Background: Topoisomerase V is a unique enzyme and the sole member of the IC subtype of type I topoisomerases.
Results: We probed the role of several amino acids comprising the topoisomerase active site to elucidate their role in catalysis.
Conclusion: The work reveals the role of several amino acids and suggests interesting parallels with type IB topoisomerases.
Significance: The results expand our understanding of this new subtype of topoisomerases.
Keywords: DNA-binding Protein, DNA Topoisomerase, DNA Topology, Enzyme Catalysis, Mutagenesis
Abstract
Topoisomerases are ubiquitous enzymes that modify the topological state of DNA inside the cell and are essential for several cellular processes. Topoisomerase V is the sole member of the type IC topoisomerase subtype. The topoisomerase domain has a unique fold among topoisomerases, and the putative active site residues show a distinct arrangement. The present study was aimed at identifying the roles of the putative active site residues in the DNA cleavage/religation process. Residues Arg-131, Arg-144, His-200, Glu-215, Lys-218, and Tyr-226 were mutated individually to a series of conservative and non-conservative amino acids, and the DNA relaxation activity at different pH values, times, and enzyme concentrations was compared with wild-type activity. The results suggest that Arg-144 is essential for protein stability because any substitution at this position was deleterious and that Arg-131 and His-200 are involved in transition state stabilization. Glu-215 reduces the DNA binding ability of topoisomerase V, especially in shorter fragments with fewer helix-hairpin-helix DNA binding motifs. Finally, Lys-218 appears to play a direct role in catalysis but not in charge stabilization of the protein-DNA intermediate complex. The results suggest that although catalytically important residues are oriented in different fashions in the active sites of type IB and type IC topoisomerases, similar amino acids play equivalent roles in both of these subtypes of enzymes, showing convergent evolution of the catalytic mechanism.
Introduction
DNA topoisomerases are found in all the three domains of life and are essential for vital cellular processes because they modify and maintain the topology of DNA (1–4). Topoisomerases are divided into two main types (Type I and Type II) based on whether they cleave single- or double-stranded DNA. Each type is further subdivided based on mechanism, sequence characteristics, and structure. Type II enzymes form dimeric or tetrameric complexes to coordinate the simultaneous cleavage of a double-stranded DNA (dsDNA) and pass an intact double strand through the resulting break (5). Type I enzymes, on the other hand, cleave a single strand of DNA and pass a single stranded DNA (ssDNA) or dsDNA through the break or rotate the broken ssDNA around the intact ssDNA (5, 6). Until recently, only two subtypes of type I topoisomerases had been identified, type IA and IB. A third subtype, type IC, was added with the discovery and characterization of Methanopyrus kandleri topoisomerase V (Topo-V)2 (7–9). The three type I subtypes are very different in sequence and structure, although type IB and type IC molecules share some overall mechanistic similarities (10).
Topoisomerases use transesterification chemistry to cleave and religate DNA during relaxation of DNA supercoils. First, a general base catalyst abstracts a proton from the active site tyrosyl oxygen and activates it for nucleophilic attack on the phosphate backbone of DNA. Upon attack, a pentavalent phosphorane transition state forms, accompanied by the buildup of negative charge on the non-bridging oxygens. The pentavalent intermediate is usually stabilized by Arg, Lys, or His residues. The transition state is resolved when a general acid makes possible the expulsion of the hydroxyl group, leading to the formation of a phosphotyrosine bond and cleavage of DNA (11, 12). DNA relaxation occurs at this stage either by strand passage or controlled rotation. The reversal of the above steps occurs when the free hydroxyl oxygen generated in the first reaction attacks the phosphorus of the covalent phosphotyrosine bond. The enzyme and the relaxed DNA molecule are released at the end of the reaction cycle (6).
Type IA topoisomerases form a covalent phosphotyrosine link with the 5′ end of the broken DNA strand, relax negatively supercoiled DNA, and require divalent metal cations and ssDNA regions for activity (5, 6). All type IA topoisomerases utilize an enzyme-bridged strand passage mechanism to change the topology of DNA and alter the linking number strictly in steps of one (6). In the crystal structures of type IA topoisomerase-DNA complexes, four conserved residues (Tyr, Lys, Arg, and Glu) form contacts at or near the scissile phosphate group (13–15). Roles for each amino acid have been proposed; Tyr is the nucleophile, Arg and Lys stabilize the pentavalent protein-DNA complex, and Glu is the proton donor for the DNA cleavage reaction (13–15). A conserved nearby histidine plays a role in proton transfer, whereas three conserved acidic residues are involved in Mg2+ binding (16–20).
Type IB enzymes form a phosphotyrosyl bond with the 3′ end of the broken DNA strand, relax either positively or negatively supercoiled DNA, and do not require divalent metal cations for activity (6). Type IB topoisomerases remove supercoils by a swiveling or “controlled rotation” mechanism (21–24), allowing the cleaved ssDNA to rotate around the uncleaved single strand and thus changing the linking number by several steps during each reaction cycle. The active site of all type IB topoisomerases contains a highly conserved pentad of residues, consisting of a Tyr, two Arg residues, a Lys, and either a His in human and viral proteins or an Asn in bacterial homologs. During catalysis, the tyrosine acts as a nucleophile and attacks the scissile phosphate, whereas the two arginines and the histidine are responsible for stabilizing the pentavalent transition state (11, 25–29). One of the arginines and the lysine are the general acid facilitating proton flow for the DNA cleavage reaction, along with a network of water molecules (27, 30).
Type IC is the newest subtype of Type I topoisomerases. Methanopyrus Topo-V is currently the sole member of this subtype. It was first identified in the hyperthermophilic archaeon M. kandleri and was later found in other Methanopyrus species. Topo-V is a two-domain protein, with an N-terminal topoisomerase domain followed by 24 helix-hairpin-helix (HhH) motifs arranged as 12 tandem (HhH)2 domains (8, 31, 32). Some of the (HhH)2 domains are involved in DNA repair, and there are at least two apurinic/apyrimidinic site-processing active sites (31, 33). Topo-V displays many characteristics similar to those of type IB enzymes; it can relax positively and negatively supercoiled DNA, forms a transient covalent bond with the 3′ end of the broken DNA, does not require magnesium for activity, and relaxes DNA by a swiveling mechanism (7, 10, 34). The most striking differences, which place Topo-V in an entirely different subtype, are the distinct domain organization with two different DNA processing activities, the unique structure of the topoisomerase domain, and the lack of sequence similarity with any other topoisomerase subtype (8, 9). The topoisomerase domain is mainly α-helical, with a helix-turn-helix motif carrying some of the important catalytic residues, and four of the (HhH)2 domains curling around the topoisomerase domain (8, 35). The active site of Topo-V contains five catalytic residues, which are identical to those found in the eukaryotic type IB active site: Tyr-226, Arg-131, Arg-144, His-200, and Lys-218 (8). Previously, Tyr-226 was shown to be the nucleophile for the topoisomerase reaction (31). Despite such close correspondence in residue identities, the overall active site structure of Topo-V differs drastically from that of the type IB enzymes, and the catalytic pentads are not superimposable (Fig. 1) (8). A superposition based on Tyr-226, Arg-131, and Arg-144 shows that the lysines in Topo-V and type IB enzymes fall on opposite sides of the active site (8). Finally, another major difference is the presence of an acidic residue, Glu-215, in Topo-V, which is located within 3–4 Å of the catalytic Tyr-226 (35). The role of this glutamate is unknown, but in type IA topoisomerases, a proximal glutamate serves as a proton donor for the DNA cleavage reaction (13). The lack of structural conservation between the active sites of Topo-V and type IB enzymes suggests that Topo-V might employ a different mechanism for DNA cleavage and religation. The aim of the present study was to test the essentiality of the catalytic pentad and glutamate in the topoisomerization reaction and identify potential roles in catalysis of these residues. The catalytic pentad and the active site glutamate were individually mutated to several amino acids with different characteristics, and the effects on DNA relaxation over a range of pH values, times, and enzyme concentrations were analyzed. The results suggest that Arg-144 is essential because mutations at this position affected protein expression and enzymatic activity. Residues Arg-131, Lys-218, His-200, and Glu-215 were successfully mutated to several amino acids with different characteristics. The mutagenesis results suggest a role for each of the active site residues. Arg-131 and His-200 are potentially involved in transition state stabilization, and Lys-218 should have a more direct role in catalysis (e.g. as a proton donor/proton relay for the reaction). Glu-215 does not contribute directly to the topoisomerase catalytic mechanism but instead affects DNA binding characteristics of Topo-V, especially in shorter fragments of Topo-V with fewer (HhH)2 domains. These observations manifest that although type IB and type IC topoisomerases have different active site architectures, they both use a similar mechanism for DNA cleavage/religation.
