Abstract
Background
There are several studies describing loss of genes through reductive evolution in microbes, but how selective forces are associated with genome expansion due to horizontal gene transfer (HGT) has not received similar attention. The aim of this study was therefore to examine how selective pressures influence genome expansion in 53 fully sequenced and assembled Escherichia coli strains. We also explored potential connections between genome expansion and the attainment of virulence factors. This was performed using estimations of several genomic parameters such as AT content, genomic drift (measured using relative entropy), genome size and estimated HGT size, which were subsequently compared to analogous parameters computed from the core genome consisting of 1729 genes common to the 53 E. coli strains. Moreover, we analyzed how selective pressures (quantified using relative entropy and dN/dS), acting on the E. coli core genome, influenced lineage and phylogroup formation.
Results
Hierarchical clustering of dS and dN estimations from the E. coli core genome resulted in phylogenetic trees with topologies in agreement with known E. coli taxonomy and phylogroups. High values of dS, compared to dN, indicate that the E. coli core genome has been subjected to substantial purifying selection over time; significantly more than the non-core part of the genome (p<0.001). This is further supported by a linear association between strain-wise dS and dN values (β = 26.94 ± 0.44, R2~0.98, p<0.001). The non-core part of the genome was also significantly more AT-rich (p<0.001) than the core genome and E. coli genome size correlated with estimated HGT size (p<0.001). In addition, genome size (p<0.001), AT content (p<0.001) as well as estimated HGT size (p<0.005) were all associated with the presence of virulence factors, suggesting that pathogenicity traits in E. coli are largely attained through HGT. No associations were found between selective pressures operating on the E. coli core genome, as estimated using relative entropy, and genome size (p~0.98).
Conclusions
On a larger time frame, genome expansion in E. coli, which is significantly associated with the acquisition of virulence factors, appears to be independent of selective forces operating on the core genome.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-882) contains supplementary material, which is available to authorized users.
Background
It has been widely documented that horizontal gene transfer (HGT) can make potentially harmless, even probiotic, bacterial species lethal [1, 2]. Considerable research has focused on how bacteria can evolve from being nonthreatening, host-independent and free-living organisms to become obligatory intracellular parasites with reduced genomes [3–9]. However, the evolutionary mechanisms explaining genome expansion due to HGT are much less documented. One reason for this is the need for a large number of fully sequenced and assembled genomes from strains of species that are particularly well suited for such analyses. The recent development of high-throughput sequencing technology has reduced sequencing costs and for many microbial species there are now multiple strains, completely sequenced and assembled, available for analyses in public databases [10]. This allowed us to explore strain-level relationships between base composition, genome size and predicted HGT in several microbial species in a recent study [11]. We found that the genome size, compared at strain-level, was predominantly correlated with genomic AT content, contrary to what has been found for prokaryotes in general [12]. Additionally, AT content correlated with predicted HGT size, which again correlated with chromosome size [11]. In this study we also analyzed the influence of selective pressures on microbial genome size using the concept of relative entropy [13, 14].
Relative entropy can be used to measure genomic distance and is computed with the Kullback–Leibler measure between observed and expected codon frequencies (see [14] for more details). The expected codon frequencies are calculated from genomic nucleotide frequencies so that decreasing distances between observed and expected codon frequencies imply increased independence between the neighboring nucleotides constituting the codons. This implies more random distributions of codon frequencies presumably due to mutations/genetic drift [13, 15]. A negative correlation between relative entropy and AT content has previously been detected in microbial genomes, implying that AT-rich genomes tend to have, on average, a more random base composition than GC-rich genomes [11, 13, 16]. The greater similarity between AT-rich genomes and random DNA sequences, with similar base compositions, is a consequence of the fact that genomic mutations are in general biased towards AT-richness [17, 18].
Horizontally transferred DNA tends to have lower relative entropy than DNA of the host chromosome. Thus, it is likely that the genomes of strains with high levels of horizontally transferred DNA will, on average, have lower relative entropy than the genomes of strains having received less HGT [13]. However, it may also suggest differences in how selective forces operate at the strain level, analogous to the general negative correlation between AT content and genome size, which appears to be largely reversed at the strain-level of bacterial species [11].
The dN /dS ratio, where dN describes the difference in non-synonymous substitutions between taxa and dS designates the difference in synonymous substitutions, has also been associated with selective pressures [19]. Indeed, a large dS relative to dN is linked to purifying selection; dN = dS is assumed to indicate neutrality of selection, while a dN greater than dS is associated with positive selection [19]. Not only does dS > dN provide an approximate quantitative measure of the selective pressures involved in purging non-synonymous substitutions resulting in reduced fitness, but the relation may also give clues about the species’ population structure [18]. Additionally, time is a central factor [19]. A recent divergence between two or more strains is often indicated by dN > dS, since such mutations are more likely to take place within a short time span [19].
It has previously been shown that purifying selection correlates with genome size for microbes above strain level [20]. In the present study we wanted to examine whether selective forces would leave a base compositional pattern in the core genomes of bacterial strains undergoing genome expansion, mediated through HGT, since such a pattern has been observed for microbial species undergoing genome reduction [3, 13]. We focused our analysis on E. coli since this particular species is renowned for extensive HGT and has many strains sequenced and fully assembled [21, 22]. Since pathogenicity has been linked with HGT [1, 2] we also wanted to test whether the pathogenic potential of the E. coli strains correlated with genomic properties such as AT content, genome size, genomic drift, and selective pressures, as estimated using relative entropy and dN/dS. To reach our aim we extracted the E. coli core genome, consisting of 1729 genes, from 53 E. coli strains and estimated dS and dN, as well as the other genomic properties mentioned above. We also generated a maximum likelihood tree based on mutations in the mutT gene, which has been associated with hyper-mutable strains [23], and compared the congruency of that tree to the trees resulting from the dS- and dN-based hierarchical cluster analyses.
