Background: EGFR lysosomal trafficking process is important for the modulation of EGF signaling.
Results: Delayed EGFR degradation in BLOS1 knockdown cells is restored by the overexpressed BLOS1 fragment that interacts with both SNX2 and TSG101.
Conclusion: BLOS1 mediates EGFR lysosomal trafficking.
Significance: Enhancement of EGFR lysosomal degradation will be effective in the treatment of cancer or lung fibrosis.
Keywords: Endosome, Epidermal Growth Factor Receptor (EGFR), Lysosome, Signaling, Trafficking
Abstract
Biogenesis of lysosome-related organelles complex-1 (BLOC-1) is a component of the molecular machinery required for the biogenesis of specialized organelles and lysosomal targeting of cargoes via the endosomal to lysosomal trafficking pathway. BLOS1, one subunit of BLOC-1, is implicated in lysosomal trafficking of membrane proteins. We found that the degradation and trafficking of epidermal growth factor receptor (EGFR) were delayed in BLOS1 knockdown cells, which were rescued through BLOS1 overexpression. A key feature to the delayed EGFR degradation is the accumulation of endolysosomes in BLOS1 knockdown cells or BLOS1 knock-out mouse embryonic fibroblasts. BLOS1 interacted with SNX2 (a retromer subunit) and TSG101 (an endosomal sorting complex required for transport subunit-I) to mediate EGFR lysosomal trafficking. These results suggest that coordination of the endolysosomal trafficking proteins is important for proper targeting of EGFR to lysosomes.
Introduction
BLOC-13 (biogenesis of lysosome-related organelles (LRO) complex-1) is a stable protein complex in eukaryotes that consists of at least eight subunits: pallidin, muted, cappuccino, dysbindin, snapin, BLOS1, BLOS2, and BLOS3 (1, 2). Recent studies have shown that KXD1 is an additional putative BLOC-1 subunit in mammals or yeast (3, 4). Two subcomplexes (pallidin-cappuccino-BLOS1 and dysbindin-snapin-BLOS2) exist in this BLOC-1 macro-complex (5). Mutations in several subunits of the BLOC-1 complex such as dysbindin, BLOS3, and pallidin (6–8) have been reported to cause a multiorganellar disorder in humans, Hermansky-Pudlak syndrome (HPS), which has features of oculocutaneous albinism, prolonged bleeding, and often death in middle age due to fibrotic lung disease and other complications. HPS features defects in melanosomes, platelet-dense granules, and several other LROs (2, 9).
BLOC-1 has been reported to function in endolysosomal transport. Specifically, BLOC-1 functions in endosomal maturation and cargo transport from early endosomes toward lysosomes or LROs (4, 10). BLOC-1 interacts physically and functionally with AP-3 to facilitate the trafficking of AP-3 cargoes such as CD63 and OCA2 for LRO biogenesis (11–13). Moreover, dysbindin or snapin mediates membrane receptor transport from endosomes to lysosomes for degradation. These proteins include dopamine receptor 2 (D2R), NMDA receptor subtype 2A (NR2A), and epidermal growth factor receptor (EGFR) (14–17). Deficiency in these BLOC-1 subunits leads to increased membrane reinsertion of these receptors to up-regulate their signaling. However, the underlying molecular mechanisms utilized by BLOC-1 in endolysosomal trafficking are still unknown. Moreover, BLOC-1 plays important roles in endosomal or lysosomal maturation by recruiting the Rab5 GTPase-activating protein Msb3 in yeast (4). Immature lysosomes accumulate, and the function of lysosomes in autophagy is impaired in snapin-deficient neurons (17). Likewise, autophagosomes accumulate in BLOS1 knock-out (KO) mouse embryonic fibroblasts (MEFs), suggesting that BLOC-1 functions in autophagy (18). It must be emphasized that the above-mentioned functions may be attributable to specific BLOC-1 subunits rather than the whole BLOC-1 complex. Phenotypic differences exist in pigmentation and hippocampal postsynaptic receptor expression and adult neurogenesis between dysbindin-1-deficient mice and muted deficient mice (19, 20).
The interacting partners of different subunits of BLOC-1 have expanded our knowledge of its biological functions (21). BLOS1, also known as GCN5L1, is a subunit of BLOC-1 with limited knowledge of its functions. BLOS1 homolog mutant flies display eye pigmentation defects due to abnormal pigment granules (22). In addition, BLOS1 participates in the vacuolar trafficking of PIN1/PIN2 to regulate root development in Arabidopsis (23). However, its roles in mammals remain largely unknown.
Lysosomal trafficking is very important for many cellular functions, such as modulation of signal transduction, biogenesis of LROs, and exocytosis (2). Several complexes have been found to mediate lysosomal trafficking, including BLOC-1, BLOC-2, BLOC-3, HOPS, AP-3, retromer, and ESCRTs (2, 24). However, how these complexes synergistically or sequentially coordinate the trafficking of a specific cargo (e.g. EGFR in particular) is unclear.
The ESCRT family includes four members, which are ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III (25, 26). ESCRTs mediate the sorting of ubiquitinylated EGFR for lysosomal degradation to regulate its signal transduction. TSG101, a subunit of ESCRT-I, interacts with HRS, a subunit of ESCRT-0, to mediate endosomal trafficking and down-regulation of EGFR (27). Another ESCRT-I subunit, UBAP1, is required for sorting EGFR to the multivesicular bodies (MVBs) and for endosomal ubiquitin homeostasis (28). Vps22/EAP30 in ESCRT-II mediates endosomal sorting of EGFR. EGFR accumulates on the limiting membranes of early endosomes and aberrantly small MVBs in Vps22-depleted cells (29). Moreover, the ESCRT-III subunit VPS24 is required for the degradation of EGFR (30).
The mammalian retromer complex contains a sorting nexin dimer composed of a combination of SNX1, SNX2, SNX5, and SNX6, and a cargo recognition trimer composed of Vps26, Vps29, and Vps35, which play very important roles in the retrograde trafficking from endosomes to the trans-Golgi network (31). It has been reported that retromer also functions in the lysosomal degradation of receptors. SNX1 is essential for lysosomal sorting of protease-activated receptor-1 (32). In addition, SNX2 plays a role in the lysosomal degradation of EGFR (33).
We previously reported that BLOS1 interacts with SNX1 to direct the membrane auxin efflux proteins PIN1/2 for vacuolar (or lysosomal) degradation, which could be mediated by ESCRTs in Arabidopsis (23). We hypothesize that this pathway may be conserved in EGFR lysosomal targeting in mammalian cells. Although the roles for BLOS1 in cargo degradation and for both SNX2 and TSG101 in EGFR degradation have been described, the role of these proteins in EGFR lysosomal trafficking remains a mystery. We report here that BLOS1 interacts with SNX2 and TSG101 to mediate the endolysosomal trafficking of EGFR for lysosomal degradation.
EXPERIMENTAL PROCEDURES
Antibodies
Mouse monoclonal anti-EEA1, anti-GM130, anti-SNX1, and anti-SNX2 antibodies were obtained from BD Transduction Laboratory (Lexington, KY). Polyclonal anti-Myc, monoclonal anti-His, anti-FLAG, anti-β-actin, and anti-LAMP1 antibodies were purchased from Sigma. Monoclonal anti-GST antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal anti-CD63 (or LAMP3) antibody was purchased from Millipore (Billerica, MA). Rabbit polyclonal anti-EGFR was obtained from Fitzgerald Industries International (Concord, MA). Mouse monoclonal anti-TSG101 antibody was obtained from Abcam (Cambridge, UK). Mouse monoclonal anti-p-Akt antibody was purchased from Cell Signaling Technology (Danvers, MA). Mouse monoclonal anti-GFP and polyclonal anti-calnexin antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX). Polyclonal snapin antibody was purchased from SYSY (Goettingen, Germany). Polyclonal anti-KXD1 (3), anti-BLOS1, and anti-BLOS2 antibodies were generated in New Zealand White rabbits against GST-tagged full-length mouse proteins.
siRNA Probes
The siRNAs against human BLOS1 were purchased from Genechem (Shanghai, China), and the sequences are as follows: 1) 5′-CAGGCCAGUGGAUCGGAAU-3′; 2) 5′-GUCUGCCCCUUCCUAGACU-3′; and 3) 5′-CCAGAGAAAGCUGGACCAU-3′.
The human TSG101 siRNA was purchased from Thermo Scientific (Hudson, NH). The sequences specific for targeting human TSG101 are 5′-CUCAAUGCCUUGAAACGAA-3′, 5′-GAACAAUCCCUGUGCCUUA-3′, 5′-CUGUCAAUGUUAUUACUCUAU-3′, and 5′-AGAGAUGGUUACCCGUUUA-3′.
The siRNAs of human SNX2 were described previously (33). A scrambled siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) was used as a negative control (GenePharma, Shanghai, China).
