Background: HIV-1 Nef inhibits activity of ABCA1 and suppresses cholesterol efflux.
Results: Nef binds to calnexin and disrupts its interaction with ABCA1 but enhances calnexin interaction with viral gp160.
Conclusion: Nef regulates activity of calnexin to favor maturation of HIV gp160 at the expense of ABCA1.
Significance: Learning how Nef regulates activity of calnexin is crucial for designing therapeutic strategies aimed at treating HIV infection and HIV-associated atherosclerosis.
Keywords: Atherosclerosis, Cholesterol Regulation, Confocal Microscopy, Endoplasmic Reticulum (ER), Human Immunodeficiency Virus (HIV)
Abstract
HIV-infected patients are at increased risk of developing atherosclerosis, in part due to an altered high density lipoprotein profile exacerbated by down-modulation and impairment of ATP-binding cassette transporter A1 (ABCA1) activity by the HIV-1 protein Nef. However, the mechanisms of this Nef effect remain unknown. Here, we show that Nef interacts with an endoplasmic reticulum chaperone calnexin, which regulates folding and maturation of glycosylated proteins. Nef disrupted interaction between calnexin and ABCA1 but increased affinity and enhanced interaction of calnexin with HIV-1 gp160. The Nef mutant that did not bind to calnexin did not affect the calnexin-ABCA1 interaction. Interaction with calnexin was essential for functionality of ABCA1, as knockdown of calnexin blocked the ABCA1 exit from the endoplasmic reticulum, reduced ABCA1 abundance, and inhibited cholesterol efflux; the same effects were observed after Nef overexpression. However, the effects of calnexin knockdown and Nef on cholesterol efflux were not additive; in fact, the combined effect of these two factors together did not differ significantly from the effect of calnexin knockdown alone. Interestingly, gp160 and ABCA1 interacted with calnexin differently; although gp160 binding to calnexin was dependent on glycosylation, glycosylation was of little importance for the interaction between ABCA1 and calnexin. Thus, Nef regulates the activity of calnexin to stimulate its interaction with gp160 at the expense of ABCA1. This study identifies a mechanism for Nef-dependent inactivation of ABCA1 and dysregulation of cholesterol metabolism.
Introduction
HIV-1 infection is associated with a high risk of developing atherosclerosis (1), and studies in animal models and in vitro demonstrated that Nef-mediated impairment of ATP-binding cassette transporter A1 (ABCA1)3 function may contribute to this side effect of HIV infection (2–4). ABCA1 mediates transport of cholesterol and phospholipids to lipid-poor apolipoprotein A-I (apoA-I), which is the initial step in the process of reverse cholesterol transport and HDL formation (5). Nef-mediated inactivation of ABCA1 causes accumulation of cholesterol in macrophages, increases the abundance of lipid rafts (sites of HIV assembly), and elevates cholesterol content of viral membranes, thus increasing HIV production and infectivity (2, 6–8). Another consequence of this activity is impairment of HDL maturation (3), which may be responsible for low HDL levels, an established risk factor for atherosclerosis in HIV-infected patients (9).
Nef down-modulates a number of cell surface transmembrane proteins, such as CD4, MHC-I, MHC-II, transferrin receptor, mannose receptor, CD80, CD86, CD8, and CCR5, by modifying their intracellular trafficking and targeting them to various degradation pathways (10, 11). The common mechanism behind this activity of Nef is binding to the target protein and connecting it to adaptor proteins, which direct the complex to degradation pathways. Nef has been shown to interact with ABCA1 (2, 12); however, this interaction was dispensable for Nef-mediated ABCA1 down-modulation and functional impairment (13), suggesting that an alternative mechanism is responsible for target protein inactivation in this case. In this study, we demonstrate that Nef affects ABCA1 activity by disrupting its interaction with calnexin.
Calnexin is an integral ER membrane calcium-binding lectin-like chaperone that binds immature glycoproteins destined for the plasma membrane, and supports folding and maturation of these proteins. Calnexin and calreticulin function as partner chaperones in the ER protein production, stabilization, and folding cycle (14). Calnexin is membrane-bound and chaperones predominantly membrane glycoproteins, whereas calreticulin is soluble and chaperones soluble proteins, although association between calreticulin and HIV-1 gp160 (a transmembrane protein) has been reported (15). It has been suggested that calnexin may stabilize glycoproteins through a dual-binding mode involving glycan and peptide binding or through either single binding mode (glycan residue or peptide binding) independently (16, 17). Although interaction between ABCA1 and calnexin has not been characterized, and the role of calnexin in ABCA1 functionality remains unknown, members of nearly every other subclass of ABC transporters, including ABCB1 (P-glycoprotein, MDR1), ABCB2 (TAP1), ABCB3 (TAP2), ABCC2, ABCC7 (CFTR), and ABCG5/G8, have been demonstrated to interact with calnexin, and among these are examples of glycan-dependent and -independent interactions (18–23). Importantly, calnexin was shown to contribute to the functionality of ABC transporters, as exemplified by CFTR and P-glycoproteins, which lost their functional activity when interaction with calnexin was impaired by mutations (24, 25).
Calnexin has also been shown to interact with HIV-1 gp160 (15, 26, 27). Interaction between gp160 and calnexin was found to be dependent on N-linked glycosylation of gp160 and was inhibited by tunicamycin (26). By analogy with its role in maturation of other glycosylated proteins in the ER, calnexin is assumed to assist folding of gp160 (28), although a direct effect of calnexin on gp160 functionality has not been demonstrated.
Here, we provide evidence that functionality of ABCA1 depends on calnexin. Nef binds calnexin and disrupts its interaction with ABCA1. This novel mechanism of action contributes to the effect of Nef on cellular cholesterol efflux.
EXPERIMENTAL PROCEDURES
Reagents
The following reagents were purchased from the indicated suppliers: mass spectrometry grade trypsin gold (Promega); Metafectene® (Biontex); rabbit anti-calnexin antibody (Assay Designs); mouse and rabbit anti-ABCA1 antibodies (Novus Biologicals); mouse monoclonal anti-calnexin-ER membrane marker antibody (Abcam); Alexa Fluor® 647 goat anti-rabbit IgG (H+L) antibody; DAPI di-lactate (Molecular Probes, Invitrogen); DyLight 550 goat anti-mouse IgG antibody (MyBiosource); goat anti-HA antibody (Genscript); mouse anti-LCB1 (SPTLC1) (BD Transduction Laboratories); control siRNA, calnexin siRNA, siRNA transfection medium, rabbit anti-MHC-I antibody, rabbit anti-calnexin (H-70) antibody, and goat anti-calnexin (C-20) antibody (Santa Cruz Biotechnology); [1,2-3H]cholesterol (PerkinElmer Life Sciences); human lipid-free apolipoprotein A-I (Biodesign); tunicamycin (Calbiochem); normal rabbit serum and ChromPure goat IgG (Jackson ImmunoResearch); cell surface protein isolation kit (Pierce); rabbit anti-dystrophin antibody (Thermo-Pierce); human calnexin cDNA (Open Biosystems); anti-FLAG M2 affinity agarose gel, EZView red protein A affinity agarose gel, murine IgG agarose gel, mouse anti-actin antibody, mouse anti-β COP antibody, rabbit anti-FLAG antibody, rabbit anti-GAPDH antibody, cycloheximide, concanavalin A (ConA)-HRP, phorbol 12-myristate 13-acetate (PMA), cholecalciferol (vitamin D3), fatty acid-free bovine serum albumin (BSA), and all other chemicals were from Sigma. Mounting medium Fluoromount-G was purchased from SouthernBiotech. Polyclonal rabbit anti-Nef antisera, mouse monoclonal anti-HIV-1 p24, and HIV-specific immunoglobulins (HIV-IG) were all from the AIDS Research and Reference Reagent Program, National Institutes of Health.
Expression Constructs
Human calnexin cDNA construct with a C-terminal HA tag was prepared by standard PCR methods from calnexin cDNA clone (Open Biosystems) in the pHCMV3 vector (Genlantis). Nef expression vectors were prepared from pT7consnefhis6 (AIDS Research and Reference Reagent Program, National Institutes of Health) in pHCMV3 by PCR cloning. All constructs were verified by sequencing.
Cells and Transfection
HeLa-ABCA1-GFP cells stably expressing GFP-tagged ABCA1 (29) were a kind gift of Dr. A. Remaley. HeLa-ABCA1 cells were cultured in DMEM-C (10% fetal calf serum (FCS), penicillin, streptomycin, and l-glutamine) containing hygromycin B and G418. HEK293T cells were grown in RPMI-C (RPMI 1640, 10% FCS, penicillin, streptomycin, and l-glutamine) and transfected with cDNA using Metafectene according to the manufacturer's (Biontex) instructions. Following transfection, cells were incubated for 48 h prior to use in experiments. A similar protocol with slight modification was used to transfect HeLa-ABCA1-GFP cells with siRNA. Following passage and overnight culture, 1 × 106 cells were transfected with control or calnexin siRNA using siRNA-Metafectene complexes (16 μl of Metafectene mixed with 8 μl of 10 μm siRNA) prepared in separate 100-μl aliquots of serum-free siRNA transfection medium (STM; Santa Cruz Biotechnology). Transfection complexes were incubated for 1 h in the dark, diluted with 800 μl of STM for a final siRNA concentration of 80 nm, and added to cells pre-washed with serum-free DMEM. Cells were incubated with transfection complexes in serum-free STM for 4 h followed by addition of DMEM-C with 2× serum and further overnight incubation. The following day, media were changed to DMEM-C, and cells were incubated for an additional 48–72 h prior to use in experiments.
THP-1 cells were transduced using lentiviral particles carrying calnexin or control shRNA sequences (Santa Cruz Biotechnology). Cells were inoculated with 50,000 infectious units of HIV particles pseudotyped with VSV-G glycoprotein in a 24-well plate setting. The following day, medium containing infectious particles was removed, and cells were cultured in RPMI-C for 2 days. Transduced cells were then selected in RPMI-C containing 4 μg/ml puromycin, and calnexin knockdown was verified by Western blot. Viable cells at the end of selection were continuously cultured in complete medium containing 2.5 μg/ml puromycin for use in experiments. Nef transfection of THP-1 cells was performed using Lipofectamine LTX reagent according to the manufacturer's recommendations, with the exception that transfected cells were cultured for 120 h prior to using in cholesterol efflux assay.