Results and discussion
Estimation of dNand dSfrom the E. colicore genome
We wanted to explore whether there was a relationship between the selective pressures that the E. coli core genome has been subjected to and genome expansion due to HGT since an association between purifying selection and genome size has previously been identified for microbial species in general [20].
We performed hierarchical cluster analyses based on dS and dN estimations from all 1729 genes belonging to the E. coli core genome. The results can be seen in Figures 1 and 2 for dS and dN estimations, respectively, and the resulting cluster groups (denoted by different colors in both Figures 1 and 2) indicate a strong association with known E. coli phylogroups [24–26]. Table 1 contains more information on the different E. coli strains and the corresponding patho-/phylo-groups resulting from the dS and dN based cluster analyses.
Figure 1.

dS -based heatmap. The heatmap demonstrates a hierarchical cluster analysis of estimated dS (the rate of synonymous distributions between taxa) of 1729 core genes from the 53 E. coli genomes. The differently colored labels designate phylogroups: D (light green), B2 (red), E (green), B1 (blue), and A (dark blue). Groups D, B2 and E consisted predominantly of pathogens; Group A was almost exclusively non-pathogenic, while Group B1 consisted of a mixture of pathogenic and non-pathogenic strains.
Figure 2.
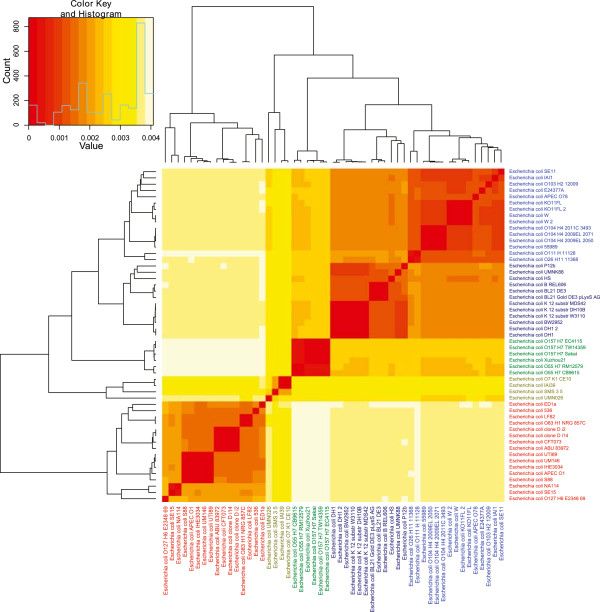
dN -based heatmap. The heatmap shows a hierarchical cluster analysis of estimated dN (the rate of non-synonymous distributions between taxa) of 1729 core genes from the 53 E. coli genomes. The differently colored labels designate phylogroups: D (light green), B2 (red), E (green), B1 (blue), and A (dark blue). Groups D, B2 and E consisted predominantly of pathogens; Group A was almost exclusively non-pathogenic, while Group B1 consisted of a mixture of pathogenic and non-pathogenic strains.
Table 1.
Information about the different E. coli strains used in the study
| Name | Pathogroup | Phylogroup |
|---|---|---|
| Escherichia coli O7:K1 CE10 | ExPEC (neonatal meningitis) | D |
| Escherichia coli IAI39 | ExPEC (Uropathogenic E. coli (UPEC)) | D |
| Escherichia coli SMS 3-5 | Multi-resistant | D |
| Escherichia coli UMN026 | ExPEC (UPEC) | D |
| Escherichia coli 536 | ExPEC (UPEC) | B2 |
| Escherichia coli ED1A | Non-pathogenic | B2 |
| Escherichia coli O83:H1 NRG 857C | AIEC (adherent-invasive E. coli) | B2 |
| Escherichia coli LF82 | AIEC | B2 |
| Escherichia coli clone D i2 | ExPEC (UPEC) | B2 |
| Escherichia coli clone D i14 | ExPEC (UPEC) | B2 |
| Escherichia coli CFT073 | ExPEC (UPEC) | B2 |
| Escherichia coli ABU 83972 | ExPEC (UPEC) | B2 |
| Escherichia coli UTI89 | ExPEC (UPEC) | B2 |
| Escherichia coli UM146 | AIEC | B2 |
| Escherichia coli IHE3034 | ExPEC (neonatal meningitis) | B2 |
| Escherichia coli APEC O1 | Avian pathogenic E. coli (APEC) | B2 |
| Escherichia coli S88 | ExPEC (neonatal meningitis) | B2 |
| Escherichia coli SE15 | Non-pathogenic | B2 |
| Escherichia coli NA114 | ExPEC (multidrug-resistant UPEC) | B2 |
| Escherichia coli E2348_69 O127:H6 | Enteropathogenic E. coli (EPEC) | B2 |
| Escherichia coli O157:H7 TW14359 | Shiga toxin-producing E. coli (STEC/EHEC) | E |
| Escherichia coli O157:H7 EC4115 | STEC/EHEC | E |
| Escherichia coli O157:H7 Sakai | STEC/EHEC | E |
| Escherichia coli Xuzhou21 | STEC/EHEC | E |
| Escherichia coli O55:H7 RM12579 | Atypical EPEC (aEPEC) | E |
| Escherichia coli O55:H7 CB9615 | aEPEC | E |
| Escherichia coli UMNK88 | Enterotoxigenic E. coli (ETEC) | A |
| Escherichia coli P12b | Non-pathogenic | A |
| Escherichia coli HS | Non-pathogenic | A |
| Escherichia coli BL21 DE3 | Lab strain – Non-pathogenic | A |
| Escherichia coli B REL606 | Lab strain – Non-pathogenic | A |
| Escherichia coli BL21 Gold DE3 pLysS AG | Lab strain – Non-pathogenic | A |
| Escherichia coli BW2952 | Lab strain – Non-pathogenic | A |
| Escherichia coli K12 substr DH10B | Lab strain – Non-pathogenic | A |
