Background: Brain inhibitory synaptic connections present challenges for molecular identification and imaging.
Results: Transgenic mice expressing tagged neuroligin-2 were used to identify associated complexes and image inhibitory synapses in multiple brain regions.
Conclusion: Neuroligin-2-associated complexes are enriched in synaptic components, and neuroligin-2 undergoes regulated dynamin-dependent endocytosis and retromer association.
Significance: New data and approaches are presented for brain inhibitory synapse proteomics and imaging.
Keywords: Adhesion, Dynamin, Endocytosis, Neuron, Synapse, Trafficking, Transgenic, GABAergic, Neurexin, Retromer
Abstract
Synapses, the basic units of communication in the brain, require complex molecular machinery for neurotransmitter release and reception. Whereas numerous components of excitatory postsynaptic sites have been identified, relatively few proteins are known that function at inhibitory postsynaptic sites. One such component is neuroligin-2 (NL2), an inhibitory synapse-specific cell surface protein that functions in cell adhesion and synaptic organization via binding to neurexins. In this study, we used a transgenic tandem affinity purification and mass spectrometry strategy to isolate and characterize NL2-associated complexes. Complexes purified from brains of transgenic His6-FLAG-YFP-NL2 mice showed enrichment in the Gene Ontology terms cell-cell signaling and synaptic transmission relative to complexes purified from wild type mice as a negative control. In addition to expected components including GABA receptor subunits and gephyrin, several novel proteins were isolated in association with NL2. Based on the presence of multiple components involved in trafficking and endocytosis, we showed that NL2 undergoes dynamin-dependent endocytosis in response to soluble ligand and colocalizes with VPS35 retromer in endosomes. Inhibitory synapses in brain also present a particular challenge for imaging. Whereas excitatory synapses on spines can be imaged with a fluorescent cell fill, inhibitory synapses require a molecular tag. We find the His6-FLAG-YFP-NL2 to be a suitable tag, with the unamplified YFP signal localizing appropriately to inhibitory synapses in multiple brain regions including cortex, hippocampus, thalamus, and basal ganglia. Altogether, we characterize NL2-associated complexes, demonstrate regulated trafficking of NL2, and provide tools for further proteomic and imaging studies of inhibitory synapses.
Introduction
Synapses are fundamental sites of communication in the brain, where neurotransmitter is released from one nerve cell to activate receptors on the target cell. Synapses are complex structural and molecular specializations containing regulated vesicle release and recycling machinery in the axon opposed to receptors and signal transduction apparatus in the dendrite. Brain function is intricately controlled by a dynamic balance of plastic excitatory and inhibitory synapses.
The molecular composition of excitatory glutamatergic postsynaptic specializations has been extensively studied and over 1000 protein constituents identified (1–6). Approaches include the purification of postsynaptic density fractions, affinity purification of glutamate receptors and associated proteins, and yeast two-hybrid screens beginning with glutamate receptor C-terminal regions. The application of similar approaches to study inhibitory synapses yielded several important regulators of GABA receptor trafficking (7, 8). However, likely due to the thin postsynaptic density of inhibitory symmetric synapses, a paucity of high affinity ligands and antibodies specific for GABA receptors, and the challenges posed by the membrane topology of inhibitory receptors for two-hybrid screens, these approaches have met with more limited success for identifying molecular components of inhibitory postsynaptic specializations.
In addition to GABA receptors, the majority of GABAergic inhibitory postsynaptic sites contain the scaffold protein gephyrin (9) and the cell surface synapse organizing protein neuroligin-2 (NL2)2 (10). NL2 directly binds to gephyrin and to the associated guanine nucleotide exchange factor collybistin (11). Presentation of NL2 to GABAergic axons is sufficient to trigger presynaptic differentiation via binding to axonal neurexin, and presentation of neurexin to dendrites is sufficient to recruit gephyrin and GABA receptors via NL2 (12, 13). Furthermore, NL2 localizes to inhibitory synapses even in the absence of gephyrin (14) or GABA receptors (15). The importance of NL2 for inhibitory synapse development is evidenced by a reduction in GABAergic and glycinergic, but not glutamatergic, miniature inhibitory postsynaptic current frequency and amplitude in genetically targeted neurons lacking NL2 (11). At the behavioral level, loss of NL2 enhanced anxiety-like behavior in dark/light and open field tests and reduced ultrasonic vocalization (16, 17). Mechanistically, loss of NL2 reduced perisomatic clustering of gephyrin and GABA receptors (11) with no change in symmetric synapse density (16). NL2 can also link through an S-SCAM scaffold to IgSF9b, which mediates adhesion at a subset of inhibitory synapses (18), and the interaction of NL2 with neurexin is the target of MDGA1 suppressor of inhibitory synapse development (19). Thus, NL2 is a central component of inhibitory synapses.