High Resolution LTQ ESI LC-MS/MS Mass Spectrometry
Total cellular protein (5 mg) from HEK293T cells co-transfected with ABCA1-FLAG and pcDNA-Nef or ABCA1-FLAG and empty control vector (pcDNA) was immunoprecipitated with M2 anti-FLAG affinity gel, and co-purifying proteins were identified by tandem MS/MS. The SeeBlue Plus2 pre-stained standard (Invitrogen) with proteins of known identity was run alongside experimental samples and processed in a similar fashion as an internal control for peptide processing and performance of the mass spectrometer. Following one-dimensional gel electrophoresis and staining with GelCode Blue dye (Thermo Pierce), unique bands were excised, and stain was removed via incubation in distilled water. Gel pieces were washed via sequential dehydration with acetonitrile and rehydration in 100 mm NH4HCO3. Gel slices were dried and hydrated on ice in Promega sequencing grade trypsin gold (12.5 ng/μl, 10–20 μl) in 50 mm NH4HCO3 for 45 min, and then a saturating volume of 50 mm ammonium bicarbonate was added, and the tubes were incubated for 16 h at 37 °C. The peptide solutions were extracted with 25 mm NH4HCO3 (25 μl, 15 min), and an equal volume of acetonitrile was then added and incubated an additional 15 min. The supernatant was collected, and the gel pieces were further extracted with two washes with 5% formic acid/acetonitrile (1:1, 30 μl), and all extracts were pooled and lyophilized. The lyophilate was dissolved in 10 μl of trifluoroacetic acid (TFA), further desalted with a C18 ZipTip (Millipore), and loaded onto a C18 trap column followed by a C18 reverse phase column using an Eksigent nano-HPLC system. Peptides were introduced into the mass spectrometer via a 10-μm silica tip (New Objective Inc., Ringoes, NJ) adapted to a nano-electrospray source (ThermoFisher Scientific). The LTQ-Orbitrap XL (ThermoFisher Scientific) was operated in data-dependent mode with one full MS (300–2000 m/z) survey scan acquired in the Orbitrap with a resolution of 30,000, and subsequently five MS/MS scans were performed in the LTQ using collision-induced dissociation with the collision gas (helium) and normalized collision energy value set at 35%. Each file was searched for protein identification using the Sequest algorithm in the Bioworks Browser software (ThermoFisher Scientific) against the Uniprot all species database indexed for fully tryptic peptides and two missed cleavages. Peptide tolerance was set at 50 ppm and fragment ion tolerance at 1.0 Da. Variable modifications were oxidation of methionine and cysteine (15.995 Da). Results were filtered based on peptide probability score with a p value <0.01.
Immunoprecipitation
HEK293T cells were transfected with ABCA1-FLAG or empty vector DNA; 48 h after transfection, cells were washed with ice-cold PBS and lysed in Nonidet P-40 Lysis Buffer (1% Nonidet P-40, 150 mm NaCl, 2 mm EDTA, 50 mm Tris-HCl, pH 7.0, protease inhibitor mixture) on ice for 30 min. Lysates were centrifuged to remove debris (10 min, 13,000 × g, 4 °C), and the supernatants were transferred to fresh 1.5-ml Eppendorf tubes, and ABCA1 was precipitated with M2 anti-FLAG affinity gel overnight at 4 °C with rotation. Murine IgG agarose gel was used in an isotype control precipitation. Precipitates were washed three times with TBS (50 mm Tris-HCl, pH 7.0, 150 mm NaCl), and FLAG-ABCA1 complexes were eluted with FLAG peptide (0.5 mg/ml, 1 h, 4 °C). Supernatants were separated on SDS-polyacrylamide gels for immunoblotting or in-gel digestion analysis and MS/MS.
For immunoprecipitation analysis in THP-1 macrophages, 1 × 107 cells were differentiated for 72 h with 100 nm PMA/100 nm vitamin D3. Cells were washed with ice-cold PBS and lysed in Nonidet P-40 Lysis Buffer on ice for 30 min. Lysates were centrifuged to remove debris (10 min, 13,000 × g, 4 °C), and the supernatants were transferred to fresh 1.5-ml Eppendorf tubes and pre-cleared by incubation with EZView red protein A in the absence of antibody. Resulting supernatants were incubated overnight with rabbit anti-ABCA1 or normal rabbit serum capture antibodies. Immune complexes were precipitated with EZView red protein A affinity gel, washed with lysis buffer, boiled in sample buffer to elute bound protein, and separated on SDS-polyacrylamide gels for immunoblotting with goat anti-calnexin antibody.
Calnexin was immunoprecipitated from transfected cells or HIV-infected macrophages using the procedure described for ABCA1, except that rabbit anti-calnexin polyclonal antibody was used. Immunoprecipitates were blotted with anti-gp160, anti-Nef, or anti-ABCA1 antibodies.
Glycosylation Analysis
HEK293T cells were transfected with empty control vector (pcDNA) or ABCA1-FLAG. Glycosylation inhibitor tunicamycin (10 μg/ml) was added immediately prior to transfection and maintained throughout the experiment to ensure that ABCA1 remained deglycosylated. 24 h after transfection, cells were lysed, and ABCA1 was precipitated with M2 anti-FLAG affinity gel as described above. Anti-FLAG immunoprecipitate was probed with rabbit anti-calnexin or rabbit anti-ABCA1 antibody. Separate anti-FLAG immunoprecipitation reactions were probed with ConA-HRP per manufacturer's instructions (Sigma).
HIV Infection
Monocyte-derived macrophages were infected with VSV-G-pseudotyped pBRNL4.3_92BR020.4(R5)nef−_IRES_EGFP or pBRNL4.3_92BR020.4(R5)nef+_IRES_EGFP (30) using virus normalized by RT activity (200,000 cpm/106 cells). Infected cultures were incubated for 3 weeks, and the level of infection was monitored by RT assay and FACS analysis.
Cholesterol Efflux
HeLa-ABCA1-GFP cells were seeded in 24-well plates at 1 × 105 cells/well and transfected using Metafectene. 72 h post-transfection, cells were labeled with [3H]cholesterol in DMEM-C for 48 h. To remove non-cell-associated cholesterol, cells were washed extensively with serum-free DMEM followed by several washes with DMEM supplemented with 1% fatty acid-free BSA. To initiate efflux, cells were incubated in DMEM, 1% fatty acid-free BSA in the presence or absence of 30 μg/ml apoA-I for 3 h as described (31). Medium was collected from the cells and cleared of cells and debris by an 800 × g spin for 10 min. Cell layers were dissolved in 0.1 n NaOH. Radioactivity in media and cells was determined by scintillation counting, and cholesterol efflux was calculated as counts in media divided by total counts (cells + media) and converted to percentage efflux.
Cholesterol efflux from THP-1 cells was measured similarly to efflux from HeLa-ABCA1 cells with the following modifications. Following labeling with [3H]cholesterol, cells were extensively washed with PBS and incubated for 18 h in complete media in the presence of 1 μm TO-901317 and 100 ng/ml PMA.
Fluorescent Microscopy
For imaging, HeLa-ABCA1-GFP cells were grown on coverslips, which were further fixed after immunolabeling and mounted on microscopic glass slides using Fluoromount G. Images were initially captured with a Carl Zeiss LSM 710 confocal microscope, using a Plan Apochromat 100×/1.46 oil immersion lens (Carl Zeiss, i.e. Fig. 4). In this case, DAPI was excited with a 405 laser line, and the emission was filtered between 410 and 495 nm; DyLight 550-encoding calnexing labeling was excited with a 561 laser line, and the emission was filtered between 575 and 640 nm, and Alexa Fluor® 647 encoding Nef labeling was excited with a 633 laser line, and the emission was filtered between 638 and 755 nm. Precise analysis was performed on a Cell Observer Spinning Disk fluorescent microscope (Carl Zeiss) equipped with Yokogawa CSU X1 spinning disk and Evolve Delta EM CCD cameras (512 × 512, Photometrics), because of its superior sensitivity. The core of the instrument is a fully automated Axio Observer microscope (Carl Zeiss). The microscope has a high precision scanning stage (ASI MS-2000, Applied Scientific Instruments) integrated with stitching software module, allowing production of high resolution virtual slides from a large scanned area of the sample. Two objective lenses were used in these experiments. Glycerine immersion lens 150×/1.35 produced a pixel size of 0.089 μm, sufficient to match diffractions at the lowest wavelength emission according to Nyquist theorem (i.e. DAPI). This lens was used to acquire images from large tile-scan regions of the culture. For achieving three-dimensional image sets from the cell cultures expressing ABCA1-GFP and labeled for nuclei (DAPI), Nef (Alexa Fluor® 647), and calnexin (DyLight 550), Plan Apochromat 100×/1.46 oil objective lens was used, which produced a voxel size of 0.133 × 0.133 × 0.2 (X/Y/Z). The optical section thickness slightly varied between the channels as a function of the variable emission recorded, being 0.82 μm for DAPI, 0.85 μm for GFP, 0.89 μm for calnexin, and 0.92 μm for Nef. The utilization of high profile objectives and imaging close to the coverslip produced images with negligible spherical aberrations, allowing for reliable quantification of the co-localization. The camera exposure time for each channel and the emission and excitation parameters were kept constant across the experiments. DAPI was excited with a 405 diode laser, and the emission was recorded with 450/50 bandpass filter. GFP was excited with a 488 diode laser, and emission was recorded with 535/30 bandpass filter. To record the calnexin immunolabeling, a 561 diode laser was used for excitation, and the emission was recorded with a 629/62 emission filter. Finally, to record the channel representing Nef immunolabeling, a 639 diode laser was used for excitation, and emission was captured with a 690/50 bandpass filter. The combination of excitation and emission produced no detectable bleed-through on the individual channels. Also, as a quality control for co-localization studies, spectral imaging with photo-bleaching was used to detect bleed-through due to FRET using a Zeiss 710 confocal instrument. Consistent with increased distance due to introduction of a two-antibodies bridge between the epitope and the fluorochrome, negligible bleed-through was detected between ABCA1-GFP and calnexin (less than 5%), and if detected by chance it affected the emissions over 560 nm, which were cut on the spinning disk confocal instrument (535/30 emission). The ABCA1-GFP signal in cultured HeLa cells was captured from live cultures to estimate the ground levels of detection. For this purpose, the microscope was equipped with integrated on-stage incubator chamber. The chamber provides a constant temperature at 37 °C and is supplied with humidified 5% CO2. The definitive focus was used when necessary, which eliminates drifts during long term experiments.
