Background: Intracellular proteins glycosylation with O-GlcNAc is able to influence cell microenvironment.
Results: O-GlcNAcylation increases hyaluronan synthase 2 (HAS2) transcription via its natural antisense transcript HAS2-AS1.
Conclusion: A novel mechanism to regulate hyaluronan synthesis via long non-coding RNA is described.
Significance: This finding highlights a new target to regulate HA synthesis, critical in many pathophysiological processes.
Keywords: Epigenetics; Glycobiology; Glycosaminoglycan; Glycosylation; Hyaluronan; Long Noncoding RNA (Long ncRNA, lncRNA); Proteoglycan
Abstract
Changes in the microenvironment organization within vascular walls are critical events in the pathogenesis of vascular pathologies, including atherosclerosis and restenosis. Hyaluronan (HA) accumulation into artery walls supports vessel thickening and is involved in many cardiocirculatory diseases. Excessive cytosolic glucose can enter the hexosamine biosynthetic pathway, increase UDP-N-acetylglucosamine (UDP-GlcNAc) availability, and lead to modification of cytosolic proteins via O-linked attachment of the monosaccharide β-N-GlcNAc (O-GlcNAcylation) from UDP-GlcNAc by the enzyme O-GlcNAc transferase. As many cytoplasmic and nuclear proteins can be glycosylated by O-GlcNAc, we studied whether the expression of the HA synthases that synthesize HA could be controlled by O-GlcNAcylation in human aortic smooth muscle cells. Among the three HAS isoenzymes, only HAS2 mRNA increased after O-GlcNAcylation induced by glucosamine treatments or by inhibiting O-GlcNAc transferase with PUGNAC (O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate). We found that the natural antisense transcript of HAS2 (HAS2-AS1) was absolutely necessary to induce the transcription of the HAS2 gene. Moreover, we found that O-GlcNAcylation modulated HAS2-AS1 promoter activation by recruiting the NF-κB subunit p65, but not the HAS2 promoter, whereas HAS2-AS1 natural antisense transcript, working in cis, regulated HAS2 transcription by altering the chromatin structure around the HAS2 proximal promoter via O-GlcNAcylation and acetylation. These results indicate that HAS2 transcription can be finely regulated not only by recruiting transcription factors to the promoter as previously described but also by modulating chromatin accessibility by epigenetic modifications.
Introduction
HA4 is an ubiquitous linear macromolecule composed of glucuronic acid and N-acetylglucosamine (GlcNAc) without any additional chemical modifications typical of the other glycosaminoglycans. HA can modulate a plethora of cellular functions in both physiological as well as pathological conditions (1), interacting with many receptors and proteins (2). HA is synthesized on the plasma membrane by three isoenzymes, HA synthase 1, 2, and 3 (HAS1, -2, and -3), that have several transmembrane domains, use cytosolic UDP-glucuronic acid, and UDP-GlcNAc as precursors and extrude the elongating chain through the plasma membrane into the extracellular matrix. HA synthesis is known to be finely regulated by several factors, including growth factors and proinflammatory cytokines, at both transcriptional and post-translational levels (3, 4).
Cardiovascular diseases are the leading causes of death in western countries and are characterized by a strong extracellular matrix remodeling with high deposition of HA in the neointima (5–7). HA has been described to have a critical role favoring vessel wall thickening and neointima formation (8–10), and inhibition of HA synthesis can be considered a new strategy to face vascular pathologies (11, 12).
Angiopathies are one of the main complications of diabetes (13), and although the molecular mechanisms involved in such diseases are poorly understood, the hexosamine biosynthetic pathway appears to have a crucial role (14). In fact, the elevated flux of nutrients that enters in the hexosamine biosynthetic pathway in hyperglycemic conditions increases the formation of UDP-GlcNAc (15). High availability of UDP-sugars induces the synthesis of HA (16–18).
Elevated levels of UDP-GlcNAc induces the activity of O-GlcNAc transferase, which catalyzes the transfer of GlcNAc from UDP-GlcNAc to serine and threonine residues (O-GlcNAcylation) (19). This intracellular glycosylation controls many cellular processes and is also involved in diabetes, cardiovascular diseases, and tumors (20, 21). O-GlcNAcylation can be easily hydrolyzed from proteins by O-GlcNAcase (OGA) (19). Several nucleo-cytoplasmic proteins are known to be regulated by O-GlcNAcylation including RNA polymerase 2 (22) and HAS2 (16).
HAS2 is mainly responsible for HA synthesis in adult mammalian tissues, and at the genetic level, HAS2 possesses a natural antisense transcript (NAT) named HAS2-AS1 (23). NAT is part of long non-coding RNA (24, 25), which has a crucial role in the modulation of gene transcription (24, 26–30).
HAS2-AS1 NAT is transcribed in the opposite strand of the HAS2 gene localized on chromosome 8 (Fig. 1). HAS2 exon 1 and HAS2-AS1 exon 2 are partially complementary. Furthermore, an alternative splicing can generate two HAS2-AS1 NAT isoforms, named long (L) and short (S), that have 257 or 174 nucleotides of perfect complementary sequence to the region starting ∼70 bp from the presumed transcription start site of human HAS2, respectively (23). This complementarity permits HAS2 mRNA·HAS2-AS1 NAT duplex formation that stabilizes the HAS2 transcript (31) as already described for other mRNA (32).
FIGURE 1.
Genomic organization of HAS2 and HAS2 genes at locus 8q24.13, localization of primers, luciferase constructs, and NF-κB sites. A, schematic representation of HAS2 (RefSeq gene NM_005328.2 on the minus DNA strand) and HAS2-AS1 (RefSeq gene NR_002835 on the plus DNA strand) genes. The scheme reports the starting and ending positions of the genes as indicated by UCSC site (Genome Bioinformatics). Light blue arrows indicate the transcription direction, and boxes are exons. The scheme is in scale. B, representation of the first exon of HAS2 gene and the exon number 1 and 2 of HAS2-AS1 gene. The scheme is not in scale, and black arrows indicate the position of the primers used in this work. The three reporter constructs used in this work containing the luciferase (Luc.) under the control of HAS2 promoter (−2118 + 43) or HAS2 promoter with the HAS2·HAS2-AS1 complementarity region (−2118 + 475; HAS2 exon 1) or HAS2-AS1 promoter (−700) are indicated. C, genomic sequence around HAS2 transcription start site that is indicated as the +1 position. Lowercase letters indicate the first 370 nucleotides of the proximal promoter region, whereas uppercase letters indicate part of HAS2 exon 1. The primer position is also indicated by arrows. D, the genomic sequence around HAS2-AS1 transcription start site is indicated as the +1 position. Lowercase letters indicate the first 444 nucleotides of the proximal promoter region, whereas uppercase letters indicate part of HAS2-AS1 exon 1. The position of primers is also indicated by arrows. The NF-κB subunit p65 binding sites are highlighted in green.
The aim of this study was to investigate whether O-GlcNAcylation could modulate HAS2 gene transcription. We found that O-GlcNAcylation induces HAS2 mRNA accumulation, and this event is strictly dependent to HAS2-AS1, which can modulate the chromatin aperture around HAS2 promoter favoring transcription.
EXPERIMENTAL PROCEDURES
Cell Cultures, Treatments, and Mammalian Samples
Primary human aortic smooth muscle cells (AoSMCs) were purchased from Lonza and grown for 5–8 passages in complete SmGm2 culture medium (Lonza) supplemented with 5% FBS as previously described (16, 33). Briefly, 3 × 105 AoSMCs were incubated with DMEM (Euroclone) supplemented with 0.2% FBS to induce quiescence. After 48 h the medium was changed to normal glucose (5 mm) DMEM-F-12 with 10% FBS, and the cells were left untreated or treated with 2 mm GlcN or 5 mm alloxan or 100 μm O-(2-acetamido-2-deoxy-d-glucopyranosylidene) amino-N-phenylcarbamate (PUGNAC) for 24 h (all from Sigma) to modulate protein O-GlcNAcylation (16). In some experiments 5 μg/ml actinomycin D (ActD) was used to inhibit RNA synthesis as previously described (34), and pyrrolidine dithiocarbamate (PDTC) was used to block NF-μB activity (35, 36).