| Escherichia coli K12 substr MDS42 | Lab strain – Non-pathogenic | A |
| Escherichia coli K12 substr W3110 | Lab strain – Non-pathogenic | A |
| Escherichia coli DH1 (AP012030.1) | Lab strain – Non-pathogenic | A |
| Escherichia coli DH1 (CP001637.1) | Lab strain – Non-pathogenic | A |
| Escherichia coli O104:H4 str. 2009EL-2050 | Enteroaggregative – EHEC (EAggEC-EHEC) | B1 |
| Escherichia coli O104:H4 str. 2009EL-2071 | EAggEC-EHEC | B1 |
| Escherichia coli O104:H4 str. 2011C-3493 | EAggEC-EHEC | B1 |
| Escherichia coli 55989 | EAggEC | B1 |
| Escherichia coli W (CP002185.1) | Lab strain | B1 |
| Escherichia coli W (CP002967.1) | Lab strain | B1 |
| Escherichia coli KO11FL_162099 (CP002516.1) | Lab strain | B1 |
| Escherichia coli KO11FL_162099 (CP002970.1) | Lab strain | B1 |
| Escherichia coli SE11 | Non pathogenic | B1 |
| Escherichia coli IAI1 | Non pathogenic | B1 |
| Escherichia coli E24377A | ETEC | B1 |
| Escherichia coli O103:H2 str. 12009 | STEC/EHEC | B1 |
| Escherichia coli O111:H- str. 11128 | STEC/EHEC | B1 |
| Escherichia coli O26:H11 str. 11368 | STEC/EHEC | B1 |
| Escherichia coli APEC O78 | APEC | B1 |
From the heatmaps in Figures 1 and 2 it can be seen that dS is considerably higher than dN implying that the E. coli core genome has been subjected to strong purifying selection [27]. Since the core genome consists of all genes common to all the strains discussed here, these genes are presumably important for the species survival and the removal of fitness-reducing mutations appears to have been of considerable importance for the evolution of the different lineages.
The similar topology of the heatmaps in Figures 1 and 2 points to corresponding differences in dS and dN for each strain and Figure 3 demonstrates that we found a strong linear association between median- dS and dN values with respect to each strain (β = 26.94 ± 0.44, R2 ~ 0.98, p < 0.001). From Table 1, as well as Figures 1 and 2, it can be seen that group B1, which includes 9 pathogens and 6 non-pathogens, is closest to group A consisting almost exclusively of non-pathogens (one pathogen and 11 non-pathogens). The predominantly pathogenic groups: B2, D and E cluster together and are further away from groups A and B1. The strong correlation between dS and dN observed in Figure 3 may indicate that the formation of the different lineages may be of a more ancient origin since both dN and dS based cluster analyses resulted in cluster groups completely congruent with the established E. coli phylogroups [24].
Figure 3.
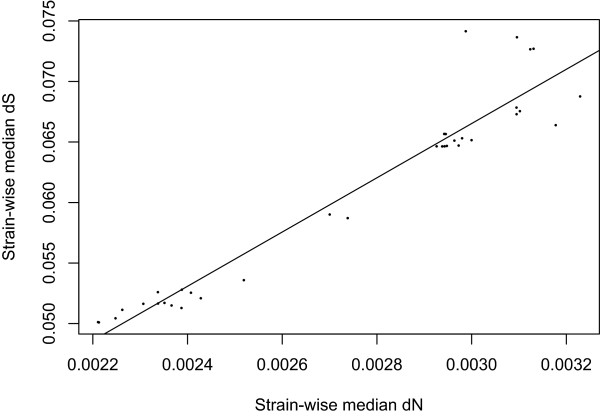
Regression plot of strain-wise median dS and dN. The figure shows median dS estimates plotted against median dN estimates for the E. coli strains in the study. The diagonal line designates the estimated regression line. All similar and clonal strains were removed for the regression analysis resulting in a sample size of 36 strains.
As mentioned above, the type of selective pressures affecting genomes can be inferred from the dN/dS ratio. To examine the selective pressures operating on the different E. coli lineages we used the dS/dN ratio instead of dN/dS for clarification [19]. The resulting dS/dN heatmap can be seen in Figure 4 and shows that although phylogroups D, B2 and E cluster together, phylogroup A is divided into two cluster groups one of which is flanked by phylogroup B1. Interestingly, a similar tree topology was observed by Didelot et al. [24] in a cluster analysis based on E. coli non-core gene content, as opposed to the core genome dS/dN ratios explored in the present study, which may suggest that the lineages represented by phylogroups A and B1 have been exposed to similar selective pressures. Indeed, Didelot et al. points out that the frequency of HGT between these two phylogroups is higher than that observed between any of the other E. coli phylogroups. Therefore it is conceivable that the strains in phylogroups A and B1 are often found within geographic proximity and in similar environments [24].
