Background: RecO anneals DNA and initiates homologous recombination.
Results: Binding of RecO to DNA/SSB is regulated by zinc during annealing and by RecR during recombination.
Conclusion: Alternative DNA repair reactions are supported by different DNA binding mechanisms of RecO.
Significance: Results explain how the same RMP supports multiple reactions during DNA repair and chromosome maintenance.
Keywords: DNA-binding Protein, DNA Repair, Homologous Recombination, Protein Structure, Protein-Protein Interaction
Abstract
Recombination mediator proteins (RMPs) are important for genome stability in all organisms. Several RMPs support two alternative reactions: initiation of homologous recombination and DNA annealing. We examined mechanisms of RMPs in both reactions with Mycobacterium smegmatis RecO (MsRecO) and demonstrated that MsRecO interacts with ssDNA by two distinct mechanisms. Zinc stimulates MsRecO binding to ssDNA during annealing, whereas the recombination function is zinc-independent and is regulated by interaction with MsRecR. Thus, different structural motifs or conformations of MsRecO are responsible for interaction with ssDNA during annealing and recombination. Neither annealing nor recombinase loading depends on MsRecO interaction with the conserved C-terminal tail of single-stranded (ss) DNA-binding protein (SSB), which is known to bind Escherichia coli RecO. However, similarly to E. coli proteins, MsRecO and MsRecOR do not dismiss SSB from ssDNA, suggesting that RMPs form a complex with SSB-ssDNA even in the absence of binding to the major protein interaction motif. We propose that alternative conformations of such complexes define the mechanism by which RMPs initiate the repair of stalled replication and support two different functions during recombinational repair of DNA breaks.
Introduction
Homologous recombination (HR)2 plays an important role in multiple aspects of chromosome maintenance and genome stability (1–5). The activity of RecA-like recombinases, which support HR, is regulated at several levels (6, 7). HR is initiated by recombinase binding to single-stranded (ss) DNA and formation of a so-called presynaptic complex (8). This step is inhibited by ssDNA binding proteins such as gp32, SSB, and replication protein A (RPA). Recombination mediator proteins (RMPs) overcome such an inhibitory effect and stimulate recombinase binding to ssDNA in response to DNA damage (9). Examples of RMPs include the phage UvsY protein, bacterial RecF, RecO, and RecR proteins (RecFOR) and eukaryotic Rad52, PALB2, and BRCA2 proteins (10–14). Mutations of BRCA2 and PALB2 are associated with cancer predisposition (15–17). RecO and RecR are implicated in drug resistance and host immune response evasion mechanisms in pathogens (18–20).
RecO, RecR, and RecF form an epistatic group involved in DNA repair and replication restart in Escherichia coli and other bacteria (19, 21–30). E. coli RecR (EcRecR) is required for EcRecO function in presynaptic complex formation in vitro, whereas EcRecF plays a regulatory role (31, 32). EcRecF stimulates efficiency of EcRecA binding to the double-stranded (ds)/ssDNA junction substrate in the presence of excess EcSSB (33, 34). RecO has a secondary recombinase-independent function of ssDNA annealing (SSA), as shown for homologs from several bacteria (35–38). As in the case of HR, SSB inhibits DNA annealing, and RecO overcomes an inhibitory effect of SSB. Neither EcRecR nor EcRecF is required for SSA by EcRecO, and EcRecR inhibits SSA activity of EcRecO (38). Eukaryotic RMPs, Rad52, and the BRCA2 homolog Brh2 also possess a similar SSA activity despite lacking sequence or structural homology with RecO and with each other (35–38). It is unclear why these proteins support two functions. One of the ideas with some experimental support in yeast suggests that SSA supports a second-end capture reaction during the post-strand invasion step of HR (39–43). How different functions are activated at different stages of HR is unknown. The function of SSA in E. coli is also not defined (44). Our recent genetic studies of Mycobacterium smegmatis RecO (MsRecO) revealed its critical role in HR and SSA pathways of dsDNA break repair (29, 45).
The mechanism by which RMPs facilitate recombinase binding to ssDNA protected by ssDNA-binding proteins is poorly understood. There are two steps in presynaptic filament formation: nucleation and extension. SSB and RPA inhibit the nucleation steps but can be beneficial for the extension (6). Rad52 and BRCA2 simultaneously bind both RPA and Rad51, thus bringing Rad51 to ssDNA proximity through protein-protein interactions (37, 46–50). Interaction with BRCA2 also alters the DNA binding properties of Rad51 (51). In contrast, EcRecOR lacks RecA binding motifs, and purified proteins do not form stable complexes with EcRecA. The interaction can potentially take place between DNA-bound proteins. For example, EcRecOR prevents dissociation of EcRecA from ssDNA (52), and EcRecFR limits the extension of EcRecA filament beyond ssDNA gap (53). However, the mechanism by which RecOR promotes initial binding of RecA to SSB-coated ssDNA is unknown.
EcRecO binds the C-terminal tail of EcSSB (EcSSB-Ct) (54). This interaction is critical for HR (27) and SSA in E. coli, but it is not conserved in other species. The SSB-Ct-binding site is significantly altered in the structure of Deinococcus radiodurans RecO (DrRecO) (55, 56), and MsRecO does not interact with SSB-Ct (29). Both Dr- and MsRecO possess a 4×Cys zinc finger motif (ZF), whereas there is only one cysteine in a structurally similar domain of EcRecO. A micromolar concentration of zinc in solution stimulates DNA binding and SSA of MsRecO (29).
In this study we have characterized the recombination function of MsRecO. Surprisingly, we found that zinc interaction with MsRecO is dispensable for the presynaptic complex formation. DNA binding of the zinc-depleted MsRecO is stimulated by MsRecR. Thus, MsRecO interacts with ssDNA via two alternative mechanisms in SSA and HR. Neither MsRecO nor MsRecOR completely displace SSB from ssDNA even in the absence of interaction with SSB-Ct. The existence of ssDNA/SSB/RMP complexes explains how two alternative reactions of the strand invasion and SSA can be supported by same RMP during consecutive steps of the recombinational repair.
EXPERIMENTAL PROCEDURES
Chemicals
Glycerol, HEPES, Mg(OAc)2, NaCl, polyethyleneimine, and Trizma base (Tris base) were purchased from Sigma, and ammonium sulfate was from Fischer. Tris(OAc)2 was made by titrating the Trizma base with glacial acetic acid purchased from EMD Millipore (Billerica, MA). Zn(OAc)2 was purchased from Hampton Research Corp (Aliso Viejo, CA). nickel-nitrilotriacetic acid resin was purchased from Molecular Cloning Laboratories (South San Francisco, CA), and 5-ml Heparin HiTrap columns were from GE Healthcare. Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) was purchased from Gold Biotechnology, Inc. (St. Louis, MO). DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA).
Protein Purification
His-tagged E. coli and M. smegmatis RecO and RecR proteins were cloned and purified as described previously (29). Mycobacterial RecO was chelated with 1 mm EDTA, reduced with 1 mm DTT, and dialyzed extensively against storage buffer (40% (v/v) glycerol, 0.5 m NaCl, 20 mm HEPES pH 7.5, 1 mm TCEP). E. coli and M. smegmatis SSB proteins were expressed in E. coli BL21(DE3)pLys without fusion to affinity tags and purified by sequential polyethyleneimine and ammonium sulfate precipitation and heparin chromatography as described previously (29). The E. coli SSB-ΔC8 expression plasmid was a gift from Dr. M. Cox (University of Wisconsin, WI) and was purified by similar protocol. E. coli RecA expression plasmid was a gift from Michael Cox (University of Wisconsin, WI). EcRecA was inserted into pMCSG7, expressed in BL21(DE3)pLys, and purified by ammonium sulfate precipitation, nickel-nitrilotriacetic acid chromatography with cleavage of the N-terminal His6 tag by tobacco etch virus, and heparin chromatography. M. smegmatis RecA was amplified from genomic DNA and was inserted into pEcoli-Cterm 6xHN (Clontech) with a stop codon preceding the His6 C-terminal tag, expressed in BL21(DE3)pLys, and purified by sequential polyethyleneimine and ammonium sulfate precipitation and anion exchange chromatography.