Eight-week-old male apolipoprotein E knock-out (apoE knock-out (KO); The Jackson Laboratory, Bar Harbor, ME) mice were fed a Western type diet containing 21% saturated fat and 0.15% cholesterol or normal chow for 4 weeks. At the end of the treatment mice were euthanized by asphyxiation, and the aortas were dissected.
cDNA from atherectomy specimens were derived from patients of both genders undergoing carotid endarterectomy with a disease severity ranging from elective, asymptomatic (stage I), and patients with transitory ischemic attack (stage II) to stroke patients (stage 4). Tissue collection and analysis were approved by the Ethics Committee of the Heinrich-Heine-University (ethics approvals number 3944) based on the patients' consent. Atherectomy specimens were directly snap-frozen in liquid nitrogen, and total RNA was extracted using peqGOLD TriFast (PEQLAB Biotechnologie GmbH, Erlangen, Germany) according to the manufacturer's instructions. 1 μg of total RNA was transcribed into cDNA using the QuantiTect Reverse Transcription kit (Qiagen, Hilden, Germany). Hearts from wild-type and OGA knock-out newborn mice (37) were obtained from newborns animals and stored at −80 °C in RNAlater.
Quantitative RT-PCR and Silencing
Total RNAs were extracted from AoSMCs or from mouse hearts with TRI reagent (Invitrogen), retro-transcribed using the High Capacity cDNA synthesis kit (Invitrogen), and amplified on an Abi Prism 7000 instrument (Applied Biosystems). Pre-developed Taqman gene expression assays (Invitrogen) were used to quantify transcripts coding for human HAS2, HAS2-AS1, HAS3, and mouse HAS2 and HAS2-AS1. β-Actin was used as a reference. The relative gene expression was determined by comparing ΔCt (38). Short interfering RNA (siRNA) from Ambion (assay ID n265529 or a scramble control) were used to abrogate the expression of HAS2-AS1 by using a nucleoporator (Nucleofector, AMAXA) (36). After 24 h of incubation, AoSMCs were treated with GlcN as described above to induce O-GlcNAcylation, and the next day, RNA was extracted to measure gene expression.
Luciferase Gene Reporter Assay
The reporter vector (based on pGL3, Promega) expressing luciferase cloned downstream from the HAS2 promoter (−2118/+43 bp) was a gift of Katri Makkonen (39), whereas the luciferase under the control of the HAS2-AS1 promoter (−700 bp) was previously described (31). The promoter of HAS2 and part of HAS2 exon 1 (−2118/+475) were synthesized by Biomatik and cloned in the pGL3 vector (Promega) by using the KpnI and SmaI restriction sites. This construct contains all the region of HAS2 exon 1 complementary to HAS2-AS1.
One million AoSMCs were nucleofected with the two HAS2 promoter-reported plasmids, split 1:2, and seeded in 35-mm dishes wells. After 24 h of incubation, AoSMCs were harvested, and 3 × 105 cells were divided in six-well plates and treated with GlcN or alloxan to modulate protein O-GlcNAcylation. CMV promoter-luciferase reporter and a pRL-TK vector (Renilla luciferase under the control of thymidine kinase promoter; Promega) were used as a positive control and nucleofection efficiency normalization, respectively. The pcDNA3-SP1 expressing vector (a gift of Professor Stephen Safe) was nucleofected in AoSMCs with the −2118 + 43 HAS2 promoter-luciferase reporter construct as an additional control. In other experiments the HAS2 gene promoter-luciferase plasmids were nucleofected with pcDNA3 vectors expressing HAS2-AS1 (see below). After 24 h of incubation, cell extracts were assayed for luciferase activity using Dual-GLO luciferase assay kit (Promega). Luciferase activities were normalized to both the protein content and the nucleofection efficiency calculated by Renilla luciferase activity as previously described (31).
Chromatin Immunoprecipitation (ChIP)
The Pierce agarose ChIP kit was used to immunoprecipitate chromatin following the manufacturer's instructions. Briefly, formaldehyde was added to 2 × 106 cells/ChIP reaction to cross-link DNA and proteins. After an incubation of 10 min at room temperature, glycine was added to quench the excess of formaldehyde. Cells were scraped into cold PBS, pelleted, and resuspended in Lysis Buffer 1 containing a 1× Halt mixture. Nuclei were isolated and resuspended in MNase Digestion Buffer. Cross-linked DNA was digested with 0.25 μl of micrococcal Nuclease (10 units/μl) at 37 °C for 15 min and centrifuged to recover the nuclei. Digested chromatin was resuspended in Lysis Buffer 2. For each IP, diluted chromatin was incubated overnight with a specific monoclonal antibody against O-GlcNAcylated proteins (HGAC85; Pierce) (40) or with anti-p65 monoclonal antibody (D14E12, Cell Signal) or with anti RNA polymerase 2 monoclonal antibody (Pierce; used as positive control) or with normal rabbit IgG (used as negative control) at 4 °C. Each immunoprecipitated complex was column-purified using the agarose resin and eluted in IP Elution Buffer. Reverse cross-linked DNA was purified by using a DNA clean-up column according to the manufacturer's instructions. 5 μl of each of the purified DNAs was used as a template for 48 cycles of quantitative PCR (qPCR) amplification using primers designed on the proximal promoter (∼−300 bp from the transcription start site) of HAS2 (5′-CTCAGGGTTCCCCAGTCCACACCTC-3′, 5′-TCTCTGGTTCAATGGGCTGCTCGAA-3′), HAS2-AS1 (5′-AGCGGCCTCACTCCTTCAGCAAAG-3′, 5′-GACCGTTGCTGCCTGTTGGGTCTC-3′), HAS2 exon 1 (5′-TTTTAAAGTGGGGAAGAATCAAACA-3′, 5′- GGCAGTTTCCAAAATTGAGGTAATA-3′) (see Fig. 1 for primer positions), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, primers provided with the ChIP kit). HAS2, HAS2-AS1, and HAS2 exon 1 primers were designed using Primer3 internet site setting parameters to have an annealing temperature of 60 °C and an amplicon size of 100 bp. qPCRs were carried out in two steps: denaturation at 95 °C for 30 s and annealing/elongation at 60 °C for 1 min using the Abi Prism 7000 instrument (Applied Biosystems) and the SYBR Green qPCR Mastermix (Invitrogen). All primers were tested on genomic DNA to avoid primer dimers and to check polymerase efficiency. Normalization was done using the ΔΔCt method. Briefly, IP samples and total input threshold cycles (Ct) for each treatment were subtracted from the Ct of the corresponding control IP (rabbit IgG). The resulting corrected value for the total input was then subtracted from the corrected experimental IP value (ΔΔCt), and these values were raised to the power of 2 (2ΔΔCt). These values were then expressed as a relative promoter binding ±S.D. as previously described (41).
Nuclease Accessibility Assay
EpiQ chromatin analysis kit (Bio-Rad) was used to perform nuclease accessibility assays following the manufacturer's protocol to measure open and closed chromatin (42). Briefly, AoSMCs were treated with reagents to modulate O-GlcNAcylation and processed in two treatment groups: undigested and digested (each in triplicate). The undigested group was not digested with the EpiQ nuclease, and the digested group was treated with limited EpiQ nuclease digestion (1 h at 37 °C) according to the kit instruction. The genomic DNA samples for both groups were isolated and subjected to qPCR to amplify the proximal promoter of HAS2, HAS2-AS1 (see sequences above), and HAS3 (5′-GCCCCACTGCGGAATTCAAAGCTAA-3′, 5′-CCCGCAAAACCTACTCAC-3′) genes by using the conditions described in ChIP section. qPCR results for all samples were normalized to an internal control, a fragment of an unexpressed rhodopsin gene (RHO) according to the kit instructions. Nuclease accessibility index was calculated as the ratio between the undigested sample and the digested sample at the same time point. The equation used was: accessibility index = (test region undigested/RHO region undigested):(test region digested/RHO region digested) as previously reported (43).
HAS2-AS1 Overexpression
The two variants of HAS2-AS1 (long (L) and short (S)) were generated by an alternative splicing within the exon 1 of HAS2-AS1. L-HAS2-AS1 and S-HAS2-AS1 have a complementary region of 257 and 174 bp with exon 2 of HAS2, respectively. L-HAS2-AS1 was synthesized by Epoch Life Science Inc. and subcloned in pcDNA3 expressing vector, whereas S-HAS2-AS1 in pcDNA3 was already available in the Bowen laboratory. Three micrograms of expressing construct were nucleofected in AoSMCs. After 24 h of incubation, the gene expression and the nuclease accessibility assays were performed as described above.