Figure 4.
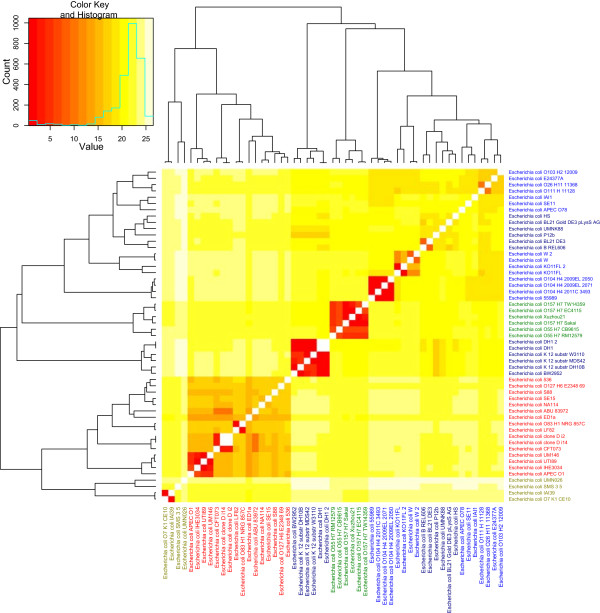
dS / dN -based heatmap. The heatmap demonstrates a hierarchical cluster analysis of estimated dS/dN (the rate of synonymous to non-synonymous substitutions between taxa) of 1729 core genes from the 53 E. coli genomes. The differently colored labels designate phylogroups: D (light green), B2 (red), E (green), B1 (blue), and A (dark blue). Groups D, B2 and E consisted predominantly of pathogens; Group A was almost exclusively non-pathogenic, while Group B1 consisted of a mixture of pathogenic and non-pathogenic strains. The horizontal axis of the color key legend indicates multiples of dS to dN, where values close to 1 designates neutrality of selection.
Phylogenetic inferences from the mutTgene
The topology of the phylogenies resulting from the dS and dN based cluster analyses (Figures 1 and 2) are congruent with the tree depicted in Figure 5, based on variants of the mutT gene, some of which are known to be associated with hyper-mutable E. coli strains [23]. The E. coli ED1a strain, one of two non-pathogens in phylogroup B2 (see Table 1), cluster outside all groups. Other genes related to the genomic mutation levels such as mutY, mutL and mutM [28] resulted in trees with topologies similar to the one we obtained with the mutT gene. However, these alignments were based on relatively large sequences with few mutations resulting in a bootstrap support that was too low for any consistent tree-topology to be inferred. Nevertheless, it is interesting to note that the mutT based-tree supported the dN and dS based phylogenies obtained above. This could mean that the different phylogroups have distinct variants of the mutT gene that coincide with both non-synonymous and synonymous substitution rates in the E. coli core genome. However, our data does not allow any conclusive statements with respect to potential effects of mutT genotypes on dN/dS values.
Figure 5.
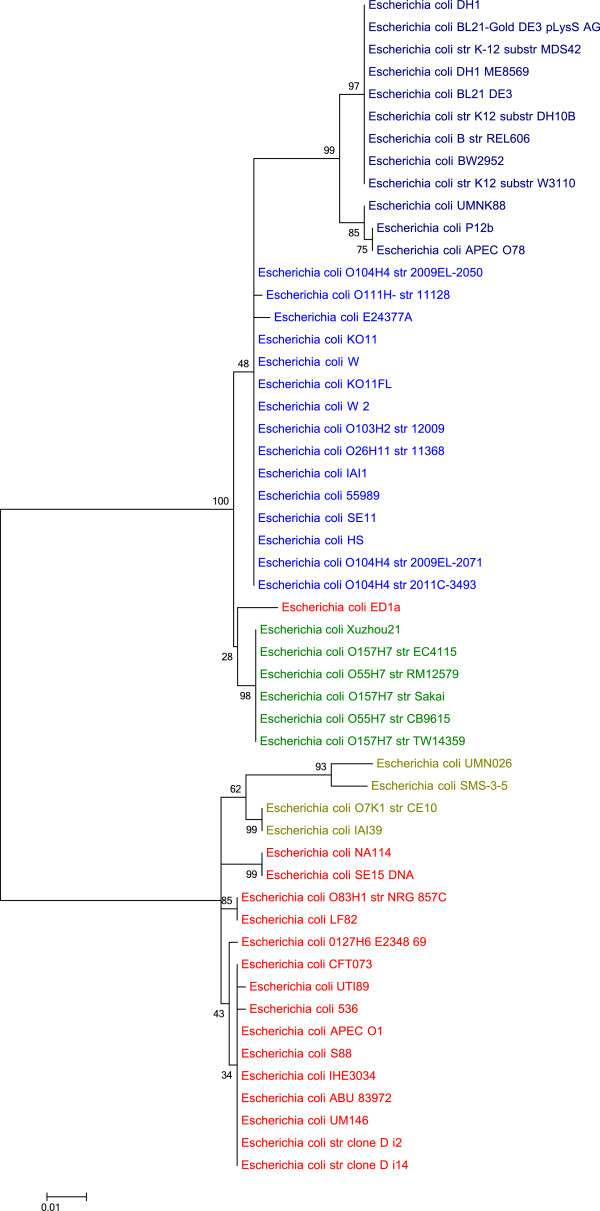
mutT based phylogenic tree. The phylogenic tree is based on alignments of the mutT gene found in the core genome of all 53 E. coli strain. The numbers close to the branches represent bootstrap support. The differently colored labels designate phylogroups: D (light green), B2 (red), E (green), B1 (dark blue), and A (blue). Groups D, B2 and E consisted predominantly of pathogens; Group A was almost exclusively non-pathogenic, while Group B1 consisted of a mixture of pathogenic and non-pathogenic strains.