DNA Binding
Proteins were diluted into buffer A (50 mm NaCl, 25% (v/v) glycerol, 50 mm HEPES pH 7.5, 1 mm TCEP) or buffer B (100 mm NaCl, 25% (v/v) glycerol, 50 mm HEPES, pH 7.5, 1 mm TCEP). Binding of DNA was assayed by titrating 5 nm fluorescein 5′-labeled dT15 oligonucleotide by RecO proteins in buffers A and B in the absence or presence of 10 μm Zn(OAc)2 or 8 μm RecR. DNA binding was assayed using fluorescence anisotropy as previously described with the excitation/emission of 485/528 nm at room temperature using a BioTek Synergy 4 plate reader (55).
Strand Exchange and ATP Hydrolysis
The entire assay and series of incubations were conducted at 37 °C. The 90-mer oligonucleotide (GCC TCT AGT CGA GGC ATC AAT ACG AAA CCT TAT TCT TTC CAG TTA CAA GCA CTT AAG GTC TTG TTC GCA GAT GGC TTA GAG CTT ATT TGC) at 25 nm (molecule concentration) was incubated with 1 μm SSB for 15 min. 1 μm RecO and 2 or 8 μm RecR were incubated for 15 min. SSB-ssDNA and RecO(R) were mixed and incubated for 15 min in Corning© 96-well Non-binding (NBSTM) clear bottom plates (product #3651). RecA (1 μm) and ATP (0.2 mm) were added to the SSB-ssDNA-RecOR mixture and incubated for 2 min. The target substrate dsDNA of 35 nucleotides long with Cy3–5′ and Cy5–3′ (/5Cy3/GCA AAT AAG CTC TAA GCC ATC TGC GAA CAA GAC CT and AGG TCT TGT TCG CAG ATG GCT TAG AGC TTA TTT GC/3Cy5Sp/) was added at 35 or 50 nm. The final volume of reaction was 100 μl. The Cy3–5′ strand is complementary to the 3′-end of the 90-mer oligonucleotide. RecA loading onto DNA was assayed by measuring DNA-dependent ATP hydrolysis with a coupled ATP regeneration colorimetric assay consisting of ATP (0.2–0.25 mm), pyruvate kinase (2–2.25 units/ml), phosphoenolpyruvate (0.2–0.25 mm), lactate dehydrogenase (2–2.25 units/ml), and NADH (0.2–0.25 mm). Decrease of NADH absorbance at 340 nm was converted to ATP hydrolysis based on a previously published protocol (33, 52). Strand exchange was measured by FRET changes with excitation at 540/25 nm (with 25-nm bandpass) and emission at 590/20 nm and 635/35 nm during 30 min. FRET was calculated using the equation, FRET = A/(A + D), where A and D are emissions from acceptor at 635 nm and donor at 590 nm. Strand exchange was normalized to 100%, corresponding to maximum possible strand exchange product formation. FRET results were verified (not shown) by analyzing the strand exchange products on Native PAGE gels by incubating the samples in 5% glycerol (v/v), 1.2% SDS, 10 mm Tris·HCl, pH 8.0, 40 mm EDTA, 0.25 mg/ml Proteinase K at 42°C for 30 min and visualizing the products and substrates by excitation/emission at 532/580, 532/670, and 633/670 nm with a Typhoon imager (GE Healthcare). 90% of maximum FRET changes were observed during the first 15 min of reaction, and this time point was selected for bar graph plots to compare activities of RMPs at different conditions. The assay was performed in buffer C (5% (v/v) glycerol, 60 mm NaCl, 25 mm Tris(OAc)2 pH 7.0, 5 mm Mg(OAc)2, 1 mm TCEP, 3 mm potassium gluconate (KGlu)) or buffer D (5% (v/v) glycerol, 60 mm NaCl, 25 mm Tris(OAc)2, 10 mm Mg(OAc)2, 1 mm DTT, 3 mm KGlu, pH 7.0, in the case of M. smegmatis or 7.5 in case of E. coli).
DNA Annealing
MsRecO annealing activity was assayed as described previously (55). 35-mer 5′-Cy3-labeled (/5Cy3/GCA AAT AAG CTC TAA GCC ATC TGC GAA CAA GAC CT) and complementary 3′-Cy5 labeled (AGG TCT TGT TCG CAG ATG GCT TAG AGC TTA TTT GC/3Cy5Sp/) oligonucleotides at 35 nm were incubated separately with MsSSB for 15 min at room temperature in buffer C in the presence or absence of 1 μm Zn(OAc)2. The MsSSB-ssDNA complexes were mixed, and 1 μm MsRecO was added. The FRET was measured by excitation at 540/25 nm and emission at 590/20 and 635/35 nm for 25 min at room temperature. Annealing was normalized to 100%, corresponding to a FRET of 0.6 for dsDNA formation. Background interaction between labeled oligonucleotides and proteins was subtracted as previously described by measuring Cy3 and Cy5 emissions of single-labeled oligonucleotides in the presence of proteins (57).
Protein Pulldown with ssDNA Immobilized on Avidin Beads
5′-Biotin-labeled 65-nucleotide-long ssDNA (/5Biosg/GCA AAT AAG CTC TAA GCC ATC TGC GAA CAA GAC CTT AAG TGC TTG TAA CTG GAA AGA ATA AGG TT) was conjugated on avidin-bound agarose resin using a previous protocol (55). Avidin-bound ssDNA (2.5 μm) was saturated with SSB (10 μm) in buffer C, incubated at 4 °C for 30 min, and spun down, and the unbound SSB was washed 3 times by the buffer. RecO (10 μm) with Zn(OAc)2 (10 μm) or with MsRecR (40 μm) in the presence or absence of MsRecA (5 μm) and ATPγS (1 mm) was added to the SSB-bound ssDNA and incubated for 30 min. Samples of the loads, supernatants, washes, and beads were incubated with denaturing buffer (10% (v/v) glycerol, 4% SDS, 10 mm Tris-HCl, 7.5, 20 mm β-mercaptoethanol) at 90 °C for 10 min and analyzed on a 12% SDS-PAGE gel. Gel photographs were processed with Adobe Photoshop CS 5.1 by normalizing the brightness across the entire gel due to irregular illumination of gel that results in dark outer wells and light inner wells.
RESULTS
MsRecO Binding to DNA Is Stimulated Independently by Zinc and RecR
Previously we demonstrated that binding of MsRecO to DNA is stimulated by the presence of a micromolar amount of zinc in solution (29). The interaction of MsRecO with ssDNA in the presence of zinc was sufficient to anneal ssDNA bound to Ms- or EcSSB under physiological conditions despite a lack of interaction between MsRecO and C-terminal tail of MsSSB. To test the recombination function of MsRecO, we first assessed the interaction of MsRecO with ssDNA in the presence of MsRecR as a required partner of RecO in RecA loading reaction. Unexpectedly, we found that MsRecR stimulated DNA binding of zinc-depleted MsRecO similarly to that of zinc (Fig. 1A). The apparent dissociation constants at 50 mm NaCl were Kd(MsRecO) = 2.08 ± 0.11 μm and Kd(MsRecO,Zn) = 0.041 ± 0.03 μm in the absence or presence of 10 μm Zn(OAc)2, correspondingly. In the presence of MsRecR, Kd(MsRecOR) = 0.028 ± 0.006 μm without zinc. At 100 mm NaCl, stimulation of DNA binding of MsRecO by MsRecR was stronger than in the case of E. coli proteins (Fig. 1B; Kd(MsRecOR) = 0.44 ± 0.06; Kd(EcRecOR) = 2.76 ± 0.27). Therefore, the interaction of MsRecO with ssDNA is regulated by two alternative mechanisms. In one case the zinc finger domain is critical for DNA binding and SSA by MsRecO alone. In the second case, the coordination of the ZF domain structure by metal is dispensable for ssDNA binding by MsRecO bound to MsRecR. To understand the role of the alternative DNA interaction mechanisms in the recombination function of MsRecOR, we cloned and purified MsSSB and MsRecA proteins and developed an assay to monitor the DNA binding and strand exchange activities of RecA.