Mouse Colonies and Gene Targeting
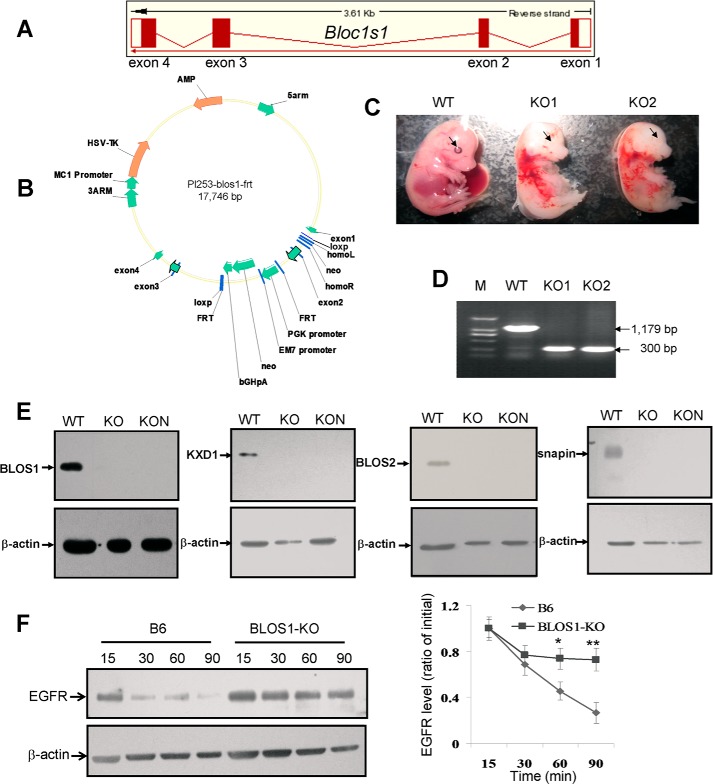
The Bloc1s1 knock-out mutant (BLOS1-KO) was generated by gene targeting in 129/J-derived ES cells according to a strategy described in Fig. 5, bred in a C57BL/6J (B6) background and crossed with EII-Cre (ubiquitous expression) or nestin-Cre (neuron specific expression) transgenic mice at the animal facility of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. To ensure the genotypes of the littermates, PCR amplifications were designed according to the targeted deletion of exon 2. All procedures were approved by the Institutional Animal Care and Use Committee of Institute of Genetics and Developmental Biology.
FIGURE 5.
Generation of Bloc1s1 knock-out mice. A, schematic diagram of exons and introns of mouse Bloc1s1 gene from the Ensemble genome database. B, schematic diagram of gene targeting vector of mouse Bloc1s1. Note that exon 2 is engineered to be deleted by inserting two flanking loxp sites in intron 1 and intron 2 and a neomycin (neo) cassette. Using this gene-targeting strategy, the obtained heterozygous Bloc1s1 knock-out mice (Bloc1s1+/−) were generated by crossing Bloc1s1loxp/+ mice with EII-Cre or nestin-Cre mice. Intercross of Bloc1s1+/− mice produced null mutant mice (Bloc1s1−/−) with exon 2 deletion. C, pictures of embryonic mice (E14.5). WT, wild-type; KO1 and KO2 are two different Bloc1s1 knock-out embryos of EII-Cre × blos1-loxp. Note that eye pigment is missing in KO mice compared with the WT mice as indicated by the arrows. D, genomic PCR verification of the deleted exon 2 of Bloc1s1. The WT allele gives rise to the PCR amplicon of 1,179 bp, whereas the mutational allele gives rise to a 300-bp amplicon. The mice were tested are shown in C by tail DNA. E, BLOS1 protein is null in both ubiquitous knock-out (KO and EII-Cre × blos1-loxp) and neuron-specific knock-out (KON and nestin-Cre × blos1-loxp) brain tissues compared with wild-type littermates (WT). Several subunits of BLOC-1 such as KXD1, BLOS2, and snapin are destabilized, supporting that BLOS1 is a subunit of BLOC-1. β-Actin is a loading control. F, degradation of EGFR is delayed in BLOS1-KO (ubiquitous) MEF cells compared with the wild-type B6 MEF cells. Quantitative data are shown on the right side.
Cell Culture and Transfection
Mouse embryonic fibroblasts were harvested at E12.5 and cultured in Dulbecco's modified Eagle's medium (DMEM, Thermo Scientific), 10% fetal bovine growth serum (FBS, Hyclone Laboratories, Logan, UT) with penicillin and streptomycin at 37 °C, 5% CO2 for a maximum of 10 passages. HeLa and HEK293T cells were cultured in DMEM supplemented with 10% FBS, including penicillin and streptomycin at 37 °C, 5% CO2.
Transfections were performed using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions. The transfection of siRNAs (final concentration, 20 nm) was performed, and the stable transfection of BLOS1 siRNA in HeLa cells was screened in DMEM supplemented with 10% FBS containing 10 μg/ml puromycin (InvivoGen, San Diego).
Immunoblotting
Cultured cells or tissue lysates were prepared in lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100 with protease inhibitor mixture (Sigma) at 4 °C for 1 h. The immunoblotting procedures were described previously (3).
Co-immunoprecipitation Assay
FLAG-tagged pCMV-2B-BLOS1 (mouse) and Myc-tagged pCMV-3B-SNX1 or SNX2 (mouse) were co-transfected into HEK293T cells cultured in DMEM, including 10% FBS (Hyclone Laboratories) for 72 h after transfection. The immunoprecipitation procedures were described previously (3).
Yeast Two-hybrid Assay
We used a protocol described in the user manual of the MATCH-MAKER 3 yeast two-hybrid system (Clontech). The constructs used in this study are listed in the figures, and the procedures were described previously (3).
GST Pulldown Assay
Full-length mouse BLOS1, N-BLOS1 (1–60 aa), and C-BLOS1 (50–125 aa) were subcloned into the pGEX-4T-1 with a GST tag. Full-length mouse SNX2 and mouse TSG101 and their truncations SNX2 (N + P) (1–268 aa), SNX2 (P + B) (136–520 aa), TSG101 (U + P) (1–215 aa), and TSG101(C + S) (215–390 aa) were subcloned into the His fusion vector pET28a. GST alone or GST fusion plasmids were transformed into Rosetta competent cells containing His fusion plasmids as indicated. The pulldown procedures were conducted as described (3).
Immunofluorescence Microscopy
For time-lapse imaging, HeLa cells were plated in 24-well cell culture plates. GFP-BLOS1 (mouse) was transfected into the cells. After 12–24 h, HeLa cells were starved in serum-free DMEM overnight. The starved cells were treated with 100 ng/ml Alexa Fluor 555-labeled EGF (Invitrogen) for 5 min at 37 °C. Then HeLa cells were internalized in serum-free DMEM at 37 °C at different time points. The HeLa cells grown on coverslips were fixed with 4% paraformaldehyde (Sigma) and permeabilized, and cells were punched by 0.4% Triton X-100 (Amresco, Cleveland, OH). Then cells were blocked with 1% BSA for 1 h and incubated with primary antibodies as indicated followed by Alexa Fluor 594- or 488-conjugated secondary antibodies (Invitrogen). The coverslips were fixed with fixing solution (PBS/glycerol, 3:1). Confocal images were obtained using a ×100 objective of C2 laser-scanning microscope (Nikon, Japan). The co-localization between EGF-Alexa Fluor 555 and the endosomal marker EEA1 was calculated for absolute intensities in 50 cells by using the Mander's overlap coefficients (34).
For subcellular localization imaging, GFP-BLOS1 (mouse) was transfected into the HeLa cells. After 24 h, the cells grown on coverslips were fixed, probed with different antibodies, and imaged by following the procedures as described above.
EGFR Degradation Assay
Control or experimental HeLa cells were cultured in serum-free DMEM overnight and then treated with 100 μg/ml cycloheximide (INALCO, Milan, Italy) and 100 ng/ml EGF (PeproTech, Rocky Hill, NJ) at different time points. The cells were washed twice with PBS at the end of each time point and then lysed in lysis buffer as described above. The lysates were subjected to SDS-PAGE and immunoblotting with corresponding antibody.
EGFR Rescue Assay
HeLa cells were cultured in DMEM containing 10% FBS. After 12–24 h, the transfection of corresponding siRNAs was performed when the cell density reached 80%. After 24 h, the siRNA-containing cells were transfected with corresponding full-length or truncated plasmids. After 24 h, cells were cultured in serum-free DMEM overnight. Then cells were stimulated with 100 ng/ml EGF together with 100 μg/ml cycloheximide for different times. Cells were collected at the end of each time point, and then lysed as described above. The lysates were subjected to SDS-PAGE and immunoblotting with corresponding antibodies.
Biotinylation and Internalization of Cell Surface EGFR
Control and stably transfected BLOS1 knockdown HeLa cells were starved in serum-free DMEM overnight. Two sets of experiments were kept unstimulated, and two sets were stimulated with 100 ng/ml EGF for 3 h. The internalization of EGF was terminated by washing with ice-cold PBS-Ca-Mg (PBS, pH 7.4, 0.1 mm CaCl2, 1 mm MgCl2). The cooling cells were incubated with biotin labeling solution including 0.5 mg/ml EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific) in PBS-Ca-Mg at 4 °C for 30 min. The reaction was stopped by washing the cells with ice-cold PBS-Ca-Mg that included 15 mm glycine. The cells were washed again with PBS and then lysed as described above. Biotinylated EGFR was purified by MagnaBindTM streptavidin beads (Thermo Scientific) according to the manufacturer's instructions. The precipitated biotin-EGFR was subjected to SDS-PAGE and immunoblotting as described above.
Electronic Microscopy
For immuno-EM, HeLa cells were grown in complete DMEM after starving for 12 h. The conjugates with anti-EGFR-gold20 were added to serum-free DMEM with 20 mm HEPES and 1% BSA (binding medium). Cells were incubated in this medium at 4 °C for 90 min to allow antibody binding. Typically, the gold conjugate was diluted 5–10 times. Then the cells were incubated at 37 °C for 5 min in fresh binding medium (without gold probes) in the presence of 100 ng/ml EGF to stimulate the internalization of the EGF receptor. Following this incubation, the cells were processed for EM examination.
