the abnormal development of the hair, skin, nails, and/or eccrine glands. One such class of ectodermal dysplasia includes the hypertrichoses syndromes, defined as excessive hair growth for a particular body site or age of an individual that is not hormone dependent, which are often associated with multiple additional anomalies (gingival hyperplasia, cardiomegaly, deafness, dental, palate, and bone defects) (Garcia-Cruz et al. 2002). Despite the pleiotrophic phenotypes of these syndromes and their reported association with large chromosomal rearrangements and copy number variants, the causative genetic mechanism underlying human hypertrichoses remained elusive.
Within the past 5 years, position effects as the underlying genetic basis of these syndromes have been reported on the TRPS1, SOX-9, and FGF-13 genes that reside ∼1 Mb to as distant as 7.3 Mb from these chromosomal rearrangements in Ambras syndrome, congenital generalized hypertrichosis terminalis (CGHT), and X-linked recessive hypertrichosis, respectively (Fantauzzo et al. 2008, 2012; DeStefano et al. 2013). Interestingly, the position effect on TRPS1 was recapitulated in the Koala mutant mouse (a model for hypertrichosis) that contains a chromosomal inversion whose proximal breakpoint maps 791 bp upstream of Trps1, leading to a marked decrease in Trps1 expression at the sites of pathology for the phenotype (Fantauzzo et al. 2008). Furthermore, TRPS1 was identified as a novel regulator of the Wnt signaling pathway and SOX-9 was found to be a direct transcriptional target gene of TRPS1, where these genes form an important feedback loop with SHH to regulate epithelial proliferation in the hair follicle (Fantauzzo and Christiano 2012; Fantauzzo et al. 2012).
In the X-linked forms of hypertrichosis, large interchromosomal insertions at the Xq27.1 extragenic palindrome site have been reported in three large kindreds, where the sequences contained within each insertion differed between the families (Zhu et al. 2011; DeStefano et al. 2013). Recently, a position effect on FGF-13, a gene that lies 1.2 Mb away from the insertion, was reported in one of these families, where FGF-13 expression was selectively and profoundly reduced throughout patient hair follicles compared to control (DeStefano et al. 2013). Furthermore, murine Fgf-13 is expressed during hair follicle induction and cycling and localizes to the bulge region (the stem cell compartment) of the postnatal hair follicle, suggesting a novel role for this noncanonical FGF in hair follicle growth and cycling (Kawano et al. 2004; DeStefano et al. 2013).
CONCLUDING REMARKS
Great progress has been made in elucidating the genetic basis of monogenic skin diseases using classical genetic techniques. Based on studies performed over the course of the past several decades, we have learned a great deal about the complexity of the skin and its constituents and the various classes of molecules and their functions that maintain the overall structural integrity of the skin. Moreover, such findings have provided novel insight into their roles and importance in other organs from the striking phenotypes observed in patients with inherited skin diseases as well as mice that harbor mutations in the same genes. Thus, the genetic approach of positional and functional cloning has been the most fruitful in driving these discoveries, and these findings have furthered research in other fields as well.
Several classes of mutations have been identified across a wide spectrum of genodermatoses, in which mutations in the same gene can lead to both dominant and recessive forms of the disease. Moreover, domains of expression (broad vs. specific) can vary for a given class of molecules (i.e., keratins), influencing the severity of the phenotype. Furthermore, comparing the diverse phenotypes between skin diseases with mutations in the same genes has taught us that the location, nature of mutations, and consequence on protein product are critical in governing the phenotypic output.
Over the past decade, great progress has also been made dissecting the genetic architecture of polygenic skin diseases, where new techniques that overcome the limitations of preexisting methods have been instrumental in defining both the genetic and epigenetic bases of these diseases. The collective findings have elucidated the transcriptional profiles at both the effector cell and end organ levels, shedding light onto the common immune basis across these skin diseases. However, because the majority of these studies have been performed on a subset of polygenic skin diseases, much work has yet to be done in the field using these new, powerful techniques. In the more well-studied polygenic skin diseases like psoriasis, a further characterization of transcriptional changes and correlations with genetic susceptibility loci (i.e., SNPs) will aid in uncovering the mechanisms behind the pathology.
To this end, it would be of interest to overlay the mutation and expression-based data sets with data from epigenetic studies (DNA methylation, deposition of histone marks, miRNAs etc) to address the potential roles of (1) SNPs in CpG-rich sequences and their influence on the methylation patterns across gene bodies as well as within regulatory elements, (2) SNPs located within miRNA seed sequences and/or complementary binding sites within the target genes, and their potential influence on the expression levels and/or stability of the transcript, (3) master regulators (i.e., immune or end-organ-related transcription factors) and potential enrichment of their binding site motifs in downstream target genes, and (4) intra- and interchromosomal interactions via 3D chromatin looping structures and their regulation of gene expression in the effector cell and end organ. Thus, an integrative approach incorporating the genetics with functional genomics and systems biology would provide a more comprehensive understanding of the complex mechanisms underlying these polygenic skin diseases.
Even further beyond the level of the gene, studies in both monogenic and complex genetic diseases have elucidated the unusual mechanisms from mosaicism to isodisomy that lead to phenotypic variation within an individual for a given disease. Similarly, other unusual genetic mechanisms, such as position effects resulting from large chromosomal abnormalities (i.e., rearrangements and CNVs), have been observed in skin diseases that are associated with additional ectodermal defects and are inherited in a Mendelian fashion. These findings have taught us that complete loss-of-function is not the only way to generate a severe phenotype, as merely altering transcript levels might be enough, shedding light onto the importance of spatiotemporal regulation of gene expression during ectodermal development.
Thus, the genetics of skin disease is quite complex, but nonetheless fascinating. Moreover, the easily accessible nature and diverse range of cell types and molecules make the skin an excellent system in which to study genetics.