FIGURE 1.
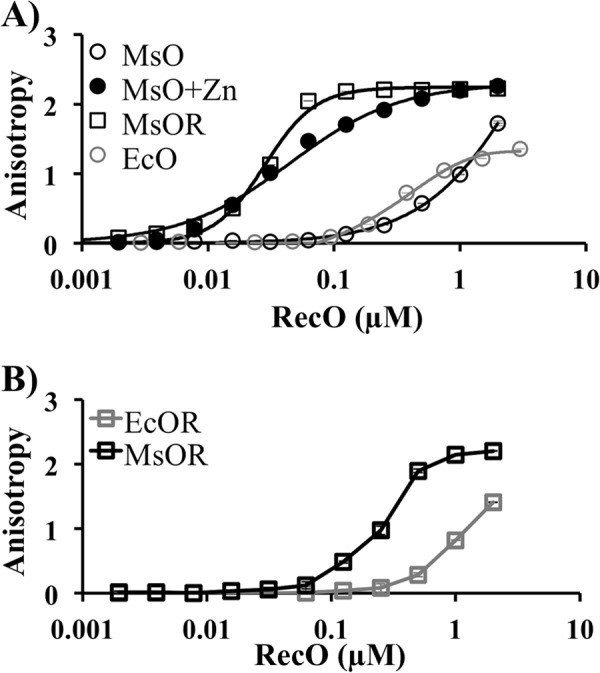
DNA binding activity of MsRecO is stimulated independently by zinc and RecR. A, isotherm showing DNA binding measured by change in fluorescence anisotropy of FAM-dT15 (5 nm) upon titration by EcRecO (EcO; gray) and MsRecO (MsO; black) in buffer A at 50 mm NaCl in the presence (square) or absence (circle) of 8 μm MsRecR without (open) and with 10 μm Zn(OAc)2 (solid). B, similar measurements of DNA binding by EcRecO (gray) and MsRecO (black) in the presence of cognate RecR (5 μm) in buffer B with 100 mm NaCl.
MsRecOR Stimulates DNA Binding and Strand Exchange Activities of MsRecA
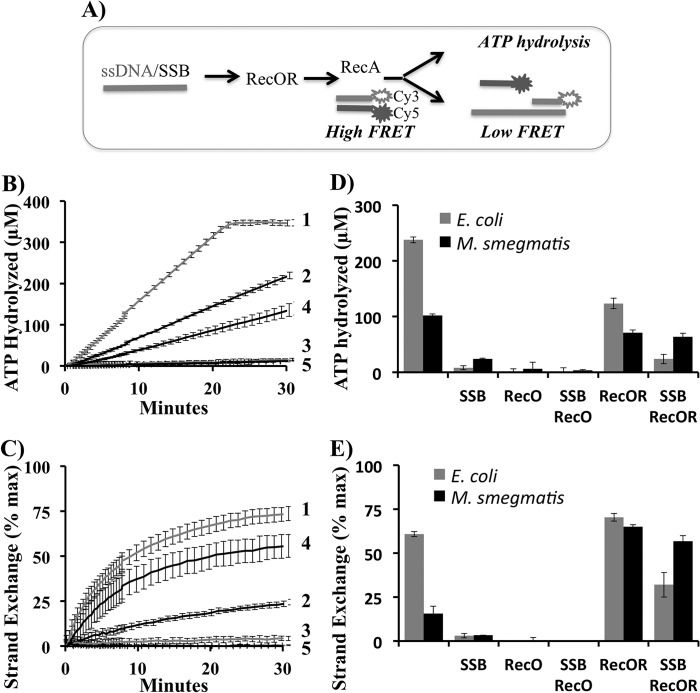
Binding of the purified MsRecA to 90-mer ssDNA and the strand exchange activity with a donor 35-bp dsDNA substrate were simultaneously monitored as described in Fig. 2. ATP hydrolysis (Fig. 2, B and D) and strand exchange activities of MsRecA alone without MsSSB (Fig. 2, C and E) were significantly weaker than those of EcRecA (Fig. 2, B–E, Table 1), in agreement with previously published data (58). The strand-exchange activity of MsRecA was observed to be particularly weak with the longer M13 DNA substrates (data not shown). SSB inhibited the DNA binding of RecA from both organisms. In both systems RecO alone did not support RecA function. MsRecOR complex stimulated the DNA binding and strand exchange activities of MsRecA in the presence of MsSSB and the strand exchange in the absence of MsSSB (Fig. 2). EcRecOR stimulated EcRecA function only in the presence of EcSSB, in agreement with previous reports (27). Thus, in the Gram-positive bacterium M. smegmatis, with a more complex array of DNA repair pathways in comparison to E. coli, activity of MsRecA is more dependent on function of RMPs.
FIGURE 2.
MsRecOR stimulates DNA binding and strand exchange activities of MsRecA. A, design of the experiment. The 90-mer ssDNA was incubated with SSB followed by incubation with RecO or RecOR. RecA was added followed by the addition of donor dsDNA 35-mer composed of a 3′-Cy5- and a 5′-Cy3-labeled strands complementary to the 3′-end of the 90-mer ssDNA. Final concentrations are 90-mer ssDNA (25 nm), 35-bp dsDNA (35 nm), RecA (1 μm), SSB (1 μm), RecO (1 μm), and RecR (2 μm). DNA-dependent ATP hydrolysis by RecA was measured by a coupled ATP regeneration colorimetric assay with NADH. Decrease of NADH absorbance at 340 nm corresponds to ATP hydrolysis. Strand exchange was measured simultaneously by a decrease of Cy3/Cy5 FRET upon strand exchange with the complementary 3′-end of ssDNA 90-mer. The decreasing FRET was inverted and normalized to reflect the percentage of maximum strand exchange. B and C, real time measurements of ATP hydrolysis (B) and strand exchange (C) reactions are shown for EcRecA (1), MsRecA (2), EcRecA with EcSSB (3), MsRecA with MsSSB and MsRecOR (4), and DNA alone (5). D and E, bar graphs representing ATP hydrolysis (D) and strand-exchange efficiency (E) by E. coli (gray) and M. smegmatis (black) RecA after a 15-min reaction time in the presence of 90-mer ssDNA and 35-bp dsDNA with cognate SSB, RecO, and RecR in buffer C and ATP regeneration system.
TABLE 1.
ATP hydrolysis rate in kcat (min−1) of E. coli and M. smegmatis RecA
DNA-dependent ATP hydrolysis rates of RecA proteins in the presence of SSB and RecOR as calculated at 15 min of the reaction time in experiment described in Fig. 2.
| ATPase rate (kcat) |
||
|---|---|---|
| E. coli | M. smegmatis | |
| min−1 | ||
| RecA | 15.9 ± 0.4 | 6.8 ± 0.2 |
| RecA, SSB | 1.1 ± 0.2 | 1.6 ± 0.1 |
| RecA, RecO | 0 | 0 |
| RecA, RecOR | 8.2 ± 0.6 | 2.9 ± 0.3 |
| RecA, SSB, RecO | 0 | 0 |
| RecA, SSB, RecOR | 1.6 ± 0.6 | 4.2 ± 0.4 |
Stimulation of MsRecA Binding to DNA by MsRecOR Does Not Depend on MsRecO Interaction with Zinc
MsRecO protein was extensively dialyzed against buffer with chelating and reducing agents and assayed for strand annealing and for recombination mediator functions. Under these conditions, MsRecOR supported the interaction of MsRecA with ssDNA and strand exchange, whereas MsRecO did not stimulate annealing (Fig. 3). The addition of zinc to the reaction buffer restored the strand annealing activity of MsRecO but not of MsRecOR. It partially inhibited MsRecA activity, probably due to zinc interference with RecA function. Nevertheless, MsRecOR did stimulate strand exchange reaction.
