Background: d-Malate dehydrogenase (DmlA) is involved in d-malate catabolism in Escherichia coli.
Results: When expressed in the presence of d-malate, DmlA allows growth of a ΔleuB strain without leucine.
Conclusion: DmlA physiologically contributes to two core metabolic reactions.
Significance: Several high level activities may coexist in one enzyme and be maintained during the course of evolution.
Keywords: Dehydrogenase, Enzyme Catalysis, Metabolism, Multifunctional Enzyme, Protein Evolution, Enzyme Promiscuity
Abstract
The enzymes of the β-decarboxylating dehydrogenase superfamily catalyze the oxidative decarboxylation of d-malate-based substrates with various specificities. Here, we show that, in addition to its natural function affording bacterial growth on d-malate as a carbon source, the d-malate dehydrogenase of Escherichia coli (EcDmlA) naturally expressed from its chromosomal gene is capable of complementing leucine auxotrophy in a leuB− strain lacking the paralogous isopropylmalate dehydrogenase enzyme. To our knowledge, this is the first example of an enzyme that contributes with a physiologically relevant level of activity to two distinct pathways of the core metabolism while expressed from its chromosomal locus. EcDmlA features relatively high catalytic activity on at least three different substrates (l(+)-tartrate, d-malate, and 3-isopropylmalate). Because of these properties both in vivo and in vitro, EcDmlA may be defined as a generalist enzyme. Phylogenetic analysis highlights an ancient origin of DmlA, indicating that the enzyme has maintained its generalist character throughout evolution. We discuss the implication of these findings for protein evolution.
Introduction
It is regularly assumed that superfamilies of enzymes evolved from ancestors featuring broad substrate specificity but low catalytic efficiency. After gene duplication, the redundant copies of the ancestor diverged and specialized to effectively accommodate one substrate and occupy a defined position in a metabolic pathway (1–5). Supporting this evolutionary scenario, modern specialized enzymes frequently retained weak activities on secondary substrates (“substrate promiscuity”) or the ability to catalyze secondary reactions (“catalytic promiscuity”), and the secondary activity of one member of the family is often the principal activity of another member of the same family (6–8). Moreover, many enzymes feature secondary activities that are unrelated to their evolutionary history (9–11). In the course of evolution, “promiscuous” activities may be improved to a physiologically relevant level by just a few mutations in the protein and are therefore considered as potential starting points for neofunctionalization (see reviews in Refs. 12–14). For example, one point mutation in the glutamyl phosphate reductase (ProA) enables the enzyme to serve both its original metabolic function and a new one in arginine biosynthesis (15). The mutation increases a promiscuous activity on N-acetylglutamyl phosphate by an order of magnitude and results in overexpression of the protein. Overexpression is necessary to simultaneously support both the original and new functions because the mutation had a significant negative effect on the original activity. In some cases, however, the original and promiscuous activities may coexist at a relatively high level in the wild-type enzyme. Genetic complementation of a functional knock-out then only requires protein overexpression (9–11, 16–18). In most of these studies, knock-out complementations were artificially achieved by overexpressing the proteins from multicopy vector systems. Recently, Nam et al. (19) estimated that 37% of Escherichia coli enzymes are generalists, i.e. they promiscuously accept more than one substrate with a relatively high efficiency. By using experimental data to model metabolic networks and fluxes in E. coli cells, they found that these generalists can contribute to the catalysis of 65% of the known metabolic reactions while expressed under the control of their natural promoter. These results are significant because enzymes with promiscuous activities on natural metabolites are believed to play an important role in the metabolic plasticity of organisms (20). However, there is no experimental evidence showing that a naturally expressed promiscuous enzyme may simultaneously contribute to more than one metabolic flux in vivo.
The superfamily of β-decarboxylating dehydrogenases gathers metabolically important enzymes catalyzing the successive oxidation and decarboxylation of substrates with a common R-malate moiety and different γ-substituents on C-3, with various sizes, polarity, or configurations (see Fig. 1). Up to now, five families have been identified: the NAD-dependent isopropylmalate dehydrogenases (IPMDHs),2 the NAD-dependent isocitrate dehydrogenases (IDHs), the NADP-dependent IDHs, the NAD-dependent d-malate/tartrate dehydrogenases (Dml or TDHs), and the NAD-dependent homoisocitrate dehydrogenases (HDHs). Crystal structures are available for several IDHs (21–23) and IPMDHs (24, 25), for the Thermus thermophilus HDH (26), and for the Pseudomonas putida TDH (PpTDH) (27). All of them present a conserved dimeric structure. The enzymes of the superfamily are Mg2+- or Mn2+-dependent and share conserved active site residues. They also have very different substrate specificities (see Fig. 1). IPMDHs are specific toward hydrophobic alkyl malate substrates (28), and IDHs exhibit high specificity toward the negatively charged isocitrate (29). In addition to homoisocitrate, the HDH of Deinococcus radiodurans also accepts isocitrate and isopropylmalate (30), whereas the HDHs of T. thermophilus and Pyrococcus horikoshii are active on isocitrate and homoisocitrate only (31, 32). Finally, although PpTDH was first identified as an enzyme supporting growth on l(+)-tartrate, it was later shown to be also active on d-malate and 3-isopropylmalate in vitro (33, 34). Although the enzyme transforms both d-malate and 3-isopropylmalate by oxidative decarboxylation, a non-decarboxylating oxidation is observed with l(+)-tartrate (Fig. 1).
FIGURE 1.
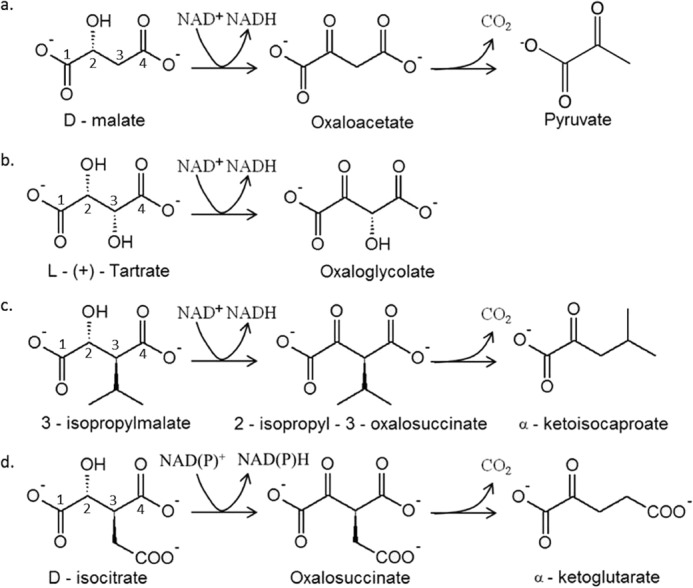
Members of the β-decarboxylating dehydrogenase family catalyze several reactions on d-malate-based substrates. a, c, and d, oxidative decarboxylation catalyzed by Dml/TDHs, IPMDHs, and NAD(P)-IDHs, respectively. b, oxidation described for PpTDH.
In E. coli, the d-malate dehydrogenase EcDmlA (dmlA gene) with high similarity to PpTDH (76% of identity) is responsible for aerobic growth on d-malate as the sole carbon source (35, 36). It coexists with a paralogous EcIPMDH (leuB gene) involved in leucine biosynthesis. It has long been known that E. coli strains harboring the leuB6 genotype produce revertants under leucine starvation conditions (37). However, the fact that most of these revertants are suppressors (i.e. no mutation could be detected in the leuB locus) indicates that the loss of EcIPMDH can be complemented by another protein (37, 38). In this work, we show that chromosomal expression of EcDmlA affords complementation of the leuB− phenotype when induced by d-malate. The EcDmlA-dependent leuB+ phenotype is not observed with glucose. These results are further supported by the biochemical characterization of the purified enzyme, and we discuss its evolutionary implications with the help of a phylogenetic analysis of the superfamily.
EXPERIMENTAL PROCEDURES
Strains and Chemicals
The ΔleuB strain HBLB1 was constructed from the HB101 strain using a standard phage P1 transduction protocol (39). The leuB6 gene was replaced with a kanamycin resistance cassette using the JW5807-2 strain (ΔleuB::kan) from the Keio Collection (40) as a donor. The cassette insertion was validated by PCR and sequencing.
Cloning
The dmlA gene was amplified from the E. coli DH12S genome by PCR (Phusion polymerase, Fisher Scientific) and cloned between the NdeI and BamHI sites of the pMal-pIII vector (New England Biolabs) in C-terminal fusion with a GSSG linker and Strep-tag II. In the final construct called phDmlA, the dmlA gene replaced the entire malE-containing ORF and was under the control of the Ptac promoter. The genetic construction was verified by sequencing.
