Background: Dysbindin-1 isoforms are selectively reduced in schizophrenic brains.
Results: Dysbindin-1C is required for the survival of hilar mossy cells and the maturation of adult newborn neurons in the dentate gyrus.
Conclusion: Dysbindin-1C, but not -1A, regulates adult hippocampal neurogenesis in a non-cell autonomous manner.
Significance: Reduced dysbindin-1C in the schizophrenic hippocampal formation likely contributes to the development of cognitive deficits.
Keywords: Apoptosis, Autophagy, Hippocampus, Neurogenesis, Schizophrenia, BLOC-1, Dysbindin-1
Abstract
DTNBP1 (dystrobrevin-binding protein 1), which encodes dysbindin-1, is one of the leading susceptibility genes for schizophrenia. Both dysbindin-1B and -1C isoforms are decreased, but the dysbindin-1A isoform is unchanged in schizophrenic hippocampal formation, suggesting dysbindin-1 isoforms may have distinct roles in schizophrenia. We found that mouse dysbindin-1C, but not dysbindin-1A, is localized in the hilar glutamatergic mossy cells of the dentate gyrus. The maturation rate of newborn neurons in sandy (sdy) mice, in which both dysbindin-1A and -1C are deleted, is significantly delayed when compared with that in wild-type mice or with that in muted (mu) mice in which dysbindin-1A is destabilized but dysbindin-1C is unaltered. Dysbindin-1C deficiency leads to a decrease in mossy cells, which causes the delayed maturation of newborn neurons. This suggests that dysbindin-1C, rather than dysbindin-1A, regulates adult hippocampal neurogenesis in a non-cell autonomous manner.
Introduction
Adult hippocampal neurogenesis, a unique form of neural network plasticity, including genesis of newborn neurons and establishment of new synapse contacts, is required for learning and memory (1, 2). Compelling evidence has shown that failing adult hippocampal neurogenesis is associated with the pathogenesis of schizophrenia, dementia, age-related cognitive decline, epilepsy, and major depression (3). However, how impaired adult hippocampal neurogenesis contributes to the pathogenesis of schizophrenia has not been extensively investigated. Remarkably, in addition to affecting prenatal and neonatal neurodevelopment, schizophrenia risk gene mutations impair adult-born neuronal development. The DISC1 (disrupted-in-schizophrenia 1) gene is a key regulator that orchestrates the tempo of functional neuronal integration in the adult mouse brain (4). Other risk genes have also been found to be involved in the regulation of adult hippocampal neurogenesis, such as NPAS3 (neuronal PAS domain protein 3) (5) and SREB2/GPR85 (G protein-coupled receptor 85) (6).
DTNBP1 (dystrobrevin-binding protein 1), which encodes dysbindin-1, is a major susceptibility gene for schizophrenia (7–9). It has shown that the sandy (sdy) mouse, which arose on the DBA/2J inbred strain and carries a spontaneously occurring large deletion (∼38 kb), which spans intron 5 to intron 7 in the Dtnbp1 gene (10), is a mouse model of schizophrenia (11–14). Neuronal differentiation of adult-born neurons in the dentate gyrus (DG)2 is impaired in sdy mice, although the underlying mechanism is unclear (15). Interestingly, synaptic dysbindin-1 protein levels in different brain regions of schizophrenia patients are reduced in an isoform-specific manner. Notably, dysbindin-1B and -1C isoforms are reduced but the dysbindin-1A isoform is unchanged in schizophrenic hippocampal formation (HF) (16). However, the contribution of the reduction of dysbindin-1 isoforms in HF to schizophrenia is unknown.
There are at least three isoforms of dysbindin-1 in primates. Dysbindin-1A is the longest isoform (351 amino acids in human and 352 amino acids in mouse). Human dysbindin-1B contains 303 amino acids with a shorter C terminus compared with dysbindin-1A, but there is no known mouse or rat ortholog of human dysbindin-1B (7). Dysbindin-1C isoform (270 amino acids in human and 271 amino acids protein in mouse) lacks the N-terminal 81 amino acids compared with the dysbindin-1A isoform (7). Our previous 5′-rapid amplification of cDNA ends assay revealed that mouse dysbindin-1C mRNA lacks exons 1–4 and contains a stretch of 5′-untranslated sequence located on intron 4 of dysbindin-1A, suggesting the usage of an alternative promoter in intron 4 (GenBank accession number AY265461) (10). This study focused on the dysbindin-1A isoform, which is a subunit of the biogenesis of lysosome-related organelles complex 1 (BLOC-1 complex) (10). BLOC-1 complex is involved in lysosomal trafficking and biogenesis of lysosome-related organelles (17). By sorting and trafficking cargo proteins, dysbindin-1 is critical for synaptic vesicle biogenesis (18–20) and postsynaptic receptor trafficking (21–25) in neurons. Dysbindin-1 likewise forms a complex with WAVE-2 (Wiskott-Aldrich syndrome protein family verprolin-homologous protein 2) and Abi-1 (Abelson interacting protein-1) to regulate dendritic spine formation (26). In addition, dysbindin-1 interacts with dystrobrevins (components of the dystrophin-associated protein complex (DPC)) in skeletal muscle and the brain, although its physiological relevance to neurodevelopment remains uncertain (27, 28). Lack of dysbindin-1 results in dysregulation of hippocampal synaptic transmission (29–31), which could contribute to the pathogenesis of schizophrenia. Thus, different dysbindin-1 isoforms may function in different pathways. However, how the dysbindin-1 isoforms are involved in adult hippocampal neurogenesis is unknown.
In this study, by using the sdy mice, in which both dysbindin-1A and -1C are deleted (1A−/1C−) (10), we explore the putative functions of dysbindin-1A and -1C in adult hippocampal neurogenesis by choosing wild-type controls (1A+/1C+) and muted (mu) mice (32), in which only dysbindin-1A is destabilized, whereas dysbindin-1C is unaltered (1A−/1C+). We found that dysbindin-1C, but not dysbindin-1A, promotes the maturation of adult newborn neurons in the DG in an isoform-specific and BLOC-1-independent manner.
EXPERIMENTAL PROCEDURES
Mice
The sdy mutant (sdy/sdy, dysbindin deficiency) and its wild-type control DBA/2J (DBA) mice, the mu mutant (mu/mu, muted deficiency) and its wild-type control CHMU/Le, and the pa mutant (pa/pa, pallidin deficiency) and its wild-type control C57BL/6J (B6) were originally obtained from The Jackson Laboratory, transferred from the Dr. Richard T. Swank laboratory, and bred in the animal facility of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. These three mutant colonies were frequently backcrossed to their parent wild-type strains during their maintenance. All procedures were approved by the Institutional Animal Care and Use Committee of Institute of Genetics and Developmental Biology (mouse protocol KYD2005-006). To ensure accurate identification of sdy, mu, and pa mutant mice, we developed PCR methods of genotyping based on the nature of the mutations in the Dtnbp1, Muted, and Pldn genes, respectively (33). Two-month-old male mice were used in all assays except where specified otherwise.
Antibodies
Polyclonal dysbindin (1:10,000 in Western blotting (WB); 1:5000 in immunofluorescence (IF) staining) and BLOS1 (WB, 1:10,000) antibodies were generated in New Zealand White rabbits against His-tagged full-length mouse proteins, respectively. Other antibodies used in this study were as follows: goat anti-WAVE2 polyclonal antibody (WB, 1:1000, sc-10394, Santa Cruz Biotechnology, Dallas, TX); goat anti-β-dystrobrevin polyclonal antibody (WB, 1:200, sc-13815, Santa Cruz Biotechnology); mouse anti-β-actin monoclonal antibody (WB, 1:10,000, A5441, Sigma); goat anti-Sox2 polyclonal antibody (IF, 1:1000, sc-17320, Santa Cruz Biotechnology); mouse anti-nestin monoclonal antibody (IF, 1:100, MAB353, Millipore, Billerica, MA); mouse anti-GFAP monoclonal antibody (IF, 1:1000, IF03L, Millipore); mouse anti-GAD67 monoclonal antibody (IF, 1:100, MAB5406, Millipore); mouse anti-calretinin monoclonal antibody (IF, 1:1000, MAB1568, Millipore); rat anti-BrdU monoclonal antibody (IF, 1:100, ab6326, Abcam, Cambridge, UK); goat anti-DCX polyclonal antibody (IF, 1:150, sc-8066, Santa Cruz Biotechnology); mouse anti-NeuN monoclonal antibody (IF, 1:800, MAB377, Millipore); rabbit anti-S100β polyclonal antibody (IF, 1:1000, ab868, Abcam); rabbit anti-phospho-CREB (Ser133) polyclonal antibody (IF, 1:200, 9198, Cell Signaling Technology, Danvers, MA), monoclonal mouse anti-Flag antibody (WB, 1:5000, Sigma); and secondary antibody Alexa Fluor 408, 488, or 594 IgG (1:2000, Molecular Probes, Eugene, OR).
Preparation of Brain Sub-synaptic Fractions
As described previously (16), a fresh whole mouse brain was homogenized in buffer A (0.1 mm CaCl2, 1 mm MgCl2, and 0.32 m sucrose) containing a protease inhibitor mixture (P8340, Sigma). The homogenate was brought to a final sucrose concentration of 1.25 m by adding 2 m sucrose and 0.1 mmol/liter CaCl2. After being overlaid with 1.0 m sucrose and 0.1 mmol/liter CaCl2, the homogenate was centrifuged at 100,000 × g for 3 h at 4 °C. The synaptosomal fraction, which forms a band at the 1.25/1.0 m sucrose interface, was collected. Next, the synaptosomal fraction was brought to a final concentration of 20 mm Tris-HCl, pH 6, 1% Triton X-100, and 0.1 mm CaCl2 by adding 40 mm Tris-HCl, pH 6, 2% Triton X-100, and 0.1 mm CaCl2. The homogenate was rotated for 30 min at 4 °C, followed by centrifugation at 40,000 × g for 30 min at 4 °C. The supernatant is the synaptic vesicle fraction. The remaining pellet was solubilized in 20 mmol/liter Tris-HCl, pH 8.0, 1% Triton X-100, and 0.1 mmol/liter CaCl2. The homogenate was rotated for 30 min at 4 °C, followed by centrifugation at 40,000 × g for 30 min at 4 °C. The resulting supernatant contains the pre-synaptic membrane fraction, and the pellet contains the postsynaptic density fraction. Supernatant fractions were precipitated by the TCA/acetone method. All fractions were dissolved in 20 mm Tris-HCl, pH 7.4, 5% SDS.