FIGURE 3.
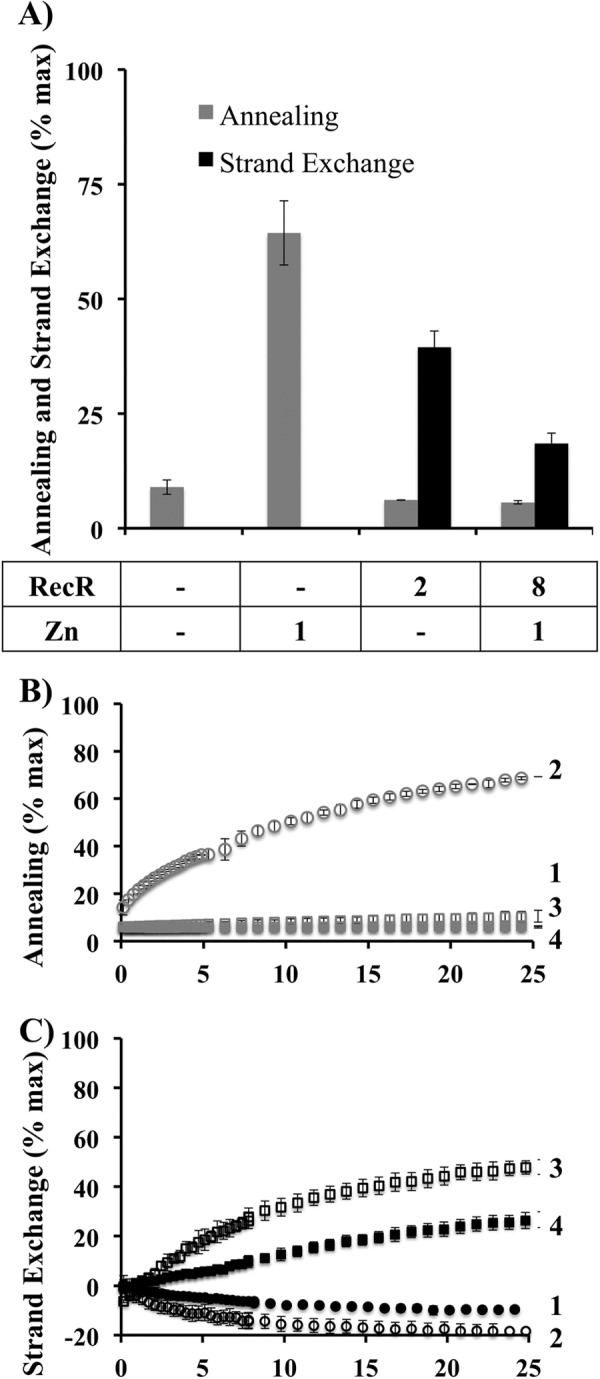
MsRecR supports strand exchange by MsRecO depleted of zinc, whereas zinc addition restores annealing activities of MsRecO. A, bar graphs showing the efficiency of annealing (gray) and strand exchange (black) by extensively dialyzing with chelating and reducing agents MsRecO (1 μm) in the presence of MsSSB (1 μm) and with or without Zn(OAc)2 and/or MsRecR (values in μm are shown under the graph) in buffer C. Annealing was performed with complementary Cy3- and Cy5-labeled 35-mer (35 nm) oligonucleotides preincubated separately with MsSSB without MsRecA. Strand exchange was assayed as described in Fig. 2 with MsRecA (1 μm). B and C, corresponding kinetic data are shown for annealing (B) and strand exchange (C) with MsRecO alone (1), in the presence of Zn(OAc)2 (2), or MsRecR (3), or Zn(OAc)2 and MsRecR (4).
MsRecR Inhibits the Annealing Activity of MsRecO
EcRecR was shown to inhibit annealing activity of EcRecO (38). Likewise, we observed that MsRecR inhibited SSA by MsRecO even in the presence of zinc under conditions favorable for annealing (Fig. 3). Ec- and MsRecOR complexes bind both ss- and dsDNA with comparative affinity to that of RecO alone (55), suggesting that the inhibitory effect is not due to incompatibility of the complex with the end product of annealing reaction. We hypothesize that the conformation of ssDNA bound to RecOR is incompatible with the strand annealing, and, consequently, that RecO and RecOR support two alternative conformations of ssDNA.
MsRecOR Function Independently of Interaction with SSB-Ct
The annealing activity of EcRecO depends on the interaction with SSB-Ct only at elevated salt concentrations. However, the presence of SSB-Ct was essential for EcRecOR function in EcRecA loading experiments even at low salt concentrations (59). We tested whether the mediator function of MsRecOR is dependent on the interaction with SSB-Ct. MsRecOR function was assayed with EcSSB as the stimulatory effect of MsRecOR on the activity of EcRecA in the presence of EcSSB was similar to that of MsRecA with MsSSB. Comparison of reactions in the presence of EcSSB and the mutant lacking last eight amino acids (EcSSB-ΔC8) demonstrated that MsRecOR supported EcRecA binding to ssDNA and strand exchange under all conditions, whereas EcRecOR activity was inhibited by EcSSB-ΔC8 (Fig. 4).
FIGURE 4.
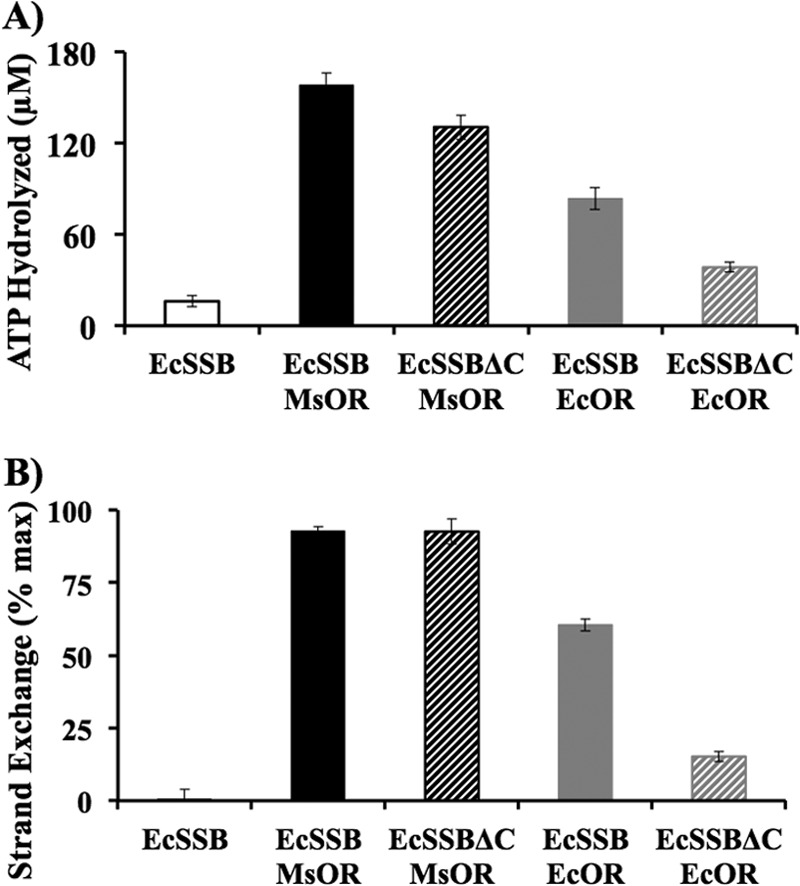
Interaction with SSB-Ct is not required for MsRecO-dependent binding of RecA to ssDNA and strand exchange. A and B, bar graphs corresponding to ATP hydrolysis rate (A) and strand-exchange efficiency (B) by EcRecA (1 μm) after 15 min of incubation time with EcSSB or EcSSB-ΔC (1 μm) and MsRecOR or EcRecOR (1 μm RecO, 8 μm RecR) in buffer D.
