Background: HUWE1 is an E3 ligase implicated in cancer and intellectual disabilities.
Results: Inducible RNAi combined with quantitative diGly proteomics was implemented to find HUWE1 substrates.
Conclusion: DDIT4 is a substrate of HUWE1.
Significance: HUWE1 is a master regulator of cell stress response proteins including DDIT4. Inducible RNAi coupled with diGly proteomics is a valuable strategy for identifying novel E3 ligase substrates.
Keywords: Protein Degradation, Proteomics, Stress Response, Ubiquitin, Ubiquitin Ligase, DDIT4, HUWE1, Quantitative diGly Proteomics, Ubiquitin-Proteasome System
Abstract
Targeted degradation of proteins through the ubiquitin-proteasome system (UPS) via the activities of E3 ubiquitin ligases regulates diverse cellular processes, and misregulation of these enzymes contributes to the pathogenesis of human diseases. One of the challenges facing the UPS field is to delineate the complete cohort of substrates for a particular E3 ligase. Advances in mass spectrometry and the development of antibodies recognizing the Lys-ϵ-Gly-Gly (diGly) remnant from ubiquitinated proteins following trypsinolysis have provided a tool to address this question. We implemented an inducible loss of function approach in combination with quantitative diGly proteomics to find novel substrates of HUWE1 (HECT, UBA, and WWE domain containing 1, E3 ubiquitin protein ligase), an E3 ligase implicated in cancer and intellectual disabilities. diGly proteomics results led to the identification of DNA damage-inducible transcript 4 (DDIT4) as a putative HUWE1 substrate. Cell-based assays demonstrated that HUWE1 interacts with and regulates ubiquitination and stability of DDIT4. Together these data suggest a model in which HUWE1 mediates DDIT4 proteasomal degradation. Our results demonstrate proof of concept that inducible knockdown of an E3 ligase in combination with diGly proteomics provides a potentially advantageous method for identifying novel E3 substrates that may help to identify candidates for therapeutic modulation in the UPS.
Introduction
Ubiquitin is a conserved, 76 amino acid-containing protein that plays an essential role in regulating protein abundance and function in organisms ranging from yeast to man (1). Through a process referred to as ubiquitination, proteins can be posttranslationally modified with ubiquitin. Typically, ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) work in concert to catalyze the formation of an isopeptide bond between the C-terminal Gly carboxyl moiety of ubiquitin and the ϵ-amino group of a Lys residue on a substrate protein (2–4). The reaction can be repeated several times with ubiquitin being conjugated to ubiquitin, generating polyubiquitin chains. In addition to other functions, polyubiquitination can serve as a signal to target the modified protein for proteasomal degradation (5). The ubiquitin ligases, classified as RING, HECT, or RING-between-RING, based on catalytic mechanisms and domain structures, are often considered to impart substrate recognition to the ubiquitination cascade (6–9). Removal of ubiquitin is carried out by a family of isopeptidases termed deubiquitinating enzymes (DUBs),3 which play a pivotal role in maintaining ubiquitination homeostasis in part by antagonizing polyubiquitination (10, 11). The ubiquitin-proteasome system (UPS) is an important regulator of many cellular processes including differentiation, division, protein quality control, apoptosis, iron homeostasis, and the immune response (12). Moreover, some DUBs and E3s are implicated as pathogenic factors in cancer, cardiovascular disorders, and neurodegeneration, making the UPS attractive as a source of therapeutic targets (13, 14).
Identifying reliable strategies for modulating E3 ubiquitin ligases would open up a novel target class for drug discovery. One of the challenges with targeting E3 ligases is to identify the full repertoire of substrates for each E3, which will inform both the potential utilities and liabilities of targeting a given enzyme. Identifying E3-substrate pairs can be difficult given the relatively weak affinities of enzyme for substrate and the low stoichiometry of ubiquitinated proteins (15). To address this gap, diverse approaches have been developed in recent years including functional genomics with targeted siRNA libraries (16–18), global protein stability profiling (19–21), extract-based functional assays (22–24), and affinity-based proteomics strategies (20, 25–27).
Quantitative Lys-ϵ-Gly-Gly (diGly) proteomics, an emerging innovative affinity-based approach, has been used to characterize the ubiquitinome (28–33) and more recently to identify potential substrates of ubiquitin ligases (34–36). Because of the Arg-Gly-Gly C-terminal sequence of ubiquitin, a diGly isopeptide bond fragment is formed when ubiquitinated proteins are trypsinized (37). Antibodies have been generated that recognize the diGly modification, allowing for enrichment of diGly peptides from cell lysates following trypsin digest (28, 29). Combining stable isotope labeling of amino acids in cell culture (SILAC) with diGly affinity capture allows not only for the quantitation and identification of ubiquitinated proteins but also the mapping of endogenously modified lysines (28, 29, 32).
HUWE1 (also known as LASU1, Mule, HECTH9, and ARF-BP1) is a large (∼482-kDa) HECT ubiquitin ligase containing ARLD, UBA, WWE, and BH3 domains (8). Conditional loss of HUWE1 in the mouse brain leads to an aberrant increase in n-MYC protein levels, inhibition of neuronal progenitor cell differentiation to mature neurons and glial cells, and neonatal death (38–40). HUWE1 also regulates proteins with important roles in cell stress response pathways such as MCL-1, TP53, c-MYC, HDAC2, and MFN2 (41–45). Not surprisingly, because of its involvement in regulating oncogenes and neurogenesis, various lines of evidence implicate HUWE1 in human disease. HUWE1 has been reported to be overexpressed in lung, stomach, colorectal, and breast carcinomas (41, 43, 46, 47). Gene deletions or inactivating mutations of HUWE1 correlate with increased n-MYC expression in high grade glioblastomas (38). In colon cancer samples, there is a direct correlation between HUWE1 expression and tumor stage (43, 46). Lastly, HUWE1 duplication or mutation is associated with X-linked intellectual disabilities (48–50).
Given the strong implications for HUWE1 in a number of human malignancies, we wished to expand the known repertoire of HUWE1 substrates to better understand the potential caveats and benefits of targeting this enzyme in disease settings. Unlike many E3s, working with HUWE1 presents a number of challenges directly attributable to its large size. Taking this into consideration, both affinity-based and quantitative diGly proteomics approaches were used for HUWE1 substrate identification, and the most interesting results were obtained with the diGly proteomics method. Here we report the identification and characterization of DDIT4 as a substrate of HUWE1.
EXPERIMENTAL PROCEDURES
Plasmid and Lentivirus Generation
Codon optimized wild-type (WT) and catalytically dead (CD; C4099A/C4341A/C4367A) (51) HUWE1 cloning vectors were obtained from GeneArt. CD HUWE1 was cloned into the pCL-N-Avi-IRES-BirA vector using Gateway cloning (Invitrogen). USP7, MCL-1, DDB1, MYH9, and DDIT4 were cloned into the NotI/BamHI, EcoRI/BamHI, EcoRV/XbaI, HindIII/XbaI, and NotI/BamHI sites, respectively, of the p3XFLAG-CMV-26 vector (Sigma). HUWE1 was cloned into the pDEST-N-SBP vector using Gateway cloning (Invitrogen). N-terminal 3×HA WT, K129R, and K0 DDIT4 vectors were obtained from Genewiz, and expression constructs were generated in the pcDNA-DEST40 vector using Gateway cloning (Invitrogen). 3×HA K129O, K155O, K188O, K218O, K219O, K220O, K129R/K155R/K188R, and K218R/K219R/K220R DDIT4 constructs were generated by site-directed mutagenesis (Stratagene). WT and CD HUWE1 were Gateway-cloned into the pcDNA3.1n/V5-DEST vector (Invitrogen). Control scrambled (5′-CAACAAGAUGAAGAGCACCAA), HUWE1 shRNA 1 (5′-UGCCGCAAUCCAGACAUAUUC), and HUWE1 shRNA 2 (5′-CGGCUCUCGGGAGAUCAACUA) shRNAs were cloned into the pLKO-Tet-On vector and packaged into lentiviruses as described previously (52). All constructs were verified by DNA sequencing.
Cell Culture and Reagents
HEK 293, HeLa, and HEK 293T cells were cultured in DMEM (Invitrogen). BT-549 cells were cultured in RPMI 1640 medium (Invitrogen), and A549 cells were cultured in F-12 medium (Invitrogen). All non-transduced cells were grown in media supplemented with 10% FBS (Hyclone). Stable cell lines inducibly expressing shRNAs were generated by transducing cells with lentiviruses at a multiplicity of infection of 0.5 and selecting with puromycin (Mediatech) at 1 μg/ml. Stable cells were grown in media containing 10% tetracycline-free FBS (Clontech), and shRNA expression was induced with addition of 50 or 100 ng/ml doxycycline (Sigma). For quantitative diGly proteomics experiments, BT-549 cells stably expressing inducible HUWE1 shRNA 1 were grown in RPMI-Flex medium (Invitrogen) supplemented with 10% dialyzed FBS, 2 mg/liter glucose, 2 mm l-glutamine, 100 μg/ml light l-lysine (K0), and 100 μg/ml light l-arginine (R0). Heavy medium was prepared as previously mentioned except that the light amino acids were replaced with 100 μg/ml heavy l-lysine (K6; [13C6]Lys) and 100 μg/ml heavy l-arginine (R10; [13C6,15N4]Arg). Transient transfections of cells with siRNAs and plasmid DNA were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Target sequences for siRNAs are as follows: non-targeting, 5′-UGGUUUACAUGUCGACUAA; USP7-1, 5′-UAAGGACCCUGCAAAUUAU; USP7-2, 5′-GUAAAGAAGUAGACUAUCG; HUWE1-1, 5′-GCAGUUGGCGGCUUUCUUA; HUWE1-2, 5′-GAGCCCAGAUGACUAAGUA; and HUWE1-3, 5′-CCGGCUUUCACCAGUCGCUUA.