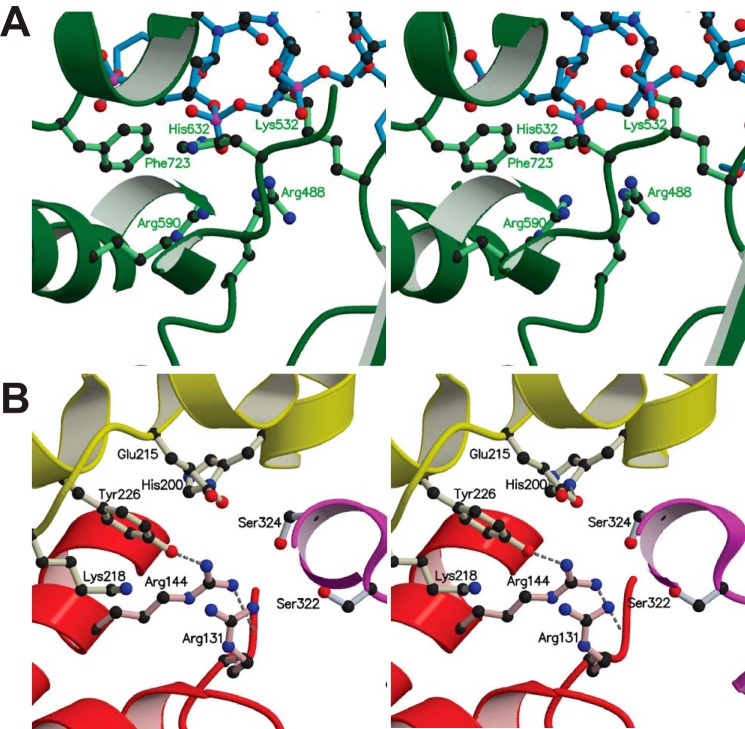
FIGURE 1.
Comparison of the topoisomerase active site of type IB and IC enzymes. A, stereo view of the active site of human topoisomerase I (Protein Data Bank entry 1A35) (41), a type IB enzyme (green) bound to DNA (blue). The catalytic pentad consists of Tyr-723 (mutated to Phe-723 in the present structure), His-632, Lys-532, Arg-488, and Arg-590. The corresponding residues of vaccinia topoisomerase are Tyr-274, His-265, Lys-167, Arg-131, and Arg-223. B, stereo view of the topoisomerase active site of an N-terminal 61-kDa fragment (Topo-61; Protein Data Bank entry 2CSB) (8) of Topo-V, a type IC enzyme. The core topoisomerase domain is in red, and the (HhH)2 domains are shown in yellow and magenta. The active site pentad consists of Tyr-226, His-200, Lys-218, Arg-131, and Arg-144. Glu-215 is also seen in the active site vicinity. A superposition of type IB and IC enzymes shows that the tyrosine and the two arginines are at similar positions in the active sites but the lysine and the histidine are not superimposable.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis and Protein Purification
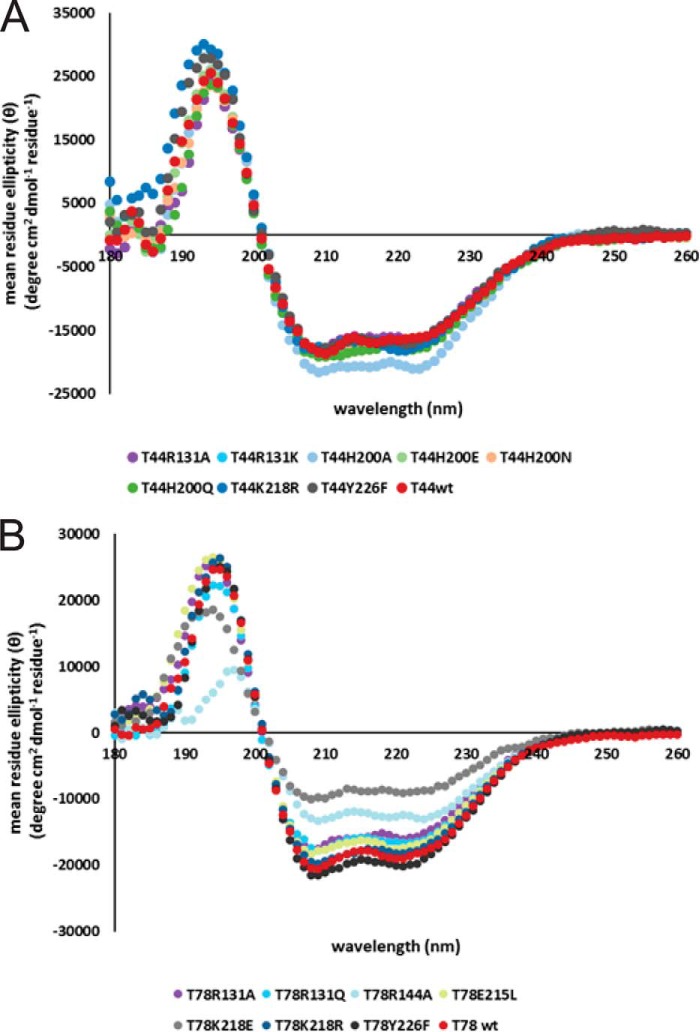
Arg-131, Arg-144, His-200, Glu-215, Lys-218, and Tyr-226 were individually mutated to the residues listed in Table 1 on the pET15bT44 and pET14bT78 expression plasmids. The pET15bT44 plasmid carries the coding sequence for the 44-kDa amino-terminal fragment of Topo-V (Topo-44) that extends to residue 380 (35). The pET14bT78 construct encodes for the 78-kDa N-terminal fragment of Topo-V (Topo-78) and spans residues 1–685 (32). For mutagenesis, DNA primers (Integrated DNA Technologies) were designed for use with the QuikChange site-directed mutagenesis kit (Stratagene). For protein production, Topo-44 mutants were transformed into Escherichia coli BL21 Rosetta (DE3) cells, whereas Topo-78 mutants were transformed into E. coli BL21 (DE3) cells. Protein induction and purification was done according to protocols reported previously (35). Pure protein was concentrated and stored in 50 mm Tris, pH 8, 250 mm NaCl, and 1 mm DTT. All of the mutants were analyzed by mass spectrometry to confirm the mutation through its molecular weight (Table 2). Topo-78 R144A was very unstable and degraded during storage, preventing accurate mass spectrometry analysis. To ensure that inactivity was not the result of folding defects, the secondary structures of the inactive mutants were tested by circular dichroism (CD) (Fig. 2).