Immunoblotting
Whole mouse brain and brain sub-regions were dissected from mice and then were homogenized in RIPA lysis buffer containing 50 mm Tris-HCl, pH 7.5, 1% (v/v) Triton X-100, 0.25% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 0.15 m NaCl, 1 mm EDTA, and protein inhibitor mixture (Sigma). The extracts were centrifuged for 15 min at 15,000 × g. The total protein concentration in the resulting supernatant was determined by the Bradford method (Bio-Rad) using fatty acid-free bovine serum albumin as a standard. Equal total protein amounts of lysates were subjected to blotting by a standard procedure, probed with the indicated antibodies, and visualized by ECL (Pierce) or ECL Plus detection reagents (Amersham Biosciences). Loading controls were probed with anti-β-actin antibody. For quantitative analysis, protein bands were scanned, and images were analyzed using ImageJ (National Institutes of Health).
Sucrose Sedimentation Velocity Analysis
Adult mouse whole brains were homogenized in a sample buffer containing 0.3 m Tris-HCl, pH 7.5, 1 mm EGTA, 1 mm dithiothreitol, 0.5 mm MgCl2, and protease inhibitor mixture (Sigma) followed by centrifugation at 5000 × g for 5 min and at 120,000 × g for 90 min at 4 °C to obtain a cytosolic extract (34). The supernatant (0.6 mg of total protein) was fractioned by ultracentrifugation on an MLA-55 rotor (Beckman, Brea, CA) at 100,000 × g for 13 h on a 5–20% (w/v) sucrose gradient (12 ml) prepared by SG 15 gradient maker (Amersham Biosciences). The total 20 fractions (∼0.5 ml each) were analyzed by immunoblotting with the indicated antibodies.
Immunohistochemistry
As described previously (35), antigen retrieval is required before using doublecortin (DCX) or nestin primary antibodies. Briefly, sections were kept in 10 mm sodium citrate buffer, pH 6.0, at 80 °C for 30 min, and then allowed to cool to room temperature. Prior to using the BrdU primary antibody, denaturation of DNA is required. Sections were incubated in 2 n HCl for 15 min at 37 °C, followed by 0.1 m borate buffer, pH 8.5, for 10 min at room temperature to neutralize the acid. Then sections were incubated with blocking solution (0.01 m PBS, pH 7.2, 0.3% Triton X-100, 5% normal goat or horse serum) for 60 min at room temperature, followed by incubation with the respective primary antibody diluted in blocking solution for 24–48 h at 4 °C. Finally, sections were incubated with fluorochrome-conjugated secondary antibody in the dark, diluted in 0.01 m PBS with 0.3% Triton X-100 solution. For double- or triple-color immunostaining, primary and secondary antibody administration was repeated sequentially. Sections were mounted with ProLong Gold antifade reagent (Molecular Probes, Eugene, OR) and kept at 4 °C for imaging.
Pattern Separation Test
To prepare the mice for behavioral studies, three generations of both the mutants and controls were bred apart before behavioral analyses. Similar to the protocol used previously (6, 36), the apparatus for this behavioral task contained an open field (84 cm long × 42 cm wide × 15 cm high) and two distinct objects measuring between 2.5 and 5 cm at the base and between 5 and 15 cm tall, which were used as stimuli in these tasks. Mice between 8 and 10 weeks of age were used for the behavioral task. In the habituation session, mice were placed in the center of an open field that contained two objects placed 20 cm apart and allowed 20 min of free exploration of the objects. The time spent exploring each object was recorded individually in 5-min intervals during the 20-min habituation stage. Then there was a 5-min inter-session interval during which the mouse was returned to its home cage, and the two objects were repositioned at 35 cm (low separation), 45 cm (medium separation), or 55 cm (high separation) apart. Next, the mouse was returned to the open field for a 5-min test session, during which it was allowed to re-explore the objects in their new locations. Also, there was a 5-min control session, during which the mice were tested when there was no change in the object location (20 cm apart). Each mouse was tested on each of the object separations, in a counter-balanced design, during different sessions. This exploratory activity was defined as the time spent exploring each object, which was recorded in 0.5-s increments. An exploration ratio was calculated as follows: (exploration time during the 5-min test session)/(exploration time during the 5-min test session + exploration time during the last 5 min of the habituation session).
BrdU Injection and Tissue Preparation
sdy, mu, and corresponding control male mice at 2 months old were used in this study. Cohort 1 mice received an intraperitoneal injection once with 200 mg/kg body weight of 5′-bromo-2′-deoxyuridine (BrdU; Sigma) dissolved in 0.9% NaCl. Two hours after the injection, the mice were anesthetized intraperitoneally with 4% pentobarbital sodium (2.5 μl/g body weight) and perfused transcardially with 0.9% NaCl, followed by 4% paraformaldehyde. Similarly, cohort 2 mice received injections daily for 5 consecutive days with 100 mg/kg BrdU; animals were perfused 30 days after the final injection. Cohort 3 mice received a single injection with 300 mg/kg BrdU; animals were perfused 14 days after the injection. Brains were postfixed overnight in 4% paraformaldehyde at 4 °C and cryoprotected with 30% sucrose. Coronal sections (40 μm thick) were cut throughout the rostrocaudal extent of the hippocampal formation using cryostat microtome (CM1900, Leica Biosystems, Germany). Sections were stored in cryoprotectant at −20 °C until use.
Imaging and Cell Counting
Images were obtained under a Nikon C2 confocal laser-scanning microscope equipped with NIS-Elements argon software (Tokyo, Japan). For cell counting, a ×10 objective (NA 0.45) was used, and a z-stack was compiled by acquiring images every 2.8 μm through the section. For co-localization analysis, a ×100 objective (NA 1.40) was used at 0.5-μm step intervals over the entire z axis. The pinhole was set as 1 airy unit. As described previously (37), the number of BrdU-labeled cells was determined from one-in-six series of sections spaced at 240 μm throughout the entire rostrocaudal extent of the hippocampal formation. Sections were coded so that observers were blind to the genotypes until all samples had been counted. All BrdU-positive cells in the granule cell layer (GCL) and subgranule zone (SGZ) (defined as a two-cell body-wide zone along the border of the granule cell layer) were counted in each section. The surface area of GCL sampled for counting was measured using the Image-Pro+ 6.0 (Media Cybernetics). The GCL sectional volume was determined as the sum of the traced areas multiplied by the thickness of sampled sections (40 μm). The number of BrdU-positive cells was then normalized by dividing the count of BrdU-positive cells by GCL sectional volume (per mm3).
To determine the phenotypic quantification of 30-day-old BrdU-positive cells, a one-in-six series of sections (240 μm spacing) through the rostrocaudal extent of the hippocampal formation was triple labeled for BrdU, NeuN, and S100β as described above. Fifty BrdU-positive cells in GCL and SGZ from each mouse were randomly selected for phenotypic analysis. BrdU-positive cells were analyzed for co-localization with either NeuN or S100β by a confocal microscope as mentioned above. The relative distribution of BrdU-positive cells co-labeled with NeuN or S100β was calculated for each genotype. The average number of cells adopting each phenotype was determined by multiplying the mean percentage for each phenotype by the total number of 30-day-old BrdU-positive cells for each mouse. Similar procedures were taken in other phenotypic analyses by in vivo immunostaining.
Statistical Analyses
Results were expressed as mean ± S.E. All comparisons were statistically tested using unpaired Student's t test or analysis of variance, two-tailed using SPSS 11.5 (IBM, Armonk, NY). In the behavioral test, for the 20-min habituation session, the time spent exploring the object was analyzed with two-way repeated measures analysis of variance with genotype as the between-group factor and exploration during the four 5-min time blocks as the repeated within-group factor. To compare ratios between both genotypes of mice, a two-way (genotype × separation) repeated measures analysis of variance with a Bonferroni's multiple comparison test was performed. All graphs were drawn in GraphPad Prism 5 (La Jolla, CA).