Phenotypic Auxotrophy Tests
All phenotypic tests were performed on amino acid-free EZ Rich defined medium. The medium was prepared as described by Neidhardt et al. (41), and the 10× EZ solution used to prepare the medium was purchased from Teknova (Hollister, California). For solid medium culture, the medium was solidified with 1.5% agar. The carbon sources were added up to 0.1% (d-glucose) or 0.15% (d-malate), and leucine was used at 1 mm if specified. Proline (1 mm) was added to the culture medium to support growth of the HBLB1 strain. Liquid cultures (100 ml) were inoculated from an overnight preculture of HBLB1 cells preliminary washed with M9 buffer. CaCO3 (10 mm) was added to the medium to reduce the lag time as described (41). Despite this step, a lag time of ∼16 h was observed. The cells were inoculated at an initial A600 of 0.05 and incubated overnight at 37 °C and 180 rpm. Growth was monitored the next day. For growth tests on solid medium, precultures grown overnight at 37 °C and 180 rpm in LB/kanamycin (50 μg/ml) were harvested by centrifugation and resuspended in 3 ml of M9 buffer. The cells were then sequentially diluted in M9 buffer up to 10−5-fold. 10 μl of each dilution was spotted on the agar. The plates were incubated at 37 °C for 24 h.
Activity Assay in Cell Extracts
50 ml of exponentially grown cell cultures (A600 = 0.2 for the culture with d-malate (0.15%) and leucine (1 mm), A600 = 0.1 for the culture with d-malate (0.15%), and A600 = 0.45 for the culture with d-glucose (0.1%) and leucine (1 mm)) were centrifuged at 4000 rpm for 10 min, and the cells were resuspended in 1 ml of KAC buffer (25 mm MOPS and 100 mm KCl, pH 7.5) (29, 42) supplemented with one tablet of Complete mini EDTA-free protease inhibitor mixture (Roche Applied Science). The samples were flash-frozen in liquid nitrogen and kept at −80 °C. For activity tests, the cells were crushed in a FastPrep-24 instrument (MP Biomedicals, Santa Ana, California) for 60 s (250 μl of glass bead suspension for 750 μl of cell suspension). The extracts were cleared by centrifugation and diluted 10 times for the activity measurements in KAC buffer. To start the reaction, 25 μl of diluted extracts was added to 125 μl of reaction mixture (500 μm d-malate or 3-isopropylmalate, 250 μm NAD+, 5 mm MnCl2, 1 mm DTT, and KAC buffer). The reaction was followed by an increase in A340. The activity data were normalized to the total concentration of protein in the extracts, measured by BCA protein assay (Pierce).
DmlA Expression and Purification
The protein was expressed from BL21 cells transformed with phDmlA. Typically, 500 ml of Terrific Broth medium was inoculated and grown at 37 °C and 180 rpm until A600 reached 0.6. Isopropyl β-d-thiogalactopyranoside was added (1 mm final concentration), and the protein was produced at 25 °C and 180 rpm for 4 h. The cells were centrifuged and resuspended in 20 ml of buffer (100 mm Tris and 150 mm NaCl, pH 8.0) supplemented with one tablet of Complete mini EDTA-free protease inhibitor mixture and flash-frozen in liquid nitrogen. Thawed cells were treated with lysozyme (1 mg/ml, 1 h), DNase (5 μg/ml) was added for an additional 20 min, and lysis was completed by 30 min of sonication (cycles of 30 s of ultrasound and 30 s of resting on ice). The protein was purified from the cleared lysate by Strep-Tactin affinity chromatography (IBA, Göttingen, Germany). The fractions containing the protein were pooled, desalted (final buffer of 10 mm Tris, pH 8.0), and concentrated by ultracentrifugation in Amicon tubes (10-kDa cutoff; Millipore). The concentration of the protein was determined by measuring the absorbance at 280 nm using a predicted extinction coefficient of 64,400 m−1 cm−1 (ProtParam tool on the ExPASy server (43)). The final concentration was typically in the range of 6 mg/ml. 25-μl aliquots were flash-frozen in liquid nitrogen and kept at −80 °C. An SDS-polyacrylamide gel of the purified DmlA protein is shown in Fig. 2.
FIGURE 2.
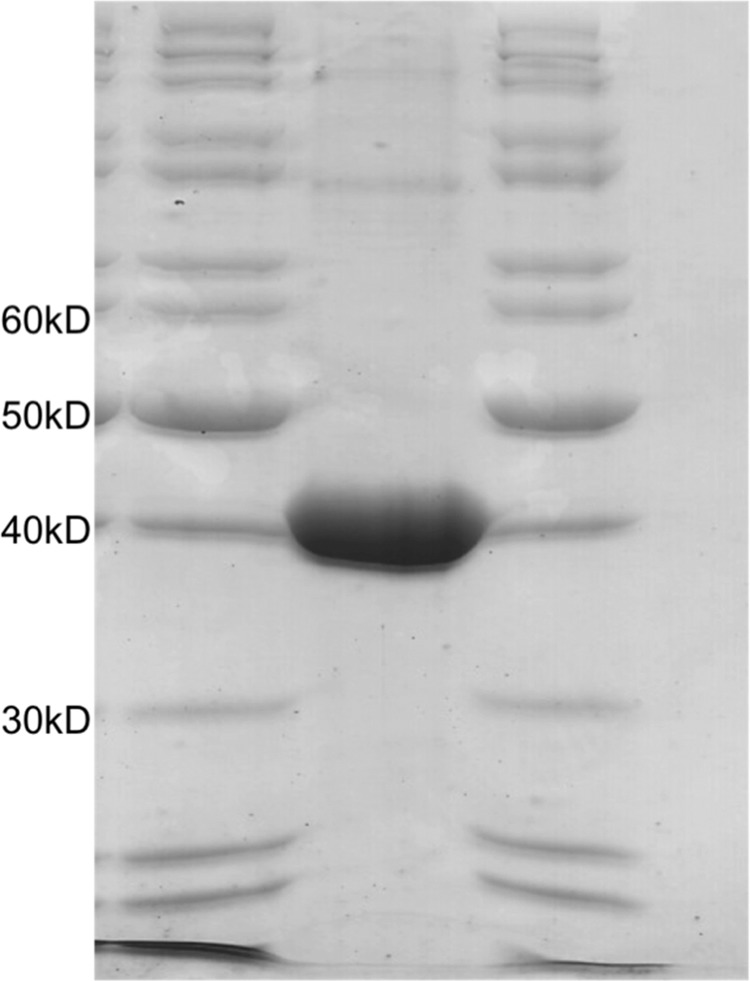
SDS-PAGE analysis of the purified DmlA protein. The protein was purified by Strep-Tactin affinity chromatography and concentrated to ∼6 mg/ml (140 μm). 20 μl was loaded on the gel. The size of the DmlA monomer is 42 kDa.
Determination of Catalytic Parameters for Different Substrates
All catalytic measurements were performed in KAC/DTT buffer (25 mm MOPS, 100 mm KCl, and 1 mm DTT, pH 7.5) at 25 °C. The pH of the substrates was adjusted to pH 7.5 with KOH. MnCl2 or MgCl2 (100 mm stock solutions in water) was added up to 5 mm to each reaction mixture. NAD+ was added up to 500 μm (for d-malate and 3-isopropylmalate assays) or 2 mm (for l(+)-tartrate assays). For the other substrates, the NAD concentration was arbitrarily fixed at 1 mm to obtain apparent kinetic parameters. The reaction was started by the addition of 25 μl of the enzyme diluted in KAC/DTT buffer. The concentration of d-malate and 3-isopropylmalate ranged from 10 to 500 μm. The concentration of l(+)-tartrate ranged from 0.1 to 4 mm. tk;2Measurements were also performed with isocitrate (0.1–5 mm), d-lactate (0.1–14 mm), and d(−)-tartrate (0.1–8 mm). The fluorescence signal (340 nm excitation, 460 nm emission) was monitored over time on a Tecan Infinite 200 PRO microplate reader. Fluorescence was converted into NADH concentration using a calibration curve. The observed kinetic constants (kobs) were calculated according to Equation 1,
 |
where [E]0 is the DmlA concentration and V0 is the initial velocity of the reaction. Nonlinear Michaelis-Menten fitting was performed with MicroCal Origin 6.0 software (OriginLab, Northampton, MA) with Equation 2,
 |
where [S] is the substrate concentration.
All measurements were performed in duplicates. The effect of pH on DmlA activity was tested by Michaelis-Menten kinetics in KAC/DTT buffer with pH adjusted to 6.0–8.5 (with steps of 0.5 pH unit) with HCl or KOH.