Examination of the base composition in the E. colicore genome
As previously mentioned, the selective pressures that the E. coli core genome has been exposed to can be analyzed using relative entropy [13]. The genomic frequencies of codons subjected to strong selective pressures are assumed to be substantially different than the corresponding products of nucleotide frequencies. Conversely, codons exposed to weak selective pressures will presumably have more similar frequencies to the corresponding product of nucleotide frequencies due to mutational bias [13, 29]. The relative entropy measure cannot separate between positive- and negative selective pressures associated with dN/dS-based methods. Therefore, with regards to relative entropy, selective pressures will denote both positive- and negative selective pressures.
We wanted to examine whether we could find base compositional differences between core- and whole genomes and whether properties deduced from the core-genomic base composition could be associated with corresponding whole genome properties. For the following statistical analyses we removed all strains that are known to be modified clones, or otherwise genetically very similar, to reduce bias. Details about the specific isolates included in these analyses can be found in Additional file 1. From Figure 6, left panel, it can be seen that relative entropy in the E. coli core genomes was significantly higher than for the corresponding whole genomes (R2 ~ 0.98, p < 0.001). This indicates that the core genome has been subjected to substantially stronger selective pressures than the non-core part of the genome. As previously shown (Figures 3 and 4) the dS estimates were substantially larger than dN estimates, which suggest strong selection of the purifying type. Additionally, it can be seen from Figure 6, right panel, that the core genome was significantly less AT rich (R2 ~ 0.98, p < 0.001) than the rest of the genome. This finding has also been linked to increased selection in other studies although of an unspecified type [30]. In the instance discussed here, relating to the E. coli core genome, there seems to be a connection between purifying selection and decreased AT content.
Figure 6.

Core genome relative entropy and AT content. The figure consists of two panels of boxplots displaying the difference between core- and whole genome relative entropy (left), and core- and whole genome fraction of AT content (right) in all 53 E. coli strains.
We also examined whether there was any association between core- and whole genome levels of both relative entropy and AT content, which could point towards similar selective pressures operating on the core- and whole genome. Our findings indicate no correlation between core- and whole genome relative entropy (p ~ 0.26) suggesting that selective pressures operating on the core genome are most likely unrelated to selective forces effective on the rest of the genome. Core- and whole genome AT content may be negatively correlated (p ~ 0.058), albeit weakly. Since this negative correlation was produced with robust regression, the result was somewhat surprising. An extra generalized additive model (GAM) [31] was therefore fitted, since such models are more capable of modeling non-linear relations, but the association between core- and whole genome AT content was no longer statistically significant (p ~ 0.23). Hence, these results seem to suggest that different selective pressures form the E. coli core and non-core genomes.
The effect of selective pressures on E. coligenome size
Since we have explored how selective pressures operate on whole and core genomes using both dN/dS and relative entropy, we have the necessary results to examine whether selective forces are associated with genome size in E. coli. Previously, a negative correlation between E. coli strains and relative entropy was established which may give the impression that an increase in genome size is a consequence of reduced selective pressures. However, foreign DNA sequences incorporated into a host genome typically have lower relative entropy. Accordingly, the negative association between genome size and relative entropy may be due to the lower relative entropy of the foreign DNA [11]. In Figure 7, it can be seen that the negative association between relative entropy and genome size still holds (R2 = 0.72, p < 0.001). In addition, we also found a significant association between the pathogenicity status of the E. coli strains and genome size (p ~ 0.001), AT content (p < 0.001), whole genome relative entropy (p < 0.001), as well as size of predicted HGT (p ~ 0.005). Hence, we find a clear association between the acquisition of virulence factors and DNA uptake as well as genome size, suggesting that the pathogenic potential of E. coli is associated with DNA uptake. However, we could not find any association between core genome relative entropy on one hand, and genome size (p ~ 0.98) or predicted GI size (p ~ 0.37) on the other. Thus, the lack of correlation between core- and whole genome relative entropy in E. coli seems to suggest that the acquisition of foreign DNA, and virulence factors in particular, is not a consequence of increased selective pressures operating on the genome, at least not on a larger time scale. In this respect, it is interesting that Shigella spp., which is more or less a distinct lineage of E. coli, has been shown to obtain its pathogenic traits through reductive evolution and relaxed selective pressures [9]. It should also be noted that due to strong bi-modality in the strain-based dN/dS estimations no reliable statistical tests could be performed between group-wise median- dN and dS and any corresponding genomic property such as AT content, genome size and relative entropy. Plotting both dN and dS or dN/dS values against the genomic properties discussed above did not reveal any indications of potential trends and were therefore excluded.
Figure 7.