Avi-tagged HUWE1-based Affinity Enrichment and Quantitative Mass Spectrometry
HEK 293T cells were transiently transfected with N-terminal Avi-tagged catalytically dead HUWE1 or an empty vector control overnight. Subsequently cells were cultured with media containing 225 nm biotin for 48 h and treated with 20 μm MG132 for 2 h prior to cell lysis. Cells were harvested by mechanical detachment, washed with excess PBS on ice, and lysed in immunoprecipitation buffer (50 mm Tris-Cl, pH 7.4, 100 mm NaCl, 5% glycerol, 1.5 mm MgCl2, 1 mm Na3VO4, 0.4% Nonidet P-40, 25 mm NaF, 10 nm calyculin A, 1 mm DTT, Complete protease inhibitor cocktail (Roche Applied Science), and 0.2 mg/ml DNase I (Sigma)) for 30 min at 4 °C. Lysates were first cleared by centrifugation and then incubated with high capacity streptavidin-agarose (Thermo) for 2 h. Beads were washed in lysis buffer without DNase and eluted by boiling for 10 min in 2× lithium dodecyl sulfate loading buffer (Invitrogen) supplemented with β-mercaptoethanol. Appropriate amounts of eluates were then resolved on a 4–12% NuPAGE gel (Invitrogen), and the gel was stained with Coomassie Brilliant Blue G (Sigma). Lanes were cut into 16 consecutive pieces. Proteins in each gel band were trypsinized and subjected to isobaric labeling with iTRAQ reagents (AB SCIEX). Corresponding samples from lanes of control and HUWE1 purifications were then pooled. Tryptic peptides were separated by online nano-high pressure liquid chromatography (Eksigent) on a C18 reversed phase column (Magic 3-mm 100-Å C18 AQ; Michrom) using an acetonitrile/water system at a flow rate of 300 nl/min prior to analysis on an LTQ Orbitrap Velos mass spectrometer (Thermo Electron). Tandem mass spectra were acquired in a data-dependent manner. Typically, 10 MS/MS measurements were performed after each high accuracy spectral acquisition range survey, and both higher-energy collisional dissociation and collision-induced dissociation tandem spectra were acquired. Raw MS files were converted to peak lists using Mascot Distiller (version 2.4.0.0) with spectrum merging enabled. The human portion (taxonomy ID, 9606) of the International Protein Index (IPI) database version 3.87 (919,491 sequences of which 810 are common contaminants) was interrogated using the Mascot search algorithm. One failed trypsin cleavage was allowed per search. The precursor and fragment ion tolerances were set to 10 ppm and 0.8 Da, respectively. Fixed modifications included the iTRAQ reagent (Lys and N terminus) and carbamidomethyl (Cys). Variable modifications included oxidation (Met), deamination (Asn and Gln), and pyroglutamic acid. Peptide and protein validation was done using Transproteomic Pipeline v3.3sqall (Institute for Systems Biology) using a false positive threshold of <1% for protein identifications. For each peptide sequence and modification state, reporter ion signal intensities from all spectral matches were summed for each reporter ion type and corrected according to the isotope correction factors given by the manufacturer. Only peptides unique to a given protein within the total data set of identified proteins were used for relative protein quantification, and only proteins with two or more unique peptides and three or more peptide-to-spectrum matches were considered for quantitation. Protein -fold changes were derived as median peptide -fold change. p values were calculated using a one-way t test (arbitrarily set to 1 for non-significant single peptide quantitations). Data were visualized for further analysis using Spotfire.
Quantitative diGly Proteomics
BT-549 cells stably transduced with an inducible HUWE1 shRNA (HUWE1 shRNA 1) and metabolically labeled with heavy or light amino acids were treated in the absence or presence of doxycycline for 72 h followed by incubation with 30 μm MG132 for 4 h. Cells were lysed in 9 m urea and mixed in a 1:1 ratio according to protein concentration determined with the modified Bradford assay (Pierce). A total protein input of ∼20 mg was used. Lysates were reduced with 10 mm DTT (Indofine Chemicals) at 30 °C for 30 min followed by alkylation with 25 mm iodoacetamide (Sigma) for 30 min at room temperature in the dark. Lysates were diluted to a final urea concentration of ∼1 m with 20 mm HEPES, pH 7.0; digested 1:100 with trypsin (Pierce) overnight at room temperature; acidified; desalted with Sep-Pak C18 cartridge (Waters); and lyophilized for 3 days.
Peptides containing the diglycyl remnant were enriched using K-ϵ-GG affinity resin (Cell Signaling Technology) according to the manufacturer's instructions. Digests were reconstituted in 1.4 ml of immunoaffinity purification buffer containing 50 mm MOPS, 10 mm Na2HPO4, and 50 mm NaCl, pH adjusted to 7.2 with NaOH. Resuspended peptides were cleared of any remaining precipitates with a 1-min 1,800 × g spin and placed on ice. One aliquot (∼40-μl packed bead volume) was washed three times with cold immunoaffinity purification buffer and mixed with the peptide sample. Incubation of sample and beads was performed with gentle end-over-end rotation at 4 °C for 2 h followed by a 1-min 2,000 × g spin to pellet the beads. The supernatant was removed and retained as the flow-through fraction. The antibody beads were washed twice with cold immunoaffinity purification buffer followed by two washes with ice-cold water. Ubiquitinated peptides were eluted from the beads with the addition of 50 μl of 0.15% trifluoroacetic acid (TFA) and allowed to stand at room temperature for 5 min. After a 1-min 2,000 × g spin, the supernatant was carefully removed and retained. A second 50-μl aliquot of 0.15% TFA was added to the beads followed by a 1-min 2,000 × g spin, and the supernatant was added to the first elution. Eluted peptides were cleared of any stray beads or potential antibody contamination using C18 StageTips (Proxeon). Tips were washed twice with 20 μl of 80% acetonitrile in 5% formic acid followed by equilibration with two aliquots of 5% formic acid. Sample was loaded to the tips in two steps (50 μl each), and flow-through was retained. A final wash with 20 μl of 30% acetonitrile in 5% formic acid was used to elute any peptides retained by the C18 membrane. Purified peptides were dried to completion and analyzed using nanospray LC-MS/MS.
A nano-LC column was prepared by creating a pulled tip with a P-2000 laser puller (Sutter Instruments) and packing the 75-μm-inner diameter fused silica with C18 material (Dr. Maisch Reprosil Pur120, C18AQ, 3 μm) to a length of 15 cm and fitted to the Eksigent nano-LC (AB SCIEX). The eluted peptides were reconstituted in 100 μl of 2% acetonitrile in 0.1% formic acid (LC Buffer A), and 25 μl was injected to the mass spectrometer (hybrid LTQ Orbitrap Velos Elite, Thermo Fisher Scientific) using a trapping column (1-cm Michrom Magic C18AQ, 5 μm), washed for 20 min, and then switched in-line with the analytical column. The peptides were eluted with a gradient of 3% Buffer B (70% acetonitrile in 0.1% formic acid) to 45% B in 80 min (0.5% B/min) delivered at a flow rate of 300 nl/min and using a top 20 collision-induced dissociation analysis method with dynamic exclusion set to 2. Data were analyzed with MaxQuant version 1.3.0.5 (Andromeda search engine with MaxQuant quantitation), searched against the UniProt Human database (V9 plus typical lab contaminants) with the addition of a MaxQuant-generated reversed database to calculate false discovery rates. Default settings were used with the exception of allowing three missed cleavages. Variable modifications of diglycyl-Lys (restricted to internal lysines only), oxidized methionine, and N-terminal acetylation were applied; carbamidomethylation of cysteine was the sole static modification. Data were visualized with Spotfire.
diGly Experiment 2 was conducted as described above with essentially the following exceptions as reported previously (53). After trypsinolysis, peptides were desalted with DSC18 tubes (Supelco). Peptides were separated by reversed phase HPLC at pH 10. Samples were separated using a Dionex 3000 fitted with a 6.4 × 150-mm Zorbax C18 Extend column with a flow rate of 1 ml/min. Eighty 1-ml fractions were collected throughout the segmented gradient (0–8% B/7 min, 8–27% B/38 min, 27–31% B/4 min, 31–39% B/8 min, 39–60% B/7 min, 60–0% B/20 min) formed with acetonitrile with a constant 4% 5 mm ammonium formate, pH 10. Fractions were pooled non-contiguously (every eighth fraction) to obtain a total of eight fractions that were individually subjected to diGly enrichment. Prior to diGly peptide enrichment, the K-ϵ-GG resin was treated with cross-linking agent as described previously (53) to minimize antibody loss particularly during peptide elution. Approximately 40 μl (packed bead volume) of cross-lined K-ϵ-GG resin was split evenly into eight aliquots for diGly enrichment.
