Background: How cortactin and WASP proteins coordinately regulate branched actin assembly is poorly understood.
Results: The Arp2/3 complex- and actin-interacting regions of cortactin and WASP proteins are functionally distinct and tailored to their regulatory roles.
Conclusion: Mechanistic distinctions between activators are important in allowing coordinate regulation of branched actin networks.
Significance: Understanding mechanistic distinctions between activators is required to understand cellular structures like lamellipodia.
Keywords: Actin, Arp2/3 Complex, Cortactin, Microscopy, Single-molecule Biophysics, WASP Proteins, Actin Bundling
Abstract
Arp2/3 complex is an important actin filament nucleator that creates branched actin filament networks required for formation of lamellipodia and endocytic actin structures. Cellular assembly of branched actin networks frequently requires multiple Arp2/3 complex activators, called nucleation promoting factors (NPFs). We recently presented a mechanism by which cortactin, a weak NPF, can displace a more potent NPF, N-WASP, from nascent branch junctions to synergistically accelerate nucleation. The distinct roles of these NPFs in branching nucleation are surprising given their similarities. We biochemically dissected these two classes of NPFs to determine how their Arp2/3 complex and actin interacting segments modulate their influences on branched actin networks. We find that the Arp2/3 complex-interacting N-terminal acidic sequence (NtA) of cortactin has structural features distinct from WASP acidic regions (A) that are required for synergy between the two NPFs. Our mutational analysis shows that differences between NtA and A do not explain the weak intrinsic NPF activity of cortactin, but instead that cortactin is a weak NPF because it cannot recruit actin monomers to Arp2/3 complex. We use TIRF microscopy to show that cortactin bundles branched actin filaments using actin filament binding repeats within a single cortactin molecule, but that N-WASP antagonizes cortactin-mediated bundling. Finally, we demonstrate that multiple WASP family proteins synergistically activate Arp2/3 complex and determine the biochemical requirements in WASP proteins for synergy. Our data indicate that synergy between WASP proteins and cortactin may play a general role in assembling diverse actin-based structures, including lamellipodia, podosomes, and endocytic actin networks.
Introduction
Control of actin filament network assembly is required for eukaryotic cells to move, divide, and differentiate (1). Arp2/3 (actin-related protein 2/3) complex is one of three major classes of actin regulators capable of de novo initiation of actin filaments and is unique among actin nucleators in that under most circumstances it creates only branched filaments (2, 3). The activity of Arp2/3 complex is tightly controlled in vivo, and there are now approximately a dozen known NPF2 proteins that bind to the complex and switch on its nucleation activity (4, 5). NPFs are responsible for regulating assembly of branched actin networks that power a broad range of cellular processes, including endocytosis (6) and protrusion of lamellipodia (7) and invadopodia; the latter of which are actin-based protrusions that allow transformed cells to degrade extracellular matrix during metastasis (8). Proper assembly of branched actin networks in these cellular structures frequently requires the coordinated action of multiple NPFs. For example, formation of invadopodia requires the activity of two distinct NPFs: cortactin and N-WASP (9–11). Similarly, multiple NPFs, including the WASP (Wiskott-Aldrich syndrome protein) family protein Wsp1, type I myosins, Pan1, Crn1, Abp1, and Dip1 contribute to Arp2/3 complex activation during endocytosis in yeast (12–15). Understanding how these distinct NPFs collectively control branched actin assembly is critical to understanding how the actin cytoskeleton mediates complex cellular processes.
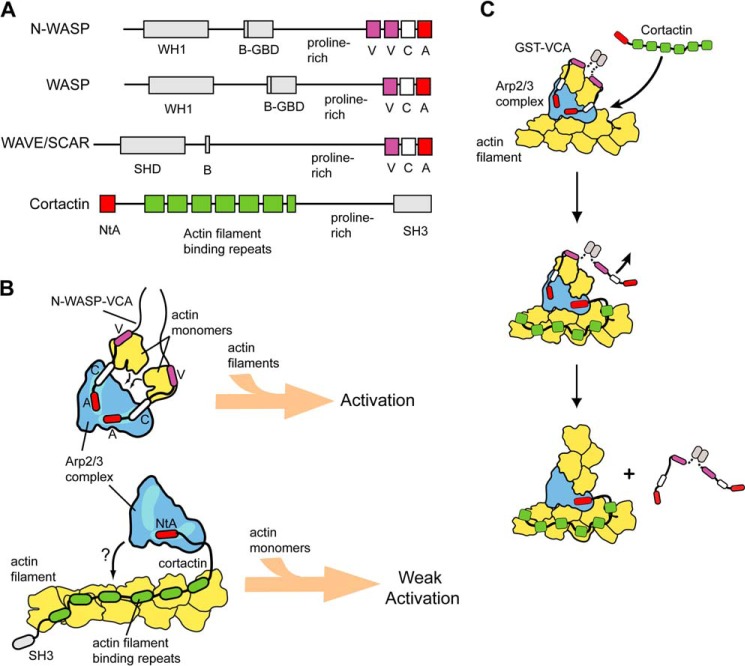
Recent evidence suggests that the biochemical mechanisms by which different NPFs turn on Arp2/3 complex influence their ability to coordinately regulate the dynamics of branched actin assembly. N-WASP and other WASP family proteins contain an acidic “A” and central “C” region that bind Arp2/3 complex, and a V (verprolin homology, also called WASP homology 2 (WH2)) region that binds monomers (16) (Fig. 1, A and B). Engineered cross-linking experiments show that actin monomer recruitment by N-WASP stimulates a conformational change in Arp2/3 complex that may be important for nucleation (17). Like WASP family proteins, cortactin contains an acidic region that interacts with Arp2/3 complex, but unlike WASP proteins, it contains an actin filament binding domain instead of a V region (18, 19) (Fig. 1, A and B). The intrinsic NPF activity of cortactin is weak compared with N-WASP, but importantly, in the presence of N-WASP, it dramatically stimulates branching nucleation (20–23) (Fig. 1, B and C). The actin filament binding repeats of cortactin potently increase synergy between these two NPFs, and single molecule TIRF microscopy experiments indicate that the repeats allow cortactin to target nascent branch junctions to displace N-WASP and stimulate nucleation (23) (Fig. 1C). These observations demonstrate that cortactin and N-WASP have distinct modes of activation of the complex that are controlled by the precise structural mechanism by which each NPF interacts with Arp2/3 complex and actin. Determining how the differences in these interactions tune regulation of the complex by NPFs, alone or in concert, is critical to understanding how branched actin networks are assembled in vivo.
FIGURE 1.
Overview of activation of Arp2/3 complex by cortactin and N-WASP. A, domain organization of WASP family proteins and cortactin. WH1, WASP homology 1; B, basic region; GBD, GTPase binding domain; V, verpolin homology (also known as WH2, WASP homology 2) domain; C, central region; A, acidic region; SHD, SCAR homology domain; NtA, N-terminal acidic region; SH3, Src homology 3. B, schematics of intrinsic NPF activities of N-WASP-VCA and cortactin. Light blue-shaded regions on Arp2/3 complex indicate NPF binding sites. C, schematic of the displacement mechanism of synergistic activation of Arp2/3 complex by cortactin and dimeric N-WASP-VCA (GST-VCA). GST-VCA must be released from the nascent branch for nucleation/elongation to proceed. Cortactin binds to the nascent branch complex at one of the two NPF sites and displaces one CA of the GST-VCA dimer from the Arp2/3 complex (reaction 1), accelerating complete release of GST-VCA (reaction 2) and allowing elongation of the newly nucleated branch filament.
The cortactin acidic region (NtA) has generally been posited to be functionally similar to the A region in WASP family proteins. Both A and NtA sequences are characterized by a stretch of multiple acidic residues surrounding a conserved tryptophan, which is required for the interaction with Arp2/3 complex (16, 19). However, the acidic regions of cortactin and WASP proteins also show several sequence differences, and evidence points to functional implications for these differences. Cross-linking assays showed that the NtA of cortactin competes with N-WASP for binding to Arp3, but unlike N-WASP, it does not bind to the Arp2 subunit (24). It is not known whether these differences tailor each NPF for its specific function in activating the complex. For instance, it is not clear whether specific structural properties of NtA are required for cortactin to synergize with N-WASP, nor whether sequence differences in NtA versus A prevent cortactin (on its own) from activating Arp2/3 complex as potently as N-WASP. Furthermore, although it is known that N-WASP can synergize with cortactin in activating Arp2/3 complex, the precise biochemical properties of N-WASP, including its oligomerization state and affinity for actin monomers, were shown to play critical roles in determining the potency of synergy (23). Therefore, it is unclear whether potent synergy with cortactin is a general feature of WASP family proteins and if so, whether the mechanism is conserved. Cortactin colocalizes with WASP family proteins such as N-WASP, WASP, and WAVE (Wiskott-Aldrich syndrome protein family verproline-homologous protein) in multiple branched actin networks, including in podosomes (25), at the leading edge of lamellipodia (26), and at sites of endocytosis (27). Determining how branched actin is assembled in each of these structures will require an understanding of how each WASP family protein coordinately regulates Arp2/3 complex with cortactin.
A key biochemical difference between cortactin and WASP family proteins is that cortactin binds filamentous actin and WASP proteins bind monomeric actin (28, 29). Because the actin monomer-binding region of WASP family proteins is required for potent Arp2/3 complex activation (16), its replacement in cortactin with a filament binding domain could explain why cortactin on its own is a weak NPF, but this has not been tested. In fact, the precise role of the actin filament binding region, which consists of 6.5 repeats of a 37-amino acid repeat sequence, is not completely understood, partially because of its multiple biochemical functions. For instance, it increases the potency of cortactin in synergizing with N-WASP and is required for the intrinsic NPF activity of cortactin (19, 23). In some studies, the actin filament binding repeats of cortactin have also been also reported to bundle actin filaments (30–32). However, bundling has not been studied in the presence of branching, despite the observation that in vivo, cortactin colocalizes with Arp2/3 complex within branched actin networks (18, 33). Therefore, the influence of the actin filament binding repeats of cortactin on the architecture of Arp2/3 complex-nucleated filament networks is not known.