RESULTS
Dysbindin-1A and -1C Display Distinct Temporal and Spatial Expression Patterns in the Mouse Brain
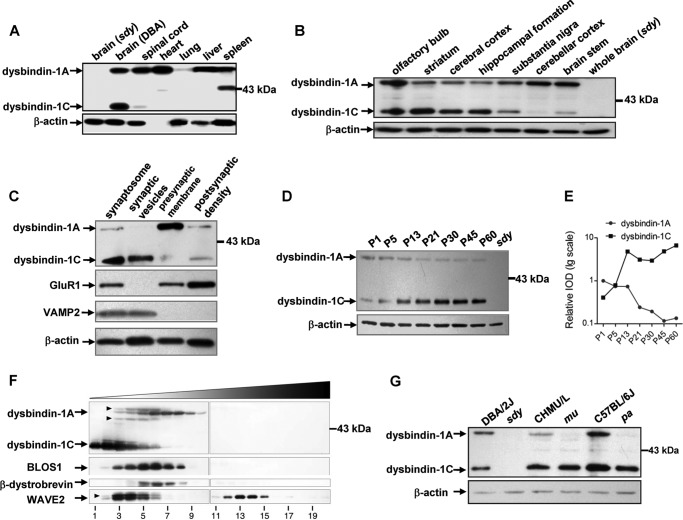
To explore whether different dysbindin-1 isoforms have distinctive spatial and temporal expression patterns in the mouse brain, we detected both dysbindin-1A (∼50 kDa) and -1C (∼35 kDa) in wild-type DBA/2J mice, but both isoforms were undetectable in sdy mice (Fig. 1A). We found that dysbindin-1A was widely expressed in multiple mouse tissues, which agrees with previous reports (10, 26), but dysbindin-1C was only detected in the brain and spinal cord (Fig. 1A). In addition to the noncentral nervous system (CNS) tissues shown in Fig. 1A, we also tested other tissues such as kidney, adrenal gland, small intestine, uterus, ovary, skeletal muscle, testis, and epididymis, in which dysbindin-1A was detectable but dysbindin-1C was not present (data not shown). In brain sub-regions, the dysbindin-1A levels were higher than dysbindin-1C in the olfactory bulb, substantia nigra, cerebellar cortex, and brain stem, but dysbindin-1C had higher expression levels than dysbindin-1A in the striatum, cerebral cortex, and HF (Fig. 1B). The expression pattern of dysbindin-1 in the HF is consistent with a previous report (7). Moreover, we found that in the synaptosome, dysbindin-1C was mainly enriched in synaptic vesicles, and dysbindin-1A was mainly localized in presynaptic membrane, although both dysbindin-1A and -1C were found in the postsynaptic density fraction (Fig. 1C). This subcellular distribution pattern is slightly different from that in human counterparts where dysbindin-1B is mainly localized in synaptic vesicles (16), suggesting that mouse dysbindin-1C may take the place of human dysbindin-1B in synaptic vesicles.
FIGURE 1.
Dysbindin-1A and -1C have distinct spatial and temporal expression patterns and dysbindin-1C is not a subunit of the BLOC-1 complex. Tissue extracts from DBA/2J mice were subjected to SDS-PAGE followed by Western blotting using anti-dysbindin-1 antibody. The brain extract from sdy was used as a negative control, and β-actin was used as a loading control. These experiments were repeated three times independently. A, dysbindin-1A is widely expressed in multiple mouse tissues, whereas dysbindin-1C is only expressed in the brain and spinal cord. B, in brain sub-regions, the dysbindin-1A levels are higher than dysbindin-1C in the olfactory bulb, substantia nigra, cerebellar cortex, and brain stem, but dysbindin-1C has higher expression levels than dysbindin-1A in the striatum, cerebral cortex, and hippocampal formation. C, dysbindin-1C is mainly enriched in the synaptic vesicles, whereas dysbindin-1A is mainly localized in the presynaptic membrane. In addition, both dysbindin-1A and -1C are found in the proportion of postsynaptic density. Successful synaptic fractionation is confirmed with VAMP2 as a marker for synaptic vesicles and GluR1 as a marker for the postsynaptic density. D and E, protein levels of dysbindin-1A in the hippocampal formation are gradually decreased. In contrast, the dysbindin-1C expression levels increase at postnatal stages. The chart in E is plotted by the relative intensities (IOD) of the bands in D. F, sedimentation velocity analyses. Mouse brain cytosol was fractioned by ultracentrifugation on a 5–20% (w/v) sucrose gradient and probed with antibodies against dysbindin-1, BLOS1, β-dystrobrevin, and WAVE2 by immunoblotting. Fractions 1 and 20 correspond to the top and bottom ends of the gradient, respectively. Dysbindin-1C does not co-sediment with subunits of the BLOC-1 complex, including dysbindin-1A and BLOS1. Moreover, dysbindin-1C does not form a stable DPC complex with β-dystrobrevin nor a stable ternary complex with WAVE2 and Abi-1. Arrowheads, nonspecific bands. G, destabilization of the dysbindin-1 in extracts of three BLOC-1 mutants (sdy, pa, and mu). Sdy is the mutant of dysbindin-1; mu is the mutant of muted; and pa is the mutant of pallidin. Inbred strain DBA/2J served as the control for sdy, CHMU/Le for mu, and C57BL/6J for pa.
We next determined the expression profile of dysbindin-1 in the mouse HF during postnatal development. From neonatal to adult stages, the protein levels of dysbindin-1A gradually decreased which was more apparent at P21. In contrast, the dysbindin-1C expression levels increased starting at P13 (Fig. 1, D and E). Taken together, mouse dysbindin-1A and -1C have distinct temporal and spatial expression patterns in the brain. Notably, dysbindin-1C is increasingly expressed in the adult HF. This implies that dysbindin-1C may play a role in the adult HF.
Dysbindin-1C Does Not Reside in the BLOC-1 Complex
To investigate whether the two isoforms dysbindin-1A and -1C reside in different complexes to function in different processes, we evaluated the co-sedimentation of dysbindin-1A/1C with different subunits of the BLOC-1 complex, DPC complex, and the WAVE-2-Abi-1 complex in mouse brain extracts. It is known that subunits of the same complex have similar sedimentation properties in sucrose gradients (34). We found that dysbindin-1A co-sedimented with a known subunit of BLOC-1, BLOS1, in fractions 3–8 as has been previously reported (38), but dysbindin-1C had a different sedimentation pattern from dysbindin-1A, which was mainly enriched in fractions 1–6 (Fig. 1F), suggesting that dysbindin-1C does not reside in the BLOC-1 complex.
Next, we explored whether dysbindin-1C resides in any of the non-BLOC-1 complexes. β-Dystrobrevin is a subunit of the DPC complex, which is the first known interactor of dysbindin-1 (27). Although β-dystrobrevin co-sedimented with the main mass of dysbindin-1A (Fig. 1F), size-exclusion chromatography revealed that dysbindin-1A and β-dystrobrevin do not co-segregate (10). We found that dysbindin-1C did not co-sediment with β-dystrobrevin (Fig. 1F). Thus, neither dysbindin-1A nor -1C is a stable subunit of the DPC complex. It has been reported that dysbindin-1A, WAVE2, and Abi-1 form a ternary complex that functions in regulating dendritic spine morphogenesis (26). Here, we showed that both dysbindin-1A and -1C sedimented differentially from WAVE2, which was enriched in fractions 3–6 and 12–15 (Fig. 1F), suggesting that dysbindin-1 may not form a stable complex with WAVE2.
In general, loss of one subunit in a heterogeneous protein complex would undermine the stability of the complex, thus leading to the degradation of other subunits. Consistently, we found that the steady-state levels of dysbindin-1A were absent in the brain lysates of mu and pa mice, in which muted and pallidin (both BLOC-1 subunits) are deficient (10). However, dysbindin-1C was not altered in the brain extracts of these mutants (Fig. 1G), consistent with the finding that dysbindin-1A, but not -1C, was reduced in mu mice (7, 39). This result further supports the notion that dysbindin-1C does not reside in the BLOC-1 complex. Thus, we concluded that dysbindin-1C functions in a BLOC-1 independent manner.
Notably, the differential destabilization of dysbindin-1A and -1C in mu or pa mice provides a unique tool for the study of functions of dysbindin-1 isoforms. In this regard, both dysbindin-1A and -1C are deleted in sdy (denoted as 1A−/1C−) when compared with the wild-type (1A+/1C+) controls (DBA/2J, CHMU/Le, or C57BL/6J). However, dysbindin-1A is absent, but dysbindin-1C is normal in mu or pa mice, which therefore can be considered as the dysbindin-1C wild-type as well as dysbindin-1A-deficient mice (1A−/1C+) (Fig. 1G).
Dysbindin-1C Is Expressed in the Glutamatergic Mossy Cells of the Hilus of the DG
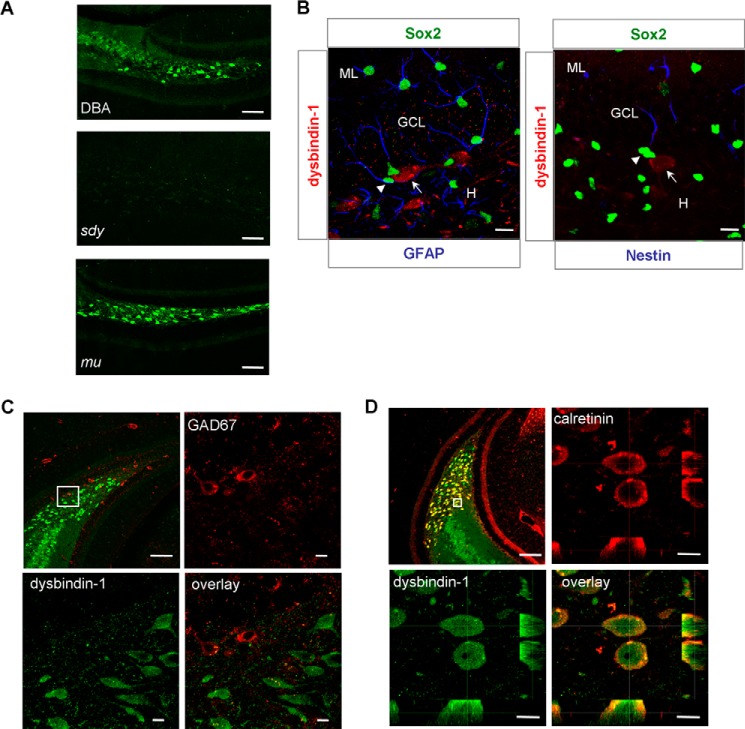
To investigate the potential role of dysbindin-1 isoforms in adult hippocampal neurogenesis, we determined the localization of dysbindin-1 isoforms in DG of HF by in vivo immunostaining. Dysbindin-1 was localized in hilar neurons and the inner molecular layer of the wild-type DG when compared with null signals in sdy mice (Fig. 2A), indicating high specificity of dysbindin-1 antibody. In addition, the immunostaining pattern of dysbindin-1 in wild-type (1A+/1C+) and mu mice (1A−/1C+) was very similar (Fig. 2A), suggesting that dysbindin-1C, but not -1A, is expressed in the DG hilus.