Immunoblot Analysis
Samples were resuspended in lithium dodecyl sulfate sample buffer and resolved on NuPAGE gels (Invitrogen) prior to immunoblotting. The following antibodies were used in immunoblots: rabbit anti-HUWE1 (Bethyl Laboratories), mouse anti-FLAG (Sigma), mouse anti-tubulin (Sigma), rabbit anti-USP7 (Cell Signaling Technology), rabbit anti-MCL-1 (Cell Signaling Technology), mouse anti-ubiquitin (Cell Signaling Technology), rabbit anti-DDIT4 (Proteintech), mouse anti-V5 (Invitrogen), and rat anti-hemagglutinin (HA) (Roche Applied Science). Immune complexes were detected by chemiluminescence using a ChemiDoc XRS imager (Bio-Rad). Immunoblots using tubulin antibodies served as loading controls.
Co-immunoprecipitation Assays
For co-immunoprecipitation assays of potential HUWE1 binding partners, cells were transfected with plasmid DNA and incubated for 24 h. Cells were washed with PBS, incubated in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, and 1× protease inhibitor cocktail (Sigma)) for 15 min on ice, and centrifuged at 17,000 × g for 15 min at 4 °C. Clarified whole cell lysates were incubated with 10 μl of FLAG affinity resin (Sigma) overnight and washed three times with lysis buffer, and the precipitates were subjected to immunoblot analysis. To validate an interaction between DDIT4-FLAG and endogenous HUWE1, a co-immunoprecipitation reaction was performed as described above. Assays demonstrating an interaction between endogenous DDIT4 and streptavidin binding peptide (SBP)-HUWE1 were carried out in a similar fashion with the exception that instead of FLAG resin high capacity streptavidin resin (Pierce) was implemented. For detection of an endogenous DDIT4-HUWE1 complex, HeLa cells were lysed and clarified as described above. Whole cell lysates were incubated with either anti-DDIT4 antibody and Protein G-Sepharose (GE Healthcare) or control IgG (Abcam) and Protein G-Sepharose. Samples were resuspended in lithium dodecyl sulfate buffer and subjected to immunoblotting.
Cell-based Ubiquitination Assays under Denaturing Conditions
HeLa cells expressing an inducible HUWE1 shRNA were transfected with an N-terminal 3×FLAG-tagged DDIT4 construct, incubated in the presence or absence of doxycycline, treated with 30 μm MG132 (Tocris) for 6 h, and washed with PBS. Cell pellets were collected by centrifugation at 17,000 × g for 15 min at 4 °C. Pellets were resuspended in 110 μl of denaturing buffer (50 mm Tris-HCl, pH 7.4, 1% SDS, and 5 mm DTT) and boiled for 10 min. Subsequently lysates were diluted with 1.2 ml of cold lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, and 1× protease inhibitor cocktail (Sigma)) and centrifuged at 17,000 × g for 15 min at 4 °C. Cleared whole cell lysate was incubated with 20 μl of FLAG resin (Sigma) overnight. Resin was washed three times with lysis buffer, and immunoprecipitated DDIT4 was eluted using 3×FLAG peptide (Sigma). Ubiquitination levels of DDIT4-FLAG were detected on a ChemiDoc XRS imager (Bio-Rad), and protein levels were quantitated using Image Lab software version 3.0 (Bio-Rad). DDIT4 ubiquitination levels were normalized to total immunoprecipitated DDIT4-FLAG.
Cycloheximide Chase Assays
HeLa cells expressing an inducible HUWE1 shRNA were incubated in the presence or absence of 50 ng/ml doxycycline for 72 h. Subsequently cells were treated with 100 μg/ml cycloheximide (Calbiochem) to block de novo protein synthesis, and cells were harvested at the indicated time points. The ChemiDoc XRS imager (Bio-Rad) was used to capture immunoblot images, and protein levels were quantitated using Image Lab software version 3.0 (Bio-Rad). DDIT4 protein levels were normalized to tubulin. Curve fitting and half-life calculations were performed using GraphPad Prism version 6.02.
Quantitative PCR
RNA was isolated with the RNeasy plus kit (Qiagen). cDNA was synthesized using TaqMan reverse transcription reagents (Applied Biosystems) according to the manufacturer's instructions. Quantitative real time PCR was performed on an Applied Biosystems ViiA 7 instrument. Relative HUWE1 and DDIT4 mRNA expression levels were normalized to β-glucuronidase mRNA levels in each sample.
RESULTS
HUWE1 Affinity Proteomics Identifies USP7 as an Interacting Partner
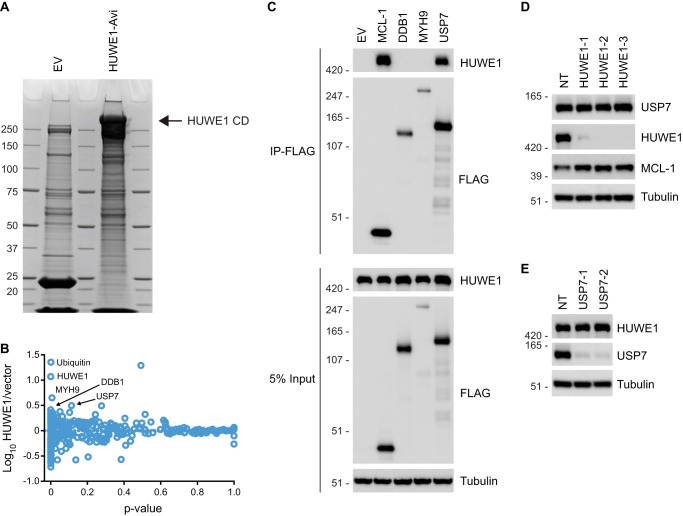
Multiple groups have linked HUWE1 to their protein of interest through affinity proteomics using their protein of interest as bait (40, 44, 54). Because we were interested in exploring HUWE1 substrates in an unbiased way, we decided to use HUWE1 as bait for affinity proteomics. HEK 293T cells were transiently transfected with an Avi-tagged CD HUWE1 expression construct or an empty vector control. Following lysis, streptavidin purification, and biotin elution, eluates were resolved by SDS-PAGE. Coomassie staining revealed a high molecular mass (>250 kDa) band in the sample from HUWE1-transfected cells, whereas in the empty vector control sample, no corresponding band was observed (Fig. 1A), suggesting that the transfection and purification steps were successful. Affinity enrichment followed by quantitative mass spectrometry analysis using isobaric labeling tags showed that among proteins enriched in the HUWE1-expressing samples were ubiquitin, DDB1, MYH9, and USP7 (Fig. 1B and supplemental Data Set S1). Considering that HUWE1 is an E3 ubiquitin ligase, is reported to be ubiquitinated at multiple lysines (GGBase), and contains a ubiquitin binding domain (UBA), it was not surprising that ubiquitin co-elutes with HUWE1. To validate the interaction between HUWE1 and other top hits from the affinity proteomics experiment, co-immunoprecipitation assays were performed. HEK 293T cells were transfected with a construct expressing FLAG-tagged DDB1, MYH9, or USP7. Cells were also transfected with a FLAG-tagged MCL-1 construct or empty vector as a positive and negative control, respectively. As expected, immunoprecipitation with FLAG affinity resin confirmed the interaction between FLAG-tagged MCL-1 and endogenous HUWE1. Among the three candidates tested, USP7-FLAG clearly precipitated endogenous HUWE1 (Fig. 1C). USP7 is a DUB that has been linked to cancers through its ability to regulate various tumor suppressors and oncogenes and has received much attention as a potential therapeutic target (55). Next, we examined whether USP7 could be a substrate for HUWE1 using RNAi. HEK 293T cells were transiently transfected with a non-targeting control siRNA or three unique HUWE1 siRNAs for 72 h. Knockdown of HUWE1 increased accumulation of the known HUWE1 substrate MCL-1 but had no effect on USP7 protein levels (Fig. 1D), suggesting that USP7 is not targeted for proteasomal degradation by HUWE1. We then tested whether USP7 could affect HUWE1 protein accumulation because HUWE1 can undergo autoubiquitination (56) and DUBs are known to regulate E3s (10). Depletion of USP7 by RNAi had no effect on HUWE1 protein expression in HEK 293T cells (Fig. 1E). Thus, USP7 is an interacting partner for HUWE1, but the functional implications for this interaction in HEK 293T cells remain to be determined.
FIGURE 1.