Here we used a mouse transgenic epitope-tagging tandem affinity purification and mass spectrometry (TAP-MS) approach to isolate and characterize complexes associated with NL2. TAP-MS involves expression of the protein of interest with multiple epitope tags that are then used sequentially to purify associated complexes and identify components by MS (20). First developed to characterize protein interactomes in yeast (21), TAP-MS is most frequently applied to mammalian cells in culture but has also been applied to transgenic and knock-in mice (22, 23). To characterize protein associations in a physiological context, we generated a transgenic mouse expressing a low level of TAP-tagged NL2 in neurons and performed TAP-MS from brain tissue, comparing with wild type mice as a negative control. Among multiple expected and novel NL2-associated proteins, we isolated many components that regulate trafficking and particularly endocytosis, and verify that NL2 can undergo regulated endocytosis. The YFP epitope tag on the NL2 transgene can also be used to image inhibitory synapses in many brain regions, without amplification in fixed tissue and potentially in live animals.
EXPERIMENTAL PROCEDURES
Thy1-HFY-NL2 Transgenic Mice
All experimental procedures with animals were conducted following the guidelines of the Canadian Council for Animal Care and the University of British Columbia Animal Care Committee. Generation of Thy1-HFY-NL2 mice was described previously (24). His6-FLAG-YFP-NL2 (HFY-NL2) contains the signal sequence of mouse neuroligin-1, hexahistidine (HHHHHH), FLAG (DYKDDDDK), and EYFP tags separated by short linkers followed by the mature coding sequence of mouse neuroligin-2. The transgenic line for analysis was chosen by the pattern and moderate level of expression of HFY-NL2 in the brain by confocal imaging. Mice were maintained on a C57BL/6 background and genotyped by PCR.
Neuron Culture and Immunocytochemistry
Hippocampal neuron culture was performed essentially as described previously (25). In brief, hippocampal neurons from embryonic day 18 rats were cultured at low density on poly-l-lysine-coated glass coverslips inverted over a feeder layer of astrocytes in neurobasal medium (Invitrogen) supplemented with B27 (Invitrogen or StemCell). Neurons were transfected using ProFection Mammalian Transfection System (Promega) at 9 days in vitro (DIV), and immunostained at DIV 12. For the localization of HFY-NL2, neurons were fixed with parafix solution (4% paraformaldehyde and 4% sucrose in PBS, pH 7.4) for 15 min followed by permeabilization with 0.2% Triton X-100 in PBS for 5 min. Neurons were then immunostained using anti-gephyrin (1:500, mAb7a; Synaptic Systems), anti-PSD-95 family (recognizes PSD-95, PSD-93, SAP102 and SAP97; 1:1000, 6G6–1C9; Thermo Fisher Scientific), anti-vesicular glutamate transporter (1:1,000, 135·304; Synaptic Systems), anti-vesicular GABA transporter (1:1,000, 131·004; Synaptic Systems), and anti-microtubule-associated protein 2 (1:4,000, IgY, ab5392; Abcam) primary antibodies, followed by Alexa dye-conjugated secondary antibodies toward the appropriate species (1:500, Alexa 488, 568, and 647; Invitrogen) and aminomethylcoumarin acetate-conjugated donkey anti-chicken IgY (1:200, 703-155-155; Jackson ImmunoResearch Laboratories, Inc.).