Standard transmission electronic microscopy fixation for HeLa cells was applied by 0.1 m cacodylate buffer with 4% formaldehyde, 0.1% glutaraldehyde, 3 μm CaCl2, and 7.5% sucrose. Cells were fixed in a 1:1 mixture of fixative liquid and cell culture medium for 15 min. Fixed cells were washed with 0.1 m cacodylate buffer before being pelleted. The cell pellet was stained in reduced osmium tetraoxide (1:1 mixture of 2% OsO4 in H2O and 3% Fe3(CN)6 in 0.1 m cacodylate) for ∼60 min. The stained cell was consequently dehydrated stepwise using 50% ethanol, 75% ethanol, 95% ethanol, and absolute ethanol. After dehydration, the cell pellet was transferred into propylene oxide and then a 1:1 mixture of propylene oxide and resin for resin infiltration. Sections about 70 nm thick were cut. The staining was conducted using 1% uranyl acetate for 15 min.
Wild-type (B6), BLOS1-KO and pallid (pa) MEFs were grown in 10-cm dishes, fixed with 2.5% glutaraldehyde, 0.1 m phosphate buffer, pH 7.4, at 4 °C for 2 h. After buffer washes, cells were post-fixed for 90 min at 4 °C in 1% OsO4. After water washes, samples were block-stained in 1% uranyl acetate at room temperature for 30 min, dehydrated in graded acetone solutions, embedded in Spurr resin, and polymerized at 70 °C for 12 h. Thin sections were stained with uranyl acetate and lead citrate and then examined on a JEM 1400 electron microscope (JEOL, Japan) at 80 KV. Digital images were captured with CCD camera.
Statistics
Endocytic organelles and EGFR-gold particles were counted in different cells according to the EM ultrastructures. Quantification of fluorescence co-localization was calculated for absolute intensities by using the Mander's overlap coefficient K1. Subcellular co-localization was calculated by Pearson's correlation. Comparisons were statistically tested using the Student's t test. Results were expressed as mean ± S.E.
RESULTS
BLOS1 Interacts with SNX2 and TSG101
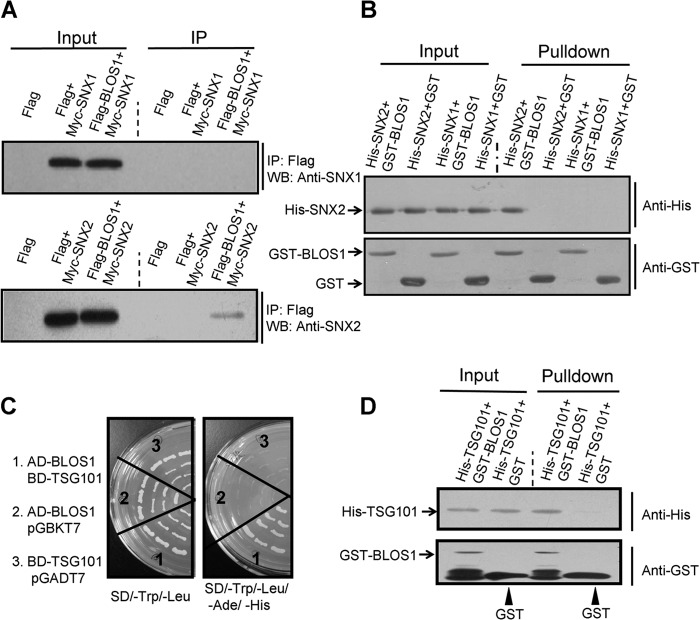
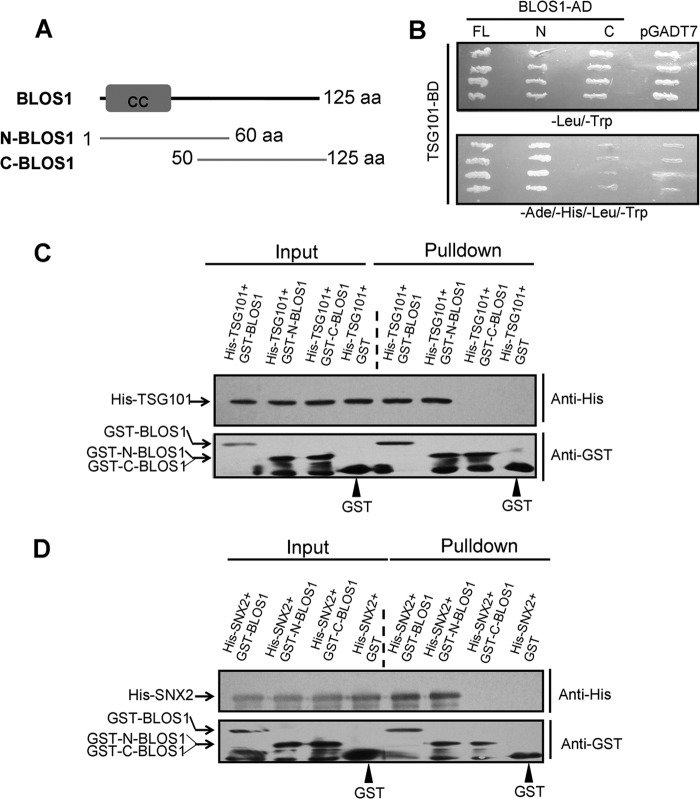
We previously showed that BLOS1 interacts with SNX1 in Arabidopsis (23). We therefore explored the interactions of their mouse counterparts. We did not find interaction between mouse BLOS1 and SNX1 by co-immunoprecipitation (co-IP) or GST pulldown assay (Fig. 1, A and B). However, we found interaction between BLOS1 and SNX2 by co-IP assay (Fig. 1A). We further confirmed the interaction between BLOS1 and SNX2 through a GST pulldown assay (Fig. 1B). This suggests that mouse SNX2, rather than SNX1, is homologous to SNX1 (At2g30330) in Arabidopsis. From our screen of a cDNA library by a yeast two-hybrid assay, we found AtELC (or At3g12400) as another interacting protein of BLOS1 in Arabidopsis, which is homologous to TSG101 in mammals. We found that BLOS1 likewise interacted with TSG101 by yeast two-hybrid and GST pulldown assays (Fig. 1, C and D).
FIGURE 1.
Interactions between BLOS1 and SNX2/TSG101. A, FLAG-tagged BLOS1 co-precipitates with Myc-SNX2 but not with Myc-SNX1. The FLAG empty vector serves as a negative control. IP, immunoprecipitation; WB, Western blotting. B, GST-BLOS1 pulls down His-SNX2 but not His-SNX1. GST alone serves as a negative control. C, BLOS1 interacts with TSG101 in the yeast two-hybrid system. Full-length TSG101 is used as a bait (with binding domain, pGBKT7) to prey BLOS1 (with activation domain, pGADT7). The left panel shows the growth of yeast on synthetic defined (SD) medium lacking leucine and tryptophan. The right panel shows the growth of yeast on SD medium lacking adenine, histidine, leucine, and tryptophan. D, GST-BLOS1 pulls down His-TSG101, although GST alone does not. The blots shown here are representative ones from three independent experiments.
EGFR Trafficking Is Delayed in BLOS1 siRNA Knockdown Cells
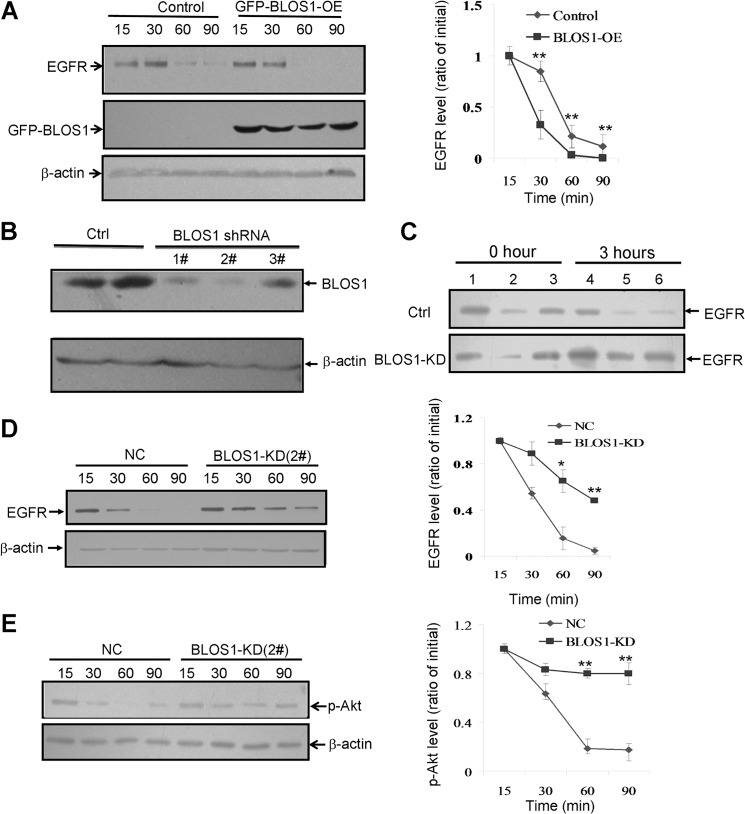
Both SNX2 and TSG101 function in EGFR trafficking (27, 33). We reasoned that the interactions between BLOS1 and SNX2/TSG101 could mediate the sorting of EGFR during its endolysosomal trafficking. We found that degradation of EGFR was accelerated in BLOS1 overexpressed cells (Fig. 2A). We then generated a stable BLOS1-knockdown (KD) HeLa cell line. The efficiency and specificity of siRNA were evaluated by Western blotting (Fig. 2B). shRNA 2# showed highest knockdown efficiency and shRNA 3# had little effect on BLOS1. The amount of EGFR on the cell surface was increased, and the degradation of EGFR was delayed in BLOS1-KD cells (Fig. 2, C and D). Consistently, the downstream target of EGF signaling, activated phospho-Akt (p-Akt), was enhanced in BLOS1-KD cells (Fig. 2E).
FIGURE 2.