Statistical Analyses
Statistical analysis of the data was done using analysis of variance (analysis of variance) followed by post hoc tests (Bonferroni) using Origin 7.5 software (OriginLab). Probability values of p < 0.01 or 0.05 were considered statistically significant. Experiments were repeated three times, each time in duplicate, and data are expressed as the means ± S.E.
RESULTS
O-GlcNAcylation Induces HAS2 mRNA and HAS2-AS1 Transcript Accumulation
As we previously reported (16), treatments with 2 mm GlcN or 100 μm PUGNAC led to an accumulation of O-GlcNAcylated proteins in AoSMCs after 24 h of incubation. The O-GlcNAcylation of HAS2 enzyme in the plasma membrane together with an increase of UDP-GlcNAc availability induced HA accumulation ∼3-fold over the controls (16). As O-GlcNAcylation is also present in many transcription factors that regulate HA metabolism (i.e. NF-κB, SP1, and YY1) (3, 44), we investigated whether expression of the HA synthases could be controlled by O-GlcNAcylation. To increase protein O-GlcNAcylation, quiescent (i.e. serum-starved) AoSMCs were incubated with DMEM-F-12 with 10% FBS, and the cells were left untreated or treated with 2 mm GlcN or 100 μm PUGNAC (an OGA inhibitor). On the other hand, to reduce protein O-GlcNAcylation, quiescent AoSMCs were incubated with DMEM-F-12 with 10% FBS, and the cells were left untreated or treated with 5 mm alloxan (an O-GlcNAc transferase inhibitor). As a further control, we treated AoSMCs with alloxan + GlcN to consider the effect augmenting UDP-GlcNAc without O-GlcNAcylation (16). After 24 h of incubation the expressions of HAS2 and HAS3 (the main HAS isoenzymes present in AoSMCs) were assayed by quantitative RT-PCR (Fig. 2A). The expression of HAS3 was not significantly modified by O-GlcNAcylation and was ∼8-fold lower than HAS2 in the control. HAS2 mRNA significantly accumulated after treatments with both GlcN and PUGNAC that increase O-GlcNAcylation. Interestingly, increasing UDP-GlcNAc without O-GlcNAcylation (alloxan + GlcN treatment) did not induce HAS2 mRNA over the control level.
FIGURE 2.
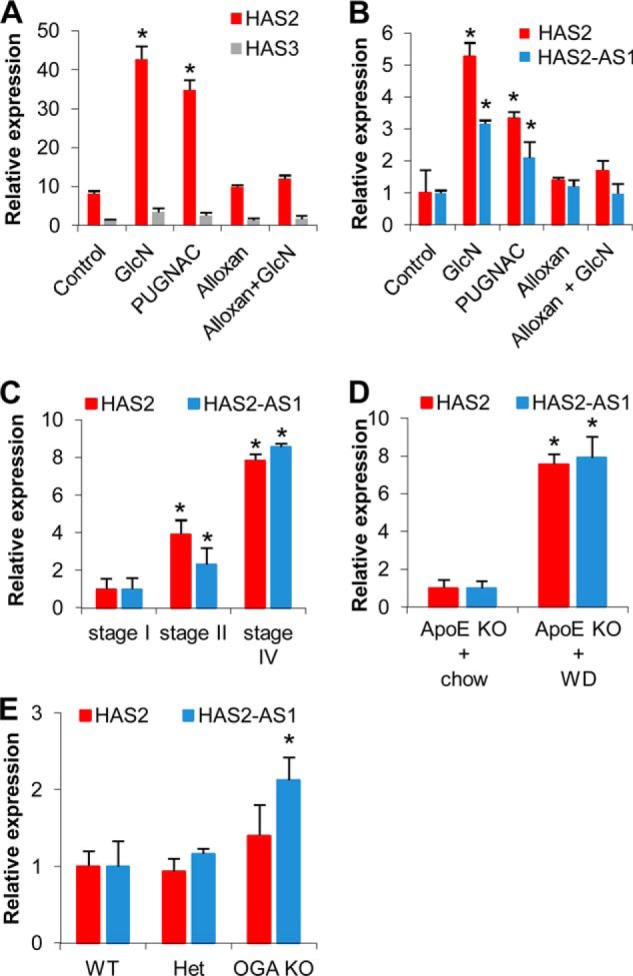
HAS2, HAS3, and HAS2-AS1 expression after modulating O-GlcNAcylation in AoSMCs and mammalian tissues. A, relative quantification of transcripts coding for HAS2 (red bars) or HAS3 (gray bars) in untreated (Control) and 48 h after treating AoSMCs with 2 mm GlcN, 100 μm PUGNAC, 5 mm alloxan, or combinations of alloxan and GlcN. The results of the relative quantification were calculated by setting the control HAS3 expression as 1, and the S.E. is shown on each bar. *, p < 0.01 control versus treated samples. B, relative quantification of HAS2 mRNA (red bars) and the HAS2-AS1 NAT (blue bars) in AoSMCs treated as described above. The results were calculated by setting the control HAS2 and HAS2-AS1 expressions as 1, and the S.E. is shown on each bar. *, p < 0.01 control versus treated samples. C, relative quantification of HAS2 mRNA and the HAS2-AS1 NAT in human atherectomy specimen at different stages of severity (Stage I, n = 22; Stage II, n = 9; Stage IV, n = 9). The results were calculated setting stage I HAS2 and HAS2-AS1 expressions as 1, and the S.E. is shown on each bar. *, p < 0.01 stage I versus the other stages. D, relative quantification of HAS2 mRNA and the HAS2-AS1 NAT in aortas dissected from ApoE KO mice fed for 4 weeks with normal chow (n = 4) or western diet (WD) (n = 7). The results were calculated by setting normal chow HAS2 and HAS2-AS1 expressions as 1, and the S.E. is shown on each bar. *, p < 0.01 chow versus western diet. E, relative quantification of HAS2 mRNA and the HAS2-AS1 NAT in hearts dissected from newborn OGA KO (n = 2), heterozygotes (n = 6), and wild-type (n = 5) mice. The results were calculated by setting normal chow HAS2 and HAS2-AS1 expressions as 1, and the S.E. is shown on each bar. *, p < 0.01 wild-type versus OGA KO.
Recently a positive effect of HAS2-AS1 NAT on HAS2 mRNA stability was described (31). Therefore, we quantified this transcript and surprisingly found that HAS2-AS1 NAT expression was also up-regulated by O-GlcNAcylation similar to HAS2 mRNA (Fig. 2B).
HAS2-AS1 NAT Is Expressed in Healthy and Pathological Mammalian Tissues
Although HAS2-AS1 NAT was reported in several tumor cell lines, we investigated whether HAS2-AS1 expression could also be detected in normal mammalian tissues (23, 31). Because of the importance of HA in the onset and progression of atherosclerosis, we analyzed the HAS2 and HAS2-AS1 expression by qRT-PCR in atherectomy samples of carotid arteries at different stages of the disease. We found that both transcripts were significantly increased in atherectomy specimens derived from more severe lesions (Fig. 2C).
To study the expression of HAS2-AS1 in an animal model of atherosclerosis, we fed apoE KO mice with normal chow or western diet (high cholesterol) for 4 weeks as this treatment is known to strongly induce plaque formation. Interestingly, we found significant increases of both HAS2 and HAS2-AS1 transcripts in aortas from atherosclerotic mice (Fig. 2D), highlighting a putative role of such a NAT in atherosclerosis.
To study the relationship of O-GlcNAcylation with the expressions of HAS2 and HAS2-AS1, we quantified their transcripts in hearts of OGA KO and wild-type newborn mice. Fig. 2E shows that HAS2-AS1 expression was significantly higher in hearts from mice with elevated levels of O-GlcNAcylation compared with hearts from wild-type or heterozygote mice. Surprisingly, in hearts from OGA KO mice, HAS2 expression was not increased relative to hearts from control animals, highlighting the specific effect of O-GlcNAcylation on HAS2-AS1 expression.