Here we use a combination of biochemical studies and TIRF microscopy to dissect how the Arp2/3 complex- and actin-interacting segments of WASP proteins and cortactin influence their roles in regulating branched actin networks. Our results show that the NtA of cortactin is tailored to synergistically activate the complex with WASP family VCAs and is biochemically and functionally distinct from the A region of WASP family proteins. The actin filament binding repeats of cortactin allow it to bundle the branched actin filaments it initiates, but bundling is antagonized by N-WASP. In addition, our data show that the intrinsic NPF activity of cortactin is weak because it lacks an actin monomer binding region. Finally, we show that synergy with cortactin is not unique to N-WASP and that other WASP family proteins, including WASP and WAVE, can cooperate with cortactin to potently activate Arp2/3 complex. Potent synergy requires dimerization of the WASP protein in all cases, suggesting that cortactin displaces each of these WASP proteins from nascent branch junctions to accelerate branching nucleation. These observations have important implications for understanding how cortactin and WASP family proteins independently and coordinately regulate the dynamics of branched actin networks.
EXPERIMENTAL PROCEDURES
Protein Purification
Rabbit skeletal muscle actin was purified and labeled with either Oregon-Green 488 or N-(1-pyrene)iodoacetamide (Molecular Probes, Eugene, OR) as previously described (17). Bovine Arp2/3 complex was purified as previously described (17). A-cortactin was generated by fusing residues 486–505 of bovine N-WASP to the N terminus of residues 46–546 of cortactin. V- and VV-cortactin constructs were constructed by adding bovine N-WASP residues 428–457 and 399–457 (V and VV, respectively) to the N terminus of a 16-residue Gly-Ser linker attached to the N terminus of full-length cortactin. The Gly-Ser linker ensures that the number of residues separating the Arp2/3 complex binding tryptophan (DDWE motif) and the C-terminal V region residues corresponds to the shortest V to A distance of any WASP sequence. Alexa 568-cortactin (residues 1–336) and Alexa 546-NtA (residues 1–48), both with a C-terminal KCK (Lys-Cys-Lys) sequence, were labeled with maleimide reactive Alexa fluorophore dyes (Molecular Probes) as previously described (23). All cortactin constructs are murine and were overexpressed and purified from Escherichia coli as previously described (23). Human WAVE1 VCA (residues 485–559), human WASP VCA (residues 429–502), human WASP CA (residues 460–502), bovine N-WASP VCA (residues 428–505), bovine N-WASP VVCA (residues 392–505), and bovine N-WASP CA (residues 461–505) were purified as dimeric (GST-tagged) or monomeric proteins as previously described (17). Synthetic peptides of human WASP C (residues 464–484) and A (residues 497–502) are as previously described (16).
Pyrene-Actin Polymerization Assays
Pyrene-actin polymerization assays were performed as previously described (15). The influence of cortactin on polymerization rates of reactions containing Arp2/3 complex and a WASP family protein was quantified by dividing the maximum polymerization rate at each cortactin concentration by the maximum polymerization rate of an identical reaction without cortactin (23). These values were reported as fold activation and plotted versus cortactin concentration. Fold activation data were fit to the following equation,
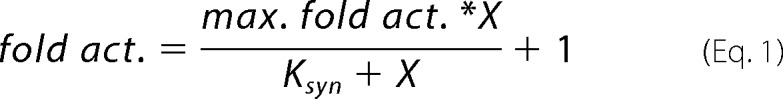 |
where X is the concentration of cortactin, and Ksyn is the concentration of cortactin at half-maximal activity. The reaction lag phase lifetime was defined as the start of the reaction to the time at which the acceleration of the reaction was the greatest. This time point was found by taking the second derivative of the raw fluorescence data that had been filtered with a five-point moving average.
Competition Binding Assays
The fluorescence anisotropy of 100 nm Alexa 546-NtA(1–48) was measured in the presence of 750 nm Arp2/3 complex and 0.2–20 μm of cortactin NtA or cortactin NtA mutants. Fluorescence anisotropy was monitored through excitation at 530 nm and emission at 570 nm in a buffered solution of 10 mm Hepes, pH 7.0, 50 mm KCl, 1 mm EGTA, 1 mm MgCl2, 1 mm DTT, and 0.2 mm ATP. Binding experiments were performed at least three times, and the data from each experiment were fit individually as previously described (34) with the probe affinity and receptor concentration held constant. The probe, Alexa 546-NtA(1–48), affinity for the Arp2/3 complex was determined by measuring the fluorescence anisotropy under conditions described above and fitting the results with a previously described equation (35). The probe affinity was found to be 0.66 ± 0.13 μm, error is 95% confidence interval. The reported affinities are the means of all individual fits.
TIRF Microscopy
Objective-based TIRF microscopy was performed on a Nikon TE2000-U microscopy body with a Nikon 100× 1.49 NA TIRF objective and a 1.5× auxiliary lens, as described previously (23). Laser light from an argon 488-nm (Dynamic Laser, Salt Lake City, UT) or solid state 561-nm (Coherent, Santa Clara, CA) laser were filtered through dual-band excitation and dichroic filters (Chroma, Bellows Falls, VT) prior to striking the sample from which the resulting fluorescent emission was filtered through dual-band dichroic and emission filters before collection on an EM-CCD camera (iXon3, Andor, or Image-EM; Hamamatsu). Actin polymerization was initiated by adding protein components to an oxygen-scavenging polymerization buffer composed of glucose-oxidase, catalase, and Trolox. The polymerization reaction was imaged after wicking the reaction into a cleaned flow chamber where the coverslip surface was passivated with polyethylene glycol. Polymerizing actin filaments were tethered to the coverslip by N-ethyl- or biotin maleimide-inactivated myosin, which was adhered to the coverslip through biotin-streptavidin interactions. The open source microscopy software Micro-Manager (36) was used to acquire 488-channel images at rates of 0.2–1 s−1 and 561-channel images at 3.6 s−1 with exposure times of 25–100 ms. Individual reaction parameters are detailed in the supplemental Movie S1 legends. Images were processed in ImageJ by background subtraction with a rolling ball radius of 10 pixels and then slightly blurred with a Gaussian filter of radius 0.6 pixels.
Image Analysis
Lifetime analysis of cortactin molecules was performed as previously described (23). Briefly, puncta in 561-channel images were automatically tracked using a nearest neighbor algorithm to determine their lifetime. Tracks were rejected if: 1) they appeared in the first or last image of the acquisition, 2) their intensity did not correspond to single molecules, or 3) their position did not correlate with actin filaments. Molecules from the tracks were manually viewed to classify them as binding to either single or bundled filaments. Molecule lifetimes were binned by 5 s, and the cumulative frequency for all molecules was determined. 1 minus cumulative frequency as a function of binned lifetimes was fit to a single exponential decay equation to yield an off rate constant and accompanying standard error.
The number of cortactin molecules per micrometer of filament was determined by manually counting cortactin molecules along a filament and measuring the length of filaments using a previously published ImageJ plugin (37). The number of filaments in the bundles were manually determined and incorporated into the filament length measurements. Binding densities were analyzed at three separate time points (105, 140, and 174 s from the start of imaging). No time-dependent difference was observed, so the average binding density reported contains measurements from all three time points.
The fraction of total filament bundled was calculated in Matlab (Mathworks Inc., Natick, MA) similar to a previously described procedure (23). Regions of interest were manually segmented to separate actin filaments from the background. Then extraneous pixels were removed using morphological opening to produce a binary image with filaments represented by positive pixels. The total length of all filaments was calculated by dividing the number of positive pixels by three (the average filament width because of the point spread function) and then the summed pixel value was converted to micrometers using 1 pixel = 0.16 or 0.017 μm (depending upon magnification used). Pixels corresponding to bundled filaments were manually marked and analyzed with the assumption that bundles contain only two filaments. Manual tracking of bundles showed that a vast majority contained only two filaments at the time points measured. Multiple nonoverlapping regions of interest were analyzed to calculate the reported fraction of total filament bundled.
Photobleaching data were collected by adhering 1 nm of NHS-biotin (Thermo-Pierce, Rockford, IL) conjugated Alexa 568-cortactin (residues 1–336) to the surface of PEGylated cover slips and acquiring 100-ms exposures continuously for 3 min. Fluorescent puncta were identified in the first image frame, and their positions were integrated across all images to produce an intensity time series for each puncta. The number of photobleaching steps was identified using a one-dimensional edge detection algorithm written in-house and implemented in Matlab.
Kinetic Modeling
Simulations were performed using previously described kinetic models (23). Briefly, time courses of actin polymerization for reactions containing GST-N-WASP-VCA, cortactin, and Arp2/3 complex were fit to a model in which cortactin can bind nascent branches and displace GST-N-WASP-VCA through an irreversible activation step (kdis), allowing a new branched filament end to form. kdis was the only parameter optimized during these fitting routines. Simulations of actin polymerization at increasing cortactin concentrations were performed using this optimized model, and the corresponding lag phase lifetimes were calculated as described above.
RESULTS
Cortactin Is a Weak NPF Because It Cannot Recruit Actin Monomers to Arp2/3 Complex
A critical difference between WASP family proteins and cortactin is that WASP proteins recruit actin monomers to Arp2/3 complex through their V regions, whereas cortactin lacks a V region and does not bind actin monomers (19). Actin monomer recruitment by WASP is required for NPF activity and has been shown to stimulate movement of Arp2 and Arp3 into a filament-like (short pitch) conformation that may template nucleation (16, 17, 38). Therefore, we hypothesized that the weak NPF activity of cortactin is due to its inability to recruit actin monomers to Arp2/3 complex. We tested this hypothesis by creating a chimeric cortactin construct in which the second V region of N-WASP was fused to the N terminus of cortactin with a 16-amino acid linker (V-cortactin; Fig. 2A). Fusion of the V region dramatically increased the activity of cortactin, increasing the maximum polymerization rate 3.1-fold compared to wild type cortactin, demonstrating that cortactin is a weak NPF because it is unable to recruit actin monomers to the Arp2/3 complex (Fig. 2, B and C). Despite its increased intrinsic NPF activity, the V-cortactin chimera had a 1.4-fold reduced maximum polymerization rate compared with N-WASP-VCA. Recent experiments show that VCA binds to two NPF binding sites on Arp2/3 complex to recruit two actin monomers and maximally stimulate nucleation (39–41). Cortactin NtA binds to only one of these two sites, so we hypothesized that V-cortactin may be less active than N-WASP because it cannot simultaneously recruit two monomers to the complex (24). To test this hypothesis, we prepared a VV-cortactin fusion, in which the tandem V region of N-WASP was appended to the N terminus of cortactin (Fig. 2A). VV-cortactin activated the Arp2/3 complex 1.6-fold more potently than V-cortactin, and to a similar extent as N-WASP-VCA, with a maximum polymerization rate at saturation of 19 nm/s (Fig. 2C). Together, these data demonstrate that differences in the actin binding properties and not the Arp2/3 complex interacting regions of cortactin and N-WASP explain their intrinsic potency as NPFs. An important question will be to determine how, in both NPFs, increasing the number of monomer binding regions from one to two increases potency, a property previously reported for N-WASP (42).