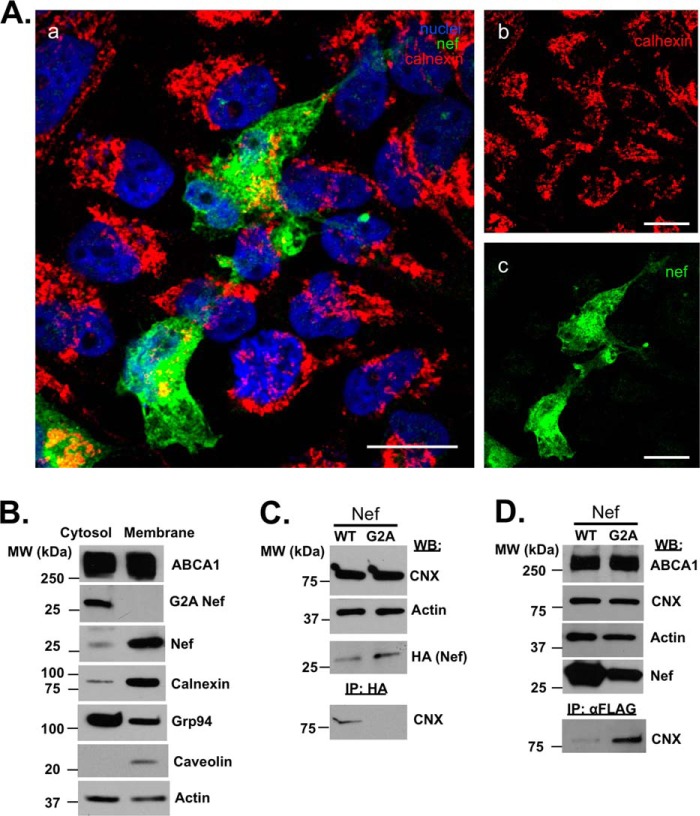
FIGURE 4.
Nef interacts with calnexin. A, confocal image representing HeLa-ABCA1-GFP cell cultures transfected with Nef. Cells were immunostained for calnexin (red channel) and Nef (green channel) using mouse monoclonal anti-calnexin/ER antibody followed by DyLight 550 goat anti-mouse IgG antibody and anti-HIV NefSF2 rabbit serum followed by Alexa Fluor® 647 goat anti-rabbit IgG antibody, respectively. Cell nuclei were stained by DAPI (blue channel). Panel a shows an overlay of calnexin/ER (red), Nef (green), and nuclei (blue). Pixels where Nef signal is overlapping with calnexin occur as orange-yellow. Single channels of calnexin/ER and Nef are presented in panels b and c, respectively. Bar sizes equal 16.6 μm. B, cytosolic and membrane fractions of HeLa-ABCA1-GFP cells transfected with WT or G2A Nef were separated by SDS-PAGE and blotted with anti-ABCA1, anti-calnexin, anti-Grp94, anti-caveolin, or anti-β actin monoclonal antibodies or anti-Nef polyclonal antibody. C, cell lysates of HEK293T cells transfected with HA-tagged wild-type (WT) Nef or G2A Nef were blotted for calnexin, actin, and HA (upper panels), and immunoprecipitates obtained using anti-HA antibody were blotted for calnexin (bottom panel). D, HEK293T cells were transfected with ABCA1-FLAG and either WT Nef or NefG2A. Cell lysates were analyzed for ABCA1, calnexin, actin, and Nef using monoclonal antibodies (upper panels), and anti-FLAG immunoprecipitates (IP) were blotted for calnexin (bottom panel). WB, Western blot.
Quantitative co-localization was assessed using Volocity software (PerkinElmer Life Sciences). The three-dimensional image sets were used for this purpose. For co-localization, the three-dimensional image sets were first subjected to intensity threshold to eliminate the dark current registered at the image, followed by extracting the product of the differences from the mean (PDM). Positive PDM was determined for a single cell, in which pixel intensities of DyLight 550 and GFP or Alexa Fluor® 647 vary synchronously, and more positive PDM indicates a stronger degree of co-localization. We used the positive PDM as an indicator for co-localizing pixels, as by definition, these pixels represent higher than the mean pixel value (over threshold) for both channels and reduce the probability of including adjacent structures in the co-localization outcome. Considering low spherical aberration in our three-dimensional image sets, the likelihood that co-localization is calculated when structures are above each other along the z axis is negligible. Additional channels representing PDM co-localization were plotted in Volocity in addition to GFP (ABCA1), DyLight 550 (CNX/ER), and Alexa Fluor® 647 (Nef) channels and were used for evaluation of spatial segregation between ABCA1, calnexin, and Nef. Volocity software was also employed for the quantitative determination of the extent of co-localization of ABCA1 with calnexin and Nef with calnexin using Manders' correlation coefficients (MCCs). Regions of interest were selected using the freehand selection tool. Two different MCC values (M1 and M2) describe the independent contributions of two selected channels to the pixels of interest; M1 and M2 coefficients provide the portion of the pixel intensity in each channel that coincides with some intensity in the other channel. MCC values range from zero (uncorrelated distributions of two probes) to one (perfect co-localization of the two images). To compute the cellular intensities for ABCA1, calnexin, and Nef, large tile scans taken with ×150 objective were used. Total field size was ∼1.16 × 570 μm, composed of adjacent stitched images taken at pixel resolution of 0.089 μm. Random cells were outlined by a hand tool, and overall cellular intensity for each channel was plotted.
RESULTS
ABCA1 Physically Interacts with Calnexin
To identify ABCA1-interacting proteins that may be affected by Nef, full-length wild-type ABCA1 with a FLAG tag at the C terminus (ABCA1-FLAG) was transfected into HEK293T cells alone or co-transfected with wild-type Nef. Mock-transfected cells served as control. Cell lysates from transfected cells were immunoprecipitated with anti-FLAG affinity gel and subjected to one-dimensional PAGE followed by MS/MS of differing bands, focusing on differences between cells transfected or not with Nef. Peptides from several previously described ABCA1-interacting proteins, including dystrophin and utrophin, were specifically identified in both Nef-positive and Nef-negative ABCA1-FLAG-transfected cells compared with mock-transfected cells (Table 1). Another protein with significant peptide coverage identified specifically in the Nef-negative ABCA1-FLAG-transfected cells was calnexin, but it was not found in Nef-positive cells (Table 1). Given that calnexin functions as an ER chaperone and contributes to the maturation of many ABC transporters, we focused on the ABCA1-calnexin interaction as a potential target of Nef.
TABLE 1.
Proteins that co-purified with ABCA1
| Co-purified protein | No. of peptide matches (control cells) | Coverage in control cells | No. of peptide matches (Nef-transfected cells) | Calculated molecular mass | Accession no. |
|---|---|---|---|---|---|
| % | |||||
| Utrophin | 66 | 26.1 | 30 | 394,247.4 | P46939 |
| Dystrophin | 1 | 0.4 | 1 | 399,153.8 | Q9VDW6 |
| Calnexin | 21 | 30.9 | 0 | 67,526.0 | P27824 |
| Filamin | 53 | 16.6 | 44 | 280,561.4 | P21333 |
| Kinesin-like protein KIF11 | 38 | 24.6 | 33 | 119,084.9 | P52732 |
To confirm physical interaction between ABCA1 and calnexin, HEK293T cells were transfected with ABCA1-FLAG, host cell factor-FLAG, or FLAG (Fig. 1A), and anti-FLAG precipitates were immunoblotted for calnexin (Fig. 1A, bottom panel). Calnexin specifically co-precipitated with ABCA1-FLAG but not with a control HCF-FLAG or FLAG. To demonstrate ABCA1-calnexin interaction in physiological conditions, we used differentiated THP-1 cells, a model of tissue macrophages. Calnexin was highly expressed in THP-1 cells regardless of differentiation status, whereas significant up-regulation of ABCA1 occurred upon differentiation of THP-1 cells into macrophage-like cells after treatment with PMA/vitamin D3 (Fig. 1B). Upon ABCA1 immunoprecipitation from lysates of differentiated THP-1 macrophages, calnexin co-immunoprecipitation could be detected by immunoblotting (Fig. 1B, bottom panel). Additional support for the close proximity of ABCA1 and calnexin came from analysis of HeLa cells stably transfected with GFP-fused ABCA1 (29) by confocal microscopy, which demonstrated co-localization of a portion of ABCA1 molecules with calnexin (orange staining in Fig. 1C, panel a). These results suggest a physical interaction between ABCA1 and calnexin in the endoplasmic reticulum.
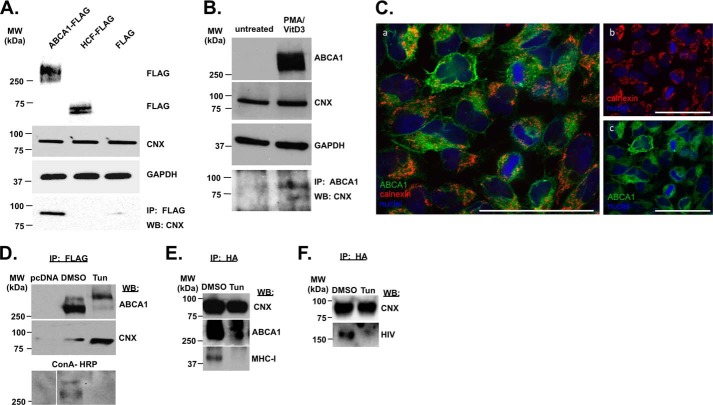
FIGURE 1.
ABCA1 interacts with calnexin. A, HEK293T cells were transfected with vectors expressing FLAG-tagged ABCA1 or HCF, or FLAG alone, lysed and blotted for FLAG, GAPDH, and CNX (upper panels). Lysates were immunoprecipitated (IP) with anti-FLAG antibody and blotted for CNX (bottom panel). B, THP-1 cells were treated or not with PMA and vitamin D3, lysed, and blotted with antibodies to ABCA1, CNX, and GAPDH (upper panels). Lysates were immunoprecipitated with anti-ABCA1 antibody and blotted for calnexin. C, HeLa-ABCA1-GFP cells were fixed and immunostained for calnexin using mouse monoclonal anti-calnexin antibody followed by DyLight 550 goat anti-mouse IgG antibody, and nuclei were stained with DAPI. Panel a shows merged layers of calnexin/ER (red), ABCA1 (green), and nuclei (blue), and layers corresponding to calnexin/ER and nuclei, or ABCA1 and nuclei are presented in panels b and c, respectively. Spots where ABCA1 is co-localized with calnexin occur as orange-yellow in panel a. Bar sizes equal 53.4 μm. D, HEK293T cells were transfected with FLAG-tagged ABCA1 or empty pcDNA vector, treated with DMSO (solvent) or tunicamycin (Tun), immunoprecipitated with anti-FLAG antibody, and blotted for CNX and ABCA1 using monoclonal antibodies (two upper panels) or for glycosylated proteins using HRP-concanavalin A (bottom panel). E, HEK293T cells were co-transfected with ABCA1 and HA-tagged calnexin, treated with DMSO or tunicamycin, immunoprecipitated with anti-HA antibody, and blotted with anti-ABCA1, anti-calnexin, and anti-MHC-I monoclonal antibodies. F, HEK293T cells were co-transfected with gp160 and HA-tagged calnexin, treated with DMSO or tunicamycin, immunoprecipitated with anti-HA antibody, and blotted with anti-HIV serum or anti-calnexin monoclonal antibody. WB, Western blot.