REFERENCES
- Aberdam D, Galliano MF, Vailly J, Pulkkinen L, Bonifas J, Christiano AM, Tryggvason K, Uitto J, Epstein EH Jr, Ortonne JP, et al. 1994. Herlitz’s junctional epidermolysis bullosa is linked to mutations in the gene (LAMC2) for the γ2 subunit of nicein/kalinin (LAMININ-5). Nat Genet 6: 299–304 [DOI] [PubMed] [Google Scholar]
- Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. 2003. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res 16: 208–214 [DOI] [PubMed] [Google Scholar]
- Allen E, Yu QC, Fuchs E. 1996. Mice expressing a mutant desmosomal cadherin exhibit abnormalities in desmosomes, proliferation, and epidermal differentiation. J Cell Biol 133: 1367–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaani N, Nagy N, Liu L, Dopping-Hepenstal PJ, Lai-Cheong JE, Clements SE, Techanukul T, Tanaka A, Mellerio JE, McGrath JA. 2010. Revertant mosaicism in recessive dystrophic epidermolysis bullosa. J Invest Dermatol 130: 1937–1940 [DOI] [PubMed] [Google Scholar]
- Anton-Lamprecht I. 1983. Genetically induced abnormalities of epidermal differentiation and ultrastructure in ichthyoses and epidermolyses: Pathogenesis, heterogeneity, fetal manifestation, and prenatal diagnosis. J Invest Dermatol 81: 149s–156s [DOI] [PubMed] [Google Scholar]
- Anton-Lamprecht I, Schnyder UW. 1982. Epidermolysis bullosa herpetiformis Dowling–Meara. Report of a case and pathomorphogenesis. Dermatologica 164: 221–235 [PubMed] [Google Scholar]
- Asano Y. 2010. Future treatments in systemic sclerosis. J Dermatol 37: 54–70 [DOI] [PubMed] [Google Scholar]
- Balada E, Ordi-Ros J, Vilardell-Tarres M. 2007. DNA methylation and systemic lupus erythematosus. Ann NY Acad Sci 1108: 127–136 [DOI] [PubMed] [Google Scholar]
- Banerjee J, Chan YC, Sen CK. 2011. MicroRNAs in skin and wound healing. Physiol Genomics 43: 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir R, Munro CS, Mason S, Stephenson A, Rees JL, Strachan T. 1993. Localisation of a gene for Darier’s disease. Hum Mol Genet 2: 1937–1939 [DOI] [PubMed] [Google Scholar]
- Baudoin C, Miquel C, Gagnoux-Palacios L, Pulkkinen L, Christiano AM, Uitto J, Tadini G, Ortonne JP, Meneguzzi G. 1994. A novel homozygous nonsense mutation in the LAMC2 gene in patients with the Herlitz junctional epidermolysis bullosa. Hum Mol Genet 3: 1909–1910 [DOI] [PubMed] [Google Scholar]
- Berlin AL, Paller AS, Chan LS. 2002. Incontinentia pigmenti: A review and update on the molecular basis of pathophysiology. J Am Acad Dermatol 47: 169–187; quiz 188–190 [DOI] [PubMed] [Google Scholar]
- Boguniewicz M, Leung DY. 2011. Atopic dermatitis: A disease of altered skin barrier and immune dysregulation. Immunol Rev 242: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifas JM, Rothman AL, Epstein EH Jr. 1991. Epidermolysis bullosa simplex: Evidence in two families for keratin gene abnormalities. Science 254: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Bowcock AM, Krueger JG. 2005. Getting under the skin: The immunogenetics of psoriasis. Nat Rev Immunol 5: 699–711 [DOI] [PubMed] [Google Scholar]
- Bowcock AM, Shannon W, Du F, Duncan J, Cao K, Aftergut K, Catier J, Fernandez-Vina MA, Menter A. 2001. Insights into psoriasis and other inflammatory diseases from large-scale gene expression studies. Hum Mol Genet 10: 1793–1805 [DOI] [PubMed] [Google Scholar]
- Bowden PE, Haley JL, Kansky A, Rothnagel JA, Jones DO, Turner RJ. 1995. Mutation of a type II keratin gene (K6a) in pachyonychia congenita. Nat Genet 10: 363–365 [DOI] [PubMed] [Google Scholar]
- Brandrup F, Hauge M, Henningsen K, Eriksen B. 1978. Psoriasis in an unselected series of twins. Arch Dermatol 114: 874–878 [PubMed] [Google Scholar]
- Brusasco A, Tadini G, Cambiaghi S, Ermacora E, Grimalt R, Caputo R. 1994. A case of congenital reticular ichthyosiform erythroderma—ichthyosis “en confettis.” Dermatology 188: 40–45 [DOI] [PubMed] [Google Scholar]
- Buchbinder LH, Lucky AW, Ballard E, Stanley JR, Stolar E, Tabas M, Bauer EA, Paller AS. 1986. Severe infantile epidermolysis bullosa simplex: Dowling–Meara type. Arch Dermatol 122: 190–198 [PubMed] [Google Scholar]
- Burge S. 1994. Darier’s disease—The clinical features and pathogenesis. Clin Exp Dermatol 19: 193–205 [DOI] [PubMed] [Google Scholar]
- Burge SM, Garrod DR. 1991. An immunohistological study of desmosomes in Darier’s disease and Hailey–Hailey disease. Br J Dermatol 124: 242–251 [DOI] [PubMed] [Google Scholar]
- Burgeson RE, Lunstrum GP, Rokosova B, Rimberg CS, Rosenbaum LM, Keene DR. 1990. The structure and function of type VII collagen. Ann NY Acad Sci 580: 32–43 [DOI] [PubMed] [Google Scholar]
- Bussmann C, Weidinger S, Novak N. 2011. Genetics of atopic dermatitis. J Dtsch Dermatol Ges 9: 670–676 [DOI] [PubMed] [Google Scholar]
- Capon F, Novelli G, Semprini S, Clementi M, Nudo M, Vultaggio P, Mazzanti C, Gobello T, Botta A, Fabrizi G, et al. 1999. Searching for psoriasis susceptibility genes in Italy: Genome scan and evidence for a new locus on chromosome 1. J Invest Dermatol 112: 32–35 [DOI] [PubMed] [Google Scholar]
- Castiglia D, Castori M, Pisaneschi E, Sommi M, Covaciu C, Zambruno G, Fischer J, Magnani C. 2009. Trisomic rescue causing reduction to homozygosity for a novel ABCA12 mutation in harlequin ichthyosis. Clin Genet 76: 392–397 [DOI] [PubMed] [Google Scholar]
- Chen M, Chen ZQ, Cui PG, Yao X, Li YM, Li AS, Gong JQ, Cao YH. 2008. The methylation pattern of p16INK4a gene promoter in psoriatic epidermis and its clinical significance. Br J Dermatol 158: 987–993 [DOI] [PubMed] [Google Scholar]