HUWE1 interacts with USP7 but does not target the protein for degradation. A, HEK 293T cells were transfected with empty vector (EV) or Avi-tagged CD HUWE1 and subsequently treated with MG132 prior to lysis. Lysates were incubated with streptavidin resin, resolved by SDS-PAGE, and stained with Coomassie Blue. B, scatter plot representing proteins identified by LC-MS/MS following pull-down of Avi-tagged CD HUWE1. Plotted on the y axis are the log10 ratios of proteins from cells expressing HUWE1 compared with a vector control sample. On the x axis is the p value indicating statistical significance of each protein. C, HEK 293T cells were transfected with FLAG-tagged candidate HUWE1-interacting partners or empty vector (EV) for 24 h. Input whole cell lysates were subjected to immunoprecipitation (IP) with FLAG affinity resin, and samples were analyzed by immunoblotting. D, immunoblot analysis of USP7 from HEK 293T cells transfected with a non-targeting (NT) control siRNA or HUWE1 siRNAs for 72 h. E, HEK 293T cells were transfected with a non-targeting (NT) control or USP7 siRNAs for 72 h and analyzed by immunoblotting.
diGly Proteomics Identifies DDIT4 as a Potential HUWE1 Substrate
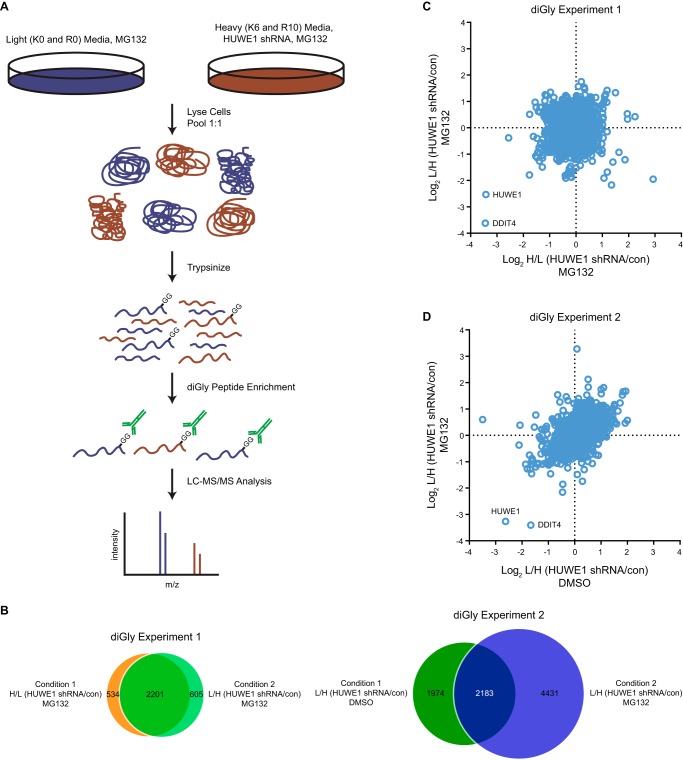
As an alternative approach to identify HUWE1 substrates in an unbiased manner, we next performed quantitative diGly proteomics. BT-549 cells, which are derived from a breast carcinoma and are sensitive to loss of HUWE1 expression (43), were stably transduced with a lentivirus encoding a doxycycline (Dox)-inducible shRNA targeting HUWE1. Cells were cultured with media containing heavy Lys and Arg isotopes (K6 and R10) or with standard light media (K0 and R0). Following several passages with light or heavy media, cells were treated with or without Dox. Dox treatment caused a pronounced reduction in HUWE1 protein levels with concomitant increase of MCL-1 in both metabolically labeled cell populations (data not shown). For quantitative diGly proteomics, cells cultured with heavy amino acids were treated with Dox to induce HUWE1 knockdown, and cells cultured with light amino acids were left untreated (Condition 1). The proteasome inhibitor MG132 was added to both cell populations to promote accumulation of ubiquitinated proteins. Subsequently, cells were lysed, and equal amounts of protein from each sample were combined and trypsinized. Using a diGly antibody, peptides were precipitated and analyzed by LC-MS/MS (Fig. 2A). A “label swap” experiment was conducted in which cells cultured with light amino acids were treated with Dox to abrogate HUWE1 expression, whereas cells cultured with heavy amino acids were not (Condition 2). Proteomics data revealed that 2,735 diGly peptides were quantitated in Condition 1, and 2,806 diGly peptides were quantitated in Condition 2 (diGly Experiment 1; Fig. 2B). Furthermore, the identities of the diGly peptides quantitated in the two conditions displayed a high degree of overlap (∼80%) (diGly Experiment 1; Fig. 2B). Analysis of SILAC ratios indicated that only two diGly peptides decreased by more than 75% following HUWE1 knockdown in both conditions. One peptide mapped to HUWE1, and the other mapped to DDIT4 (also known as REDD1, Dig2, and Rtp801) (Fig. 2C and supplemental Data Set S2). Because HUWE1 expression was ablated via RNAi, it was expected that levels of all HUWE1 peptides irrespective of ubiquitination levels would be lower. In contrast, DDIT4 appeared to be a candidate protein whose ubiquitination is strongly regulated by HUWE1.
FIGURE 2.
Quantitative diGly proteomics implicates DDIT4 as an HUWE1 substrate. A, schematic illustrating a quantitative diGly proteomics experiment to identify possible HUWE1 substrates. B, Venn diagram of all diGly peptides identified and quantitated in the two quantitative diGly proteomics experiments. C and D, scatter plot representing peptides identified by LC-MS/MS following diGly affinity purification in BT-549 cells. C, log2 ratios of H/L (HUWE1 shRNA/con) on the x axis and L/H (HUWE1 shRNA/con) conditions on the y axis from cells treated with MG132. D, log2 ratios of L/H (HUWE1 shRNA/con) from cells treated with DMSO on the x axis and cells treated with MG132 on the y axis. con, control, L, light, and H, heavy.
To test whether general stabilization of UPS substrates by MG132 treatment affected our ability to detect significant relative -fold changes for ubiquitinated peptides upon HUWE1 knockdown, we performed an additional diGly proteomics experiment with a modified protocol providing additional analytical depth (see “Experimental Procedures”). BT-549 cells cultured with light amino acids expressing an inducible HUWE1 shRNA were treated with Dox to induce HUWE1 knockdown, and cells cultured with heavy amino acids were not treated. Both cell populations were incubated with DMSO (Condition 1). A parallel experiment was also performed where both light and heavy labeled cells were incubated with MG132 (Condition 2). When cells were treated with MG132, 6,614 diGly peptides were quantitated compared with 4,157 diGly peptides quantitated from cells treated with DMSO (diGly Experiment 2; Fig. 2B) in agreement with previous reports that proteasome inhibition increases the number of peptides identified by quantitative diGly proteomics (53). Moreover, only ∼52.5% of diGly peptides quantitated in the DMSO condition were also identified and quantitated in the MG132-treated condition (diGly Experiment 2; Fig. 2B), suggesting a certain degree of qualitative difference in the ubiquitinome in the presence and absence of proteasome perturbation. Peptides mapping to HUWE1 and DDIT4 demonstrated the greatest reduction across both conditions, consistent with results obtained in the first diGly proteomics experiment, further suggesting that DDIT4 may be a substrate of HUWE1 (Fig. 2D and supplemental Data Set S3).
HUWE1 Interacts with DDIT4 and Regulates Its Polyubiquitination Levels
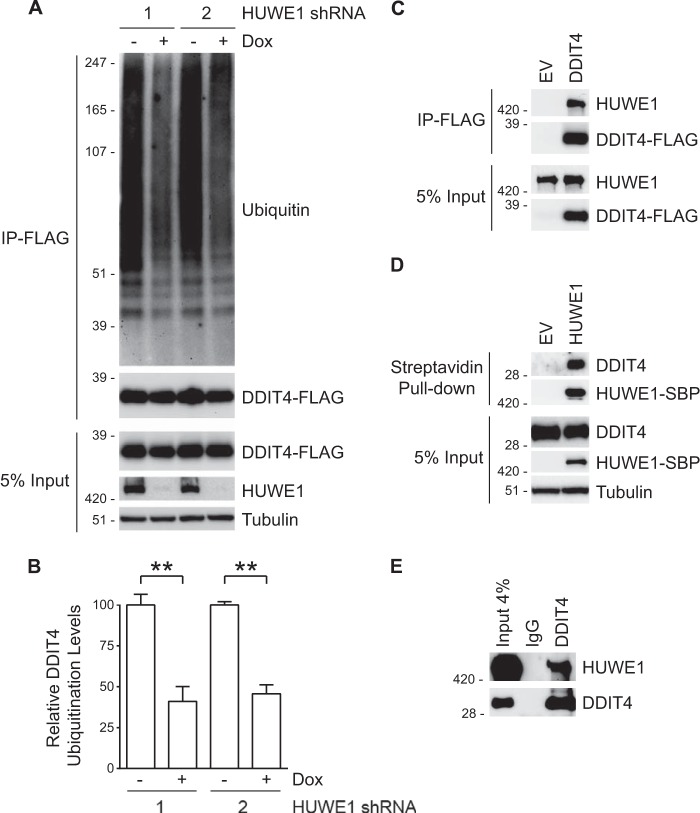
To confirm the results of the diGly proteomics indicating that reduction of HUWE1 reduces ubiquitination of DDIT4, we carried out a cell-based ubiquitination assay. HeLa cells expressing Dox-inducible HUWE1 shRNAs were transfected with a DDIT4-FLAG construct, incubated in the absence or presence of Dox to control HUWE1 expression, and treated with MG132. Following lysis under denaturing conditions (57), DDIT4-FLAG was immunoprecipitated with FLAG affinity resin. Ubiquitination levels were assessed by immunoblotting. When HUWE1 expression was unperturbed (− Dox), accumulation of high molecular weight ubiquitinated species was observed. However, when HUWE1 expression was abrogated with either of two unique HUWE1 shRNAs (+ Dox) the ubiquitin signal decreased ∼55–60% (Fig. 3, A and B).