HAS2-AS1 NAT Is Required for HAS2 mRNA Accumulation after GlcN Treatment
A previous study revealed that HAS2-AS1 NAT is necessary for HAS2 mRNA accumulation in renal proximal tubular epithelial cells after cytokine treatments (31). To assess whether HAS2-AS1 NAT was involved in HAS2 mRNA accumulation after O-GlcNAcylation induction, we efficiently abrogated HAS2-AS1 NAT in AoSMCs by siRNA relative to the control and the siSCR treatment (Fig. 3A). The abrogation of HAS2-AS1 also eliminated the NAT augment after the treatment with GlcN, in contrast with the siSCR plus GlcN treatment. As shown in Fig. 3B, the HAS2-AS1 NAT silencing prevented the HAS2 mRNA increase due to GlcN treatment. The abrogation of HAS2-AS1 NAT reduced the HAS2 mRNA expression by ∼50%, indicating that it could also be involved in HAS2 basal transcription. The effects of HAS2-AS1 NAT knockdown were specific, as the treatment with a scrambled siRNA did not alter the expected results and because the HAS3 expression was not influenced by HAS2-AS1 NAT silencing.
FIGURE 3.
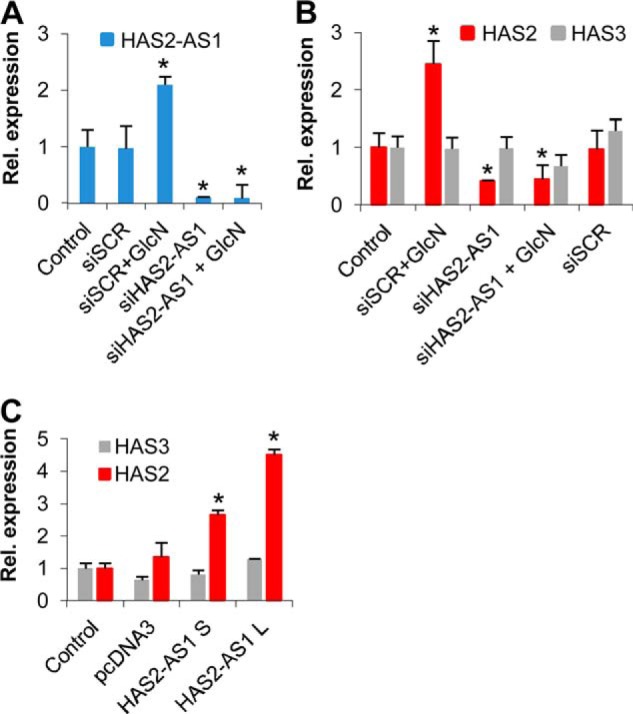
HAS2-AS1 NAT is necessary to increase HAS2 mRNA transcription after O-GlcNAcylation. A, relative expression of HAS2-AS1 after HAS2-AS1 abrogation by siRNA (siHAS2-AS1). AoSMCs were nucleofected with siHAS2-AS1 or a scrambled control (siSCR) and after 24 h treated with or without 2 mm GlcN. After 24 h, total RNA was extracted, retrotranscribed, and subjected to qRT-PCR. The results of the relative quantification are calculated by setting the HAS2-AS1 expression in control samples as 1, and the S.E. is shown on each bar. *, p < 0.01 siSCR+GlcN- versus siHAS2-AS1-treated samples. B, relative expression of HAS2 (red bars) and HAS3 (gray bars) mRNAs after HAS2-AS1 abrogation by siRNA. AoSMCs were treated as described above, and the results were calculated setting HAS2 and HAS3 expression in control samples as 1. The S.E. is shown on each bar. *, p < 0.01 siSCR+GlcN- versus siHAS2-AS1-treated samples. C, HAS2-AS1 overexpression increases expression of HASs mRNA. AoSMCs were nucleofected with no DNA (control) or with 3 μg of pcDNA3 vector or with 3 μg of pcDNA3 that expresses the splice variant of HAS2-AS1 short (S) the splice variant long (L). After 48 h the relative expressions of HAS2 and HAS3 were assayed by qRT-PCR. The results are calculated setting HAS2 and HAS3 expression in control samples as 1, and the S.E. is shown on each bar. *, p < 0.01 control versus treated samples.
To confirm the positive role of HAS2-AS1 NAT, we nucleofected pcDNA3 based vectors that drive the expression of the two splice variants HAS2-AS1 short (S) and long (L), which have a complementarity region of 174 and 257 bp, respectively, with the HAS2 exon 1. Overexpression of both HAS2-AS1 S and L NATs induced a significant increase of HAS2 mRNA (Fig. 3C) confirming the positive effects on HAS2 mRNA favoring its accumulation probably through stabilization of HAS2 messenger or facilitating its transcription.
mRNA Stabilization Is Not Involved in HAS2 Messenger Accumulation
NATs can form duplexes with their cognate mRNAs, and it has been demonstrated that such complexes are protected from ribonuclease degradation (31, 32). To investigate this possibility, we treated AoSMCs with GlcN and with ActD, a common inhibitor of transcription that has already been used in these cells (34). AoSMCs were treated without GlcN or with GlcN for 24 h (Fig. 4A, first and second bars, respectively). ActD was then added or not to the cultures, and the expression of HAS2 mRNA was assayed after 60 and 120 min (Fig. 4A, red bars). Interestingly, the inhibition of transcription by ActD prevented the HAS2 messenger increase at 120 min, suggesting that RNA stabilization was not involved in HAS2 mRNA accumulation after GlcN treatment. Similarly, the increase of HAS2 mRNA due to HAS2-AS1 L overexpression was strictly dependent on transcription as shown by the ability of ActD to block the increase of HAS2 transcription after 24 h of GlcN treatment of AoSMCs nucleofected with the HAS2-AS1 L coding vector (Fig. 4B).
FIGURE 4.
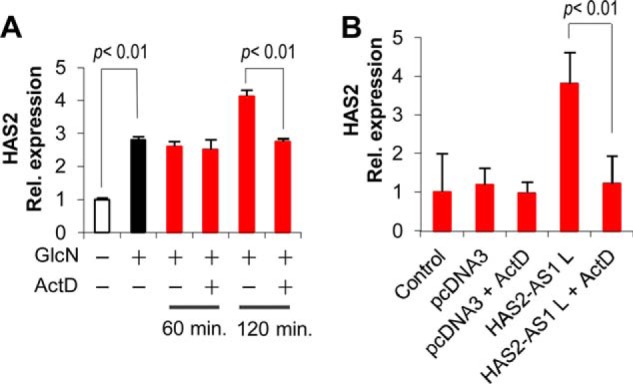
HAS2 expression induced by O-GlcNAcylation is not due to mRNA stability. A, AoSMCs were incubated with (black bar) or without (white bar) 2 mm GlcN for 24 h and then left untreated or treated with 5 μg/ml ActD. After 60 or 120 min (red bars) of incubation total RNA was extracted, retrotranscribed, and subjected to qRT-PCR. The results were calculated by setting HAS2 expression in untreated samples as 1, and the S.E. is shown on each bar. B, HAS2 expression induced by HAS2-AS1 L overexpression is not due to mRNA stability. AoSMCs were nucleofected with no DNA (control) or with 3 μg of pcDNA3 vector or with 3 μg of pcDNA3 expressing the HAS2-AS1 long (L) isoform. After 24 h cells were treated with ActD (5 μg/ml final concentration) or with vehicle alone and incubated for 120 min. The results are calculated setting HAS2 expression in untreated samples as 1, and the S.E. is shown on each bar.
HAS2 and HAS2-AS1 Promoters Are Activated by O-GlcNAcylation
As mRNA stability was not involved in HAS2 messenger accumulation after O-GlcNAcylation, we investigated whether the GlcN treatments could activate HAS2 and HAS2-AS1 promoters. Luciferase reporter assays were performed by using the two HAS2 promoters (−2118/+43 and −2118 + 475 from the transcription start site, respectively) (39) and the HAS2-AS1 promoter (−700 from the transcription start site) (31). The first HAS2 promoter construct has only a small part of exon 1, whereas the second possesses all the regions of HAS2 exon 1 complementary to the HAS2-AS1 exon 2 (see the schemes in Fig. 1). The vectors were nucleofected in AoSMCs, and the cells were treated with GlcN with or without alloxan to modulate O-GlcNAcylation. As shown in Fig. 5A, the −2118/+43 HAS2 promoter was not able to induce luciferase activity when the nucleofected cells were treated with GlcN. As SP1 is known to activate the HAS2 promoter as a positive control, AoSMCs were nucleofected with a pcDNA3 plasmid coding for SP1 together with the −2118/+43 HAS2 promoter/luciferase vector and, as expected, found an increase of luciferase activity. These data provide evidence that the −2118/+43 HAS2 promoter was not directly involved in HAS2 transcription after O-GlcNAcylation.
FIGURE 5.
