Unique cell wall lipids play key roles in the physiology and pathogenicity of Mycobacterium tuberculosis. Drug discovery efforts aim at targeting their biosynthesis, export, and regulation.
Abstract
Mycobacterium tuberculosis (Mtb) lipids are indelibly imprinted in just about every key aspect of tuberculosis (TB) basic and translational research. Although the interest in these compounds originally stemmed from their abundance, structural diversity, and antigenicity, continued research in this field has been driven by their important contribution to TB pathogenesis and their interest from the perspective of drug, vaccine, diagnostic, and biomarker development. This article summarizes what is known of the roles of lipids in the physiology and pathogenicity of Mtb and the exciting developments that have occurred in recent years in identifying new lead compounds targeting their biogenesis.
The mycobacterial cell envelope is the dominant feature of the biology of Mycobacterium tuberculosis (Mtb) and other mycobacteria, based on sugars and lipids of exceptional structure. Cell envelope lipids constitute ∼40% of the cell dry mass, although this percentage may vary depending on species/isolate and growth conditions (Goren and Brennan 1979; Minnikin et al. 1982). The particular chemical structures and organization of these lipids between the inner and outer membrane of the mycobacterial cell envelope account for much of its impermeability to biocides and for the unique staining properties of mycobacteria that aid in the diagnosis of infected specimens (Goren and Brennan 1979; Jarlier and Nikaido 1994; Brennan and Nikaido 1995). Being located at the interface between the bacterium and the host, the outer membrane and capsular lipids of Mtb also play important roles in directing host–pathogen interactions. Several lipid components of Mtb (e.g., isoprenoid lipids, glycerophospholipids, phosphatidylinositol mannosides, lipoarabinomannan, mycolic acids, and trehalose mycolates) are produced by all mycobacterial species, consistent with their requirement for growth. The biosynthesis of some of these lipids is the site of action of anti-TB drugs such as isoniazid (INH) and ethionamide (ETH), as well as multiple other lead compounds under development. Other lipids (e.g., long-chain polymethyl-branched fatty acid–containing acyltrehaloses and long-chain diols) are not essential for growth but are unique to pathogenic mycobacteria and have been involved in pathogenesis.
Studies initiated in 1927 by R.J. Anderson and colleagues mark the beginning of the pioneering work that has led to the discovery and early structural definition of some of the most biologically relevant lipids of Mtb, including phosphatidylinositol mannosides, mycolic acids, and branched fatty acids and polyols (Goren and Brennan 1979). Studies conducted for decades to follow by generations of outstanding biochemists in France, the United Kingdom, and the United States, in particular, have resulted in a thorough understanding of the chemical structures of these lipids. Their detailed descriptions have been compiled in several recent books and reviews and the reader is referred to these for further reading on this topic (Brennan 1988; Barry et al. 1998; Kaur et al. 2009; Daffé et al. 2014). Developments in the genetic manipulation of mycobacteria in the 1990s and the release of the first complete genome sequence of Mtb in 1998 have provided a major impetus to the study of the biosynthesis of these compounds with the result that many of the enzymes and transporters involved in their biogenesis have now been identified. Following the classification of Mtb into six major genetic lineages associated with specific geographical regions (Gagneux and Small 2007), recent years have also seen a return of interest in comparative analyses of the lipid content of clinical isolates from different regions of the world in the hope of identifying correlates of pathogenicity. Interestingly, not only are these studies providing new insights into the biochemical diversity of Mtb and the putative roles of some lipids in host–pathogen interactions (Daffé et al. 1991; Constant et al. 2002; Reed et al. 2004, 2007; Tsenova et al. 2005; Sinsimer et al. 2008; Huet et al. 2009; Krishnan et al. 2011), they are also shedding light on the genetic basis of observations made some 20 to 40 years ago by early investigators in the field (Goren et al. 1974a; Daffé et al. 1991; Krishnan et al. 2011; Supply et al. 2013). What is known of the biosynthesis of Mtb lipids has been reviewed recently (Daffé et al. 2014). This review focuses on our current understanding of the roles of cell wall lipids in the physiology and pathogenicity of Mtb and describes ongoing drug discovery efforts aimed at targeting their biosynthesis, export, and regulation.
PHYSICAL ORGANIZATION AND PHYSIOLOGICAL ROLES OF Mtb CELL ENVELOPE LIPIDS
Architecture of the Mtb Cell Envelope
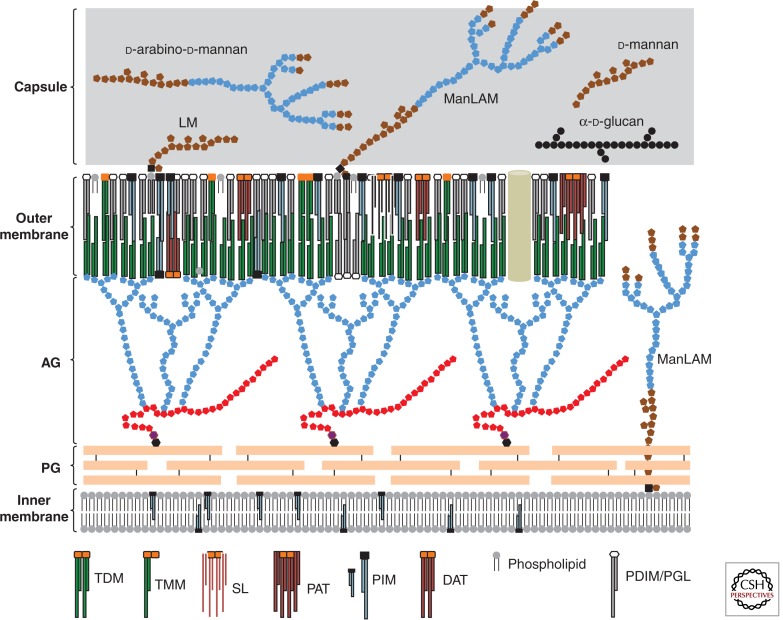
A schematic representation of the cell envelope’s architecture of Mtb is presented in Figure 1. The innermost layer is the plasma membrane that seems typical of bacterial membranes. Outside the plasma membrane is a massive cell wall “core” comprised of peptidoglycan (PG) covalently attached to the heteropolysaccharide arabinogalactan (AG), in turn esterified at its nonreducing ends to α-alkyl, β-hydroxy long-chain (C60-C90) mycolic acids. Intercalated within this lipid environment are the “free” (noncovalently linked) lipids and lipoglycans that have intrigued researchers for more than eight decades: the phosphatidylinositol mannosides (PIMs), the phthiocerol dimycocerosates (PDIMs), the phenolic glycolipids (PGLs), a variety of acyltrehaloses, mannose-capped lipoarabinomannan (ManLAM), etc. (Figs. 2 and 3). The covalently bound mycolic acids of the cell wall core and the free lipids form the inner and outer leaflets, respectively, of a highly impermeable asymmetrical bilayer (known as the “mycomembrane” or outer membrane) that confers to mycobacteria their characteristic resistance to many therapeutic agents (Minnikin et al. 1982; Jarlier and Nikaido 1994; Hoffmann et al. 2008; Zuber et al. 2008) (Fig. 1). Finally, a loosely attached capsular-like structure outside the outer membrane of Mtb was shown to mainly consist of polysaccharides and proteins with only minor amounts of lipids (2%–3% of the material) (Lemassu and Daffé 1994; Ortalo-Magné et al. 1995; Sani et al. 2010) (Fig. 1). Thus, in spite of being Gram-positive bacteria, mycobacteria possess a cell envelope characterized by the presence of an outer membrane and a periplasmic space.
Figure 1.
Schematic representation of the Mtb cell envelope. Many of the classes of (glyco)lipids discussed in the text are represented schematically and are shown in probable locations in the cell envelope. Light blue symbols represent arabinose residues, red symbols represent galactose residues, brown symbols represent mannose residues, and black circles represent glucose residues. d-arabino-d-mannan, d-glucan, and d-mannan are capsular polysaccharides. Mycolic acid chains are shown in dark green. LM, lipomannan; ManLAM, mannose-capped lipoarabinomannan; AG, arabinogalactan; PG, peptidoglycan.
Figure 2.
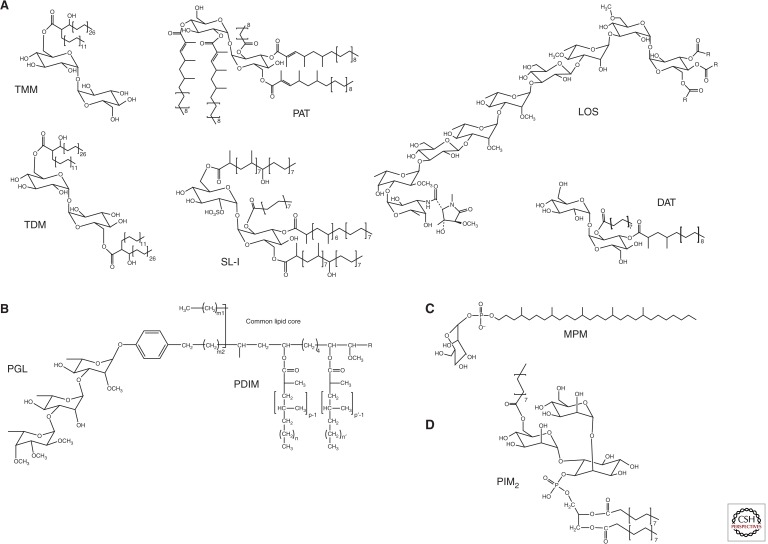
Structures of Mtb lipids. (A) Structures of acyltrehaloses: trehalose monomycolate (TMM); trehalose dimycolate (TDM); sulfolipid (SL-I); diacyltrehalose (DAT); polyacyltrehalose (PAT); and the lipooligosaccharide (LOS) of Mtb Canettii (R=Ac). The major sulfolipid SL-I (2,3,6,6′-tetraacyl α-α′-trehalose-2′-sulfate) is represented. In SL-I, trehalose is sulfated at the 2′ position and esterified with palmitic acid and the multimethyl-branched phthioceranic and hydroxyphthioceranic acids. In DAT (2,3-di-O-acyltrehalose), trehalose is esterified with palmitic acid and the multimethyl-branched mycosanoic acid. In PAT, trehalose is esterified with palmitic acid and the multimethyl-branched mycolipenic acids. (B) Structures of the phthiocerol dimycocerosate (PDIM) and phenolic glycolipid (PGL) of Mtb. p, p′ = 3–5; n, n′ = 16–18; m1 = 20–22; m2 = 15–17; R = CH2-CH3 or CH3. (C) Structure of the predominant mannosyl-β-1-phosphomycoketide (MPM) from Mtb H37Rv. (D) Structure of a triacylated phosphatidylinositol dimannoside (PIM2), one of the major forms of PIMs produced by Mtb.
Figure 3.