To determine whether the interaction between ABCA1 and calnexin occurs through the glucosyl groups on ABCA1 (32, 33), we used tunicamycin, which inhibits an early step in formation of N-glycans on nascent glycoproteins. Following treatment of ABCA1-FLAG-transfected HEK293T cells with tunicamycin, immunoprecipitated ABCA1-FLAG lost a major ABCA1-reactive band (presumably glycosylated ABCA1) but preserved (and even increased) the slower migrating band likely corresponding to alternatively modified or multimerized protein (Fig. 1D, upper panel). This interpretation was supported by glycosylation analysis using the carbohydrate-binding protein ConA. ConA-HRP detected ABCA1 precipitated from DMSO-treated cells, but no reactivity was detected in anti-FLAG IPs from tunicamycin-treated cells, indicating that ABCA1 in these cells was not glycosylated (Fig. 1D, bottom panel). Surprisingly, blotted anti-FLAG IPs remained positive for calnexin. Thus, despite the reduction in ABCA1 protein and the lack of glycosylation observed upon tunicamycin treatment, calnexin still co-precipitated with ABCA1 (Fig. 1D, middle panel), suggesting that the calnexin-ABCA1 interaction does not require glycosylation. Similar results were obtained in cells co-transfected with HA-calnexin and ABCA1-FLAG, when IP was performed with anti-HA antibody, and immunoblots were stained for calnexin and ABCA1; ABCA1 still co-precipitated with HA-calnexin from cells treated with tunicamycin (Fig. 1E, top two panels). Again, ABCA1 that immunoprecipitated with calnexin from tunicamycin-treated cells had reduced amounts of faster migrating bands likely representing glycosylated species. Importantly, MHC-I, which has been shown to interact with calnexin in a glycan-dependent fashion (34, 35), failed to precipitate with calnexin upon treatment of the cells with tunicamycin (Fig. 1E, bottom panel).
Maturation of HIV-1 envelope precursor gp160 depends on calnexin (26). We transfected HEK293T cells with HA-calnexin and gp160-expressing vector and analyzed the effect of tunicamycin on calnexin-gp160 interaction. Consistent with a previously published report (26), tunicamycin inhibited interaction between calnexin and gp160 (Fig. 1F). Thus, calnexin interaction with ABCA1 and gp160 may involve two different mechanisms as follows: N-linked glycosylation-dependent binding for gp160, and binding dependent on peptide recognition for ABCA1. An alternative, although less likely, explanation of this result is a conformational change in calnexin due to possible deglycosylation; N-linked glycosylation has been shown for the yeast homolog of calnexin, Cne1p (36). However, this mechanism also implies different requirements for calnexin interactions with ABCA1 and gp160.
Nef Reduces Physical Interaction between Calnexin and ABCA1 but Enhances Interaction of Calnexin with gp160
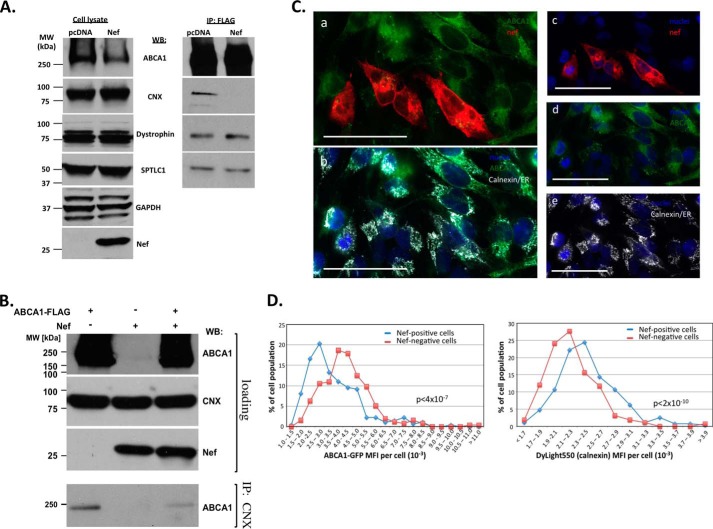
To determine the effect of Nef on ABCA1 interaction with calnexin, we co-transfected HEK293T cells with ABCA1-FLAG and Nef (or empty vector) and assessed the relative amounts of proteins in Nef-positive and Nef-negative cell lysates (Fig. 2A, left panels) and ABCA1-FLAG immunoprecipitates (Fig. 2A, right panels). We analyzed proteins identified in our MS/MS analysis (calnexin and dystrophin) and another previously identified ABCA1-interacting protein, SPTLC1 (37). Co-transfection with Nef caused the disappearance of calnexin from ABCA1-FLAG immunoprecipitates, but it did not affect ABCA1 interaction with other partners (Fig. 2A, right panels), indicating that Nef specifically disrupts the interaction between ABCA1 and calnexin. A similar result was obtained when calnexin was immunoprecipitated from these lysates and probed with an anti-ABCA1 antibody, although in this case Nef greatly reduced but did not entirely eliminate the detection of ABCA1 (Fig. 2B).
FIGURE 2.
Nef disrupts ABCA1 interaction with calnexin. A, HEK293T cells were co-transfected with FLAG-tagged ABCA1 and Nef or empty pcDNA vector, lysed, and blotted for ABCA1, calnexin, dystrophin, serine palmitoyltransferase (SPTLC1), GAPDH, and Nef (left panels). Following immunoprecipitation with anti-FLAG antibody, immunoprecipitates were blotted for calnexin, dystrophin, and SPLTC1 (right panels). B, cell lysates of HEK293T cells transfected with FLAG-tagged ABCA1, Nef, empty pcDNA vector, or co-transfected with FLAG-tagged ABCA1 and Nef were immunoprecipitated (IP) with anti-calnexin antibody and blotted for ABCA1. C, HeLa-ABCA1-GFP cells were transfected with Nef, and 48 h post-transfection, cells were fixed and immunostained for calnexin and Nef using mouse monoclonal anti-calnexin/ER marker antibody revealed with DyLight 550 goat anti-mouse IgG antibody (gray channel) and anti-HIV NefSF2 rabbit serum revealed with Alexa Fluor® 647 goat anti-rabbit IgG antibody (red channel), respectively, and ABCA1-GFP was present in the green channel and nuclear stain DAPI in the blue channel. For clarity, panels a, c, d, and e show a combination of two channels representing Nef and ABCA1 (panel a), nuclei and Nef (panel c), nuclei and ABCA1 (panel d), and nuclei and calnexin (panel e), whereas panel b shows overlay of nuclei, ABCA1, and calnexin channels. Bar sizes equal 53.4 μm. D, confocal images of individual cells were analyzed using Volocity software. Histograms of frequency distribution of MFI of ABCA1-GFP (left panel) and calnexin (stained with DyLight 550-labeled antibody, right panel) in individual cells with (blue line) and without (red line) Nef expression (271 Nef-positive and 257 Nef-negative cells were analyzed; p values are shown for t test comparison of MFI in Nef-positive and Nef-negative cells). WB, Western blot.
Analysis by confocal microscopy of individual cells expressing and not expressing Nef in cultures of Nef-transfected HeLa-ABCA1-GFP cells is shown in Fig. 2, C and D. The effect of Nef on ABCA1 is shown in Fig. 2C, where nuclei are stained blue (DAPI); ABCA1 is stained green (GFP); Nef is stained red (Alexa Fluor® 647); and calnexin/ER is stained gray (DyLight 550). The area containing four Nef-positive and eight Nef-negative cells was selected for single-cell analysis (Fig. 2C, panel a, layers corresponding to nuclei and calnexin were removed). Cells positive for Nef had reduced levels of ABCA1 (Fig. 2C, panel d, Nef and calnexin layers were removed for better visibility). In addition, the remaining ABCA1 in Nef-positive cells localized to intracellular regions enriched in calnexin (Fig. 2C, panels b, d, and e), indicating ER association, whereas in Nef-negative cells ABCA1 was well represented on the periphery of the cells. Reduction of cell surface ABCA1 is likely due both to prevention of ABCA1 delivery from ER to the plasma membrane and internalization and degradation of ABCA1 that had been delivered (7). Histograms showing frequency distribution of mean fluorescence intensities (MFI) in 271 Nef-positive and 257 Nef-negative randomly selected cells are shown in Fig. 2D. Fig. 2D, left panel, represents MFI of ABCA1-GFP and demonstrates that the presence of Nef was associated with lower ABCA1 protein levels, and the difference was highly statistically significant (p < 4 × 10−7). Fig. 2D, right panel, represents MFI of DyLight 550 (encoding calnexin) on the same cells; a significant increase in calnexin was observed in Nef-positive cells (p < 2 × 10−10).
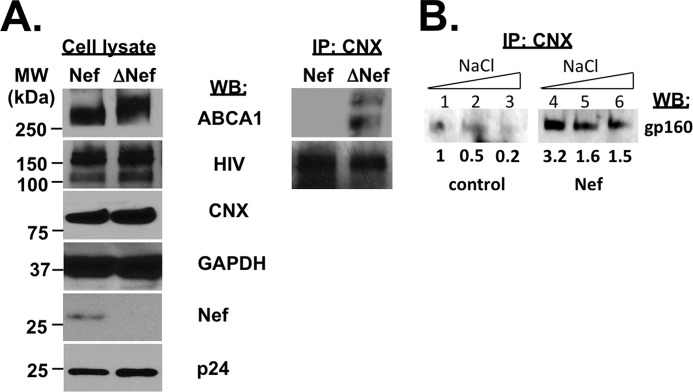
To determine whether ABCA1-calnexin interaction is impaired during HIV-1 infection, we infected monocyte-derived macrophages with wild-type or Nef-deficient HIV-1 and tested calnexin interaction with ABCA1 and HIV-1 Env by pulling down calnexin and blotting the immunoprecipitate with antibody to ABCA1 or immunoglobulins from an HIV-positive patient (Fig. 3A, right panels). ABCA1 co-immunoprecipitated with calnexin from cells infected with Nef-deficient HIV-1; however, no co-immunoprecipitation of ABCA1 and calnexin could be detected from cells infected with Nef-positive virus. Interestingly, interaction between HIV-1 gp160 and calnexin was not reduced by the presence of Nef but seemed to be enhanced (Fig. 3A, right panels).