FIGURE 3.
DDIT4 interacts with HUWE1, and its ubiquitination status is regulated by the E3. A, HeLa cells stably expressing two unique Dox-inducible HUWE1 shRNAs were treated in the absence and presence of Dox for 48 h, transfected with FLAG-tagged DDIT4 for 24 h, and incubated with MG132 for an additional 6 h. Input whole cell lysates were subjected to immunoprecipitation (IP) with FLAG resin, and samples were analyzed by immunoblotting. B, quantification of ubiquitinated DDIT4-FLAG from three assays as described in A. Data are represented as the means ± S.E. with p values determined using Student's unpaired t test (**, p < 0.005). Error bars represent S.E. C, HeLa cells were transfected with DDIT4-FLAG or an empty vector (EV) control for 24 h. Following immunoprecipitation with FLAG resin, samples were analyzed by immunoblotting. D, immunoblot analysis of lysates from HeLa cells transfected with SBP-tagged HUWE1 or empty vector (EV) for 24 h and subjected to streptavidin pull-down. E, endogenous HUWE1 can be immunoprecipitated with anti-DDIT4 resin from HeLa cell extracts. Rabbit IgG served as a control.
Although quantitative proteomics and cell-based ubiquitination assays are consistent with HUWE1 modulating levels of DDIT4 ubiquitination, these approaches do not address whether HUWE1 directly regulates DDIT4. To demonstrate that HUWE1 and DDIT4 interact in cells, HeLa cells were transfected with DDIT4-FLAG. Co-immunoprecipitation experiments demonstrated that endogenous HUWE1 interacts with DDIT4-FLAG (Fig. 3C). Reciprocal experiments confirmed that SBP-tagged HUWE1 precipitates endogenous DDIT4 (Fig. 3D). Furthermore, immunoprecipitation of DDIT4 indicates that HUWE1 and DDIT4 form an endogenous complex (Fig. 3E). Taken together these data suggest that HUWE1 interacts with DDIT4 and regulates DDIT4 polyubiquitination.
DDIT4 Stability Is Controlled via HUWE1-dependent Proteasomal Degradation
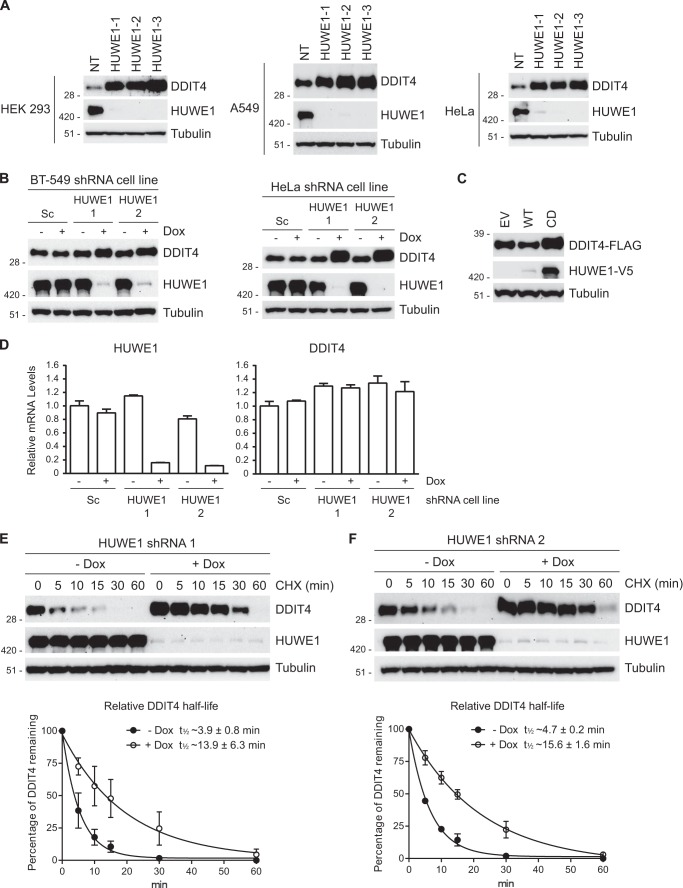
Posttranslational modification with ubiquitin can have myriad effects on protein function including regulation of interactions, localization, activity, and stability (9). Consistent with published observations (58, 59), steady-state protein levels of endogenous DDIT4 were elevated in cells treated with MG132 compared with cells treated with DMSO, indicating that DDIT4 stability is regulated by the proteasome (data not shown). To test whether HUWE1 regulates the stability of DDIT4, we conducted a series of RNAi experiments. Knockdown of HUWE1 using shRNAs and siRNAs increased accumulation of DDIT4 in multiple human cell lines (Fig. 4, A and B). Co-transfection of DDIT4-FLAG and WT HUWE1-V5 reduced DDIT4-FLAG levels. Conversely, co-expression of DDIT4-FLAG and CD HUWE1-V5 caused increased accumulation of DDIT4-FLAG (Fig. 4C) ostensibly through a dominant-negative mechanism, suggesting that the ubiquitin ligase activity of HUWE1 is required for its regulation of DDIT4. Consistent with HUWE1 regulating DDIT4 stability through a posttranslational mechanism, mRNA levels of DDIT4 were unchanged with loss of HUWE1 expression (Fig. 4D). Moreover, DDIT4 ectopically expressed from a plasmid that does not contain its endogenous promoter or untranslated regions displayed enhanced accumulation following HUWE1 knockdown (see below).
FIGURE 4.
HUWE1 regulates the accumulation and stability of DDIT4. A, several human cells lines were transfected with non-targeting (NT) control or three HUWE1 siRNAs for 72 h and analyzed by immunoblotting. B, HeLa and BT-549 cells stably expressing Dox-inducible scrambled (Sc) or two unique HUWE1 shRNAs were treated in the absence or presence of Dox for 72 h. Immunoblotting demonstrates that silencing of HUWE1 increases steady-state levels of DDIT4. C, HEK 293T cells were co-transfected with plasmids expressing DDIT4-FLAG and either empty vector (EV), WT, or CD HUWE1-V5 proteins for 24 h. DDIT4-FLAG levels were assessed by immunoblot analysis. D, HeLa cells stably expressing Dox-inducible scrambled (Sc) or two unique HUWE1 shRNAs were treated in the absence or presence of Dox for 72 h. Relative HUWE1 and DDIT4 mRNA levels were measured by quantitative PCR. Assays were performed in triplicate with data represented as the means ± S.E. E and F, HeLa cells stably expressing a Dox-inducible HUWE1 shRNA were treated in the absence or presence of Dox for 72 h. Cells were incubated with cycloheximide (CHX) and harvested at the indicated time points. Top panels, DDIT4 levels as measured by immunoblotting. Bottom panels, half-life of DDIT4 based on the quantitation of three independent immunoblots. Error bars represent S.E.
To further test whether HUWE1 regulates the stability of DDIT4, we carried out a cycloheximide chase assay. A previous report estimated the half-life of DDIT4 at ∼5 min or less (60). In HeLa cells expressing a Dox-inducible HUWE1 shRNA (HUWE1 shRNA 1), DDIT4 had a relative half-life of ∼3.9 min in the absence of Dox. Addition of Dox resulted in an ∼3.6-fold increase in DDIT4 half-life (∼13.9 min; Fig. 4E). Analogously, knockdown of HUWE1 with a second shRNA (HUWE1 shRNA 2) caused a ∼3.3-fold increase in DDIT4 half-life (∼4.7 versus ∼15.6 min; Fig. 4F).