TABLE 1.
Mutants that were designed in both the Topo-44 and Topo-78 fragments
The first column lists the six residues involved in the present study, the second column lists all of the mutations that were designed for each of the active site residues, and the third and fourth columns list the mutants that were successfully purified and assayed in the Topo-44 and Topo-78 backbones, respectively.
| Residue | Designed mutations | Purified in Topo-44 backbone | Purified in Topo-78 backbone |
|---|---|---|---|
| Arginine 131 | Ala, Lys, Gln | Ala, Lys | Ala, Gln |
| Arginine 144 | Ala, Lys, Gln | Ala | |
| Histidine 200 | Ala, Arg, Asn, Gln, Glu | Ala, Arg, Asn, Gln, Glu | Ala, Arg, Asn, Glu |
| Glutamate 215 | Ala, His, Leu, Gln, Arg | Ala, His, Gln, Arg | Ala, His, Leu, Gln |
| Lysine 218 | Ala, Glu, Gln, Arg | Arg | Ala, Glu, Gln, Arg |
| Tyrosine 226 | Phe | Phe | Phe |
TABLE 2.
MS analysis of the different proteins used in the experiments
T78R144A was not analyzed by MS because it degraded during storage for MS analysis. T44H200A is degraded by 2,627 Da, but it was active in the time course and protein gradient experiments. For this mutant, a C-terminal deletion of 27 amino acids (corresponding to residues 1–353) has a predicted Mr of 41,256.9, which is close to the observed Mr by MS analysis. Other than these two samples, the molecular weights of all of the mutants are in excellent agreement with the expected values.
| Name | Mr from MS | Difference in Mr compared with WT | Expected difference in Mr |
|---|---|---|---|
| T44wt | 43,884.09 | ||
| T44R131A | 43,799.11 | 85 | 85 |
| T44R131K | 43,856 | 28 | 28 |
| T44H200R | 43,903 | 19 | 19 |
| T44H200A | 41,257.14 | −2626.95 | 66.05 |
| T44H200Q | 43,875 | 9 | 9 |
| T44H200N | 43,861 | 23 | 23 |
| T44H200E | 43,876.16 | 8 | 8 |
| T44E215A | 43,826.04 | 58 | 58 |
| T44E215R | 43,911.21 | 27 | 27 |
| T44E215Q | 43,883.3 | 1 | 1 |
| T44E215H | 43,892.25 | 8 | 8 |
| T44K218R | 43,918.83 | 34.74 | 16 |
| T44Y226F | 43,868.01 | 16 | 16 |
| T78wt | 78,426.66 | ||
| T78R131A | 78,341.19 | 85.47 | 85.11 |
| T78H200A | 78,360.45 | 66.21 | 66.16 |
| T78H200R | 78,445.84 | 19.18 | 19.04 |
| T78H200N | 78,403.75 | 22.91 | 23.04 |
| T78H200E | 78,419.36 | 7.3 | 8.03 |
| T78E215A | 78,368.26 | 58.4 | 58.03 |
| T78E215Q | 78,426.52 | 0.14 | 0.99 |
| T78E215L | 78,410.34 | 16.32 | 15.96 |
| T78E215H | 78,434.35 | 7.69 | 8.02 |
| T78K218A | 78,368.9 | 57.76 | 57.19 |
| T78K218E | 78,427.67 | 1.01 | 0.94 |
| T78K218R | 78,454.73 | 28.07 | 28.01 |
| T78Y226F | 78,410.26 | 16.4 | 16 |
FIGURE 2.
CD spectra of selected Topo-44 (A) and Topo-78 (B) mutants. Proteins that were inactive in relaxation assays were analyzed by CD to confirm that the mutants were folded. The spectra of the Topo-44 mutants overlap well with that of the wild-type fragment, whereas the spectra of some Topo-78 mutants show minor differences. The spectra for Topo-78 R144A and Topo-78 K218E appear to be different from that of the wild-type fragment. Arg-144 may be essential for stability of protein because most of the Arg-144 mutants could not be purified. In the crystal structure of the Topo-44 fragment (Protein Data Bank entry 3M7D), Arg-144 makes contact with Arg-131 and Tyr-226 residues of the active site (35). Even after extensive purification, Topo-78 K218E is probably bound to DNA, as observed by the 260/280 ratio of 1.55. This might account for the difference in the spectrum of this protein.
DNA Relaxation Assays
DNA relaxation assays were carried out as described previously (35). Briefly, the enzyme was incubated with 250 ng of negatively supercoiled pUC19 plasmid in 15 μl of reaction mix containing a 50 mm concentration of the appropriate buffer, 30 mm NaCl, and 0.2 mm EDTA for 15 min. Activity was measured at 65 °C, covering a pH range from 4 to 10. The different buffers used were sodium acetate for pH 4 and 5, MES for pH 6, HEPES for pH 7, Tris for pH 8, CHES for pH 9, and CAPS for pH 10. The pH of all buffers was adjusted at 65 °C. For Topo-78 mutants, 0.15 μg of enzyme and for Topo-44 mutants, 0.75 μg of pure enzyme were used in each reaction unless otherwise noted. Protein concentration gradients and time course reactions were also carried out with the different mutants to analyze the effect of varying enzyme concentration and incubation times on the DNA relaxation activity. For both time and concentration gradient experiments, the buffer was at pH 5. For concentration gradient and time course experiments, the relaxation was quantified using ImageQuant (version 5.2) based on the appearance of relaxed or partially relaxed DNA in each lane. Two important assumptions were made: 1) the amount of nicked DNA in a plasmid is constant, and 2) relaxed and nicked DNA run identically under the experimental conditions used. Based on the fact that on a given lane, DNAtot = DNAn + DNAsc + DNArel, where DNAtot is the total amount of DNA in the reaction, DNAn is the amount of nicked DNA, DNAsc is amount of fully supercoiled DNA, and DNArel is the amount of relaxed and partially relaxed DNA (topoisomers), it is possible to write the fraction of relaxed DNA as freli = 1 − fn0 − fsci, where the superscript refers to either a time point or a concentration condition, and fn0 refers to the fraction of nicked DNA either at the starting time or in the absence of protein. In practice, first, a large rectangular box was drawn around each lane and covering both the supercoiled and relaxed DNA, and the total intensity inside of this box was measured (BB). Next, a smaller box was drawn covering the supercoiled plasmid in each lane, and the intensity was measured (SB). To correct for variations in the background, BB and SB were selected in an empty lane, and the intensity was measured. The background values were subtracted from the corresponding boxes in the sample lanes to obtain BBc and SBc. From the initial conditions, it is possible to obtain fnic = 1 − SBc0/BBc0. At any given point, freli = 1 − fnic − SBci/BBci or freli = SBc0/BBc0 − SBci/BBci. Finally, freli can be expressed as the fraction of relaxed DNA by correcting for the presence of nicked DNA as freli* = (1 − fn0 − fsci)/(1 − fi0) or freli* = (1 − SBciBBc0/SBc0BBci). The latter values were used in plots of either time or protein concentration versus relaxation activity.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA experiments were carried out as described previously (35). The sequence of the forward strand for 39-mer DNA is as follows: 5′-GCGACGCGAGGCTGGATGGCCTTCCCCATTATGATTCTT-3′. Some of the mutants made in the Topo-44 backbone were used in these assays.