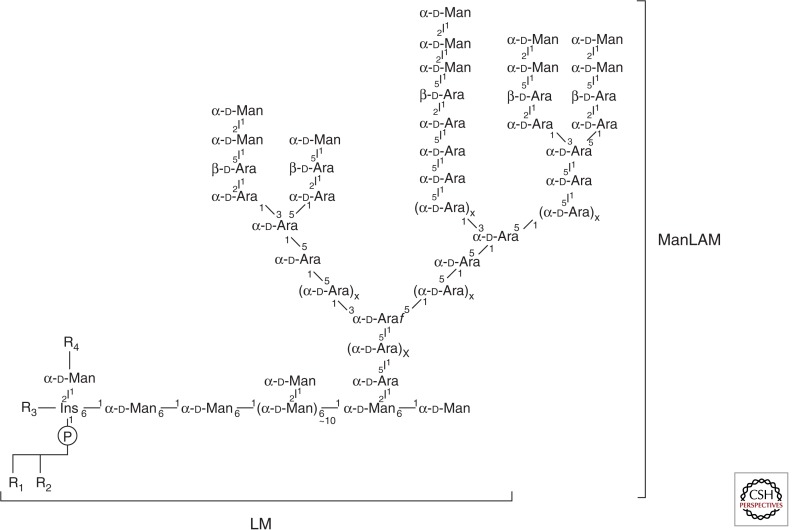
Structures of Mtb lipoglycans. Mannose-capped lipoarabinomannan (ManLAM); the mannan moiety of ManLAM consists of ∼20–30 Manp residues and the arabinan polymers of ∼60 Araf units; the precise number of arabinan chain(s) attached to mannan is still uncertain. Lipomannan (LM) is devoid of the arabinan chains of ManLAM. R1, R2, R3, R4 are tuberculostearic acid, oleic acid, or palmitic acid.
Mycolic Acids
The interest in drugs targeting the biosynthesis of mycolic acids is clearly illustrated by the therapeutic efficacy of INH and ETH (Table 1). This essentially owes to their role as key structural elements of the outer membrane. The mycolic acids covalently linked to AG represent about half of the weight of the cell wall core (Draper 1998). Mycolic acids also occur as acyl substituents esterifying a variety of outer membrane (glyco)lipids including trehalose mono- and dimycolates (Fig. 2), glycerol monomycolate, and glucose monomycolate. They play key roles in the permeability of the cell envelope, the ability of Mtb to form biofilms (Ojha et al. 2005, 2010; Sambandan et al. 2013), and the pathogenicity of the bacterium during the actively replicating and persistent stages of the infection (Barry et al. 1998; Daffé and Draper 1998; Yuan et al. 1998; Dubnau et al. 2000; Glickman et al. 2000; Sugawara et al. 2002; Vander Beken et al. 2011).
Table 1.
TB drug discovery pipeline showing the inhibitors targeting lipid biosynthesis
| Pathway targeted | (Proposed) target protein | Inhibitor | ||
|---|---|---|---|---|
| Discovery | Preclinical development | Clinical development | ||
| Mycolic acids | ||||
| InhA | CD117 (Vilchèze et al. 2011) Pyridomycin (Hartkoorn et al. 2012) Triclosan derivatives (Pan and Tonge 2012; North et al. 2013) Novel structural classes (GlaxoSmithKline) |
|||
| HadABC | ISO and TAC analogs (Phetsuksiri et al. 1999; Bhowruth et al. 2006; Alahari et al. 2007; Shahab et al. 2010; Coxon et al. 2013) NAS-21, NAS-91 and analogs (Bhowruth et al. 2008; Gratraud et al. 2008) |
|||
| KasA/KasB | Thiolactomycin and analogs (Pan and Tonge 2012; North et al. 2013) |
|||
| Pks13 | Thiophene compounds; Compound 3′ (Ioerger et al. 2013; Wilson et al. 2013) |
|||
| FadD32 | 4,6-diaryl-5,7-dimethyl coumarins (Stanley et al. 2013) |
|||
| Antigen 85 A, B, and C | Ebselen (Favrot et al. 2013) I3-AG85 (Warrier et al. 2012) |
|||
| MmpL3 | Adamantyl ureas (Grzegorzewicz et al. 2012a) BM212 and analogs (La Rosa et al. 2012; Poce et al. 2013) C215 (Stanley et al. 2012) SPIRO2 and THPP1 (Remuinan et al. 2013) Compound 2 (Ioerger et al. 2013) |
SQ109 (Tahlan et al. 2012) (Sequella) |
||
| Unknown (Inhibition of oxygenated mycolic acid biosynthesis) |
PA-824 (Stover et al. 2000) OPC-67683 (Matsumoto et al. 2006) |
|||
| EthR | ETH “boosters”: BDM 31343 and analogs (Willand et al. 2009; Villemagne et al. 2012) | |||
| Decaprenyl phosphate | ||||
| IspC | Fosfomidomycin analogs (Obiol-Pardo et al. 2011; Uh et al. 2011) |
|||
| Isoprenoid-based biosynthetic precursors | ||||
| PG biosynthesis | MraY | Capuramycin analogs (Sequella) |
SQ641 (lead capuramycin analog) (Sequella) |
|
| PG biosynthesis | WecA | Caprazamycin derivatives (CPZEN-45) (Ishizaki et al. 2013) |
||
| LAM (and AG) biosynthesis |
DprE1 | DNB1 (Christophe et al. 2009) VI-9376 (Magnet et al. 2010) 377790 (Stanley et al. 2012) TCA-1 (Wang et al. 2013) Several synthetic compounds (TB Alliance, Scripps) |
Benzothiazinones (BTZ043) (Makarov et al. 2009) |
|
| Menaquinones | ||||
| MenA | Aurachin RE, Ro 48-8071 and analogs (Dhiman et al. 2009; Debnath et al. 2012) |
|||
| MenB | 1,4-benzoxazines; 4-oxo-4-phenylbut-2-enoates (Li et al. 2010; 2011) |
|||
| MenE | Sulfonyladenosine analogs (Lu et al. 2008; 2012) |
|||
| PGL | ||||
| FadD22 | 5′-O-[N-(4-hydroxybenzoyl)sulfamoyl]-adenosine (Ferreras et al. 2008) |
|||
| Ser/Thr kinases | ||||
| PknA PknB PknG |
Several chemical scaffolds (Vertex Pharmaceuticals Inc.) (Magnet et al. 2010; Danilenko et al. 2011; Lougheed et al. 2011; Chapman et al. 2012) |
|||
See text and the Working Group for New Drugs, Stop TB Partnership (http://www.newtbdrugs.org) for details.
Inner and Outer Membrane Extractable Lipids
Glycerophospholipids, PIM, LM, and ManLAM. Glycerophospholipids (phosphatidylinositol [PI], phosphatidylglycerol, phosphatidylserine [PS], phosphatidylethanolamine [PE], cardiolipin [CL]), and mannosylated forms of PI known collectively as PIMs (Fig. 2) are the major lipid constituents of the plasma membrane of Mtb. In addition, the plasma membrane of Mtb contains major lipoglycans known as lipomannan (LM) and ManLAM (Fig. 3) (Pitarque et al. 2008), and a number of isoprenoids that play various critical functions in the cell. Among them, decaprenyl phosphate (Dec-P) serves as a carrier of activated sugars in the synthesis of essential cell envelope polymers including PG, AG, LM, and ManLAM (Kaur et al. 2009), and menaquinones are essential components of the respiratory chain (Weinstein et al. 2005; Dhiman et al. 2009). Thus, as in other bacteria, plasma membrane lipids play a vital role in Mtb, providing a selective permeability barrier around the cell and participating in a number of critical physiological processes, including electron transport, ATP formation, DNA replication, and the biosynthesis of a variety of cell envelope components.
Glycerophospholipids (particularly PE), PIM, LM, and ManLAM are also found in the outermost layers of the cell envelope of Mtb and other Mycobacterium species where they may interact with the host (Ortalo-Magné et al. 1996; Pitarque et al. 2008; Dhiman et al. 2011). Whether Mtb requires phosphatidylglycerol, CL, PS, and PE for growth is at present not known. PI, PIM, LM, and ManLAM, in contrast, are all required for the viability of Mtb in vitro (Kaur et al. 2009; Guerin et al. 2010). Emerging data indicate that PIM play important roles in the permeability of the cell envelope, inner membrane integrity, and regulation of cell septation and division (Parish et al. 1997; Korduláková et al. 2002; Patterson et al. 2003; Morita et al. 2005, 2006). Likewise, structural defects in LM and ManLAM were shown to affect the β-lactam susceptibility and acid-fast staining properties of mycobacterial cells, indicating an equally important role of these molecules in cell envelope integrity (Fukuda et al. 2013).
Trehalose esters, PDIM, and PGL. Other outer membrane/surface-exposed lipids include trehalose monomycolates (TMM) and trehalose dimycolates (TDM), PDIM, sulfolipids (SL), diacyltrehaloses (DAT), polyacyltrehaloses (PAT), phenolic glycolipids (PGL), and lipooligosaccharides (LOS) (Fig. 2). In addition to serving as a mycolic acid donor in the formation of TDM and the cell wall core, TMM was recently identified as the likely form under which mycolic acids, which are synthesized in the cytoplasm, translocate to the periplasm or outer membrane (Grzegorzewicz et al. 2012a). Consistently, compounds that prevent TMM translocation are potent inhibitors of Mtb (Grzegorzewicz et al. 2012a; Tahlan et al. 2012; Stanley et al. 2012; La Rosa et al. 2012; Remuinan et al. 2013). In contrast to TDM and TMM, which are considered essential lipid components of the cell envelope of all mycobacteria, SL, DAT, PAT, and PDIM are more restricted in their distribution and are not essential for the growth of Mtb in vitro, although they have been involved in pathogenicity (see the section Roles of Mtb Lipids in Pathogenicity). PDIM and closely related phthiocerol diphthioceranates are essentially confined to pathogenic Mycobacterium species (Daffé and Lanéelle 1988). Consistent with their important roles in the permeability of the cell envelope (Camacho et al. 2001) and pathogenicity, PDIMs are apparently produced by all virulent clinical isolates of Mtb. DAT and PAT are confined to virulent isolates of the Mycobacterium tuberculosis complex (Goren and Brennan 1979; Jackson et al. 2007), and SL are exclusively found in the human pathogen Mtb (Goren 1990). Structural similarities between SL, DAT, and PAT and, to some extent PDIM, particularly the fact that they all are esterified with long-chain polymethyl-branched fatty acids, is suggestive of at least partially redundant functions in the cells. One of them is to alleviate the propionate-mediated stress undergone by Mtb when the bacterium switches to host cholesterol and fatty acids as major carbon sources (Singh et al. 2009; Lee et al. 2013). The propionyl-CoA generated upon β-oxidation of cholesterol is converted to methylmalonyl-CoA by the propionyl-CoA carboxylase and then used by dedicated polyketide synthases in the elongation of the polymethyl-branched fatty acids esterifying PDIM, SL, DAT, and PAT. Independent from this metabolic function and suggestive of their collective role in the permeability of the cell envelope, Mtb mutants deficient in the synthesis of more than one polymethyl-branched fatty acid–containing lipid (either PDIM and SL or DAT, PAT, and SL) but not mutants lacking only one lipid species, lose the ability to stain with neutral red (Cardona et al. 2006). Finally, DAT and PAT have been involved in the retention of the capsular material at the surface of Mtb (Rousseau et al. 2003a).