- Christiano AM, Greenspan DS, Hoffman GG, Zhang X, Tamai Y, Lin AN, Dietz HC, Hovnanian A, Uitto J. 1993. A missense mutation in type VII collagen in two affected siblings with recessive dystrophic epidermolysis bullosa. Nat Genet 4: 62–66 [DOI] [PubMed] [Google Scholar]
- Christiano AM, Anhalt G, Gibbons S, Bauer EA, Uitto J. 1994a. Premature termination codons in the type VII collagen gene (COL7A1) underlie severe, mutilating recessive dystrophic epidermolysis bullosa. Genomics 21: 160–168 [DOI] [PubMed] [Google Scholar]
- Christiano AM, Greenspan DS, Lee S, Uitto J. 1994b. Cloning of human type VII collagen. Complete primary sequence of the α1(VII) chain and identification of intragenic polymorphisms. J Biol Chem 269: 20256–20262 [PubMed] [Google Scholar]
- Christiano AM, Ryynanen M, Uitto J. 1994c. Dominant dystrophic epidermolysis bullosa: Identification of a Gly→Ser substitution in the triple-helical domain of type VII collagen. Proc Natl Acad Sci 91: 3549–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiano AM, Hoffman GG, Zhang X, Xu Y, Tamai Y, Greenspan DS, Uitto J. 1997. Strategy for identification of sequence variants in COL7A1 and a novel 2-bp deletion mutation in recessive dystrophic epidermolysis bullosa. Hum Mutat 10: 408–414 [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Hutton ME, Letai A, Hebert A, Paller AS, Fuchs E. 1991. Point mutations in human keratin 14 genes of epidermolysis bullosa simplex patients: Genetic and functional analyses. Cell 66: 1301–1311 [DOI] [PubMed] [Google Scholar]
- Craddock N, Dawson E, Burge S, Parfitt L, Mant B, Roberts Q, Daniels J, Gill M, McGuffin P, Powell J, et al. 1993. The gene for Darier’s disease maps to chromosome 12q23-q24.1. Hum Mol Genet 2: 1941–1943 [DOI] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. 1994. Purification of CpG islands using a methylated DNA binding column. Nat Genet 6: 236–244 [DOI] [PubMed] [Google Scholar]
- Cui Y, Sheng Y, Zhang X. 2013. Genetic susceptibility to SLE: Recent progress from GWAS. J Autoimmun 41: 25–33 [DOI] [PubMed] [Google Scholar]
- Czuwara-Ladykowska J, Shirasaki F, Jackers P, Watson DK, Trojanowska M. 2001. Fli-1 inhibits collagen type I production in dermal fibroblasts via an Sp1-dependent pathway. J Biol Chem 276: 20839–20848 [DOI] [PubMed] [Google Scholar]
- Dang N, Murrell DF. 2008. Mutation analysis and characterization of COL7A1 mutations in dystrophic epidermolysis bullosa. Exp Dermatol 17: 553–568 [DOI] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN, Giardina E, Stuart PE, Nair R, Helms C, et al. 2009. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet 41: 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V, Rogers GR, Hamrock DJ, Marekov LN, Steinert PM, Compton JG, Markova N, Rizzo WB. 1996. Sjogren–Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nat Genet 12: 52–57 [DOI] [PubMed] [Google Scholar]
- DeStefano GM, Fantauzzo KA, Petukhova L, Kurban M, Tadin-Strapps M, Levy B, Warburton D, Cirulli ET, Han Y, Sun X, et al. 2013. Position effect on FGF13 associated with X-linked congenital generalized hypertrichosis. Proc Natl Acad Sci 110: 7790–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL, Spelman LS, Martin NG. 1993. Psoriasis in Australian twins. J Am Acad Dermatol 29: 428–434 [DOI] [PubMed] [Google Scholar]
- Eichner R, Bonitz P, Sun TT. 1984. Classification of epidermal keratins according to their immunoreactivity, isoelectric point, and mode of expression. J Cell Biol 98: 1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JA, Stebbing M, Harrap SB. 2001. Polymorphism of the androgen receptor gene is associated with male pattern baldness. J Invest Dermatol 116: 452–455 [DOI] [PubMed] [Google Scholar]
- Epstein EH Jr. 1996. The genetics of human skin diseases. Curr Opin Genet Dev 6: 295–300 [DOI] [PubMed] [Google Scholar]
- Fantauzzo KA, Christiano AM. 2012. Trps1 activates a network of secreted Wnt inhibitors and transcription factors crucial to vibrissa follicle morphogenesis. Development 139: 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo KA, Tadin-Strapps M, You Y, Mentzer SE, Baumeister FA, Cianfarani S, Van Maldergem L, Warburton D, Sundberg JP, Christiano AM. 2008. A position effect on TRPS1 is associated with Ambras syndrome in humans and the Koala phenotype in mice. Hum Mol Genet 17: 3539–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo KA, Kurban M, Levy B, Christiano AM. 2012. Trps1 and its target gene Sox9 regulate epithelial proliferation in the developing hair follicle and are associated with hypertrichosis. PLoS Genet 8: e1003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber EM, Nall ML, Watson W. 1974. Natural history of psoriasis in 61 twin pairs. Arch Dermatol 109: 207–211 [PubMed] [Google Scholar]
- Fassihi H, Wessagowit V, Ashton GH, Moss C, Ward R, Denyer J, Mellerio JE, McGrath JA. 2005. Complete paternal uniparental isodisomy of chromosome 1 resulting in Herlitz junctional epidermolysis bullosa. Clin Exp Dermatol 30: 71–74 [DOI] [PubMed] [Google Scholar]
- Fassihi H, Lu L, Wessagowit V, Ozoemena LC, Jones CA, Dopping-Hepenstal PJ, Foster L, Atherton DJ, Mellerio JE, McGrath JA. 2006. Complete maternal isodisomy of chromosome 3 in a child with recessive dystrophic epidermolysis bullosa but no other phenotypic abnormalities. J Invest Dermatol 126: 2039–2043 [DOI] [PubMed] [Google Scholar]