FIGURE 2.
Dysbindin-1C is expressed in glutamatergic mossy cells of the DG hilus. A, dysbindin-1 is mainly localized in the hilar neurons and the inner molecular layer of hippocampal DG in wild-type DBA/2J (DBA) mice (upper panel). Immunostaining of dysbindin-1 in the sdy mice served as a negative control (middle panel). The localization of dysbindin-1C in mu mice is similar to dysbindin-1 in DBA mice (lower panel). Scale bars, 100 μm. B, dysbindin-1-positive neurons (arrow, red) are not co-localized with GFAP (blue) and Sox2 (green) double-positive adult neuronal progenitor cells (arrowhead) in SGZ (left panel). Likewise, dysbindin-1-positive neurons (arrow, red) are not co-localized with nestin (blue) and Sox2 (green) double-positive neuronal progenitor cells (arrowhead) in SGZ (right panel). ML, molecular layer; GCL, granule cell layer; H, hilus. Scale bars, 10 μm. C, double staining of dysbindin-1 (green) and GAD67 (red) reveals that dysbindin-1 is not present in GAD67 + GABAergic hilar interneurons. Upper left panel, 10× objective, scale bars, 100 μm. Other panels show different channels of the area from upper left panel under 100× objective magnification, scale bars, 10 μm. D, dysbindin-1 (green) and calretinin (red) are co-localized in glutamatergic mossy cell body of hilus and mossy cell axon of inner molecular layer. Upper left panel, 10× objective, scale bars, 100 μm. Other panels show different channels of the area from upper left panel under 100× objective magnification, scale bars, 10 μm.
To further clarify which type of hilar neurons in which dysbindin-1 is expressed, we performed immunostaining with the antibodies against Sox2, GFAP, and nestin to label the adult neural progenitor cells that localized in the SGZ. We found that dysbindin-1 was not expressed in adult neuronal progenitor cells of SGZ (Fig. 2B). Hilar neurons are mainly divided into two types as follows: one is a GABAergic interneuron marked with GAD67 (glutamate decarboxylase 67-kDa isoform), and the other is a glutamatergic mossy cell marked with calretinin (1). We found that dysbindin-1 did not co-reside with GAD67, which labeled the GABAergic interneurons (Fig. 2C), but co-labeled mainly with calretinin-positive mossy cell bodies and the inner molecular layer where mossy cell axons are localized (Fig. 2D). These results suggest that dysbindin-1C is expressed in the glutamatergic mossy cells of the DG hilus.
Dysbindin-1C Promotes the Maturation of Adult-born Neurons Non-cell Autonomously
Proliferation and survival of progenitor cells were normal, but the percentage of BrdU-positive cells that co-labeled with NeuN was significantly lower in sdy mice than in wild-type controls, suggesting that the maturation of newborn neurons was impaired (15). However, the underlying mechanism and the contribution of different dysbindin-1 isoforms are unclear. To investigate the role of different dysbindin-1 isoforms in adult hippocampal neurogenesis, we adopted a slightly different method using the progenitor cell tracer BrdU together with different cell lineage-specific markers (40, 41). Adult neural progenitor cells (BrdU+) in the SGZ undergo asymmetric division and give rise to astrocytes expressing S100β and immature neurons expressing DCX. Only a small proportion of newborn neurons survive beyond 4 weeks and go through morphological and physiological maturation, migrating into the granule cell layer of the DG. Eventually, these 30-day-old new granule neurons integrate into local circuits and express NeuN as mature neurons.
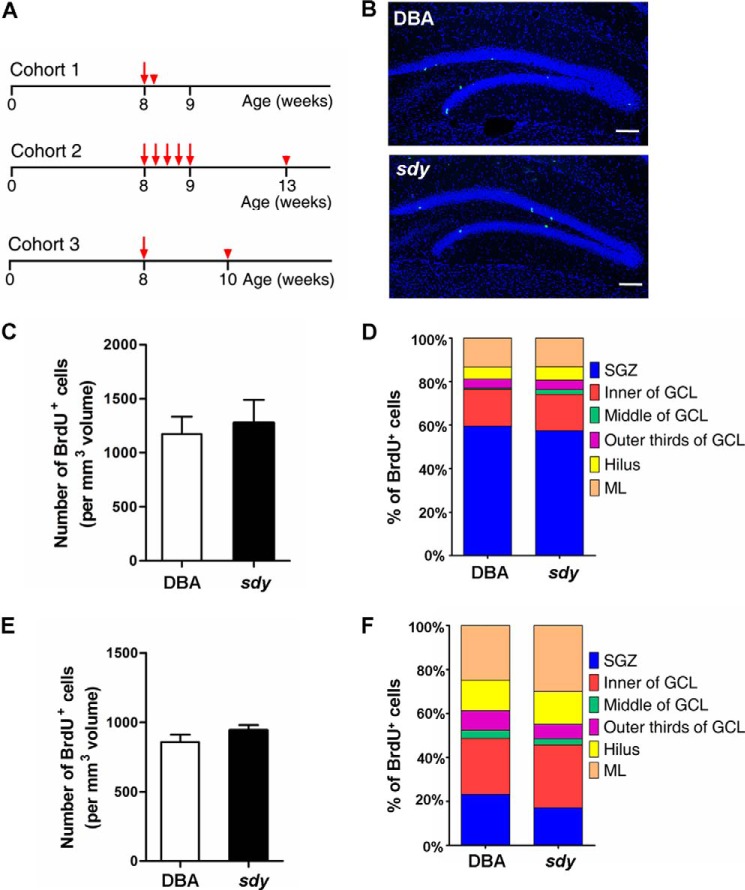
By using the cohort 1 method shown in Fig. 3A, we found that the number and the percentile distribution of BrdU-positive cells within each sub-region of DG were similar between wild-type and sdy mice (Fig. 3, B–D), indicating that proliferation and localization of neuronal progenitor cells were not affected in sdy mice. Likewise, when choosing the cohort 2 method (Fig. 3A), there was no significant difference in the number and the percentile distribution of BrdU-positive cells within each sub-region of the DG between wild-type and sdy mice, suggesting that the survival and migration of adult-born cells in the DG were not altered in sdy mice (Fig. 3, E and F).
FIGURE 3.
Proliferation of neuronal progenitor cells and survival of adult-born cells in the DG are not affected in sdy mice. A, experimental schemes of BrdU injection for assessing cell proliferation (cohort 1) and survival (cohort 2) in the adult hippocampal formation. Cohort 3 represents the experimental scheme of BrdU injection for assessing phosphorylation of CREB of newborn neurons. B, representative DBA/2J (DBA, WT) and sdy brain sections stained with an antibody against BrdU (green) and DAPI (blue) for in vivo neurogenesis analyses. Scale bars, 100 μm. C, 2 h after BrdU injection, the number of BrdU+ cells per mm3 volume in the granule cell layer (GCL) and subgranule zone (SGZ) of DG was counted for each genotype (mean ± S.E.; n = 4 for each group). The number of BrdU+ cells is similar between wild-type and sdy mice (DBA, 1173 ± 80 versus sdy, 1275 ± 108, p > 0.05). D, positioning or percentile distribution of BrdU+ cells in the DG of the adult hippocampal formation of sdy mice is similar to WT (p > 0.05). The localization of BrdU+ cells is defined as six sub-regions in the DG: hilus (H), SGZ, inner of GCL, middle of GCL, outer-third of GCL, molecular layer (ML). E, 30 days after a 5-day consecutive BrdU injection, the number of BrdU+ cells per mm3 volume in the GCL and SGZ of DG was counted for each genotype (mean ± S.E.; n = 4 for each group). The number of 30-day-old BrdU+ cells is similar between wild-type and sdy mice (DBA, 858 ± 27 versus sdy, 942 ± 22, p > 0.05). F, similar to D, positioning of surviving BrdU+ cells in the DG of sdy mice has no significant difference from that in WT (p > 0.05).
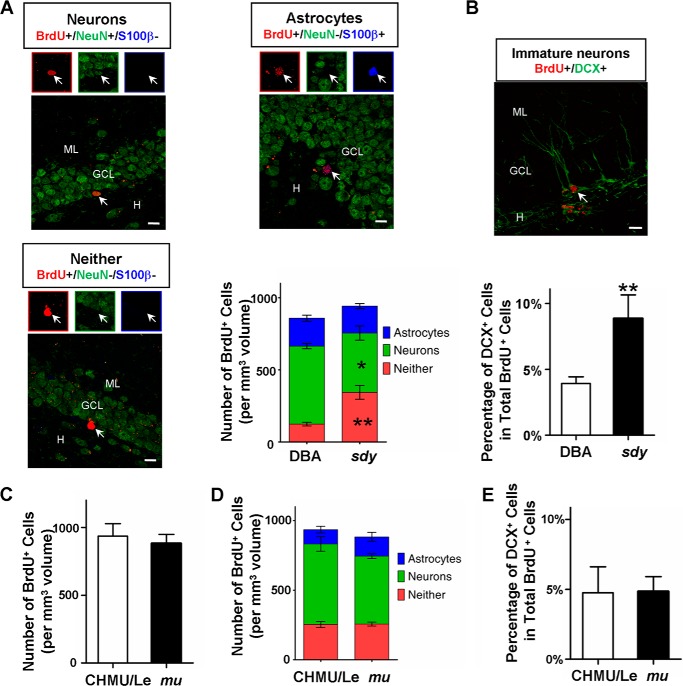
We further assessed neuronal differentiation fates by counting different cell types with the triple labeling method, 30 days after the final injection of BrdU (cohort 2, Fig. 3A). We found that sdy mice had about a 24% reduction in the number of mature neurons (BrdU+/NeuN+/S100β−) compared with wild-type DBA/2J controls (Fig. 4A). However, we found no significant difference in the number of surviving newborn astrocytes (BrdU+/NeuN−/S100β+) in sdy mice compared with wild-type mice. In contrast, in sdy mice, there was a significantly higher number of immature neurons expressing only BrdU (BrdU+/NeuN−/S100β−) than in DBA/2J controls (Fig. 4A), suggesting that the maturation of adult-born neurons is impaired in sdy mice. To further confirm the neuronal maturation defect in sdy mice, we performed double BrdU/DCX immunostaining to measure the fraction of immature neurons at 30 days post-injection (cohort 2, Fig. 3A) (42). We found that sdy mice had a significantly increased percentage of BrdU+ cells co-labeled with DCX compared with wild-type controls (Fig. 4B). Collectively, in agreement with a previous finding (15), the increased number of immature neurons and decreased number of mature neurons in sdy mice (1A−/1C−) suggest that dysbindin-1 is required for adult-born neuronal maturation in the DG.