MsRecO and MsRecOR Do Not Displace MsSSB from ssDNA
Interaction of EcRecO with EcSSB led to the hypothesis that RecO does not completely dismiss SSB from ssDNA upon binding (22). Accordingly, we demonstrated that EcSSB retains on dT70 immobilized on avidin beads after incubation with excess of EcRecO or EcRecOR (54). Interestingly, similar results were obtained at low salt concentration with SSB-ΔC8 construct, where the interaction with SSB-Ct is dispensable for annealing. Because MsRecO does not bind SSB-Ct, we addressed the same question using mycobacterial proteins. We assayed MsSSB displacement using a similar pulldown assay with 65-mer ssDNA immobilized on beads. MsSSB was incubated with beads, and the unbound protein was removed. Next, the beads were incubated with the excess of MsRecO or MsRecOR in the absence of unbound MsSSB. Similar to the E. coli system, no significant dissociation of MsSSB was observed upon incubation with MsRecO, and comparable amounts of both MsRecO and MsSSB were retained on beads (Fig. 5A). In the case of MsRecOR, a partial dissociation of MsSSB from DNA was observed. However, the amount of MsSSB retained on the beads was also comparable with that of MsRecOR (Fig. 5B). Moreover, the addition of MsRecA resulted in the retention of all four proteins on ssDNA. The results together with previous analysis of E. coli proteins suggest that RecO(R) can form a complex with ssDNA and SSB regardless of interaction with SSB-Ct.
FIGURE 5.
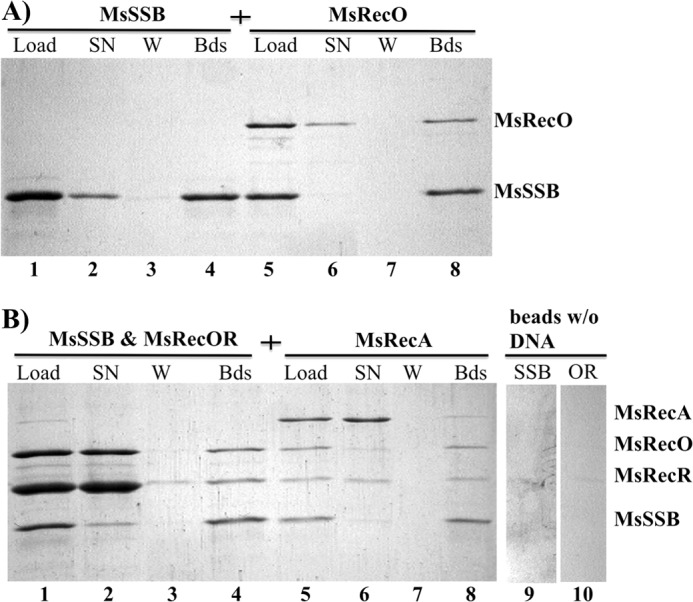
MsRecO(R) and MsRecA share ssDNA with MsSSB. A, SDS-PAGE analysis showing proteins pulled down on avidin-immobilized ssDNA. Lane 1, initial mixture of MsSSB (10 μm) with avidin-bound 65-mer ssDNA (load); lane 2, unbound MsSSB in supernatant (SN) after incubation with ssDNA beads; lane 3, proteins in solution after the third round of beads washing (W); lane 4, MsSSB retained on beads after the third wash (Bds); lane 5, ssDNA beads prebound to MsSSB mixed with MsRecO (10 μm); lane 6, unbound protein in initial mixture; lane 7, unbound protein after the third wash; lane 8, proteins remaining on beads after three washes. B, similarly prepared ssDNA immobilized on beads and bound to MsSSB were incubated with MsRecO (10 μm) and MsRecR (40 μm) (lanes 1–4) and, consequently, with MsRecA (5 μm) and ATPγS (1 mm) (lanes 5–8) in buffer C. Lanes 9 and 10 contain negative controls for beads without DNA incubated with MsSSB (SSB) or MsRecOR (OR), respectively, after extensive wash.
MsSSB was previously reported to interact with MsRecA and stimulate its function (58). In our experiments, MsSSB inhibited ATPase and strand exchange activities of MsRecA. However, in the presence of MsRecOR the titration of the reaction by MsSSB revealed a partial stimulatory effect of MsSSB (Fig. 6, A and B). Such an increase was not observed with E. coli proteins, in line with the previously reported results (Fig. 6, C and D) (59). These data further support a functional interaction between RecOR and SSB on ssDNA.
FIGURE 6.
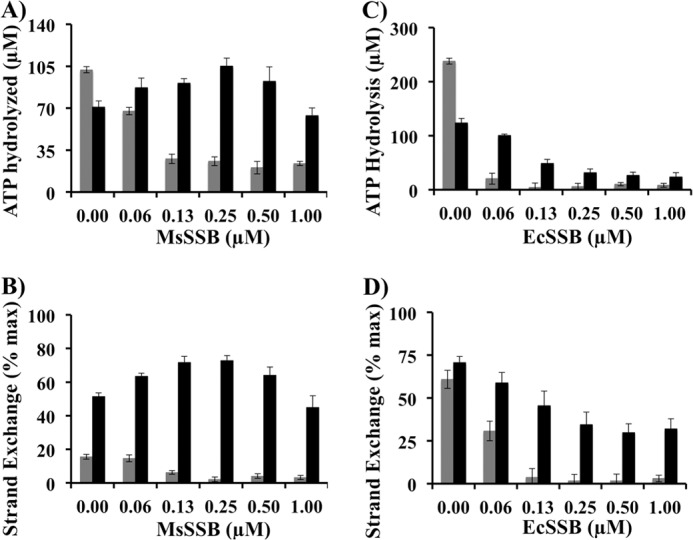
MsSSB partially promotes MsRecA activity in the presence of MsRecOR. A and B, shown are bar graphs representing values of ATP hydrolysis (A) and strand-exchange efficiency (B) by MsRecA (1 μm) at 15 min with ssDNA preincubated with different amounts of MsSSB in buffer C in the absence (gray) or presence (black) of MsRecO (1 μm) and MsRecR (2 μm). C and D, similar bar graphs showing ATP hydrolysis (C) and strand-exchange efficiency (D) by EcRecA (1 μm) upon titration with EcSSB in buffer C in the absence (gray) and presence (black) of EcRecO (1 μm) and EcRecR (2 μm).
DISCUSSION
RecO and RecOR Support Two Distinct Conformations of ssDNA during SSA and HR
The data presented in this work demonstrate that different structural elements of MsRecO are involved in ssDNA binding in two alternative DNA repair pathways. During annealing, ssDNA binding depends on the presence of zinc in solution likely due to structural requirements of the ZF domain. The depletion of zinc by chelating agents results in an annealing-inactive protein with significantly lower affinity to ssDNA. However, DNA binding is restored in the presence of MsRecR, which by itself has a very weak affinity to DNA in the millimolar range of concentration.
RecR has a highly conserved amino acid sequence (60, 61). Despite its essential role in RecA binding, RecR does not form stable complexes with RecA. A weak DNA-dependent interaction between RecOR and RecA is still a possibility. Alternatively, we hypothesize that the specific conformation of ssDNA bound to RecOR is favorable for interaction with RecA. Interestingly, the excess of RecOR in solution does not inhibit RecA function. An inhibitory property of RecR in DNA annealing further supports the importance of specific ssDNA conformation. For example, the RecOR complex interacts with comparable affinities with both ds- and ssDNA. Thus, the inhibition can be explained by a conformation of ssDNA incompatible with annealing. Therefore, we suggest that RecO can support two alternative conformations of ssDNA; one favorable for annealing and another, in the presence of RecR, favorable for RecA binding to ssDNA. Likewise, DNA conformational changes supported by gp32/UvsY proteins were suggested to play a major functional role in homologous recombination (62).