- Fuchs E, Green H. 1980. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell 19: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Fuchs E, Esteves RA, Coulombe PA. 1992. Transgenic mice expressing a mutant keratin 10 gene reveal the likely genetic basis for epidermolytic hyperkeratosis. Proc Natl Acad Sci 89: 6906–6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ. 1993. Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: Evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci 90: 10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cruz D, Figuera LE, Cantu JM. 2002. Inherited hypertrichoses. Clin Genet 61: 321–329 [DOI] [PubMed] [Google Scholar]
- Giehl KA, Eckstein GN, Pasternack SM, Praetzel-Wunder S, Ruzicka T, Lichtner P, Seidl K, Rogers M, Graf E, Langbein L, et al. 2012. Nonsense mutations in AAGAB cause punctate palmoplantar keratoderma type Buschke–Fischer–Brauer. Am J Hum Genet 91: 754–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik KH, Bornholdt D, Oeffner F, Konig A, del Carmen Boente M, Enders H, Fritz B, Hertl M, Grasshoff U, Hofling K, et al. 2007. Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nat Genet 39: 833–835 [DOI] [PubMed] [Google Scholar]
- Gudjonsson JE, Ding J, Li X, Nair RP, Tejasvi T, Qin ZS, Ghosh D, Aphale A, Gumucio DL, Voorhees JJ, et al. 2009. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J Invest Dermatol 129: 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happle R. 1985. Lyonization and the lines of Blaschko. Hum Genet 70: 200–206 [DOI] [PubMed] [Google Scholar]
- Happle R. 1991. Somatic recombination may explain linear psoriasis. J Med Genet 28: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happle R. 1999. Loss of heterozygosity in human skin. J Am Acad Dermatol 41: 143–164 [DOI] [PubMed] [Google Scholar]
- Happle R. 2007. Superimposed segmental manifestation of polygenic skin disorders. J Am Acad Dermatol 57: 690–699 [DOI] [PubMed] [Google Scholar]
- Hillmer AM, Hanneken S, Ritzmann S, Becker T, Freudenberg J, Brockschmidt FF, Flaquer A, Freudenberg-Hua Y, Jamra RA, Metzen C, et al. 2005. Genetic variation in the human androgen receptor gene is the major determinant of common early-onset androgenetic alopecia. Am J Hum Genet 77: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl D, Huber M, Frenk E. 1993. Analysis of the cornified cell envelope in lamellar ichthyosis. Arch Dermatol 129: 618–624 [PubMed] [Google Scholar]
- Hovnanian A, Duquesnoy P, Blanchet-Bardon C, Knowlton RG, Amselem S, Lathrop M, Dubertret L, Uitto J, Goossens M. 1992. Genetic linkage of recessive dystrophic epidermolysis bullosa to the type VII collagen gene. J Clin Invest 90: 1032–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovnanian A, Pollack E, Hilal L, Rochat A, Prost C, Barrandon Y, Goossens M. 1993. A missense mutation in the rod domain of keratin 14 associated with recessive epidermolysis bullosa simplex. Nat Genet 3: 327–332 [DOI] [PubMed] [Google Scholar]
- Hovnanian A, Hilal L, Blanchet-Bardon C, de Prost Y, Christiano AM, Uitto J, Goossens M. 1994. Recurrent nonsense mutations within the type VII collagen gene in patients with severe recessive dystrophic epidermolysis bullosa. Am J Hum Genet 55: 289–296 [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Bonifas JM, Beech J, Bench G, Shigihara T, Ogawa H, Ikeda S, Mauro T, Epstein EH Jr. 2000. Mutations in ATP2C1, encoding a calcium pump, cause Hailey–Hailey disease. Nat Genet 24: 61–65 [DOI] [PubMed] [Google Scholar]
- Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SP, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D. 1995. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science 267: 525–528 [DOI] [PubMed] [Google Scholar]
- Hunt DM, Rickman L, Whittock NV, Eady RA, Simrak D, Dopping-Hepenstal PJ, Stevens HP, Armstrong DK, Hennies HC, Kuster W, et al. 2001. Spectrum of dominant mutations in the desmosomal cadherin desmoglein 1, causing the skin disease striate palmoplantar keratoderma. Eur J Hum Genet 9: 197–203 [DOI] [PubMed] [Google Scholar]
- Hwang LY, Lee JB, Richard G, Uitto JJ, Hsu S. 2003. Type 1 segmental manifestation of Hailey–Hailey disease. J Am Acad Dermatol 49: 712–714 [DOI] [PubMed] [Google Scholar]
- Ikeda S, Welsh EA, Peluso AM, Leyden W, Duvic M, Woodley DT, Epstein EH Jr. 1994. Localization of the gene whose mutations underlie Hailey–Hailey disease to chromosome 3q. Hum Mol Genet 3: 1147–1150 [DOI] [PubMed] [Google Scholar]
- Ikeda S, Shigihara T, Mayuzumi N, Yu X, Ogawa H. 2001. Mutations of ATP2C1 in Japanese patients with Hailey–Hailey disease: Intrafamilial and interfamilial phenotype variations and lack of correlation with mutation patterns. J Invest Dermatol 117: 1654–1656 [DOI] [PubMed] [Google Scholar]
- Jabbari A, Suarez-Farinas M, Dewell S, Krueger JG. 2012. Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol 132: 246–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, Cole JB, Gowan K, Holland PJ, Bennett DC, et al. 2012. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet 44: 676–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman MF, Scheffer H, Stulp R, Pas HH, Nijenhuis M, Heeres K, Owaribe K, Pulkkinen L, Uitto J. 1997. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell 88: 543–551 [DOI] [PubMed] [Google Scholar]
- Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, Zhang W, Bowcock AM. 2011. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet 20: 4025–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari-Dehaghi M, Anhalt GJ, Camilleri MJ, Chernyavsky AI, Chun S, Felgner PL, Jasinskas A, Leiferman KM, Liang L, Marchenko S, et al. 2013. Pemphigus vulgaris autoantibody profiling by proteomic technique. PLoS ONE 8: e57587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Suzuki S, Suzuki M, Oki J, Imamura T. 2004. Bulge- and basal layer-specific expression of fibroblast growth factor-13 (FHF-2) in mouse skin. J Invest Dermatol 122: 1084–1090 [DOI] [PubMed] [Google Scholar]
- Kelsell DP, Norgett EE, Unsworth H, Teh MT, Cullup T, Mein CA, Dopping-Hepenstal PJ, Dale BA, Tadini G, Fleckman P, et al. 2005. Mutations in ABCA12 underlie the severe congenital skin disease harlequin ichthyosis. Am J Hum Genet 76: 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimonis V, DiGiovanna JJ, Yang JM, Doyle SZ, Bale SJ, Compton JG. 1994. A mutation in the V1 end domain of keratin 1 in non-epidermolytic palmar-plantar keratoderma. J Invest Dermatol 103: 764–769 [DOI] [PubMed] [Google Scholar]
- Kivirikko S, McGrath JA, Baudoin C, Aberdam D, Ciatti S, Dunnill MG, McMillan JR, Eady RA, Ortonne JP, Meneguzzi G, et al. 1995. A homozygous nonsense mutation in the α3 chain gene of laminin 5 (LAMA3) in lethal (Herlitz) junctional epidermolysis bullosa. Human Molecular Genetics 4: 959–962 [DOI] [PubMed] [Google Scholar]
- Kleinjan DJ, van Heyningen V. 1998. Position effect in human genetic disease. Hum Mol Genet 7: 1611–1618 [DOI] [PubMed] [Google Scholar]
- Kljuic A, Bazzi H, Sundberg JP, Martinez-Mir A, O’Shaughnessy R, Mahoney MG, Levy M, Montagutelli X, Ahmad W, Aita VM, et al. 2003. Desmoglein 4 in hair follicle differentiation and epidermal adhesion: Evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell 113: 249–260 [DOI] [PubMed] [Google Scholar]
- Koch PJ, Mahoney MG, Ishikawa H, Pulkkinen L, Uitto J, Shultz L, Murphy GF, Whitaker-Menezes D, Stanley JR. 1997. Targeted disruption of the pemphigus vulgaris antigen (desmoglein 3) gene in mice causes loss of keratinocyte cell adhesion with a phenotype similar to pemphigus vulgaris. J Cell Biol 137: 1091–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Fuchs E. 1989. The use of retinoic acid to probe the relation between hyperproliferation-associated keratins and cell proliferation in normal and malignant epidermal cells. J Cell Biol 109: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai-Cheong JE, McGrath JA, Uitto J. 2011. Revertant mosaicism in skin: Natural gene therapy. Trends Mol Med 17: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen FS, Holm NV, Henningsen K. 1986. Atopic dermatitis. A genetic-epidemiologic study in a population-based twin sample. J Am Acad Dermatol 15: 487–494 [PubMed] [Google Scholar]
- Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, et al. 2012. Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet 8: e1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang K, Chen H, Sun F, Xu J, Wu Z, Li P, Zhang L, Du Y, Luan H, et al. 2013. A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjögren’s syndrome at 7q11.23. Nat Genet 45: 1361–1365 [DOI] [PubMed] [Google Scholar]
- Luo Y, Wang Y, Wang Q, Xiao R, Lu Q. 2013. Systemic sclerosis: Genetics and epigenetics. J Autoimmun 41: 161–167 [DOI] [PubMed] [Google Scholar]
- Makris C, Godfrey VL, Krahn-Senftleben G, Takahashi T, Roberts JL, Schwarz T, Feng L, Johnson RS, Karin M. 2000. Female mice heterozygous for IKK γ/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol Cell 5: 969–979 [DOI] [PubMed] [Google Scholar]
- Matthews D, Fry L, Powles A, Weber J, McCarthy M, Fisher E, Davies K, Williamson R. 1996. Evidence that a locus for familial psoriasis maps to chromosome 4q. Nat Genet 14: 231–233 [DOI] [PubMed] [Google Scholar]
- Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, Kowal-Bielecka O, Gay RE, Michel BA, Distler JH, et al. 2010. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 62: 1733–1743 [DOI] [PubMed] [Google Scholar]
- McGrath JA, Kivirikko S, Ciatti S, Moss C, Dunnill GS, Eady RA, Rodeck CH, Christiano AM, Uitto J. 1995a. A homozygous nonsense mutation in the α3 chain gene of laminin 5 (LAMA3) in Herlitz junctional epidermolysis bullosa: Prenatal exclusion in a fetus at risk. Genomics 29: 282–284 [DOI] [PubMed] [Google Scholar]
- McGrath JA, McMillan JR, Dunnill MG, Pulkkinen L, Christiano AM, Rodeck CH, Eady RA, Uitto J. 1995b. Genetic basis of lethal junctional epidermolysis bullosa in an affected fetus: Implications for prenatal diagnosis in one family. Prenat Diagn 15: 647–654 [DOI] [PubMed] [Google Scholar]
- McGrath JA, McMillan JR, Shemanko CS, Runswick SK, Leigh IM, Lane EB, Garrod DR, Eady RA. 1997. Mutations in the plakophilin 1 gene result in ectodermal dysplasia/skin fragility syndrome. Nat Genet 17: 240–244 [DOI] [PubMed] [Google Scholar]
- McGrath JA, Hoeger PH, Christiano AM, McMillan JR, Mellerio JE, Ashton GH, Dopping-Hepenstal PJ, Lake BD, Leigh IM, Harper JI, et al. 1999. Skin fragility and hypohidrotic ectodermal dysplasia resulting from ablation of plakophilin 1. Br J Dermatol 140: 297–307 [DOI] [PubMed] [Google Scholar]
- McLean WH, Rugg EL, Lunny DP, Morley SM, Lane EB, Swensson O, Dopping-Hepenstal PJ, Griffiths WA, Eady RA, Higgins C, et al. 1995. Keratin 16 and keratin 17 mutations cause pachyonychia congenita. Nat Genet 9: 273–278 [DOI] [PubMed] [Google Scholar]