Degradation of EGFR in BLOS1 overexpression and knockdown HeLa cells. Experimental conditions are described under “Experimental Procedures.” The blots shown here are representative ones from three independent experiments. *, p < 0.05; **, p < 0.01. A, overexpression of GFP-BLOS1 (GFP-BLOS1-OE) promotes the degradation of EGFR compared with normal control cells. Quantitative data are shown on the right side. B, efficiency and specificity of BLOS1 siRNA interference compared with the scrambled siRNA (Ctrl). Note that the 2# probe is used for the following assays. C, level of cell surface biotinylated EGFR is increased in BLOS1-KD cells after 3 h of treatment of EGF (100 ng/ml) compared with no treatment (Ctrl) at 0 h. Lanes 1 and 4, input supernatant at 0 and 3 h; lanes 2 and 5, supernatant after binding to beads at 0 and 3 h; lanes 3 and 6, cell surface biotinylated EGFR at 0 and 3 h. D, degradation of EGFR is delayed in BLOS1 knockdown (BLOS1-KD) HeLa cells compared with normal control cells (NC). Quantitative data are shown on the right side. E, downstream EGF signaling activator, phosphorylation of Akt (p-Akt), is enhanced in BLOS1-KD cells compared with normal control cells (NC). Quantitative data are shown on the right side.
BLOS1 Is Partially Localized to the Endosomal Compartments
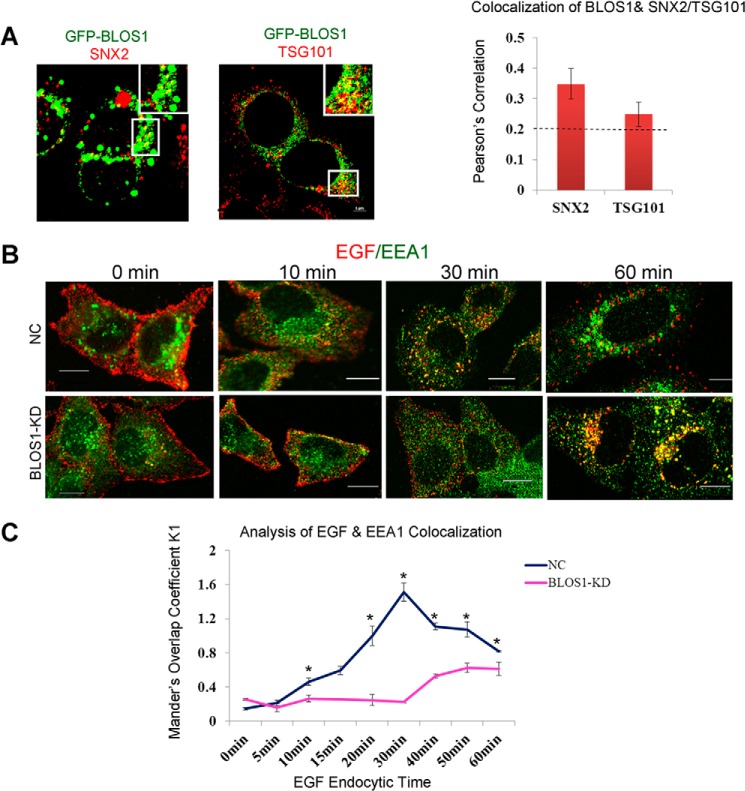
It has been reported that SNX2 is mainly localized at early endosomes, whereas TSG101 is mainly localized at MVBs (33, 35). To understand the cellular process mediated by BLOS1 interactions, we next tested for co-localization between mouse BLOS1 and SNX2 or TSG101 by immunofluorescence microscopy. We found that GFP-BLOS1 partially (>20%) co-localized with SNX2 and TSG101, respectively (Fig. 3A). These data indicate that BLOS1 is likely co-localized with SNX2 and TSG101 at endosomal compartments, which is consistent with findings in plant cells (23).
FIGURE 3.
Co-localization of BLOS1 with SNX2/TSG101 and endosomal targeting of EGF is delayed in stable BLOS1 knockdown HeLa cells. A, GFP-BLOS1 partially co-localizes with endogenous SNX2 and TSG101. The co-localization ratios were calculated by Pearson's correlation. B, representative images at different time points upon EGF (red) treatment in BLOS1-KD cells and normal control cells (NC). Endogenous EEA1 is labeled in green. Scale bar, 10 μm. C, co-localizations of EGF and EEA1 are represented by Mander's overlap coefficient K1. Data collected are from 50 cells and calculated as mean ± S.E. *, p < 0.05.
We then investigated subcellular co-localization with an early endosome marker EEA1 in the time course of EGF endocytic trafficking. We found that co-localization of EGF-Alexa Fluor 555 with EEA1 was delayed in BLOS1-KD cells compared with control cells (Fig. 3, B and C). Taken together, these results suggest that EGFR is delayed at endosomal compartments when BLOS1 is deficient.
Endolysosomes Are Increased in Stable BLOS1-KD cells or BLOS1-KO MEFs
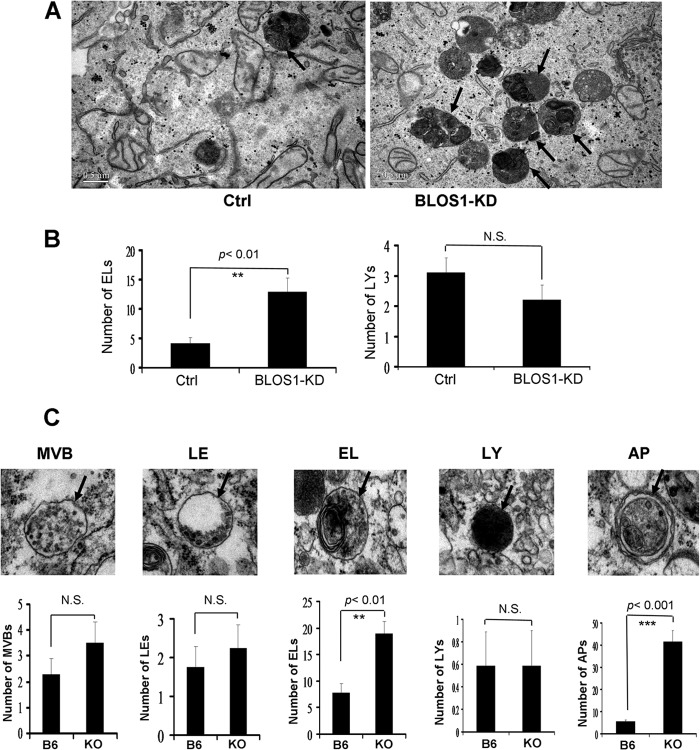
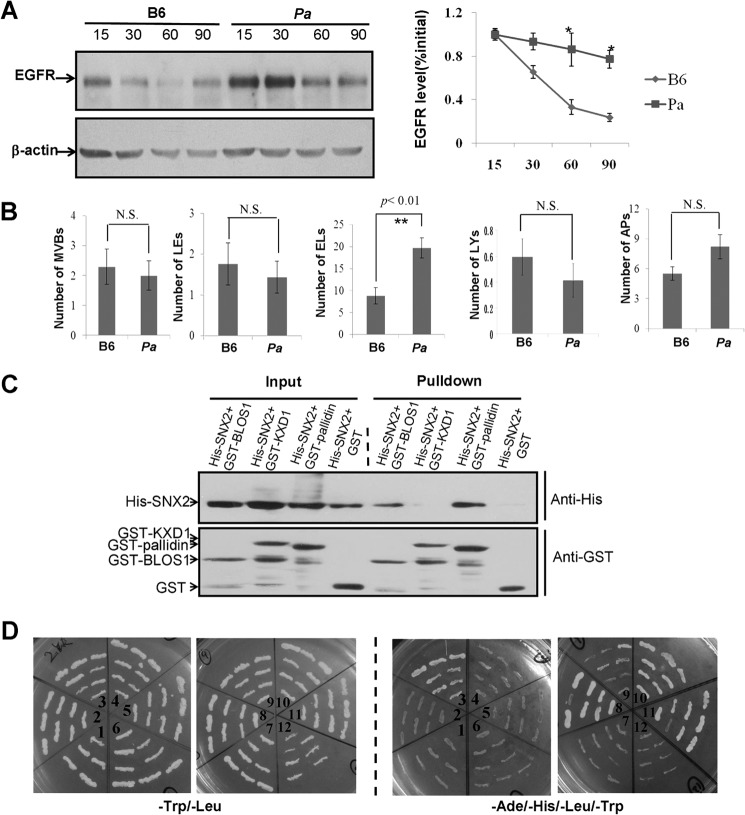
To investigate why EGFR trafficking to lysosomes is delayed, we first examined the ultrastructures of endocytic compartments by transmission electronic microscopy. We found drastic accumulation of endolysosomes in stable BLOS1-KD HeLa cells but no significant change in the number of lysosomes, although a decline tendency was noted (Fig. 4, A and B). The residual BLOS1 in BLOS1-KD cells could make some difference from the cells with a complete loss of BLOS1. Similarly, we found a significant increase of endolysosomes in BLOS1-KO MEFs (Fig. 4C). In addition, autophagosomes were significantly increased in BLOS1-KO MEFs (Fig. 4C), which is consistent with findings in the MEFs from another conventional BLOS1-KO mouse line (18).
FIGURE 4.