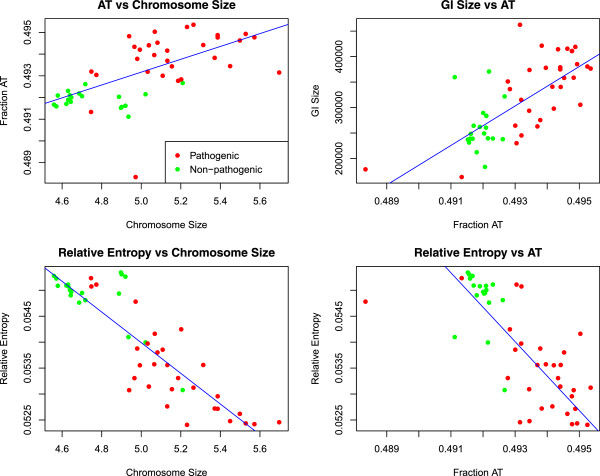
Statistical analyses of genomic properties in 53 E. coli strains. The figure consists of 4 panels showing different associations between selected genomic properties of pathogenic (red dots) and non-pathogenic (green dots) E. coli strains. The blue line denotes the estimated regression line, which was significant for all panels (p < 0.05). Top left panel shows genomic AT content versus chromosome size, while the top right panel depicts estimated HGT size versus genomic AT content. Bottom right panel designates whole genome relative entropy versus genome size, and bottom left panel shows whole genome relative entropy plotted against genomic fraction of AT.
Genome expansion and genome reduction in E. coli
To our knowledge, there are no previous studies of evolutionary forces responsible for genome expansion due to HGT. A recent study discusses evolutionary aspects of recombination in recently emerged clonal Staphylococcus aureus and Clostridium botulinum isolates by examining dN and dS of SNPs in core-, non-core- and recombined DNA, but does not deal with genome expansion as such [32]. Our findings suggest that the E. coli core genome has been subjected to substantial selective pressures over time compared to the genome as a whole. The linear association between median dS and dN estimations for all strains indicates that purifying selection has been directing E. coli lineage evolution and the comparably low rates of non-synonymous substitutions (dN) may indicate that the core genome has remained intact for a longer time span [19]. It should also be noted that all E. coli strains examined in this study are publicly available whole genome sequences, and the fact that they have been selected for sequencing may be due to some special traits not commonly observed in wild-type E. coli.
Conclusions
Our results support previous studies arguing that acquisition of traits through HGT may be a consequence of “spandrel”-like evolutionary processes [33] where the functions of acquired genes are formed through positive selection over time or eventually lost [34, 35]. Hence, increase of selective pressures appears not to be the driving force behind chromosome expansion and acquisition of new traits in E. coli, which is consistent with related findings, also those pointing to an analogous evolutionary trail for gene duplications [36, 37]. Pathogenic E. coli may thus have evolved as a consequence of a hostile environment, where virulence associated genes are abundant. We anticipate that a lot more will be said about this in the future.
Methods
59 E. coli genomes, with their annotated coding genes and corresponding proteins, were downloaded from NCBI/Genbank [10]. In six of the genomes we discovered a lack of correspondence between the coding genes and their listed proteins, and these six genomes were discarded from the downstream analysis. See Additional file 1 for more information on the different E. coli strains used in the study. Genomic properties such as genome size, AT content and relative entropy were estimated using in-house scripts that are available upon request. All statistical analyses were performed with R [38].
Extraction of the core genome
All proteins from every genome were BLASTed (blastp) [39] against all proteins of all other genomes, and a distance was computed between all protein pairs as described in [40]. Based on these distances, proteins were clustered using hierarchical clustering with complete linkage, and divided into clusters by cutting the dendrogram tree at distance 0,1. Loosely speaking, this means any two proteins in the same cluster share 90% similarity. Using this rather strict cutoff resulted in a set of 1729 core clusters, i.e. clusters with at least one protein for each of the 53 genomes in the study. Next, paralogs were eliminated from each cluster using the same procedure as described in [40]. The 53 orthologs in each cluster were aligned using the MCoffee software [41] and the protein-alignments were back-translated to DNA-alignments using the TranslatorX software [42].
Estimation of core genome dNand dS
To calculate dN and dS we followed the method first described by Li et al. [43]. Briefly, we sequentially performed gene-wise multiple alignments as described above on all 1729 core genes from the 53 strains used in the study. The alignment ends were trimmed manually so that the sequences within the alignments were all of the same length. We then used the seqinr package [44] in R to read the alignments, and subsequently calculated gene-by-strain dN and dS values using the kaks() command. For strain-wide assessments, the dN and dS estimates for individual genes were added up and weighted according to gene length. dN and dS for each strain were based on the median from all versus all comparisons.
Due to the bimodal distribution of the core genome-based dN and dS values, heatmaps based on hierarchical clustering with Euclidean distance were created for each of the dS, dN and dS/dN estimated distance matrices so that potential differences between the strains could be examined. These matrices are included in an R-file (see Additional file 2).
Relative entropy
Relative entropy measures the Kullback–Leibler divergence between observed and expected codon frequencies in coding regions [13], i.e.:
where the sum is taken over all 64 possible codons XYZ consisting of nucleotides X, Y and Z, respectively. Fi is a function returning the frequency of codon XYZ, or nucleotides X, Y and Z, from genome i. A low DKL indicates that the observed codon frequencies are, on average, similar to the individual nucleotide frequencies, signifying that the codon frequencies are more random, presumably due to relaxation of the selective forces operating on the genome [13].
The mutTbased phylogenetic tree
The phylogenic tree based on the mutT gene was created, after sequence alignment, using maximum likelihood estimation and 500 bootstraps using the package Mega 6 [45]. Based on statistical analyses carried out with the “Ape” package in R [46], we found that a nucleotide substitution model based on the Tamura-Nei 93 model [47], which assumes equal transversion rates and unequal transition rates, with Gamma-distributed among-site rate variation was the model with the lowest AIC [48] and therefore chosen. The Gamma distribution was discretized into 6 categories, which is the default number of categories; changes to this number did not notably affect the tree topology. The DNA sequences which the mutT based phylogenetic tree is based on are included in FASTA-format (Additional file 3).