FIGURE 3.
Effects of Nef on calnexin interaction with ABCA1 and gp160. A, monocyte-derived macrophages were infected with Nef-positive or Nef-deficient HIV-1 (ΔNef), cultured for 2 weeks, lysed, and immunoblotted using anti-ABCA1 antibody (ABCA1), pooled anti-HIV immunoglobulins from HIV-positive patients (HIV), and antibodies to calnexin (CNX), GAPDH, Nef and p24 (left panels). Cell lysates were immunoprecipitated (IP) using anti-calnexin antibody and immunoblotted using anti-ABCA1 antibody (ABCA1) and anti-HIV immunoglobulins (HIV) (right panels). B, HEK293T cells were co-transfected with gp160 and Nef or empty vector (control). Cells were lysed with buffer containing NaCl at 150 mm (lanes 1 and 4), 600 mm (lanes 2 and 5), or 1.2 m (lanes 3 and 6), and immunoprecipitation was carried out with anti-CNX antibody. Immunoprecipitates were blotted for gp160 using monoclonal antibody. Density of the bands was quantified using ImageJ program and is presented below the bands as relative units. WB, Western blot.
A possible mechanism for enhancing the interaction between gp160 and calnexin by Nef is to increase affinity of this interaction. Higher affinity should make an interaction resistant to higher salt concentration in the interaction buffer. Therefore, to further substantiate the effect of Nef on interaction between calnexin and gp160, we transfected HEK293T cells (these cells do not express ABCA1) with gp160 and Nef (or empty vector) and performed calnexin immunoprecipitation in increasing salt concentrations (Fig. 3B). In the absence of Nef, the amount of gp160 co-precipitating with calnexin was decreased by 50% in 600 mm NaCl (Fig. 3B, lane 2) and by 80% in 1.2 m NaCl (lane 3). In the presence of Nef, the amount of gp160 co-precipitating with calnexin in normal salt concentration (150 mm) was increased 3.2-fold (Fig. 3B, compare lanes 4 and 1). In 600 mm salt, the amount of gp160 co-precipitating with calnexin in the presence of Nef was decreased by 50%, and in 1.2 m salt by 53% (Fig. 3B), indicating that Nef increases the affinity of calnexin-gp160 interaction.
These results support the notion that Nef specifically disrupts interaction between calnexin and ABCA1, but it promotes interaction between calnexin and gp160.
Nef Physically Interacts with Calnexin
Calnexin is an essential protein of the endoplasmic reticulum and is largely confined to the ER due to ER-retention signals. It is anchored to the membranes of ER by its transmembrane domain. Given the observed effect of Nef on calnexin interactions with target proteins, it appeared likely that Nef regulates calnexin by physically interacting with it. However, ER localization of Nef has not been reported. Our analysis of HeLa-ABCA1-GFP cells transfected with Nef demonstrated that a proportion of Nef co-localized with calnexin (Fig. 4A, orange staining in panel a). Fig. 4A, panel a, shows a merged image of calnexin/ER (red), Nef (green), and nuclei (blue). Fig. 4A,, yellow/orange pixels depict areas in panel a with relatively close proximity of ER and Nef. Panels b and c in Fig. 4A display single channels of the same image set representing calnexin and Nef, respectively. To test whether Nef associates with calnexin, HEK293T cells were transfected with HA-tagged wild-type or mutant Nef (G2A mutation), precipitated with agarose-immobilized anti-HA antibody, and blotted for calnexin (Fig. 4B). The NefG2A mutant was used as a negative control, as this mutation abrogates Nef myristoylation and membrane association (38), and thus is expected to disrupt association with calnexin. Indeed, although the IP from cells transfected with HA-tagged Nef clearly identified a band corresponding to calnexin, no such band was observed in IPs from HA-NefG2-transfected cells, suggesting that membrane association of Nef was required for the Nef-calnexin interaction (Fig. 4C). Given that fractionation data demonstrated predominantly membrane localization of Nef (Fig. 4B), this result indicates that a proportion of Nef localizes to ER membranes.
Having observed that Nef physically interacts with calnexin and that the NefG2A mutant fails to associate with calnexin, we sought to determine whether association of Nef with calnexin was required for Nef interference with the ABCA1-calnexin interaction. To address this question, HEK293T cells were co-transfected with ABCA1-FLAG and either WT Nef or NefG2A, and anti-FLAG IPs were analyzed for calnexin. According to this analysis, ABCA1 showed negligible interaction with calnexin in the presence of WT Nef, although in the presence of NefG2A, ABCA1 co-precipitated with calnexin (Fig. 4D). Combined with the observation that the NefG2A mutant was consistently defective in impairing ABCA1-mediated cholesterol efflux activity (2), this result suggests that Nef interaction with calnexin is required for Nef-mediated disruption of ABCA1-calnexin interaction and functional impairment of ABCA1.
Areas of Calnexin Interaction with Nef Are Spatially Separated in ER from Calnexin-ABCA1 Interaction
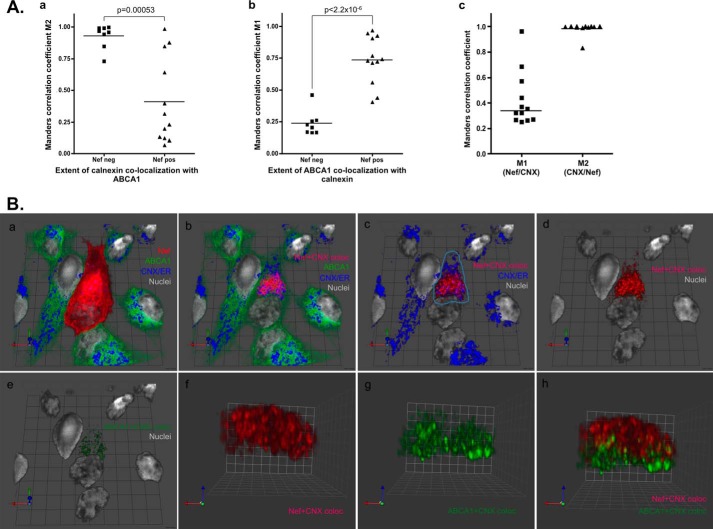
Using three-dimensional image sets taken at high resolution (×150/1.35) and subsequent analysis with Volocity software, we next compared co-localization of ABCA1 and calnexin within eight fully reconstructed cells lacking Nef but strongly positive for ABCA1 to that within 12 cells selected for strong Nef expression. Although 93 ± 3.3% (n = 8) of calnexin co-localized with ABCA1 in the absence of Nef, this proportion dropped to about 41 ± 9.7% (n = 12) in the presence of Nef. This difference was highly statistically significant (p = 0.00053) (Fig. 5A, panel a). Although we found a decrease in the amount of calnexin co-localizing with ABCA1 in the Nef-expressing cells, the median proportion of ABCA1 co-localizing with ER increased 3-fold (p < 2.2 × 10−6), consistent with ABCA1 retention in ER (Fig. 5A, panel b). When the same 12 cells strongly expressing Nef were analyzed for co-localization of Nef and calnexin, we found that almost all calnexin/ER (median 99.8 ± 1.4%; range 83.1–100%) was co-localizing with approximately one-third (median 33.9 ± 6.2%; range 25.1–96.3%) of Nef present in the cell (Fig. 5A, panel c).
FIGURE 5.
Effect of Nef on co-localization of ABCA1, Nef, and calnexin/ER. A, co-localization of ABCA1 and calnexin within Nef-negative but ABCA1 strongly positive cells (n = 8) and Nef strongly positive cells (n = 12) taken from Nef-transfected cultures of HeLa-ABCA1-GFP cells. Three-dimensional image sets were collected with ×100/1.46 lens at voxel resolution of 0.133 × 0.133 × 0.2 (x/y/z) and used for this analysis. Extent of calnexin co-localization with ABCA1 was determined by MCC M2 (panel a) using Volocity software. Extent of ABCA1 co-localization with calnexin is plotted as MCC M1 (panel b). Panel c shows the extent of Nef co-localization with calnexin (M1) and the extent of calnexin co-localization with Nef (M2) in cells strongly expressing Nef. Bars indicate mean MCCs. B, three-dimensional image reconstruction of ABCA1 (green), Nef (red), calnexin/ER (blue), and nuclei (gray) within HeLa ABCA1-GFP cells. Panel a shows all four channels overlapped, with Nef signal obscuring the underlying structures due to its high cell surface expression. Panel b, the Nef channel was stripped from the three-dimensional rendering to allow visualization of the underlying calnexin (blue) and ABCA1 (green). Also, the voxels representing Nef + calnexin and ABCA1 + calnexin-positive co-localization were rendered in red and green, respectively. Panel c, ABCA1 channel was also removed. Panel d, both ABCA1 and calnexin layers were removed. Panel e shows nuclei and ABCA1 co-localized with calnexin in a Nef-positive cell. Panels f–h represent the volume from the endoplasmic reticulum image of the area of co-localization of calnexin, ABCA1, and Nef in the Nef-expressing cell, color encoding as in panel c. Co-localization of calnexin and Nef is in red (panel f), and co-localization of calnexin and ABCA1 is in green (panel g). Overlay of both co-localizations is shown in panel h. Spatial distribution of the voxels encoding ABCA1 + calnexin and Nef + calnexin is as follows: 1 unit = 4.57 μm in panels a–e; 1 unit = 1.17 μm in panels f–h.
To determine the mutual co-localization of Nef, ABCA1, and calnexin, we performed analysis of three-dimensional image sets of the 12 cells with high Nef expression. Images of one representative cell are shown in Fig. 5B. Fig. 5B, panel a, shows nuclei in gray, ABCA1 in green, calnexin in blue, and Nef in red. In Fig. 5B, panel b, the red color encodes voxels representing Nef co-localization with calnexin. In Fig. 5B, panel c, the ABCA1-encoding green channel is removed to illustrate the calnexin and Nef interactions. In Fig. 5B, panel d, the calnexin layer is removed as well. Fig. 5B, panel e, shows nuclei and the voxels encoding two positive co-localization entities as follows: voxels representing ABCA1 co-localized with calnexin are presented in green, and voxels representing Nef co-localization with calnexin are presented in red. Fig. 5B, panels f–h, represents a detailed image of the area of co-localization of calnexin, ABCA1, and Nef in the Nef-expressing cell marked in panel c. Co-localization of calnexin and Nef is colored red (Fig. 5B, panel f), and co-localization of calnexin and ABCA1 is colored green (panel g). Overlay of both co-localizations is shown in Fig. 5B, panel h. In agreement with the immunoprecipitation data, this analysis demonstrated that ABCA1 and Nef co-localization with calnexin is for the most part mutually exclusive despite the fact that they co-localized in generally the same area of the cell (ER).