The HA-NL2 vector was a gift from P. Scheiffele (26). For the endocytosis analysis of HA-NL2, the initial surface HA-NL2 was labeled with FITC-conjugated rat anti-HA Fab fragment (1 μg/ml, 11988506001; Roche Applied Science). Soluble neurexin 1β-alkaline phosphatase (AP) or AP as a negative control were prepared as described (27) and added at 10 nm in conditioned medium at 37 °C for 60 min. To block the activity of dynamin, dynasore (80 μm; Abcam) or DMSO as a negative control was added in the conditioned medium. Neurons were then fixed with parafix, and the pre-labeled NL2 still present on the cell surface was labeled with Alexa 568-conjugated anti-rat secondary antibody (1:500; Invitrogen). The ratio of the intensity of green to red fluorescence was used to assess endocytosis of HA-NL2. For colocalization analysis with VPS35, following cell surface labeling with FITC-conjugated rat anti-HA Fab for HA-NL2 and incubation with neurexin 1β-AP or AP as described above, neurons were incubated an additional hour and then fixed in parafix, permeabilized with 0.2% Triton X-100, and co-labeled with anti-VPS35 (H00055737-M02, Abnova) and appropriate secondary antibody. Images for Fig. 6 were acquired on an Axioplan2 microscope (Carl Zeiss) with an oil immersion objective (40× 1.3 numerical aperture or 63× 1.4 numerical aperture) and cooled CCD camera (Sensys; Photometrics) using MetaMorph software (Molecular Devices) and customized filter sets. Images for Fig. 7 were obtained on a Zeiss LSM 700 confocal microscope with a 40× 1.4 numerical aperture oil objective and sequential scanning with individual lasers and optimized filters. For quantification, cells were stained simultaneously and imaged with identical settings. All imaging and analysis was done blind.
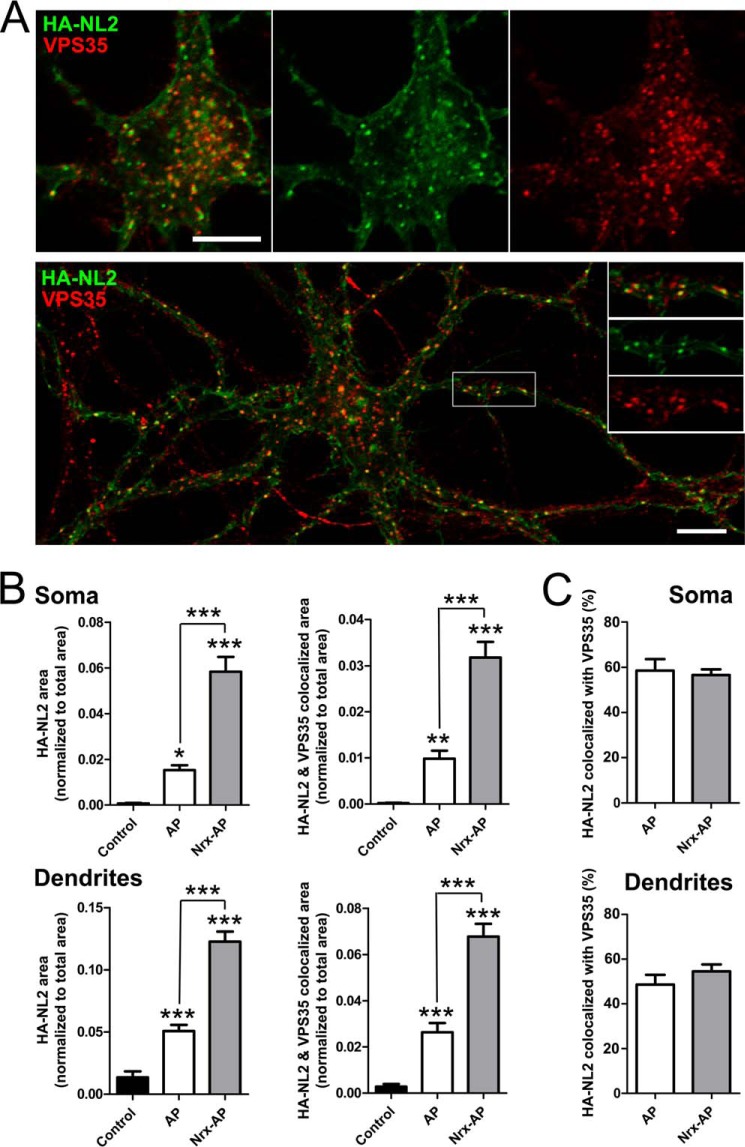
FIGURE 6.