Ultrastructure of endocytic compartments in BLOS1 stable knockdown HeLa cells and BLOS1 knock-out MEFs. A, more endolysosomes or pre-lysosomes are shown in BLOS1-KD cells as shown by the arrows compared with control (Ctrl) HeLa cells. Scale bar, 0.5 μm. B, average number of endolysosomes (ELs) in BLOS1-KD cells is significantly higher than that in control cells (12.89 ± 2.41 in 9 KD cells versus 4.13 ± 1.06 in 8 control cells. **, p < 0.01). However, the average number of mature lysosomes (LYs) tends to lower but the change is not significant (N.S.) (2.2 ± 0.42 in 10 KD cells versus 3.1 ± 0.48 in 10 control cells. p > 0.05). C, upper panel, representative pictures of endomembrane compartments in BLOS1-KO MEFs are shown. MVB, multivesicular bodies; LE, late endosomes; EL, endolysosomes; LY, lysosomes; AP, autophagosomes. In the lower panel, average numbers of the endocytic compartments are compared between the wild-type (B6) and KO MEFs. The number of endolysosomes (18.88 ± 2.31 in 17 KO cells versus 7.79 ± 1.77 in 16 control cells. **, p < 0.01) and the number of autophagosomes (41.41 ± 5.13 in 17 KO cells versus 5.53 ± 0.68 in 17 control cells. ***, p < 0.001) are significantly increased in KO MEFs, although there are no significant (N.S.) changes in the numbers of MVBs, late endosomes, and lysosomes. The definition of these endomembrane structures is based on the criteria defined by others (18, 42, 43). Briefly, MVBs contain evenly distributed numerous ILVs. In contrast, late endosomes contain fused or enlarged ILVs in additional to a few regular ILVs. Endolysosomes contain partial electron dense areas with single-layer limiting membrane. In contrast, lysosomes contain an electron-dense lumen. Autophagosomes contain structures with a bi-layer limiting membrane.
In this study, we generated a BLOS1-KO mouse line by using the conditional Cre/loxp system (Fig. 5, A and B). Both the BLOS1EII-Cre/loxp-KO (ubiquitous) and the BLOS1nestin-Cre/loxp-KO (neuron-specific) mice were embryonic lethal starting from E12.5. In addition, eye pigment was not visible in the BLOS1EII-Cre/loxp-KO embryos (Fig. 5, C and D), and several other BLOC-1 subunits such as KXD1, BLOS2, and snapin were destabilized in brain tissues of both BLOS1EII-Cre/loxp-KO and the BLOS1nestin-Cre/loxp-KO mice (Fig. 5E). These features suggest that BLOS1 resides in the BLOC-1 complex and functions in the LRO (e.g. melanosome) biogenesis. We then examined whether EGFR degradation is impaired in the BLOS1-KO cells. Similarly, EGFR degradation was delayed in the BLOS1EII-Cre/loxp-KO MEFs as seen in the BLOS1-KD cells (Figs. 2D and 5F). Taken together, the increase of endolysosomes is an indication of increased premature lysosomes, which is similar to snapin deficiency (17), and therefore, it may result in impaired EGFR degradation.
EGFR Accumulates in Endolysosomes in Stable BLOS1-KD Cells
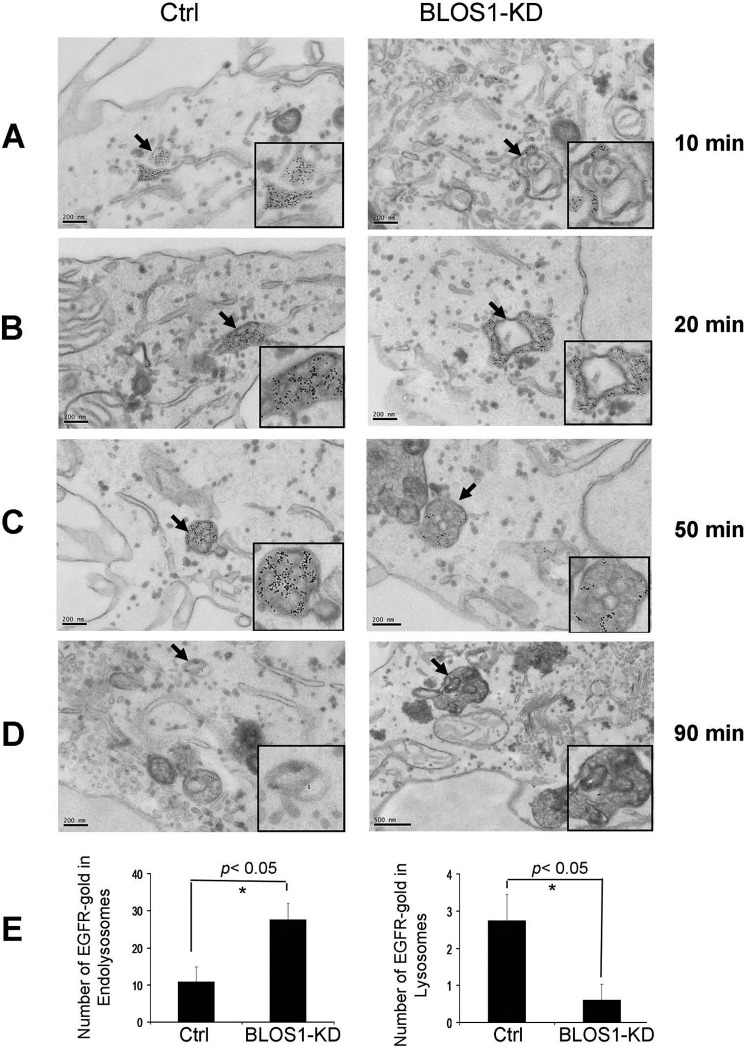
We next examined the distribution and accumulation of EGFR in endosomal and lysosomal compartments by immunoelectronic microscopy. Upon treatment of EGF, gold-labeled EGFR mainly accumulated in the tubular structures of early endosomes at 10 or 20 min (Fig. 6, A and B), with less distribution on the membrane of intralumenal vacuoles (ILVs) of MVBs at 50 min (Fig. 6C), and more accumulated in the endolysosomes at 90 min (Fig. 6D) in the stable BLOS1-KD cells. We confirmed the accumulation of EGFR-gold-labeled particles in endolysosomes and less distribution in lysosomes in the BLOS1-KD cells by counting the number of particles on these organelles (Fig. 6E). This suggests that the altered distribution of EGFR at endosomes and the accumulation of EGFR at endolysosomes may lead to delay of EGFR degradation.
FIGURE 6.
EGFR distribution upon EGF treatment in BLOS1 stable knockdown (KD) HeLa cells. A and B, cells were treated with 100 ng/ml EGF for 10 min (B) or 20 min (C). Gold-labeled EGFR particles are distributed throughout the early endosomes in the control cells but are localized to tubular structures of early endosomes in the BLOS1-KD cells. Indicated early endosomes structures by arrows are shown in the insets. Scale bar, 200 nm. C, cells were treated with 100 ng/ml EGF for 50 min. Gold-labeled EGFR particles are distributed mainly on membrane structures of ILVs within the MVBs in the control cells but were localized to both ILVs and limiting membrane of MVBs in the BLOS1-KD cells. Indicated MVBs by arrows are shown in the insets. Scale bar, 200 nm. D, cells were treated with 100 ng/ml EGF for 90 min. Gold-labeled EGFR particles are localized to lysosomes in control cells but are localized to endolysosomes in the BLOS1-KD cells. Indicated lysosomal structures are shown in the insets. Scale bar, 200 nm. E, EGFR-gold-labeled particles in endolysosomes and lysosomes as shown in D were counted. The average number of EGFR particles localized on endolysosomes is significantly increased in BLOS1-KD cells (27.6 ± 4.41 in 10 KD cells versus 10.88 ± 3.96 in 8 control (Ctrl) cells. *, p < 0.05), whereas the average number of EGFR particles localized on lysosomes is significantly lower in BLOS1-KD cells compared with the control cells (0.6 ± 0.43 in 10 KD cells versus 2.75 ± 0.70 in 8 control cells. *, p < 0.05). The definitions of endocytic compartments are described in the figure legends of Fig. 4.
N-terminal BLOS1, Which Interacts with Both TSG101 and SNX2, Is Required for Degradation of EGFR
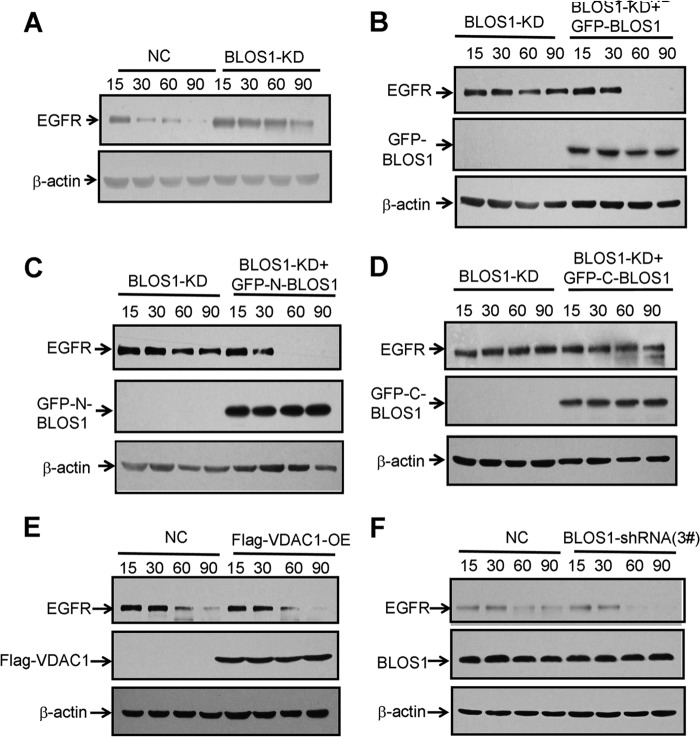
To understand the underlying molecular mechanism for regulating EGFR trafficking, we next investigated whether the interactions between BLOS1 and SNX2/TSG101 is required for the degradation of EGFR. We generated two overlapping truncations of BLOS1, N-BLOS1 (1–60 aa), and C-BLOS1 (50–125 aa) (Fig. 7A). N-BLOS1 was necessary for the interaction with both TSG101 and SNX2 through yeast two-hybrid and GST pulldown assays (Fig. 7, B–D). Full-length BLOS1 (denoted as BLOS1), N-BLOS1, and C-BLOS1 were overexpressed in BLOS1-KD cells. We found that the delayed degradation of EGFR was rescued in BLOS1-KD cells by overexpressed full-length BLOS1 or N-BLOS1 but not by C-BLOS1 (Fig. 8, A–D). In contrast, overexpression of an irrelevant protein, a mitochondrial membrane protein VDAC1, and the transfection of an inefficient shRNA 3#, as shown in Fig. 2B, did not show any effects on the EGFR degradation compared with the controls (Fig. 8, E and F). These results suggest that N-BLOS1, which contains the interacting domains for both SNX2 and TSG101, is required for the degradation of EGFR.