HGT predictions
HGT predictions based on the SIGI-HMM method were downloaded and computed using the Islandviewer webpage [49–51].
Statistical analyses
The statistical analyses were carried out using an iterative robust MM-type regression (M-type estimator with Tukey’s biweight and initial coefficient estimates provided from an S-type estimator) [52] with significance estimates (p-values) obtained from t-statistics. All similar strains were discarded before these statistical analyses so that the sample size was reduced to 36 strains (see Additional file 4 and Additional file 5). Robust regression was used where there were several outlying residuals resulting in moderately skewed distributions, otherwise standard ordinary least squares regression was used, which additionally includes a goodness-of-fit estimate (R2). The association between core- and whole genome AT content was also tested using a generalized additive model (GAM), where the predictor (whole genome AT content) was modeled using a spline-function [53]. Additional file 5 contains all estimates resulting from the statistical analyses.
Electronic supplementary material
Additional file 1: An Excel file containing information about the strains used in the study. (XLSX 15 KB)
Additional file 2: An R-file with the dN/dS estimates used in the cluster analyses. (ZIP 30 KB)
Additional file 3: FASTA file with aligned sequences used to make mutT-based tree. (ZIP 1 KB)
Additional file 4: An Excel file with data used for regression analyses. (XLSX 18 KB)
Additional file 5: An Excel file containing data from the regression analyses. (XLSX 11 KB)
Acknowledgements
JB was supported by Norwegian Institute of Public Health, CS by The Norwegian Veterinary Institute. OB and LS were both funded by The Norwegian University of Life Sciences. We thank Dr. Christine L. Parr for help with language and grammar.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JB: Designed the study, performed analyses, wrote the paper. OB: Carried out dS/dN calculations. CS: E. coli based analyses. LS: Computed the E. coli core genome. OB, CS and LS all contributed to the writing of the manuscript. All authors have read and approved this submission.
Contributor Information
Jon Bohlin, Email: jon.bohlin@fhi.no.
Ola B Brynildsrud, Email: ola.bronstad.brynildsrud@nmbu.no.
Camilla Sekse, Email: camilla.sekse@vetinst.no.
Lars Snipen, Email: lars.snipen@nmbu.no.
References
- 1.Fournier PE, Drancourt M, Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infect Dis. 2007;7(11):711–723. doi: 10.1016/S1473-3099(07)70260-8. [DOI] [PubMed] [Google Scholar]
- 2.Pallen MJ, Wren BW. Bacterial pathogenomics. Nature. 2007;449(7164):835–842. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- 3.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10(1):13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 4.Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108(5):583–586. doi: 10.1016/S0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 5.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science (New York, NY) 2009;323(5912):379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 6.Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16(2):149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wernegreen JJ. Reduced selective constraint in endosymbionts: elevation in radical amino acid replacements occurs genome-wide. PLoS One. 2011;6(12):e28905. doi: 10.1371/journal.pone.0028905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yus E, Maier T, Michalodimitrakis K, Van Noort V, Yamada T, Chen WH, Wodke JA, Guell M, Martinez S, Bourgeois R, Kühner S, Raineri E, Letunic I, Kalinina OV, Rode M, Herrmann R, Gutiérrez-Gallego R, Russell RB, Gavin AC, Bork P, Serrano L. Impact of genome reduction on bacterial metabolism and its regulation. Science (New York, NY) 2009;326(5957):1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 9.Hershberg R, Tang H, Petrov DA. Reduced selection leads to accelerated gene loss in Shigella. Genome Biol. 2007;8(8):R164. doi: 10.1186/gb-2007-8-8-r164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2014;42(1):D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohlin J, Sekse C, Skjerve E, Brynildsrud O. Positive correlations between genomic% AT and genome size within strains of bacterial species. Environ Microbiol Rep. 2014;6(3):278–286. doi: 10.1111/1758-2229.12145. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell D. GC content and genome length in Chargaff compliant genomes. Biochem Biophys Res Commun. 2007;353(0006–291; 1):207–210. doi: 10.1016/j.bbrc.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Bohlin J, Van Passel MW, Snipen L, Kristoffersen AB, Ussery D, Hardy SP. Relative entropy differences in bacterial chromosomes, plasmids, phages and genomic islands. BMC Genomics. 2012;13:66-2164-2113-2166. doi: 10.1186/1471-2164-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cover TM, Thomas JA. Elements of Information Theory. New York: John Wiley & Sons, Inc; 1991. [Google Scholar]
- 15.Bohlin J, Brynildsrud O, Vesth T, Skjerve E, Ussery DW. Amino acid usage is asymmetrically biased in AT- and GC-rich microbial genomes. PLoS One. 2013;8(7):e69878. doi: 10.1371/journal.pone.0069878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohlin J, Skjerve E, Ussery DW. Investigations of oligonucleotide usage variance within and between prokaryotes. PLoS Comput Biol. 2008;4(4):e1000057. doi: 10.1371/journal.pcbi.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershberg R, Petrov DA. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6(9):e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocha EP, Feil EJ. Mutational patterns cannot explain genome composition: Are there any neutral sites in the genomes of bacteria? PLoS Genet. 2010;6(9):e1001104. doi: 10.1371/journal.pgen.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocha EP, Smith JM, Hurst LD, Holden MT, Cooper JE, Smith NH, Feil EJ. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239(2):226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 20.Novichkov PS, Wolf YI, Dubchak I, Koonin EV. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol. 2009;191(1):65–73. doi: 10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5(1):e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Passel MW, Marri PR, Ochman H. The emergence and fate of horizontally acquired genes in Escherichia coli. PLoS Comput Biol. 2008;4(4):e1000059. doi: 10.1371/journal.pcbi.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wielgoss S, Barrick JE, Tenaillon O, Wiser MJ, Dittmar WJ, Cruveiller S, Chane-Woon-Ming B, Medigue C, Lenski RE, Schneider D. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc Natl Acad Sci U S A. 2013;110(1):222–227. doi: 10.1073/pnas.1219574110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Didelot X, Meric G, Falush D, Darling AE. Impact of homologous and non-homologous recombination in the genomic evolution of Escherichia coli. BMC Genomics. 2012;13:256. doi: 10.1186/1471-2164-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monk JM, Charusanti P, Aziz RK, Lerman JA, Premyodhin N, Orth JD, Feist AM, Palsson BO. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc Natl Acad Sci U S A. 2013;110(50):20338–20343. doi: 10.1073/pnas.1307797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeves PR, Liu B, Zhou ZM, Li D, Guo D, Ren Y, Clabots C, Lan RT, Johnson JR, Wang L. Rates of Mutation and Host Transmission for an Escherichia coli Clone over 3 Years. Plos One. 2011;6(10):e26907. doi: 10.1371/journal.pone.0026907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balbi KJ, Feil EJ. The rise and fall of deleterious mutation. Res Microbiol. 2007;158(10):779–786. doi: 10.1016/j.resmic.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Gonzalez A, Rivera-Rivera RJ, Massey SE. The Presence of the DNA Repair Genes mutM, mutY, mutL, and mutS is Related to Proteome Size in Bacterial Genomes. Front Genet. 2012;3:3. doi: 10.3389/fgene.2012.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. Codon usage between genomes is constrained by genome-wide mutational processes. Proc Natl Acad Sci U S A. 2004;101(10):3480–3485. doi: 10.1073/pnas.0307827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hildebrand F, Meyer A, Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6(9):e1001107. doi: 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hastie TJ, Tibshirani RJ. Generalized additive models. New York: Chapman and Hall/CRC Press; 1990. [Google Scholar]
- 32.Castillo-Ramirez S, Harris SR, Holden MT, He M, Parkhill J, Bentley SD, Feil EJ. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog. 2011;7(7):e1002129. doi: 10.1371/journal.ppat.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fall S, Mercier A, Bertolla F, Calteau A, Gueguen L, Perriere G, Vogel TM, Simonet P. Horizontal gene transfer regulation in bacteria as a "spandrel" of DNA repair mechanisms. PLoS One. 2007;2(10):e1055. doi: 10.1371/journal.pone.0001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chattopadhyay S, Weissman SJ, Minin VN, Russo TA, Dykhuizen DE, Sokurenko EV. High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection. Proc Natl Acad Sci U S A. 2009;106(30):12412–12417. doi: 10.1073/pnas.0906217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Treangen TJ, Rocha EP. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7(1):e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci. 2012;279(1749):5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paauw A, Leverstein-van Hall MA, Verhoef J, Fluit AC. Evolution in quantum leaps: multiple combinatorial transfers of HPI and other genetic modules in Enterobacteriaceae. PLoS One. 2010;5(1):e8662. doi: 10.1371/journal.pone.0008662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Core Team . R: A language and environment for statistical computing. 2012. [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Snipen L, Wassenaar TM, Altermann E, Olson J, Kathariou S, Lagesen K, Takamiya M, Knochel S, Ussery DW, Meinersmann RJ. Analysis of evolutionary patterns of genes in Campylobacter jejuni and C. coli. Microb Inform Exp. 2012;2(1):8. doi: 10.1186/2042-5783-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34(6):1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38(Web Server issue):W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2(2):150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 44.Charif D, Thioulouse J, Lobry JR, Perriere G. Online synonymous codon usage analyses with the ade4 and seqinR packages. Bioinformatics. 2005;21(4):545–547. doi: 10.1093/bioinformatics/bti037. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 48.Akaike H. A New Look at the Statistical Model Identification. IEEE Trans Auto Contrl. 1974;AC-19(6):716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 49.Langille MG, Brinkman FS. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics (Oxford, England) 2009;25(5):664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langille MG, Hsiao WW, Brinkman FS. Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol. 2010;8(5):373–382. doi: 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- 51.Waack S, Keller O, Asper R, Brodag T, Damm C, Fricke WF, Surovcik K, Meinicke P, Merkl R. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yohai VJ, Stahel WA, Zamar RH. A procedure for robust estimation and inference in linear regression. Directions in robust statistics and diagnostics. New York: Springer; 1991. pp. 365–374. [Google Scholar]
- 53.Wood SN. Generalized Additive Models: An Introduction with R. Boca Raton: Chapman & Hall/CRC Press; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: An Excel file containing information about the strains used in the study. (XLSX 15 KB)
Additional file 2: An R-file with the dN/dS estimates used in the cluster analyses. (ZIP 30 KB)
Additional file 3: FASTA file with aligned sequences used to make mutT-based tree. (ZIP 1 KB)
Additional file 4: An Excel file with data used for regression analyses. (XLSX 18 KB)
Additional file 5: An Excel file containing data from the regression analyses. (XLSX 11 KB)