Differential Scanning Fluorometry
The protein was diluted in KAC buffer and used at a concentration of 2 μm in the unfolding assay. The substrates of the enzyme were added at a concentration of 1 mm. SYPRO Orange dye was added (5000-fold dilution of dimethyl sulfoxide stock solution, Thermo Fisher Scientific). Using an Applied Biosciences StepOnePlus RT-PCR system (Thermo Fisher Scientific), a temperature gradient of 1 °C/min (ranging from 25 to 90 °C) was applied to the samples, and dye fluorescence was monitored (emission filter at 580 nm). The unfolding data were corrected with the blank carried out under the same conditions without protein and were fitted to the Boltzmann sigmoid equation (Equation 3) describing the fluorescence intensity (y) as a function of temperature (x) (as described in Ref. 44) using MicroCal Origin 6.0 software.
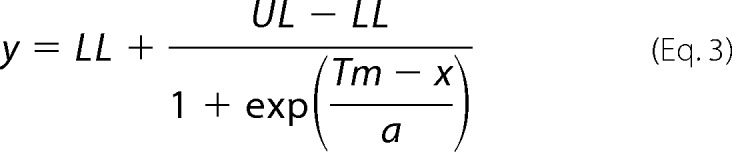 |
LL and UL are the minimum and maximum fluorescence intensities of the transition, respectively. Tm is the melting temperature. The constant a is the slope of the curve at Tm. The measurements were done in duplicates.
Phylogenetic Analysis
We constructed a data set of ∼250 homologous sequences after performing a BLAST search with PpTDH as bait. The sequences were aligned, and those containing mutations of conserved active site residues were discarded. As most of the sequences obtained were from α-, β-, and γ-Proteobacteria, we kept only the sequences from these classes of organisms in the final data set for a more clear analysis. Sequences from fungi, archaea, and other Proteobacteria were discarded, as well as two HDH sequences. Finally, the data set was screened for highly similar sequences. We tried to keep Dml/TDHs and IPMDHs from the same organisms in the data set when it was possible. The resulting final data set contains 95 different IDH, Dml/TDH, IPMDH, and TtuC sequences (see supplemental material). The annotation attributed to some proteins did not correspond to the conserved motif in the substrate recognition structure (45). These sequences were properly re-annotated. Alignments were done with the Clustal algorithm. The best fit model for amino acid replacement was determined using the ProtTest 3 server (46). The LG +I +G model was selected. The phylogenetic tree was calculated with the PhyML program, and 100 rounds of bootstrapping analysis were performed (47). The trees were generated using the TreeDyn algorithm, available online at the Phylogeny.fr web site (48). Finally, to evaluate the robustness of the tree, similar analyses from initial Muscle and T-Coffee alignments and from the Clustal alignment using the WAG substitution model were performed. Similar conclusions could be drawn from all constructed trees.
RESULTS
Expression of DmlA under the Control of Its Native Promoter Complements the ΔleuB Phenotype
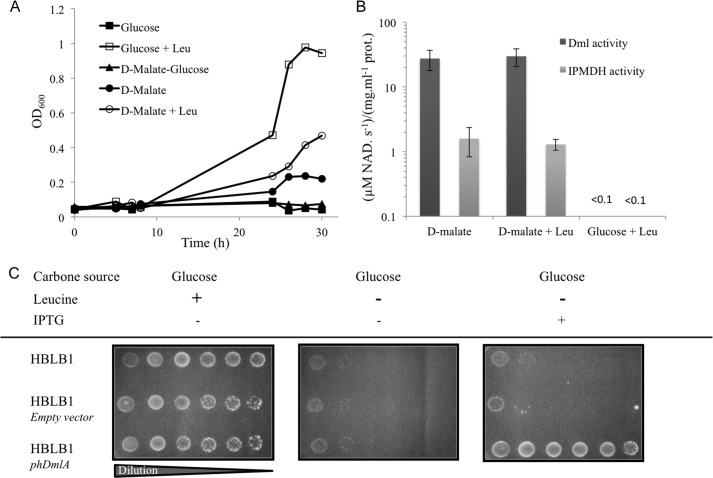
Given the high similarity of EcDmlA to PpTDH and the activity of PpTDH detected on 3-isopropylmalate, we hypothesized that EcDmlA could be responsible for suppressive leuB knock-out complementation. To study the IPMDH function of EcDmlA in vivo and to avoid direct leuB− reversion, we constructed the HBLB1 strain, in which the leuB6 gene of the HB101 strain was deleted (ΔleuB strain). Leucine auxotrophy was tested in chemically defined medium with different carbon sources. As shown in Fig. 3a, with d-glucose as the carbon source, leucine supplementation was necessary to support HBLB1 growth. With d-malate, however, HBLB1 cells grew without leucine, and growth was inhibited when glucose was added as an alternative carbon source to d-malate. The same result was obtained on solid medium (data not shown). As EcDmlA expression is induced by d-malate and repressed by glucose (49), this experiment supports our initial hypothesis. In liquid medium, A600 reached ∼0.4 on d-malate and leucine and 0.2 on d-malate at stationary phase. The lower plateau reached by the cells in the absence of leucine indicates that the fitness of the strain is limited by either the d-malate catabolism or the synthesis of leucine, as both pathways are supported by the same enzyme and compete with each other. We collected samples of exponentially growing cells and tested the activities on d-malate (Dml) and 3-isopropylmalate (IPMDH) in the extracts. Both activities were present in cells grown on d-malate (with and without leucine) and absent from cells grown on d-glucose medium supplemented with leucine (Fig. 3b). No difference in the level of Dml or IPMDH activity was detected between cells grown on d-malate in the presence or absence of leucine. Altogether, these results indicate that normal induction of EcDmlA by d-malate is sufficient for leuB− complementation and that no overexpression mechanism is involved. To confirm the role of EcDmlA in leucine prototrophy, its gene was cloned in an expression plasmid under the control of the Ptac promoter. Competent HBLB1 cells were transformed with the resulting phDmlA vector. With glucose as the carbon source, leucine prototrophy of the phDmlA-HBLB1 strain depended on DmlA induction by isopropyl β-d-thiogalactopyranoside (IPTG) (Fig. 3c). Besides the selection of a few revertants, no growth was observed for the control strain transformed with the empty vector.
FIGURE 3.
DmlA complements the ΔleuB phenotype. A, HBLB1 (ΔleuB strain) growth in liquid EZ Rich defined medium. ■, d-glucose as the carbon source (0.1%) without leucine; □, d-glucose as the carbon source (0.1%) with leucine (1 mm); ▴, d-glucose (0.1%) and d-malate (0.15%) as carbon sources; ●, d-malate as the carbon source (0.15%) without leucine; ○, d-malate as the carbon source (0.15%) with leucine (1 mm). B, d-malate (dark gray bars) and 3-isopropylmalate (light gray bars) dehydrogenase activities in extracts prepared from exponentially growing HBLB1 cells collected under d-malate, d-malate + leucine, and d-glucose + leucine conditions. The measured activities were normalized to the total protein concentrations. C, EcDmlA expressed from HBLB1 cells transformed with the phDmlA vector. Complementation of leucine auxotrophy on EZ Rich defined medium depends on the presence of the isopropyl β-d-thiogalactopyranoside (IPTG) inducer (1 mm).