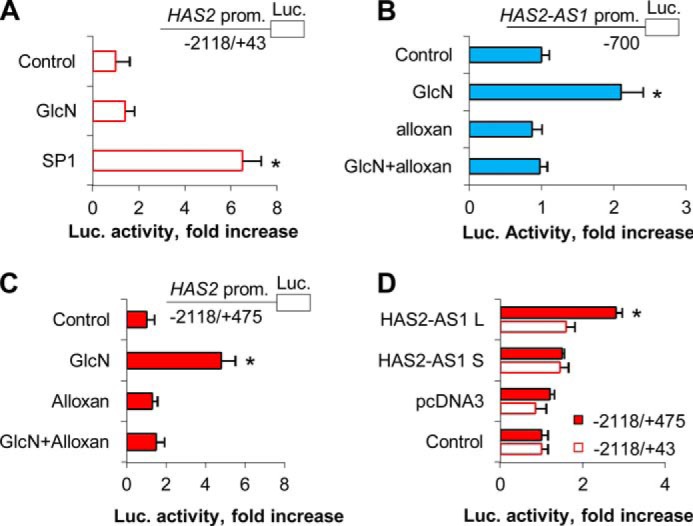
O-GlcNAcylation induces HAS2-AS1 and HAS2 promoter activities. A, AoSMCs were nucleofected with the −2118/+43 HAS2 promoter-luciferase reporter construct and incubated for 24 h. The cells were then plated in six-well plates, treated with or without 2 mm GlcN, and incubated for 24 h before determining luciferase activity. Normalized luciferase activity is expressed as -fold increase with respect to untreated control samples (set as 1). As a positive control, AoSMCs were conucleofected with an SP1 overexpression vector and the −2118/+43 HAS2 promoter-luciferase construct. After 48 h of incubation, the activity of luciferase was assayed. *, p < 0.01 control versus transfected samples. B, AoSMCs were nucleofected with a −700 HAS2-AS1 promoter-luciferase reporter construct. After 24 h of incubation the cells were plated in six-well plates, treated with or without 2 mm GlcN or 5 mm alloxan, and incubated for 24 h before determining luciferase (Luc.) activity. Results are expressed as the mean ± S.E. in three different determinations. *, p < 0.01 control versus treatment, respectively. C, HAS2·HAS2-AS1 complementarity region (i.e. HAS2 exon 1) is critical to modulate transcription. AoSMCs were nucleofected with a −2118/+475 HAS2 promoter-luciferase reporter construct, and after 24 h the cells were plated in six-well plates treated with or without GlcN or alloxan. After 24 h, the luciferase activity was assayed as described above. *, p < 0.01 control versus treated samples. D, AoSMCs were nucleofected with either the −2118/+43 or the −2118/+475 HAS2 promoter-luciferase reporter constructs (Control) or conucleofected with the −2118/+43 or −2118/+475 constructs with 3 μg of pcDNA3 vector or pcDNA3 expressing either the splice variant of HAS2-AS1 short (S) or long (L). After 24 h, the activity of luciferase was measured as described above. *, p < 0.01 control versus transfected samples.
Surprisingly, the promoter of HAS2-AS1 significantly induced luciferase activity after GlcN treatment (Fig. 5B), and the addition of alloxan prevented the induction. These results clearly indicate that O-GlcNAcylation can directly regulate the transcription of HAS2-AS1 by modulating its promoter activity.
We then determined whether the HAS2 exon 1 could have a role in the activation of transcription after inducing O-GlcNAcylation as it is partially complementary to the HAS2-AS1 exon 2 (see Fig. 1). AoSMCs nucleofected with the −2118 + 475 HAS2 promoter-luciferase construct increased luciferase activity significantly after inducing O-GlcNAcylation with GlcN treatment (Fig. 5C). To further demonstrate the critical role of exon 1 in the regulation of HAS2 gene transcription, we conucleofected the −2118 + 475 or the −2118/+43 HAS2 promoter-luciferase plasmids with the pcDNA3 vectors expressing HAS2-AS1 S or L and found that only the HAS2 promoter construct containing the exon 1 was able to drive luciferase activity when nucleofected with the HAS2-AS1 L expressing vector (Fig. 5D).
p65 and O-GlcNAcylated Factor(s) Binds to HAS2-AS1 and HAS2 Promoters
As HAS2-AS1 promoter was activated by GlcN, whereas the HAS2 (−2118/+43) promoter was not able to drive luciferase activity after GlcN treatment, we hypothesized that a transcription factor(s) could interact with HAS2-AS1 but not with −2118/+43 HAS2 promoters after O-GlcNAcylation. To test this we performed ChIP analyses using or the HGAC85 monoclonal antibody that is specific for O-GlcNAc modifications on proteins or anti-p65 (also known as RelA, one of the major components of the NF-κB complex) or anti RNA polymerase 2 antibodies. The cross-linked and digested chromatin was immunoprecipitated with the antibodies or rabbit IgG (control), and the amount of promoter DNA associated with the IP chromatin was quantified by quantitative PCR with primers specific to the human HAS2-AS1 or HAS2 proximal promoter regions. After GlcN treatments, we found significant increases of O-GlcNAcylated proteins associated to the HAS2-AS1 proximal promoter, whereas no significant changes were observable in O-GlcNAcylated proteins bound to the HAS2 proximal promoter (Fig. 6A). Interestingly, we found that a significant increase of O-GlcNAcylated proteins associated with the overlapping region between HAS2 exon 1 and HAS2-AS1 exon 2 after GlcN treatment (Fig. 6A), highlighting the crucial role of this genomic region in the regulation of HAS2 transcription.
FIGURE 6.
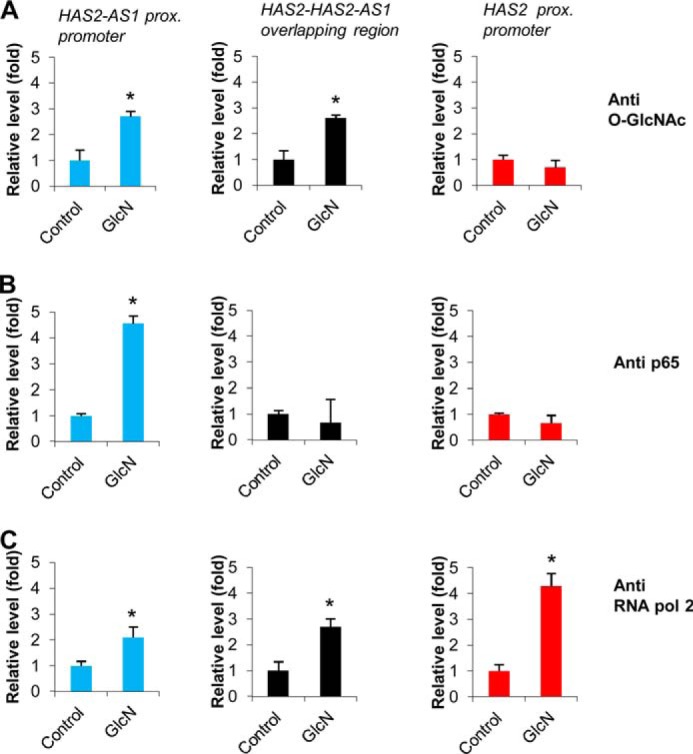
O-GlcNAcylated factor(s) and p65 are associated with HAS2-AS1 proximal promoter. AoSMCs were left untreated (Control) or treated with 2 mm GlcN. After 48 h, chromatin was purified, and ChIP experiments were performed by using specific monoclonal antibodies against O-GlcNAc (A), p65 (B), RNA polymerase 2 (RNA pol 2) (C) or rabbit IgG. PCR conducted on DNA derived from the input chromatin template served as a positive control, and that of the IgG-precipitated template served as a specificity control. The amount of proximal promoter DNAs (−300 bp with respect to the transcription start site) associated with the IP chromatin was quantified by qPCR with specific primers for HAS2-AS1 proximal promoter or HAS2·HAS2-AS1 complementarity region or HAS2 proximal promoter. ChIP promoter binding in control AoSMCs was set at 1 ± S.D. Results are expressed as the mean ± S.E. in three different determinations. *, p < 0.01 untreated versus treatment, respectively. The IgG control ChIP assays produced only 0.05× of the antibody binding levels observed with untreated as well as treated AoSMCs.