- Meneguzzi G, Marinkovich MP, Aberdam D, Pisani A, Burgeson R, Ortonne JP. 1992. Kalinin is abnormally expressed in epithelial basement membranes of Herlitz’s junctional epidermolysis bullosa patients. Exp Dermatol 1: 221–229 [DOI] [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. 1982. The catalog of human cytokeratins: Patterns of expression in normal epithelia, tumors and cultured cells. Cell 31: 11–24 [DOI] [PubMed] [Google Scholar]
- Monreal AW, Zonana J, Ferguson B. 1998. Identification of a new splice form of the EDA1 gene permits detection of nearly all X-linked hypohidrotic ectodermal dysplasia mutations. Am J Hum Genet 63: 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton NE. 1955. Sequential tests for the detection of linkage. Am J Hum Genet 7: 277–318 [PMC free article] [PubMed] [Google Scholar]
- Nair RP, Henseler T, Jenisch S, Stuart P, Bichakjian CK, Lenk W, Westphal E, Guo SW, Christophers E, Voorhees JJ, et al. 1997. Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet 6: 1349–1356 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Sekigawa I, Ogasawara H, Mitsuishi K, Hira K, Ikeda S, Ogawa H. 2006. Expression of DNMT-1 in patients with atopic dermatitis. Arch Dermatol Res 298: 253–256 [DOI] [PubMed] [Google Scholar]
- Natsuga K, Nishie W, Arita K, Shinkuma S, Nakamura H, Kubota S, Imakado S, Akiyama M, Shimizu H. 2010. Complete paternal isodisomy of chromosome 17 in junctional epidermolysis bullosa with pyloric atresia. J Invest Dermatol 130: 2671–2674 [DOI] [PubMed] [Google Scholar]
- Neldner KH. 1988. Pseudoxanthoma elasticum. Int J Dermatol 27: 98–100 [DOI] [PubMed] [Google Scholar]
- Parente MG, Chung LC, Ryynanen J, Woodley DT, Wynn KC, Bauer EA, Mattei MG, Chu ML, Uitto J. 1991. Human type VII collagen: cDNA cloning and chromosomal mapping of the gene. Proc Natl Acad Sci 88: 6931–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmooij AM, Pas HH, Deviaene FC, Nijenhuis M, Jonkman MF. 2005. Multiple correcting COL17A1 mutations in patients with revertant mosaicism of epidermolysis bullosa. Am J Hum Genet 77: 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmooij AM, Pas HH, Bolling MC, Jonkman MF. 2007. Revertant mosaicism in junctional epidermolysis bullosa due to multiple correcting second-site mutations in LAMB3. J Clin Invest 117: 1240–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternoster L, Standl M, Chen CM, Ramasamy A, Bonnelykke K, Duijts L, Ferreira MA, Alves AC, Thyssen JP, Albrecht E, et al. 2012. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet 44: 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RW, Spargo B. 1961. Electron microscope studies of dermal-epidermal separation in human skin. J Invest Dermatol 36: 213–224 [DOI] [PubMed] [Google Scholar]
- Peluso AM, Bonifas JM, Ikeda S, Hu Z, Devries S, Waldman F, Badura M, O’Connell P, Damen L, Epstein E, et al. 1995. Narrowing of the Hailey–Hailey disease gene region on chromosome 3q and identification of one kindred with a deletion in this region. Genomics 30: 77–80 [DOI] [PubMed] [Google Scholar]
- Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, Kim H, Singh P, Lee A, Chen WV, et al. 2010. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani M, Ruocco V. 1984. “Twin” psoriasis in monozygotic twins. Arch Dermatol 120: 1418–1419 [PubMed] [Google Scholar]
- Pohler E, Mamai O, Hirst J, Zamiri M, Horn H, Nomura T, Irvine AD, Moran B, Wilson NJ, Smith FJ, et al. 2012. Haploinsufficiency for AAGAB causes clinically heterogeneous forms of punctate palmoplantar keratoderma. Nat Genet 44: 1272–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen L, Christiano AM, Airenne T, Haakana H, Tryggvason K, Uitto J. 1994a. Mutations in the γ2 chain gene (LAMC2) of kalinin/laminin 5 in the junctional forms of epidermolysis bullosa. Nat Genet 6: 293–297 [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Christiano AM, Gerecke D, Wagman DW, Burgeson RE, Pittelkow MR, Uitto J. 1994b. A homozygous nonsense mutation in the β3 chain gene of laminin 5 (LAMB3) in Herlitz junctional epidermolysis bullosa. Genomics 24: 357–360 [DOI] [PubMed] [Google Scholar]
- Reed MJ, Purohit A, Woo LW, Newman SP, Potter BV. 2005. Steroid sulfatase: Molecular biology, regulation, and inhibition. Endocr Rev 26: 171–202 [DOI] [PubMed] [Google Scholar]
- Reis A, Kuster W, Eckardt R, Sperling K. 1992. Mapping of a gene for epidermolytic palmoplantar keratoderma to the region of the acidic keratin gene cluster at 17q12-q21. Hum Genet 90: 113–116 [DOI] [PubMed] [Google Scholar]
- Reis A, Hennies HC, Langbein L, Digweed M, Mischke D, Drechsler M, Schrock E, Royer-Pokora B, Franke WW, Sperling K, et al. 1994. Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK). Nat Genet 6: 174–179 [DOI] [PubMed] [Google Scholar]
- Richard G, Korge BP, Wright AR, Mazzanti C, Harth W, Annicchiarico-Petruzzelli M, Compton JG, Bale SJ. 1995. Hailey–Hailey disease maps to a 5 cM interval on chromosome 3q21-q24. J Invest Dermatol 105: 357–360 [DOI] [PubMed] [Google Scholar]
- Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. 1990. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum 33: 1665–1673 [DOI] [PubMed] [Google Scholar]
- Rickman L, Simrak D, Stevens HP, Hunt DM, King IA, Bryant SP, Eady RA, Leigh IM, Arnemann J, Magee AI, et al. 1999. N-terminal deletion in a desmosomal cadherin causes the autosomal dominant skin disease striate palmoplantar keratoderma. Hum Mol Genet 8: 971–976 [DOI] [PubMed] [Google Scholar]
- Ringpfeil F, Lebwohl MG, Christiano AM, Uitto J. 2000. Pseudoxanthoma elasticum: Mutations in the MRP6 gene encoding a transmembrane ATP-binding cassette (ABC) transporter. Proc Natl Acad Sci 97: 6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo WB. 1993. Sjogren–Larsson syndrome. Semin Dermatol 12: 210–218 [PubMed] [Google Scholar]