FIGURE 4.
Dysbindin-1C is required for the maturation of newborn neurons. A, co-expression of NeuN (green) and S100β (blue) in BrdU+ cells (red) from DBA/2J (DBA, WT) and sdy mice. Arrows in the merged images and enlarged boxes indicate BrdU+/NeuN+ neuron (upper left, yellow), BrdU+/S100β+ astrocyte (upper right, purple), and BrdU+/NeuN-/S100β-immature neurons (lower left, red), respectively. H, hilus, GCL, granule cell layer, M, molecular layer. Scale bars, 10 μm. The lower right panel shows the distribution of BrdU+ cells in the GCL and SGZ 30 days after BrdU injection (mean ± S.E.; n = 4 for each group). Compared with WT, there is a significantly lower number of BrdU+/NeuN+ newborn neurons (green boxes) in the DG of sdy mice (DBA, 541 ± 18 versus sdy, 411 ± 49 *, p < 0.05). In contrast, the number of BrdU+ immature neurons (red boxes) is significantly increased compared with WT (DBA, 124 ± 13 versus sdy, 344 ± 48 **, p < 0.01), although the number of BrdU+/S100β+ newborn astrocytes (blue boxes) is unaltered in sdy mice (DBA, 194 ± 21 versus sdy, 187 ± 17 p > 0.05). B, co-expression of BrdU (red) and DCX (green) in newborn cells from DBA and sdy mice (upper panel). The arrow in the image indicates a BrdU and DCX double-positive immature neuron. Scale bar, 10 μm. The distribution of BrdU+/DCX+ immature neurons in all BrdU+ cells 30 days after BrdU injection (mean ± S.E.; n = 3 for each group in total of ∼400 neurons) (lower panel). Compared with wild type, a significantly increased fraction of BrdU+/DCX+ immature neurons in GCL and SGZ of sdy mice (DBA, 3.92 ± 0.26% versus sdy, 8.87 ± 0.89% **, p < 0.01). C, 30 days after the 5-day consecutive BrdU injection, the number of BrdU+ cells per mm3 volume in the DG was counted for each genotype (mean ± S.E.; n = 4 for each group). The number of 30-day-old BrdU+ cells is similar between wild-type (CHMU/Le) and mu mice (CHMU/Le, 936 ± 46 versus mu, 883 ± 33, p > 0.05). D, as described in A, the distribution of BrdU+ cells in the GCL and SGZ 30 days after BrdU injection (mean ± S.E.; n = 4 for each group) was determined in wild-type and mu mice. There are no significant differences between wild-type and mu mice in the number of BrdU+ immature neurons (red boxes) (CHMU/Le, 254 ± 21 versus mu 257 ± 14, p > 0.05), BrdU+/NeuN+ neurons (green boxes) (CHMU/Le, 579 ± 53 versus mu, 488 ± 17, p > 0.05), and BrdU+/S100β+ astrocytes (blue boxes) (CHMU/Le, 103 ± 24 versus mu, 138 ± 32, p > 0.05), respectively. E, as described in B, the distribution of BrdU+/DCX+ immature neurons in all BrdU+ cells 30 days after BrdU injection (mean ± S.E.; n = 3 for each group in total of ∼400 neurons) was determined. The percentile of BrdU+/DCX+ immature neurons in GCL and SGZ of mu mice is similar to wild type (CHMU/Le, 4.76% ± 0.93 versus sdy 4.85% ± 0.53, p > 0.05).
Similarly, we examined the proliferation, survival, and maturation of adult newborn neurons in mu mice (1A−/1C+) in which dysbindin-1C functions normally, compared with the wild-type controls (1A+/1C+). We found that proliferation and localization, survival and migration, and neuronal maturation and differentiation of the neuronal progenitors were not altered in the DG of mu mice (Fig. 4, C–E). Taken together, these results suggest that dysbindin-1C, but not dysbindin-1A, plays an important role in the maturation of adult-born neurons.
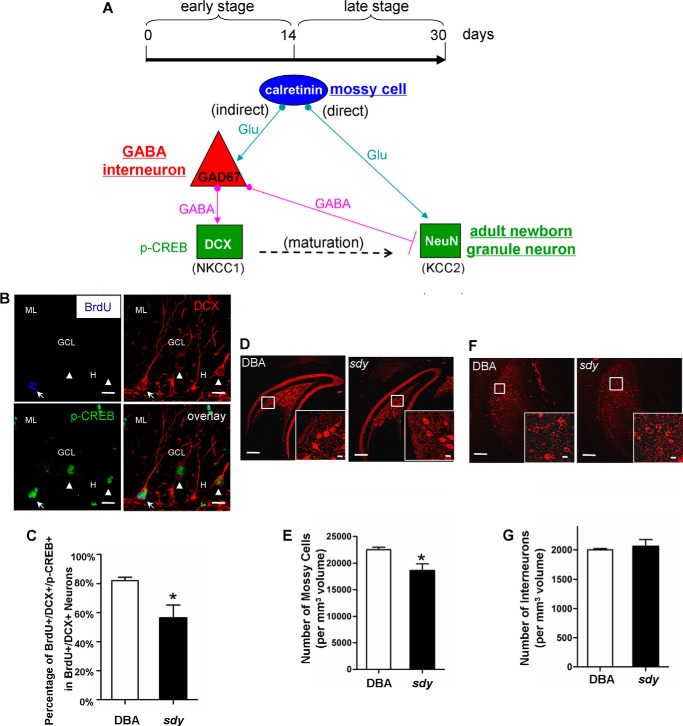
As dysbindin-1C is not localized in adult neuronal progenitor cells, but in hilar mossy cells where dysbindin-1A is not expressed, we speculated that dysbindin-1C regulates adult-born neuronal maturation non-cell autonomously, requiring extrinsic signals from the neuronal stem cell niche. Based on the timing of depolarizing GABA (indirect pathway) and glutamate-induced excitation (direct pathway) of newborn neurons (Fig. 5A), defects in GABA-induced depolarization or glutamatergic inputs would contribute to the delayed maturation of newborn neurons in sdy mice, leading to the increase of immature neurons.
FIGURE 5.
Loss of dysbindin-1 diminishes the phosphorylation of CREB in 14-day-old newborn DCX-positive immature neurons and decreases the number of mossy cells in the DG hilus. A, schematic of direct and indirect pathway during the maturation of adult newborn neurons (1). Hilar mossy cell and GABA interneuron, immature, and mature newborn granule neuron are marked with calretinin and GAD67, DCX, and NeuN, respectively. In the early stage and the indirect pathway, hilar glutamatergic mossy cells input excitatory action on GABA interneurons (53). The latter exerts initially excitatory actions on immature neurons, which are depolarized by GABA due to high expression levels of the chloride importer NKCC1 (Na+/K+/2Cl− co-transporter), to activate phosphorylated CREB (p-CREB) signaling pathway (54) and thus to promote early maturation of newborn neurons. In the late stage, GABA becomes inhibitory as a result of the down-regulation of NKCC1 and the up-regulation of the chloride exporter KCC2 (K+-coupled Cl− transporter 2) (55–57). Meanwhile, glutamatergic mossy cell inputs begin to promote further maturation of newborn granule neurons through the direct pathway after 14 days (58, 59). B, co-expression of BrdU (red), DCX (green), and p-CREB (blue) in newborn cells from DBA/2J (DBA, WT) and sdy mice. Arrow in the images indicates a 14-day-old newborn BrdU+/DCX+/p-CREB+ immature neuron; arrowhead indicates an adjacent BrdU-/DCX+/p-CREB+ immature neuron. GCL, granule cell layer; ML, molecular layer; H, hilus. Scale bars, 10 μm. C, phenotypic percentile of BrdU+/DCX+/p-CREB+ immature neurons in all BrdU+/DCX+ immature neurons 14 days after BrdU injection (mean ± S.E.; n = 3 for each group in total of ∼300 neurons). Compared with wild type, the fraction of BrdU+/DCX+/p-CREB+ newborn neurons was significantly decreased in the GCL and SGZ of sdy mice (DBA, 82.07% ±1.35 versus sdy, 56.27% ± 5.20 *, p < 0.05). D, representative DBA/2J and sdy brain sections stained with the anti-calretinin antibody, which is the marker of mossy cells. Insets show the enlarged view of calretinin+ mossy cells in the DG hilus. Scale bars, 100 μm; scale bars in insets, 10 μm. E, number of mossy cells per mm3 volume in the DG hilus was calculated for each genotype (mean ± S.E.; n = 3 for each group). The number of mossy cells is modestly but significantly decreased in sdy mice compared with wild type (DBA, 22,510 ± 303 versus sdy, 18,590 ± 741, *, p < 0.05). F, representative DBA and sdy brain sections stained with the anti-GAD67 antibody, which is the marker of GABAergic interneurons. Insets show the enlarged view of GAD67+ interneurons in the DG hilus. Scale bars, 100 μm; scale bars in insets, 10 μm. G, number of interneurons per mm3 volume in the GCL and hilus of DG was counted for each genotype (mean ± S.E.; n = 3 for each group). The number of GABAergic interneurons is not altered in sdy mice compared with controls (DBA, 2003 ± 11 versus sdy, 2056 ± 70, p > 0.05).