PGL and LOS are other lipids acylated by polymethyl-branched fatty acids produced by Mtb and related tubercle bacilli, albeit not all lineages (Fig. 2). LOS have thus far only been found in Mycobacterium canettii (Daffé et al. 1991), a representative of smooth tubercle bacilli that seem to have originated from the same pool of ancestors as Mtb but rarely causes human disease (Supply et al. 2013). Based on studies performed in Mycobacterium marinum (Ren et al. 2007), the presence of LOS in the cell envelope of M. canettii may account for the particular smooth colonial morphotype of this strain. The ability to synthesize PGL, on the other hand, has only been retained by M. canettii and some Mtb isolates of the East Asian/Beijing lineage (Daffé et al. 1987; Constant et al. 2002; Reed et al. 2004; Huet et al. 2009). PGL have been involved in pathogenicity (see the section Roles of Mtb Lipids in Pathogenicity). In an attempt to correlate the lipid content with the virulence of Mtb isolates, Goren et al. characterized a methoxylated phenolphthiocerol (a nonglycosylated variant of PGL), the so-called “attenuation indicator lipid” (Goren et al. 1974a). This lipid and its unmethylated form were detected in East Asian/Beijing isolates and found to accumulate in all Indo-Oceanic Mtb strains examined (Krishnan et al. 2011). Similarly, Beijing strains were reported to accumulate variants of PDIM and eventually PGL, known as phthiotriol and glycosylated phenolphthiotriol dimycocerosates (Huet et al. 2009). The correlation between the occurrence of these lipids and variations in virulence remains, however, unclear (Huet et al. 2009; Krishnan et al. 2011). PGL and LOS are clearly not essential for growth but they are potent B-cell antigens making them potentially useful, albeit lineage-specific, serodiagnostics (Brennan 1988; He et al. 1991; Vera-Cabrera et al. 1994; Simonney et al. 1995, 1996; Constant et al. 2002; Julian et al. 2002).
Other Lipids
Another family of lipids known as the mannosyl-β-1-phosphomycoketides (MPM) are produced by slow growing mycobacteria including Mtb, Mycobacterium avium, and M. marinum (Fig. 2). They are found inside the cells and released in the culture medium. They are not essential for the growth of Mtb but are thought to be involved in pathogenicity (Matsunaga and Sugita 2012).
Triglycerides (TAG) are considered intracellular lipids of Mtb. They represent the main apolar lipid when glycerol serves as the major carbon source in the medium (Brennan 1988). TAGs are thought to serve as an energy reserve for the long-term survival of Mtb during the persistence phase of infection (Brennan 1988; Daniel et al. 2004), as well as a means by which free fatty acids are detoxified. Mtb Beijing family isolates accumulate large quantities of TAG, possibly as a result of the up-regulation of the TAG synthase, Rv3130c (Reed et al. 2007).
Regulation of Lipid Synthesis
The physiological status of mycobacterial bacilli (i.e., age of the culture, actively replicating vs. persistent) and a number of environmental factors including temperature, oxygen tension, and nitrogen and carbon sources are known to impact their lipid composition, particularly phospholipids, TAG, PIM, LM, ManLAM, mycolic acid–based glycolipids, and polymethyl-branched fatty acid–containing lipids (Brennan 1988; Barry et al. 1998; Daffé and Draper 1998; Singh et al. 2009; Dhiman et al. 2011; Lee et al. 2013; Yang et al. 2013). Less is known of the regulation of lipid synthesis during host infection. PDIM and mycolic acids are apparently produced in abundant quantities by in vivo–grown tubercle bacilli (Kanai et al. 1970; Bhamidi et al. 2012). Yet, variations in the acid-fast characteristics of Mtb during mouse and guinea pig infection suggest that the bacterium undergoes drastic adaptative changes of its lipid composition in vivo (Ryan et al. 2010). Increased production of SL and DAT during host infection is supported by the up-regulation of biosynthetic genes on phagocytosis of Mtb by human primary macrophages or during progressive pulmonary TB infection in mice (Graham and Clark-Curtiss 1999; Rodriguez et al. 2013). The virulence-associated two-component regulator PhoP-PhoR may be involved in this up-regulation as it has been found to coordinately and positively regulate the synthesis of SL, DAT, and PAT in Mtb (Gonzalo-Asencio et al. 2006; Walters et al. 2006). The metabolic switching of Mtb to host lipids as carbon sources during infection and the associated increase in the production of PDIM, SL, DAT, and PAT were shown to be facilitated by the regulatory protein WhiB3 (Singh et al. 2009). In recent years, PknA, PknB, and other members of the Ser/Thr kinase family were found to regulate the activity of multiple enzymes and transporters involved in the biosynthesis of mycolic acids, ManLAMs, and PDIMs (Molle and Kremer 2010). It is thus apparent that lipid synthesis in Mtb is tightly controlled and that regulation occurs both at the transcriptional and posttranslational levels.
ROLES OF Mtb LIPIDS IN PATHOGENICITY
Several excellent reviews have been published about the biological activities of Mtb lipids, including very recently (Daffé and Draper 1998; Russell et al. 2002; Karakousis et al. 2004; Bertozzi and Schelle 2008; Britton and Triccas 2008; Gilleron et al. 2008; Glickman 2008; Guilhot et al. 2008; Guenin-Macé et al. 2009; Neyrolles and Guilhot 2011; Russell 2011; Daffé et al. 2014). Thus, only a brief overview of what is known and remains to be investigated about the involvement of lipids in the pathogenesis of TB will be presented here.
The existence of a possible correlation between the lipid content of Mtb clinical isolates and their pathogenicity was noted early on by Middlebrook and Goren among others. These studies led to the identification of SL, TDM, and PDIM and derivatives as lipids of potential relevance to TB immunopathogenesis (Bloch 1950; Goren et al. 1974a,b; Daffé and Draper 1998). Further work on the biological activities of purified lipids in vitro and in vivo followed, in the postgenomic era, by an examination of the virulence phenotype and immune responses induced by Mtb mutants deficient in various aspects of their biosynthesis lent further support to the involvement of lipids in host–pathogen interactions. Their contribution to the infection process accompanies every step of the life cycle of the bacterium. The entry of Mtb inside host macrophages and dendritic cells is essentially mediated by specialized phagocytic receptors including complement receptors, scavenger receptors, and C-type lectins, such as the mannose receptor, DC-SIGN, and Mincle. PIM, TDM, and ManLAM are among the predominant glycolipids mediating Mtb’s interactions with these receptors (Villeneuve et al. 2005; Schlesinger et al. 2008; Ishikawa et al. 2009; Neyrolles and Guilhot 2011). Once inside phagocytic cells, Mtb resides in a phagosome that fails to fuse with lysozymes. The isoprenoid-derived compounds, isotuberculosinol and tuberculosinol, ManLAM, TDM, PDIM, SL, and DAT all have been implicated in the impairment of phagosome maturation, although the mechanisms involved and amount of supporting data varies for each compound (Daffé and Draper 1998; Brodin et al. 2010; Neyrolles and Guilhot 2011; Russell 2011; Mann and Peters 2012). Inside the cells, Mtb releases significant amounts of proteins and lipids, among them TMM, TDM, PIM, PGL, PE, phosphatidylglycerol, CL, and ManLAM, that traffic within the cell and may be released through exocytosis (Russell et al. 2002; Russell 2011). These molecules are not only taken up by bystander antigen-presenting cells, they also act as modulators of the function of the host cell and surrounding tissue, impacting the secretion of pro- and anti-inflammatory cytokines (e.g., PGL, TDM, mycolic acids, SL, PIM, LM, ManLAM, and lipoproteins), apoptosis (lipoproteins, PIM, ManLAM, and TDM), T-cell functions (ManLAM, DAT, and PAT), the induction of foamy macrophages (oxygenated mycolates and TDM), and granuloma formation (TMM, TDM, and ManLAM) (Daffé and Draper 1998; Jackson et al. 2007; Gilleron et al. 2008; Peyron et al. 2008; Guenin-Macé et al. 2009; Neyrolles and Guilhot 2011; Rajni et al. 2011; Russell 2011; Vander Beken et al. 2011; Li et al. 2012; Sakamoto et al. 2013; Vir et al. 2013). Importantly, several Mtb lipids, including diacylated forms of SL, PIM, ManLAM, MPM, and mycolyl lipids, represent themselves as antigens for immune recognition and are presented to T lymphocytes by MHC-I-like molecules of the CD1 family (Matsunaga and Sugita 2012; Arora et al. 2013; Ly et al. 2013; Van Rhijn et al. 2013). Some of these lipids may show potential as subunit vaccines (Guiard et al. 2009; Arora et al. 2013). Finally, ManLAM, SL, TDM, PGL, LOS, DAT, and lipoproteins are also potent inducers of humoral immune responses.
Although the critical involvement of PDIM and TDM in TB pathogenesis is now supported by a wealth of in vitro and in vivo data (Guilhot et al. 2008; Neyrolles and Guilhot 2011; Rajni et al. 2011; Li et al. 2012), the role(s) played by other prominent (glyco)lipids including (iso)tuberculosinol, SL, DAT, PAT, PGL, some PDIM variants, and ManLAM during host infection are far from being as clear. Their roles in pathogenicity typically come into question when their demonstrated biological activities in vitro or in vivo (e.g., inhibition of phagosome maturation and immunomodulatory activities), usually as purified molecules, fail to translate into a clear “virulence” phenotype in Mtb strains genetically engineered to produce or, on the contrary, lose the ability to produce them. Classical examples of this include, for instance, SL, whose plethora of biological activities (Goren and Brennan 1979; Goren 1990; Daffé and Draper 1998; Bertozzi and Schelle 2008) fail to translate into a clear replication or persistence advantage in infected mice or guinea pigs (Rousseau et al. 2003b); DAT and PAT, whose absence from Mtb H37Rv does not hamper the ability of this strain to grow in mice (Rousseau et al. 2003a); or ManLAM, the suppression of the bioactive mannose caps of which fails to translate into any detectable phenotype in macrophages and animal models of infection (Afonso-Barroso et al. 2012). A combination of factors may account for this. First, no pathogenic trait of Mtb is attributable to a single virulence factor and so compensatory mechanisms within certain lipid families (e.g., polymethyl-branched fatty acid–containing lipids) or cell ligands (e.g., ManLAMs and mannosylated glycoproteins) are known to exist (Pitarque et al. 2005; Passemar et al. 2014). Second, an inherent flaw associated with studies that use purified molecules is that they may not accurately mimic qualitatively and/or quantitatively the way these molecules are presented to the host when carried by whole bacilli. Furthermore, the genetic background of the Mtb strain in which they are produced is likely to affect the way they interact with the host and their subsequent contribution to pathogenicity (e.g., PGL) (Sinsimer et al. 2008). Third, lipids such as SL and (iso)tuberculosinol are restricted to the human pathogen, Mtb, and may only show a clear phenotype in this specific host. The facts that the ability of Mtb to produce some PDIM variants and that PGL is relatively restricted to East-Asian isolates (Constant et al. 2002; Reed et al. 2004; Huet et al. 2009) suggest that the different Mtb lineages may even have evolved regionally to tailor their lipid composition to the genetic background of their human host (Neyrolles and Guilhot 2011). From a mechanistic standpoint, concluding to the direct effect of a lipid on a specific host function can be challenging. This owes to the roles that lipids often play in the structure and permeability of the cell envelope in addition to potentially interacting with the host. Thus, a deficiency in their production may not only cause Mtb to become more susceptible (i.e., more permeable) to host defense mechanisms (e.g., PDIMs and their impact on the resistance of Mtb to oxidative stress) (Rousseau et al. 2004), it may also profoundly alter the way other immunomodulatory antigens are presented at the cell surface and interact with host functions.