FIGURE 2.
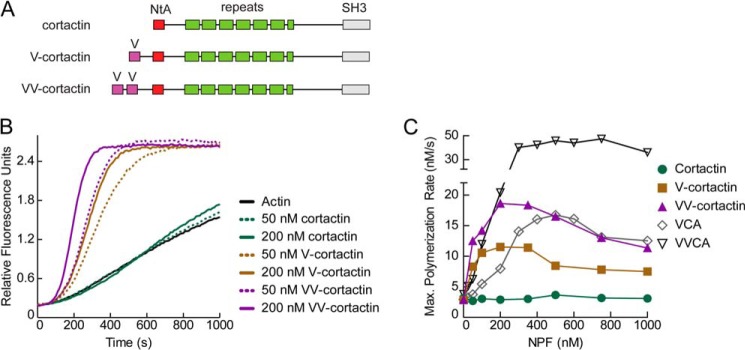
Cortactin is a weak NPF because it cannot recruit actin monomers to the Arp2/3 complex. A, domain organization of wild type cortactin and N-terminal V region fusions. B, pyrene-actin polymerization time courses showing Arp2/3 complex activation by cortactin, V-cortactin, or VV-cortactin. Reactions contain 3 μm 15% pyrene-actin, 40 nm Arp2/3 complex, and the indicated concentrations of cortactin constructs. C, maximum polymerization rate versus NPF concentration for N-WASP VVCA, N-WASP VCA, cortactin, V-cortactin, and VV-cortactin. Reactions conditions were as described in B. Maximum polymerization rates were calculated as previously described (15).
Cortactin Bundles Branched Actin Filaments to Create Actin Networks Distinct from N-WASP-activated Arp2/3 Complex
Low speed copelleting assays, negative staining electron microscopy, and small angle x-ray scattering showed that cortactin bundles spontaneously nucleated (unbranched) actin filaments (30–32). Cortactin localizes with Arp2/3 complex in branched actin networks in vivo (33) and specifically targets branch junctions in vitro (23), but it is uncertain whether cortactin can bundle branched actin filaments or how its nucleation-promoting and bundling activities might together influence branched actin filament architectures. Therefore, we used TIRF microscopy to directly visualize GST-N-WASP-VCA- or cortactin-mediated branching nucleation. GST-N-WASP-VCA-activated Arp2/3 complex created both primary branches that grew from spontaneously nucleated filaments and secondary or higher order branches from Arp2/3-nucleated filaments, creating dense dendritic networks (Fig. 3A, top panel, and supplemental Movie S1). In contrast, cortactin-mediated branching formed dramatically different networks, in which branching was less dendritic, and instead consisted of a few daughter filaments elongating from mother filaments that were frequently bundled. In many instances, daughter filaments growing away from the mother filament (or mother filament bundle) collided with a nearby “sister filament” to form daughter filament bundles (Fig. 3A, bottom panel, and supplemental Movie S2). These data show that the bundling and branching activation activities of cortactin together create a network of interconnected bundles distinct from N-WASP-mediated branching.
FIGURE 3.
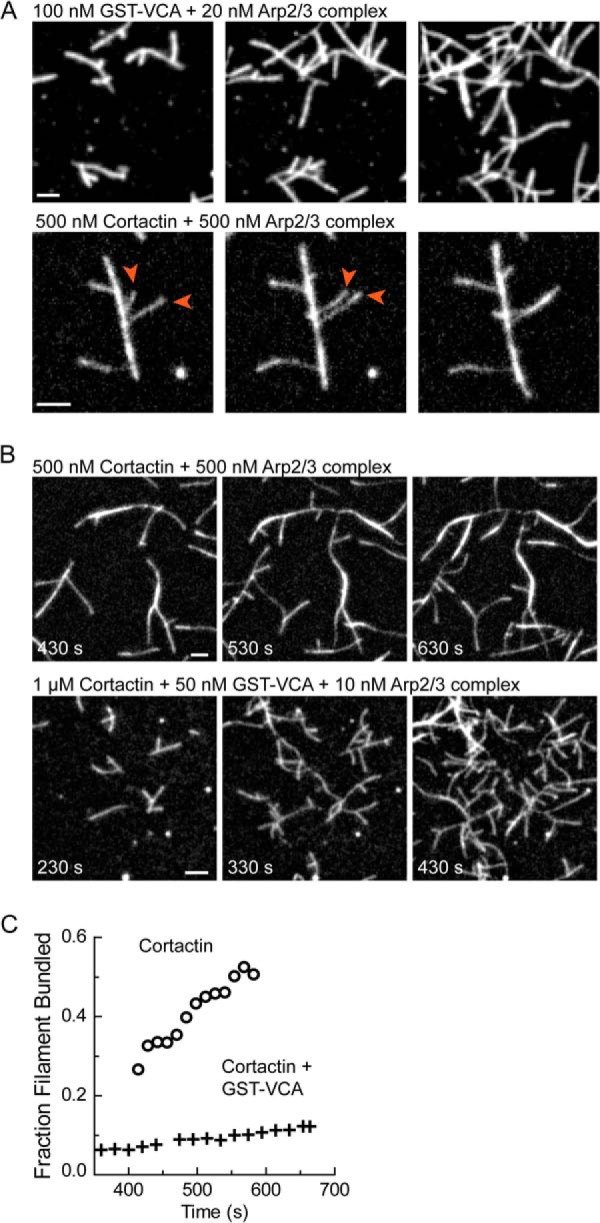
Cortactin bundles branched actin filaments to form actin networks distinct from N-WASP. A, time lapse TIRF microscopy images of branched networks nucleated by Arp2/3 complex activated by either GST-N-WASP-VCA (top panels) or cortactin (residues 1–336) (bottom panel) at the indicated concentrations. Arrowheads indicate an example of daughter filament bundling. B, time lapse TIRF microscopy images of branched networks nucleated by Arp2/3 complex activated by cortactin alone (residues 1–336) (top panel) or cortactin and GST-N-WASP-VCA (bottom panel). C, quantification of the fraction of actin filaments in bundles from reactions in B as a function of time. All TIRF reactions contain 1.5 μm 33% Oregon-Green actin. Scale bars are 2 μm.
Electron micrographs show actin filaments in lamellipodia and at sites of endocytosis are dendritic and more similar to the N-WASP-initiated networks we observed in our TIRF images than the bundled cortactin-mediated networks (43, 44). This presents an apparent discrepancy, because cortactin localizes to both of these actin-based structures (28, 45). To resolve this apparent discrepancy, we examined the architecture of branched networks created in the presence of both N-WASP (GST-N-WASP-VCA) and cortactin. Reactions containing both NPFs showed highly dendritic networks, similar to those created with N-WASP-VCA alone (Fig. 3B). Even at high concentrations of cortactin, which normally cause dramatic bundling (supplemental Movie S3), very little bundling was observed when GST-N-WASP-VCA was present (Fig. 3, B and C, and supplemental Movie S4). These data suggest that high rates of branching nucleation antagonize bundling and that the architectures of cortactin-containing actin networks in vivo will be modulated by the activities of the other NPFs present.
Cortactin Monomers Bundle Parallel and Antiparallel Actin Filaments
To better understand the bundling mechanism, we directly observed bundling of unbranched elongating filaments by cortactin using two-color TIRF microscopy. For these assays, we used an Alexa 568-labeled C-terminally truncated construct of cortactin that lacks the proline-rich region and the Src homology 3 domain. This construct has the same NPF activity as full-length cortactin (23). Multiple pathways to bundling were observed. In some events, actin filaments with bound fluorescent cortactin puncta collided at an acute angle with a second filament (Fig. 4A and supplemental Movie S5). Either the two filaments then zippered together, or the shorter end of the Y-shaped structure bound to the side of the longer filament, creating a gap that closed from each side. In other cases, filaments aligned along their lengths collided and immediately snapped together, and in others, two filaments attached to the surface elongated into each other (Fig. 4A and supplemental Movies S6 and S7). Using fiduciary marks to identify barbed ends, we observed both parallel and anti-parallel bundling events (Fig. 4A). We hypothesized that bundling could occur by cortactin oligomerization, which would allow the actin filament binding region from each cortactin molecule to contact a separate filament. Alternatively, a cortactin monomer could utilize a single set of filament binding repeats to bridge two actin filaments (Fig. 4B). To distinguish between these possibilities, we measured the oligomerization state of cortactin by attaching it to the imaging surface through biotinylation and observing its photobleaching behavior. The vast majority of fluorescent puncta bleached in a single step, demonstrating that cortactin is a monomer and supporting a model in which monomers of cortactin mediate bundling using a single set of repeats to bridge two filaments (Fig. 4C). To determine how bundling influences the interactions of cortactin with actin, we measured the lifetimes of cortactin bound to single filaments versus bundles. The off rate of cortactin for filaments was nearly identical in each scenario (Fig. 4D). In addition, the number of cortactin molecules bound per filament length was nearly identical for cortactin bound to single versus bundled filaments (Fig. 4E). These observations indicate that bundling does not dramatically alter the interactions between cortactin and actin filaments.
FIGURE 4.