As previous analyses revealed the presence of NF-κB binding sites on the HAS2-AS1 promoter (31) (see Fig. 1), we investigated whether p65 was present on the HAS2-AS1 promoter after GlcN treatments. As shown in Fig. 6B, ChIP analyses revealed that GlcN treatment induced a strong and specific recruitment of p65 on HAS2-AS1 proximal promoter, whereas p65 was not detected on the HAS2 proximal promoter or in the overlapping region. As the positive control experiment, we performed ChIP with anti-RNA polymerase 2 antibody and found an increased recruitment of this enzyme on HAS2-AS1 and HAS2 promoters as well as on the overlapping region as expected.
NF-κB Is Required after GlcN Treatment to Induce HAS2-AS1 NAT Accumulation
To demonstrate the role of NF-κB subunit p65 in the accumulation of HAS2-AS1 NAT after induction of O-GlcNAcylation with GlcN treatments, quiescent AoSMCs were untreated or treated with GlcN or treated with GlcN and PDTC, an inhibitor of NF-κB, at 0.5 and 2 μm as final concentrations. PDTC specifically prevented the GlcN-induced HAS2-AS1 NAT accumulation in a dose-response manner without altering HAS3 expression (Fig. 7) and, therefore, highlighted the involvement of NF-κB in the regulation of HAS2-AS1 NAT.
FIGURE 7.
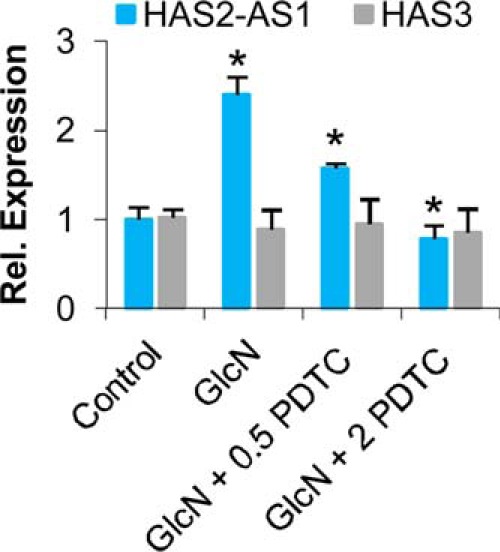
HAS2-AS1 expression is mediated by NF-κB. Relative quantification of HAS2-AS1 (blue bars) and HAS3 (gray bars) in untreated (Control) and after 48 h of treatment of AoSMCs with 2 mm GlcN or with 2 mm GlcN plus 0.5 or 2 μm PDTC (an NF-κB inhibitor). The results of the relative quantification were calculated by setting control HAS2-AS1 and HAS3 expression as 1, and the S.E. is shown on each bar. *, p < 0.01 GlcN- versus PDTC-treated samples.
HAS-AS1 Induces Chromatin Remodeling in the HAS2 Proximal Promoter
NATs can regulate their cognate transcript in several ways (26). We focused on the possibility that HAS2-AS1 NAT could interact in cis with the HAS2 gene and modulate chromatin structure. To investigate this point, we performed nuclease accessibility assays on AoSMCs treated with reagents that modulate O-GlcNAcylation. The nuclease accessibility assay can be used to determine the sensitivity of DNA regions to nucleases revealing the location of open or closed chromatin. The method is based on the more rapid nuclease-catalyzed hydrolysis observed with open chromatin structures than with closed chromatin structures. As shown in Fig. 8A, the induction of O-GlcNAcylation by GlcN or PUGNAC treatments increased the nuclease accessibility index of the proximal promoters of HAS2 and HAS2-AS1, suggesting an open chromatin structure. Conversely, the chromatin structure around the HAS3 promoter, which shares no complementarity with HAS2-AS1 NAT, was not influenced by O-GlcNAcylation.
FIGURE 8.
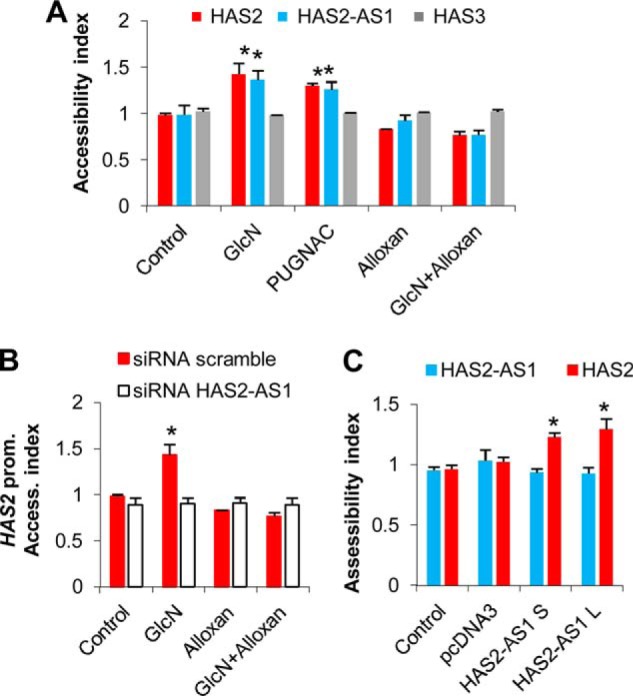
O-GlcNAcylation and HAS2-AS1 induce chromatin remodeling. A, AoSMCs were left untreated or treated with 2 mm GlcN, 100 μm PUGNAC, 40 μm DON, 5 mm alloxan, or combinations of alloxan and GlcN. After 48 h, the nuclease accessibility index was calculated for the proximal promoter of HAS2 (red bars), HAS2-AS1 (blue bars), and HAS3 (gray bars). The nuclease accessibility index was set at 1 in control samples. Results are expressed as the mean ± S.E. in three different determinations. *, p < 0.01 untreated versus treatment, respectively. B, HAS2-AS1 abrogation prevented the aperture of chromatin in proximal promoter (Prom.) of HAS2. AoSMCs were nucleofected with siHAS2-AS1 (white bars) or a scrambled control (red bars). After 24 h of incubation, the cells were left untreated (Control) or treated with 2 mm GlcN or 5 mm alloxan or a combination of alloxan and GlcN. After 24 h nuclease accessibility assays were performed as described above. Results are expressed as the mean ± S.E. in three different determinations. *, p < 0.01 untreated versus treatment, respectively. C, overexpression of HAS2-AS1 S and L induced aperture of chromatin around HAS2 proximal promoter. AoSMCs were nucleofected with no DNA (Control) or with 3 μg of pcDNA3 vector or with 3 μg of pcDNA3 expressing either splice variant of HAS2-AS1 short (S) or long (L). After 48 h the accessibility of nuclease to HAS2 (red bars) or HAS2-AS1 (blue bars) proximal promoters was determined. Results are expressed as the mean ± S.E. in three different determinations. *, p < 0.01 control versus treatment, respectively.
To determine whether the compaction status of chromatin around the HAS2 proximal promoter could depend on HAS2-AS1 NAT, we silenced HAS2-AS1 by siRNA in AoSMCs and treated the cells with GlcN and alloxan to modulate protein O-GlcNAcylation. Abrogation of HAS2-AS1 did not permit the opening of chromatin around the HAS2 proximal promoter after inducing O-GlcNAcylation (Fig. 8B). To confirm the role of HAS2-AS1 in modulating the chromatin structure, we nucleofected pcDNA3 vectors expressing HAS2-AS1 S or L variants and found that overexpression of these NATs was sufficient to increase the aperture of the region around the HAS2 proximal promoter (Fig. 8C). This change in chromatin structure was specific for the HAS2 gene as the region around the HAS2-AS1 proximal promoter was not altered by HAS2-AS1.
DISCUSSION
O-GlcNAcylation is an intracellular glycosylation involved in many pathologies as diabetic vascular diseases (21). At the nuclear level, O-GlcNAcylation has been already described to modify histones as well as several transcription factors and RNA polymerase 2 (46). Therefore, the alteration in chromatin structure could be due to histone glycosylation and, in this study we found that O-GlcNAcylation increased the accessibility index of the proximal promoters of both HAS2 and HAS2-AS1. Furthermore, the chromatin opening of the HAS2 promoter required the HAS2-AS1 NAT, and the overexpression of such a NAT was able to induce similar chromatin remodeling specifically at HAS2 promoter.
Although all histones can by modified by O-GlcNAc (47), GlcN treatment is known to induce histone H3 O-GlcNAcylation with a reduction of H3 phosphorylation. As O-GlcNAcylated H3 was found in association with both H3K4me3 or H3K9me3, which are active or inactive histone marks, respectively, the role of histone glycosylation in chromatin regulation is still under investigation (48).