Mtb LIPIDS IN THE CONTEXT OF DRUG DISCOVERY
General Considerations
The previous sections have highlighted the diversity, unique structures, and essential roles played by cell envelope lipids in the physiology and pathogenicity of Mtb. These features account for the specificity and therapeutic efficacy of first- and second-line TB drugs such as INH and ETH, which inhibit the biosynthesis of mycolic acids (Banerjee et al. 1994; Vilchèze and Jacobs 2007; Grzegorzewicz et al. 2012b) (Fig. 4), and those of ethambutol (EMB), which inhibits the formation of the arabinan domains of ManLAM (Fig. 3) and AG (Mikušova et al. 1995). In addition, pyrazinamide was reported to be an inhibitor of the fatty acid synthase I (FAS-I) (Zimhony et al. 2000). With multidrug resistance on the increase, recent years have seen a marked intensification of TB drug discovery efforts with the result that many new lead compounds are now at various stages of the drug discovery and preclinical development pipeline. Interestingly, these efforts keep pointing at cell envelope lipids as one of the Achille’s heels of Mtb (Table 1) (Jackson et al. 2013). As importantly, they are leading the TB field to revisit earlier impressions that drug targeting the biogenesis of the cell envelope lipids may not be synergistic with other drugs, useful against MRD-Mtb isolates, or active against persistent bacilli. This section summarizes recent developments that have occurred in the discovery of drugs targeting lipid synthesis in Mtb.
Figure 4.
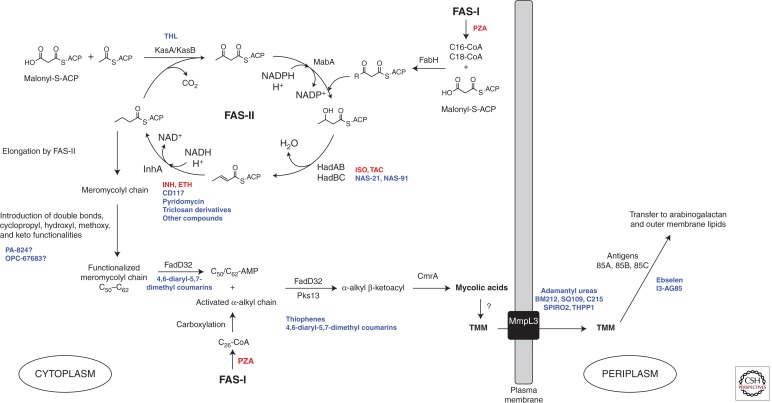
Biosynthetic pathway of mycolic acids in Mtb and site of action of anti-TB drugs. The saturated α-alkyl chain (C24 or C26 in Mtb) and the meromycoloyl chain (C50–C62) are synthesized by fatty-acid synthase-I (FAS-I) and fatty-acid synthase-II (FAS-II), respectively. The initial substrates of FAS-II are medium length keto-acyl-ACP resulting from the condensation by the Mtb FabH protein of the acyl-CoA products of FAS-I with malonyl-ACP. After reduction by the β-keto-acyl-ACP reductase MabA, dehydration by the (3R)-hydroxyl-dehydratases HadAB and HadBC and reduction by the enoyl-CoA reductase InhA, either the β-ketoacyl-ACP synthase KasA or KasB catalyzes the condensation of the resulting product with malonyl-ACP units, thereby initiating the next round of elongation. Methyltransferases and other as yet unknown enzymes modify the meromycolyl chain introducing double bonds, cyclopropyl, hydroxy, methoxy, and keto functionalities. These two chains are coupled together via Claisen condensation by the acyl-AMP ligase FadD32 and the polyketide synthase Pks13. Upon release from Pks13, reduction by CmrA yields mycolic acids, which are then translocated to the periplasm under the form of TMM by partially elucidated translocation machinery involving the essential integral membrane transporter, MmpL3, and attached to AG or another molecule of TMM to form TDM, by the antigen 85 complex. The sites of action of Mtb inhibitors are indicated. Drugs presently or formerly used in the clinical treatment of TB are in red; inhibitors under development are in blue (see Table 1). THL, thiolactomycin; PZA, pyrazinamide.
Drugs Targeting Mycolic Acid Biosynthesis and Export
The biosynthesis of mycolic acids is shown schematically in Figure 4. In the past 10 years, both target-based and whole cell–based approaches have been used toward the identification and development of inhibitors acting on virtually all portions of this biosynthetic pathway (Fig. 4 and Table 1). A comprehensive review of these efforts was recently published by North et al. (2013), and we will here only briefly describe some of the most exciting developments that have occurred in the last few years. Target-based screening uses biochemical and structural approaches to design, screen, and further evaluate a compound’s activity against a specific molecular target. This type of approach has been used extensively to rationally optimize inhibitors of the FAS-II elongation cycle—in particular, analogs of thiolactomycin as inhibitors of KasA/KasB and triclosan derivatives as inhibitors of InhA (Pan and Tonge 2012). The screening of inhibitors showing activity against the Plasmodium falciparum enoyl reductase and β-hydroxyacyl-ACP dehydratase, FabZ, identified compounds that appear to target the corresponding FAS-II enzymes in Mtb and display an MIC against whole mycobacterial cells (Bhowruth et al. 2008; Gratraud et al. 2008; Vilchèze et al. 2011). Pyridomycin, a cyclic depsipeptide isolated from Streptomyces with an MIC value of 0.4 µg/mL against Mtb, is now known to also target InhA (Haatkoorn et al. 2012), as are novel structural classes of inhibitors in the lead-optimization phase reported by the Working Group on New TB Drugs (Table 1). Incidentally, the recent elucidation of the mode of action of isoxyl (ISO) and thiacetazone (TAC), two prodrugs formerly used in the clinical treatment of Mtb, revealed that they both inhibit the dehydration step of the FAS-II cycle catalyzed by HadAB and HadBC (Grzegorzewicz et al. 2012b). The finding that relatively simple structural modifications of the parent ISO and TAC compounds can increase the potency of analogs more than 20-fold is encouraging in that it suggests that it may be possible to identify compounds that could be administered at a lower dose, thereby reducing side effects (Phetsuksiri et al. 1999; Bhowruth et al. 2006; Alahari et al. 2007; Shahab et al. 2010; Coxon et al. 2013).
Because of their pivotal roles in the final assembly of mycolic acids and their transfer to cell envelope acceptors, Pks13, FadD32, and the mycoloyltransferases of the antigen 85 complex are also receiving a lot of attention from the perspective of drug development (Fig. 4). High-throughput screening assays were reported for FadD32 (Galandrin et al. 2013) and the antigens 85 (Boucau et al. 2009; Elamin et al. 2009; Sanki et al. 2009; Favrot et al. 2013). The latter assays led to the identification of a number of inhibitors of antigens 85, one of them, known as ebselen, also shows activity against Mtb in culture (MIC of 20 µg/mL). The molecular mechanism of inhibition of antigens 85 by ebselen was elucidated and found to be particularly interesting in the sense that it is unlikely to lead to the selection of drug-resistant isolates (Favrot et al. 2013). Small molecule binders of antigen 85C identified by magnetic resonance spectroscopy also showed activity against Mtb in vitro and inside macrophages in the 100-µm range (Warrier et al. 2012). The return of interest in whole cell–based screening that the TB field has witnessed in recent years has led to the identification of thiophenes and other compounds active against Pks13 (Ioerger et al. 2013; Wilson et al. 2013), diaryl coumarin–based compounds active against FadD32 (Stanley et al. 2013), and diverse chemotypes active against the TMM transporter, MmpL3 (Grzegorzewicz et al. 2012a; Stanley et al. 2012; Ioerger et al. 2013; Remuinan et al. 2013). BM212 and derivatives (La Rosa et al. 2012; Poce et al. 2013) and SQ109, a drug candidate originally designed to be an EMB analog and currently undergoing phase II clinical trials (Sacksteder et al. 2012; Tahlan et al. 2012), were also shown to kill Mtb through the inhibition of MmpL3. The reason why so many different chemical scaffolds apparently inhibit the MmpL3-mediated translocation of TMM is unclear. Establishing a validated assay for the direct measurement of MmpL3 inhibition is a high priority, because it would greatly facilitate the further development of these inhibitors and could potentially reveal new scaffolds for development.
In summary, recent drug discovery efforts have yielded a number of chemotypes inhibiting various aspects of the synthesis of mycolic acid, some of which are promising candidates for further development. Inhibiting the FAS-II system and MmpL3 currently appear to be the most attractive/druggable targets for TB drug discovery in this pathway.
Isoprenoids and Related Lipids
Inhibitors of the MEP pathway. As a lipid carrier of activated saccharide subunits, polyisoprenyl phosphate (Dec-P) is involved in the biosynthesis of the arabinan portion of AG, the mannan and arabinan domains of arabinomannan, LM, and ManLAM, the lipid I and lipid II precursors of PG, and the biosynthesis of the “linker unit” between AG and PG (Kaur et al. 2009; Daffé et al. 2014) (Fig. 1). The discovery that Dec-P synthesis in Mtb proceeds through the methylerythritol phosphate pathway, which has no homolog in humans, has provided stimulus for the characterization and identification of inhibitors of the relevant enzymes. Fosmidomycin, which is currently in clinical studies for the treatment of malaria, is a competitive inhibitor of the second enzyme of the pathway, 1-deoxy-d-xylulose-5-phosphate reductoisomerase (IspC). Several derivatives of this antibiotic have been synthesized, some of which have shown promising results against Mtb (Jackson et al. 2013) (Table 1).