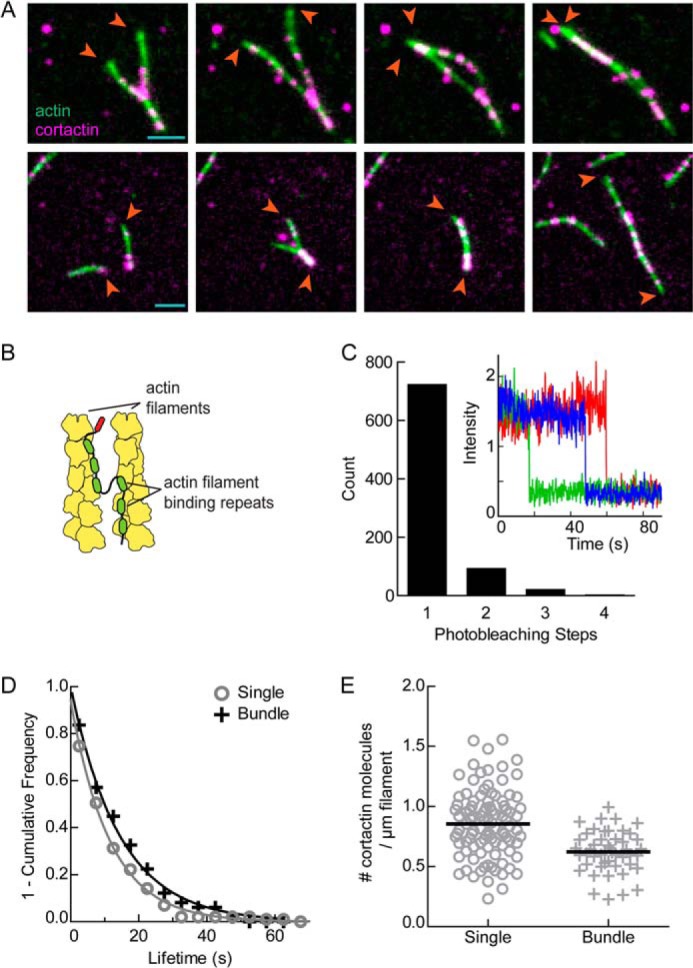
Cortactin monomers bundle parallel and antiparallel actin filaments. A, two-color time lapse TIRF microscopy images of cortactin molecules (magenta) bundling actin filaments (green) in either parallel (top panels) or antiparallel (bottom panels) orientations with respect to their barbed ends (arrowheads). Colocalized signals are white. Reaction contains 1.5 μm 33% Oregon-Green actin and 10 nm Alexa 568-cortactin (1–336). B, graphic representation of proposed cortactin bundling mechanism. C, count of fluorescent puncta corresponding to the indicated number of observed photobleaching steps from a reaction of 1 nm biotinylated Alexa 568-cortactin attached to a streptavidin-coated coverslip. Inset, plot of fluorescence intensity versus time for three different monomeric Alexa 568-cortactin molecules during photobleaching experiment. D, frequency plot of tracked cortactin lifetimes for molecules bound to single (gray circles, n = 99) or bundled (black crosses, n = 49) filaments from the reaction in A. The data were fit with a single exponential decay function to yield off rates of 0.09 ± 0.01 and 0.07 ± 0.01 s−1 for single and bundled filaments, respectively. Errors are 95% confidence interval from fit. E, number of cortactin molecules bound per micrometer of actin filament for molecules bound to either single (circles, n = 99) or bundled (crosses, n = 48) filaments from reaction in A. Black bars represent means; single = 0.85 ± 0.28 and bundled = 0.62 ± 0.17 cortactin molecules per micrometer of actin filament. Errors are S.D.
The Arp2/3 Complex Binding Region of Cortactin Has Unique Biochemical Properties Required for Synergistic Activation
To better understand how functional segments of Arp2/3 complex activators influence branching nucleation, we next focused on the Arp2/3 complex interacting regions of N-WASP and cortactin. Specifically, we asked if features of Arp2/3 complex binding regions in these NPFs influenced their ability to coordinately regulate the complex. Cortactin and N-WASP have been shown to synergistically activate the complex using a displacement mechanism, in which cortactin increases the nucleation rate by binding to the complex at nascent branch junctions and displacing N-WASP-VCA (23) (Fig. 1C). This model is consistent with the observation that the NtA region of cortactin is sufficient for synergy with N-WASP and competes with N-WASP-CA for binding to the Arp3 subunit (23, 24). It is also consistent with the observation that N-WASP must be released from the complex at nascent branch junctions before nucleation, whereas cortactin can remain bound throughout nucleation and elongation (23, 46). Structural modeling suggests the V region in WASP-VCA blocks the barbed ends of recruited actin monomers from interacting with actin monomers, explaining why VCA must be released (47). These results suggest that any interaction that can promote displacement of VCA from the nascent branch junction without blocking incoming actin monomers might synergistically activate the Arp2/3 complex. Therefore, we asked whether the A region of WASP, which can compete with VCA for binding to Arp2/3 complex (16), can synergistically activate the complex with GST-N-WASP-VCA. Unexpectedly, the A region of WASP was not synergistic with GST-N-WASP-VCA, even at concentrations where NtA-mediated synergy was saturated (Fig. 5A). These data demonstrate that NtA is functionally distinct from A and harbors unique structural features required for synergy. The failure of A to synergize with GST-N-WASP-VCA cannot be explained by its relatively low affinity (KD = ∼9 μm) compared with NtA (KD = 1.6 μm), because neither WASP-CA nor N-WASP-CA (KD = ∼1 μm) synergistically activated the complex with GST-N-WASP-VCA (16, 48) (Fig. 5A). Consistent with this observation, the WASP C region also failed to synergize with GST-N-WASP-VCA (Fig. 5A).
FIGURE 5.
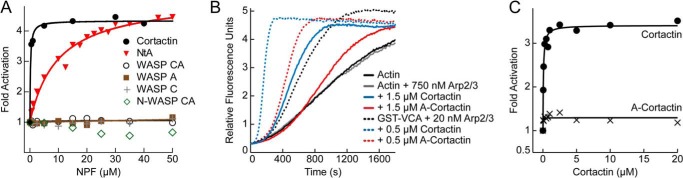
Cortactin NtA has unique biochemical properties compared with the CA region of WASP. A, fold Arp2/3 complex activation versus NPF concentration for pyrene-actin polymerization reactions containing either cortactin, NtA, WASP CA, WASP A, WASP C, or N-WASP CA. All reactions contained 2 μm 15% pyrene-actin, 20 nm Arp2/3 complex, 250 nm GST-N-WASP-VCA, and the indicated NPF. Fold activation is defined as the maximum polymerization rate from a reaction with both GST-N-WASP-VCA and either cortactin or WASP proteins divided by the maximum polymerization rate of the reaction with just GST-N-WASP-VCA. B, pyrene-actin polymerization time courses showing intrinsic NPF activity and synergy with 150 nm GST-N-WASP-VCA (GST-VCA) by cortactin and N-WASP acidic cortactin (A-cortactin) for reactions containing 3 μm 15% pyrene-actin and the indicated protein concentrations. C, plot of fold activation versus cortactin concentration for cortactin and A-cortactin. Reactions include 2 μm 15% pyrene-actin, 20 nm Arp2/3 complex, and 150 nm GST-N-WASP-VCA.
In some contexts CA inhibits WASP-mediated activation of the complex (49, 50). Our data here show that monomeric CA constructs are not effective inhibitors of GST-N-WASP-VCA. Although we cannot eliminate the possibility that CA-mediated inhibition is masked by weak synergy between CA and GST-N-WASP-VCA, the observation that WASP proteins do not bind to branch junctions indicates that CA lacks a key biochemical property for displacement-mediated synergy (23, 46). Therefore, we speculate the weak inhibitory potency of CA we measured is due to its weak binding to isolated Arp2/3 complex compared with dimeric N-WASP constructs (16, 48).
Because the actin filament binding domain of cortactin increases its potency in synergizing with N-WASP (Fig. 5A), we next replaced the NtA region of cortactin with the acidic (A) region of N-WASP and asked whether this chimeric construct (A-cortactin) could synergize with GST-N-WASP-VCA. A-cortactin showed dramatically reduced synergy with GST-N-WASP-VCA compared with wild type cortactin (Fig. 5, B and C). The chimeric construct increased the maximum polymerization rate 1.4-fold over the rate with GST-N-WASP-VCA alone, compared with the 3.5-fold increase of wild type cortactin. These data show that even in the context of the actin filament binding repeats, NtA is functionally distinct from WASP family A regions.