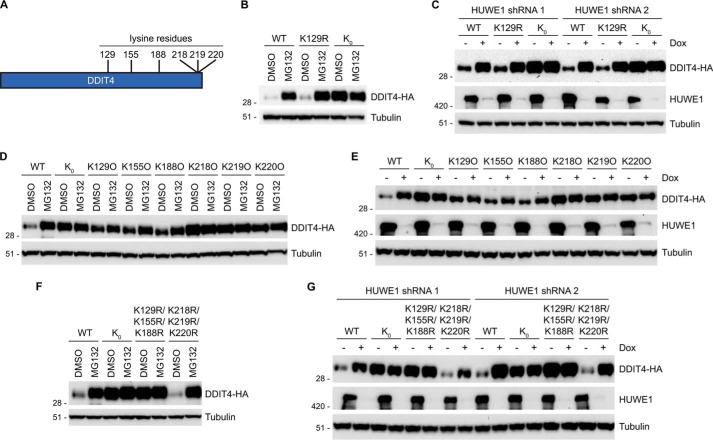
Mapping of Lysine Residues Involved in Regulation of DDIT4
DDIT4 is a 232 amino acid-containing protein with six lysines at positions 129, 155, 188, 218, 219, and 220 (Fig. 5A). A DDIT4 tryptic peptide carrying a diGly modification at Lys-129 showed down-regulation upon HUWE1 knockdown (supplemental Data Sets S2 and S3), suggesting that Lys-129 may be a physiological site of HUWE1-dependent ubiquitination and hence required for DDIT4 regulation. To investigate the requirement of Lys-129 modification in the regulation of DDIT4, we generated a HA-tagged DDIT4 construct in which Lys-129 was mutated to Arg (K129R). Both HA-tagged WT and K129R DDIT4 displayed marked accumulation when cells were treated with MG132 compared with vehicle, indicating that modification of Lys-129 is not required for DDIT4 stabilization (Fig. 5B). We speculated that DDIT4 has additional ubiquitination sites not identified in the proteomics experiment and that HUWE1 is sufficiently flexible to recognize additional ubiquitination sites. We generated a lysine-less HA-tagged DDIT4 construct in which all six lysine residues were mutated to arginine (K0), to remove all possible Lys-based ubiquitination sites. In contrast to WT and K129R, K0 DDIT4 appeared to be constitutively stable in the absence and presence of proteasome inhibition (Fig. 5B). Consistently, WT and K129R DDIT4 showed preferentially higher levels when HUWE1 expression was abrogated via RNAi. In contrast, K0 DDIT4 had constitutively high levels in both control (− Dox) and HUWE1 knockdown (+ Dox) cells (Fig. 5C). These data suggest that multiple Lys residues are required for HUWE1-mediated proteasomal degradation of DDIT4.
FIGURE 5.
Multiple lysine residues are required for DDIT4 regulation. A, schematic demonstrating positions of lysine residues on DDIT4. B, 24 h after transfection HeLa cells were treated with either MG132 or DMSO for 6 h. WT and variant DDIT4-HA levels were assessed by immunoblot analysis. C, HeLa cells stably expressing two unique HUWE1 shRNAs were treated in the absence and presence of Dox for 48 h, transfected with WT and variant DDIT4-HA constructs for 24 h, and incubated with MG132 or DMSO for an additional 6 h. DDIT4-HA levels were determined by immunoblotting. D, DDIT4-HA constructs containing a single lysine residue were assayed as described in B. E, HeLa cells stably expressing inducible HUWE1 shRNA 1 were transfected with DDIT4-HA constructs containing a single lysine residue and analyzed as described in C. F and G, K129R/K155R/K188R and K218R/K219R/K220R DDIT4 constructs were analyzed as in B and C, respectively.
To further investigate potential lysines required for DDIT4 regulation, six DDIT4 constructs were generated in which five lysine residues were mutated to arginine, leaving only one of the six lysines present on the protein (K129O, K155O, K188O, K218O, K219O, and K220O). For example, the K129O construct contained Lys to Arg mutations at residues Lys-155, Lys-188, Lys-218, Lys-219, and Lys-220, leaving only Lys-129 available for ubiquitin modification. These six constructs were transfected into HeLa cells to assess their responsiveness to proteasome inhibition. K129O, K155O, and K188O DDIT4 showed at least a partial increase in expression when cells were treated with MG132, although the effects were less robust than with WT DDIT4. By comparison, K218O, K219O, and K22O DDIT4 appeared constitutively stable as their expression did not increase with MG132 treatment, phenocopying K0 DDIT4 (Fig. 5D). In agreement with MG132 treatment, similar results were observed with loss of HUWE1 expression (Fig. 5E). Thus, Lys-129, Lys-155, and Lys-188 are sufficient to confer some degree of regulation, suggesting that they might have a role in controlling DDIT4 levels.
To test this idea, a DDIT4 construct was generated in which Lys-129, Lys-155, and Lys-188 were mutated to Arg (K129R/K155R/K188R). K129R/K155R/K188R DDIT4 was insensitive to both proteasome inhibition and loss of HUWE1 expression akin to K0 DDIT4. However, a DDIT4 construct in which Lys-218, Lys-219, and Lys-220 were mutated to Arg (K218R/K219R/K220R) showed enhanced expression with MG132 treatment and HUWE1 knockdown comparable with WT DDIT4 (Fig. 5, F and G). Hence, these data strongly suggest that, like many E3 ligase substrates, DDIT4 contains multiple lysines that are required for its modification and proteasomal degradation. In this case, a combination of lysine residues, likely including Lys-129, Lys-155, and Lys-188, are modified to target DDIT4 for degradation. Collectively, the data presented here invoke a model in which HUWE1 regulates the stability of DDIT4 via polyubiquitination of multiple lysines, targeting DDIT4 for proteasomal degradation.
DISCUSSION
One of the biggest challenges facing the ubiquitin field is the elucidation of the complete cohort of substrates modified by each of the ∼600 E3 ubiquitin ligases encoded in the human genome (15). To find substrates of the E3 ubiquitin ligase HUWE1, affinity purification proteomics was performed. These data identified the DUB USP7 as a candidate interacting partner. Although immunoprecipitation assays indicated that HUWE1 interacts with USP7 in agreement with other studies (61, 62), the E3 does not appear to regulate USP7 stability via the UPS. It has been reported that USP7 can interact with and deubiquitinate HUWE1 presumably to protect the E3 ligase from autoubiquitination-mediated degradation in HeLa cells (61). However, loss of USP7 expression did not markedly alter HUWE1 protein levels in HEK 293T cells, suggesting that this regulation may not occur in all cell types.
Of note, various proteasomal subunits were identified in the HUWE1 affinity proteomics (supplemental Data Set S1) data, consistent with previous reports suggesting that HUWE1 can interact with the proteasome (63, 64). However, the significance of these potential interactions requires further investigation.
Because HUWE1-based affinity proteomics did not lead to the identification of HUWE1 substrates, we aimed to address this question with quantitative diGly proteomics. Two diGly proteomics experiments strongly hinted that DDIT4 is a substrate of HUWE1, and several cell-based assays confirmed the E3-substrate relationship. Immunoblotting data showed that repressing HUWE1 expression decreases DDIT4 ubiquitination levels in human cells. DDIT4 stability appears to be regulated by HUWE1-mediated polyubiquitination and subsequent proteasomal degradation. It was recently reported that knockdown of HUWE1 has no effect on DDIT4 half-life (59). One explanation for this difference may be the knockdown efficiency of HUWE1 in the two experiments. The shRNAs used here appear to cause a more pronounced reduction of HUWE1 expression compared with the siRNAs in the previous report. Regardless, we demonstrated that HUWE1 knockdown by two unique shRNAs led to an ∼3.5-fold increase in the apparent half-life of DDIT4. These same shRNAs decreased DDIT4 ubiquitination and caused aberrant accumulation of the protein.
Reduction of HUWE1 expression increased the half-life of DDIT4 from ∼4.3 to ∼14.8 min. Interestingly, even with blunted HUWE1 levels, DDIT4 is rapidly turned over compared with most proteins. This suggests either that residual HUWE1 remaining after shRNA knockdown is sufficient to continue to drive DDIT4 degradation or that other regulatory factors in the UPS or additional proteolytic mechanisms such as lysosomes or other proteases may be involved in DDIT4 regulation. The fact that HUWE1 knockdown and MG132 treatment have similar effects on the different lysine mutants of DDIT4 could suggest that HUWE1 is the key E3 ligase responsible for DDIT4 degradation via the proteasome under the conditions of our experiments.
diGly peptides mapping to previously identified HUWE1 substrates TP53, MFN2, c-MYC, and HDAC2 were found in our quantitative diGly proteomics experiments. However, in comparing HUWE1 knockdown with control samples, none of these diGly peptides showed a >2-fold decrease under any condition in either of the two quantitative diGly proteomics experiments (supplemental Data Sets S2 and S3). Although it was somewhat surprising that known HUWE1 substrates did not score higher in our assays, several factors may have contributed to these results. The cell line used for the diGly proteomics might affect the results of the experiments. For example, HUWE1 regulation of TP53 and c-MYC is not observed in every cell line (40), and n-MYC is generally expressed at high levels in human cell lines of neuronal origin (data not shown). Our screening approach examined diGly levels under basal conditions. HUWE1 substrates like MFN2 are preferentially regulated by the E3 under conditions of cell stress (44), and as a result, the corresponding ubiquitinated peptides might have been below the detection limit given the experimental design of the presented study. Conversely, greater coverage of the ubiquitinome in our diGly proteomics experiments might have led to identification of peptides (corresponding to both known and unknown HUWE1 substrates) that change significantly with HUWE1 reduction. We and others are working on further optimizing the underlying experimental workflows to increase the analytical depth of diGly proteomics. Nevertheless, the identification of DDIT4 as a potential HUWE1 substrate is compelling based on the magnitude and consistency in which the same DDIT4 diGly peptide scored in all quantitative diGly proteomics samples regardless of whether or not the proteasome was inhibited.