Calnexin Knockdown Mimics the Effects of Nef on ABCA1 Down-regulation
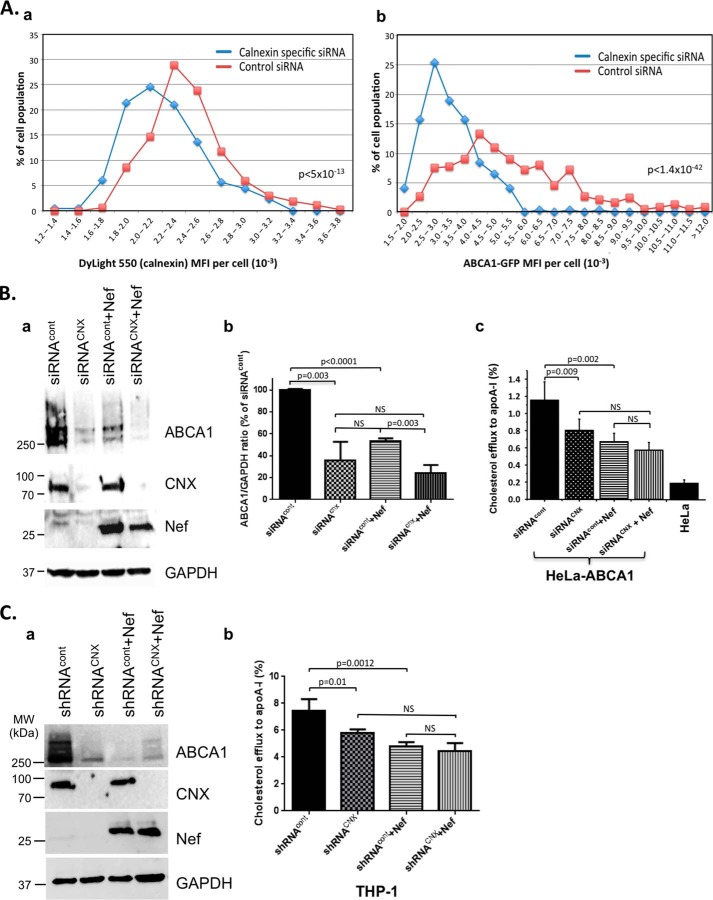
Calnexin has been shown to retain immature and misfolded proteins in the ER thus preventing their exit from the ER into the secretory pathway to reach their functional cellular domains at the plasma membrane, as exemplified by ABC transporter CFTR mutants defective in maturation (25, 39). To test the effect of calnexin knockdown on ABCA1 abundance and localization, we monitored ABCA1-GFP in cells transfected with control or anti-CNX siRNA. Quantification of calnexin staining on a large number of cells confirmed down-regulation of calnexin in cultures transfected with CNX siRNA relative to cells transfected with control siRNA (Fig. 6A, panel a). Importantly, abundance of ABCA1 was also reduced in cells transfected with CNX siRNA (Fig. 6A, panel b). Comparison of MFI of HeLa-ABCA1-GFP cells in cultures transfected with control siRNA (375 cells analyzed) and calnexin-specific siRNA (248 cells analyzed) showed that the down-regulation of both calnexin and ABCA-1 after transfection with calnexin-specific siRNA was statistically significant (p < 5 × 10−13 and p < 1.4 × 10−42, respectively). Cells were selected for analysis based on a large tile scanned region from the cell culture captured at high magnification and resolution (data not shown). For consistency, only cells where the optical section represents the cell nucleus (to avoid the contribution of ABCA1 localized on the cellular surface) were sampled.
FIGURE 6.
Effects of Nef and calnexin knockdown on ABCA1 abundance and functionality. A, HeLa-ABCA1-GFP cells transfected with either control siRNA or calnexin-specific siRNA were immunostained for calnexin using mouse monoclonal anti-calnexin/ER antibody visualized by DyLight 550 goat anti-mouse IgG antibody. Histograms obtained by analysis of confocal images using Volocity software show comparison in frequency distribution of MFI of calnexin (DyLight 550, panel a) and ABCA1 (GFP, panel b) in cells from cultures transfected with control siRNA or calnexin-specific siRNA. Analysis was done on 248 randomly selected cells from cultures transfected with calnexin-specific siRNA and 375 randomly selected cells from cultures transfected with control siRNA. Differences in frequency of MFI distribution were statistically highly significant for both calnexin and ABCA1 (p < 5 × 10−13 and p < 1.4 × 10−42, respectively). B, HeLa-ABCA1-GFP cells were co-transfected with Nef and control siRNA or siRNA targeting calnexin and analyzed by Western blotting for expression of ABCA1, calnexin, Nef, and GAPDH (panel a). Results were quantified by densitometry and are presented as mean ± S.D. of three independent experiments (panel b). Statistical analysis was done by Student's unpaired two-tailed t test. NS indicates nonsignificant values (p > 0.05). Panel c shows results of analysis of cholesterol efflux to apoA-I performed with the same cells. Cholesterol efflux to apoA-I from HeLa cells is shown as control. C, ABCA1, CNX, Nef, and GAPDH were analyzed in THP-1 cells stably expressing control (shRNAcont) or calnexin-specific shRNA (shRNACNX) and transfected with Nef or empty vector (panel a). Cholesterol efflux analysis with these cells is shown in panel b. Results show apoA-I-specific efflux presented as mean ± S.D. of four replicates. Statistical analysis was done by Student's unpaired two-tailed t test. NS, indicates nonsignificant values (p > 0.05).
As independent proof of the effect of calnexin down-regulation on ABCA1 abundance, we analyzed ABCA1 by Western blotting in HeLa-ABCA1-GFP cells transfected with siRNA to calnexin and/or the Nef-expressing construct (Fig. 6B, panel a). Quantification of the bands by densitometry demonstrated that ABCA1 abundance (normalized to GAPDH) was reduced by over 50% in cells transfected with calnexin siRNA relative to cells transfected with control siRNA (Fig. 6B, panel b), consistent with results in Fig. 6A (panel b). Transfection of the cells with Nef also resulted in significant reduction of ABCA1 abundance (Fig. 6B, panel b). Importantly, co-transfection of Nef and calnexin siRNA together did not produce an additive effect on ABCA1 abundance; in fact, ABCA1 abundance in co-transfected cells was similar to the level in cells transfected with siRNACNX alone, supporting the notion that Nef-mediated reduction of ABCA1 abundance depends on calnexin. There was significant difference between ABCA1 abundance in cells transfected with Nef and cells transfected with Nef plus siRNACNX, likely due to differences in expression levels of these two factors.
To determine the effect of calnexin and Nef on ABCA1 functionality, we analyzed apoA-I-specific cholesterol efflux in HeLa-ABCA1-GFP cells transfected with calnexin siRNA and/or Nef. Consistent with previously published findings (2, 6–8, 13) and results in Fig. 6B (panel b), siRNACNX and Nef individually and significantly reduced cholesterol efflux (Fig. 6B, panel c). The effect of calnexin knockdown on cholesterol efflux was slightly smaller than the effect of Nef transfection but was still highly significant. However, the combined effect of Nef transfection and calnexin knockdown on cholesterol efflux was not statistically different from the effect of each of these treatments alone and in particular from the effect of calnexin knockdown (Fig. 6B, panel c). Of note, cholesterol efflux from HeLa-ABCA1 cells transfected with Nef or siRNACNX (or co-transfected with both) was not reduced to the level observed in parent HeLa cells (Fig. 6B, panel c), which do not express ABCA1 and do not support cholesterol efflux to apoA-I (2, 40), indicating that down-regulation was not saturated and did not reach a maximal level, likely due to an insufficient expression level of inhibitory molecules. There was good correlation between cholesterol efflux (Fig. 6B, panel c) and ABCA1 abundance (Fig. 6B, panel b), although a significant difference in ABCA1 levels between cells transfected with Nef alone and Nef together with siRNACNX did not translate to a difference in cholesterol efflux. A likely reason for this discrepancy is different populations of ABCA1 molecules assessed by these two assays: Western blot examines all ABCA1 present in the cell and cholesterol efflux analysis reflects activity of functional ABCA1, represented predominantly by cell surface molecules. Lack of close correlation between the levels of ABCA1 and cholesterol efflux was also observed in another study using the same cell line (29).
To demonstrate that the effects observed in cells where ABCA1 was introduced by transfection are valid for cells expressing endogenous ABCA1, we analyzed cholesterol efflux from THP-1 cells (a monocytic cell line) transfected with Nef. For this analysis, we used THP-1 cells transduced with lentiviruses expressing control or calnexin shRNA and differentiated into macrophage-like cells by PMA treatment. Western blot analysis in Fig. 6C (panel a) shows that calnexin expression was effectively suppressed in cells transduced with shRNACNX, and ABCA1 was down-regulated in cells transduced with shRNACNX and/or transfected with Nef. Results of cholesterol efflux analysis from these cells are shown in Fig. 6C, panel b. Similar to results in HeLa-ABCA1 cells (Fig. 6B, panel c), knockdown of calnexin or transfection with Nef significantly reduced cholesterol efflux. Most importantly, the combined effect of calnexin knockdown and expression of Nef was not significantly different from the effect of individual treatments (and calnexin knockdown in particular), indicating that Nef does not inhibit cholesterol efflux when calnexin is down-regulated.
Taken together, these results demonstrate that calnexin down-regulation has a similar effect on ABCA1 abundance as expression of HIV-1 Nef, and calnexin is essential for ABCA1 functionality. Most importantly, they support the notion that the blockade of the interaction between ABCA1 and calnexin is an essential component of the effect of Nef on ABCA1 abundance and apoA-I-specific cholesterol efflux.
DISCUSSION
In this report, we describe a novel mechanism by which Nef interferes with activity of a host cell protein ABCA1, which is essential for the first step of reverse cholesterol transport, cholesterol efflux. We demonstrate that Nef binds to endoplasmic reticulum chaperone calnexin and disrupts its interaction with ABCA1, leading to retention of this transporter in the ER and its eventual degradation. Importantly, Nef does not indiscriminately disrupt all calnexin interactions, as interaction between calnexin and HIV-1 Env, which is critical for proper Env maturation and functionality (28), was actually stimulated by Nef.