RESULTS
Design of Protein Mutants and Purification
The putative catalytic residues were mutated individually both to similar and dissimilar residues. A list of the substitutions made is shown in Table 1. Conservative substitutions retained at least one of the chemical characteristics of the wild-type residue. Sterics, electrostatics, and acid/base properties were also taken into account when selecting the substitutions. Replacements to alanine were introduced to determine essentiality. Mutations were introduced into two different fragments of Topo-V: Topo-44 (an N-terminal 44-kDa fragment of Topo-V) and Topo-78 (an N-terminal 78-kDa fragment of Topo-V). The decision to conduct the mutagenesis experiments using protein fragments of Topo-V as opposed to full-length protein was due to several reasons. The protein expression of full-length Topo-V is low compared with that of Topo-V fragments, and this is not ideal for expressing some of the topoisomerase active site mutants. Full-length Topo-V is highly insoluble and requires 1 m NaCl for solubility. It has been observed previously that as the number of (HhH)2 domains increases, the relaxation activity of Topo-V fragments also increases, mainly due to the increased binding affinity of the protein to DNA (35). On the other hand, the isolated topoisomerase domain of Topo-V (Topo-31) has very limited DNA relaxation activity because it cannot bind to DNA in the absence of any (HhH)2 domains (35). Thus, the Topo-44 fragment, which contains the topoisomerase domain and one full and one partial (HhH)2 domain, is the best minimal fragment for analyzing the role of the catalytic residues. Experiments were also done with mutants made in the Topo-78 fragment. Topo-78 has seven full (HhH)2 domains and is a fragment with both topoisomerase and DNA repair activities (31, 32). The Topo-78 backbone helped to assess the effect of mutations in instances where the corresponding Topo-44 mutant was inactive, presumably due to protein-DNA binding effects mediated by the (HhH)2 domains. There were also instances where a particular mutant could not be purified in the Topo-44 backbone but could be purified in the Topo-78 backbone (e.g. T78R144A). Thus, studies of the putative active site residues in both the Topo-44 and Topo-78 backbones are complementary and gave a better understanding of the roles of these residues in catalysis. Protein expression and purification followed the general procedure described previously (35). Some of the substitutions resulted in an undetectable amount of soluble protein, and hence these mutants could not be purified. The list of proteins successfully purified and assayed is shown in Table 1. The results presented are representative of two independent experiments.
Effect of Mutations on DNA Relaxation Activity
The role of each putative catalytic residue (Tyr-226, Arg-131, Arg-144, His-200, Glu-215, and Lys-218) in the topoisomerization reaction was tested by performing DNA relaxation assays with various mutants over a wide pH range (pH 4–10). The pH range was varied because it was expected that, similar to type IA and IB enzymes, Topo-V employs general base catalysis to facilitate the attack of the catalytic tyrosine on the scissile phosphodiester bond and general acid catalysis to expel the 5′-hydroxyl of the leaving group. In addition, previous studies have shown that various Topo-V fragments differ in the optimal pH for DNA relaxation (35); Topo-44 relaxes DNA optimally around pH 5, whereas Topo-78 is equally active in the pH 5–9 range. In the case of type IB topoisomerases, some of the work characterizing the reaction mechanism took advantage of the strong sequence specificity of the viral topoisomerases, which assisted in accurate quantification of the effect of the mutations (36, 37). Topo-V, unlike viral topoisomerases, binds nonspecifically to DNA. In addition, DNA binding experiments using phosphorothioate-modified DNA showed that Topo-V avoids modified nucleotides and binds to non-modified sites in the modified oligonucleotide (data not shown). This characteristic precludes the use of suicide cleavage assays to measure rate constants under steady-state or single-turnover conditions, as in the case of the vaccinia enzyme. For pH analysis, the different mutants were compared qualitatively by checking how DNA relaxation varied over different pH when compared with the wild-type protein. None of the active site mutants showed a marked shift in the optimal pH for activity.
In addition to pH analysis, DNA relaxation activity of each mutant was analyzed using different protein concentrations and incubation times. For these experiments, the reaction buffer was at pH 5 because both Topo-44 and Topo-78 fragments are active at this pH. The time course and concentration gradient experiments show whether longer incubations or higher protein concentrations can improve the activity of a mutant compared with the regular reaction conditions. DNA relaxation activity was quantified by measuring the appearance of relaxed or partially relaxed DNA when compared with the control lane (no protein) as explained under “Experimental Procedures.” It was noted that at high protein concentrations, the protein had an inhibitory effect. The reason for this is not clear, but for this reason, data at high protein concentrations were not included in the final analyses.
The final assessment of the role of each amino acid was made based on pH profile, time, and concentration gradients and comparison with similar residues in type IB enzymes. The results for each of the putative active site residues are described below.
Tyrosine 226
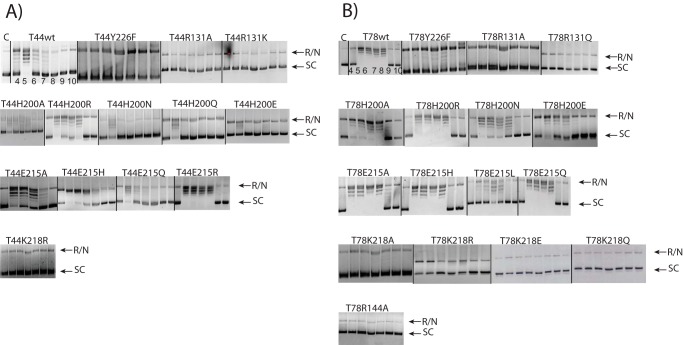
Tyr-226, previously identified as the catalytic tyrosine through phenylalanine scans, was mutated to Phe in both the Topo-44 (T44) and Topo-78 (T78) backbones. As expected, replacement of tyrosine with phenylalanine abolished activity at all pH values in T44Y226F (Fig. 3) and showed only minimal activity at pH 8 in the case of T78Y226F. It was also noted that in the Topo-44 backbone, the Y226F mutation abolished DNA binding in EMSA experiments (Fig. 4). In the EMSA experiments, a putative faint cleaved DNA band was observed for the wild-type and Glu-215 mutants but not for the other mutants. The shorter DNA band could be due to the protein cleaving the DNA near the 5′ end of the oligonucleotide, forming a covalent complex with the shorter DNA fragment, and releasing a longer oligonucleotide with a short unpaired region at one end.
FIGURE 3.
DNA relaxation activity pH profile of different mutants. A, DNA relaxation activity of Topo-44 mutants. For the Topo-44 mutants, 0.75 μg of protein were incubated with 250 ng of supercoiled pUC19 plasmid. B, DNA relaxation activity of Topo-78 mutants. For the Topo-78 mutants, 0.15 μg of protein were incubated with 250 ng of supercoiled pUC19 plasmid. Each mutant is labeled above the gel showing the relaxation assay. In general, the relaxation activities of the various active site mutants agree well in both the Topo-44 and Topo-78 backbones, although there is a higher activity associated with the Topo-78 backbone. The mutants that are completely inactive in the Topo-44 backbone are also inactive in the Topo-78 backbone, showing the essentiality of these residues in the catalytic reaction. The pH of the reaction varied from 4 to 10 from left to right of the panel for each mutant. The leftmost panel in the top row corresponds to control DNA alone (C). R/N, represents relaxed or nicked DNA; SC, supercoiled DNA. The bands between R/N and SC represent different DNA topoisomers.
FIGURE 4.