NtA Harbors Non-WASP-like Sequences That Contribute to Synergy
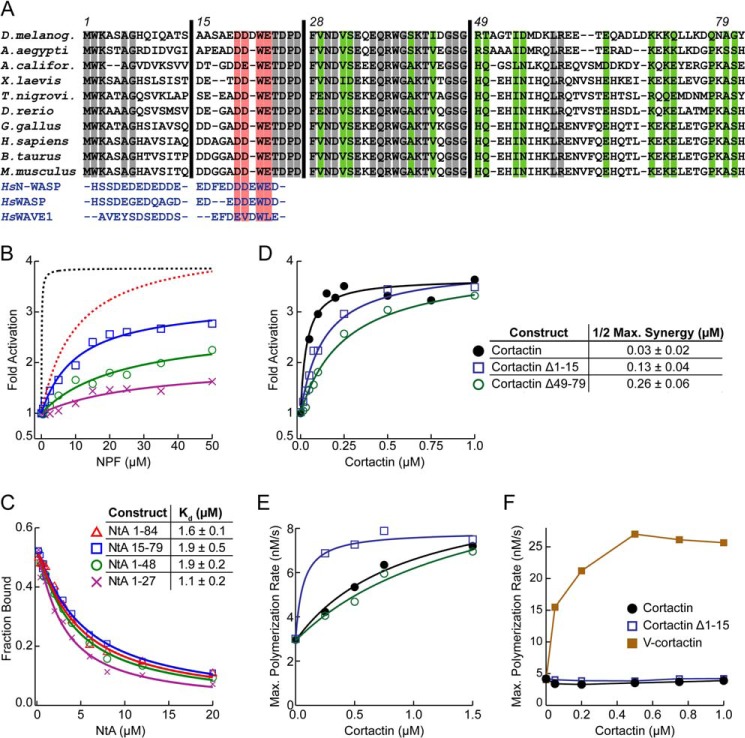
To determine the basis of functional differences between cortactin NtA and N-WASP A, we aligned cortactin sequences from multiple species to the A region of WASP family proteins. This analysis revealed that the acidic motif of cortactin (the DDWE motif) is embedded within a ∼60-amino acid stretch of residues highly conserved among cortactin sequences but not present in WASP family proteins (Fig. 6A). To determine the importance of each of these conserved NtA segments, we tested the influence of truncations of NtA on its ability to synergistically activate the complex. We first asked whether the conserved region C-terminal to the DDWE motif is required for synergy. We made a construct (NtA(1–27)) that contains the short conserved stretch N-terminal to the DDWE motif plus the DDWE motif but lacks the C-terminal region of NtA. NtA(1–27) showed a dramatic reduction in synergy with GST-N-WASP-VCA. The maximum polymerization rate at 50 μm NtA(1–27) was 2.2-fold less than the full-length NtA construct (NtA(1–84)). A construct lacking only the C-terminal 36 amino acids of the NtA (NtA(1–48)) was 1.6-fold defective compared with NtA(1–84), whereas a construct lacking 14 residues at the N terminus (NtA(15–79)) showed a 1.3-fold decrease in synergy (Fig. 6B). These data indicate that the conserved non-WASP-like sequences in NtA region contribute to the ability of cortactin to synergistically activate Arp2/3 complex with N-WASP.
FIGURE 6.
All regions of the cortactin NtA contribute to synergy. A, sequence alignment of cortactin NtA and WASP family acidic regions. WASP family proteins are human (Hs) N-WASP (residues 482–505), WASP (residues 481–502), and WAVE1 (residues 540–559). Gray- and green-shaded NtA residues are 100 and 80% conserved, respectively. Red-shaded residues represent the WASP-like DDWE motif. Residue numbering is based on Mus musculus cortactin. Vertical black lines indicate truncations of the NtA. B, plot of fold activation versus concentrations of cortactin (black dashed line), NtA(1–84) (red dashed line), NtA(15–79), NtA(1–48), and NtA(1–27). Dashed lines indicate previously published data (23). C, fluorescence anisotropy binding assay showing competition between 100 nm Alexa 546-labeled NtA(1–48) and unlabeled NtA constructs for 750 nm Arp2/3 complex. Plot shows representative competition binding titration data for each construct with accompanying fit. Reported affinities (inset) are averages of three separate titrations. Errors are S.D. D, plot of fold Arp2/3 complex activation for a range of concentrations of cortactin in pyrene-actin polymerization reactions containing the indicated cortactin construct and 250 nm GST-N-WASP-VCA, 2 μm 15% pyrene-actin, and 20 nm Arp2/3 complex. The right panel contains best fit values with 95% confidence intervals of concentration to half-maximum saturation of synergistic activation. Fits were performed as described under “Experimental Procedures.” E, plot of maximum polymerization rate versus cortactin concentration for actin polymerization reactions containing 3 μm 15% pyrene-actin and 0.5 μm Arp2/3 complex, but no GST-N-WASP-VCA. F, plot of maximum polymerization rate versus cortactin concentration for actin polymerization reactions containing 3 μm 15% pyrene-actin, 40 nm Arp2/3 complex, and the indicated cortactin construct.
To determine how regions outside the DDWE motif contribute to synergy, we asked whether the NtA truncations influenced interactions with Arp2/3 complex. We measured the affinity of the NtA constructs for free Arp2/3 complex using a fluorescence anisotropy competition binding assay (Fig. 6C). Unexpectedly, all NtA constructs bound Arp2/3 complex with a similar affinity to the full-length NtA. The displacement model of synergistic activation predicts that cortactin binds Arp2/3 complex at nascent branch junctions, where interactions of the complex with N-WASP, actin monomers, and actin filaments could influence the NtA binding surface (Fig. 1C). Therefore, the mutations in NtA might decrease interactions with Arp2/3 complex specifically at nascent branch junctions. To test this, we measured the concentrations of mutant NtA constructs required to saturate synergy with GST-N-WASP-VCA. For these assays, we introduced the NtA mutations in the context of full-length cortactin to enable us to saturate the reactions. The concentrations required for half-maximal synergy for the full-length NtA mutants were 4–8-fold greater than wild type cortactin, whereas the potency of each mutant at saturation was identical to wild type cortactin (Fig. 6D). These results are consistent with a model in which mutations in the NtA decrease the affinity of cortactin for the nascent branch junctions rather than slowing a post-binding first order reaction rate.
The Distinct Modes of Activation of Arp2/3 Complex by Cortactin Are Partially Separable and Encoded within the NtA
The intrinsic NPF activity of cortactin requires both the DDWE motif of the NtA and the actin filament binding repeats, leading to the hypothesis that cortactin recruits free Arp2/3 complex to a pre-existing filament to activate nucleation (24) (Fig. 1B). In the recruitment pathway to activation, the interactions of NtA with the free Arp2/3 complex could influence rates of activation. This is in contrast to the displacement mechanism of synergistic activation, in which interaction of NtA with nascent branch junctions (and not the isolated Arp2/3 complex) define activation rates. Therefore, we hypothesized that NtA residues outside of the DDWE motif, which are important for synergistic activation but not for binding to the isolated complex, might be dispensable for the intrinsic NPF activity of cortactin. Consistent with this hypothesis, we found that deleting the C-terminal portion of NtA had no effect on the intrinsic NPF activity of cortactin (Fig. 6E). Similarly, truncating the N terminus of NtA also failed to reduce cortactin's intrinsic NPF activity. Instead, this mutant showed increased activity compared with wild type cortactin, indicating that the N terminus of NtA antagonizes the intrinsic NPF activity of cortactin (Fig. 6E). The stimulatory effect of the N-terminal truncation was insignificant when directly compared with potent stimulation caused by the N-terminal V-cortactin fusions (Fig. 6F). Together, these observations indicate that the intrinsic versus synergistic activation activities of cortactin are mechanistically distinct and that mutations in the NtA allow functional separation of these activities.
Cortactin Synergistically Activates Arp2/3 Complex to Different Extents with WAVE, WASP, and N-WASP
Cortactin regulates branched actin assembly in multiple cellular structures, including podosomes (51), invadopodia (52), sites of endocytosis (45), and at the leading edge of the cell (33). Each of these structures contains both cortactin and a WASP family protein, suggesting these two NPFs might synergize to assemble the branched actin filaments in each of these structures. However, N-WASP is the only WASP family protein demonstrated to potently synergize with cortactin, and few experiments have explored synergy with other WASP family proteins (20–23). Dissection of the mechanism revealed strict biochemical requirements for N-WASP, making it uncertain whether potent synergy with cortactin is a general feature of WASP family proteins. To answer this question, we tested combinations of cortactin and different WASP family proteins for their ability to synergistically activate Arp2/3 complex.
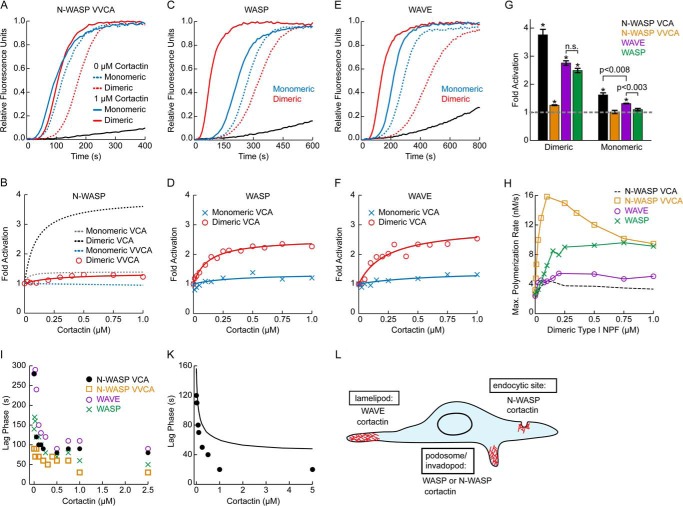
Cortactin and N-WASP colocalize in ventral actin-based protrusions in transformed cells called invadopodia (53). We previously showed that GST-N-WASP-VCA and cortactin synergistically activate Arp2/3 complex in vitro (23), potentially explaining why both NPFs are required for assembly of actin filaments in invadopodia (10). Potent synergy required dimerization of N-WASP-VCA; monomeric N-WASP-VCA containing either one or both of the two native V regions (N-WASP-VCA or N-WASP-VVCA) showed little or no synergy, respectively (23). This does not rule out the possibility that N-WASP synergizes with cortactin in invadopodia, because there is building evidence that WASP family proteins act as oligomers in vivo (48, 54–56). However, experiments to measure the synergy of dimerized N-WASP have all used a construct of N-WASP that includes only one of its two native V regions (20, 21). Because the additional V region increases the intrinsic NPF activity of N-WASP (Fig. 7H), and we observed that intrinsic and synergistic activity are inversely correlated (23, 42) (see “Discussion”), we reasoned that the additional V region may influence synergy with cortactin. Therefore, we tested the ability of GST-N-WASP-VVCA to synergize with cortactin. The addition of cortactin to GST-N-WASP-VVCA increased the maximum polymerization only 1.3-fold over GST-N-WASP-VVCA alone, whereas its addition to GST-N-WASP-VCA increased the polymerization rate 3.8-fold (Fig. 7, A, B, and G). These data demonstrate that the presence of an additional V region in N-WASP significantly decreases synergy with cortactin. Therefore, synergy between these two NPFs may not contribute significantly to nucleation of branched filaments in invadopodia.
FIGURE 7.