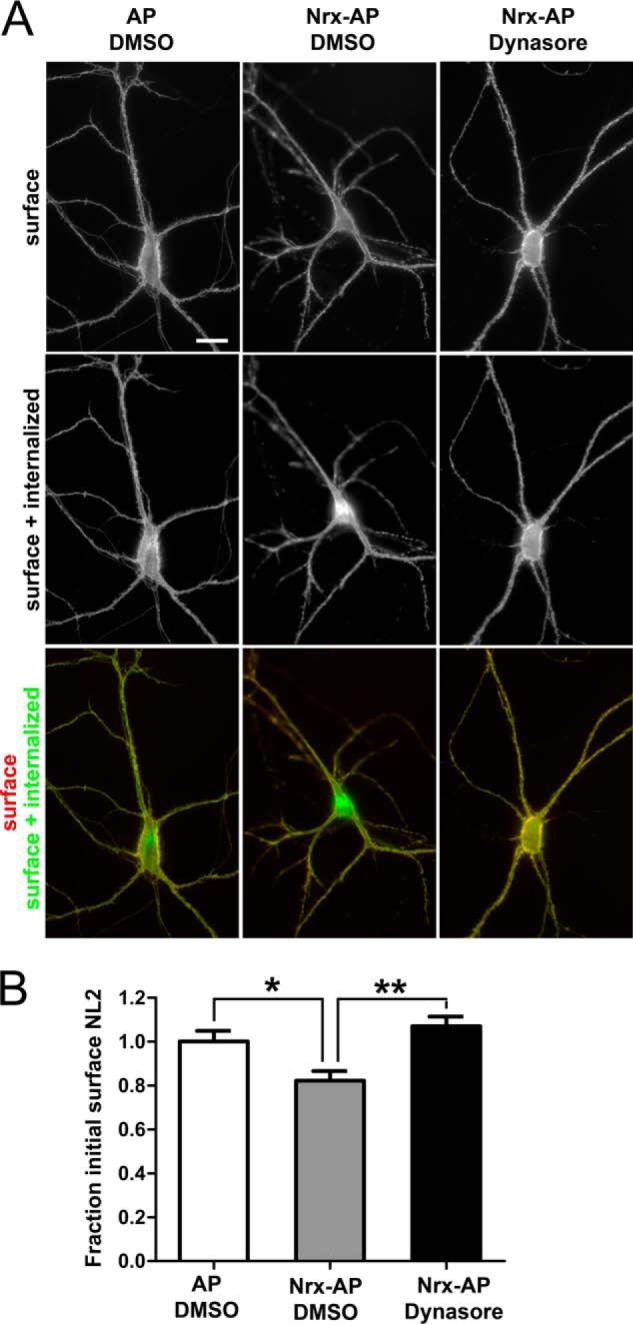
NL2 can undergo dynamin-dependent regulated endocytosis in neurons. Cultured hippocampal neurons were transfected to express HA-NL2 at 9 DIV and assayed at 12 DIV. Live neurons were incubated with FITC-conjugated rat anti-HA Fab fragment for 10 min, then for 1 h with soluble NL2 ligand neurexin Nrx-AP or AP control in the presence of the dynamin inhibitor dynasore or vehicle DMSO control. Cells were fixed without permeabilization and incubated with Alexa 568-conjugated anti-rat antibody. Thus, green FITC indicates surface plus internalized HA-NL2, whereas red Alexa 568 indicates surface HA-NL2 remaining. This HA-NL2 was expressed at a higher level than the HFY-NL2 in Fig. 1, hence there is a higher level at extrasynaptic sites along dendrites, but this high expression was needed for sufficient sensitivity for the internalization assay. A, Nrxn1β-AP relative to AP control induced an obvious increase in HA-NL2 internalization. Scale bar, 20 μm. B, the fraction of initial surface HA-NL2 remaining, measured as the ratio of Alexa 568 to FITC signal normalized to the AP + DMSO control condition, differed significantly among groups (analysis of variance, p = 0.0006, *, p < 0.05; **, p < 0.001 by Tukey's multiple comparison test, n = 34–37 from two independent experiments).