EcDmlA Is a Generalist Enzyme
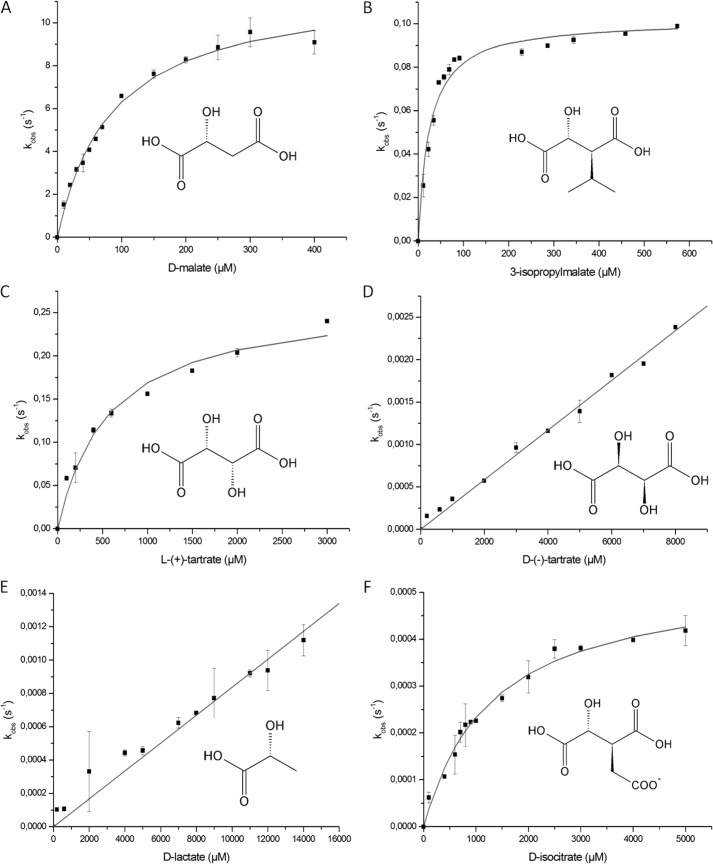
Strep-tag II-fused EcDmlA was expressed, purified, and biochemically characterized. Similar to other β-decarboxylating dehydrogenases, EcDmlA activity depended on the presence of divalent metal ion (Mn2+ or Mg2+) (data not shown). As several TDHs and HDHs are activated by K+ or NH4+ and are less active in the presence of Na+ (33, 50, 51), we tested the effect of these monovalent cations on the activity of EcDmlA and found a similar behavior (Fig. 4). Enzyme kinetics (50) and structural data (27) suggest that the K+ or NH4+ ion interacts with the NAD and helps in cofactor positioning. The activity-pH profile has a bell shape with an optimum at pH 7.0–7.5 (data not shown). As predicted by amino acid sequence, EcDmlA preferred NAD to NADP by ∼500-fold (data not shown). The activity of the enzyme on d-malate (kcat/Km(malate) = 105 m−1 s−1) (Table 1) corresponded to that reported for a moderately efficient enzyme in energy metabolism (52). Strikingly, the enzyme exhibited high activity on 3-isopropylmalate, with kcat/Km(IPM) = 4 × 103 m−1 s−1. This is only 50 times lower than the constant for EcIPMDH at the same temperature (kcat/Km(IPM) = 2 × 105 m−1 s−1) (28), supporting leuB− complementation. Finally, significant activity on l(+)-tartrate was also detected (kcat/Km(l-(+)-tartrate) = 5 × 102 m−1 s−1). The impact of divalent metal ion on substrate specificity was investigated by replacing Mn2+ with Mg2+ for activity measurements. With the Mg2+ metal ion, all three activities decreased by 2–5-fold, but their relative levels remained roughly unchanged (data not shown).
FIGURE 4.
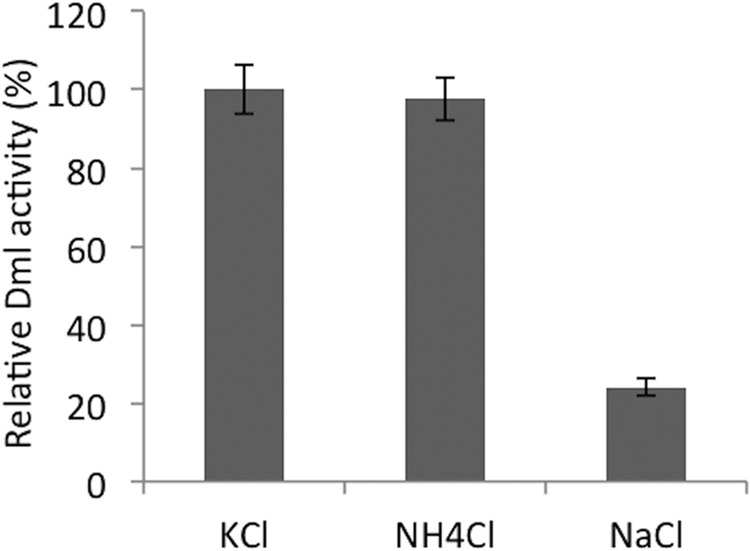
Monovalent cations modulate the catalytic activity of DmlA. The activity of DmlA on d-malate was tested in 25 mm MOPS buffer, pH 7.5, supplemented with 150 mm K+, NH4+, or Na+ chloride salts. The activity of the enzyme with KCl was set as the reference.
TABLE 1.
Apparent kinetic parameters determined for EcDmlA on different substrates and in the presence of different divalent metal ions
The concentration of Mn2+ was fixed at 5 mm for all measurements. The measurements were done at 25 °C and pH 7.5 in the presence of 150 mm KCl. The rate versus substrate concentration data are presented in Fig. 6. ND, not determined.
|
EcDmlA |
PpTDHc | |||||
|---|---|---|---|---|---|---|
| kcat | Km(substrate)a | kcat/Km(substrate) | Km(NAD)b | kcat/Km(NAD) | kcat/Km(substrate) | |
| s−1 | μm | m−1 s−1 | μm | m−1 s−1 | m−1 s−1 | |
| d-Malated | 11.7 ± 0.3 | 96 ± 7 | 1 × 105 | 94 ± 8 | 1 × 105 | 2 × 105 |
| 3-Isopropylmalated | 0.102 ± 0.003 | 24 ± 3 | 4 × 103 | 70 ± 3 | 2 × 103 | 1 × 104 |
| l(+)-Tartrated | 0.26 ± 0.01 | 567 ± 84 | 5 × 102 | 600 ± 200 | 5 × 102 | 4 × 102 |
| d-Isocitratef | (5.4 ± 0.2) × 10−4 | 1300 ± 125 | 4 × 10−1 | ND | ND | ND |
| d(−)-Tartratef,g | ND | ≥8000 | 3 × 10−1 | ND | ND | ND |
| d-Lactatef,g | ND | ≥14,000 | 1 × 10−1 | ND | ND | ND |
a Apparent Km for the specified substrate with a fixed concentration of NAD.
b Apparent Km for NAD with a fixed concentration of the specified substrate.
c Data are taken from Ref. 33.
d For these substrate, the concentration of NAD was fixed at 500 μm for the Km(substrate) measurements, and the concentration of substrates was fixed at 500 μm for the Km(NAD) measurements.
e For l(+)-tartrate, the concentration of NAD was fixed at 2 mm for the Km(substrate) measurements, and the concentration of substrates was fixed at 5 mm for the Km(NAD) measurements.
f For these substrates, the concentration of NAD was fixed at 1 mm to obtain apparent kcat/Km(substrate).
g Because of the high Km(substrate), the kinetic parameters could not be obtained for these substrates, and the kcat/Km(substrate) values were estimated from the linear part of the Michaelis-Menten function.
The kcat/Km parameters obtained for EcDmlA are very similar to the data reported for PpTDH at the same temperature for d-malate, 3-isopropylmalate, and l(+)-tartrate (Table 1; data from Ref. 33). However, overexpressed EcDmlA did not afford any growth of the HBLB1 strain on l(+)-tartrate in the presence of 1 mm leucine (data not shown). In a similar experiment, Lukas et al. (49) reported a very slow growth but only when the medium was enriched with amino acids. The turnover rate constant (kcat) of DmlA with l(+)-tartrate was twice as high as that with 3-isopropylmalate. However, the Km for l(+)-tartrate and NAD was 5–20-fold higher than the Km for all previously tested substrates. Additionally, a low affinity of EcDmlA for l(+)-tartrate was suggested by thermal unfolding of the protein in a differential scanning fluorometry experiment (Fig. 5). In a differential scanning fluorometry assay, the temperature at which a protein unfolds in the presence of a ligand is compared with its unfolding temperature in the apo form. The stabilizing effect increases with binding affinity (53, 54). In the presence of 1 mm d-malate or 3-isopropylmalate, the protein unfolded at higher temperature compared with its apo form (1.5–2 °C higher Tm). In contrast, no significant Tm shift was observed in the presence of the same concentration of l(+)-tartrate, indicating that the interaction between EcDmlA and l(+)-tartrate is weaker than the interaction between EcDmlA and d-malate or 3-isopropylmalate. Notably, l(+)-tartrate is the only substrate in Fig. 1 with C-3 in the R-configuration, whereas all other substrates feature the S-configuration. The increase in Km for both l(+)-tartrate and NAD may be explained by steric hindrance between the substrate and cofactor or between the substituent on C-3 of l(+)-tartrate and a structural part of the active site.
FIGURE 5.