Mycobacterial RMPs Co-localizes with SSB on ssDNA
At physiological conditions, interaction with SSB-Ct is critical for function of EcRecO and EcRecOR, which do not displace EcSSB from ssDNA during the initial steps of SSA and HR. MsRecO does not bind MsSSB-Ct. However, we observed a co-localization of MsRecO and MsRecOR with MsSSB on ssDNA. A partial displacement of MsSSB cannot be ruled out by this experiment. Nevertheless, the amount of the protein retained on beads-immobilized ssDNA is comparable with that of MsRecO or MsRecOR. Moreover, MsSSB slightly facilitates MsRecOR-mediated strand-exchange activity of MsRecA in a limited concentration range. Likewise, previously we demonstrated that at low salt concentrations EcRecO co-localizes with EcSSB-ΔC8 mutant on ssDNA beads. These data support a conserved ssDNA “sharing” mechanism by RMPs and SSB. Thermus thermophilus proteins also do not dismiss SSB from ssDNA (55, 63). T4 phage UvsY (64, 65) and eukaryotic Rad52 (57, 66) and PALB2 (67) do not dismiss gp32 and RPA, respectively.
The mechanism of DNA sharing by SSB and RMPs remains to be investigated. 65-nucleotide-long ssDNA is wrapped around SSB tetramer in a so-called (SSB)65 mode (68–71) with some nucleotides forming extensive contacts with the protein, whereas others are more solvent-exposed. RecO and RecOR can interact initially with such less tightly bound regions. Alternatively, RecO(R) can alter the DNA binding mode of SSB from (SSB)65 to (SSB)35, where half of ssDNA wrapped around SSB tetramer can become available for RecO(R) binding (72). In either case, RecO and RecOR alter the conformation of ssDNA bound to SSB to promote annealing or RecA recruitment.
The Role of SSB-RMP-ssDNA Complexes in DNA Repair
RecA binding to ssDNA occurs at an early stage of DNA repair as RecA-ssDNA complexes initiate such events as the SOS response in bacteria. Thus, RMPs, which stimulate RecA binding to ssDNA, can function as initial sensors of DNA damage or stalled replication. The existence of SSB-RMP-ssDNA complexes observed in our studies, in studies of UvsY/gp32 (73), and of Rad52-RPA (47, 74) can explain how RMP initiates recombinase loading in response to DNA damage. We speculate that SSA and HR reactions proceed through several reversible steps of RecO(R) interaction with SSB-ssDNA as depicted in Fig. 7. Such a mechanism will have two important features. First, a sequential formation of reversible complexes can lead to relatively slow kinetics, e.g. compared to that of replication. Second, the completion of each reaction depends on the availability of an appropriate substrate, e.g. complementary ssDNA in case of SSA. Otherwise, the complexes can dissociate to free SSB for interaction with other proteins. These features can explain how such RMP-SSB complexes will load RecA on ssDNA upon replication stalling but will not interfere during replication (34, 75–77). Accordingly, the lethality of a UvrD/Rep E. coli mutant due to slow replication and lack of anti-recombinase activity is bypassed by deletion of either of the RecFOR proteins (78, 79).
FIGURE 7.
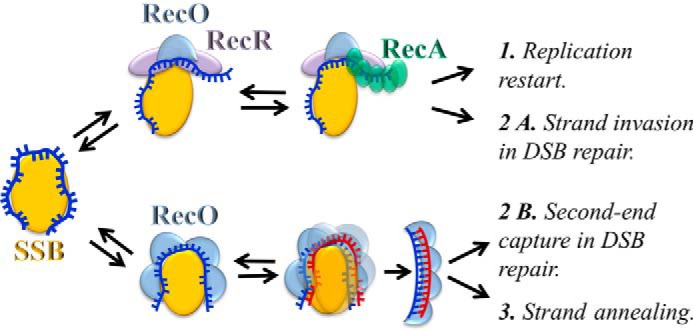
Hypothetical model of strand exchange and annealing reactions mediated by RecO(R). 1, RecOR binds SSB-ssDNA and promotes conformational changes of ssDNA favorable for recruitment of RecA to ssDNA at the site of stalled replication or (2A) to the recessed ssDNA end at dsDNA break (DSB). Alternately, (2B) RecO forms intermediate complexes with SSB-ssDNA and facilitates SSA when complementary ssDNA is available during a second-end capture reaction or (3) during annealing of repetitive ends. Reversibility of each intermediate step eliminates the inhibitory effect of RMPs on other functions of SSB-ssDNA complexes and permits selective activation of each pathway only under kinetically favorable conditions.
In the case of dsDNA break repair, the proposed model will explain why RMPs initially support only presynaptic complex formation on a resected DNA end but not SSA due to the lack of the complementary ssDNA substrate. In the post-strand invasion step, upon generation of complementary ssDNA, RMPs can support the second-end capture reaction as was previously hypothesized (66, 80). The existence of reversible Rad51-DNA complexes and Rad52-RPA complexes was previously proposed to control recombination initiation in eukaryotes (81).
Regulation of RecO Function in Mycobacteria
MsRecO retains weak SSA activity for a limited time immediately after purification under reducing conditions without zinc in solvent (data not shown). Estimation of zinc binding affinity by MsRecO revealed an apparent dissociation constant of 0.1 μm (29). Such moderate to low affinity, as compared with other zinc finger domains (82), can serve as a regulatory mechanism of SSA activity of MsRecO in mycobacteria. Zinc is an important cofactor in all organisms, and mycobacteria possess several mechanisms that regulate the intracellular zinc concentration (83, 84). An overall concentration of zinc in bacteria is estimated at 0.2 mm, although the amount of unbound zinc is in the femtomolar concentration range (85). Therefore, zinc binding by MsRecO depends on competition for zinc with other zinc binding molecules in the cell. The ZF domain of MsRecO can also be sensitive to oxidation as it contains four cysteines. Therefore, the annealing function of mycobacterial RecO can be inhibited upon zinc depletion or oxidative conditions, whereas a recombination mediator function should be supported under such conditions.
The structural role of the ZF domain in RecO remains to be investigated. Despite low homology, structures of Dr- and EcRecO are similar including the folding of ZF domain (54, 56). RecO is composed of an N-terminal OB-fold (oligonucleotide/oligosaccharide binding-fold) domain, a central α-helical domain and a ZF domain (56, 86). The N-terminal domain likely serves as a primary ssDNA-binding site and also interacts with RecR (87, 88). ZF domain is located at the opposite side of the globular structure to the N-terminal domain and was suggested to serve as a secondary DNA-binding site based on limited mutagenesis studies of D. radiodurans RecO (86). Alternatively, it can be involved in protein-protein interactions, e.g. in the case when annealing function requires formation of an oligomeric RecO structure on ssDNA similar to that of Rad52 oligomer (57, 89). Neither of the studied RecO proteins form oligomeric structures in solution. At the same time, interaction of EcRecO with dT70 is several orders of magnitude stronger than with dT16 (see the supplemental data in Ref. 90), suggesting formation of specific complexes or cooperativity during DNA binding. MsRecO oligomerization in the presence of zinc with or without ssDNA is currently under investigation. These studies are complicated by the low solubility of MsRecO, particularly of cysteine mutants, and formation of high molecular weight aggregates in the presence of zinc.
Specific structural requirements of MsRecO during annealing but not recombination can be efficiently utilized for function separation and comprehensive analysis of the interplay between two functions in vitro and in vivo in mycobacteria in the future. Such studies will be important for understanding the defense mechanism of Mycobacterium tuberculosis due to the key role of RecO in multiple DNA repair pathways in mycobacteria.