FIGURE 7.
Endocytosed NL2 colocalizes with VPS35 retromer. Cultured hippocampal neurons were transfected to express HA-NL2 at 9 DIV and assayed at 12 DIV. Live neurons were incubated with FITC-conjugated rat anti-HA Fab fragment for 10 min and either fixed immediately (control) or subsequently incubated for 1 h with soluble NL2 ligand neurexin Nrx-AP or AP control then an additional hour in conditioned media, and fixed. Thus, green FITC indicates surface plus internalized HA-NL2. Fixed cells were permeabilized and immunolabeled for VPS35 (red). A, confocal images focusing on the soma (top) and dendrites (bottom) of labeled neurons treated with Nrx-AP for 1 h show colocalization of endocytosed punctate HA-NL2 with VPS35. B and C, for quantification, central regions of the soma and large dendrites were outlined interior to the plasma membrane. Fractional area occupied by punctate pre-labeled HA-NL2 and HA-NL2 colocalized with VPS35 was measured (B, analysis of variance, p < 0.0001; *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Tukey's multiple comparison test, comparisons are with control group except where indicated otherwise, n = 30 from two independent experiments). The percentage of HA-NL2 punctate area colocalized with VPS35 did not differ between Nrx-AP and AP groups (C). Scale bars, 10 μm.
Cell Surface Binding Assays
COS7 cells were cultured in DMEM supplemented with 10% fetal bovine serum, and transfected with HFY-NL2 or YFP-CD4 (74) using TransIT-LT1 transfection reagent (Mirus). For the binding assay, transfected live cells were washed with extracellular solution (ECS: 168 mm NaCl, 2.6 mm KCl, 10 mm Hepes, pH 7.2, 2 mm CaCl2, 2 mm MgCl2, and 10 mm d-glucose) with 100 μg/ml of BSA (ECS/BSA), and then incubated with 20 nm purified neurexin 1β-Myc-AP or plain Myc-AP protein for 1 h at 4 °C. These cells were then fixed with parafix solution and immunostained with anti-Myc (1:1000, ab32; Abcam) and anti-GFP (1:1,000, A11122; Invitrogen) to visualize bound AP fusion proteins and surface YFP, respectively.
Brain Immunofluorescence and Imaging
For YFP imaging, mice between 2 and 4 months old were transcardially perfused with 2% paraformaldehyde in PBS using a low-flow peristaltic pump. Brains collected were post-fixed with 2% paraformaldehyde in PBS for 1 h, cryoprotected in 20%, then 30% (w/v) sucrose in PBS at 4 °C, and rapidly frozen. Cryostat sections (20 μm) were incubated in blocking solution (5% normal goat serum, 5% BSA, and 0.25% Triton X-100, 0.1 m PBS) for 1 h at room temperature then incubated overnight at 4 °C with anti-gephyrin (1:1000; mouse IgG1; mAb7a, Synaptic Systems) and in some cases anti-Synapsin1 (1:1000; rabbit; AB1543P, EMD Millipore) primary antibodies followed by appropriate Alexa-conjugated secondary antibodies (Invitrogen). For GAD65 labeling (1:100; mouse IgG2a, GAD-6-c, Developmental Studies Hybridoma Bank), perfused brains required longer post-fixation, which greatly reduced the YFP signal, so sections were co-labeled with anti-GFP (1:500; rabbit, Invitrogen A11122). For fluorescent labeling of nuclei, 4′,6-diamidino-2-phenylindole (DAPI, 100 ng/ml) was included with the secondary antibodies. Coverslips were mounted in Elvanol (Tris-HCl, glycerol, polyvinyl alcohol, 2% 1,4-diazabicyclo[2,2,2]octane). Confocal images were captured sequentially on an Olympus Fluoview FV500 confocal on a BX61W microscope with a 60× 1.42 numerical aperture oil-immersion lens and customized filter sets.