Cortactin synergistically activates Arp2/3 complex to different extents with WAVE, WASP, and N-WASP. A, C, and E, time courses of pyrene-actin polymerization for reactions containing indicated WASP family proteins with or without 1 μm cortactin, plus 20 nm Arp2/3 complex, 2 μm 15% pyrene-actin, and either 750 nm N-WASP-VVCA, 100 nm GST-N-WASP-VVCA, 750 nm WASP-VCA, 250 nm GST-WASP-VCA, 750 nm WAVE-VCA, or 250 nm GST-WAVE-VCA. B, D, and F, fold activation for cortactin-mediated synergistic activation of Arp2/3 complex with either monomeric or dimeric WASP family proteins. Reactions conditions are the same as above but with varying concentrations of cortactin. The data were fit as described under “Experimental Procedures.” Dashed lines indicate previously published data (23). G, average fold activation of Arp2/3 complex by cortactin for reactions containing 1 μm cortactin, 20 nm Arp2/3 complex, 2 μm 15% pyrene-actin, and a WASP family protein at concentrations listed above. Dimeric N-WASP VCA, monomeric N-WASP VCA, and monomeric N-WASP VVCA were previously published (23). The dashed gray line indicates no synergy. p values were calculated by two-tailed Student's t test. Error bars indicate S.E. n.s., not significant. H, plot of maximum polymerization rate versus dimeric NPF concentration for actin polymerization reactions performed with 3 μm 15% pyrene-actin, 20 nm Arp2/3 complex, and the indicated concentrations of either GST-N-WASP-VCA (black dashed line; previously published (23)), GST-N-WASP-VVCA, GST-WAVE-VCA, or GST-WASP-VCA. I, plot of lag phase lifetime versus cortactin concentration for actin polymerization reactions containing 2 μm 15% pyrene-actin, 20 nm Arp2/3 complex, indicated concentrations of cortactin and either 250 nm GST-N-WASP-VCA, 100 nm GST-N-WASP-VVCA, 250 nm GST-WAVE-VCA, or 250 nm GST-WASP-VCA. K, plot of lag phase lifetime versus cortactin concentration for either actin polymerization reactions performed with 3 μm 15% pyrene-actin, 50 nm Arp2/3 complex, 100 nm GST-N-WASP-VCA, and the indicated concentrations of cortactin (black closed circles) or simulations of actin polymerization reactions (black line) from a kinetic model of cortactin mediated synergistic displacement. Kinetic model fits and parameter optimization were published previously (23). L, schematic indicating the actin based cellular structures were cortactin and a corresponding WASP family protein have been observed to colocalize.
WASP is a hematopoietic cell-specific WASP family protein required for the assembly of actin filaments in podosomes, protrusive structures similar to invadopodia that occur in nontransformed cells (57). WASP has a single V region and has increased intrinsic dimeric NPF activity compared with GST-N-WASP-VCA (Fig. 7H). Dimeric WASP-VCA was recently shown by TIRF microscopy to increase Arp2/3 complex-mediated branching in the presence of cortactin (22). However, it is unknown whether the biochemical differences between WASP and N-WASP influence the potency or mechanism of synergy. To test this, we added cortactin to reactions containing Arp2/3 complex and dimeric or monomeric WASP-VCA. We found that dimeric WASP (GST-WASP-VCA), but not monomeric WASP (WASP-VCA), synergistically activated Arp2/3 complex with cortactin (Fig. 7, C, D, and G). Therefore, like N-WASP, the oligomerization state of WASP-VCA is a critical determinant of synergy, suggesting N-WASP and WASP use a similar mechanism to synergize with cortactin. The addition of cortactin to GST-WASP-VCA increased the maximum polymerization rate 2.6-fold over GST-WASP-VCA alone. This value was significantly less than dimeric N-WASP-VCA, indicating that sequence differences in the VCA of distinct WASP proteins tune the potency of their synergy with cortactin (Fig. 7G).
WAVE and cortactin colocalize with Arp2/3 complex in lamellipodia, and RNAi knockdown experiments indicate that both NPFs contribute to nucleation of actin filaments in these structures (26, 58, 59). WAVE contains one V region and as a dimer was found to have slightly higher activity than dimeric N-WASP-VCA but less activity than dimeric WASP VCA (Fig. 7H). Dimeric WAVE-VCA (GST-WAVE-VCA) showed a similar potency as dimeric WASP-VCA in cortactin-mediated synergistic activation of Arp2/3 complex (Fig. 7, E–G).
In addition to increasing the polymerization rate, synergistic activation of Arp2/3 complex by WASP family VCAs and cortactin significantly decreased the lag phase of the polymerization time courses (Fig. 7I). To determine whether the displacement mechanism can account for the decreased lag phase, we simulated time courses of reactions of cortactin, GST-N-WASP-VCA, and Arp2/3 complex using a previously optimized kinetic model of cortactin-mediated displacement (23). In our simulations, increasing cortactin concentrations decreased the length of the lag phase almost 3-fold, similar to the decreases measured in the reactions (Fig. 7K). Although the duration of the lag phase simulated at saturating cortactin was greater than the observed value, it was also overestimated at 0 μm cortactin. This indicates that that the reactions describing spontaneous and/or GST-N-WASP-VCA-activated nucleation may be oversimplified, creating the observed differences between predicted and measured lag phase in reactions containing cortactin.
Together, these results show that the ability to synergize with cortactin is a general feature of WASP family proteins. Therefore, synergistic activation of Arp2/3 complex by cortactin and a WASP family protein may be important for assembly of branched actin filaments in diverse actin-based structures, including podosomes, endocytic sites, and lamellipodia (Fig. 7L).
DISCUSSION
Cortactin Is a Weak NPF Because It Cannot Recruit Actin Monomers to the Arp2/3 Complex
Here we showed that differences between the cortactin NtA and the WASP A region cannot explain the weak intrinsic NPF activity of cortactin. Instead, we showed that cortactin is a weak NPF because it cannot recruit actin monomers to Arp2/3 complex. The absence of an actin monomer binding domain may ensure the NPF activity of cortactin is suppressed until a WASP family protein is activated, and branched networks can be assembled by the two NPFs synergistically. Alternatively, the NPF activity of cortactin may be stimulated when it binds proteins that allow it to indirectly tether monomers to Arp2/3 complex. For instance, WIP (WASP interacting protein) contains both a proline-rich region that binds to the Src homology 3 domain of cortactin and two V regions that bind actin monomers. In vitro, WIP increases cortactin-mediated activation of Arp2/3 complex, and coexpression of WIP and cortactin stimulates membrane protrusion (60). Several other NPFs, including type I myosins and Abp1 (actin binding protein 1) (5), share the sequence characteristics of cortactin, in that they contain Arp2/3 complex interacting and actin filament binding regions, but no actin monomer binding segments. Indirect recruitment of actin monomers by these NPFs may be a general mechanism to control their activity. Indeed, myosin 1, like cortactin, shows weak intrinsic Arp2/3 activation, but its NPF activity is potently stimulated by a direct interaction with WIP (61).
The observation that fusing a V region onto the N terminus of cortactin potently activated its NPF activity was unexpected given that cortactin lacks a canonical C region (Fig. 6A). The C region of WASP VCA is thought to be critical for stimulating an activating conformational change in Arp2/3 complex (62). Why the C region is required for activation in the context of VCA but not in V-cortactin is unclear. One possibility is that the cortactin NtA harbors sequences unique from the WASP C region that stimulate conformational changes in the complex. In support of this, single particle reconstructions of electron microscopy images of cortactin-bound Arp2/3 complex show Arp2 and Arp3 in a filament-like conformation distinct from the splayed conformation observed in crystal structures of inactive Arp2/3 complex (63). FRET experiments by Goley et al. (64) showed that unlike CA, an NtA construct containing residues 1–39 does not stimulate conformation changes in the complex, indicating that NtA residues important for these changes may be C-terminal to residue 39. Determining precisely how V-cortactin and WIP-bound cortactin activate Arp2/3 complex without a C region will be critical for understanding the structural mechanisms by which NPFs activate the complex.
The activity of V-cortactin was also unexpected because NtA binds to a single NPF site on the complex (24). In contrast, multiple lines of evidence, including analytical ultracentrifugation, isothermal titration calorimetry, and chemical cross-linking, indicate that WASP CA binds two sites on the complex, one on Arp3 and the other on ARPC1 and Arp2 (15, 39–41). Furthermore, mutational studies showed that recruitment of two actin monomers through engagement of VCA to both NPF sites is required for maximal activation (39). Assuming that the conserved tryptophan in NtA binds to the same surface on Arp3 as WASP (24), our structural modeling suggests the V-cortactin construct can deliver an actin monomer to the barbed end of Arp3. V-cortactin could potentially also deliver an actin monomer to the barbed end of Arp2, but the linear distance is much greater (107 versus 50 angstroms) and is unlikely to be spanned by the 43 amino acids between the end of the V to the tryptophan, especially considering that a fully extended conformation of this region would occupy excluded volume in the model. Therefore, these data indicate that V-cortactin activates the complex by recruiting a single actin monomer to Arp3. This is consistent with the results of Padrick et al. (39), who showed that recruitment of actin monomers to Arp3 is more important for activation than monomer recruitment to Arp2. However, as with WASP-VCA, recruitment of actin monomers to both sites may contribute to activation, because we found that fusion of the tandem V of N-WASP to the N terminus of cortactin (VV-cortactin) further stimulated NPF activity.
Cortactin Bundles Branched Actin Filaments to Create Actin Networks Distinct from N-WASP
Recent observations indicate that filament bundling by fimbrin plays a role in modulating the function of Arp2/3-nucleated branched networks in vivo (65). These observations suggest that other bundlers, including cortactin, may also influence branched filament networks in vivo. However, we showed that the bundling potency of cortactin is reduced in the presence of N-WASP, presumably because increased nucleation rates create shorter branches that cannot bundle. Therefore, if cortactin-mediated bundling is important in vivo, its bundling activity is likely tuned by the presence of other actin regulators that influence branch density and filament lengths, including capping protein (66). During endocytosis, bundling may also be influenced by dynamin, which is thought to oligomerize cortactin, thereby directly regulating its bundling activity (32, 67).