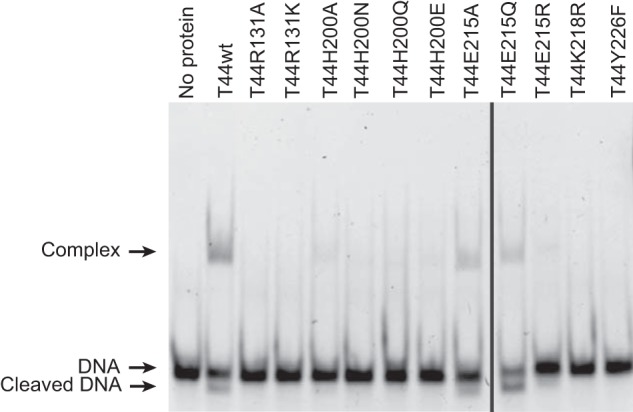
EMSA experiments with 39-mer dsDNA and various Topo-44 active site mutants. 4 μm DNA was incubated with 10 μm protein at 65 °C for 30 min, and the products were separated on a 6% native polyacrylamide gel. The wild-type protein and some of the Glu-215 mutants form clear protein-DNA complexes or a cleaved product. The His-200 mutants show a faint gel shift, demonstrating an effect on DNA binding by the mutations. The Lys-218, Arg-131, and Y226F mutants show no visible shift. The gel was cropped to remove a failed assay.
Arginine 131
A panel of substitutions (Table 1) was designed based on the fact that Arg-131 is a positively charged residue that could be involved in general acid catalysis or could stabilize interactions with the negatively charged DNA backbone. The R131A mutant was purified successfully in both the Topo-44 and Topo-78 backbones and showed no detectable DNA relaxation activity in either fragment (Fig. 5). R131K is a conservative substitution that retains both the electrostatic and acid/base properties, except for the loss of the bidentate coordination possible through the guanidinium group of the arginine. Whereas the R131K mutant was purified successfully in the Topo-44 backbone, it resulted in degradation when expressed in the Topo-78 backbone. DNA relaxation assays with the Topo-44 R131K protein at different pH values showed no activity for this mutant. Finally, replacing Arg-131 with glutamine prevented detectable protein expression in the Topo-44 backbone and ablated activity in the Topo-78 backbone across all pH values tested (Fig. 3). Concentration gradient and time course experiments in the Topo-44 and the Topo-78 backbones (Fig. 5) also show that none of the Arg-131 mutants are active under any conditions tested. Overall, the results suggest that Arg-131 plays an essential role in DNA relaxation. Because a similar charge substitution, to a lysine residue, was inactive, Arg-131 could be involved in stabilizing a protein-DNA intermediate through bidentate coordination through its side chain.
FIGURE 5.
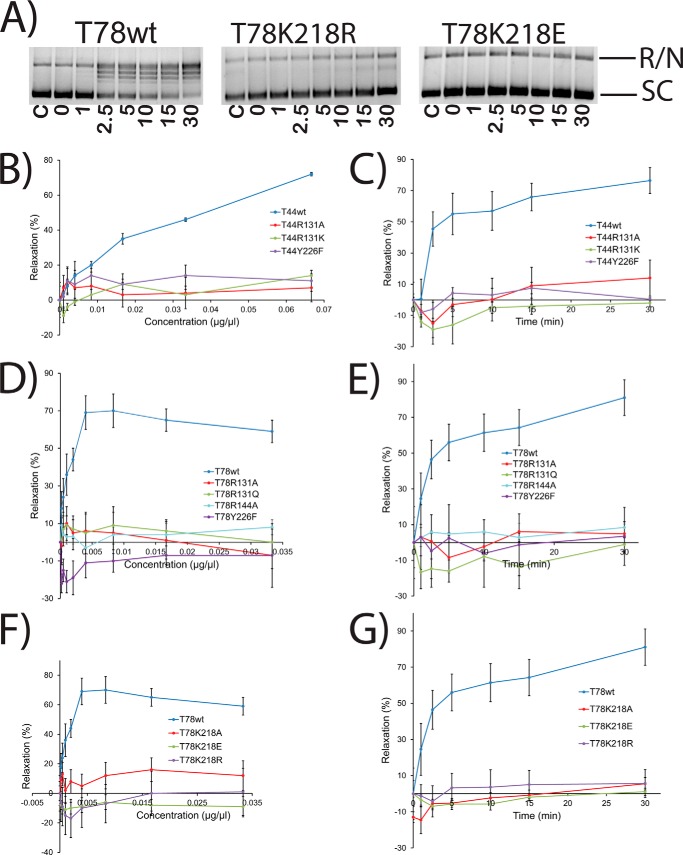
Enzyme concentration gradient and time course experiments with several inactive mutants. A, representative gels showing time course of supercoiled DNA relaxation activity by wild-type Topo-78 and two inactive mutants. The bands corresponding to fully relaxed or nicked (R/N) and supercoiled (SC) DNA are marked. The time points (min) corresponding to each lane are shown. C, control with no protein. In all cases, the reaction buffer was at pH 5, and the final volume was 15 μl. B–G, plots showing the percentage relaxation activity versus protein concentration or incubation times for different inactive mutants in both the Topo-44 and Topo-78 backbones. The activity of an inactive mutant stayed flat and identical to the activity at the starting point. The experiments were repeated two times for each mutant. Error bars, S.E. Activity is inhibited at high protein concentrations, and these points were omitted from the analyses. See “Experimental Procedures” for details about how the plots were obtained.
Arginine 144
Similar to the analyses undertaken for Arg-131, a panel of substitutions, including Ala, Lys, and Gln, was designed to probe whether Arg-144 acts as a transition state stabilizer or a general acid. Most Arg-144 mutants showed low or no protein solubility in both backbones. Only Topo-78 R144A could be purified, and this protein was very sensitive and degraded very quickly after purification. Relaxation assays at different pH values, protein concentrations, and time points showed no activity for this mutant (Figs. 3 and 5). The fact that change of one amino acid dramatically affected protein expression shows that Arg-144 may be essential for protein stability. R144A in Topo-78 is completely inactive in all of the conditions tested, suggesting that Arg-144 plays an important role in the relaxation process.
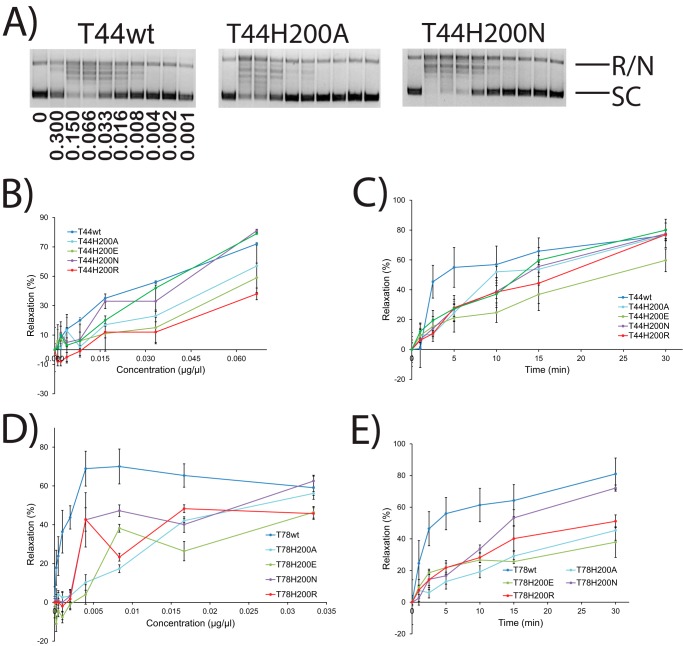
Histidine 200
Because the optimal pH for DNA relaxation in Topo-44 is ∼5 and the theoretical side chain pKa of histidine is ∼6, His-200 was a plausible candidate for the role of general acid. To test these possibilities, a panel of substitutions, including Ala, Arg, Asn, Gln, and Glu, was studied. H200A, H200R, H200N, and H200E mutants were successfully purified in both the Topo-78 and Topo-44 backbones, whereas H200Q was purified only in the Topo-44 backbone. Relaxation assays with these mutants at different pH conditions showed that T44H200A, T44H200Q, T44H200N, and T44H200E mutants have low but detectable DNA relaxation activity at pH 5, whereas T44H200R had relaxation over a pH range similar to the wild-type enzyme. In the Topo-78 backbone, qualitatively, H200R is as active as the wild type, H200A and H200N are slightly less active than wild type, and H200E shows a more marked reduction in activity across all of the pH values tested. In all cases, there is no shifting of the optimal pH for activity (Fig. 3). The concentration gradient and varying incubation time experiments show (Fig. 6) that although His-200 mutants are not as active as wild type at lower protein concentration or earlier time points, many of the mutants reach the same DNA relaxation level as the wild-type enzyme with higher protein concentration and longer incubations. Based on the pH dependence analysis, especially the results with the larger fragment, it is likely that His-200 is involved in charge stabilization of the protein-DNA intermediate. Finally, substitutions of His by Gln or Asn, which provide the necessary hydrogen bonding capabilities but do not have ionizable side chains, do not abolish activity, as would be expected if the histidine were acting as a proton donor/acceptor.