Western Blot Analysis
For Western blots, protein of equal amounts from Thy1-HFY-NL2 or wild type mice was loaded per lane for SDS-PAGE, run on 8% polyacrylamide gels, and transferred to Immobilon P membrane. Membranes were blocked with 5% skim milk and 0.05% Tween 20 in Tris-buffered saline, and incubated with primary antibodies as follows: anti-GFP (1:1000; rabbit, A11122, Invitrogen), anti-neuroligin-2 (1:2000 (28)), anti-GABAARγ2 (1:1000; rabbit, NB300-151, Novus), anti-GABAARα2 (1:1000; rabbit, Alomone Labs), anti-gephyrin (1:1000; mouse, SC25311, Santa Cruz), anti-synapsin1 (1:1000; rabbit, AB1543P, EMD Millipore), anti-PSD-95 family (recognizes PSD-95, PSD-93, SAP102, and SAP97; 1:1000; mouse IgG2a, 6G6-1C9, Thermo Scientific), and anti-β-actin (1:2500; rabbit, Abcam). Following incubation in appropriate HRP-conjugated secondary antibodies, the signal was detected using the Immobilon Western Chemiluminescent HRP Substrate kit (WBKLS0100, Millipore) and imaged with the Bio-Rad Molecular Imager VersaDoc MP Imaging System (Bio-Rad). For silver staining, the PageSilver kit (K0681, Fermentas) was used.
Tandem Affinity Purification
All of the protein work was performed at 4 °C. Mice between 2 and 4 months of age were sacrificed. Seven brains each from sex- and age-matched transgenic or wild type mice were pooled in homogenization buffer (50 mm NaH2PO4, 300 mm NaCl, pH 8.0, with Mini Protease Inhibitor Mixture, 04693159001, Roche Applied Science). The whole brain tissue was briefly sonicated for two 10-s bursts with a microtip using a Sonic Dismembrator (FB120110, Fisher Scientific), then homogenized using a Dounce homogenizer (099C K54, Thomas Scientific). The brain homogenate was centrifuged for 10 min at 1500 × g to remove nuclei and cellular debris. The supernatant was centrifuged for 1 h at 100,000 × g and the pellet resuspended in homogenization buffer and further homogenized using the Dounce homogenizer. The homogenate was then lysed with 0.5% (w/v) n-dodecyl-β-d-maltoside (DDM, D4641, Sigma) in homogenization buffer rotating slowly for 16 h. Cytoskeletal debris was removed by ultracentrifugation (100,000 × g for 60 min). Protein concentration was determined using the Bio-Rad RC/DC Protein Assay Kit (number 500-0121, Bio-Rad).
Nickel-nitrilotriacetic acid (Ni-NTA)-agarose (30230, Qiagen) resin was equilibrated with homogenization buffer and incubated with brain lysate prepared as described above for 16 h rotating at 200 rpm on a rotary shaker. The resin was washed twice with 10 mm imidazole in homogenization buffer, collected by centrifugation for 10 s at 15,000 × g. The proteins bound to Ni-NTA resin were eluted three times with 200 mm imidazole in homogenization buffer by centrifugation for 30 s at 15,000 × g between each elution step. Pooled eluate was incubated with biotin-conjugated anti-GFP antibody (0.4 μg/mg of brain lysate; 600-106-215, Rockland) for 1 h, followed by incubation with streptavidin-conjugated Sepharose 4B beads (1 μl/mg of brain lysate; S7973-89E, US Biological) for 16 h. The immune complexes were washed twice with 0.1% DDM in homogenization buffer, and purified proteins were eluted three times with SDS sample buffer (0.0626 m Tris-HCl, 2% SDS, 0.01% bromphenol blue, 100 mm dithiothreitol, 10% glycerol, pH 6.8) pooling the supernatant.