This study provides the first characterization of the ABCA1-calnexin interaction. In fact, nothing is known about the role of calnexin in maturation of ABCA family members. Our data suggest that the interaction between ABCA1 and calnexin is glycan-independent and likely involves a peptide-binding mechanism of calnexin interaction with target proteins. In fact, it appears that calnexin interaction with nonglycosylated ABCA1 is stronger than with glycosylated protein (Fig. 1C), suggesting that affinity of calnexin-ABCA1 interaction is reduced by glycosylation. Glycan-independent interaction with calnexin is not unique to ABCA1, as another ABC transporter, ABCB1/P-glycoprotein, has been reported to interact with calnexin via a lectin-independent mechanism that involves the transmembrane regions of P-glycoprotein (18, 25). It is possible that the ABCA1-calnexin interaction also involves transmembrane regions of ABCA1. In contrast, interaction of calnexin with gp160 depends on glycosylation (this study and Ref. 26), suggesting that Nef binding to calnexin may induce a conformational change that promotes calnexin interaction with glycans but inhibits peptide recognition. Alternatively, Nef may compete with ABCA1 for binding to calnexin, although this mechanism appears less likely given that Nef has not been reported to enter the lumen of the ER, where calnexin N- and P-domains responsible for most of interactions of this protein are located (41). The extensive list of ABC transporters interacting with calnexin, some via a glycan-independent mechanism, indicates that calnexin functions as a prototypical chaperone for membrane glycoproteins of the ABC superfamily and suggests that these proteins may represent heretofore unappreciated targets of HIV-1 Nef.
Calnexin knockdown or Nef expression increased ABCA1 association with ER, reduced ABCA1 abundance, and inhibited ABCA1-dependent cholesterol efflux. This result suggests that calnexin functions as an ABCA1 quality-control chaperone that increases ABCA1 cholesterol efflux activity. Interestingly, similar retention of ABCA1 in the ER was described for one of the ABCA1 mutants responsible for Tangier disease (33, 42). HIV-1 Nef exploits this requirement for ABCA1 interaction with calnexin to reduce ABCA1 function. The essential role of Nef-mediated disruption of calnexin-ABCA1 interaction in the overall effect of Nef on cholesterol efflux is demonstrated by the lack of significant difference between the effects of Nef and Nef + siRNACNX, whereas the individual effects of both Nef and siRNACNX were highly significant (Fig. 6, B and C). The overall effect of Nef is to retain ABCA1 in the ER with subsequent delivery to proteasomes (13).
Intriguingly, while disrupting calnexin interaction with ABCA1, Nef stimulates the calnexin-gp160 interaction. Functional relevance of this effect of calnexin for HIV-1 replication remains to be determined, but previous reports indicate that calnexin is essential for gp160 maturation (28). We propose the following model for calnexin-related effects of Nef (Fig. 7). In uninfected cells, calnexin can interact with proteins undergoing post-translational modifications within the ER, either through peptide-dependent binding or through glycan recognition. Newly synthesized ABCA1 interacts with calnexin via a peptide-dependent mechanism. Calnexin exerts a rigorous quality control facilitating folding of a functional ABCA1, which is directed to the plasma membrane. Dysfunctional ABCA1 (misfolded or improperly glycosylated molecules) is directed to degradation in proteasomes (43). In HIV-infected cells, Nef binds to calnexin and changes its interaction affinities, promoting interaction with gp160 while impairing interaction with ABCA1. This prevents calnexin from exercising its chaperone function on ABCA1 but enhances its activity on gp160. As a result, Env maturation may be optimized while that of ABCA1 is impaired, leading to retention of ABCA1 in the ER and its subsequent degradation. Nef may also stimulate internalization and lysosomal degradation of cell surface ABCA1 (7). The proposed model is in part speculative, as not all of its elements have been proven; however, it is consistent with our findings and those of others.
FIGURE 7.
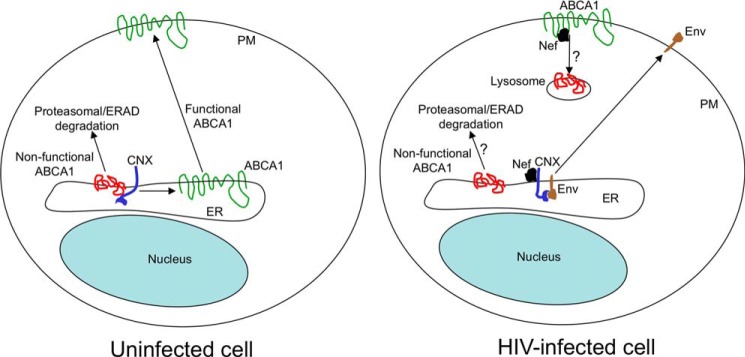
Proposed model of calnexin-mediated Nef effects. In uninfected cells, CNX interacts with ABCA1 in the ER in a glycosylation-independent fashion, facilitating folding and maturation of ABCA1. Matured ABCA1 is delivered to the plasma membrane (PM), and incorrectly folded nonfunctional protein is retained in the ER and degraded via the proteasomal/ERAD pathway. In HIV-infected cells, Nef binds to calnexin and disrupts the glycosylation-independent calnexin-ABCA1 interaction, resulting in ABCA1 retention in the ER and subsequent degradation, presumably by proteasomes (13). Glycosylation-dependent interaction of calnexin with HIV-1 gp160 is enhanced by Nef, promoting maturation of viral envelope proteins. Nef may also contribute to internalization of surface ABCA1 and its degradation, presumably in lysosomes (7). Question marks indicate pathways that have been shown for ABCA1 degradation, but not for specific events described in the figure.
A possible mechanism for such a differential effect of Nef on calnexin interactions is a conformational change in calnexin, which stimulates recognition of glycans but inhibits peptide recognition. Ongoing structural studies will test this hypothesis. Calnexin is a particularly good target for Nef because interfering with the function of this chaperone would allow Nef to modulate membrane expression of a variety of host cell proteins. This effect of Nef on calnexin interactions with cellular proteins may be part of the viral strategy to ensure the availability of calnexin and to optimize its function to assist in the maturation and folding of viral Env proteins.
In summary, the results presented in this report demonstrate that Nef stimulates interaction between calnexin and gp160 at the expense of the physiological target of calnexin, ABCA1, and identify a novel mechanism by which HIV-1 Nef influences activity of viral and host cell proteins by modulating their interaction with calnexin. The effect of Nef on calnexin provides a mechanism for the previously described Nef-mediated impairment of cellular cholesterol efflux, which leads to disruption of HDL maturation, retention of cellular cholesterol, increased abundance of lipid rafts, and enhanced virus production and fusion capacity of the virions.
Acknowledgments
The following reagents were obtained through the AIDS Reagent Program, Division of AIDS, NIAID, National Institutes of Health; HIV-1 Nef antiserum was from Dr. Ronald Swanstrom (44); HIV-IG was from NABI and NHLBI; monoclonal antibody to HIV-1 p24 (AG3.0) was from Dr. Jonathan Allan (45); HIV-1 gp160 antiserum was from Refs. 46, 47; pT7consnefhis6 was from Dr. Ron Swanstrom (44). HeLa-ABCA1-GFP cells stably expressing GFP-tagged ABCA1 were the kind gift of Dr. A. Remaley (29). Clones of Nef-positive (pBRNL4.3_92BR020.4(R5)nef+_IRES_EGFP) and Nef-deficient (pBRNL4.3_92BR020.4(R5)nef-_IRES_EGFP) macrophage-tropic HIV-1 were obtained from Dr. Frank Kirchhoff (30).
This work was supported, in whole or in part, by National Institutes of Health Grants HL093818, HL101274, and AI108533 and by Program Grant 5P30AI087714 (to District of Columbia Developmental Center for AIDS Research). This work is part of a dissertation to be presented by R. H. to the Institute for Biomedical Sciences, George Washington University, in partial fulfillment of the requirements for the Ph.D. degree.
- ABCA1
- ATP-binding cassette A1
- ER
- endoplasmic reticulum
- CNX
- calnexin
- CFTR
- cystic fibrosis transmembrane conductance regulator
- PDM
- product of the differences from the mean
- MFI
- mean fluorescence intensity
- PMA
- phorbol 12-myristate 13-acetate
- MCC
- Manders' correlation coefficient
- ConA
- concanavalin-A
- IP
- immunoprecipitation
- Env
- envelope.