Inhibitors of lipid-linked biosynthetic precursors. The phospho-N-acetylmuramyl pentapeptide translocase (MraY) catalyzes the exchange of UDP-N-acetylmuramic acid pentapeptide from UDP to Dec-P thereby generating lipid I in PG biosynthesis. The synthesis of capuramycin analogs as inhibitors of this enzyme has led to the identification of several compounds with 2- to 4-µg/mL MICs against drug-susceptible and MDR Mtb isolates (Jackson et al. 2013). Although its mode of action has yet to be confirmed in whole Mtb cells, one of them, SQ641, is at the stage of preclinical development (Table 1).
Very recently, the caprazamycin derivative CPZEN-45 was shown to be an inhibitor of Mtb’s decaprenyl-phosphate-GlcNAc-1-phosphate transferase, WecA, in the biosynthesis of AG (Ishizaki et al. 2013).
Decaprenylphosphoryl arabinose (DPA) is the only known arabinose donor in the building of the arabinan domains of the two essential cell envelope (lipo)polysaccharides, AG, and ManLAM. In the last 4 years, whole cell–based screening of compounds against Mtb has produced several inhibitors of the essential epimerase DprE1 required for the formation of DPA (Table 1). The molecular mechanism of action of some of these compounds has been thoroughly investigated and several DPA inhibitors are now reported to be in the hit-to-lead or preclinical development phases (Table 1) (Jackson et al. 2013).
Menaquinones. The lipoquinones involved in the respiratory chains of mycobacteria consist of menaquinones (2-methyl-3-polyprenyl-1,4-naphthoquinones) (Daffé et al. 2014). Evidence based on transcriptomics profiling and the inhibitory activity of menaquinone inhibitors against Mtb in culture has highlighted the requirement of this bacterium for electron transport and, therefore, menaquinone synthesis both during active replication and microaerophilic persistence (Dhiman et al. 2009). Accordingly, three key enzymes of the menaquinone biosynthetic pathway of Mtb, MenB (1,4-dihydroxy-2-napthoic acid synthase), MenE (O-succinylbenzoyl-CoA synthase), and MenA (1,4-dihydroxy-2-napthoic acid octaprenyl transferase) have been studied as potential drug targets and a number of promising inhibitors synthesized (Table 1).
Inhibition of PIM, LM, and ManLAM Biosynthesis
The essential character of PIM, LM, and ManLAM, their restricted distribution to mycobacteria and closely related Actinomycetes, and their demonstrated impact on the structure and permeability of the cell envelope of Mtb make the biosynthetic enzymes of these molecules attractive candidates for the development of specific Mtb inhibitors with the potential to synergize with or potentiate the activity of other drugs used in combination. Accordingly, target-to-drug approaches are pursuing various essential enzymes acting both at early (e.g., PimA, PimB’) and late (e.g., Emb proteins and other lipid-linked sugar-utilizing glycosyltransferases) stages of the pathway. Innovative high-throughput screening assays have been developed for some of these enzymes (Zhang et al. 2010, 2011). The most advanced ManLAM inhibitors to date are those targeting DPA synthesis, as described in the previous section.
Targeting Lipids Involved in the Replication and Persistence of Mtb In Vivo
Targeting TAG synthesis through inhibition of the primary TAG synthase of Mtb, Rv3130c (Daniel et al. 2004; Sirakova et al. 2006), may represent a viable approach to the eradication of persistent bacilli for the key role TAG is likely to play as an energy reserve for the long-term survival of Mtb during the persistence phase of infection.
As detailed in the previous section, PDIMs are critical components of the permeability barrier surrounding Mtb and contribute to a significant extent to the survival and ability of the bacterium to replicate in vivo. To the best of our knowledge, no specific PDIM inhibitors have yet been reported but promising compounds targeting the Ser/Thr kinases of Mtb that regulate their biosynthesis (among other physiological processes) are under development (Jackson et al. 2013) (Table 1). Another regulatory system of interest in the context of drug development is the two-component transcriptional regulator PhoP-PhoR, which regulates multiple virulence-associated processes in Mtb, including the biosynthesis of SL, DAT, and PAT (Gonzalo-Asencio et al. 2006). Finally, a compound inhibiting the production of PGL in whole Mtb cells has been reported (Ferreras et al. 2008).
CONCLUDING REMARKS
Mtb lipids have been for the last eight decades and still remain today the object of much research activity. One of the highlights of this research in the last five years is undoubtedly the discovery of a variety of novel chemical scaffolds inhibiting various aspects of lipid synthesis, particularly mycolic acids, ManLAM, and AG. Beyond their therapeutic applications, lipids involved in the immunopathogenesis of TB may also represent useful targets for future preventive interventions, as illustrated by current studies aimed at investigating the vaccine potential of CD1-restricted lipid antigens (G Puzo, pers. comm.) and that of an attenuated phoP-based Mtb knockout mutant (Nambiar et al. 2012). Although the interest in developing TB serodiagnostics based on Mtb lipids seems to have somewhat waned in the last decade, important efforts are currently focusing on their potential as biomarkers to monitor TB reactivation, as well as the efficacy of treatment and vaccination (Shui et al. 2012; Wallis et al. 2013; Chan et al. 2013).
ACKNOWLEDGMENTS
Vicki Jones is gratefully acknowledged for her help with the preparation of the figures. The author’s research on cell envelope lipids and inhibitors is supported through the National Institutes of Health/National Institute of Allergy and Infectious Diseases grants AI064798 and AI063054.
REFERENCES
- Afonso-Barroso A, Clark SO, Williams A, Rosa GT, Nobrega C, Silva-Gomes S, Vale-Costa S, Ummels R, Stoker N, Movahedzadeh F, et al. 2012. Lipoarabinomannan mannose caps do not affect mycobacterial virulence or the induction of protective immunity in experimental animal models of infection and have minimal impact on in vitro inflammatory responses. Cell Microbiol 15: 660–674 [DOI] [PubMed] [Google Scholar]
- Alahari A, Trivelli X, Guerardel Y, Dover LG, Besra GS, Sacchettini JC, Reynolds RC, Coxon GD, Kremer L. 2007. Thiacetazone, an antitubercular drug that inhibits cyclopropanation of cell wall mycolic acids in mycobacteria. PLoS ONE 2: e1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P, Foster EL, Porcelli SA. 2013. CD1d and natural killer T cells in immunity to Mycobacterium tuberculosis. Adv Exp Med Biol 783: 199–223 [DOI] [PubMed] [Google Scholar]
- Banerjee A, Dubnau E, Quémard A, Balasubramanian V, Um KS, Wilson T, Collins D, Lisle G, Jacobs WR Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263: 227–230 [DOI] [PubMed] [Google Scholar]
- Barry CE III, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y. 1998. Mycolic acids: Structure, biosynthesis and physiological functions. Prog Lipid Res 37: 143–179 [DOI] [PubMed] [Google Scholar]
- Bertozzi CR, Schelle MW. 2008. Sulfated metabolites from Mycobacterium tuberculosis: Sulfolipid-1 and beyond. In The mycobacterial cell envelope (ed. Daffé M, Reyrat J-M), pp. 291–304. ASM, Washington, DC [Google Scholar]
- Bhamidi S, Shi L, Chatterjee D, Belisle JT, Crick DC, McNeil MR. 2012. A bioanalytical method to determine the cell wall composition of Mycobacterium tuberculosis grown in vivo. Anal Biochem 421: 240–249 [DOI] [PubMed] [Google Scholar]
- Bhowruth V, Brown AK, Reynolds RC, Coxon GD, Mackay SP, Minnikin DE, Besra GS. 2006. Symmetrical and unsymmetrical analogues of isoxyl; active agents against Mycobacterium tuberculosis. Bioorg Med Chem Lett 16: 4743–4747 [DOI] [PubMed] [Google Scholar]
- Bhowruth V, Brown AK, Besra GS. 2008. Synthesis and biological evaluation of NAS-21 and NAS-91 analogues as potential inhibitors of the mycobacterial FAS-II dehydratase enzyme Rv0636. Microbiology 154: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch H. 1950. Studies on the virulence of tubercle bacilli; isolation and biological properties of a constituent of virulent organisms. J Exp Med 91: 197–218, pl [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucau J, Sanki AK, Voss BJ, Sucheck SJ, Ronning DR. 2009. A coupled enzymatic assay measuring Mycobacterium tuberculosis antigen 85C enzymatic activity. Anal Biochem 385: 120–127 [DOI] [PubMed] [Google Scholar]
- Brennan PJ. 1988. Mycobacterium and other actinomycetes. In Microbial lipids (ed. Ratledge C, Wilkinson SG), pp. 203–298. Academic, London [Google Scholar]
- Brennan PJ, Nikaido H. 1995. The envelope of mycobacteria. Annu Rev Biochem 64: 29–63 [DOI] [PubMed] [Google Scholar]
- Britton WJ, Triccas JA. 2008. The constituents of the cell envelope and their impact on the host immune system. In The mycobacterial cell envelope (ed. Daffé M, Reyrat J-M), pp. 249–270. ASM, Washington, DC [Google Scholar]
- Brodin P, Poquet Y, Levillain F, Peguillet I, Larrouy-Maumus G, Gilleron M, Ewann F, Christophe T, Fenistein D, Jang J, et al. 2010. High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog 6: e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho LR, Constant P, Raynaud C, Lanéelle MA, Triccas JA, Gicquel B, Daffé M, Guilhot C. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem 276: 19845–19854 [DOI] [PubMed] [Google Scholar]
- Cardona P-J, Soto CY, Martin C, Gicquel B, Agusti G, Guirado E, Sirakova TD, Kolattukudy PE, Julian E, Luquin M. 2006. Neutral red reaction is related to virulence and cell wall methyl-branched lipids in Mycobacterium tuberculosis. Microbes Infect 8: 183–190 [DOI] [PubMed] [Google Scholar]
- Chan CE, Zhao BZ, Cazenave-Gassiot A, Pang SW, Bendt AK, Wenk MR, Macary PA, Hanson BJ. 2013. Novel phage display–derived mycolic acid–specific antibodies with potential for tuberculosis diagnosis. J Lipid Res 54: 2924–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman TM, Bouloc N, Buxton RS, Chugh J, Lougheed KE, Osborne SA, Saxty B, Smerdon SJ, Taylor DL, Whalley D. 2012. Substituted aminopyrimidine protein kinase B (PknB) inhibitors show activity against Mycobacterium tuberculosis. Bioorg Med Chem Lett 22: 3349–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, Genovesio A, Carralot JP, Ewann F, Kim EH, et al. 2009. High content screening identifies decaprenyl-phosphoribose 2′ epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog 5: e1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant P, Perez E, Malaga W, Lanéelle MA, Saurel O, Daffé M, Guilhot C. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J Biol Chem 277: 38148–38158 [DOI] [PubMed] [Google Scholar]