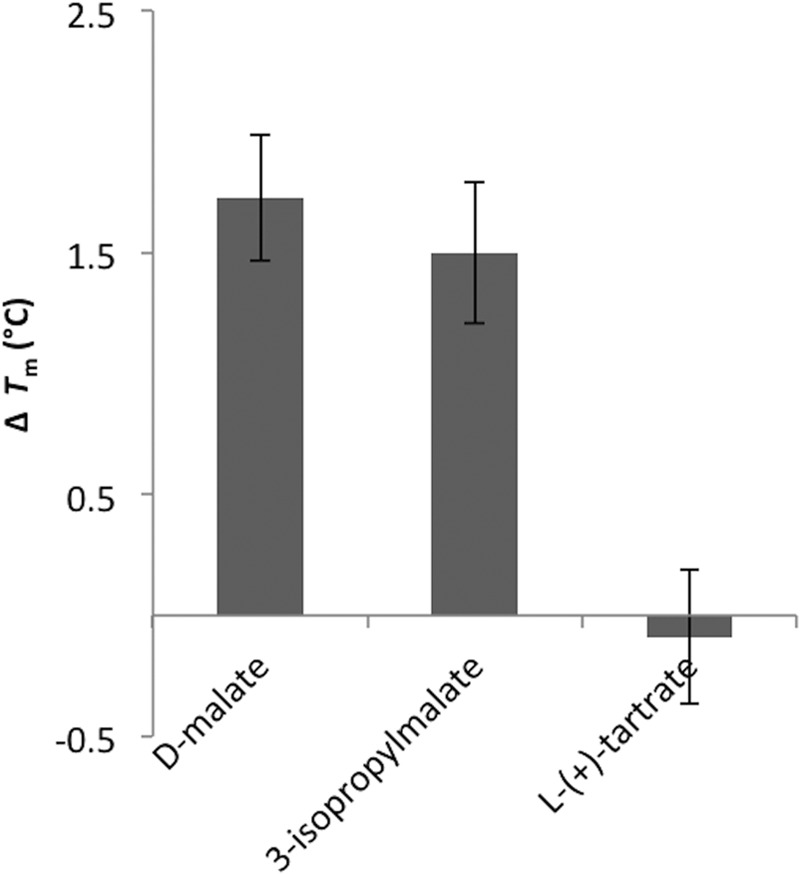
DmlA unfolds at a higher temperature in the presence of d-malate and 3-isopropylmalate, but not l-(+)-tartrate. Tm was measured by differential scanning fluorometry of the apo form and in the presence of 1 mm d-malate, 3-isopropylmalate, and l(+)-tartrate. The ΔTm values represent the difference in Tm between the protein in the presence of the specified substrate and the apoprotein.
In the results presented above, we showed that EcDmlA was active on a substrate with a large hydrophobic γ-substituent (3-isopropylmalate), as well as a substrate with hydrogen in that position (d-malate) and a substrate with a polar hydroxyl as a γ-substituent in an inverted configuration (l(+)-tartrate). We further observed a weak activity of the enzyme on isocitrate (kcat/Km(isocitrate) = 4 × 10−1 m−1 s−1), a molecule with a negatively charged γ-substituent. d-Lactate, a minimalist substrate without the C-4 carboxyl, was also a poor substrate for EcDmlA (kcat/Km(lactate) = 1 × 10−1 m−1 s−1). Finally, the enzyme was also active on d(−)-tartrate (kcat/Km(d-(−)-tartrate) = 3 × 10−1 m−1 s−1), which has C-3 in the preferred S-configuration but features an inverted configuration of C-2 involved in hydride transfer with the cofactor. Hence, EcDmlA is a highly versatile enzyme and accepts substrates featuring different sizes and polarities of the γ-substituent, a carboxyl or a hydrogen as the other substituent of C-3, and both configurations of C-2 and C-3. The rate versus substrate concentration data for all tested substrates are shown in Fig. 6.
FIGURE 6.
Kinetic characterization of DmlA on various substrates. The kobs values were measured for six substrates at different concentrations with fixed concentrations of NAD+ (see “Experimental Procedures” for concentrations) and Mn2+ (5 mm). The kinetic parameters were calculated from the data by fitting to the Michaelis-Menten equation (Equation 2) (A–C and F) and by estimating the kcat/Km parameter from the linear part of the activity-substrate curve (D and E). The calculated parameters are reported in Table 1.
Phylogenetic Analysis Suggests an Ancient Origin for Dml/TDH Enzymes
d-Malate/tartrate dehydrogenases are metabolic enzymes supporting aerobic growth either on d-malate (Dml activity) or on l(+)-tartrate (TDH and TtuC activities). It is important to emphasize that the current data on the Dml/TDH family are limited, especially at the metabolic level. Hence, the enzymes hereafter are annotated according to their initial characterization (Dml or TDH), which may not precisely reflect their actual metabolic role(s). So far, biochemical data are available for PpTDH, Pseudomonas fluorescens TDH (55), and Rhodobacter sphaeroides TDH (51), as well as EcDmlA (this study). Additionally, the metabolic role of two other proteins was experimentally confirmed: d-malate growth for Enterobacter aerogenes Dml (55) and l(+)-tartrate growth for Agrobacterium vitis TtuC (56).
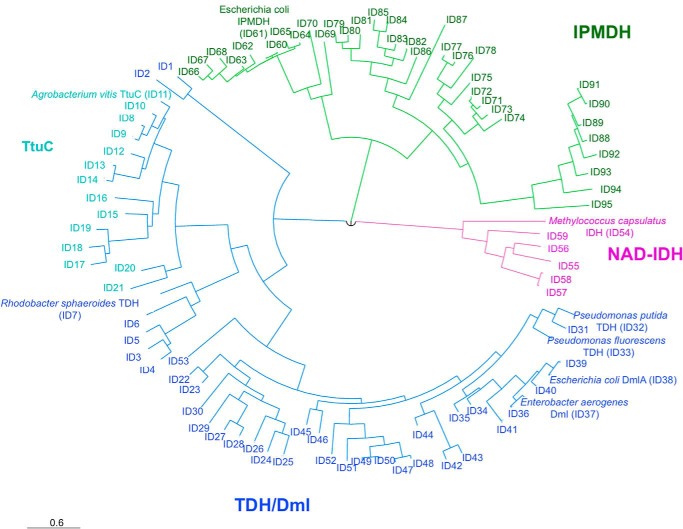
The phylogenetic analysis of a data set of sequences homologous to PpTDH and EcDmlA (minimum 35% identity) showed that the sequences cluster into three groups: the IPMDHs, NAD-dependent IDHs, and Dml/TDHs (Fig. 7). In the data set, the function of the proteins was assigned using the conserved motifs in the substrate recognition region identified by Chen and Jeong (45). The motif SXN is conserved in IDHs, whereas the motifs EX2–3(L/I)L and LX3L are proper to 3-isopropylmalate and d-malate/tartrate dehydrogenases, respectively (45, 57).
FIGURE 7.
Phylogenetic relationship between Dml/TDHs, IPMDHs, and IDHs. The unrooted phylogenetic tree shows the evolutionary relationship between IPMDHs (green), NAD-dependent IDHs (pink), and Dml/TDH/TtuC enzymes (Dml/TDHs are blue, and TtuC enzymes are cyan) from α-, β-, and γ-Proteobacteria. The protein names are indicated for those proteins that have been characterized in vivo and/or in vitro (28, 51, 55, 56, 66). The other proteins are designed with an ID number referring to the complete data set presented in the supplemental material.
The evolutionary trajectory that was hypothesized for explaining the low activity of PpTDH on l(+)-tartrate and its activity on isopropylmalate starts by a recent duplication of an IPMDH gene, followed by the evolution of the enzyme into a generalist, and ends with the partial re-specialization for the new catalytic function (33). However, the topology of our phylogenetic tree suggests that the Dml/TDH activity is more ancient than previously thought. Instead of branching to the IPMDH cluster, the Dml/TDHs are monophyletic and diverge more or less from the same point as the NAD-dependent IDHs and IPMDHs. Hence, Dml/TDH proteins likely evolved directly from the common ancestor of the β-decarboxylating dehydrogenases that was proposed to be a generalist enzyme (58) rather than from a duplicated IPMDH gene. Another interesting observation is that the generalist character seems to be a common feature of the Dml/TDH enzymes. Activity on d-malate, l(+)-tartrate, and 3-isopropylmalate was reported for PpTDH and EcDmlA, sharing 76% identity along the entire sequence length. d-Malate and tartrate activities were also reported for R. sphaeroides TDH, sharing 60 and 65% identities, respectively, with the two others and located in a different phylogenetic subcluster. The highest activity of the purified R. sphaeroides TDH, the Dml activity, is only 2 orders of magnitude higher than its TDH activity (in terms of kcat/Km) (51). Moreover, R. sphaeroides TDH supports the growth of R. sphaeroides on l(+)-tartrate. The activity of this enzyme toward 3-isopropylmalate was not tested. The promiscuous properties of these three divergent enzymes suggest that the generalist trait was maintained through the evolution of Dml/TDHs.