To alter chromatin structure many factors can be involved, but epigenetic modifications represent the most common mechanism that allows the aperture of nucleosomes, permitting a more efficient interaction between DNA and the transcription machinery (49, 50). Recently, O-GlcNAc transferase has been found associated with promoters where it regulates ten-eleven translocation (TET) family enzymes that have a role in DNA demethylation process (22, 51–53).
Based on the observation that many histone-modifying enzymes lack specific DNA binding domain (54) and that long non-coding RNAs and NATs might interact with ubiquitously expressed histone modifying enzymes providing the required level of binding specificity (24), we speculated that HAS2-AS1 could bind an uncharacterized factor(s) able to modify histones and mediate chromatin structure change (Fig. 9). Several epigenetic modification proteins have been described to interact with long non-coding RNA as the histone methyltransferase mixed-lineage leukemia that is able to activate transcription at HOXA locus (55).
FIGURE 9.

Working model of HAS2-AS1 mechanism of action. In basal conditions the accessibility of both HAS2 and HAS2-AS1 promoters are low allowing a limited production of transcripts. After induction of O-GlcNAcylation or in hyperglycemic conditions in vivo, O-GlcNAcylated p65 binds to the HAS2-AS1 promoter, increasing HAS2-AS1 NAT accumulation. HAS2-AS1, pairing with the complementarity region on HAS2 exon 1, could bring the enzyme(s) (circle with question mark) involved in epigenetic modification of histones (indicated by the triangle flags on cylinders), permitting chromatin aperture on HAS2 promoter and allowing a more efficient transcription.
The specific activity of HAS2-AS1 NAT to induce HAS2 expression could be due to the complementarity region that is close to HAS2 promoter. HAS2-AS1 could interact in cis with HAS2 exon 1 and permit the modification of chromatin specifically on HAS2 promoter as outlined in the scheme of Fig. 9. How such a complementary region between HAS2 and HAS2-AS1 controls transcription is still under investigation, but as the simple overexpression of HAS-AS1 (in particular the long isoform that has an extended complementarity region respect to the short isoform) without any treatment is able to induce transcriptional activity, we hypothesize a RNA-DNA interaction between HAS2-AS1 NAT and HAS2 gene at exon 1 level. An intriguing point to investigate would be whether HAS2 could control HAS2-AS1 expression in an opposite regulation.
The importance of HAS-AS1 is supported by the fact that HAS2-AS1 promoter is finely tuned by SP1 And SMAD2/3 (31). Our results show that this promoter is activated by O-GlcNAcylation and that NF-κB subunit p65 is associated with the HAS2-AS1 promoter after GlcN treatment. Interestingly, p65 is known to increase its transcriptional activity after O-GlcNAcylation (45, 56). On the other hand, although HAS2 promoter is known to be finely regulated by several transcription factors (3), we found that the activity of HAS2 promoter was not altered by O-GlcNAcylation, and no O-GlcNAcylated factor(s) was detectable bound with it. Interestingly, the addition of the complementarity region between HAS2 and HAS2-AS1 to the HAS2 promoter (construct −2218/+475) led to an increment of luciferase activity after induction of O-GlcNAcylation, highlighting a crucial role of this region (i.e. HAS2 exon 1) in transcription regulation. This issue is also supported by the fact that this region was able to interact with the O-GlcNAcylated factor(s).
As the overexpression of HAS2 and HA accumulation can be detrimental in vivo in the case of malignancies and vascular pathologies (10), we found that the expression of HAS2-AS1 NAT is augmented in human atheromatous plaque with an increased grade of severity known as the proatherosclerotic effect of HA. Furthermore, in a mouse model of atherosclerosis induced by high cholesterol, the accumulation of both HAS2 and HAS2-AS1 is evident highlighting the potential role in the pathology of such NAT. Although our in vivo data cannot explore the molecular mechanism of HAS2-AS1 action, the expression of HAS2-AS1 in mice with a high level of O-GlcNAcylation was double with respect to wild-type or heterozygous animals, confirming the role such glycosylation to control the NAT expression.
In conclusion, our data identify HAS2-AS1 as new important player in the regulation of HAS2 gene transcription and highlight the coordinated expression of such NAT and its sense counterpart in healthy and atherosclerotic human carotids as well as in murine model of atherosclerosis.
Acknowledgments
We thank Stephen Safe (Texas A&M University, for pcDNA3-SP1 vector), Katri Makkonen (University of Eastern Finland, for HAS2 promoter luciferase vectors), Stuart Schreiber (Howard Hughes Medical Institute, for pcDNA3-HADC1 vector), Hendrik Stunnenberg (Radboud University, The Netherlands, for pcDNA3-p300 vector). We acknowledge the “Centro Grandi Attrezzature per la Ricerca Biomedica,” Università degli Studi dell'Insubria, for instrument availability.
This work was supported by FAR, PRIN, Fondazione Comunitaria del Varesotto-ONLUS, and European Union Grant IRSES INFLAMA (to A. P.).
- HA
- hyaluronan
- O-GlcNAcylation
- O-linked N-acetylglucosamine
- OGA
- O-GlcNAcase
- HAS
- hyaluronan synthase
- AoSMCs
- human aortic smooth muscle cells
- NAT
- natural antisense transcript
- PUGNAC
- O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate
- PDTC
- pyrrolidine dithiocarbamate
- ActD
- actinomycin D
- qPCR
- quantitative PCR
- Ct
- threshold cycle
- IP
- immunoprecipitation
- L
- long
- S
- short.
REFERENCES
- 1. Jiang D., Liang J., Noble P. W. (2011) Hyaluronan as an immune regulator in human diseases. Physiol. Rev. 91, 221–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vigetti D., Karousou E., Viola M., Deleonibus S., De Luca G., Passi A. (2014) Hyaluronan: biosynthesis and signaling. Biochim. Biophys. Acta 1840, 2452–2459 [DOI] [PubMed] [Google Scholar]
- 3. Tammi R. H., Passi A. G., Rilla K., Karousou E., Vigetti D., Makkonen K., Tammi M. I. (2011) Transcriptional and post-translational regulation of hyaluronan synthesis. FEBS J. 278, 1419–1428 [DOI] [PubMed] [Google Scholar]
- 4. Vigetti D., Viola M., Karousou E., De Luca G., Passi A. (2014) Metabolic control of hyaluronan synthases. Matrix Biol. 35, 8–13 [DOI] [PubMed] [Google Scholar]
- 5. Ross R. (1999) Atherosclerosis: an inflammatory disease. N. Engl. J. Med. 340, 115–126 [DOI] [PubMed] [Google Scholar]
- 6. Vigetti D., Moretto P., Viola M., Genasetti A., Rizzi M., Karousou E., Pallotti F., De Luca G., Passi A. (2006) Matrix metalloproteinase 2 and tissue inhibitors of metalloproteinases regulate human aortic smooth muscle cell migration during in vitro aging. FASEB J. 20, 1118–1130 [DOI] [PubMed] [Google Scholar]
- 7. Riessen R., Wight T. N., Pastore C., Henley C., Isner J. M. (1996) Distribution of hyaluronan during extracellular matrix remodeling in human restenotic arteries and balloon-injured rat carotid arteries. Circulation 93, 1141–1147 [DOI] [PubMed] [Google Scholar]
- 8. Chai S., Chai Q., Danielsen C. C., Hjorth P., Nyengaard J. R., Ledet T., Yamaguchi Y., Rasmussen L. M., Wogensen L. (2005) Overexpression of hyaluronan in the tunica media promotes the development of atherosclerosis. Circ. Res. 96, 583–591 [DOI] [PubMed] [Google Scholar]
- 9. Cuff C. A., Kothapalli D., Azonobi I., Chun S., Zhang Y., Belkin R., Yeh C., Secreto A., Assoian R. K., Rader D. J., Puré E. (2001) The adhesion receptor CD44 promotes atherosclerosis by mediating inflammatory cell recruitment and vascular cell activation. J. Clin. Invest. 108, 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vigetti D., Viola M., Karousou E., Genasetti A., Rizzi M., Clerici M., Bartolini B., Moretto P., De Luca G., Passi A. (2008) Vascular pathology and the role of hyaluronan. ScientificWorldJournal 8, 1116–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vigetti D., Rizzi M., Moretto P., Deleonibus S., Dreyfuss J. M., Karousou E., Viola M., Clerici M., Hascall V. C., Ramoni M. F., De Luca G., Passi A. (2011) Glycosaminoglycans and glucose prevent apoptosis in 4-methylumbelliferone-treated human aortic smooth muscle cells. J. Biol. Chem. 286, 34497–34503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vigetti D., Rizzi M., Viola M., Karousou E., Genasetti A., Clerici M., Bartolini B., Hascall V. C., De Luca G., Passi A. (2009) The effects of 4-methylumbelliferone on hyaluronan synthesis, MMP2 activity, proliferation, and motility of human aortic smooth muscle cells. Glycobiology 19, 537–546 [DOI] [PubMed] [Google Scholar]