FIGURE 6.
Enzyme concentration gradient and time course experiments with His-200 mutants. A, representative gels showing concentration gradient supercoiled DNA relaxation activity by wild-type Topo-44 and two His-200 mutants. The bands corresponding to fully relaxed or nicked (R/N) and supercoiled (SC) DNA are marked. The concentration of protein (μg/μl) used in each lane is shown in the wild type panel. 0, control with no protein. In all cases, the reaction buffer was at pH 5, and the final volume was 15 μl. B–E, plots showing the percentage relaxation activity versus either protein concentration or incubation times for different His-200 mutants in both the Topo-44 and Topo-78 backbones. The results show that amino acids with different characteristics can be tolerated at position 200, with a preference for positive or neutral residues compared with negatively charged amino acids. The experiments were repeated two times for each mutant. Error bars, S.E. Activity is inhibited at high protein concentrations, and these points were omitted from the analyses. See “Experimental Procedures” for details about how the plots were obtained.
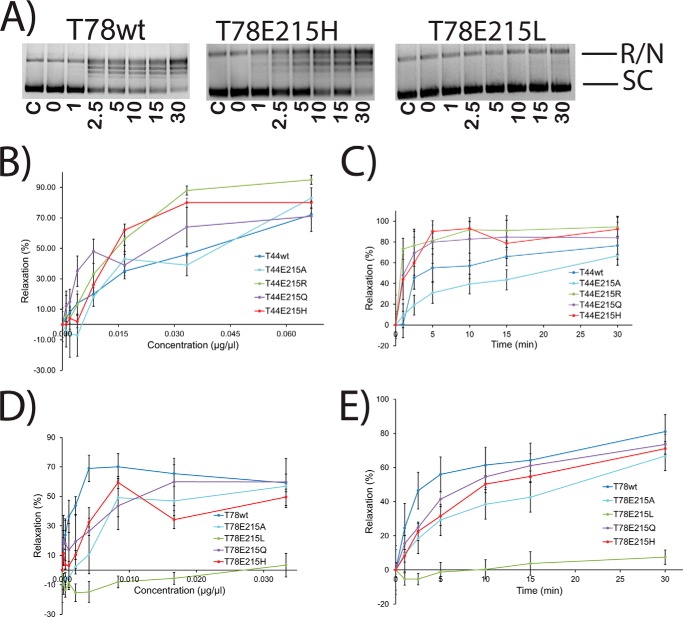
Glutamic Acid 215
The proximity of glutamate 215 to the active site of Topo-V suggests that it might play a role in catalysis, although substitution of Glu-215 to alanine does not have a noticeable effect on DNA relaxation activity (8). Additionally, because wild-type Topo-44 relaxes DNA at pH 5 and the theoretical pKa of glutamic acid is ∼4.3, Glu-215 is a prime candidate for proton donor. To test this hypothesis, Glu-215 was mutated to Ala, His, Leu, Gln, and Arg in the Topo-44 and Topo-78 backbones. E215A, E215H, E215Q, and E215R were purified in the Topo-44 backbone, whereas E215A, E215H, E215L, and E215Q were purified in the Topo-78 backbone. The pH profiles of all of the mutants (Fig. 3) as well as protein concentration gradients and time courses (Fig. 7) were tested. The results of the pH analyses show that in the Topo-44 fragment, all of the Glu-215 mutants were active over a wider range of pH values, compared with wild-type Topo-44. In Topo-78, all Glu-215 mutants show the same pH profile except T78E215L. This suggests that a hydrophobic residue at this position is not tolerated. Glutamine is isosteric with glutamate but not ionizable, and this mutant shows a similar level of activity in both backbones, confirming that Glu-215 is not a proton donor. Mutations of Glu-215 to His, with a pKa of 6.1, and Arg, with a pKa of 12.48, were tested because these residues provide a positive charge in place of the negative charge of glutamic acid. Surprisingly, both of these mutations showed slightly enhanced activity (Fig. 3) over a wider range of pH values, especially in the Topo-44 backbone. Protein concentration and time course experiments (Fig. 7) again agree with the results from the pH analysis; some of the Glu-215 mutants in the Topo-44 backbone exhibit slightly higher activity than the wild-type enzyme, whereas no difference in activity is noted in the mutants made in the Topo-78 backbone. Another observation is that in EMSA experiments, although wild-type, His-200, and Glu-215 mutants in the Topo-44 fragment can bind to DNA, none of the other Topo-44 active site mutants can bind to DNA (Fig. 4).
FIGURE 7.
Enzyme concentration gradient and time course experiments with Glu-215 mutants. A, representative gels showing time course supercoiled DNA relaxation activity by wild-type Topo-78 and two mutants. The bands corresponding to fully relaxed or nicked (R/N) and supercoiled (SC) DNA are marked. The time points (min) corresponding to each lane are shown. C, control with no protein. In all cases, the reaction buffer was at pH 5, and the final volume was 15 μl. B–E, the plots show the percentage relaxation activity versus either protein concentration or incubation times for different Glu-215 mutants in both the Topo-44 and Topo-78 backbones. The effects of the mutations are more pronounced in the Topo-44 backbone, where changing Glu-215 to an Arg, His, or Gln results in slightly higher activity than in the wild-type protein, even at low protein concentration or early time points. The presence of Ala at position 215 reduces the activity of the protein, whereas the presence of a hydrophobic residue (T78E215L) renders the protein inactive. The experiments were repeated two times for each mutant. Error bars, S.E. Activity is inhibited at high protein concentrations, and these points were omitted from the analyses. See “Experimental Procedures” for details about how the plots were obtained.
Lysine 218
In order to probe the role of Lys-218, particularly focusing on its possible contribution to acid/base catalysis and/or transition state stabilization during transesterification, Lys-218 was mutated to Ala, Glu, Gln, and Arg in both backbones. All the mutants were successfully purified in the Topo-78 backbone, whereas only K218R was purified in the Topo-44 backbone. The results for pH gradient, concentration gradient, and time variation (Figs. 3 and 5) are shown. Most mutants made in both backbones are inactive. The fact that K128R substitution shows very minimal activity at pH 8 rules out a possible role of Lys-218 in charge stabilization through its positively charged side chain. T78K218Q is inactive in all of the pH conditions tested. The low yield of T78K218Q protein hindered concentration gradient and time course experiments with this mutant. Taken together, these results suggest that Lys-218 plays an important role in catalysis, probably as a proton donor or as part of a proton relay mechanism and is unlikely to be involved in charge stabilization.