Protein Digestion and LC-MS/MS
Tandem affinity purified material from each anti-GFP immunoprecipitation following Ni-NTA elution was run into a 10% SDS-PAGE gel. Proteins were visualized by colloidal Coomassie (29) and split into 6 fractions where proteins in each fraction were digested out of the gel as described (30). Peptide samples were purified by solid phase extraction on C-18 STAGE tips (31). Purified peptides were analyzed using a linear-trapping quadrupole, Orbitrap mass spectrometer (LTQ-Orbitrap Velos; ThermoFisher Scientific) on-line coupled to an Agilent 1290 Series HPLC using a nanospray ionization source (ThermoFisher Scientific) including a 2-cm long, 100-μm inner diameter fused silica trap column, 50-μm inner diameter fused silica-fritted analytical column, and a 20-μm inner diameter fused silica gold-coated spray tip (6-μm diameter opening, pulled on a P-2000 laser puller from Sutter Instruments, coated on Leica EM SCD005 Super Cool Sputtering Device). The trap column is packed with 5-μm diameter Aqua C-18 beads (Phenomenex), whereas the analytical column is packed with a 3.0-μm diameter Reprosil-Pur C-18-AQ beads (Dr. Maisch). Buffer A consisted of 0.5% aqueous acetic acid, and buffer B consisted of 0.5% acetic acid and 80% acetonitrile in water. Samples were resuspended in buffer A and loaded with the same buffer. Standard 90-min gradients were run from 10 to 32% B over 51 min, then from 32 to 40% B in the next 5 min, then increased to 100% B over a 2-min period, held at 100% B for 2.5 min, and then dropped to 0% B for another 20 min to recondition the column. For a 60-min run the gradient was from 10 to 40% B over 32 min, then to 100% B over 0.5 min, held at 100% B for 1.5 min and dropped to 0% B for 15 min. For 120 min (180 min) methods buffer B was from 10 to 25% over 60 min (120 min), from 25 to 60% over 20 min, from 60 to 100% B over 7 min, kept at 100% for 2.5 min and then the column was reconditioned for 20 min with buffer A. The HPLC system included Agilent 1290 series Pump and Autosampler with Thermostat. The thermostat temperature was set at 6 °C. The sample was loaded on the trap column at 5 μl/min and the analysis was performed at 0.1 μl/min. The LTQ-Orbitrap was set to acquire a full-range scan at 60,000 resolution from 350,000 to 1,600,000 in the Orbitrap to simultaneously fragment the top 10 peptide ions by CID in each cycle in the LTQ (minimum intensity 1000 counts). Parent ions were then excluded from MS/MS for the next 30 s. Singly charged ions were excluded because in ESI mode peptides usually carry multiple charges. The Orbitrap was continuously recalibrated using the lock-mass function.
Protein Identification
Centroided fragment peak lists were processed with Proteome Discoverer version 1.2 (ThermoFisher Scientific). The search was performed with Mascot algorithm version 2.4 against the mouse Uniprot database (August 2013 version) with common contaminants added (50863 total sequences) using the following parameters: peptide mass accuracy 10 parts per million; fragment mass accuracy 0.6 Da; trypsin enzyme specificity with one missed cleavage, fixed modifications, carbamidomethyl; variable modifications, methionine oxidation; deamidated N, Q, and N-acetylpeptides, ESI-TRAP fragment characteristics. Only those peptides with IonScores exceeding the individually calculated 99% confidence limit (as opposed to the average limit for the whole experiment) were considered as accurately identified. Proteome Discoverer parameters were: event detector: mass precision 4 ppm (corresponds to extracted ion chromatograms at ± 12 ppm max error), S/N threshold 1; quantitation method − ratio calculation − replace missing quantitation values with minimum intensity, yes; use single peak quantitation channels, yes; protein quantification, use all peptides, yes.
Selection of NL2-specific Interactors
From all the proteins identified across five biological replicates, those that might be specifically interacting with HFY-NL2 were selected by applying the following criteria: (a) reproducible detection in the sample purified from transgenic (TG) mice (at least 2 unique peptides in two experiments or 1 unique peptide in three experiments); and (b) selective purification from TG mice relative to WT controls (a peptide spectrum match (PSM) ratio of ≥ 4:1 TG:WT in at least two experiments, a PSM ratio ≤1:2 TG:WT in no more than one experiment, and cumulative total PSM ratio >2:1 TG:WT). These criteria resulted in a list of 76 putative HFY-NL2-associated proteins (Table 1, categorized by manual curation from PubMed and GeneCards).