Dynamic Multivalent Contacts with Actin Filaments Can Explain Bundling by Cortactin
Our data demonstrate that multiple actin binding regions are harbored within the 6.5 repeats of cortactin. We propose a model in which a cortactin molecule binds to the side of a filament, with individual repeats dynamically binding and releasing the filament. When an adjacent filament collides with the bound cortactin, one or more of the filament binding repeats release from the parent filament and interact with the bundling partner to bridge the filaments, leaving at least one repeat bound to the parent filament (Fig. 4B). Several lines of evidence support this model. First, chemical cross-linking and circular dichroism experiments demonstrate that the repeats are dynamic and unstructured in the absence of actin (68). Flexibility within the repeat region would allow it to dynamically switch between single and multiple filament binding configurations. Second, the binding stoichiometry of cortactin to actin filaments is approximately five actin subunits to one cortactin, consistent with the idea that multiple actin subunits are contacted by a single filament binding repeat region and that each repeat contacts approximately one subunit (69). Third, we showed that the off rate of cortactin for single filaments is identical to its off rate for bundles, and that the apparent affinity of cortactin for single filaments is similar to its apparent affinity for bundles. This suggests that the strength of contacts made by the repeat region is similar whether cortactin is bound to a single or bundled filament. This could occur if repeats act as flexibly connected binding modules, and the total strength of the interaction is additive and tuned by the total number of repeats attached, whether to a single or bundled filaments. This mode of interaction may allow both the actin filament binding and bundling activities of cortactin to be regulated by alternative splicing, which generates isoforms of cortactin that have between 2.5 and 6.5 repeats (70). We note that our data are inconsistent with experiments suggesting that the fourth repeat is entirely responsible for the actin binding affinity of cortactin (18) and with small angle x-ray scattering studies and modeling exercises that led to the hypothesis that the repeats fold into a stable globular domain (31).
The Arp2/3 Complex Binding Regions of Cortactin and WASP Are Functionally Distinct
NtA versus A sequence differences likely contribute to the distinct binding site specificities of NtA versus WASP for the complex, providing one explanation for the functional differences we observed here. Biochemical data suggest that WASP binds more tightly to the NPF site that spans Arp2 and ARPC1, with an ∼100-fold weaker affinity for the Arp3 NPF site, which is the only site bound by the cortactin NtA (24, 40, 41). However, simple competition with dimeric WASP for binding to the Arp3 site cannot explain synergy, because at high concentrations CA and A can bind to both sites, yet we did not observe synergistic activation of the Arp2/3 complex even at very high concentrations of CA or A. Understanding the molecular bases for the functional differences between NtA and A will likely require high resolution structural information showing how both WASP-A and NtA bind to the complex.
Our comparisons of WASP A versus NtA segments have implications for understanding diverse regulators of Arp2/3 complex. In addition to NPFs like WASP and cortactin, the A region is found in several inhibitors of Arp2/3 complex, including PICK1 (71), Gadkin (72), and Arpin (73). These proteins block branching nucleation by competing with WASP family proteins for binding to the complex. However, unlike cortactin, none of these proteins have been observed to synergize with WASP family proteins in activation of Arp2/3 complex. Our data suggest that the A regions of these proteins may lack specific features present in cortactin NtA required for potent synergy.
The Presence of Tandem V Regions in N-WASP Hinders Its Synergy with Cortactin
We showed that the ability to synergistically activate Arp2/3 complex with cortactin is a general feature of WASP family proteins, but that the potency of different WASP family proteins varies widely. Native N-WASP had the weakest synergistic activity, whether monomeric or dimerized. The weak synergy of native N-WASP is due to the presence of two V regions, because deletion of one V dramatically increased the potency of dimeric N-WASP in synergistic activation of Arp2/3 complex (23) (Fig. 7, A, B, and G). How tandem actin monomer recruitment regions could influence synergy with cortactin can be understood in the context of the displacement mechanism. In this mechanism, the rate of WASP release from the nascent branch junction limits or partially limits the rate of branching nucleation (23, 46). By displacing WASP from the nascent branch junction, cortactin increases branching rates. Therefore, the displacement mechanism predicts an inverse correlation between the intrinsic activity of WASP family proteins at saturation (which likely reflects the limiting release rate) and the fold increase in polymerization rate because of addition of cortactin. We previously observed this correlation for point mutations within the V region of GST-N-WASP-VCA (23). Here we observe this correlation when comparing diverse WASP family proteins. For instance, GST-N-WASP-VVCA shows a much higher intrinsic NPF activity and a much lower synergy with cortactin compared with GST-N-WASP-VCA (23) (Fig. 7, A, B, and G). These observations suggest that GST-N-WASP-VVCA shows decreased synergistic activation because it is released more rapidly from the nascent branch than GST-N-WASP-VCA. Although we do not know how the number of V regions could influence the WASP release rate from nascent branches, one possibility is that recruitment of additional actin monomers helps stimulate a conformational change that accelerates WASP release and nucleation. Finally, we note that although we observed an inverse correlation between intrinsic NPF activity of WASP proteins to their synergy with cortactin, it was unexpected that the synergy of cortactin with GST-WAVE-VCA was not significantly greater than with GST-WASP-VCA. This observation may indicate that the intrinsic NPF activity at saturation is a relatively crude indicator of potential synergy with cortactin for reasons that we do not currently understand.
All WASP Family Proteins May Use a Similar Mechanism to Synergistically Activate Arp2/3 Complex with Cortactin
Our previous data indicate that release of dimeric but not monomeric N-WASP from the nascent branch junction is rate-limiting, explaining why dimerization of WASP proteins is required for synergy with cortactin. Here we showed that multiple WASP family proteins synergize with cortactin to activate Arp2/3 complex, but that in each case potent synergy required dimerization of the WASP protein. These data suggest that all WASP family proteins use the displacement mechanism to synergize with cortactin. This finding has important implications for understanding the role of cortactin in assembling branched actin networks in diverse cellular structures. WASP, N-WASP, and WAVE are frequently attached to cellular membranes, allowing them to initiate branched actin networks that provide pushing forces to move or remodel cellular membranes (74). WASP proteins are thought to provide a transient connection between membranes and the polymerizing actin networks by binding the barbed ends of polymerizing filaments, nascent branch junctions, or both (75, 76). Using the displacement mechanism to synergistically activate Arp2/3 complex with WASP family proteins, cortactin can not only increase rates of branching nucleation but may also decrease the lifetime of WASP-mediated connections between the polymerizing network and the membrane (22). The importance of WASP-mediated connections to polymerizing actin networks has been demonstrated using in vitro bead motility assays (75), and an important question to resolve will be the role of these connections in modulating actin network-membrane interactions in vivo.
Supplementary Material
Acknowledgments
We thank Byron Hetrick and Su Ling-Liu for help with N-WASP and Arp2/3 complex purifications, as well as Bruce Bowerman and Chris Doe for use of their microscope and Raghuveer Parthasarathy for the particle tracking script.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM092917 (to B. J. N.) and Predoctoral Training Grant GM007759 (to L. A. H.). This work was also supported by American Heart Association Grant 10SDG2610189 (to B. J. N.) and the NICHD Summer Research Program at the University of Oregon through National Institutes of Health Grant R25HD070817 (to J. G. P.).

This article contains supplemental Movies S1–S7.
- NPF
- nucleation promoting factor
- N-WASP
- neural WASP
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1. Pollard T. D., Cooper J. A. (2009) Actin, a central player in cell shape and movement. Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blanchoin L., Pollard T. D., Mullins R. D. (2000) Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10, 1273–1282 [DOI] [PubMed] [Google Scholar]
- 3. Wagner A. R., Luan Q., Liu S.-L., Nolen B. J. (2013) Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr. Biol. 23, 1990–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Higgs H. N., Pollard T. D. (2001) Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70, 649–676 [DOI] [PubMed] [Google Scholar]
- 5. Goley E. D., Welch M. D. (2006) The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726 [DOI] [PubMed] [Google Scholar]
- 6. Kaksonen M., Toret C. P., Drubin D. G. (2006) Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7, 404–414 [DOI] [PubMed] [Google Scholar]
- 7. Stradal T. E., Rottner K., Disanza A., Confalonieri S., Innocenti M., Scita G. (2004) Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 14, 303–311 [DOI] [PubMed] [Google Scholar]
- 8. Nürnberg A., Kitzing T., Grosse R. (2011) Nucleating actin for invasion. Nat. Rev. Cancer 11, 177–187 [DOI] [PubMed] [Google Scholar]
- 9. Mizutani K., Miki H., He H., Maruta H., Takenawa T. (2002) Essential role of neural Wiskott-Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Res. 62, 669–674 [PubMed] [Google Scholar]
- 10. Ayala I., Baldassarre M., Giacchetti G., Caldieri G., Tetè S., Luini A., Buccione R. (2008) Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J. Cell Sci. 121, 369–378 [DOI] [PubMed] [Google Scholar]
- 11. Oser M., Yamaguchi H., Mader C. C., Bravo-Cordero J. J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A. J., Condeelis J. (2009) Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186, 571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sirotkin V., Berro J., Macmillan K., Zhao L., Pollard T. D. (2010) Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol. Biol. Cell 21, 2894–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galletta B. J., Chuang D. Y., Cooper J. A. (2008) Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 6, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Basu R., Chang F. (2011) Characterization of dip1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr. Biol. 21, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu S.-L., Needham K. M., May J. R., Nolen B. J. (2011) Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J. Biol. Chem. 286, 17039–17046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marchand J.-B., Kaiser D. A., Pollard T. D., Higgs H. N. (2001) Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 3, 76–82 [DOI] [PubMed] [Google Scholar]