- Coxon GD, Craig D, Corrales RM, Vialla E, Gannoun-Zaki L, Kremer L. 2013. Synthesis, antitubercular activity and mechanism of resistance of highly effective thiacetazone analogues. PloS ONE 8: e53162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffé M, Lanéelle M-A. 1988. Distribution of phthiocerol diester, phenolic mycosides and related compounds in mycobacteria. J Gen Microbiol 134: 2049–2055 [DOI] [PubMed] [Google Scholar]
- Daffé M, Draper P. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol 39: 131–203 [DOI] [PubMed] [Google Scholar]
- Daffé M, Lacave C, Lanéelle MA, Lanéelle G. 1987. Structure of the major triglycosyl phenol-phthiocerol of Mycobacterium tuberculosis (strain Canetti). Eur J Biochem 167: 155–160 [DOI] [PubMed] [Google Scholar]
- Daffé M, McNeil MR, Brennan PJ. 1991. Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30: 378–388 [DOI] [PubMed] [Google Scholar]
- Daffé M, Crick DC, Jackson M. 2014. Genetics of capsular polysaccharides and cell envelope (glyco)lipids. Microbiol Spectrum 2: MGM2-0021–2013 [DOI] [PubMed] [Google Scholar]
- Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 186: 5017–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilenko VN, Osolodkin DI, Lakatosh SA, Preobrazhenskaya MN, Shtil AA. 2011. Bacterial eukaryotic type serine-threonine protein kinases: From structural biology to targeted anti-infective drug design. Curr Top Med Chem 11: 1352–1369 [DOI] [PubMed] [Google Scholar]
- Debnath J, Siricilla S, Wan B, Crick DC, Lenaerts AJ, Franzblau SG, Kurosu M. 2012. Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J Med Chem 55: 3739–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman RK, Mahapatra S, Slayden RA, Boyne ME, Lenaerts A, Hinshaw JC, Angala SK, Chatterjee D, Biswas K, Narayanasamy P, et al. 2009. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol Microbiol 72: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman RK, Dinadayala P, Ryan GJ, Lenaerts AJ, Schenkel AR, Crick DC. 2011. Lipoarabinomannan localization and abundance during growth of Mycobacterium smegmatis. J Bacteriol 193: 5802–5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper P. 1998. The outer parts of the mycobacterial envelope as permeability barriers. Frontiers Biosci 3: D1253–D1261 [DOI] [PubMed] [Google Scholar]
- Dubnau E, Chan J, Raynaud C, Mohan VP, Lanéelle M-A, Yu K, Quémard A, Smith I, Daffé M. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol Microbiol 36: 630–637 [DOI] [PubMed] [Google Scholar]
- Elamin AA, Stehr M, Oehlmann W, Singh M. 2009. The mycolyltransferase 85A, a putative drug target of Mycobacterium tuberculosis: Development of a novel assay and quantification of glycolipid-status of the mycobacterial cell wall. J Microbiol Method 79: 358–363 [DOI] [PubMed] [Google Scholar]
- Favrot L, Grzegorzewicz AE, Lajiness DH, Marvin RK, Boucau J, Isailovic D, Jackson M, Ronning DR. 2013. Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen. Nat Commun 4: 2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras JA, Stirrett KL, Lu X, Ryu JS, Soll CE, Tan DS, Quadri LE. 2008. Mycobacterial phenolic glycolipid virulence factor biosynthesis: Mechanism and small-molecule inhibition of polyketide chain initiation. Chem Biol 15: 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Matsumura T, Ato M, Hamasaki M, Nishiuchi Y, Murakami Y, Maeda Y, Yoshimori T, Matsumoto S, Kobayashi K, et al. 2013. Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. mBio 4: e00472–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S, Small PM. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis 7: 328–337 [DOI] [PubMed] [Google Scholar]
- Galandrin S, Guillet V, Rane RS, Leger M, Radha N, Eynard N, Das K, Balganesh TS, Mourey L, Daffé M, et al. 2013. Assay development for identifying inhibitors of the mycobacterial FadD32 activity. J Biomol Screen. 18: 576–587 [DOI] [PubMed] [Google Scholar]
- Gilleron M, Jackson M, Nigou J, Puzo G. 2008. Structure, activities and biosynthesis of the phosphatidyl-myo-inositol-based lipoglycans. In The mycobacterial cell envelope (ed. Daffé M, Reyrat J-M), pp. 75–105. ASM, Washington, DC [Google Scholar]
- Glickman MS. 2008. Cording, cord factors and trehalose dimycolate. In The mycobacterial cell envelope (ed. Daffé M, Reyrat J-M), pp. 63–73. ASM, Washington DC [Google Scholar]
- Glickman MS, Cox JS, Jacobs WR Jr, 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell 5: 717–727 [DOI] [PubMed] [Google Scholar]
- Gonzalo Asensio J, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffe M, Gicquel B, Martin C, et al. 2006. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem 281: 1313–1316 [DOI] [PubMed] [Google Scholar]
- Goren MB. 1990. Mycobacterial fatty acid esters of sugars and sulfosugars. In handbook of lipid research glycolipids, phosphoglycolipids and sulfoglycolipids (ed. Kates M), pp. 363–461. Plenum, New York [Google Scholar]
- Goren MB, Brennan PJ. 1979. Mycobacterial lipids: Chemistry and biologic activities. In Tuberculosis (ed. Youmans GP), pp. 63–193. W.B. Saunders, Philadelphia [Google Scholar]
- Goren MB, Brokl O, Schaefer WB. 1974a. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: Phthiocerol dimycocerosate and the attenuation indicator lipid. Infect Immun 9: 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren MB, Brokl O, Schaefer WB. 1974b. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: Correlation of virulence with elaboration of sulfatides and strongly acidic lipids. Infect Immun 9: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JE, Clark-Curtiss JE. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc Natl Acad Sci 96: 11554–11559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratraud P, Surolia N, Besra GS, Surolia A, Kremer L. 2008. Antimycobacterial activity and mechanism of action of NAS-91. Antimicrob Agents Chemother 52: 1162–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorzewicz AE, Pham H, Gundi VAKB, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SEM, Korduláková J, et al. 2012a. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorzewicz AE, Korduláková J, Jones V, Born SE, Belardinelli JM, Vaquié A, Gundi VA, Madacki J, Slama N, Laval F, et al. 2012b. A common mechanism of inhibition of the Mycobacterium tuberculosis mycolic acid biosynthetic pathway by isoxyl and thiacetazone. J Biol Chem 287: 38434–38441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenin-Macé L, Simeone R, Demangel C. 2009. Lipids of pathogenic Mycobacteria: Contributions to virulence and host immune suppression. Transbound Emerg Dis 56: 255–268 [DOI] [PubMed] [Google Scholar]
- Guerin ME, Korduláková J, Alzari PM, Brennan PJ, Jackson M. 2010. Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. J Biol Chem 285: 33577–33583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiard J, Collmann A, Garcia-Alles LF, Mourey L, Brando T, Mori L, Gilleron M, Prandi J, De Libero G, Puzo G. 2009. Fatty acyl structures of Mycobacterium tuberculosis sulfoglycolipid govern T cell response. J Immunol 182: 7030–7037 [DOI] [PubMed] [Google Scholar]
- Guilhot C, Chalut C, Daffé M. 2008. Biosynthesis and roles of phenolic glycolipids and related molecules in Mycobacterium tuberculosis. In The mycobacterial cell envelope (ed. Daffé M, Reyrat J-M), pp. 273–289. ASM, Washington, DC [Google Scholar]
- Hartkoorn RC, Sala C, Neres J, Pojer F, Magnet S, Mukherjee R, Uplekar S, Boy-Rottger S, Altmann KH, Cole ST. 2012. Towards a new tuberculosis drug: Pyridomycin—Nature’s isoniazid. EMBO Mol Med 4: 1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Oka S, Han YK, Yamamura Y, Kusunose E, Kusunose M, Yano I. 1991. Rapid serodiagnosis of human mycobacteriosis by ELISA using cord factor (trehalose-6,6′-dimycolate) purified from Mycobacterium tuberculosis as antigen. FEMS Microbiol Immunol 3: 201–204 [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. 2008. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci 105: 3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet G, Constant P, Malaga W, Lanéelle MA, Kremer K, van Soolingen D, Daffé M, Guilhot C. 2009. A lipid profile typifies the Beijing strains of Mycobacterium tuberculosis: Identification of a mutation responsible for a modification of the structures of phthiocerol dimycocerosates and phenolic glycolipids. J Biol Chem 284: 27101–27113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, O’Malley T, Liao R, Guinn KM, Hickey MJ, Mohaideen N, Murphy KC, Boshoff HI, Mizrahi V, Rubin EJ, et al. 2013. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PloS ONE 8: e75245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. 2009. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med 206: 2879–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y, Hayashi C, Inoue K, Igarashi M, Takahashi Y, Pujari V, Crick DC, Brennan PJ, Nomoto A. 2013. Inhibition of the first step in synthesis of the mycobacterial cell wall core, catalyzed by the GlcNAc-1-phosphate transferase WecA, by the novel caprazamycin derivative CPZEN-45. J Biol Chem 288: 30309–30319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Stadthagen G, Gicquel B. 2007. Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: Biosynthesis, transport, regulation and biological activities. Tuberculosis 87: 78–86 [DOI] [PubMed] [Google Scholar]
- Jackson M, McNeil MR, Brennan PJ. 2013. Progress in targeting cell envelope biogenesis in Mycobacterium tuberculosis. Future Microbiol 8: 855–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarlier V, Nikaido H. 1994. Mycobacterial cell wall: Structure and role in natural resistance to antibiotics. FEMS Microbiol Lett 123: 11–18 [DOI] [PubMed] [Google Scholar]
- Julian E, Matas L, Perez A, Alcaide J, Laneelle MA, Luquin M. 2002. Serodiagnosis of tuberculosis: Comparison of immunoglobulin A (IgA) response to sulfolipid I with IgG and IgM responses to 2,3-diacyltrehalose, 2,3,6-triacyltrehalose, and cord factor antigens. J Clin Microbiol 40: 3782–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai K, Wiegeshaus E, Smith DW. 1970. Demonstration of mycolic acid and phthiocerol dimycocerosate in “in vivo grown tubercle bacilli”. Jpn J Med Sci Biol 23: 327–333 [PubMed] [Google Scholar]
- Karakousis PC, Bishai WR, Dorman SE. 2004. Mycobacterium tuberculosis cell envelope lipids and the host immune response. Cell Microbiol 6: 105–116 [DOI] [PubMed] [Google Scholar]
- Kaur D, Guerin ME, Skovierova H, Brennan PJ, Jackson M. 2009. Chapter 2. Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol 69: 23–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korduláková J, Gilleron M, Mikušová K, Puzo G, Brennan PJ, Gicquel B, Jackson M. 2002. Definition of the first mannosylation step in phosphatidylinositol synthesis: PimA is essential for growth of mycobacteria. J Biol Chem 277: 31335–31344 [DOI] [PubMed] [Google Scholar]