Inducible silencing of an E3 ligase or other component of the UPS in combination with quantitative diGly proteomics to identify substrate proteins presents several potential advantages over ectopic or overexpression methods. First, inhibiting an endogenously expressed E3 ligase may lead to the identification of more physiologically relevant substrates. Second, the ability to inducibly regulate expression of an E3 ligase reduces the chances of having a cell line adapt to the perturbation of the E3. Lastly, for large E3s like HUWE1 or other E3s that are tightly regulated by autoubiquitination or other mechanisms, overexpression can be difficult to accomplish even when transcription is driven from strong constitutive promoters. Using a lentivirus-based approach presents additional options as lentiviruses can be used to infect cells both in vitro (two-dimensional and three-dimensional models) and in vivo providing a broader context for knockdown of UPS components. Thus, when overexpression of an E3 of interest is not possible or a chemical inhibitor is not available, inducible gene silencing methods such as RNAi provide alternative strategies for pairing E3s with their cognate substrates. We also note that under the conditions used for HUWE1, diGly proteomics appeared superior to standard affinity proteomics for novel substrate identification.
DDIT4 is believed to function in multiple biological settings such as inhibiting mTORC1 signaling and regulating production of reactive oxygen species (58, 65–67). How DDIT4 exerts its effect in various signaling pathways remains incompletely understood. DDIT4 contains no predicted functional motifs, and a crystal structure of the C terminus revealed an α/β sandwich fold with no significant homology to any previously solved protein structures (68). In mice challenged with an environmental stress, reduced expression of DDIT4 has a protective effect. Apoptosis and pathological neovascularization are attenuated in the retina of Ddit4−/− mice compared with WT littermates subjected to an oxygen-induced retinopathy model (69). Chronic exposure to cigarette smoke causes emphysema in WT mice, whereas Ddit4−/− mice are largely shielded from the disease (70). In cancer cells, DDIT4 function appears to be context-dependent. In some cancer cell lines and mouse models, DDIT4 appears to act as an oncogene in a prosurvival role in part by blocking apoptosis (71, 72), whereas in other cell-based and in vivo experiments, it functions as a tumor suppressor (67, 73). When considering whether HUWE1 is an appropriate target for cancer or other disease indications, it should be noted that inhibition of HUWE1 with a concomitant increase in DDIT4 expression may have beneficial or detrimental effects for the therapeutic strategy.
Mechanistically, DDIT4 is transcriptionally up-regulated in response to a variety of cell perturbations including hypoxia (65, 74), DNA damage (58), and endoplasmic reticulum stress (72). Our data suggest an added posttranslational regulatory mechanism for DDIT4 via HUWE1-mediated proteasomal degradation. Thus, DDIT4 is the newest member of a cadre of proteins such as MFN2, MCL-1, TP53, and HDAC2 involved in cell stress response pathways whose stability is modulated by HUWE1.
We and others have demonstrated that DDIT4 has a half-life of ∼5 min. Unstable proteins are energetically unfavorable as they require constant synthesis. However, this class of proteins offers advantageous flexibility to a cell, enabling rapid responses to biological cues through a quick fine-tuning of their concentration (75). Hence, many short lived proteins like transcription factors, cyclins, and metabolic enzymes have critical regulatory functions. In the case of HUWE1-DDIT4, the biological or pathobiological function for this regulatory axis remains to be elucidated. Of particular interest is identifying conditions that trigger loss of HUWE1-dependent DDIT4 proteasomal degradation and the resulting phenotypic consequences. It is tempting to speculate that with the high energy investment due to the rapid turnover of DDIT4 an important regulatory payout awaits identification.
Overall, the observations reported here contribute to an expanding body of studies directed at using quantitative diGly proteomics to match ubiquitin ligases with their cognate substrates. Our results demonstrate that inducible knockdown of an E3 ligase in combination with diGly proteomics provides a potentially advantageous method for identifying novel substrates even for previously characterized E3s like HUWE1. As technologies for substrate identification continue to improve, we hope to further expand our understanding of the relevant substrates for E3 ubiquitin ligases to provide guidance for selecting promising drug target candidates.
Supplementary Material
Acknowledgments
We thank J. DaSilva and A. Carlson for technical assistance.

This article contains supplemental Data Sets S1–S3.
- DUB
- deubiquitinating enzyme
- HUWE1
- HECT, UBA, and WWE domain-containing 1, E3 ubiquitin protein ligase
- DDIT4
- DNA damage-inducible transcript 4
- UPS
- ubiquitin-proteasome system
- SILAC
- stable isotope labeling of amino acids in cell culture
- diGly
- Lys-ϵ-Gly-Gly
- CD
- catalytically dead
- SBP
- streptavidin binding peptide
- Dox
- doxycycline
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Pickart C. M. (2004) Back to the future with ubiquitin. Cell 116, 181–190 [DOI] [PubMed] [Google Scholar]
- 3. Schulman B. A., Harper J. W. (2009) Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ye Y., Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schrader E. K., Harstad K. G., Matouschek A. (2009) Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 7. Eisenhaber B., Chumak N., Eisenhaber F., Hauser M. T. (2007) The ring between ring fingers (RBR) protein family. Genome Biol. 8, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 9. Komander D., Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 10. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 11. Ventii K. H., Wilkinson K. D. (2008) Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bedford L., Lowe J., Dick L. R., Mayer R. J., Brownell J. E. (2011) Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 10, 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nalepa G., Rolfe M., Harper J. W. (2006) Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 5, 596–613 [DOI] [PubMed] [Google Scholar]
- 15. Williamson A., Werner A., Rape M. (2013) The Colossus of ubiquitylation: decrypting a cellular code. Mol. Cell 49, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin L., Pahuja K. B., Wickliffe K. E., Gorur A., Baumgärtel C., Schekman R., Rape M. (2012) Ubiquitin-dependent regulation of COPII coat size and function. Nature 482, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cano F., Bye H., Duncan L. M., Buchet-Poyau K., Billaud M., Wills M. R., Lehner P. J. (2012) The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J. 31, 3596–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salahudeen A. A., Thompson J. W., Ruiz J. C., Ma H. W., Kinch L. N., Li Q., Grishin N. V., Bruick R. K. (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yen H. C., Elledge S. J. (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322, 923–929 [DOI] [PubMed] [Google Scholar]
- 20. Emanuele M. J., Elia A. E., Xu Q., Thoma C. R., Izhar L., Leng Y., Guo A., Chen Y. N., Rush J., Hsu P. W., Yen H. C., Elledge S. J. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu G., Middleton R. E., Sun H., Naniong M., Ott C. J., Mitsiades C. S., Wong K. K., Bradner J. E., Kaelin W. G., Jr. (2014) The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Merbl Y., Kirschner M. W. (2009) Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc. Natl. Acad. Sci. U.S.A. 106, 2543–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Persaud A., Alberts P., Amsen E. M., Xiong X., Wasmuth J., Saadon Z., Fladd C., Parkinson J., Rotin D. (2009) Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4–2 using proteome arrays. Mol. Syst. Biol. 5, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Merbl Y., Refour P., Patel H., Springer M., Kirschner M. W. (2013) Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell 152, 1160–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tan M. K., Lim H. J., Bennett E. J., Shi Y., Harper J. W. (2013) Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol. Cell 52, 9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mark K. G., Simonetta M., Maiolica A., Seller C. A., Toczyski D. P. (2014) Ubiquitin ligase trapping identifies an SCF(Saf1) pathway targeting unprocessed vacuolar/lysosomal proteins. Mol. Cell 53, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis M. A., Larimore E. A., Fissel B. M., Swanger J., Taatjes D. J., Clurman B. E. (2013) The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 27, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu G., Paige J. S., Jaffrey S. R. (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Danielsen J. M., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., Nielsen M. L. (2011) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10, M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Udeshi N. D., Mani D. R., Eisenhaure T., Mertins P., Jaffe J. D., Clauser K. R., Hacohen N., Carr S. A. (2012) Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics 11, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 34. Krönke J., Udeshi N. D., Narla A., Grauman P., Hurst S. N., McConkey M., Svinkina T., Heckl D., Comer E., Li X., Ciarlo C., Hartman E., Munshi N., Schenone M., Schreiber S. L., Carr