Acknowledgments
We are thankful to O. Koroleva for cloning and support of biochemical experiments and to K. Malley for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants AI064693 (to M. G.) and GM073837 (to S. K.). This work was also supported by President's Research Fund, Saint Louis University (to S. K.).
- HR
- homologous recombination
- RPA
- replication protein A
- ssDNA
- single-stranded DNA
- SSB
- ssDNA-binding protein
- RMP
- recombination mediator protein
- SSA
- ssDNA annealing
- Ms-
- M. smegmatis
- -Ct
- C-terminal tail
- Dr-
- D. radiodurans
- ZF
- zinc finger motif
- TCEP
- Tris (2-carboxyethyl) phosphine hydrochloride
- Ec-
- E. coli
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Cox M. M. (1991) The RecA protein as a recombinational repair system. Mol. Microbiol. 5, 1295–1299 [DOI] [PubMed] [Google Scholar]
- 2. Kuzminov A. (2001) DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8461–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li X., Heyer W. D. (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18, 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thompson L. H., Schild D. (1999) The contribution of homologous recombination in preserving genome integrity in mammalian cells. Biochimie 81, 87–105 [DOI] [PubMed] [Google Scholar]
- 5. Venkitaraman A. R. (2014) Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 343, 1470–1475 [DOI] [PubMed] [Google Scholar]
- 6. Cox M. M. (2007) Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 42, 41–63 [DOI] [PubMed] [Google Scholar]
- 7. San Filippo J., Sung P., Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 8. Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58, 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beernink H. T., Morrical S. W. (1999) RMPs: recombination/replication mediator proteins. Trends Biochem. Sci. 24, 385–389 [DOI] [PubMed] [Google Scholar]
- 10. Umezu K., Kolodner R. D. (1994) Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 269, 30005–30013 [PubMed] [Google Scholar]
- 11. Yang H., Li Q., Fan J., Holloman W. K., Pavletich N. P. (2005) The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433, 653–657 [DOI] [PubMed] [Google Scholar]
- 12. Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 [DOI] [PubMed] [Google Scholar]
- 13. New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 [DOI] [PubMed] [Google Scholar]
- 14. Buisson R., Dion-Côté A. M., Coulombe Y., Launay H., Cai H., Stasiak A. Z., Stasiak A., Xia B., Masson J. Y. (2010) Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17, 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stratton M. R., Wooster R. (1996) Hereditary predisposition to breast cancer. Curr. Opin. Genet. Dev. 6, 93–97 [DOI] [PubMed] [Google Scholar]
- 16. Couch F. J., Nathanson K. L., Offit K. (2014) Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science 343, 1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Southey M. C., Teo Z. L., Winship I. (2013) PALB2 and breast cancer: ready for clinical translation! Appl. Clin. Genet. 6, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sechman E. V., Rohrer M. S., Seifert H. S. (2005) A genetic screen identifies genes and sites involved in pilin antigenic variation in Neisseria gonorrhoeae. Mol. Microbiol. 57, 468–483 [DOI] [PubMed] [Google Scholar]
- 19. Wang G., Lo L. F., Maier R. J. (2011) The RecRO pathway of DNA recombinational repair in Helicobacter pylori and its role in bacterial survival in the host. DNA Repair 10, 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robinson K., Loughlin M. F., Potter R., Jenks P. J. (2005) Host adaptation and immune modulation are mediated by homologous recombination in Helicobacter pylori. J. Infect. Dis. 191, 579–587 [DOI] [PubMed] [Google Scholar]
- 21. Mahdi A. A., Lloyd R. G. (1989) Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol. Gen. Genet. 216, 503–510 [DOI] [PubMed] [Google Scholar]
- 22. Umezu K., Chi N. W., Kolodner R. D. (1993) Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl. Acad. Sci. U.S.A. 90, 3875–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lovett S. T., Clark A. J. (1983) Genetic analysis of regulation of the RecF pathway of recombination in Escherichia coli K-12. J. Bacteriol. 153, 1471–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alonso J. C., Stiege A. C. (1991) Molecular analysis of the Bacillus subtilis recF function. Mol. Gen. Genet. 228, 393–400 [DOI] [PubMed] [Google Scholar]
- 25. Asai T., Kogoma T. (1994) The RecF pathway of homologous recombination can mediate the initiation of DNA damage-inducible replication of the Escherichia coli chromosome. J. Bacteriol. 176, 7113–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Courcelle J., Carswell-Crumpton C., Hanawalt P. C. (1997) recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 94, 3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sakai A., Cox M. M. (2009) RecFOR and RecOR as distinct RecA loading pathways. J. Biol. Chem. 284, 3264–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Satoh K., Kikuchi M., Ishaque A. M., Ohba H., Yamada M., Tejima K., Onodera T., Narumi I. (2012) The role of Deinococcus radiodurans RecFOR proteins in homologous recombination. DNA Repair 11, 410–418 [DOI] [PubMed] [Google Scholar]
- 29. Gupta R., Ryzhikov M., Koroleva O., Unciuleac M., Shuman S., Korolev S., Glickman M. S. (2013) A dual role for mycobacterial RecO in RecA-dependent homologous recombination and RecA-independent single-strand annealing. Nucleic Acids Res. 41, 2284–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Courcelle J. (2005) Recs preventing wrecks. Mutat. Res. 577, 217–227 [DOI] [PubMed] [Google Scholar]
- 31. Sandler S. J., Clark A. J. (1994) RecOR suppression of recF mutant phenotypes in Escherichia coli K-12. J. Bacteriol. 176, 3661–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Korolev S. (2011) ATP Binding Cassette Properties of Recombination Mediator Protein RecF. In DNA Repair (Kruman I. ed.) InTech pp. 1–23, www.intechopen.com/books/dna-repair/atp-binding-cassette-properties-of-recombination-mediator-protein-recf
- 33. Morimatsu K., Kowalczykowski S. C. (2003) RecFOR Proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 34. Morimatsu K., Wu Y., Kowalczykowski S. C. (2012) RecFOR proteins target RecA protein to a DNA gap with either DNA or RNA at the 5′ terminus: implication for repair of stalled replication forks. J. Biol. Chem. 287, 35621–35630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sugiyama T., New J. H., Kowalczykowski S. C. (1998) DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 95, 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mazloum N., Zhou Q., Holloman W. K. (2007) DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry 46, 7163–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mortensen U. H., Bendixen C., Sunjevaric I., Rothstein R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. U.S.A. 93, 10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kantake N., Madiraju M. V., Sugiyama T., Kowalczykowski S. C. (2002) Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: a common step in genetic recombination. Proc. Natl. Acad. Sci. U.S.A. 99, 15327–15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mazloum N., Holloman W. K. (2009) Second-end capture in DNA double-strand break repair promoted by Brh2 protein of Ustilago maydis. Mol. Cell 33, 160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McIlwraith M. J., West S. C. (2008) DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol. Cell 29, 510–516 [DOI] [PubMed] [Google Scholar]
- 41. Nimonkar A. V., Sica R. A., Kowalczykowski S. C. (2009) Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc. Natl. Acad. Sci. U.S.A. 106, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi I., Hallwyl S. C., Seong C., Mortensen U., Rothstein R., Sung P. (2009) Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J. Biol. Chem. 284, 33275–33284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lao J. P., Oh S. D., Shinohara M., Shinohara A., Hunter N. (2008) Rad52 promotes postinvasion steps of meiotic double-strand break repair. Mol. Cell 29, 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manfredi C., Suzuki Y., Yadav T., Takeyasu K., Alonso J. C. (2010) RecO-mediated DNA homology search and annealing is facilitated by SsbA. Nucleic Acids Res. 38, 6920–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gupta R., Barkan D., Redelman-Sidi G., Shuman S., Glickman M. S. (2011) Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 79, 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Park M. S., Ludwig D. L., Stigger E., Lee S. H. (1996) Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 271, 18996–19000 [DOI] [PubMed] [Google Scholar]