DISCUSSION
Role of the Putative Active Site Residues
In the present study, our aim was to identify the residues involved in the transesterification reaction catalyzed by Topo-V using a series of activity assays covering different pH values, protein concentration gradients, and incubation times.
It was previously observed that an R131A substitution in the 61-kDa N-terminal fragment of topoisomerase V (Topo-61) led to a severe reduction in activity, indicating that it was critical for DNA relaxation (8). Substituting Lys for Arg at position 131 led to loss of activity, suggesting that the bidentate coordination of the Arg side chain with the DNA backbone is important. Interestingly, in type IB enzymes, an arginine (Arg-130, a residue corresponding to Arg-131 of Topo-V) has also been implicated in transition state stabilization (26–28, 30, 38). Mutational studies of Arg-130 in vaccinia virus topoisomerase suggested that this residue makes a bidentate contact with DNA (26), which was confirmed by crystal structures showing that Arg-130 hydrogen bonds to a nonbridging oxygen of the pentavalent transition state and to the 5′ leaving group oxygen (27, 28). This second contact suggested that Arg-130 may play an additional role in proton relay to the 5′ hydroxyl of the leaving DNA strand (27, 30). Although the active site of Topo-V is different from the type IB active site, the two arginines (Arg-130 of vaccinia viral topoisomerase and Arg-131 of Topo-V) are located in nearly identical positions with respect to the tyrosine. In the Topo-44 structure (Protein Data Bank entry 3M7D), the side chain of Arg-131 is only 4 Å away from the hydroxyl group of Tyr-226 (35). Furthermore, the R131Q mutant is inactive, suggesting that it plays a role not just confined to binding. Taken together, these facts suggest that Arg-131 may be essential for bidentate contacts with the highly charged phosphorane transition state and its stabilization and might play a role in the proton relay mechanism, as in type IB enzymes.
The difficulty in purifying Arg-144 mutants suggests that either this residue has a profound effect on protein stability or that the Arg-144 mutants are bound to DNA, and the protein is pelleting along with the insoluble cell fraction. The only mutant purified, T78R144A, was minimally expressed compared with other mutants, was highly unstable, and had a high level of bound DNA. The CD spectrum of this mutant is shown in Fig. 2 and shows that although the protein has the signature of a folded protein, it looks different from that of the wild-type protein. The equivalent residue in type IB enzymes, Arg-223 in vaccinia topoisomerase I, makes a monodentate contact with a phosphate (11, 39). Additional studies, including structural analyses of variola viral topoisomerase I and biochemical studies with modified substrates, also implicated Arg-223 in simple electrostatic transition state stabilization (12, 27, 28). The crystal structure of Topo-44 shows that Arg-144 forms a 2.8-Å hydrogen bond to the Tyr-226 hydroxyl and also that there is a water network connecting Arg-144 and His-200, suggesting that Arg-144 could easily approach the phosphate backbone during the cleavage/religation reaction (Protein Data Bank entry 3M7D) (35). These observations suggest that Arg-144 could play a role in transition state stabilization.
Mutations at His-200 are tolerated, and there is no change in the optimal pH of the enzyme when the histidine is replaced, suggesting that it cannot be the essential proton donor. Instead, the results suggest that His-200 is involved in transition state stabilization because a positively charged residue is favored over a negatively charged one at this position. In vaccinia topoisomerase I, the histidine of the catalytic pentad contacts the nonbridging phosphate oxygens to stabilize the buildup of negative charge on the pentavalent transition state, and there is no contribution to acid/base catalysis (27–29, 40). The present mutational studies suggest that His-200 of Topo-V might play a similar role.
The role of Glu-215 could not be determined unambiguously. It was initially hypothesized that Glu-215 might act as a general acid catalyst (8). The present mutational analyses instead suggest that Glu-215 has a negative effect on the relaxation reaction because a positively charged residue at this position has increased activity over a wider pH range. Because the mutants made in Topo-78 do not show a drastic change in activity, we conclude that Glu-215 is negatively regulating the binding of protein to DNA in the presence of limited number of (HhH)2 domains. It has been observed that Asp-168 of viral topoisomerases inhibits DNA binding, reduces the rate of DNA cleavage and ligation, and also promotes catalytic equilibrium to favor the ligation reaction (28). Asp-168 forms a bridging interaction between the sugars of both DNA strands and, along with Lys-167, promotes the correct geometry of the sugar to attack the phosphotyrosine linkage (28). The positions of Asp-168 in the type IB active site and that of Glu-215 in the Topo-V active site are not equivalent, so it is unclear whether Glu-215 plays a role in Topo-V equivalent to that of Asp-168 in type IB enzymes. DNA binding studies with the Glu-215 mutants in Topo-44 backbone showed that the mutants form stable complexes with DNA oligonucleotides, whereas most other active site mutants disrupt DNA binding (Fig. 4).
Mutations of Lys-218 show that no change is tolerated at this position. The inactivity of the K218R mutant suggests that the positive charge is not the sole determinant of activity, although it is possible that the slightly bulkier Arg side chain is not properly positioned, rendering the mutant protein inactive. T78K218Q mutagenesis results indicate that Lys-218 plays a direct role in catalysis, probably in proton transfer, similar to the equivalent lysine residue in type IB topoisomerases.
Conclusions
This is the first systematic study aimed at understanding the role of the topoisomerase active site residues in a type IC enzyme. Previous studies have shown that both type IB and IC enzymes use a constrained rotation mechanism for DNA relaxation, although the details, such as the mean change in linking number and the relaxation velocity, are different in both cases (10). Nonetheless, due to the overall structural differences, it was not clear whether the two subtypes of enzymes employ similar chemical strategies. From the experiments described here, the role of five residues found in the active site is now clearer. One of the arginines, Arg-144, appears to be essential for protein stability. The other arginine, Arg-131, and a histidine, His-200, clearly play a role in charge stabilization. The histidine does not appear to play a role in proton relay. The sole lysine is unlikely to be involved in charge stabilization and is more likely to be involved in proton transfer. Finally, the glutamate at the active site might play a role in electrostatic interaction with DNA because replacement with positively charged residues enhances DNA relaxation. Additional studies, such as crystal structures of Topo-V bound to DNA, are essential to confirm some of the roles assigned to specific residues and also to find additional functions for these residues. In the case of type IA and type IB topoisomerases, a combination of approaches was required to understand the reaction mechanism employed by these proteins, and this is likely to be the case for type IC enzymes. Nevertheless, it is clear that despite the structural differences between type IB and IC enzymes, they share a similar DNA relaxation mechanism, pointing to convergent evolution in their mechanisms.
Acknowledgments
We acknowledge staff and instrumentation support from the Keck Biophysics Facility for CD and the Integrated Molecular Structure Education and Research Center for mass spectrometry analysis. Support from the R. H. Lurie Comprehensive Cancer Center of Northwestern University for the Structural Biology Facility is also acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM51350 (to A. M.). This work was also supported by American Heart Association Grant 10POST2600325 (to R. R.).
- Topo-V
- topoisomerase V
- HhH
- helix-hairpin-helix
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid.