FIGURE 7.
Interacting domain between BLOS1 and TSG101/SNX2. A, schematic structure of BLOS1 and its N- and C-terminal truncations. B, both full-length BLOS1 and N-BLOS1 interact with TSG101 in yeast two-hybrid assays. Full-length TSG101 was used as a bait (with binding domain) to test different preys (with activation domain). pGADT7 was used as a negative prey control. The upper panel shows the growth of yeast on SD medium lacking leucine and tryptophan. The lower panel shows the growth of yeast on SD medium lacking adenine, histidine, leucine, and tryptophan. C, both GST-BLOS1 and GST-N-BLOS1 pull down His-TSG101, but GST-C-BLOS1 and GST alone do not pull down His-TSG101. D, GST-BLOS1 and GST-N-BLOS1 pull down His-SNX2 but GST-C-BLOS1 and GST alone do not pull down His-SNX2. The blots shown here are representative ones from three independent experiments.
FIGURE 8.
N-terminal BLOS1, which interacts with both SNX2 and TSG101, is required for the degradation of EGFR. A, degradation of EGFR is delayed in BLOS1 knockdown (BLOS1-KD) HeLa cells compared with normal control cells (NC). B, full-length GFP-BLOS1 rescues the delayed degradation of EGFR in BLOS1-KD cells. C, GFP-N-BLOS1 rescues the delayed degradation of EGFR in BLOS1-KD cells. D, GFP-C-BLOS1 does not rescue the delayed degradation of EGFR in BLOS1-KD cells. E, degradation of EGFR is unaltered in HeLa cells overexpressed with an irrelevant mitochondrial protein VDAC1 (VDAC1-OE) compared with normal control cells. F, degradation of EGFR is unaltered in HeLa cells transfected with an inefficient shRNA (3#) as shown in Fig. 2B compared with normal control cells. β-Actin is a loading control. The blots shown here are representative ones from three independent experiments.
C-terminal SNX2, Which Interacts with BLOS1, Is Required for the Degradation of EGFR
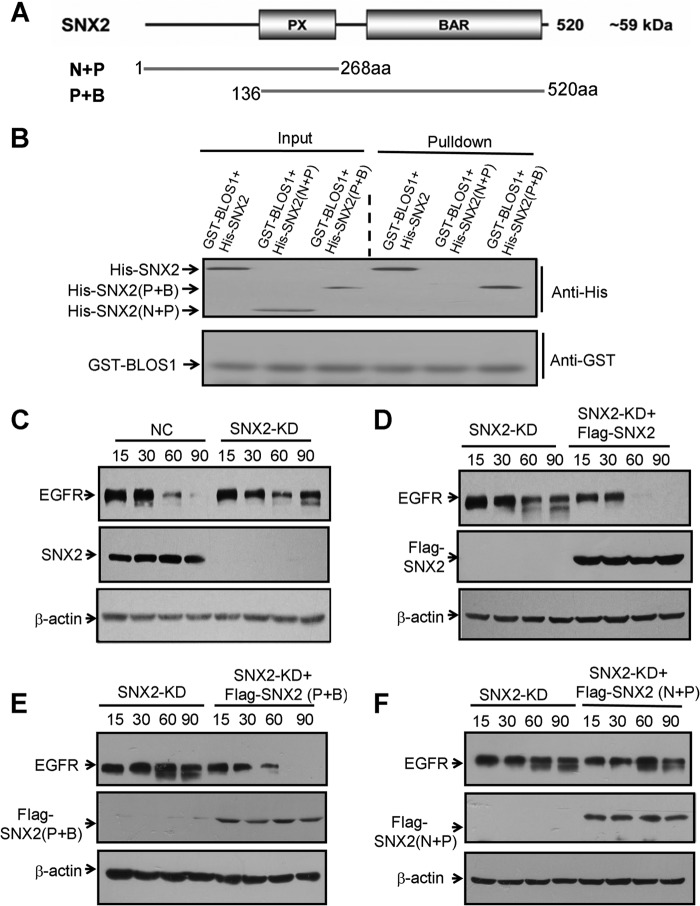
Similarly, we mapped the interacting domain in SNX2 for its interaction with BLOS1. We constructed two overlapping truncations of SNX2, N + P fragment with PX domain (1–268 aa) and P + B fragment with both PX and BAR domains (136–520 aa) (Fig. 9A). The P + B truncation mediated the interaction with BLOS1 by GST pulldown assays (Fig. 9B). Full-length SNX2 (denoted as SNX2), P + B, and N + P truncations were overexpressed in SNX2-KD cells. We found that degradation of EGFR was delayed in SNX2-KD cells, in agreement with a previous report (Fig. 9C) (33). Furthermore, the delayed degradation of EGFR was rescued in SNX2-KD cells by overexpressing full-length SNX2 and P + B truncation but not by N + P truncation (Fig. 9, D–F). These results suggest that the C-terminal SNX2 with the BAR domain interacts with BLOS1, which is required for the degradation of EGFR.
FIGURE 9.
C-terminal SNX2 (P + B), which interacts with BLOS1, is required for the degradation of EGFR. A, schematic structure of SNX2 and its N- and C-terminal truncations, N + P, and P + B. PX domain (P), binding PI3P. BAR domain (B), curvature and dimerization. B, GST-BLOS1 pulls down both full-length and C-terminal SNX2 (P + B) but not the N-terminal SNX2 (N + P). C, degradation of EGFR is delayed in SNX2 knockdown cells compared with normal control (NC) HeLa cells. D, full-length FLAG-SNX2 rescues the delayed degradation of EGFR in SNX2-KD cells. E, FLAG-SNX2(P + B) rescues the delayed degradation of EGFR in SNX2-KD cells. F, FLAG-SNX2(N + P) does not rescue the delayed degradation of EGFR in SNX2-KD cells. β-Actin in C–F is a loading control. Blots shown here are representative ones from three independent experiments.
C-terminal TSG101, Which Interacts with BLOS1, Is Required for Degradation of EGFR
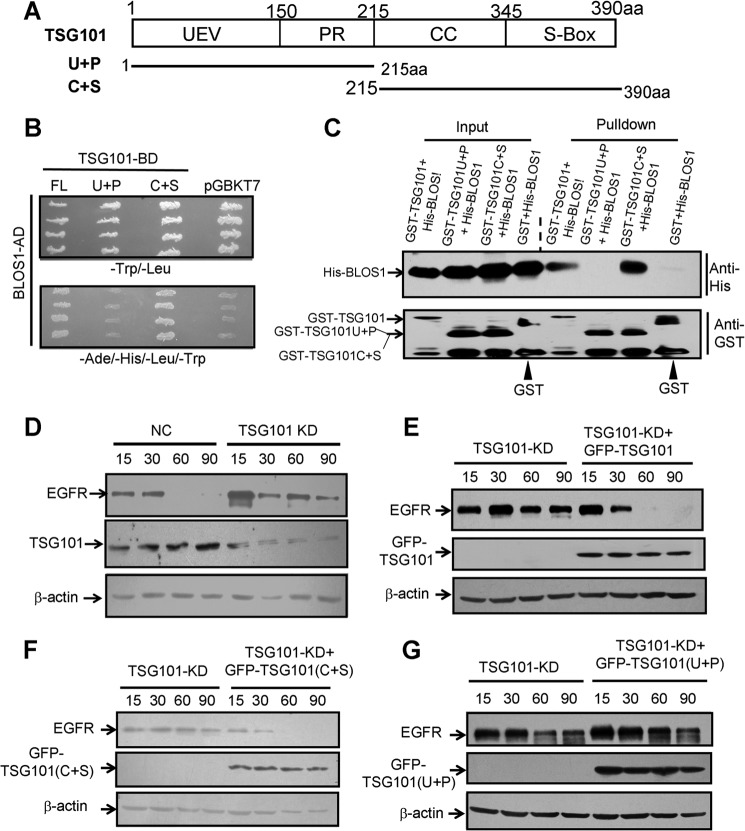
To map the interacting domain of TSG101 with BLOS1, we constructed two truncations of TSG101, U + P fragment with the UEV and PR domains (1–215 aa) and C fragment with the coiled-coil and S-box domains (215–390 aa) (Fig. 10A). The C + S truncation mediated the interaction with BLOS1 through yeast two-hybrid and GST pulldown assays (Fig. 10, B and C). Full-length TSG101 (denoted as TSG101), U + P, and C + S truncations were overexpressed in TSG101-KD cells. We found that degradation of EGFR was delayed in TSG101-KD cells, in agreement with a previous report (Fig. 10D) (27). Likewise, the delayed degradation of EGFR was rescued in TSG101-KD cells by overexpressing full-length TSG101 and the C + S truncation, but not by the U + P truncation (Fig. 10, E–G). These results suggest that the C-terminal TSG101, with the coiled-coil and S-box domains, interacts with BLOS1, which is required for the degradation of EGFR.