- 47. Seong C., Sehorn M. G., Plate I., Shi I., Song B., Chi P., Mortensen U., Sung P., Krejci L. (2008) Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J. Biol. Chem. 283, 12166–12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu J., Ehmsen K. T., Heyer W. D., Morrical S. W. (2011) Presynaptic filament dynamics in homologous recombination and DNA repair. Crit. Rev. Biochem. Mol. Biol. 46, 240–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Holloman W. K., Schirawski J., Holliday R. (2008) The homologous recombination system of Ustilago maydis. Fungal Genet. Biol. 45, S31–S39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang H., Jeffrey P. D., Miller J., Kinnucan E., Sun Y., Thoma N. H., Zheng N., Chen P. L., Lee W. H., Pavletich N. P. (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297, 1837–1848 [DOI] [PubMed] [Google Scholar]
- 51. Carreira A., Hilario J., Amitani I., Baskin R. J., Shivji M. K., Venkitaraman A. R., Kowalczykowski S. C. (2009) The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136, 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bork J. M., Cox M. M., Inman R. B. (2001) The RecOR proteins modulate RecA protein function at 5′ ends of single- stranded DNA. EMBO J. 20, 7313–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Webb B. L., Cox M. M., Inman R. B. (1997) Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 91, 347–356 [DOI] [PubMed] [Google Scholar]
- 54. Ryzhikov M., Korolev S. (2012) Structural studies of SSB interaction with RecO. Methods Mol. Biol. 922, 123–131 [DOI] [PubMed] [Google Scholar]
- 55. Ryzhikov M., Koroleva O., Postnov D., Tran A., Korolev S. (2011) Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 39, 6305–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Makharashvili N., Koroleva O., Bera S., Grandgenett D. P., Korolev S. (2004) A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure 12, 1881–1889 [DOI] [PubMed] [Google Scholar]
- 57. Grimme J. M., Honda M., Wright R., Okuno Y., Rothenberg E., Mazin A. V., Ha T., Spies M. (2010) Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 38, 2917–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ganesh N., Muniyappa K. (2003) Characterization of DNA strand transfer promoted by Mycobacterium smegmatis RecA reveals functional diversity with Mycobacterium tuberculosis RecA. Biochemistry 42, 7216–7225 [DOI] [PubMed] [Google Scholar]
- 59. Hobbs M. D., Sakai A., Cox M. M. (2007) SSB protein limits RecOR binding onto single-stranded DNA. J. Biol. Chem. 282, 11058–11067 [DOI] [PubMed] [Google Scholar]
- 60. Aravind L., Leipe D. D., Koonin E. V. (1998) Toprim: a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26, 4205–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rocha E. P., Cornet E., Michel B. (2005) Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 1, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu J., Qian N., Morrical S. W. (2006) Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions. J. Biol. Chem. 281, 26308–26319 [DOI] [PubMed] [Google Scholar]
- 63. Inoue J., Honda M., Ikawa S., Shibata T., Mikawa T. (2008) The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins. Nucleic Acids Res. 36, 94–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sweezy M. A., Morrical S. W. (1997) Single-stranded DNA binding properties of the uvsY recombination protein of bacteriophage T4. J. Mol. Biol. 266, 927–938 [DOI] [PubMed] [Google Scholar]
- 65. Jiang H., Giedroc D., Kodadek T. (1993) The role of protein-protein interactions in the assembly of the presynaptic filament for T4 homologous recombination. J. Biol. Chem. 268, 7904–7911 [PubMed] [Google Scholar]
- 66. Sugiyama T., Kantake N. (2009) Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates. J. Mol. Biol. 390, 45–55 [DOI] [PubMed] [Google Scholar]
- 67. Genois M. M., Mukherjee A., Ubeda J. M., Buisson R., Paquet E., Roy G., Plourde M., Coulombe Y., Ouellette M., Masson J. Y. (2012) Interactions between BRCA2 and RAD51 for promoting homologous recombination in Leishmania infantum. Nucleic Acids Res. 40, 6570–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. George N. P., Ngo K. V., Chitteni-Pattu S., Norais C. A., Battista J. R., Cox M. M., Keck J. L. (2012) Structure and cellular dynamics of Deinococcus radiodurans SSB/DNA complexes. J. Biol. Chem. 287, 22123–221232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Raghunathan S., Kozlov A. G., Lohman T. M., Waksman G. (2000) Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 7, 648–652 [DOI] [PubMed] [Google Scholar]
- 70. Antony E., Weiland E. A., Korolev S., Lohman T. M. (2012) Plasmodium falciparum SSB tetramer wraps single-stranded DNA with similar topology but opposite polarity to E. coli SSB. J. Mol. Biol. 420, 269–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yadav T., Carrasco B., Myers A. R., George N. P., Keck J. L., Alonso J. C. (2012) Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 40, 5546–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bujalowski W., Lohman T. M. (1986) Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry 25, 7799–7802 [DOI] [PubMed] [Google Scholar]
- 73. Sweezy M. A., Morrical S. W. (1999) Biochemical interactions within a ternary complex of the bacteriophage T4 recombination proteins uvsY and gp32 bound to single-stranded DNA. Biochemistry 38, 936–944 [DOI] [PubMed] [Google Scholar]
- 74. Plate I., Hallwyl S. C., Shi I., Krejci L., Müller C., Albertsen L., Sung P., Mortensen U. H. (2008) Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 283, 29077–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. (2011) Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Petermann E., Orta M. L., Issaeva N., Schultz N., Helleday T. (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lia G., Rigato A., Long E., Chagneau C., Le Masson M., Allemand J. F., Michel B. (2013) RecA-promoted, RecFOR-independent progressive disassembly of replisomes stalled by helicase inactivation. Mol. Cell 49, 547–557 [DOI] [PubMed] [Google Scholar]
- 78. Lestini R., Michel B. (2008) UvrD and UvrD252 counteract RecQ, RecJ and RecFOR in the rep mutant. J. Bacteriol. 190, 5995–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Michel B., Flores M. J., Viguera E., Grompone G., Seigneur M., Bidnenko V. (2001) Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8181–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sugiyama T., Kantake N., Wu Y., Kowalczykowski S. C. (2006) Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 25, 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jackson D., Dhar K., Wahl J. K., Wold M. S., Borgstahl G. E. (2002) Analysis of the human replication protein A:Rad52 complex: evidence for crosstalk between RPA32, RPA70, Rad52 and DNA. J. Mol. Biol. 321, 133–148 [DOI] [PubMed] [Google Scholar]
- 82. Hanas J. S., Larabee J. L., Hocker J. R. (2005) Zinc finger interactions with metals and other mall molecules. In Zinc Finger Proteins: From Atomic Contact to Cellular Function (Iuchi S., Kuldell N., eds.) pp. 39–46, Landes Bioscience and Kluwer Academic/Plenum Publishers [Google Scholar]
- 83. Maciag A., Dainese E., Rodriguez G. M., Milano A., Provvedi R., Pasca M. R., Smith I., Palù G., Riccardi G., Manganelli R. (2007) Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189, 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Serafini A., Pisu D., Palù G., Rodriguez G. M., Manganelli R. (2013) The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS ONE 8, e78351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Outten C. E., O'Halloran T. V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 [DOI] [PubMed] [Google Scholar]
- 86. Leiros I., Timmins J., Hall D. R., McSweeney S. (2005) Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J. 24, 906–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Radzimanowski J., Dehez F., Round A., Bidon-Chanal A., McSweeney S., Timmins J. (2013) An “open” structure of the RecOR complex supports ssDNA binding within the core of the complex. Nucleic Acids Res. 41, 7972–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Timmins J., Leiros I., McSweeney S. (2007) Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding. EMBO J. 26, 3260–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Singleton M. R., Wentzell L. M., Liu Y., West S. C., Wigley D. B. (2002) Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. U.S.A. 99, 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhou R., Kozlov A. G., Roy R., Zhang J., Korolev S., Lohman T. M., Ha T. (2011) SSB functions as a sliding platform that migrates on DNA via repetition. Cell 146, 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]