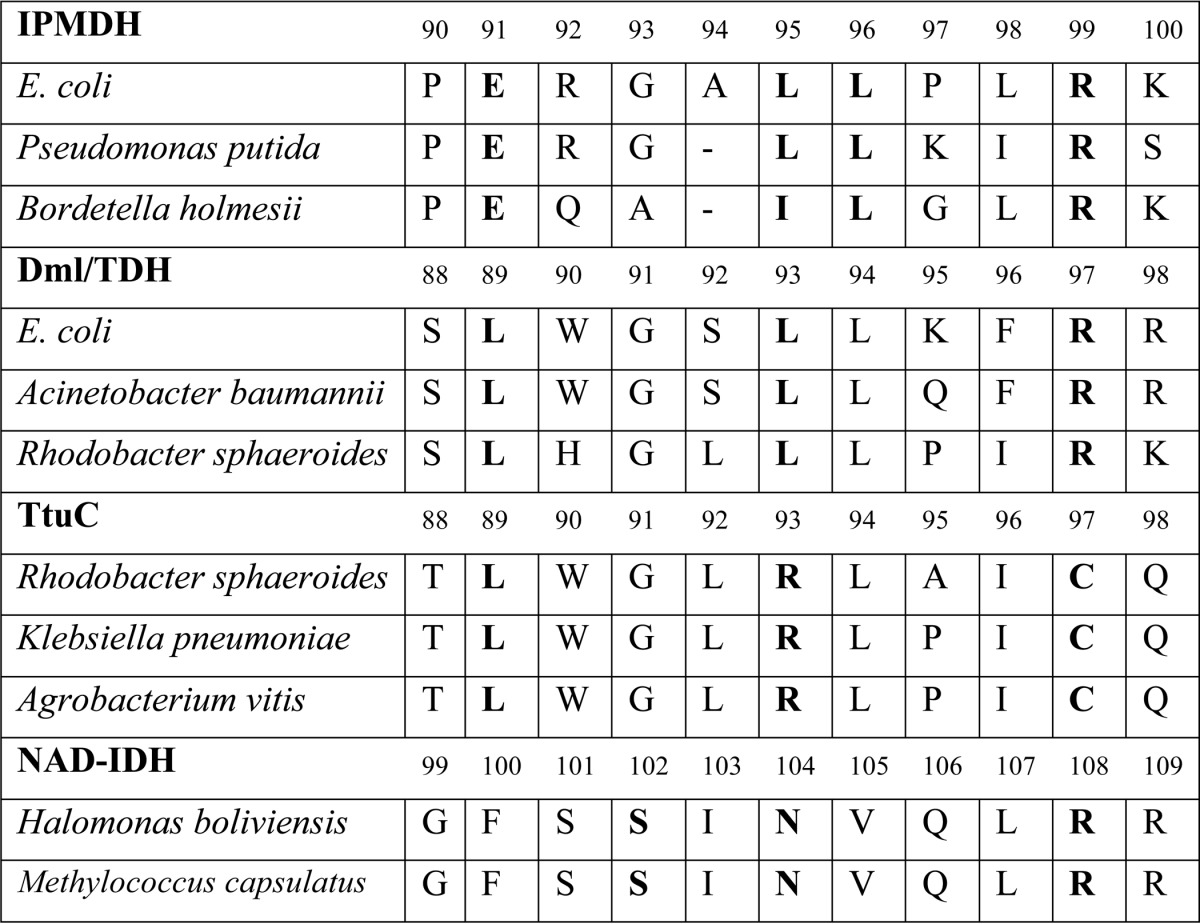
The phylogenetic tree also shows that TtuC-like proteins form a subcluster among the d-malate/tartrate dehydrogenases (Fig. 7). Surprisingly, some active site residues that are usually conserved in the enzymes of the β-decarboxylating dehydrogenase superfamily are modified in TtuC proteins (Table 2). First, the last leucine of the LXXXL motif specific to the substrate recognition loop of the Dml/TDH enzymes is replaced with arginine. Second, Arg-97 (EcDmlA numbering), which is conserved in all β-decarboxylating dehydrogenases and interacts with both carboxylates of the common d-malate moiety of the substrates, is mutated to cysteine in TtuC proteins. Despite these mutations, A. vitis TtuC is essential for growth on l(+)-tartrate (56). This suggests a unique substrate-binding mechanism among the β-decarboxylating dehydrogenases. As many organisms, such as R. sphaeroides, Klebsiella pneumoniae, and Agrobacterium tumefaciens, contain both TtuC and TDH, the enzymes may have different metabolic roles or functions.
TABLE 2.
Alignment of several IPMDH, Dml/TDH, TtuC, and NAD-dependent IDH proteins in the substrate recognition region
The numbers indicate the amino acid numbering of the first sequence from each group of enzymes. The conserved residues, identified as conserved motifs, are shown in boldface. The complete set of sequences used for the phylogenetic analysis and the alignment of the substrate recognition regions is provided in the supplemental material.
DISCUSSION
In this work, we have shown that the promiscuous activity of wild-type and naturally expressed EcDmlA on 3-isopropylmalate is high enough to complement the knock-out of the paralogous LeuB enzyme from the leucine biosynthesis pathway. Moreover, EcDmlA is simultaneously active in its original d-malate catabolism pathway and in the leucine biosynthesis pathway. The bifunctionality is significant because relatively high fluxes through these two pathways are essential to the bacteria under the applied growth conditions. To our knowledge, this result provides the first experimental example of a wild-type generalist enzyme supporting two core metabolic fluxes in vivo in the absence of protein overexpression. The capacity of EcDmlA to function in both metabolic pathways is explained by its high catalytic activity on d-malate and isopropylmalate. The enzyme also has a relatively high activity on l(+)-tartrate, although our results indicate a weak affinity of the protein for this alternative substrate compared with d-malate and 3-isopropylmalate. This can be linked to the work on PpTDH done by Serfozo and Tipton (59, 60), who suggested a common binding mode for substrates featuring the S-configuration of C-3 (d-malate, 3-isopropylmalate) and an alternative binding mode for substrates featuring the R-configuration of C-3 (l(+)-tartrate). The alternative R-configuration binding mode has not been characterized yet and was proposed to explain the release of the oxidized intermediate without further decarboxylation. As the kinetic values obtained for EcDmlA are very similar to the kinetic values reported for PpTDH on all three substrates (33), the catalytic mechanisms of both enzymes should be similar, and the inability of EcDmlA overexpression to support growth of the bacteria on l(+)-tartrate is more likely due to a lack of membrane transporters for l(+)-tartrate or to metabolic differences between E. coli and P. putida.
Enzymes are generally described in textbooks as remarkable specialists. However, the number of promiscuous enzymes reported in the literature is rapidly growing, suggesting that most enzymes promiscuously accept alternative substrates (13). EcDmlA is a remarkable example because it features relatively high activities on three different substrates and may therefore be defined as a generalist. Moreover, the topology of our phylogenetic tree suggests that Dml/TDHs are ancient and emerged about the time when the core metabolic pathways of leucine biosynthesis and the Krebs cycle diverged. Because the three Dml/TDHs that have been biochemically characterized accept multiple substrates and are well dispersed in the tree, the whole family most probably shares the generalist trait, although a more systematic study would be necessary to confirm this hypothesis.
Why was EcDmlA maintained as a generalist in the course of evolution? Was it selected for several activities, or is it just the result of fortuitous enzyme infidelity (i.e. promiscuity (61))? The later hypothesis is supported by the regulation mechanism of the dmlA gene. The expression of the protein is determined by the carbon source used, independent of the presence or absence of leucine, because no overexpression could be detected under leucine starvation conditions. This indicates that its IPMDH activity is a consequence of enzyme infidelity rather than a selected alternative route for leucine biosynthesis. We propose two hypotheses to explain the evolutionary retention of the generalist trait.
(i) The selection pressure for specialization may be low. In other words, promiscuity may be maintained as long as it does not interfere with any metabolic function. Although EcDmlA will have to support a high metabolic flux when d-malate is used as the carbon source, these conditions may be very occasional (49). Moreover, when DmlA is expressed in a wild-type strain with an active leucine biosynthesis pathway, the two pathways will probably not interfere significantly. Indeed, as an intermediate metabolite, 3-isopropylmalate will probably be maintained at a low concentration. In contrast, if a source of exogenous leucine is present in the medium, the expression of the genes encoded in the leu operon will be down-regulated (62), and the intracellular concentration of the intermediate metabolites will be even lower.
(ii) The full specialization may be difficult to achieve in this particular active site scaffold. Indeed, the positioning of the cofactor relative to the substrate is a critical aspect for catalysis by the β-decarboxylating dehydrogenases (63, 64). The cofactor may be constrained to its optimal conformation by active site residues (as for IPMDHs (63)) or by the γ-substituent of the substrate (as for IDHs (64)). For PpTDH, structural data suggest that cofactor positioning probably involves an interaction with the monovalent cation (K+ or NH4+) that is necessary for catalysis (27, 33). However, the interactions between K+ ions and their ligands are dominantly electrostatic, and a wider range of distances and coordination numbers are then acceptable (65). An active site geometry shaped by a K+ ion would probably allow more conformational changes to accommodate different substrates. This hypothesis is supported by the biochemical data obtained for other β-decarboxylating dehydrogenases. Indeed, among the biochemically characterized HDHs, the cation-dependent HDH of D. radiodurans features broader substrate specificity than the HDHs of T. thermophilus and P. horikoshii, which do not require any monovalent cation (30–32). The IDHs and IPMDHs, which are cation-independent, are more specific to their respective natural substrates.