FIGURE 10.
C-terminal TSG101 (C + S), which interacts with BLOS1, is required for the degradation of EGFR. A, schematic structure of TSG101 and its N- and C-terminal truncations, U + P, and C + S. UEV, ubiquitin E2 variant (U). PR, proline-rich domain (P). CC, coiled-coil (C). S-Box, steadiness box (S). B, full-length TSG101 and C-terminal TSG101(C + S) interact with BLOS1 in yeast two-hybrid assays. Full-length BLOS1 is used as a prey (with activation domain) to test different baits (with binding domain). pGBKT7 is used as a negative bait control. The upper panel shows the growth of yeast on SD medium lacking leucine and tryptophan. The lower panel shows the growth of yeast on SD medium lacking adenine, histidine, leucine, and tryptophan. C, GST-TSG101 and GST-TSG101(C + S) pull down His-BLOS1, but the N-terminal GST-TSG101 (U + P) does not. D, degradation of EGFR is delayed in TSG101 knockdown cells compared with the normal control (NC) HeLa cells. E, full-length GFP-TSG101 rescues the delayed degradation of EGFR in TSG101-KD cells. F, GFP-TSG101(C + S) rescues the delayed degradation of EGFR in TSG101-KD cells. G, GFP-TSG101(U + P) does not rescue the delayed degradation of EGFR in TSG101-KD cells. β-Actin in D–G is a loading control. The blots shown here are representative ones from three independent experiments.
EGFR Degradation in Other BLOC-1 Subunit-deficient Cells
To address whether the delayed EGFR degradation in BLOS1-deficient cells is a representation of BLOC-1 function, we tested another BLOC-1 subunit, pallidin. Our results showed that EGFR degradation was significantly slowed in pallidin-null (pa) MEFs (Fig. 11A). In addition, we examined the ultrastructure of endocytic organelles of the MEFs from this pa mutant (Fig. 11B). Interestingly, both BLOS1-KO and pa MEFs that exhibit impaired EGFR trafficking showed significantly increased endolysosomes, but autophagosomes significantly accumulated only in BLOS1-KO cells, which may distinguish it from viable BLOC-1 mutants.
FIGURE 11.
Other BLOC-1 subunits in EGFR trafficking and in the interactions with SNX2/TSG101. A, degradation of EGFR is delayed in pa MEF cells compared with the wild-type B6 MEF cells. Quantitative data are shown on the right side. B, numbers of endolysosomes (19.67 ± 2.31 in 20 pa cells versus 8.82 ± 1.89 in 20 B6 cells. **, p < 0.01) are significantly increased in KO MEFs, although there are no significant (N.S.) changes in the numbers of MVBs, late endosomes, lysosomes, and autophagosomes. The definition of endocytic compartments is described in Fig. 4 legend. C, GST-BLOS1 and GST-pallidin pull down His-SNX2, whereas GST-KXD1 and GST alone do not pull down His-SNX2. D, TSG101 as a bait with binding domain (BD) interacts with muted (3), snapin (8), and BLOS1 (11) as preys with activation domain (AD) in yeast two-hybrid assays. The interaction between BLOS1 and KXD1 (10) is a positive control. pGBKT7 (in 1 and 12) is a negative bait control. The left two panels show the growth of yeast on SD medium lacking leucine and tryptophan. The right two panels show the growth of yeast on SD medium lacking adenine, histidine, leucine, and tryptophan. 1, AD-dysbindin/BD-pGBKT7; 2, AD-dysbindin/BD-TSG101; 3, AD-muted/BD-TSG101; 4, AD-BLOS2/BD-TSG101; 5, AD-BLOS3/BD-TSG101; 6, AD-KXD1/BD-TSG101; 7, AD-cappuccino/BD-TSG101; 8, AD-snapin/BD-TSG101; 9, AD-pallidin/BD-TSG101; 10, AD-KXD1/BD-BLOS1; 11, AD-BLOS1/BD-TSG101; 12, AD-TSG101/BD-pGBKT7.
We tested whether other BLOC-1 subunits interact with SNX2 or TSG101, which could be involved in the endocytic trafficking of EGFR. Indeed, our GST pulldown and yeast two-hybrid assays showed that SNX2 interacted with pallidin (Fig. 11C), and TSG101 interacted with both muted and snapin (Fig. 11D). Taken together, our results suggest that BLOC-1 is involved in the endocytic trafficking of EGFR that is mediated by SNX2 and TSG101.
DISCUSSION
EGFR lysosomal degradation plays a pivotal role in the modulation of EGF signaling. EGF stimulation of the EGFR promotes receptor dimerization, autophosphorylation, and protein-tyrosine kinase activity, triggering signal cascades involved in multiple cellular processes, including cell proliferation, migration, differentiation, and survival (36). Progression of EGFR through the endocytic pathway is critical for its activation and is regulated by sorting constituents. Components from diverse protein trafficking complexes such as retromer, BLOC-1, and ESCRTs have been involved in the sorting of EGFR in the endocytic pathway (17, 27, 33). Our results suggest that BLOC-1 relays two well known sorting complexes, retromer and ESCRT-1, in the endocytic trafficking of EGFR. The discovery of this missing link between the early endosomal sorting complex retromer and the MVB sorting complex ESCRT-I is important for understanding of cargo transport between these two morphologically distinct compartments.
BLOC-1 consists of nine subunits (pallidin, muted, cappuccino, dysbindin, snapin, BLOS1, BLOS2, BLOS3, and KXD1), which function in endolysosomal trafficking (2). Snapin deficiency leads to accumulation of pre-lysosomes in cultured cortical neurons (17). Similarly, we found the accumulation of endolysosomes (or pre-lysosomes) in BLOS1- or pallidin-deficient cells. This may lead to the common impairment of lysosomal functions and the delay of EGFR degradation. However, autophagosomes accumulate in both snapin-KO (17) and BLOS1-KO cells but not in pallidin-deficient cells. The snapin-KO and BLOS1-KO mice are embryonic or neonatal-lethal (Fig. 5) (37), which is distinct from other viable BLOC-1-deficient mice such as sandy (dysbindin-deficient), muted (muted-deficient), pallid (pallidin-deficient), cappuccino (cappuccino-deficient), reduced pigmentation (BLOS3-deficient) (38), and KXD1-KO mice (3). In fact, the impaired autophagy in BLOS1-KO mice could contribute to developmental defects or a lethal phenotype, which is documented in the snapin-KO mice (39). The variable phenotypes of different BLOC-1 subunit deficiencies suggest that different subunits may play different roles in the complex (intra-complex roles) or in other processes through their interacting proteins (extra-complex roles) (19–21).
In our immunoelectronic microscopy pictures, EGFR is mislocalized to sub-regions of endosomal compartments. The localization of EGFR to tubular structures in the BLOS1-KD cells may facilitate its recycling to the plasma membrane (Fig. 2C). This is in agreement with the enhanced reinsertion of dopamine receptor 2 (15) and glutamate receptor NR2A (16) in dysbindin-deficient mice. Likewise, plasma membrane EGFR is increased in snapin-deficient mice (17). EGFR could be sorted into the ILVs of the MVBs in the BLOS1-KD cells (Fig. 6C), which suggests that the structure and sorting function of MVBs are apparently normal. Our results reveal that BLOC-1 plays dual roles in both lysosomal maturation and endocytic trafficking of EGFR.
Mutation of several HPS genes causes lung fibrosis in some types of HPS (9). Increased expression of EGFR has been found in different types of lung fibrosis (40). It is unknown whether the lack of some HPS proteins would cause the impaired lysosomal degradation of EGFR in these patients, which may contribute to the development of lung fibrosis in their middle age. However, down-regulation of EGFR signaling is a therapeutic target to ameliorate lung fibrosis (41). Therefore, intervention of EGFR lysosomal trafficking to facilitate its degradation could be an alternative target for treating HPS patients with lung fibrosis as well as patients with cancers.
Acknowledgments
We are in debt to Dr. Richard Swank who provided the HPS mutants and proofread this manuscript. We thank Dr. Xiang Gao for support in generating the BLOS1 knock-out mice and providing the nestin-Cre transgenic mice. We thank Dr. Xiao Yang for providing the EII-Cre transgenic mice. We thank Dr. Qing Yang for the preparation of the anti-KXD1 antibody and Wenwen Zhou for preparation of the anti-BLOS2 antibody.
This work was supported in part by National Natural Science Foundation of China Grants 31230046, 31471333, and 91332116 and by Chinese Academy of Sciences Grant KJZD-EW-L08.
- BLOC
- lysosome-related organelle complex
- BLOS1
- BLOC-1 subunit 1
- EGFR
- epidermal growth factor receptor
- ESCRT
- endosomal sorting complex required for transport
- HPS
- Hermansky-Pudlak syndrome
- LRO
- lysosome-related organelle
- MEF
- mouse embryonic fibroblast
- aa
- amino acid
- KD
- knockdown
- pa
- pallidin-null mutant
- ILV
- intralumenal vacuole
- MVB
- multivesicular body
- AD
- activation domain
- BD
- binding domain
- SD
- synthetic defined.