- Krishnan N, Malaga W, Constant P, Caws M, Tran TH, Salmons J, Nguyen TN, Nguyen DB, Daffé M, Young DB, et al. 2011. Mycobacterium tuberculosis lineage influences innate immune response and virulence and is associated with distinct cell envelope lipid profiles. PloS ONE 6: e23870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa V, Poce G, Canseco JO, Buroni S, Pasca MR, Biava M, Raju RM, Porretta GC, Alfonso S, Battilocchio C, et al. 2012. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob Agents Chemother 56: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Vanderven BC, Fahey RJ, Russell DG. 2013. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem 288: 6788–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemassu A, Daffé M. 1994. Structural features of the exocellular polysaccharides of Mycobacterium tuberculosis. Biochem J 297: 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu N, Zhang H, Knudson SE, Slayden RA, Tonge PJ. 2010. Synthesis and SAR studies of 1,4-benzoxazine MenB inhibitors: Novel antibacterial agents against Mycobacterium tuberculosis. Bioorg Med Chem Lett 20: 6306–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu N, Zhang H, Knudson SE, Li HJ, Lai CT, Simmerling C, Slayden RA, Tonge PJ. 2011. CoA adducts of 4-oxo-4-phenylbut-2-enoates: Inhibitors of MenB from the M. tuberculosis menaquinone biosynthesis pathway. ACS Med Chem Lett 2: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Du Q, Deng W, Xie J. 2012. The biology of mycobacterium cord factor and roles in pathogen–host interaction. Crit Rev Eukaryot Gene Expr 22: 289–297 [DOI] [PubMed] [Google Scholar]
- Lougheed KE, Osborne SA, Saxty B, Whalley D, Chapman T, Bouloc N, Chugh J, Nott TJ, Patel D, Spivey VL, et al. 2011. Effective inhibitors of the essential kinase PknB and their potential as anti-mycobacterial agents. Tuberculosis (Edinb) 91: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhang H, Tonge PJ, Tan DS. 2008. Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis. Bioorg Med Chem Lett 18: 5963–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhou R, Sharma I, Li X, Kumar G, Swaminathan S, Tonge PJ, Tan DS. 2012. Stable analogues of OSB-AMP: Potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis. Chem Biochem 13: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly D, Kasmar AG, Cheng TY, de Jong A, Huang S, Roy S, Bhatt A, van Summeren RP, Altman JD, Jacobs WR Jr, et al. 2013. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med 210: 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Hartkoorn RC, Szekely R, Pato J, Triccas JA, Schneider P, Szantai-Kis C, Orfi L, Chambon M, Banfi D, et al. 2010. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis (Edinb) 90: 354–360 [DOI] [PubMed] [Google Scholar]
- Makarov V, Manina G, Mikušová K, Mollmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, et al. 2009. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324: 801–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann FM, Peters RJ. 2012. Isotuberculosinol: The unusual case of an immunomodulatory diterpenoid from Mycobacterium tuberculosis. Med Chem Comm 3: 899–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, Shimokawa Y, Komatsu M. 2006. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med 3: e466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga I, Sugita M. 2012. Mycoketide: A CD1c-presented antigen with important implications in mycobacterial infection. Clin Dev Immunol 2012: 981821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikušová K, Slayden RA, Besra GS, Brennan PJ. 1995. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob Agents Chemother 39: 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin DE. 1982. Lipids: Complex lipids, their chemistry, biosynthesis and roles. In The biology of mycobacteria (ed. Ratledge C, Stanford J), pp. 95–184. Academic, London [Google Scholar]
- Molle V, Kremer L. 2010. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol Microbiol 75: 1064–1077 [DOI] [PubMed] [Google Scholar]
- Morita YS, Velasquez R, Taig E, Waller RF, Patterson JH, Tull D, Williams SJ, Billman-Jacobe H, McConville MJ. 2005. Compartmentalization of lipid biosynthesis in mycobacteria. J Biol Chem 280: 21645–21652 [DOI] [PubMed] [Google Scholar]
- Morita YS, Sena CCB, Waller RF, Kurokawa K, Sernee MF, Nakatani F, Haites RE, Billman-Jacobe H, McConville MJ, Maeda Y, et al. 2006. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J Biol Chem 281: 25143–25155 [DOI] [PubMed] [Google Scholar]
- Nambiar JK, Pinto R, Aguilo JI, Takatsu K, Martin C, Britton WJ, Triccas JA. 2012. Protective immunity afforded by attenuated, PhoP-deficient Mycobacterium tuberculosis is associated with sustained generation of CD4+ T-cell memory. Eur J Immunol 42: 385–392 [DOI] [PubMed] [Google Scholar]
- Neyrolles O, Guilhot C. 2011. Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis (Edinb) 91: 187–195 [DOI] [PubMed] [Google Scholar]
- North EJ, Jackson M, Lee RE. 2013. New approaches to target the mycolic acid biosynthesis pathway for the development of tuberculosis therapeutics. Curr Pharm Des (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiol-Pardo C, Rubio-Martinez J, Imperial S. 2011. The methylerythritol phosphate (MEP) pathway for isoprenoid biosynthesis as a target for the development of new drugs against tuberculosis. Curr Med Chem 18: 1325–1338 [DOI] [PubMed] [Google Scholar]
- Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR Jr, Hatfull GF. 2005. GroEL1: A dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123: 861–873 [DOI] [PubMed] [Google Scholar]
- Ojha AK, Trivelli X, Guerardel Y, Kremer L, Hatfull GF. 2010. Enzymatic hydrolysis of trehalose dimycolate releases free mycolic acids during mycobacterial growth in biofilms. J Biol Chem 285: 17380–17389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortalo-Magné A, Dupont MA, Lemassu A, Andersen AB, Gounon P, Daffé M. 1995. Molecular composition of the outermost capsular material of the tubercle bacillus. Microbiol 141: 1609–1620 [DOI] [PubMed] [Google Scholar]
- Ortalo-Magné A, Lemassu A, Lanéelle MA, Bardou F, Silve G, Gounon P, Marchal G, Daffé M. 1996. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacteriol 178: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan P, Tonge PJ. 2012. Targeting InhA, the FASII enoyl-ACP reductase: SAR studies on novel inhibitor scaffolds. Curr Top Med Chem 12: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish T, Liu J, Nikaido H, Stoker NG. 1997. A Mycobacterium smegmatis mutant with a defective inositol monophosphate phosphatase gene homolog has altered cell envelope permeability. J Bacteriol 179: 7827–7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passemar C, Arbues A, Malaga W, Mercier I, Moreau F, Lepourry L, Neyrolles O, Guilhot C, Astarie-Dequeker C. 2014. Multiple deletions in the polyketide synthase gene repertoire of Mycobacterium tuberculosis reveal functional overlap of cell envelope lipids in host-pathogen interactions. Cell Microbiol 16: 195–213 [DOI] [PubMed] [Google Scholar]
- Patterson JH, Waller RF, Jeevarajah D, Billman-Jacobe H, McConville MJ. 2003. Mannose metabolism is required for mycobacterial growth. Biochem J 372: 77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffé M, Emile JF, Marchou B, Cardona PJ, et al. 2008. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog 4: e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phetsuksiri B, Baulard AR, Cooper A, Minnikin DE, Douglas JD, Besra GS, Brennan PJ. 1999. Antimycobacterial activities of isoxyl and new derivatives through the inhibition of mycolic acid synthesis. Antimicrob Agents Chemother 43: 1042–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitarque S, Herrmann JL, Duteyrat JL, Jackson M, Stewart GR, Lecointe F, Payre B, Schwartz O, Young DB, Marchal G, et al. 2005. Deciphering the molecular bases of Mycobacterium tuberculosis binding to the lectin DC-SIGN reveals an underestimated complexity. Biochem J 392: 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitarque S, Larrouy-Maumus G, Payré B, Jackson M, Puzo G, Nigou J. 2008. The immunomodulatory lipoglycans, lipoarabinomannan and lipomannan, are exposed at the mycobacterial cell surface. Tuberculosis 88: 560–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poce G, Bates RH, Alfonso S, Cocozza M, Porretta GC, Ballell L, Rullas J, Ortega F, De Logu A, Agus E, et al. 2013. Improved BM212 MmpL3 inhibitor analogue shows efficacy in acute murine model of tuberculosis infection. PloS ONE 8: e56980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajni, Rao N, Meena LS. 2011. Biosynthesis and virulent behavior of lipids produced by Mycobacterium tuberculosis: LAM and Cord Factor: An overview. Biotechnol Res Int 2011: 274693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431: 84–87 [DOI] [PubMed] [Google Scholar]
- Reed MB, Gagneux S, Deriemer K, Small PM, Barry CE 3rd. 2007. The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J Bacteriol 189: 2583–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remuinan MJ, Perez-Herran E, Rullas J, Alemparte C, Martinez-Hoyos M, Dow DJ, Afari J, Mehta N, Esquivias J, Jimenez E, et al. 2013. Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N-benzyl-6',7'-dihydrospiro [piperidine-4,4'-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PloS ONE 8: e60933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Dover LG, Islam ST, Alexander DC, Chen JM, Besra GS, Liu J. 2007. Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol Microbiol 63: 1345–1359 [DOI] [PubMed] [Google Scholar]
- Rodriguez JE, Ramirez AS, Salas LP, Helguera-Repetto C, Gonzalez-y-Merchand J, Soto CY, Hernandez-Pando R. 2013. Transcription of genes involved in sulfolipid and polyacyltrehalose biosynthesis of Mycobacterium tuberculosis in experimental latent tuberculosis infection. PloS ONE 8: e58378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Neyrolles O, Bordat Y, Giroux S, Sirakova TD, Prevost M-C, Kolattukudy PE, Gicquel B, Jackson M. 2003a. Deficiency in mycolipenate- and mycosanoate-derived acyltrehaloses enhances early interactions of Mycobacterium tuberculosis with host cells. Cell Microbiol 5: 405–415 [DOI] [PubMed] [Google Scholar]
- Rousseau C, Turner OC, Rush E, Bordat Y, Sirakova TD, Kolattukudy PE, Ritter S, Orme IM, Gicquel B, Jackson M. 2003b. Sulfolipid deficiency does not affect the virulence of Mycobacterium tuberculosis H37Rv in mice and guinea pigs. Infect Immun 71: 4684–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Winter N, Pivert E, Bordat Y, Neyrolles O, Ave P, Huerre M, Gicquel B, Jackson M. 2004. Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell Microbiol 6: 277–287 [DOI] [PubMed] [Google Scholar]