S. A., Ebert B. L. (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee K. A., Hammerle L. P., Andrews P. S., Stokes M. P., Mustelin T., Silva J. C., Black R. A., Doedens J. R. (2011) Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J. Biol. Chem. 286, 41530–41538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sarraf S. A., Raman M., Guarani-Pereira V., Sowa M. E., Huttlin E. L., Gygi S. P., Harper J. W. (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldknopf I. L., Busch H. (1977) Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. U.S.A. 74, 864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao X., D' Arca D., Lim W. K., Brahmachary M., Carro M. S., Ludwig T., Cardo C. C., Guillemot F., Aldape K., Califano A., Iavarone A., Lasorella A. (2009) The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev. Cell 17, 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. D'Arca D., Zhao X., Xu W., Ramirez-Martinez N. C., Iavarone A., Lasorella A. (2010) Huwe1 ubiquitin ligase is essential to synchronize neuronal and glial differentiation in the developing cerebellum. Proc. Natl. Acad. Sci. U.S.A. 107, 5875–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao X., Heng J. I., Guardavaccaro D., Jiang R., Pagano M., Guillemot F., Iavarone A., Lasorella A. (2008) The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 10, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. (2005) ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 42. Zhong Q., Gao W., Du F., Wang X. (2005) Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121, 1085–1095 [DOI] [PubMed] [Google Scholar]
- 43. Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Kogel U., Scheffner M., Helin K., Eilers M. (2005) The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123, 409–421 [DOI] [PubMed] [Google Scholar]
- 44. Leboucher G. P., Tsai Y. C., Yang M., Shaw K. C., Zhou M., Veenstra T. D., Glickman M. H., Weissman A. M. (2012) Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol. Cell 47, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang J., Kan S., Huang B., Hao Z., Mak T. W., Zhong Q. (2011) Mule determines the apoptotic response to HDAC inhibitors by targeted ubiquitination and destruction of HDAC2. Genes Dev. 25, 2610–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoon S. Y., Lee Y., Kim J. H., Chung A. S., Joo J. H., Kim C. N., Kim N. S., Choe I. S., Kim J. W. (2005) Over-expression of human UREB1 in colorectal cancer: HECT domain of human UREB1 inhibits the activity of tumor suppressor p53 protein. Biochem. Biophys. Res. Commun. 326, 7–17 [DOI] [PubMed] [Google Scholar]
- 47. Confalonieri S., Quarto M., Goisis G., Nuciforo P., Donzelli M., Jodice G., Pelosi G., Viale G., Pece S., Di Fiore P. P. (2009) Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene 28, 2959–2968 [DOI] [PubMed] [Google Scholar]
- 48. Froyen G., Corbett M., Vandewalle J., Jarvela I., Lawrence O., Meldrum C., Bauters M., Govaerts K., Vandeleur L., Van Esch H., Chelly J., Sanlaville D., van Bokhoven H., Ropers H. H., Laumonnier F., Ranieri E., Schwartz C. E., Abidi F., Tarpey P. S., Futreal P. A., Whibley A., Raymond F. L., Stratton M. R., Fryns J. P., Scott R., Peippo M., Sipponen M., Partington M., Mowat D., Field M., Hackett A., Marynen P., Turner G., Gécz J. (2008) Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am. J. Hum. Genet. 82, 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Turner G., Gedeon A., Mulley J. (1994) X-linked mental retardation with heterozygous expression and macrocephaly: pericentromeric gene localization. Am. J. Med. Genet. 51, 575–580 [DOI] [PubMed] [Google Scholar]
- 50. Froyen G., Belet S., Martinez F., Santos-Rebouças C. B., Declercq M., Verbeeck J., Donckers L., Berland S., Mayo S., Rosello M., Pimentel M. M., Fintelman-Rodrigues N., Hovland R., Rodrigues dos Santos S., Raymond F. L., Bose T., Corbett M. A., Sheffield L., van Ravenswaaij-Arts C. M., Dijkhuizen T., Coutton C., Satre V., Siu V., Marynen P. (2012) Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am. J. Hum. Genet. 91, 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Love K. R., Pandya R. K., Spooner E., Ploegh H. L. (2009) Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem. Biol. 4, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wiederschain D., Wee S., Chen L., Loo A., Yang G., Huang A., Chen Y., Caponigro G., Yao Y. M., Lengauer C., Sellers W. R., Benson J. D. (2009) Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 [DOI] [PubMed] [Google Scholar]
- 53. Udeshi N. D., Svinkina T., Mertins P., Kuhn E., Mani D. R., Qiao J. W., Carr S. A. (2013) Refined preparation and use of anti-diglycine remnant (K-ϵ-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yin L., Joshi S., Wu N., Tong X., Lazar M. A. (2010) E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation of the circadian heme receptor Rev-erbα. Proc. Natl. Acad. Sci. U.S.A. 107, 11614–11619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nicholson B., Suresh Kumar K. G. (2011) The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem. Biophys. 60, 61–68 [DOI] [PubMed] [Google Scholar]
- 56. Pandya R. K., Partridge J. R., Love K. R., Schwartz T. U., Ploegh H. L. (2010) A structural element within the HUWE1 HECT domain modulates self-ubiquitination and substrate ubiquitination activities. J. Biol. Chem. 285, 5664–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bloom J., Pagano M. (2005) Experimental tests to definitively determine ubiquitylation of a substrate. Methods Enzymol. 399, 249–266 [DOI] [PubMed] [Google Scholar]
- 58. Ellisen L. W., Ramsayer K. D., Johannessen C. M., Yang A., Beppu H., Minda K., Oliner J. D., McKeon F., Haber D. A. (2002) REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell 10, 995–1005 [DOI] [PubMed] [Google Scholar]
- 59. Tan C. Y., Hagen T. (2013) mTORC1 dependent regulation of REDD1 protein stability. PLoS One 8, e63970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kimball S. R., Do A. N., Kutzler L., Cavener D. R., Jefferson L. S. (2008) Rapid turnover of the mTOR complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following inhibition of protein synthesis. J. Biol. Chem. 283, 3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Khoronenkova S. V., Dianov G. L. (2013) USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic Acids Res. 41, 1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Poulsen E. G., Steinhauer C., Lees M., Lauridsen A. M., Ellgaard L., Hartmann-Petersen R. (2012) HUWE1 and TRIP12 collaborate in degradation of ubiquitin-fusion proteins and misframed ubiquitin. PLoS One 7, e50548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tai H. C., Besche H., Goldberg A. L., Schuman E. M. (2010) Characterization of the brain 26S proteasome and its interacting proteins. Front. Mol. Neurosci. 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reiling J. H., Hafen E. (2004) The hypoxia-induced paralogs scylla and charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 18, 2879–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brugarolas J., Lei K., Hurley R. L., Manning B. D., Reiling J. H., Hafen E., Witters L. A., Ellisen L. W., Kaelin W. G., Jr. (2004) Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Horak P., Crawford A. R., Vadysirisack D. D., Nash Z. M., DeYoung M. P., Sgroi D., Ellisen L. W. (2010) Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 4675–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vega-Rubin-de-Celis S., Abdallah Z., Kinch L., Grishin N. V., Brugarolas J., Zhang X. (2010) Structural analysis and functional implications of the negative mTORCl regulator REDD1. Biochemistry 49, 2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brafman A., Mett I., Shafir M., Gottlieb H., Damari G., Gozlan-Kelner S., Vishnevskia-Dai V., Skaliter R., Einat P., Faerman A., Feinstein E., Shoshani T. (2004) Inhibition of oxygen-induced retinopathy in RTP801-deficient mice. Invest. Ophthalmol. Vis. Sci. 45, 3796–3805 [DOI] [PubMed] [Google Scholar]
- 70. Yoshida T., Mett I., Bhunia A. K., Bowman J., Perez M., Zhang L., Gandjeva A., Zhen L., Chukwueke U., Mao T., Richter A., Brown E., Ashush H., Notkin N., Gelfand A., Thimmulappa R. K., Rangasamy T., Sussan T., Cosgrove G., Mouded M., Shapiro S. D., Petrache I., Biswal S., Feinstein E., Tuder R. M. (2010) Rtp801, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke-induced pulmonary injury and emphysema. Nat. Med. 16, 767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schwarzer R., Tondera D., Arnold W., Giese K., Klippel A., Kaufmann J. (2005) REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. Oncogene 24, 1138–1149 [DOI] [PubMed] [Google Scholar]
- 72. Wang Z., Malone M. H., Thomenius M. J., Zhong F., Xu F., Distelhorst C. W. (2003) Dexamethasone-induced gene 2 (dig2) is a novel pro-survival stress gene induced rapidly by diverse apoptotic signals. J. Biol. Chem. 278, 27053–27058 [DOI] [PubMed] [Google Scholar]
- 73. DeYoung M. P., Horak P., Sofer A., Sgroi D., Ellisen L. W. (2008) Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 22, 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shoshani T., Faerman A., Mett I., Zelin E., Tenne T., Gorodin S., Moshel Y., Elbaz S., Budanov A., Chajut A., Kalinski H., Kamer I., Rozen A., Mor O., Keshet E., Leshkowitz D., Einat P., Skaliter R., Feinstein E. (2002) Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 22, 2283–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kristensen A. R., Gsponer J., Foster L. J. (2013) Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 9, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.