How can the generalist traits of an enzyme impact the metabolic plasticity of the organism? Enzyme promiscuous activities are believed to be good starting points for evolution by providing possibilities of innovation before or after a gene duplication event. As discussed in the Introduction, McLoughlin and Copley (15) reported an example of an enzyme featuring one unique mutation that affords bifunctionality but at the expense of a significant negative trade-off between the new and old functions. Hence, at the early stage of evolution, a compromise between the newly evolved and original activities has to be found to allow the survival of the organism. The particular example of EcDmlA shows that a secondary high level activity may coexist with the original one without any mutation-associated compromise. Despite the fact that the activity of the enzyme on 3-isopropylmalate is high (kcat/Km ∼ 103 m−1 s−1) and sufficient to affect the phenotype of the organism, its principal activity on d-malate remains compatible with an efficient enzyme from secondary metabolism (52). There is, however, a compromise imposed by gene sharing but at the metabolic level when both activities are simultaneously required. Indeed, the growth of the ΔleuB strain on d-malate is lower when exogenous leucine is absent, indicating a competition between the two metabolic pathways sustained by the same enzyme. If the selection pressure for one or both activities increases because higher or more constant metabolic fluxes are required, the fitness will probably improve upon a simple overexpression of the protein by mutation in the promoter region or by gene duplication. Interestingly, sequence alignments suggest that TtuC enzymes form a distinct cluster among Dml/TDHs and feature a different substrate-binding motif. The gene coding for TtuC of A. vitis (a grapevine pathogen) belongs to the tartrate operon (ttu), suggesting that TtuC-like enzymes could have arisen from the specialization of a tdh gene for tartrate utilization (56). In A. tumefaciens, the ttuC gene is encoded in a similar tartrate ttu operon and coexists with a tdh gene (see supplemental material), suggesting a gene duplication scenario. If TtuC enzymes evolved to efficiently catabolize tartrate, then this specialization required consequent mutations of otherwise conserved residues. Hence, a deeper study of substrate specificities among Dml/TDHs regarding the potential existence of structural features promoting enzyme promiscuity or specialization and the trade-offs between the different activities in the course of evolution would be very informative.
Are these results relevant in the context of natural evolution? The experimental design applied in this study is artificial, as a metabolic limitation was created by the knock-out of a metabolically important gene. However, it mimics a new requirement for neofunctionalization that could arise naturally due to environmental changes. It is yet unclear if other proteins have secondary activities close to a physiologically relevant level. In particular, it is important to emphasize that all of the systematic screenings for enzyme generalists were realized under overexpression conditions using multicopy vectors (9, 10). However, some of the enzymes selected in these studies may be sufficiently active on the alternative substrates to afford a phenotype without overexpression. Previous studies (11, 15) showed that a single mutation in a chromosomal gene might increase a promiscuous activity at a level sufficient to complement a metabolic knock-out. Hence, the development of new techniques for directed evolution of chromosome-encoded proteins is a key challenge for gaining further insight into the mechanisms governing natural evolution of enzymes.
Supplementary Material
Acknowledgments
We acknowledge Prof. Jean-François Collet (Université catholique de Louvain) for providing the strain from the Keio Collection. We also thank Dr. Florian Hollfelder (University of Cambridge, Cambridge, United Kingdom) for comments on the manuscript.
This work was supported by a research assistant fellowship from the Belgian Fonds National de la Recherche Scientifique (FNRS) and by Belgian “Interuniversity Attraction Poles” Program Project P7/44 iPROS.

This article contains supplemental material.
- IPMDH
- isopropylmalate dehydrogenase
- IDH
- isocitrate dehydrogenase
- TDH
- tartrate dehydrogenase
- HDH
- homoisocitrate dehydrogenase
- Pp
- P. putida
- Ec
- E. coli.
REFERENCES
- 1. Ohno S. (1970) Evolution by Gene Duplication, Springer-Verlag, New York [Google Scholar]
- 2. Voordeckers K., Brown C. A., Vanneste K., van der Zande E., Voet A., Maere S., Verstrepen K. J. (December 11, 2012) Reconstitution of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PloS Biol. 10, e1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang R., Hippauf F., Rohrbeck D., Haustein M., Wenke K., Feike J., Sorrelle N., Piechulla B., Barkman T. J. (2012) Enzyme functional evolution through improved catalysis of ancestrally nonpreferred substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 2966–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jensen R. A. (1976) Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30, 409–425 [DOI] [PubMed] [Google Scholar]
- 5. Copley S. D. (2012) Toward a systems biology perspective on enzyme evolution. J. Biol. Chem. 287, 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glasner M. E., Gerlt J. A., Babbitt P. C. (2006) Evolution of enzyme superfamilies. Curr. Opin. Chem. Biol. 10, 492–497 [DOI] [PubMed] [Google Scholar]
- 7. Afriat L., Roodveldt C., Manco G., Tawfik D. S. (2006) The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 45, 13677–13686 [DOI] [PubMed] [Google Scholar]
- 8. van Loo B., Jonas S., Babtie A. C., Benjdia A., Berteau O., Hyvönen M., Hollfelder F. (2010) An efficient, multiply promiscuous hydrolase in the alkaline phosphatase superfamily. Proc. Natl. Acad. Sci. U.S.A. 107, 2740–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patrick W. M., Quandt E. M., Swartzlander D. B., Matsumura I. (2007) Multicopy suppression underpins metabolic evolvability. Mol. Biol. Evol. 24, 2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soo V. W., Hanson-Manful P., Patrick W. M. (2011) Artificial gene amplification reveals an abundance of promiscuous resistance determinants in E. coli. Proc. Natl. Acad. Sci. U.S.A. 108, 1484–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yip S. H., Matsumura I. (2013) Substrate ambiguous enzymes within the E. coli proteome offer different evolutionary solutions to the same problem. Mol. Biol. Evol. 30, 2001–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khersonsky O., Roodveldt C., Tawfik D. S. (2006) Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10, 498–508 [DOI] [PubMed] [Google Scholar]
- 13. Khersonsky O., Tawfik D. S. (2010) Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 [DOI] [PubMed] [Google Scholar]
- 14. Kaltenbach M., Tokuriki N. (2014) Dynamics and constraints of enzyme evolution. J. Exp. Zool. B Mol. Dev. Evol. 9999, 1–20 [DOI] [PubMed] [Google Scholar]
- 15. McLoughlin S. Y., Copley S. D. (2008) A compromise required by gene sharing enables survival: implications for evolution of new enzyme activities. Proc. Natl. Acad. Sci. U.S.A. 105, 13497–13502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller B. G., Raines R. T. (2004) Identifying latent enzymes activities: substrate ambiguity within modern bacteria sugar kinases. Biochemistry 43, 6387–6392 [DOI] [PubMed] [Google Scholar]
- 17. Miller B. G., Raines R. T. (2005) Reconstitution of a defunct glycolytic pathway via recruitment of ambiguous sugar kinases. Biochemistry 44, 10776–10783 [DOI] [PubMed] [Google Scholar]
- 18. Yang K., Metcalf W. W. (2004) A new activity for an older enzyme: Escherichia coli bacterial alkaline phosphatase is a phosphite-dependent dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 101, 7919–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nam H., Lewis N. E., Lerman J. A., Lee D. H., Chang R. L., Kim D., Palsson B. O. (2012) Network context and selection in the evolution to enzyme specificity. Science 337, 1101–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D'Ari R., Casadesús J. (1998) Underground metabolism. BioEssays 20, 181–186 [DOI] [PubMed] [Google Scholar]
- 21. Fedøy A. E., Yang N., Martinez A., Leiros H. K., Steen I. H. (2007) Structural and functional properties of isocitrate dehydrogenase from the psychrophilic bacterium Desulfotalea psychrophila reveal a cold-active enzyme with an unusual high thermal stability. J. Mol. Biol. 371, 130–149 [DOI] [PubMed] [Google Scholar]