REFERENCES
- 1. Desvarieux M., Boccara F., Meynard J. L., Bastard J. P., Mallat Z., Charbit B., Demmer R. T., Haddour N., Fellahi S., Tedgui A., Cohen A., Capeau J., Boyd A., Girard P. M. (2013) Infection duration and inflammatory imbalance are associated with atherosclerotic risk in HIV-infected never-smokers independent of antiretroviral therapy. AIDS 27, 2603–2614 [DOI] [PubMed] [Google Scholar]
- 2. Mujawar Z., Rose H., Morrow M. P., Pushkarsky T., Dubrovsky L., Mukhamedova N., Fu Y., Dart A., Orenstein J. M., Bobryshev Y. V., Bukrinsky M., Sviridov D. (2006) Human immunodeficiency virus impairs reverse cholesterol transport from macrophages. PLoS Biol. 4, e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asztalos B. F., Mujawar Z., Morrow M. P., Grant A., Pushkarsky T., Wanke C., Shannon R., Geyer M., Kirchhoff F., Sviridov D., Fitzgerald M. L., Bukrinsky M., Mansfield K. G. (2010) Circulating Nef induces dyslipidemia in simian immunodeficiency virus-infected macaques by suppressing cholesterol efflux. J. Infect. Dis. 202, 614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dubrovsky L., Van Duyne R., Senina S., Guendel I., Pushkarsky T., Sviridov D., Kashanchi F., Bukrinsky M. (2012) Liver X receptor agonist inhibits HIV-1 replication and prevents HIV-induced reduction of plasma HDL in humanized mouse model of HIV infection. Biochem. Biophys. Res. Commun. 419, 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Navab M., Reddy S. T., Van Lenten B. J., Fogelman A. M. (2011) HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat. Rev. Cardiol. 8, 222–232 [DOI] [PubMed] [Google Scholar]
- 6. Morrow M. P., Grant A., Mujawar Z., Dubrovsky L., Pushkarsky T., Kiselyeva Y., Jennelle L., Mukhamedova N., Remaley A. T., Kashanchi F., Sviridov D., Bukrinsky M. (2010) Stimulation of the liver X receptor pathway inhibits HIV-1 replication via induction of ATP-binding cassette transporter A1. Mol. Pharmacol. 78, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui H. L., Grant A., Mukhamedova N., Pushkarsky T., Jennelle L., Dubrovsky L., Gaus K., Fitzgerald M. L., Sviridov D., Bukrinsky M. (2012) HIV-1 Nef mobilizes lipid rafts in macrophages through a pathway that competes with ABCA1-dependent cholesterol efflux. J. Lipid Res. 53, 696–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang H., Badralmaa Y., Yang J., Lempicki R., Hazen A., Natarajan V. (2012) Retinoic acid and liver X receptor agonist synergistically inhibit HIV infection in CD4+ T cells by up-regulating ABCA1-mediated cholesterol efflux. Lipids Health Dis. 11, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giannarelli C., Klein R. S., Badimon J. J. (2011) Cardiovascular implications of HIV-induced dyslipidemia. Atherosclerosis 219, 384–389 [DOI] [PubMed] [Google Scholar]
- 10. Roeth J. F., Collins K. L. (2006) Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol. Mol. Biol. Rev. 70, 548–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leonard J. A., Filzen T., Carter C. C., Schaefer M., Collins K. L. (2011) HIV-1 Nef disrupts intracellular trafficking of major histocompatibility complex class I, CD4, CD8, and CD28 by distinct pathways that share common elements. J. Virol. 85, 6867–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jacob D., Hunegnaw R., Sabyrzyanova T. A., Pushkarsky T., Chekhov V. O., Adzhubei A. A., Kalebina T. S., Bukrinsky M. (2014) The ABCA1 domain responsible for interaction with HIV-1 Nef is conformational and not linear. Biochem. Biophys. Res. Commun. 444, 19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mujawar Z., Tamehiro N., Grant A., Sviridov D., Bukrinsky M., Fitzgerald M. L. (2010) Mutation of the ATP cassette binding transporter A1 (ABCA1) C terminus disrupts HIV-1 Nef binding but does not block the Nef enhancement of ABCA1 protein degradation. Biochemistry 49, 8338–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caramelo J. J., Parodi A. J. (2008) Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283, 10221–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Papandréou M. J., Barbouche R., Guieu R., Rivera S., Fantini J., Khrestchatisky M., Jones I. M., Fenouillet E. (2010) Mapping of domains on HIV envelope protein mediating association with calnexin and protein-disulfide isomerase. J. Biol. Chem. 285, 13788–13796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cabral C. M., Liu Y., Sifers R. N. (2001) Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem. Sci. 26, 619–624 [DOI] [PubMed] [Google Scholar]
- 17. Leach M. R., Cohen-Doyle M. F., Thomas D. Y., Williams D. B. (2002) Localization of the lectin, ERp57 binding, and polypeptide binding sites of calnexin and calreticulin. J. Biol. Chem. 277, 29686–29697 [DOI] [PubMed] [Google Scholar]
- 18. Loo T. W., Clarke D. M. (1995) P-glycoprotein. Associations between domains and between domains and molecular chaperones. J. Biol. Chem. 270, 21839–21844 [DOI] [PubMed] [Google Scholar]
- 19. Suh W. K., Mitchell E. K., Yang Y., Peterson P. A., Waneck G. L., Williams D. B. (1996) MHC class I molecules form ternary complexes with calnexin and TAP and undergo peptide-regulated interaction with TAP via their extracellular domains. J. Exp. Med. 184, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagmann W., Schubert J., König J., Keppler D. (2002) Reconstitution of transport-active multidrug resistance protein 2 (MRP2; ABCC2) in proteoliposomes. Biol. Chem. 383, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 21. Amaral M. D. (2004) CFTR and chaperones: processing and degradation. J. Mol. Neurosci. 23, 41–48 [DOI] [PubMed] [Google Scholar]
- 22. Okiyoneda T., Kono T., Niibori A., Harada K., Kusuhara H., Takada T., Shuto T., Suico M. A., Sugiyama Y., Kai H. (2006) Calreticulin facilitates the cell surface expression of ABCG5/G8. Biochem. Biophys. Res. Commun. 347, 67–75 [DOI] [PubMed] [Google Scholar]
- 23. Papadopoulos M., Momburg F. (2007) Multiple residues in the transmembrane helix and connecting peptide of mouse tapasin stabilize the transporter associated with the antigen-processing TAP2 subunit. J. Biol. Chem. 282, 9401–9410 [DOI] [PubMed] [Google Scholar]
- 24. Pind S., Riordan J. R., Williams D. B. (1994) Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 269, 12784–12788 [PubMed] [Google Scholar]
- 25. Loo T. W., Clarke D. M. (1994) Prolonged association of temperature-sensitive mutants of human P-glycoprotein with calnexin during biogenesis. J. Biol. Chem. 269, 28683–28689 [PubMed] [Google Scholar]
- 26. Otteken A., Moss B. (1996) Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperone function similar to that of calnexin. J. Biol. Chem. 271, 97–103 [DOI] [PubMed] [Google Scholar]
- 27. Dettenhofer M., Yu X. F. (2001) Characterization of the biosynthesis of human immunodeficiency virus type 1 Env from infected T-cells and the effects of glucose trimming of Env on virion infectivity. J. Biol. Chem. 276, 5985–5991 [DOI] [PubMed] [Google Scholar]
- 28. Land A., Braakman I. (2001) Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie 83, 783–790 [DOI] [PubMed] [Google Scholar]
- 29. Neufeld E. B., Remaley A. T., Demosky S. J., Stonik J. A., Cooney A. M., Comly M., Dwyer N. K., Zhang M., Blanchette-Mackie J., Santamarina-Fojo S., Brewer H. B., Jr. (2001) Cellular localization and trafficking of the human ABCA1 transporter. J. Biol. Chem. 276, 27584–27590 [DOI] [PubMed] [Google Scholar]
- 30. Schindler M., Münch J., Kirchhoff F. (2005) Human immunodeficiency virus type 1 inhibits DNA damage-triggered apoptosis by a Nef-independent mechanism. J. Virol. 79, 5489–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Low H., Hoang A., Sviridov D. (2012) Cholesterol efflux assay. J. Vis. Exp. 61, e3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanaka A. R., Ikeda Y., Abe-Dohmae S., Arakawa R., Sadanami K., Kidera A., Nakagawa S., Nagase T., Aoki R., Kioka N., Amachi T., Yokoyama S., Ueda K. (2001) Human ABCA1 contains a large amino-terminal extracellular domain homologous to an epitope of Sjogren's syndrome. Biochem. Biophys. Res. Commun. 283, 1019–1025 [DOI] [PubMed] [Google Scholar]
- 33. Tanaka A. R., Abe-Dohmae S., Ohnishi T., Aoki R., Morinaga G., Okuhira K., Ikeda Y., Kano F., Matsuo M., Kioka N., Amachi T., Murata M., Yokoyama S., Ueda K. (2003) Effects of mutations of ABCA1 in the first extracellular domain on subcellular trafficking and ATP binding/hydrolysis. J. Biol. Chem. 278, 8815–8819 [DOI] [PubMed] [Google Scholar]
- 34. Vassilakos A., Cohen-Doyle M. F., Peterson P. A., Jackson M. R., Williams D. B. (1996) The molecular chaperone calnexin facilitates folding and assembly of class I histocompatibility molecules. EMBO J. 15, 1495–1506 [PMC free article] [PubMed] [Google Scholar]
- 35. Danilczyk U. G., Williams D. B. (2001) The lectin chaperone calnexin utilizes polypeptide-based interactions to associate with many of its substrates in vivo. J. Biol. Chem. 276, 25532–25540 [DOI] [PubMed] [Google Scholar]
- 36. Parlati F., Dominguez M., Bergeron J. J., Thomas D. Y. (1995) Saccharomyces cerevisiae CNE1 encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functions as a constituent of the ER quality control apparatus. J. Biol. Chem. 270, 244–253 [DOI] [PubMed] [Google Scholar]
- 37. Tamehiro N., Zhou S., Okuhira K., Benita Y., Brown C. E., Zhuang D. Z., Latz E., Hornemann T., von Eckardstein A., Xavier R. J., Freeman M. W., Fitzgerald M. L. (2008) SPTLC1 binds ABCA1 to negatively regulate trafficking and cholesterol efflux activity of the transporter. Biochemistry 47, 6138–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geyer M., Fackler O. T., Peterlin B. M. (2001) Structure-function relationships in HIV-1 Nef. EMBO Rep. 2, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Okiyoneda T., Harada K., Takeya M., Yamahira K., Wada I., Shuto T., Suico M. A., Hashimoto Y., Kai H. (2004) ΔF508 CFTR pool in the endoplasmic reticulum is increased by calnexin overexpression. Mol. Biol. Cell 15, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu Y., Mukhamedova N., Ip S., D'Souza W., Henley K. J., DiTommaso T., Kesani R., Ditiatkovski M., Jones L., Lane R. M., Jennings G., Smyth I. M., Kile B. T., Sviridov D. (2013) ABCA12 regulates ABCA1-dependent cholesterol efflux from macrophages and the development of atherosclerosis. Cell Metab. 18, 225–238 [DOI] [PubMed] [Google Scholar]
- 41. Schrag J. D., Bergeron J. J., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. (2001) The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell 8, 633–644 [DOI] [PubMed] [Google Scholar]
- 42. Tanaka A. R., Kano F., Ueda K., Murata M. (2008) The ABCA1 Q597R mutant undergoes trafficking from the ER upon ER stress. Biochem. Biophys. Res. Commun. 369, 1174–1178 [DOI] [PubMed] [Google Scholar]
- 43. Feng B., Tabas I. (2002) ABCA1-mediated cholesterol efflux is defective in free cholesterol-loaded macrophages. Mechanism involves enhanced ABCA1 degradation in a process requiring full NPC1 activity. J. Biol. Chem. 277, 43271–43280 [DOI] [PubMed] [Google Scholar]
- 44. Shugars D. C., Smith M. S., Glueck D. H., Nantermet P. V., Seillier-Moiseiwitsch F., Swanstrom R. (1993) Analysis of human immunodeficiency virus type 1 Nef gene sequences present in vivo. J. Virol. 67, 4639–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simm M., Shahabuddin M., Chao W., Allan J. S., Volsky D. J. (1995) Aberrant Gag protein composition of a human immunodeficiency virus type 1 vif mutant produced in primary lymphocytes. J. Virol. 69, 4582–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsushita S., Robert-Guroff M., Rusche J., Koito A., Hattori T., Hoshino H., Javaherian K., Takatsuki K., Putney S. (1988) Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J. Virol. 62, 2107–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rusche J. R., Lynn D. L., Robert-Guroff M., Langlois A. J., Lyerly H. K., Carson H., Krohn K., Ranki A., Gallo R. C., Bolognesi D. P. (1987) Humoral immune response to the entire human immunodeficiency virus envelope glycoprotein made in insect cells. Proc. Natl. Acad. Sci. U.S.A. 84, 6924–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]