To ascertain whether the indirect pathway is defective in sdy mice, we administered BrdU to wild-type and sdy mice, and we performed triple BrdU/DCX/p-CREB immunostaining (Fig. 5B) at 14 days post-injection (cohort 3, Fig. 3A). Notably, the inputs to adult newborn neurons are from the hilar GABA interneurons rather than GABA interneurons from the outer molecular layer (43). We therefore only counted the GABAergic cells in the hilar region rather than those in the outer molecular layer. We found that the fraction of immature neurons co-labeled with p-CREB (BrdU+/DCX+/p-CREB+) from the total population of immature neurons (BrdU+/DCX+) was significantly decreased (31.44% reduction) in sdy mice compared with wild-type controls (Fig. 5C). This suggests that depolarizing GABA-CREB signaling is impaired in the DG of sdy mice due to loss of dysbindin-1C, via the indirect pathway in the early stage. As shown in Fig. 4A, the number of newborn mature neurons is lower in sdy mice when counted at 30 days post-injection (late stage). It is difficult to completely rule out the possibility of the dysfunction of the direct pathway due to the existing early stage effect on newborn neurons. Taken together, this suggests that dysbindin-1C in mossy cells plays a role in the early maturation of newborn neurons, likely through the indirect pathway to depolarize GABA signaling.
Loss of Dysbindin-1C Causes Mossy Cell Loss
To investigate how dysbindin-1C deficiency could down-regulate the GABA-CREB pathway in immature neurons, we performed immunostaining using an antibody against the mossy cell marker calretinin and quantified the number of hilar mossy cells of wild-type and sdy mice. We found that the number of mossy cells in sdy mice is modestly (17.41% reduction) but significantly decreased compared with wild-type controls (Fig. 5, D and E). In contrast, using the interneuron marker GAD67, no significant differences in the number of hilar GABA interneurons between sdy mice and controls were shown (Fig. 5, F and G). This result indicates that loss of dysbindin-1C compromises the survival of mossy cells. However, the underlying mechanism of mossy cell loss awaits further investigation.
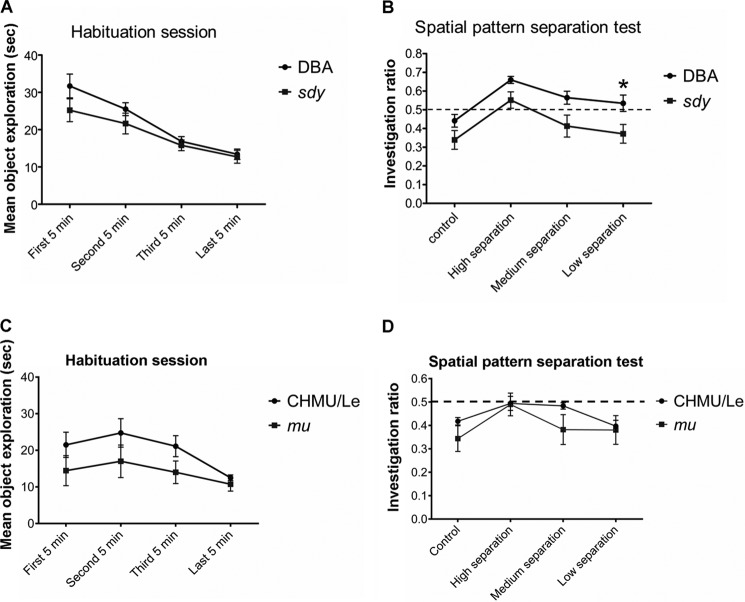
Dysbindin-1C Deficiency Alters the Behaviors of Lower Pattern Separation
It is well known that impaired adult hippocampal neurogenesis leads to the impairment of pattern separation (2, 6). To investigate whether the impaired adult neurogenesis in sdy mice is associated with abnormal behaviors in pattern separation, we adopted methods dedicated to the pattern recognition test (6, 36). We found that the interaction between the genotype and exploration time was not significant in the habituation session (Fig. 6A). However, sdy mice had retarded pattern recognition with increased difficulties in discrimination especially in the low separation design compared with wild-type controls in the test session (p < 0.05, Fig. 6B). Using similar settings, we found that there was no significant difference in either the habituation session or the test session in mu mice (with normal dysbindin-1C) compared with the wild-type controls (p > 0.05, Fig. 6, C and D). This suggests that the impaired adult neurogenesis in sdy mice, due to the lack of dysbindin-1C, likely contributes to the impaired cognitive deficits that mimic the symptoms in schizophrenic patients.
FIGURE 6.
sdy mice displayed impaired spatial pattern separation capacity. A, for the habituation period, both wild-type (WT, DBA/2J) and sdy mice display adaptability to the stimulus objects as demonstrated by a decrease in the time spent exploring the objects during the 20-min interval. The interaction between the genotype and exploration time is not statistically significant (F1, 54 = 3.20, p = 0.09, n = 12). B, under higher difficulty discrimination conditions, the sdy mice showed impaired spatial pattern separation capacity. An exploration ratio of >0.5 indicates the ability of a mouse to discriminate a change in spatial orientation. As a control, when there was no change in the object location between habituation and test sessions (20 cm apart), all of the mice showed shorter investigation durations. During the test session, the genotypes (WT and sdy) affects pattern separation performance significantly during various separation conditions (F1, 22 = 7.58, p = 0.01, n = 12). Both sdy and control mice discriminate a high spatial separation (55 cm apart) between the stimulus objects (WT, 0.66 ± 0.02 versus sdy, 0.55 ± 0.04, post hoc t = 1.76, p = 0.33, n = 12). At the medium (45 cm apart) and the low (35 cm apart) spatial separation, only the wild-type mice are able to discriminate the difference in spatial orientation of the objects, but sdy mice show a lack of spatial pattern separation ability. Notably, the performance of sdy at low separation is significantly impaired compared with the wild-type mice (medium, WT 0.56 ± 0.03 versus sdy 0.41 ± 0.06; post hoc t = 2.48, p = 0.06, n = 12; low, WT 0.53 ± 0.04 versus sdy 0.37 ± 0.05, post hoc t = 2.66, *, p = 0.037, n = 12). Data are expressed as mean ± S.E. C, using the same method described in A, during the habituation period both wild-type (WT, CHMU/Le) and mu mice display adaptability to the stimulus objects as demonstrated by a decrease in time spent exploring the objects during the 20-min interval. The interaction between the genotypes and exploration time is not statistically significant (F3, 42 = 0.60, p = 0.62, n = 12). D, using the same method described in B, when there was no change in the object location between habituation and test sessions (20 cm apart), all of the mice showed shorter investigation durations. During the test session, both WT and mu mice show similar pattern separation capacity during various separation conditions (F1, 14 = 1.72, p = 0.21, n = 12).
DISCUSSION
In this study, we adopted adult hippocampal neurogenesis in mice as a cellular model to investigate whether dysbindin-1 regulates neurodevelopment in an isoform-specific manner. First, in the adult mouse brain, dysbindin-1C has a distinctive spatial and temporal expression pattern in contrast to dysbindin-1A. Second, dysbindin-1C does not reside in the BLOC-1 complex. Third, dysbindin-1C is localized in glutamatergic mossy cells of the hilus in the DG for the maintenance of their survival. Finally, dysbindin-1C promotes the early maturation of adult-born neurons most likely via the excitatory actions of GABA-CREB signaling. Taken together, our study provides new insights into the isoform-specific and BLOC-1-independent function of dysbindin-1, highlighting the relationship between defects in adult hippocampal neurogenesis and the pathogenesis of schizophrenia. Our results support the importance of decreased dysbindin-1C found in the schizophrenic hippocampal formation for the development of cognitive deficits (16).
Previous studies have focused on the neuronal functions of dysbindin-1A residing in the BLOC-1 complex in cargo sorting into synaptic vesicles and postsynaptic receptor trafficking (17, 44). Little attention was paid to the function of other dysbindin-1 isoforms, except for their expression and distribution (7). Intriguingly, if the development of schizophrenic symptoms is attributable to dysbindin-1 in the BLOC-1 complex, one would expect similar effects of other BLOC-1 subunits (44). Linkage and association studies have shown strong evidence for the involvement of the DTNBP1 gene in schizophrenia (45). However, except for BLOC1S3, genes encoding other BLOC-1 subunits have no significant association with schizophrenia risk although an epistatic interaction between DTNBP1 and MUTED may exist (46). Currently, there is no support for MUTED as a susceptibility gene for schizophrenia (47). A recent study has shown the distinct effects on the expression of postsynaptic receptors in the HF between sdy and mu mice (39), supporting the notion that lack of different BLOC-1 subunits may lead to variable phenotypes, due to their different interactors or isoforms. Furthermore, our pattern separation tests did not support the involvement of the MUTED gene in the development of cognitive deficits. This highlights a unique role of dysbindin-1 in the pathogenesis of schizophrenia with the speculation that dysbindin-1 may have additional roles, mediated through its isoforms, in the brain via the BLOC-1-independent pathway.