- Rodriguez TA, Fernandes KE, Dresser KL, Duvic M. 2010. Concordance rate of alopecia areata in identical twins supports both genetic and environmental factors. J Am Acad Dermatol 62: 525–527 [DOI] [PubMed] [Google Scholar]
- Roop DR, Huitfeldt H, Kilkenny A, Yuspa SH. 1987. Regulated expression of differentiation-associated keratins in cultured epidermal cells detected by monospecific antibodies to unique peptides of mouse epidermal keratins. Differentiation 35: 143–150 [DOI] [PubMed] [Google Scholar]
- Ruchusatsawat K, Wongpiyabovorn J, Shuangshoti S, Hirankarn N, Mutirangura A. 2006. SHP-1 promoter 2 methylation in normal epithelial tissues and demethylation in psoriasis. J Mol Med (Berl) 84: 175–182 [DOI] [PubMed] [Google Scholar]
- Russell TJ, Schultes LM, Kuban DJ. 1972. Histocompatibility (HL-A) antigens associated with psoriasis. N Engl J Med 287: 738–740 [DOI] [PubMed] [Google Scholar]
- Russell LJ, DiGiovanna JJ, Hashem N, Compton JG, Bale SJ. 1994. Linkage of autosomal recessive lamellar ichthyosis to chromosome 14q. Am J Hum Genet 55: 1146–1152 [PMC free article] [PubMed] [Google Scholar]
- Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, Bale SJ. 1995. Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet 9: 279–283 [DOI] [PubMed] [Google Scholar]
- Ryynanen M, Knowlton RG, Parente MG, Chung LC, Chu ML, Uitto J. 1991. Human type VII collagen: Genetic linkage of the gene (COL7A1) on chromosome 3 to dominant dystrophic epidermolysis bullosa. Am J Hum Genet 49: 797–803 [PMC free article] [PubMed] [Google Scholar]
- Sakuntabhai A, Ruiz-Perez V, Carter S, Jacobsen N, Burge S, Monk S, Smith M, Munro CS, O’Donovan M, Craddock N, et al. 1999. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat Genet 21: 271–277 [DOI] [PubMed] [Google Scholar]
- Sakuntabhai A, Dhitavat J, Burge S, Hovnanian A. 2000. Mosaicism for ATP2A2 mutations causes segmental Darier’s disease. J Invest Dermatol 115: 1144–1147 [DOI] [PubMed] [Google Scholar]
- Sarig O, Bercovici S, Zoller L, Goldberg I, Indelman M, Nahum S, Israeli S, Sagiv N, Martinez de Morentin H, Katz O, et al. 2012. Population-specific association between a polymorphic variant in ST18, encoding a pro-apoptotic molecule, and pemphigus vulgaris. J Invest Dermatol 132: 1798–1805 [DOI] [PubMed] [Google Scholar]
- Schultz Larsen F. 1993. Atopic dermatitis: A genetic-epidemiologic study in a population-based twin sample. J Am Acad Dermatol 28: 719–723 [DOI] [PubMed] [Google Scholar]
- Smahi A, Courtois G, Vabres P, Yamaoka S, Heuertz S, Munnich A, Israel A, Heiss NS, Klauck SM, Kioschis P, et al. 2000. Genomic rearrangement in NEMO impairs NF-κB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature 405: 466–472 [DOI] [PubMed] [Google Scholar]
- Smith FJ, Morley SM, McLean WH. 2004. Novel mechanism of revertant mosaicism in Dowling–Meara epidermolysis bullosa simplex. J Invest Dermatol 122: 73–77 [DOI] [PubMed] [Google Scholar]
- Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, Liao H, Evans AT, Goudie DR, Lewis-Jones S, et al. 2006. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet 38: 337–342 [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, et al. 2010. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol 126: 581–589; e581–520 [DOI] [PubMed] [Google Scholar]
- Spritz RA. 2008. The genetics of generalized vitiligo. Curr Dir Autoimmun 10: 244–257 [DOI] [PubMed] [Google Scholar]
- Spritz RA. 2011. Recent progress in the genetics of generalized vitiglio. J Genet Genomics 38: 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Pispa J, Hartung AJ, Du Y, Ezer S, Jenks T, Shimada T, Pekkanen M, Mikkola ML, Ko MS, et al. 1997. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc Natl Acad Sci 94: 13069–13074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley JR. 1993. Cell adhesion molecules as targets of autoantibodies in pemphigus and pemphigoid, bullous diseases due to defective epidermal cell adhesion. Adv Immunol 53: 291–325 [DOI] [PubMed] [Google Scholar]
- Suarez-Farinas M, Lowes MA, Zaba LC, Krueger JG. 2010. Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA). PLoS ONE 5: e10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya RD, Coda AB, Sinha AA. 2010. Transcriptional profiling in alopecia areata defines immune and cell cycle control related genes within disease-specific signatures. Genomics 96: 146–153 [DOI] [PubMed] [Google Scholar]
- Sudbrak R, Brown J, Dobson-Stone C, Carter S, Ramser J, White J, Healy E, Dissanayake M, Larregue M, Perrussel M, et al. 2000. Hailey–Hailey disease is caused by mutations in ATP2C1 encoding a novel Ca2+ pump. Hum Mol Genet 9: 1131–1140 [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, et al. 2009. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum 60: 1065–1075 [DOI] [PubMed] [Google Scholar]
- Tidman MJ, Eady RA. 1985. Evaluation of anchoring fibrils and other components of the dermal-epidermal junction in dystrophic epidermolysis bullosa by a quantitative ultrastructural technique. J Invest Dermatol 84: 374–377 [DOI] [PubMed] [Google Scholar]
- Tidman MJ, Eady RA. 1986. Hemidesmosome heterogeneity in junctional epidermolysis bullosa revealed by morphometric analysis. J Invest Dermatol 86: 51–56 [DOI] [PubMed] [Google Scholar]