- 13. Nigro J., Osman N., Dart A. M., Little P. J. (2006) Insulin resistance and atherosclerosis. Endocr. Rev. 27, 242–259 [DOI] [PubMed] [Google Scholar]
- 14. Brownlee M. (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 414, 813–820 [DOI] [PubMed] [Google Scholar]
- 15. Copeland R. J., Bullen J. W., Hart G. W. (2008) Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 295, E17–E28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vigetti D., Deleonibus S., Moretto P., Karousou E., Viola M., Bartolini B., Hascall V. C., Tammi M., De Luca G., Passi A. (2012) Role of UDP-N-acetylglucosamine (GlcNAc) and O-GlcNAcylation of hyaluronan synthase 2 in the control of chondroitin sulfate and hyaluronan synthesis. J. Biol. Chem. 287, 35544–35555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vigetti D., Ori M., Viola M., Genasetti A., Karousou E., Rizzi M., Pallotti F., Nardi I., Hascall V. C., De Luca G., Passi A. (2006) Molecular cloning and characterization of UDP-glucose dehydrogenase from the amphibian Xenopus laevis and its involvement in hyaluronan synthesis. J. Biol. Chem. 281, 8254–8263 [DOI] [PubMed] [Google Scholar]
- 18. Jokela T. A., Makkonen K. M., Oikari S., Kärnä R., Koli E., Hart G. W., Tammi R. H., Carlberg C., Tammi M. I. (2011) Cellular content of UDP-N-acetylhexosamines controls hyaluronan synthase 2 expression and correlates with O-GlcNAc modification of transcription factors YY1 and SP1. J. Biol. Chem. 286, 33632–33640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hart G. W., Housley M. P., Slawson C. (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 20. Buse M. G. (2006) Hexosamines, insulin resistance, and the complications of diabetes: current status. Am. J. Physiol. Endocrinol. Metab. 290, E1–E8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Slawson C., Copeland R. J., Hart G. W. (2010) O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanover J. A., Krause M. W., Love D. C. (2012) Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13, 312–321 [DOI] [PubMed] [Google Scholar]
- 23. Chao H., Spicer A. P. (2005) Natural antisense mRNAs to hyaluronan synthase 2 inhibit hyaluronan biosynthesis and cell proliferation. J. Biol. Chem. 280, 27513–27522 [DOI] [PubMed] [Google Scholar]
- 24. Magistri M., Faghihi M. A., St Laurent G., 3rd, Wahlestedt C. (2012) Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 28, 389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rinn J. L., Chang H. Y. (2012) Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang K. C., Chang H. Y. (2011) Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoon J. H., Abdelmohsen K., Gorospe M. (2013) Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 425, 3723–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mercer T. R., Mattick J. S. (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307 [DOI] [PubMed] [Google Scholar]
- 29. Lee J. T. (2012) Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439 [DOI] [PubMed] [Google Scholar]
- 30. Faghihi M. A., Wahlestedt C. (2009) Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 10, 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Michael D. R., Phillips A. O., Krupa A., Martin J., Redman J. E., Altaher A., Neville R. D., Webber J., Kim M. Y., Bowen T. (2011) The human hyaluronan synthase 2 (HAS2) gene and its natural antisense RNA exhibit coordinated expression in the renal proximal tubular epithelial cell. J. Biol. Chem. 286, 19523–19532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Faghihi M. A., Modarresi F., Khalil A. M., Wood D. E., Sahagan B. G., Morgan T. E., Finch C. E., St Laurent G., 3rd, Kenny P. J., Wahlestedt C. (2008) Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vigetti D., Clerici M., Deleonibus S., Karousou E., Viola M., Moretto P., Heldin P., Hascall V. C., De Luca G., Passi A. (2011) Hyaluronan synthesis is inhibited by adenosine monophosphate-activated protein kinase through the regulation of HAS2 activity in human aortic smooth muscle cells. J. Biol. Chem. 286, 7917–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raman P., Krukovets I., Marinic T. E., Bornstein P., Stenina O. I. (2007) Glycosylation mediates up-regulation of a potent antiangiogenic and proatherogenic protein, thrombospondin-1, by glucose in vascular smooth muscle cells. J. Biol. Chem. 282, 5704–5714 [DOI] [PubMed] [Google Scholar]
- 35. Schreck R., Meier B., Männel D. N., Dröge W., Baeuerle P. A. (1992) Dithiocarbamates as potent inhibitors of nuclear factor κB activation in intact cells. J. Exp. Med. 175, 1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vigetti D., Genasetti A., Karousou E., Viola M., Moretto P., Clerici M., Deleonibus S., De Luca G., Hascall V. C., Passi A. (2010) Proinflammatory cytokines induce hyaluronan synthesis and monocyte adhesion in human endothelial cells through hyaluronan synthase 2 (HAS2) and the nuclear factor-κB (NF-κB) pathway. J. Biol. Chem. 285, 24639–24645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang Y. R., Song M., Lee H., Jeon Y., Choi E. J., Jang H. J., Moon H. Y., Byun H. Y., Kim E. K., Kim D. H., Lee M. N., Koh A., Ghim J., Choi J. H., Lee-Kwon W., Kim K. T., Ryu S. H., Suh P. G. (2012) O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448 [DOI] [PubMed] [Google Scholar]
- 38. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 39. Makkonen K. M., Pasonen-Seppänen S., Törrönen K., Tammi M. I., Carlberg C. (2009) Regulation of the hyaluronan synthase 2 gene by convergence in cyclic AMP response element-binding protein and retinoid acid receptor signaling. J. Biol. Chem. 284, 18270–18281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Love D. C., Ghosh S., Mondoux M. A., Fukushige T., Wang P., Wilson M. A., Iser W. B., Wolkow C. A., Krause M. W., Hanover J. A. (2010) Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 7413–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang I. C., Chen Y. J., Hughes D., Petrovic V., Major M. L., Park H. J., Tan Y., Ackerson T., Costa R. H. (2005) Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25, 10875–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rao S., Procko E., Shannon M. F. (2001) Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 167, 4494–4503 [DOI] [PubMed] [Google Scholar]
- 43. Yuan W., Wu T., Fu H., Dai C., Wu H., Liu N., Li X., Xu M., Zhang Z., Niu T., Han Z., Chai J., Zhou X. J., Gao S., Zhu B. (2012) Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337, 971–975 [DOI] [PubMed] [Google Scholar]
- 44. Ozcan S., Andrali S. S., Cantrell J. E. (2010) Modulation of transcription factor function by O-GlcNAc modification. Biochim. Biophys. Acta 1799, 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allison D. F., Wamsley J. J., Kumar M., Li D., Gray L. G., Hart G. W., Jones D. R., Mayo M. W. (2012) Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-κB acetylation and transcription. Proc. Natl. Acad. Sci. U.S.A. 109, 16888–16893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Slawson C., Hart G. W. (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sakabe K., Wang Z., Hart G. W. (2010) β-N-Acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang S., Roche K., Nasheuer H. P., Lowndes N. F. (2011) Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J. Biol. Chem. 286, 37483–37495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chi P., Allis C. D., Wang G. G. (2010) Covalent histone modifications: miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bannister A. J., Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen Q., Chen Y., Bian C., Fujiki R., Yu X. (2013) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., Urh M., Daniels D. L., Fuks F. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bernstein E., Allis C. D. (2005) RNA meets chromatin. Genes Dev. 19, 1635–1655 [DOI] [PubMed] [Google Scholar]
- 55. Du Z., Fei T., Verhaak R. G., Su Z., Zhang Y., Brown M., Chen Y., Liu X. S. (2013) Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 20, 908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang W. H., Park S. Y., Nam H. W., Kim do H., Kang J. G., Kang E. S., Kim Y. S., Lee H. C., Kim K. S., Cho J. W. (2008) NFκB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. U.S.A. 105, 17345–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]