The distinct spatial and temporal expressions of mouse dysbindin-1A and -1C are likely due to the alternative usage of different promoters. Dysbindin-1C results from a downstream start codon on exon 5 of dysbindin-1A thus lacking the N-terminal 81 residues. The 72–140-amino acid region of dysbindin-1 contains the binding domain for BLOC-1 subunits such as pallidin and snapin (28). It is unknown whether the difference in the N termini of dysbindin-1A and -1C contributes to the residency in the BLOC-1 complex. Interestingly, a default nucleus localization signal may lie on the N terminus of dysbindin-1A and -1B, and lack of this sequence in dysbindin-1C may lead to its cytosolic retention (48). This agrees with the observation that the DNA-dependent protein kinase complex interacts only with dysbindin-1A and -1B in the nucleus but not with dysbindin-1C in cytosol (49). Although the physiological relevance of DNA-PK phosphorylation of dysbindin-1A/-1B, but not -1C, is unknown, it is unlikely that this process is involved in regulating the maturation of DG adult-born neurons in which dysbindin-1C is required.
Our findings that dysbindin-1C regulates the GABA-CREB signaling through inputs from mossy cells suggest an interplay between the GABA signaling and DISC1, another leading susceptibility gene of schizophrenia, in regulating adult hippocampal neurogenesis. During early postnatal hippocampal neurogenesis, DISC1 restricts GABA-mediated Akt-mTOR signaling and exerts an inhibitory role on the maturation of adult-born neurons (50). Importantly, several schizophrenia susceptibility genes such as DTNBP1, DISC1, GAD67, NKCC1, and AKT1 converge in this pathway (45, 50, 51).
The delayed maturation of adult-born neurons is expected to affect the hippocampal CA3-CA1 functionality and synaptic plasticity. Indeed, an enhancement in long term potentiation and reduced NMDA receptor-dependent synaptic potentiation was found in sdy hippocampal neurons (30, 31). Moreover, altered hippocampal activity was revealed by magnetic resonance imaging of the sdy mice (52). These structural and functional endophenotypes in dysbindin-1-deficient mice suggest an association with impaired cognition, learning, and memory. In humans, DTNBP1 SNPs and haplotypes have been associated with the cognitive deficits or negative symptoms (45), although its relationship with dysbindin-1 isoforms is unknown. Similarly, in sdy mice, social withdrawal, recognition deficits, altered memory capacities, or contextual fear conditioning have been reported, which may be related to dysfunction in adult hippocampal neurogenesis (11, 14, 31). Our additional tests on pattern separation in this study further suggest that impaired adult hippocampal neurogenesis plays an important role in the development of these cognitive deficits.
Acknowledgments
We are in debt to Dr. Richard T. Swank who provided the mouse mutants for this study. We thank Dr. John R. Speakman for his proofreading of the manuscript. We are grateful for the critical comments from Dr. Zuhang Sheng.
This work was supported in part by National Natural Science Foundation of China Grants 91332116, 31230046, and 30900777, by National Basic Research Program of China Grant 2014CB942803, and Chinese Academy of Sciences Grant KJZD-EW-L08.
This article was selected as a Paper of the Week.
- DG
- dentate gyrus
- BLOC-1
- biogenesis of lysosome-related organelles complex 1
- DPC
- dystrophin-associated protein complex
- DTNBP1
- dystrobrevin-binding protein 1
- HF
- hippocampal formation
- SGZ
- subgranule zone
- WB
- Western blotting
- DCX
- doublecortin
- IF
- immunofluorescence
- GCL
- granule cell layer
- CREB
- cAMP response element-binding protein
- GFAP
- glial fibrillary acidic protein.
REFERENCES
- 1. Ming G. L., Song H. (2011) Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deng W., Aimone J. B., Gage F. H. (2010) New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat. Rev. Neurosci. 11, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kempermann G., Krebs J., Fabel K. (2008) The contribution of failing adult hippocampal neurogenesis to psychiatric disorders. Curr. Opin. Psychiatry 21, 290–295 [DOI] [PubMed] [Google Scholar]
- 4. Duan X., Chang J. H., Ge S., Faulkner R. L., Kim J. Y., Kitabatake Y., Liu X. B., Yang C. H., Jordan J. D., Ma D. K., Liu C. Y., Ganesan S., Cheng H. J., Ming G. L., Lu B., Song H. (2007) Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130, 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pieper A. A., Wu X., Han T. W., Estill S. J., Dang Q., Wu L. C., Reece-Fincanon S., Dudley C. A., Richardson J. A., Brat D. J., McKnight S. L. (2005) The neuronal PAS domain protein 3 transcription factor controls FGF-mediated adult hippocampal neurogenesis in mice. Proc. Natl. Acad. Sci. U.S.A. 102, 14052–14057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Q., Kogan J. H., Gross A. K., Zhou Y., Walton N. M., Shin R., Heusner C. L., Miyake S., Tajinda K., Tamura K., Matsumoto M. (2012) SREB2/GPR85, a schizophrenia risk factor, negatively regulates hippocampal adult neurogenesis and neurogenesis-dependent learning and memory. Eur. J. Neurosci. 36, 2597–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Talbot K., Ong W. Y., Blake D. J., Tang J., Louneva N., Carlson G. C., Arnold S. E. (2009) in Handbook of Neurochemistry and Molecular Neurobiology, Schizophrenia (Javitt D., Kantrowitz J., eds) 3rd Ed., pp. 107–241, Springer Science, New York [Google Scholar]
- 8. Straub R. E., Jiang Y., MacLean C. J., Ma Y., Webb B. T., Myakishev M. V., Harris-Kerr C., Wormley B., Sadek H., Kadambi B., Cesare A. J., Gibberman A., Wang X., O'Neill F. A., Walsh D., Kendler K. S. (2002) Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am. J. Hum. Genet. 71, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ayalew M., Le-Niculescu H., Levey D. F., Jain N., Changala B., Patel S. D., Winiger E., Breier A., Shekhar A., Amdur R., Koller D., Nurnberger J. I., Corvin A., Geyer M., Tsuang M. T., Salomon D., Schork N. J., Fanous A. H., O'Donovan M. C., Niculescu A. B. (2012) Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol. Psychiatry 17, 887–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li W., Zhang Q., Oiso N., Novak E. K., Gautam R., O'Brien E. P., Tinsley C. L., Blake D. J., Spritz R. A., Copeland N. G., Jenkins N. A., Amato D., Roe B. A., Starcevic M., Dell'Angelica E. C., Elliott R. W., Mishra V., Kingsmore S. F., Paylor R. E., Swank R. T. (2003) Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 35, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feng Y. Q., Zhou Z. Y., He X., Wang H., Guo X. L., Hao C. J., Guo Y., Zhen X. C., Li W. (2008) Dysbindin deficiency in sandy mice causes reduction of snapin and displays behaviors related to schizophrenia. Schizophr. Res. 106, 218–228 [DOI] [PubMed] [Google Scholar]
- 12. Hattori S., Murotani T., Matsuzaki S., Ishizuka T., Kumamoto N., Takeda M., Tohyama M., Yamatodani A., Kunugi H., Hashimoto R. (2008) Behavioral abnormalities and dopamine reductions in sdy mutant mice with a deletion in Dtnbp1, a susceptibility gene for schizophrenia. Biochem. Biophys. Res. Commun. 373, 298–302 [DOI] [PubMed] [Google Scholar]
- 13. Takao K., Toyama K., Nakanishi K., Hattori S., Takamura H., Takeda M., Miyakawa T., Hashimoto R. (2008) Impaired long-term memory retention and working memory in sdy mutant mice with a deletion in Dtnbp1, a susceptibility gene for schizophrenia. Mol. Brain 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhardwaj S. K., Baharnoori M., Sharif-Askari B., Kamath A., Williams S., Srivastava L. K. (2009) Behavioral characterization of dysbindin-1 deficient sandy mice. Behav. Brain Res. 197, 435–441 [DOI] [PubMed] [Google Scholar]
- 15. Nihonmatsu-Kikuchi N., Hashimoto R., Hattori S., Matsuzaki S., Shinozaki T., Miura H., Ohota S., Tohyama M., Takeda M., Tatebayashi Y. (2011) Reduced rate of neural differentiation in the dentate gyrus of adult dysbindin null (sandy) mouse. PLoS One 6, e15886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Talbot K., Louneva N., Cohen J. W., Kazi H., Blake D. J., Arnold S. E. (2011) Synaptic dysbindin-1 reductions in schizophrenia occur in an isoform-specific manner indicating their subsynaptic location. PLoS One 6, e16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei A. H., Li W. (2013) Hermansky-Pudlak syndrome: pigmentary and non-pigmentary defects and their pathogenesis. Pigment Cell Melanoma Res. 26, 176–192 [DOI] [PubMed] [Google Scholar]