- Russell DG. 2011. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev 240: 252–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DG, Mwandumba HC, Rhoades EE. 2002. Mycobacterium and the coat of many lipids. J Cell Biol 158: 421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan GJ, Hoff DR, Driver ER, Voskuil ML, Gonzalez-Juarrero M, Basaraba RJ, Crick DC, Spencer JS, Lenaerts AJ. 2010. Multiple M. tuberculosis phenotypes in mouse and guinea pig lung tissue revealed by a dual-staining approach. PloS ONE 5: e11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder KA, Protopopova M, Barry CE III, Andries K, Nacy CA. 2012. Discovery and development of SQ109: A new antitubercular drug with a novel mechanism of action. Future Microbiol 7: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Kim MJ, Rhoades ER, Allavena RE, Ehrt S, Wainwright HC, Russell DG, Rohde KH. 2013. Mycobacterial trehalose dimycolate reprograms macrophage global gene expression and activates matrix metalloproteinases. Infect Immun 81: 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan D, Dao DN, Weinrick BC, Vilchèze C, Gurcha SS, Ojha A, Kremer L, Besra GS, Hatfull GF, Jacobs WR Jr. 2013. Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. mBio 4: e00222–00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M, Houben ENG, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jimenez CR, Daffé M, et al. 2010. Direct visualization by cryo-EM of the mycobacterial capsular layer: A labile structure containing ESX-1-secreted proteins. PLoS Pathog 6: e1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanki AK, Boucau J, Ronning DR, Sucheck SJ. 2009. Antigen 85C-mediated acyl-transfer between synthetic acyl donors and fragments of the arabinan. Glycoconj J 26: 589–596 [DOI] [PubMed] [Google Scholar]
- Schlesinger LS, Azad AK, Torrelles JB, Roberts E, Vergne I, Deretic V. 2008. Determinants of phagocytosis, phagosome biogenesis and autophagy for Mycobacterium tuberculosis. In Handbook of tuberculosis: Immunology and cell biology (ed. Kaufmann SHE, Britton WJ), pp. 1–22. Wiley-VCH, Darmstadt, Germany [Google Scholar]
- Shahab FM, Kobarfard F, Shafaghi B, Dadashzadeh S. 2010. Preclinical pharmacokinetics of KBF611, a new antituberculosis agent in mice and rabbits, and comparison with thiacetazone. Xenobiotica 40: 225–234 [DOI] [PubMed] [Google Scholar]
- Shui G, Bendt AK, Jappar IA, Lim HM, Laneelle M, Herve M, Via LE, Chua GH, Bratschi MW, Zainul Rahim SZ, et al. 2012. Mycolic acids as diagnostic markers for tuberculosis case detection in humans and drug efficacy in mice. EMBO Mol Med 4: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonney N, Molina JM, Molimard M, Oksenhendler E, Perronne C, Lagrange PH. 1995. Analysis of the immunological humoral response to Mycobacterium tuberculosis glycolipid antigens (DAT, PGLTb1) for diagnosis of tuberculosis in HIV-seropositive and -seronegative patients. Eur J Clin Microbiol Infect Dis 14: 883–891 [DOI] [PubMed] [Google Scholar]
- Simonney N, Molina JM, Molimard M, Oksenhendler E, Lagrange PH. 1996. Comparison of A60 and three glycolipid antigens in an ELISA test for tuberculosis. Clin Microbiol Infect 2: 214–222 [DOI] [PubMed] [Google Scholar]
- Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. 2009. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog 5: e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsimer D, Huet G, Manca C, Tsenova L, Koo MS, Kurepina N, Kana B, Mathema B, Marras SA, Kreiswirth BN, et al. 2008. The phenolic glycolipid of Mycobacterium tuberculosis differentially modulates the early host cytokine response but does not in itself confer hypervirulence. Infect Immun 76: 3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirakova TD, Dubey VS, Deb C, Daniel J, Korotkova TA, Abomoelak B, Kolattukudy PE. 2006. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology 152: 2717–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, et al. 2012. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem Biol 7: 1377–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Kawate T, Iwase N, Shimizu M, Clatworthy AE, Kazyanskaya E, Sacchettini JC, Ioerger TR, Siddiqi NA, Minami S, et al. 2013. Diarylcoumarins inhibit mycolic acid biosynthesis and kill Mycobacterium tuberculosis by targeting FadD32. Proc Natl Acad Sci 110: 11565–11570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover KC, Warrener P, VanDevanter DR, Sherman DR, Arain TM, Langhorne MH, Anderson SW, Towell JA, Yuan Y, McMurray DN, et al. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405: 962–966 [DOI] [PubMed] [Google Scholar]
- Sugawara I, Udagawa T, Hua SC, Reza-Gholizadeh M, Otomo K, Saito Y, Yamada H. 2002. Pulmonary granulomas of guinea pigs induced by inhalation exposure of heat-treated BCG Pasteur, purified trehalose dimycolate and methyl ketomycolate. J Med Microbiol 51: 131–137 [DOI] [PubMed] [Google Scholar]
- Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, et al. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45: 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE 3rd, et al. 2012. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsenova L, Ellison E, Harbacheuski R, Moreira AL, Kurepina N, Reed MB, Mathema B, Barry CE III, Kaplan G. 2005. Virulence of selected Mycobacterium tuberculosis clinical isolates in the rabbit model of meningitis is dependent on phenolic glycolipid produced by the bacilli. J Infect Dis 192: 98–106 [DOI] [PubMed] [Google Scholar]
- Uh E, Jackson ER, San Jose G, Maddox M, Lee RE, Lee RE, Boshoff HI, Dowd CS. 2011. Antibacterial and antitubercular activity of fosmidomycin, FR900098, and their lipophilic analogs. Bioorg Med Chem Lett 21: 6973–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rhijn I, Ly D, Moody DB. 2013. CD1a, CD1b, and CD1c in immunity against mycobacteria. Adv Exp Med Biol 783: 181–197 [DOI] [PubMed] [Google Scholar]
- Vander Beken S, Al Dulayymi JR, Naessens T, Koza G, Maza-Iglesias M, Rowles R, Theunissen C, De Medts J, Lanckacker E, Baird MS, et al. 2011. Molecular structure of the Mycobacterium tuberculosis virulence factor, mycolic acid, determines the elicited inflammatory pattern. Eur J Immunol 41: 450–460 [DOI] [PubMed] [Google Scholar]
- Vera-Cabrera L, Handzel V, Laszlo A. 1994. Development of an enzyme-linked immunosorbent assay (ELISA) combined with a streptavidin-biotin and enzyme amplification method to detect anti-2,3-di-o-acyltrehalose (DAT) antibodies in patients with tuberculosis. J Immunol Methods 177: 69–77 [DOI] [PubMed] [Google Scholar]
- Vilchèze C, Jacobs WR Jr, 2007. The mechanism of isoniazid killing: Clarity through the scope of genetics. Annu Rev Microbiol 61: 35–50 [DOI] [PubMed] [Google Scholar]
- Vilchèze C, Baughn AD, Tufariello J, Leung LW, Kuo M, Basler CF, Alland D, Sacchettini JC, Freundlich JS, Jacobs WR Jr. 2011. Novel inhibitors of InhA efficiently kill Mycobacterium tuberculosis under aerobic and anaerobic conditions. Antimicrob Agents Chemother 55: 3889–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne B, Crauste C, Flipo M, Baulard AR, Deprez B, Willand N. 2012. Tuberculosis: The drug development pipeline at a glance. Eur J Med Chem 51: 1–16 [DOI] [PubMed] [Google Scholar]
- Villeneuve C, Gilleron M, Maridonneau-Parini I, Daffé M, Astarie-Dequeker C, Etienne G. 2005. Mycobacteria use their surface-exposed glycolipids to infect human macrophages through a receptor-dependent process. J Lipid Res 46: 475–483 [DOI] [PubMed] [Google Scholar]
- Vir P, Gupta D, Agarwal R, Verma I. 2013. Immunomodulation of alveolar epithelial cells by Mycobacterium tuberculosis phosphatidylinositol mannosides results in apoptosis. APMIS 122: 268–282 [DOI] [PubMed] [Google Scholar]
- Wallis RS, Kim P, Cole S, Hanna D, Andrade BB, Maeurer M, Schito M, Zumla A. 2013. Tuberculosis biomarkers discovery: Developments, needs, and challenges. Lancet Infect Dis 13: 362–372 [DOI] [PubMed] [Google Scholar]
- Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffé M, Smith I. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol 60: 312–330 [DOI] [PubMed] [Google Scholar]
- Wang F, Sambandan D, Halder R, Wang J, Batt SM, Weinrick B, Ahmad I, Yang P, Zhang Y, Kim J, et al. 2013. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc Natl Acad Sci 110: E2510–E2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrier T, Tropis M, Werngren J, Diehl A, Gengenbacher M, Schlegel B, Schade M, Oschkinat H, Daffé M, Hoffner S, et al. 2012. Antigen 85C inhibition restricts Mycobacterium tuberculosis growth through disruption of cord factor biosynthesis. Antimicrob Agents Chemother 56: 1735–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein EA, Yano T, Li LS, Avarbock D, Avarbock A, Helm D, McColm AA, Duncan K, Lonsdale JT, Rubin H. 2005. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc Natl Acad Sci 102: 4548–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willand N, Dirie B, Carette X, Bifani P, Singhal A, Desroses M, Leroux F, Willery E, Mathys V, Deprez-Poulain R, et al. 2009. Synthetic EthR inhibitors boost antituberculous activity of ethionamide. Nat Med 15: 537–544 [DOI] [PubMed] [Google Scholar]
- Wilson R, Kumar P, Parashar V, Vilchèze C, Veyron-Churlet R, Freundlich JS, Barnes SW, Walker JR, Szymonifka MJ, Marchiano E, et al. 2013. Antituberculosis thiophenes define a requirement for Pks13 in mycolic acid biosynthesis. Nat Chem Biol 9: 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Sinha T, Carlson TK, Keiser TL, Torrelles JB, Schlesinger LS. 2013. Changes in the major cell envelope components of Mycobacterium tuberculosis during in vitro growth. Glycobiology 23: 926–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhu Y, Crane DD, Barry CE III. 1998. The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol Microbiol 29: 1449–1458 [DOI] [PubMed] [Google Scholar]
- Zhang J, Amin AG, Holemann A, Seeberger PH, Chatterjee D. 2010. Development of a plate-based scintillation proximity assay for the mycobacterial AftB enzyme involved in cell wall arabinan biosynthesis. Bioorg Med Chem 18: 7121–7131 [DOI] [PubMed] [Google Scholar]
- Zhang J, Angala SK, Pramanik PK, Li K, Crick DC, Liav A, Jozwiak A, Swiezewska E, Jackson M, Chatterjee D. 2011. Reconstitution of functional mycobacterial arabinosyltransferase aftc proteoliposome and assessment of decaprenylphosphorylarabinose analogues as arabinofuranosyl donors. ACS Chem Biol 6: 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimhony O, Cox JS, Welch JT, Vilchèze C, Jacobs WR Jr. 2000. Pyrazinamide inhibits the eukaryotic-like fatty acid synthetase I (FASI) of Mycobacterium tuberculosis. Nat Med 6: 1043–1047 [DOI] [PubMed] [Google Scholar]
- Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffé M. 2008. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bact 190: 5672–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]