- 17. Hetrick B., Han M. S., Helgeson L. A., Nolen B. J. (2013) Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 20, 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. (2000) Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 151, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uruno T., Liu J., Zhang P., Fan Y., Egile C., Li R., Mueller S. C., Zhan X. (2001) Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 3, 259–266 [DOI] [PubMed] [Google Scholar]
- 20. Weaver A. M., Karginov A. V., Kinley A. W., Weed S. A., Li Y., Parsons J. T., Cooper J. A. (2001) Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370–374 [DOI] [PubMed] [Google Scholar]
- 21. Uruno T., Liu J., Li Y., Smith N., Zhan X. (2003) Sequential interaction of Actin-related proteins 2 and 3 (Arp2/3) complex with neural Wiscott-Aldrich syndrome protein (N-WASP) and cortactin during branched actin filament network formation. J. Biol. Chem. 278, 26086–26093 [DOI] [PubMed] [Google Scholar]
- 22. Siton O., Ideses Y., Albeck S., Unger T., Bershadsky A. D., Gov N. S., Bernheim-Groswasser A. (2011) Cortactin releases the brakes in actin-based motility by enhancing WASP-VCA detachment from Arp2/3 branches. Curr. Biol. 21, 2092–2097 [DOI] [PubMed] [Google Scholar]
- 23. Helgeson L. A., Nolen B. J. (2013) Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. Elife 2, e00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weaver A. M., Heuser J. E., Karginov A. V., Lee W. L., Parsons J. T., Cooper J. (2002) Interaction of cortactin and N-WASp with Arp2/3 complex. Curr. Biol. 12, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 25. Tehrani S., Faccio R., Chandrasekar I., Ross F. P., Cooper J. A. (2006) Cortactin has an essential and specific role in osteoclast actin assembly. Mol. Biol. Cell 17, 2882–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martinez-Quiles N., Ho H.-Y., Kirschner M. W., Ramesh N., Geha R. S. (2004) Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell Biol. 24, 5269–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taylor M. J., Perrais D., Merrifield C. J. (2011) A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9, e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu H., Parsons J. T. (1993) Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J. Cell Biol. 120, 1417–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Machesky L. M., Insall R. H. (1998) Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347–1356 [DOI] [PubMed] [Google Scholar]
- 30. Huang C., Ni Y., Wang T., Gao Y., Haudenschild C. C., Zhan X. (1997) Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J. Biol. Chem. 272, 13911–13915 [DOI] [PubMed] [Google Scholar]
- 31. Cowieson N. P., King G., Cookson D., Ross I., Huber T., Hume D. A., Kobe B., Martin J. L. (2008) Cortactin adopts a globular conformation and bundles actin into sheets. J. Biol. Chem. 283, 16187–16193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamada H., Abe T., Satoh A., Okazaki N., Tago S., Kobayashi K., Yoshida Y., Oda Y., Watanabe M., Tomizawa K., Matsui H., Takei K. (2013) Stabilization of actin bundles by a dynamin 1/cortactin ring complex is necessary for growth cone filopodia. J. Neurosci. 33, 4514–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaksonen M., Peng H. B., Rauvala H. (2000) Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J. Cell Sci. 113, 4421–4426 [DOI] [PubMed] [Google Scholar]
- 34. Wang Z. (1995) An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 360, 111–114 [DOI] [PubMed] [Google Scholar]
- 35. Liu S.-L., May J. R., Helgeson L. A., Nolen B. J. (2013) Insertions within the actin core of actin-related protein 3 (Arp3) modulate branching nucleation by Arp2/3 complex. J. Biol. Chem. 288, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. (2010) Computer control of microscopes using μManager. Curr. Protoc. Mol. Biol. Unit 14.20.1–14.20.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuhn J. R., Pollard T. D. (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boczkowska M., Rebowski G., Petoukhov M. V., Hayes D. B., Svergun D. I., Dominguez R. (2008) X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure 16, 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Padrick S. B., Doolittle L. K., Brautigam C. A., King D. S., Rosen M. K. (2011) Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. U.S.A. 108, E472–E479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ti S.-C., Jurgenson C. T., Nolen B. J., Pollard T. D. (2011) Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 108, E463–E471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boczkowska M., Rebowski G., Kast D. J., Dominguez R. (2014) Structural analysis of the transitional state of Arp2/3 complex activation by two actin-bound WCAs. Nat. Commun. 5, 3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamaguchi H., Miki H., Suetsugu S., Ma L., Kirschner M. W., Takenawa T. (2000) Two tandem verprolin homology domains are necessary for a strong activation of Arp2/3 complex-induced actin polymerization and induction of microspike formation by N-WASP. Proc. Natl. Acad. Sci. U.S.A. 97, 12631–12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Svitkina T. M., Borisy G. G. (1999) Organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Collins A., Warrington A., Taylor K. A., Svitkina T. (2011) Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr. Biol. 21, 1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cao H., Orth J. D., Chen J., Weller S. G., Heuser J. E., McNiven M. A. (2003) Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol. Cell Biol. 23, 2162–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith B. A., Padrick S. B., Doolittle L. K., Daugherty-Clarke K., Corrêa I. R., Jr., Xu M.-Q., Goode B. L., Rosen M. K., Gelles J. (2013) Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife 2, e01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Padrick S. B., Cheng H.-C., Ismail A. M., Panchal S. C., Doolittle L. K., Kim S., Skehan B. M., Umetani J., Brautigam C. A., Leong J. M., Rosen M. K. (2008) Hierarchical regulation of WASP/WAVE proteins. Mol. Cell 32, 426–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. (1999) The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97, 221–231 [DOI] [PubMed] [Google Scholar]
- 50. Hüfner K., Higgs H. N., Pollard T. D., Jacobi C., Aepfelbacher M., Linder S. (2001) The verprolin-like central (vc) region of Wiskott-Aldrich syndrome protein induces Arp2/3 complex-dependent actin nucleation. J. Biol. Chem. 276, 35761–35767 [DOI] [PubMed] [Google Scholar]
- 51. Destaing O., Saltel F., Geminard J.C., Jurdic P., Bard F. (2003) Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol. Biol. Cell 14, 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bowden E. T., Barth M., Thomas D., Glazer R. I., Mueller S. C. (1999) An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene 18, 4440–4449 [DOI] [PubMed] [Google Scholar]
- 53. Desmarais V., Yamaguchi H., Oser M., Soon L., Mouneimne G., Sarmiento C., Eddy R., Condeelis J. (2009) N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells. Cell Motil. Cytoskeleton 66, 303–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q.-X., Nixon B. T., Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suetsugu S. (2013) Activation of nucleation promoting factors for directional actin filament elongation: allosteric regulation and multimerization on the membrane. Semin. Cell Dev. Biol. 24, 267–271 [DOI] [PubMed] [Google Scholar]
- 56. Gohl C., Banovic D., Grevelhörster A., Bogdan S. (2010) WAVE forms hetero- and homo-oligomeric complexes at integrin junctions in Drosophila visualized by bimolecular fluorescence complementation. J. Biol. Chem. 285, 40171–40179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Murphy D. A., Courtneidge S. A. (2011) The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bryce N. S., Clark E. S., Leysath J. L., Currie J. D., Webb D. J., Weaver A. M. (2005) Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 15, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 59. Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. (2003) Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kinley A. W., Weed S. A., Weaver A. M., Karginov A. V., Bissonette E., Cooper J. A., Parsons J. T. (2003) Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr. Biol. 13, 384–393 [DOI] [PubMed] [Google Scholar]
- 61. Sirotkin V., Beltzner C. C., Marchand J.-B., Pollard T. D. (2005) Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J. Cell Biol. 170, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Panchal S. C., Kaiser D. A., Torres E., Pollard T. D., Rosen M. K. (2003) A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat. Struct. Biol. 10, 591–598 [DOI] [PubMed] [Google Scholar]
- 63. Xu X.-P., Rouiller I., Slaughter B. D., Egile C., Kim E., Unruh J. R., Fan X., Pollard T. D., Li R., Hanein D., Volkmann N. (2012) Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 31, 236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goley E. D., Rodenbusch S. E., Martin A. C., Welch M. D. (2004) Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol. Cell 16, 269–279 [DOI] [PubMed] [Google Scholar]
- 65. Skau C. T., Courson D. S., Bestul A. J., Winkelman J. D., Rock R. S., Sirotkin V., Kovar D. R. (2011) Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J. Biol. Chem. 286, 26964–26977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu J., Casella J. F., Pollard T. D. (1999) Effect of capping protein, CapZ, on the length of actin filaments and mechanical properties of actin filament networks. Cell Motil. Cytoskeleton 42, 73–81 [DOI] [PubMed] [Google Scholar]
- 67. Mooren O. L., Kotova T. I., Moore A. J., Schafer D. A. (2009) Dynamin2 GTPase and cortactin remodel actin filaments. J. Biol. Chem. 284, 23995–24005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shvetsov A., Berkane E., Chereau D., Dominguez R., Reisler E. (2009) The actin-binding domain of cortactin is dynamic and unstructured and affects lateral and longitudinal contacts in F-actin. Cell Motil. Cytoskeleton 66, 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. MacGrath S. M., Koleske A. J. (2012) Arg/Abl2 modulates the affinity and stoichiometry of binding of cortactin to F-actin. Biochemistry 51, 6644–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Katsube T., Togashi S., Hashimoto N., Ogiu T., Tsuji H. (2004) Filamentous actin binding ability of cortactin isoforms is responsible for their cell-cell junctional localization in epithelial cells. Arch. Biochem. Biophys. 427, 79–90 [DOI] [PubMed] [Google Scholar]
- 71. Rocca D. L., Martin S., Jenkins E. L., Hanley J. G. (2008) Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat. Cell Biol. 10, 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maritzen T., Zech T., Schmidt M. R., Krause E., Machesky L. M., Haucke V. (2012) Gadkin negatively regulates cell spreading and motility via sequestration of the actin-nucleating ARP2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 109, 10382–10387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dang I., Gorelik R., Sousa-Blin C., Derivery E., Guérin C., Linkner J., Nemethova M., Dumortier J. G., Giger F. A., Chipysheva T. A., Ermilova V. D., Vacher S., Campanacci V., Herrada I., Planson A.-G., Fetics S., Henriot V., David V., Oguievetskaia K., Lakisic G., Pierre F., Steffen A., Boyreau A., Peyriéras N., Rottner K., Zinn-Justin S., Cherfils J., Bièche I., Alexandrova A. Y., David N. B., Small J. V., Faix J., Blanchoin L., Gautreau A. (2013) Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503, 281–284 [DOI] [PubMed] [Google Scholar]
- 74. Takenawa T., Suetsugu S. (2007) The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 75. Co C., Wong D. T., Gierke S., Chang V., Taunton J. (2007) Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell 128, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Akin O., Mullins R. D. (2008) Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.