- 18. Larimore J., Tornieri K., Ryder P. V., Gokhale A., Zlatic S. A., Craige B., Lee J. D., Talbot K., Pare J. F., Smith Y., Faundez V. (2011) The schizophrenia susceptibility factor dysbindin and its associated complex sort cargoes from cell bodies to the synapse. Mol. Biol. Cell 22, 4854–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newell-Litwa K., Salazar G., Smith Y., Faundez V. (2009) Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol. Biol. Cell 20, 1441–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newell-Litwa K., Chintala S., Jenkins S., Pare J. F., McGaha L., Smith Y., Faundez V. (2010) Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus. J. Neurosci. 30, 820–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iizuka Y., Sei Y., Weinberger D. R., Straub R. E. (2007) Evidence that the BLOC-1 protein dysbindin modulates dopamine D2 receptor internalization and signaling but not D1 internalization. J. Neurosci. 27, 12390–12395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeans A., Malins R., Padamsey Z., Reinhart M., Emptage N. (2011) Increased expression of dysbindin-1A leads to a selective deficit in NMDA receptor signaling in the hippocampus. Neuropharmacology 61, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 23. Ji Y., Yang F., Papaleo F., Wang H. X., Gao W. J., Weinberger D. R., Lu B. (2009) Role of dysbindin in dopamine receptor trafficking and cortical GABA function. Proc. Natl. Acad. Sci. U.S.A. 106, 19593–19598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marley A., von Zastrow M. (2010) Dysbindin promotes the post-endocytic sorting of G protein-coupled receptors to lysosomes. PLoS One 5, e9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang T. T., Yang F., Chen B. S., Lu Y., Ji Y., Roche K. W., Lu B. (2009) Dysbindin regulates hippocampal LTP by controlling NMDA receptor surface expression. Proc. Natl. Acad. Sci. U.S.A. 106, 21395–21400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito H., Morishita R., Shinoda T., Iwamoto I., Sudo K., Okamoto K., Nagata K. (2010) Dysbindin-1, WAVE2 and Abi-1 form a complex that regulates dendritic spine formation. Mol. Psychiatry 15, 976–986 [DOI] [PubMed] [Google Scholar]
- 27. Benson M. A., Newey S. E., Martin-Rendon E., Hawkes R., Blake D. J. (2001) Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J. Biol. Chem. 276, 24232–24241 [DOI] [PubMed] [Google Scholar]
- 28. Nazarian R., Starcevic M., Spencer M. J., Dell'Angelica E. C. (2006) Reinvestigation of the dysbindin subunit of BLOC-1 (biogenesis of lysosome-related organelles complex-1) as a dystrobrevin-binding protein. Biochem. J. 395, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen X. W., Feng Y. Q., Hao C. J., Guo X. L., He X., Zhou Z. Y., Guo N., Huang H. P., Xiong W., Zheng H., Zuo P. L., Zhang C. X., Li W., Zhou Z. (2008) DTNBP1, a schizophrenia susceptibility gene, affects kinetics of transmitter release. J. Cell Biol. 181, 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Orozco I. J., Koppensteiner P., Ninan I., Arancio O. (2013) The schizophrenia susceptibility gene DTNBP1 modulates AMPAR synaptic transmission and plasticity in the hippocampus of juvenile DBA/2J mice. Mol. Cell. Neurosci. 58, 76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glen W. B., Jr., Horowitz B., Carlson G. C., Cannon T. D., Talbot K., Jentsch J. D., Lavin A. (2014) Dysbindin-1 loss compromises NMDAR-dependent synaptic plasticity and contextual fear conditioning. Hippocampus 24, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Q., Li W., Novak E. K., Karim A., Mishra V. S., Kingsmore S. F., Roe B. A., Suzuki T., Swank R. T. (2002) The gene for the muted (mu) mouse, a model for Hermansky-Pudlak syndrome, defines a novel protein which regulates vesicle trafficking. Hum. Mol. Genet. 11, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li W., He M., Zhou H., Bourne J. W., Liang P. (2006) Mutational data integration in gene-oriented files of the Hermansky-Pudlak Syndrome database. Hum. Mutat. 27, 402–407 [DOI] [PubMed] [Google Scholar]
- 34. Falcón-Pérez J. M., Starcevic M., Gautam R., Dell'Angelica E. C. (2002) BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J. Biol. Chem. 277, 28191–28199 [DOI] [PubMed] [Google Scholar]
- 35. Wojtowicz J. M., Kee N. (2006) BrdU assay for neurogenesis in rodents. Nat. Protoc. 1, 1399–1405 [DOI] [PubMed] [Google Scholar]
- 36. Hunsaker M. R., Wenzel H. J., Willemsen R., Berman R. F. (2009) Progressive spatial processing deficits in a mouse model of the fragile X permutation. Behav. Neurosci. 123, 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verret L., Jankowsky J. L., Xu G. M., Borchelt D. R., Rampon C. (2007) Alzheimer's-type amyloidosis in transgenic mice impairs survival of newborn neurons derived from adult hippocampal neurogenesis. J. Neurosci. 27, 6771–6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Starcevic M., Dell'Angelica E. C. (2004) Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279, 28393–28401 [DOI] [PubMed] [Google Scholar]
- 39. Larimore J., Zlatic S. A., Gokhale A., Tornieri K., Singleton K. S., Mullin A. P., Tang J., Talbot K., Faundez V. (2014) Mutations in the BLOC-1 subunits dysbindin and muted generate divergent and dosage-dependent phenotypes. J. Biol. Chem. 289, 14291–14300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kempermann G., Jessberger S., Steiner B., Kronenberg G. (2004) Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 27, 447–452 [DOI] [PubMed] [Google Scholar]
- 41. Zhao C., Deng W., Gage F. H. (2008) Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660 [DOI] [PubMed] [Google Scholar]
- 42. Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M. C., Friocourt G., McDonnell N., Reiner O., Kahn A., McConnell S. K., Berwald-Netter Y., Denoulet P., Chelly J. (1999) Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron 23, 247–256 [DOI] [PubMed] [Google Scholar]
- 43. Song J., Zhong C., Bonaguidi M. A., Sun G. J., Hsu D., Gu Y., Meletis K., Huang Z. J., Ge S., Enikolopov G., Deisseroth K., Luscher B., Christian K. M., Ming G. L., Song H. (2012) Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature 489, 150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mullin A. P., Gokhale A., Larimore J., Faundez V. (2011) Cell biology of the BLOC-1 complex subunit dysbindin, a schizophrenia susceptibility gene. Mol. Neurobiol. 44, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balu D. T., Coyle J. T. (2011) Neuroplasticity signaling pathways linked to the pathophysiology of schizophrenia. Neurosci. Biobehav. Rev. 35, 848–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris D. W., Murphy K., Kenny N., Purcell S. M., McGhee K. A., Schwaiger S., Nangle J. M., Donohoe G., Clarke S., Scully P., Quinn J., Meagher D., Baldwin P., Crumlish N., O'Callaghan E., Waddington J. L., Gill M., Corvin A. P. (2008) Dysbindin (DTNBP1) and the biogenesis of lysosome-related organelles complex 1 (BLOC-1): main and epistatic gene effects are potential contributors to schizophrenia susceptibility. Biol. Psychiatry 63, 24–31 [DOI] [PubMed] [Google Scholar]
- 47. Gerrish A., Williams H., Moskvina V., Owen M. J., O'Donovan M. C., Williams N. M. (2009) An examination of MUTED as a schizophrenia susceptibility gene. Schizophr. Res. 107, 110–111 [DOI] [PubMed] [Google Scholar]
- 48. Fei E., Ma X., Zhu C., Xue T., Yan J., Xu Y., Zhou J., Wang G. (2010) Nucleocytoplasmic shuttling of dysbindin-1, a schizophrenia-related protein, regulates synapsin I expression. J. Biol. Chem. 285, 38630–38640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oyama S., Yamakawa H., Sasagawa N., Hosoi Y., Futai E., Ishiura S. (2009) Dysbindin-1, a schizophrenia-related protein, functionally interacts with the DNA-dependent protein kinase complex in an isoform-dependent manner. PLoS One 4, e4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim J. Y., Liu C. Y., Zhang F., Duan X., Wen Z., Song J., Feighery E., Lu B., Rujescu D., St Clair D., Christian K., Callicott J. H., Weinberger D. R., Song H., Ming G. L. (2012) Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell 148, 1051–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hyde T. M., Lipska B. K., Ali T., Mathew S. V., Law A. J., Metitiri O. E., Straub R. E., Ye T., Colantuoni C., Herman M. M., Bigelow L. B., Weinberger D. R., Kleinman J. E. (2011) Expression of GABA signaling molecules KCC2, NKCC1, and GAD1 in cortical development and schizophrenia. J. Neurosci. 31, 11088–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lutkenhoff E., Karlsgodt K. H., Gutman B., Stein J. L., Thompson P. M., Cannon T. D., Jentsch J. D. (2012) Structural and functional neuroimaging phenotypes in dysbindin mutant mice. Neuroimage 62, 120–129 [DOI] [PubMed] [Google Scholar]
- 53. Chancey J. H., Poulsen D. J., Wadiche J. I., Overstreet-Wadiche L. (2014) Hilar mossy cells provide the first glutamatergic synapses to adult-born dentate granule cells. J. Neurosci. 34, 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jagasia R., Steib K., Englberger E., Herold S., Faus-Kessler T., Saxe M., Gage F. H., Song H., Lie D. C. (2009) GABA-cAMP response element-binding protein signaling regulates maturation and survival of newly generated neurons in the adult hippocampus. J. Neurosci. 29, 7966–7977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ge S., Goh E. L., Sailor K. A., Kitabatake Y., Ming G. L., Song H. (2006) GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature 439, 589–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Overstreet Wadiche L., Bromberg D. A., Bensen A. L., Westbrook G. L. (2005) GABAergic signaling to newborn neurons in dentate gyrus. J. Neurophysiol. 94, 4528–4532 [DOI] [PubMed] [Google Scholar]
- 57. Tozuka Y., Fukuda S., Namba T., Seki T., Hisatsune T. (2005) GABAergic excitation promotes neuronal differentiation in adult hippocampal progenitor cells. Neuron 47, 803–815 [DOI] [PubMed] [Google Scholar]
- 58. Espósito M. S., Piatti V. C., Laplagne D. A., Morgenstern N. A., Ferrari C. C., Pitossi F. J., Schinder A. F. (2005) Neuronal differentiation in the adult hippocampus recapitulates embryonic development. J. Neurosci. 25, 10074–10086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tashiro A., Sandler V. M., Toni N., Zhao C., Gage F. H. (2006) NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature 442, 929–933 [DOI] [PubMed] [Google Scholar]