- Tiepolo L, Zuffardi O, Fraccaro M, di Natale D, Gargantini L, Muller CR, Ropers HH. 1980. Assignment by deletion mapping of the steroid sulfatase X-linked ichthyosis locus to Xp223. Hum Genet 54: 205–206 [DOI] [PubMed] [Google Scholar]
- Tomfohrde J, Silverman A, Barnes R, Fernandez-Vina MA, Young M, Lory D, Morris L, Wuepper KD, Stastny P, Menter A, et al. 1994. Gene for familial psoriasis susceptibility mapped to the distal end of human chromosome 17q. Science 264: 1141–1145 [DOI] [PubMed] [Google Scholar]
- Toumi A, Abida O, Ben Ayed M, Masmoudi A, Turki H, Masmoudi H. 2013. Cytokine gene polymorphisms in Tunisian endemic pemphigus foliaceus: A possible role of il-4 variants. Hum Immunol 74: 658–665 [DOI] [PubMed] [Google Scholar]
- Traupe H. 1999. Functional X-chromosomal mosaicism of the skin: Rudolf Happle and the lines of Alfred Blaschko. Am J Med Genet 85: 324–329 [DOI] [PubMed] [Google Scholar]
- Trembath RC, Clough RL, Rosbotham JL, Jones AB, Camp RD, Frodsham A, Browne J, Barber R, Terwilliger J, Lathrop GM, et al. 1997. Identification of a major susceptibility locus on chromosome 6p and evidence for further disease loci revealed by a two stage genome-wide search in psoriasis. Hum Mol Genet 6: 813–820 [DOI] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, et al. 2012. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 44: 1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, Christiano AM. 1992. Molecular genetics of the cutaneous basement membrane zone. Perspectives on epidermolysis bullosa and other blistering skin diseases. J Clin Invest 90: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, Chung-Honet LC, Christiano AM. 1992. Molecular biology and pathology of type VII collagen. Exp Dermatol 1: 2–11 [DOI] [PubMed] [Google Scholar]
- Uitto J, Pulkkinen L, Christiano AM. 1994. Molecular basis of the dystrophic and junctional forms of epidermolysis bullosa: Mutations in the type VII collagen and kalinin (laminin 5) genes. J Invest Dermatol 103: 39S–46S [DOI] [PubMed] [Google Scholar]
- Vailly J, Verrando P, Champliaud MF, Gerecke D, Wagman DW, Baudoin C, Aberdam D, Burgeson R, Bauer E, Ortonne JP. 1994. The 100-kDa chain of nicein/kalinin is a laminin B2 chain variant. Eur J Biochem 219: 209–218 [DOI] [PubMed] [Google Scholar]
- Vassar R, Coulombe PA, Degenstein L, Albers K, Fuchs E. 1991. Mutant keratin expression in transgenic mice causes marked abnormalities resembling a human genetic skin disease. Cell 64: 365–380 [DOI] [PubMed] [Google Scholar]
- Verrando P, Blanchet-Bardon C, Pisani A, Thomas L, Cambazard F, Eady RA, Schofield O, Ortonne JP. 1991. Monoclonal antibody GB3 defines a widespread defect of several basement membranes and a keratinocyte dysfunction in patients with lethal junctional epidermolysis bullosa. Lab Invest 64: 85–92 [PubMed] [Google Scholar]
- Wada T, Shirakata Y, Takahashi H, Murakami S, Iizuka H, Suzuki H, Hashimoto K. 2003. A Japanese case of segmental Darier’s disease caused by mosaicism for the ATP2A2 mutation. Br J Dermatol 149: 185–188 [DOI] [PubMed] [Google Scholar]
- Wang Y, Fan PS, Kahaleh B. 2006. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum 54: 2271–2279 [DOI] [PubMed] [Google Scholar]
- Webster D, France JT, Shapiro LJ, Weiss R. 1978. X-linked ichthyosis due to steroid-sulphatase deficiency. Lancet 1: 70–72 [DOI] [PubMed] [Google Scholar]
- Weidinger S, Illig T, Baurecht H, Irvine AD, Rodriguez E, Diaz-Lacava A, Klopp N, Wagenpfeil S, Zhao Y, Liao H, et al. 2006. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol 118: 214–219 [DOI] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT. 1995. Heterochromatin and gene expression in Drosophila. Annu Rev Genet 29: 577–605 [DOI] [PubMed] [Google Scholar]
- Yamanishi K. 1996. Gene analysis of human skin and skin diseases. J Dermatol Sci 11: 169–176 [DOI] [PubMed] [Google Scholar]
- Yan L, Wang JM, Zeng K. 2012. Association between HLA-DRB1 polymorphisms and pemphigus vulgaris: A meta-analysis. Br J Dermatol 167: 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Broady R, Huang Y, Wang Y, Yu J, Gao M, Levings M, Wei S, Zhang S, Xu A, et al. 2012. Transcriptome analysis reveals markers of aberrantly activated innate immunity in vitiligo lesional and non-lesional skin. PLoS ONE 7: e51040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. 2012. Genome-wide association study of skin complex diseases. J Dermatol Sci 66: 89–97 [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang R, Li X, Yin G, Niu X, Hou R. 2007. The mRNA expression and promoter methylation status of the p16 gene in colony-forming cells with high proliferative potential in patients with psoriasis. Clin Exp Dermatol 32: 702–708 [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Huang W, Yang S, Sun LD, Zhang FY, Zhu QX, Zhang FR, Zhang C, Du WH, Pu XM, et al. 2009. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet 41: 205–210 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lu Q. 2008. DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmun Rev 7: 376–383 [DOI] [PubMed] [Google Scholar]
- Zhou X, Krueger JG, Kao MC, Lee E, Du F, Menter A, Wong WH, Bowcock AM. 2003. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics 13: 69–78 [DOI] [PubMed] [Google Scholar]
- Zhu H, Shang D, Sun M, Choi S, Liu Q, Hao J, Figuera LE, Zhang F, Choy KW, Ao Y, et al. 2011. X-linked congenital hypertrichosis syndrome is associated with interchromosomal insertions mediated by a human-specific palindrome near SOX3. Am J Hum Genet 88: 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]