- 22. Hurley J. H., Thorsness P. E., Ramalingam V., Helmers N. H., Koshland D. E., Jr., Stroud R. M. (1989) Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 86, 8635–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stokke R., Karlström M., Yang N., Leiros I., Ladenstein R., Birkeland N. K., Steen I. H. (2007) Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure analysis and engineering of chimers. Extremophiles 11, 481–493 [DOI] [PubMed] [Google Scholar]
- 24. Gráczer É., Merli A., Singh R. K., Karuppasamy M., Závodszky P., Weiss M. S., Vas M. (2011) Atomic level description of the domain closure in a dimeric enzyme: Thermus thermophilus 3-isopropylmalate dehydrogenase. Mol. Biosyst. 7, 1646–1659 [DOI] [PubMed] [Google Scholar]
- 25. Wallon G., Kryger G., Lovett S. T., Oshima T., Ringe D., Petsko G. A. (1997) Crystal structure of E. coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus. J. Mol. Biol. 266, 1016–1031 [DOI] [PubMed] [Google Scholar]
- 26. Miyazaki J., Asada K., Fushinobu S., Kuzuyama T., Nishiyama M. (2005) Crystal structure of the tetrameric homoisocitrate dehydrogenase from an extreme thermophile, Thermus thermophilus: involvement of hydrophobic dimer-dimer interaction in extremely high thermotolerance. J. Bacteriol. 187, 6779–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malik R., Viola R. E. (2010) Structural characterization of tartrate dehydrogenase: a versatile enzyme catalyzing multiple reactions. Acta Crystallogr. D Biol. Crystallogr. 66, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyazaki K., Kakinuma K., Terasawa H., Oshima T. (1993) Kinetic analysis on the substrate specificity of 3-isopropylmalate dehydrogenase. FEBS Lett. 332, 35–36 [DOI] [PubMed] [Google Scholar]
- 29. Dean A. M., Shiau A. K., Koshland D. E., Jr. (1996) Determinants of performance in the isocitrate dehydrogenase of Escherichia coli. Protein Sci. 5, 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyazaki K. (2005) Identification of a novel trifunctional homoisocitrate dehydrogenase and modulation of the broad substrate specificity through site-directed mutagenesis. Biochem. Biophys. Res. Commun. 336, 596–602 [DOI] [PubMed] [Google Scholar]
- 31. Miyazaki J., Kobashi N., Nishiyama M., Yamane H. (2003) Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic bacterium, Thermus thermophilus HB27, and evolutionary implication of β-decarboxylating dehydrogenase. J. Biol. Chem. 278, 1864–1871 [DOI] [PubMed] [Google Scholar]
- 32. Miyazaki K. (2005) Bifunctional isocitrate-homoisocitrate dehydrogenase: a missing link in the evolution of β-decarboxylating dehydrogenase. Biochem. Biophys. Res. Commun. 331, 341–346 [DOI] [PubMed] [Google Scholar]
- 33. Tipton P. A., Beecher B. S. (1994) Tartrate dehydrogenase, a new member of the family of metal-dependent decarboxylating R-hydroxyacid dehydrogenases. Arch. Biochem. Biophys. 313, 15–21 [DOI] [PubMed] [Google Scholar]
- 34. Tipton P. A., Peisach J. (1990) Characterization of the multiple catalytic activities of tartrate dehydrogenase. Biochemistry 29, 1749–1756 [DOI] [PubMed] [Google Scholar]
- 35. Stern J. R., Hegre C. S. (1966) Inducible d-malic enzyme in E. coli. Nature 212, 1611–1612 [DOI] [PubMed] [Google Scholar]
- 36. Reed J. L., Patel T. R., Chen K. H., Joyce A. R., Applebee M. K., Herring C. D., Bui O. T., Knight E. M., Fong S. S., Palsson B. O. (2006) Systems approach to refining genome annotation. Proc. Natl. Acad. Sci. U.S.A. 103, 17480–17484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wright B. E., Minnick M. F. (1997) Reversion rates in a leuB auxotroph of E. coli K-12 correlate with ppGpp levels during exponential growth. Microbiology 143, 847–854 [DOI] [PubMed] [Google Scholar]
- 38. Jin J., Gao P., Mao Y. (2002) Occurence of leu+ revertants under starvation cultures in E. coli is growth-dependent. BMC Genet. 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomason L. C., Costantino N., Court D. L. (2007) E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 79, 1.17.1–1.17.8 [DOI] [PubMed] [Google Scholar]
- 40. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (February 21, 2006) Construction of E. coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neidhardt F. C., Bloch P. L., Smith D. F. (1974) Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dean A. M., Lee M. H., Koshland D. E., Jr. (1989) Phosphorylation inactivates E. coli isocitrate dehydrogenase by preventing isocitrate binding. J. Biol. Chem. 264, 20482–20486 [PubMed] [Google Scholar]
- 43. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) Protein identification and analysis tools on the ExPASy server. in The Proteomics Protocols Handbook (Walker J. M., ed), pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 44. Niesen F. H., Berglund H., Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 45. Chen R., Jeong S. S. (2000) Functional prediction: identification of protein orthologs and paralogs. Protein Sci. 9, 2344–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Darriba D., Taboada G. L., Doallo R., Posada D. (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 48. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lukas H., Reimann J., Kim O. B., Grimpo J., Unden G. (2010) Regulation of aerobic and anaerobic d-malate metabolism of E. coli by the LysR-type regulator DmlR (YeaT). J. Bacteriol. 192, 2503–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin Y., West A. H., Cook P. F. (2008) Potassium is an activator of homoisocitrate dehydrogenase from Saccharomyces cerevisiae. Biochemistry 47, 10809–10815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Giffhorn F., Kuhn A. (1983) Purification and characterization of a bifunctional l-(+)-tartrate dehydrogenase-d-(+)-malate dehydrogenase (decarboxylating) from Rhodopseudomonas sphaeroides Y. J. Bacteriol. 155, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bar-Even A., Noor E., Savir Y., Liebermeister W., Davidi D., Tawfik D. S., Milo R. (2011) The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 [DOI] [PubMed] [Google Scholar]
- 53. Matulis D., Kranz J. K., Salemme F. R., Todd M. J. (2005) Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 44, 5258–5266 [DOI] [PubMed] [Google Scholar]
- 54. Senisterra G. A., Markin E., Yamazaki K., Hui R., Vedadi M., Awrey D. E. (2006) Screening for ligands using a generic and high-throughput light-scattering-based assay. J. Biomol. Screen. 11, 940–948 [DOI] [PubMed] [Google Scholar]
- 55. Knichel W., Radler F. (1982) d-Malic enzyme of Pseudomonas fluorescens. Eur. J. Biochem. 123, 547–552 [DOI] [PubMed] [Google Scholar]
- 56. Crouzet P., Otten L. (1995) Sequence and mutational analysis of a tartrate utilization operon from Agrobacterium vitis. J. Bacteriol. 177, 6518–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yaoi T., Miyazaki K., Oshima T. (1997) Substrate recognition of isocitrate dehydrogenase and 3-isopropylmalate dehydrogenase from Thermus thermophilus. J. Biochem. 121, 77–81 [DOI] [PubMed] [Google Scholar]
- 58. Zhu G., Golding G. B., Dean A. M. (2005) The selective cause of an ancient adaptation. Science 307, 1279–1282 [DOI] [PubMed] [Google Scholar]
- 59. Tipton P. A. (1993) Intermediate partitioning in the tartrate dehydrogenase-catalyzed oxidative decarboxylation of d-malate. Biochemistry 32, 2822–2827 [DOI] [PubMed] [Google Scholar]
- 60. Serfozo P., Tipton P. A. (1995) Substrate determinants of the course of tartrate dehydrogenase-catalyzed reactions. Biochemistry 34, 7517–7524 [DOI] [PubMed] [Google Scholar]
- 61. Tawfik D. S. (2010) Messy biology and the origins of evolutionary innovations. Nat. Chem. Biol. 6, 692–996 [DOI] [PubMed] [Google Scholar]
- 62. Wessler S. R., Calvo J. M. (1981) Control of leu operon expression in E. coli by transcription attenuation mechanism. J. Mol. Biol. 149, 579–597 [DOI] [PubMed] [Google Scholar]
- 63. Dean A. M., Dvorak L. (1995) The role of the glutamate 87 in the kinetic mechanism of Thermus thermophilus isopropylmalate dehydrogenase. Protein Sci. 4, 2156–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gonçalves S., Miller S. P., Carrondo M. A., Dean A. M., Matias P. M. (2012) Induced fit and the catalytic mechanism of isocitrate dehydrogenase. Biochemistry 51, 7098–7115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harding M. M. (2002) Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr. D Biol. Crystallogr. 58, 872–874 [DOI] [PubMed] [Google Scholar]
- 66. Stokke R., Madern D., Fedøy A. E., Karlsen S., Birkeland N. K., Steen I. H. (2007) Biochemical characterization of isocitrate dehydrogenase from Methylococcus capsulatus reveals a unique NAD+-dependent homotetrameric enzyme. Arch. Microbiol. 187, 361–370 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.