REFERENCES
- 1. Pommier Y., Leo E., Zhang H., Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schoeffler A. J., Berger J. M. (2008) DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101 [DOI] [PubMed] [Google Scholar]
- 3. Vos S. M., Tretter E. M., Schmidt B. H., Berger J. M. (2011) All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang J. C. (2009) A journey in the world of DNA rings and beyond. Annu. Rev. Biochem. 78, 31–54 [DOI] [PubMed] [Google Scholar]
- 5. Champoux J. J. (2001) DNA Topoisomerases: Structure, Function, and Mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 6. Baker N. M., Rajan R., Mondragón A. (2009) Structural studies of type I topoisomerases. Nucleic Acids Res. 37, 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slesarev A. I., Stetter K. O., Lake J. A., Gellert M., Krah R., Kozyavkin S. A. (1993) DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature 364, 735–737 [DOI] [PubMed] [Google Scholar]
- 8. Taneja B., Patel A., Slesarev A., Mondragón A. (2006) Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 25, 398–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forterre P. (2006) DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 24, 245–247 [DOI] [PubMed] [Google Scholar]
- 10. Taneja B., Schnurr B., Slesarev A., Marko J. F., Mondragón A. (2007) Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc. Natl. Acad. Sci. U.S.A. 104, 14670–14675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krogh B. O., Shuman S. (2000) Catalytic mechanism of DNA topoisomerase IB. Mol. Cell 5, 1035–1041 [DOI] [PubMed] [Google Scholar]
- 12. Tian L., Claeboe C. D., Hecht S. M., Shuman S. (2005) Mechanistic plasticity of DNA topoisomerase IB: phosphate electrostatics dictate the need for a catalytic arginine. Structure 13, 513–520 [DOI] [PubMed] [Google Scholar]
- 13. Changela A., DiGate R. J., Mondragón A. (2001) Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature 411, 1077–1081 [DOI] [PubMed] [Google Scholar]
- 14. Changela A., DiGate R. J., Mondragón A. (2007) Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J. Mol. Biol. 368, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Cheng B., Tse-Dinh Y. C. (2011) Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 108, 6939–6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Domanico P. L., Tse-Dinh Y. C. (1991) Mechanistic studies on E. coli DNA topoisomerase I: divalent ion effects. J. Inorg. Biochem. 42, 87–96 [DOI] [PubMed] [Google Scholar]
- 17. Zhu C. X., Roche C. J., Tse-Dinh Y. C. (1997) Effect of Mg(II) binding on the structure and activity of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 272, 16206–16210 [DOI] [PubMed] [Google Scholar]
- 18. Zhu C. X., Roche C. J., Papanicolaou N., DiPietrantonio A., Tse-Dinh Y. C. (1998) Site-directed mutagenesis of conserved aspartates, glutamates and arginines in the active site region of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 273, 8783–8789 [DOI] [PubMed] [Google Scholar]
- 19. Zhu C. X., Tse-Dinh Y. C. (2000) The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J. Biol. Chem. 275, 5318–5322 [DOI] [PubMed] [Google Scholar]
- 20. Perry K., Mondragón A. (2002) Biochemical characterization of an invariant histidine involved in Escherichia coli DNA topoisomerase I catalysis. J. Biol. Chem. 277, 13237–13245 [DOI] [PubMed] [Google Scholar]
- 21. Koster D. A., Croquette V., Dekker C., Shuman S., Dekker N. H. (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434, 671–674 [DOI] [PubMed] [Google Scholar]
- 22. Koster D. A., Czerwinski F., Halby L., Crut A., Vekhoff P., Palle K., Arimondo P. B., Dekker N. H. (2008) Single-molecule observations of topotecan-mediated TopIB activity at a unique DNA sequence. Nucleic Acids Res. 36, 2301–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stewart L., Redinbo M. R., Qiu X., Hol W. G., Champoux J. J. (1998) A model for the mechanism of human topoisomerase I. Science 279, 1534–1541 [DOI] [PubMed] [Google Scholar]
- 24. Seol Y., Zhang H., Pommier Y., Neuman K. C. (2012) A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 109, 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petersen B. O., Wittschieben J., Shuman S. (1996) Mutations within a conserved region of vaccinia topoisomerase affect the DNA cleavage-religation equilibrium. J. Mol. Biol. 263, 181–195 [DOI] [PubMed] [Google Scholar]
- 26. Wittschieben J., Shuman S. (1997) Mechanism of DNA transesterification by vaccinia topoisomerase: catalytic contributions of essential residues Arg-130, Gly-132, Tyr-136 and Lys-167. Nucleic Acids Res. 25, 3001–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perry K., Hwang Y., Bushman F. D., Van Duyne G. D. (2006) Structural basis for specificity in the poxvirus topoisomerase. Mol. Cell 23, 343–354 [DOI] [PubMed] [Google Scholar]
- 28. Perry K., Hwang Y., Bushman F. D., Van Duyne G. D. (2010) Insights from the structure of a smallpox virus topoisomerase-DNA transition state mimic. Structure 18, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petersen B. O., Shuman S. (1997) Histidine 265 is important for covalent catalysis by vaccinia topoisomerase and is conserved in all eukaryotic type I enzymes. J. Biol. Chem. 272, 3891–3896 [DOI] [PubMed] [Google Scholar]
- 30. Krogh B. O., Shuman S. (2002) Proton relay mechanism of general acid catalysis by DNA topoisomerase IB. J. Biol. Chem. 277, 5711–5714 [DOI] [PubMed] [Google Scholar]
- 31. Belova G. I., Prasad R., Kozyavkin S. A., Lake J. A., Wilson S. H., Slesarev A. I. (2001) A type IB topoisomerase with DNA repair activities. Proc. Natl. Acad. Sci. U.S.A. 98, 6015–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belova G. I., Prasad R., Nazimov I. V., Wilson S. H., Slesarev A. I. (2002) The domain organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J. Biol. Chem. 277, 4959–4965 [DOI] [PubMed] [Google Scholar]
- 33. Rajan R., Prasad R., Taneja B., Wilson S. H., Mondragón A. (2013) Identification of one of the apurinic/apyrimidinic lyase active sites of topoisomerase V by structural and functional studies. Nucleic Acids Res. 41, 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slesarev A. I., Lake J. A., Stetter K. O., Gellert M., Kozyavkin S. A. (1994) Purification and characterization of DNA topoisomerase V. An enzyme from the hyperthermophilic prokaryote Methanopyrus kandleri that resembles eukaryotic topoisomerase I. J. Biol. Chem. 269, 3295–3303 [PubMed] [Google Scholar]
- 35. Rajan R., Taneja B., Mondragón A. (2010) Structures of minimal catalytic fragments of topoisomerase V reveals conformational changes relevant for DNA binding. Structure 18, 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stivers J. T., Shuman S., Mildvan A. S. (1994) Vaccinia DNA topoisomerase I: single-turnover and steady-state kinetic analysis of the DNA strand cleavage and ligation reactions. Biochemistry 33, 327–339 [DOI] [PubMed] [Google Scholar]
- 37. Stivers J. T., Shuman S., Mildvan A. S. (1994) Vaccinia DNA topoisomerase I: kinetic evidence for general acid-base catalysis and a conformational step. Biochemistry 33, 15449–15458 [DOI] [PubMed] [Google Scholar]
- 38. Wittschieben J., Shuman S. (1994) Mutational analysis of vaccinia DNA topoisomerase defines amino acid residues essential for covalent catalysis. J. Biol. Chem. 269, 29978–29983 [PubMed] [Google Scholar]
- 39. Cheng C., Wang L. K., Sekiguchi J., Shuman S. (1997) Mutational analysis of 39 residues of vaccinia DNA topoisomerase identifies Lys-220, Arg-223, and Asn-228 as important for covalent catalysis. J. Biol. Chem. 272, 8263–8269 [DOI] [PubMed] [Google Scholar]
- 40. Yang Z., Champoux J. J. (2001) The role of histidine 632 in catalysis by human topoisomerase I. J. Biol. Chem. 276, 677–685 [DOI] [PubMed] [Google Scholar]
- 41. Redinbo M. R., Stewart L., Kuhn P., Champoux J. J., Hol W. G. (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279, 1504–1513 [DOI] [PubMed] [Google Scholar]