REFERENCES
- 1. Starcevic M., Dell'Angelica E. C. (2004) Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279, 28393–28401 [DOI] [PubMed] [Google Scholar]
- 2. Wei A. H., Li W. (2013) Hermansky-Pudlak syndrome: pigmentary and non-pigmentary defects and their pathogenesis. Pigment Cell Melanoma Res. 26, 176–192 [DOI] [PubMed] [Google Scholar]
- 3. Yang Q., He X., Yang L., Zhou Z., Cullinane A. R., Wei A., Zhang Z., Hao Z., Zhang A., He M., Feng Y., Gao X., Gahl W. A., Huizing M., Li W. (2012) The BLOS1-interacting protein KXD1 is involved in the biogenesis of lysosome-related organelles. Traffic 13, 1160–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. John Peter A. T., Lachmann J., Rana M., Bunge M., Cabrera M., Ungermann C. (2013) The BLOC-1 complex promotes endosomal maturation by recruiting the Rab5 GTPase-activating protein Msb3. J. Cell Biol. 201, 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee H. H., Nemecek D., Schindler C., Smith W. J., Ghirlando R., Steven A. C., Bonifacino J. S., Hurley J. H. (2012) Assembly and architecture of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 287, 5882–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li W., Zhang Q., Oiso N., Novak E. K., Gautam R., O'Brien E. P., Tinsley C. L., Blake D. J., Spritz R. A., Copeland N. G., Jenkins N. A., Amato D., Roe B. A., Starcevic M., Dell'Angelica E. C., Elliott R. W., Mishra V., Kingsmore S. F., Paylor R. E., Swank R. T. (2003) Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 35, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cullinane A. R., Curry J. A., Carmona-Rivera C., Summers C. G., Ciccone C., Cardillo N. D., Dorward H., Hess R. A., White J. G., Adams D., Huizing M., Gahl W. A. (2011) A BLOC-1 Mutation screen reveals that PLDN is mutated in Hermansky-Pudlak syndrome type 9. Am. J. Hum. Genet. 88, 778–787 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Morgan N. V., Pasha S., Johnson C. A., Ainsworth J. R., Eady R. A., Dawood B., McKeown C., Trembath R. C., Wilde J., Watson S. P., Maher E. R. (2006) A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8). Am. J. Hum. Genet. 78, 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huizing M., Helip-Wooley A., Westbroek W., Gunay-Aygun M., Gahl W. A. (2008) Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu. Rev. Genomics Hum. Genet. 9, 359–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Setty S. R., Tenza D., Truschel S. T., Chou E., Sviderskaya E. V., Theos A. C., Lamoreux M. L., Di Pietro S. M., Starcevic M., Bennett D. C., Dell'Angelica E. C., Raposo G., Marks M. S. (2007) BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol. Biol. Cell 18, 768–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sitaram A., Dennis M. K., Chaudhuri R., De Jesus-Rojas W., Tenza D., Setty S. R., Wood C. S., Sviderskaya E. V., Bennett D. C., Raposo G., Bonifacino J. S., Marks M. S. (2012) Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol. Biol. Cell 23, 3178–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salazar G., Craige B., Styers M. L., Newell-Litwa K. A., Doucette M. M., Wainer B. H., Falcon-Perez J. M., Dell'Angelica E. C., Peden A. A., Werner E., Faundez V. (2006) BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol. Biol. Cell 17, 4014–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Pietro S. M., Falcón-Pérez J. M., Tenza D., Setty S. R., Marks M. S., Raposo G., Dell'Angelica E. C. (2006) BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell 17, 4027–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marley A., von Zastrow M. (2010) Dysbindin promotes the post-endocytic sorting of G protein-coupled receptors to lysosomes. PLoS One 5, e9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ji Y., Yang F., Papaleo F., Wang H. X., Gao W. J., Weinberger D. R., Lu B. (2009) Role of dysbindin in dopamine receptor trafficking and cortical GABA function. Proc. Natl. Acad. Sci. U.S.A. 106, 19593–19598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang T. T., Yang F., Chen B. S., Lu Y., Ji Y., Roche K. W., Lu B. (2009) Dysbindin regulates hippocampal LTP by controlling NMDA receptor surface expression. Proc. Natl. Acad. Sci. U.S.A. 106, 21395–21400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cai Q., Lu L., Tian J. H., Zhu Y. B., Qiao H., Sheng Z. H. (2010) Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron 68, 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scott I., Webster B. R., Chan C. K., Okonkwo J. U., Han K., Sack M. N. (2014) GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J. Biol. Chem. 289, 2864–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H., Yuan Y., Zhang Z., Yan H., Feng Y., Li W. (August 25, 2014) Dysbindin-1C is required for the survival of hilar mossy cells and the maturation of adult newborn neurons in dentate gyrus. J. Biol. Chem. 10.1074/jbc.M114.590927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Larimore J., Zlatic S. A., Gokhale A., Tornieri K., Singleton K. S., Mullin A. P., Tang J., Talbot K., Faundez V. (2014) Mutations in the BLOC-1 subunits dysbindin and muted generate divergent and dosage-dependent phenotypes. J. Biol. Chem. 289, 14291–14300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W., Feng Y., Hao C., Guo X., Cui Y., He M., He X. (2007) The BLOC interactomes form a network in endosomal transport. J. Genet. Genomics 34, 669–682 [DOI] [PubMed] [Google Scholar]
- 22. Cheli V. T., Daniels R. W., Godoy R., Hoyle D. J., Kandachar V., Starcevic M., Martinez-Agosto J. A., Poole S., DiAntonio A., Lloyd V. K., Chang H. C., Krantz D. E., Dell'Angelica E. C. (2010) Genetic modifiers of abnormal organelle biogenesis in a Drosophila model of BLOC-1 deficiency. Hum. Mol. Genet. 19, 861–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cui Y., Li X., Chen Q., He X., Yang Q., Zhang A., Yu X., Chen H., Liu N., Xie Q., Yang W., Zuo J., Palme K., Li W. (2010) BLOS1, a putative BLOC-1 subunit, interacts with SNX1 and modulates root growth in Arabidopsis. J. Cell Sci. 123, 3727–3733 [DOI] [PubMed] [Google Scholar]
- 24. Luzio J. P., Parkinson M. D., Gray S. R., Bright N. A. (2009) The delivery of endocytosed cargo to lysosomes. Biochem. Soc. Trans. 37, 1019–1021 [DOI] [PubMed] [Google Scholar]
- 25. Schmidt O., Teis D. (2012) The ESCRT machinery. Curr. Biol. 22, R116–R120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raiborg C., Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 27. Lu Q., Hope L. W., Brasch M., Reinhard C., Cohen S. N. (2003) TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. U.S.A. 100, 7626–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stefani F., Zhang L., Taylor S., Donovan J., Rollinson S., Doyotte A., Brownhill K., Bennion J., Pickering-Brown S., Woodman P. (2011) UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr. Biol. 21, 1245–1250 [DOI] [PubMed] [Google Scholar]
- 29. Malerød L., Stuffers S., Brech A., Stenmark H. (2007) Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic 8, 1617–1629 [DOI] [PubMed] [Google Scholar]
- 30. Bache K. G., Stuffers S., Malerød L., Slagsvold T., Raiborg C., Lechardeur D., Wälchli S., Lukacs G. L., Brech A., Stenmark H. (2006) The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell 17, 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bonifacino J. S., Hurley J. H. (2008) Retromer. Curr. Opin. Cell Biol. 20, 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gullapalli A., Wolfe B. L., Griffin C. T., Magnuson T., Trejo J. (2006) An essential role for SNX1 in lysosomal sorting of protease-activated receptor-1: evidence for retromer-, Hrs-, and Tsg101-independent functions of sorting nexins. Mol. Biol. Cell 17, 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gullapalli A., Garrett T. A., Paing M. M., Griffin C. T., Yang Y., Trejo J. (2004) A role for sorting nexin 2 in epidermal growth factor receptor down-regulation: evidence for distinct functions of sorting nexin 1 and 2 in protein trafficking. Mol. Biol. Cell 15, 2143–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McDonald J. H., Dunn K. W. (2013) Statistical tests for measures of co-localization in biological microscopy. J. Microsc. 252, 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bishop N., Horman A., Woodman P. (2002) Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 157, 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lemmon M. A., Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tian J. H., Wu Z. X., Unzicker M., Lu L., Cai Q., Li C., Schirra C., Matti U., Stevens D., Deng C., Rettig J., Sheng Z. H. (2005) The role of Snapin in neurosecretion: snapin knock-out mice exhibit impaired calcium-dependent exocytosis of large dense-core vesicles in chromaffin cells. J. Neurosci. 25, 10546–10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Swank R. T., Novak E. K., McGarry M. P., Rusiniak M. E., Feng L. (1998) Mouse models of Hermansky Pudlak syndrome: a review. Pigment Cell Res. 11, 60–80 [DOI] [PubMed] [Google Scholar]
- 39. Zhou B., Zhu Y. B., Lin L., Cai Q., Sheng Z. H. (2011) Snapin deficiency is associated with developmental defects of the central nervous system. Biosci. Rep. 31, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tzouvelekis A., Ntolios P., Karameris A., Vilaras G., Boglou P., Koulelidis A., Archontogeorgis K., Kaltsas K., Zacharis G., Sarikloglou E., Steiropoulos P., Mikroulis D., Koutsopoulos A., Froudarakis M., Bouros D. (2013) Increased expression of epidermal growth factor receptor (EGF-R) in patients with different forms of lung fibrosis. Biomed. Res. Int. 654354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Madala S. K., Schmidt S., Davidson C., Ikegami M., Wert S., Hardie W. D. (2012) MEK-ERK pathway modulation ameliorates pulmonary fibrosis associated with epidermal growth factor receptor activation. Am. J. Respir. Cell Mol. Biol. 46, 380–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peng J., Zhang R., Cui Y., Liu H., Zhao X., Huang L., Hu M., Yuan X., Ma B., Ma X., Takashi U., Masaaki K., Liang X., Yu L. (2014) Atg5 regulates late endosome and lysosome biogenesis. Sci. China Life Sci. 57, 59–68 [DOI] [PubMed] [Google Scholar]
- 43. Huotari J., Helenius A. (2011) Endosome maturation. EMBO J. 30, 3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]