TABLE 1.
Proteins associated with HFY-NL2
Two-step purification of complexes using the His6 and YFP tags was performed as described in the legend to Fig. 5 from five sets of adult age-matched and sex-matched Thy1-HFY-NL2 (TG) and WT mice. After removal of human keratins, all components are listed that passed the identity criteria of at least 2 unique peptides in two experiments or 1 unique peptide in three experiments in TG samples, and the selection criteria of PSM ratio of ≥4:1 TG:WT in at least two experiments and PSM ratio ≤1:2 TG:WT in no more than one experiment and cumulative total PSM ratio >2:1 TG:WT. Listed here are the cumulative total PSM from TG and WT samples from the five experiments; italics indicates uncertainty in PSM allocation among closely related proteins due to some common peptides. Protein categories were defined by manual curation from PubMed and GeneCards.
| Accession | Gene symbol | Protein name | TG total PSM | WT total PSM |
|---|---|---|---|---|
| Synaptic adhesion | ||||
| IPI00468605.4 | Nlgn2 | Neuroligin 2 (transgene) | 192 | 2 |
| IPI00885337.1 | Nlgn3 | Neuroligin 3 | 30 | 1 |
| IPI00858277.1 | Nlgn4 | Neuroligin 4 | 44 | 0 |
| IPI00930898.1 | Nfasc | Neurofascin | 8 | 2 |
| Neurotransmitter receptor | ||||
| IPI00113772.1 | Gabra1 | GABAAR α1 | 16 | 2 |
| IPI00323554.3 | Gabrb2 | GABAAR β2 | 11 | 0 |
| IPI00874658.1 | Gabrg2 | GABAAR γ2 | 11 | 0 |
| Trafficking | ||||
| IPI00420426.1 | Ap3b2 | AP-3 complex subunit β-2 | 7 | 0 |
| IPI00221754.3 | Atl1 | atlastin 1 | 8 | 1 |
| IPI00670114.1 | Cadps | Calcium-dependent secretion activator 1 isoform 4 | 15 | 3 |
| IPI00650037.2 | Dnajc6 | DnaJ (Hsp40) homolog, subfamily C, member 6 isoform 2 | 6 | 0 |
| IPI00874710.1 | Dnm1l | Dynamin 1-like | 9 | 3 |
| IPI00625738.3 | Dnm3 | Dynamin 3 | 24 | 9 |
| IPI00122399.1 | Glg1 | Golgi apparatus protein 1 | 8 | 1 |
| IPI00323881.1 | Kpnb1 | Importin subunit beta-1 | 6 | 1 |
| IPI00776187.1 | Myo6 | Myosin VI | 5 | 0 |
| IPI00311515.2 | Napb | N-Ethylmaleimide-sensitive factor attachment protein β | 34 | 2 |
| IPI00889265.1 | Ncdn | Neurochondrin | 19 | 2 |
| IPI00337980.5 | Rab21 | Rab21 | 5 | 2 |
| IPI00622482.2 | Rufy3 | RUN and FYVE domain containing 3 isoform 1 | 8 | 1 |
| IPI00123349.2 | Sec23a | Sec23A | 4 | 0 |
| IPI00653617.1 | Snap91 | Synaptosomal-associated protein, 91kDa | 14 | 4 |
| IPI00885385.1 | Stx1a | Syntaxin 1a | 7 | 1 |
| IPI00874893.1 | Stx1b | Syntaxin 1b | 31 | 9 |
| IPI00125267.4 | Vapa | Vesicle-associated membrane protein-associated protein A | 4 | 0 |
| IPI00135655.3 | Vapb | Vesicle-associated membrane protein-associated protein B | 5 | 0 |
| IPI00111181.1 | Vps35 | Vacuolar protein sorting-associated protein 35 | 6 | 1 |
| IPI00395038.3 | XpoI | exportin-1 | 7 | 0 |
| Kinases | ||||
| IPI00119762.4 | Dclk1 | Double cortin and calcium/calmodulin-dependent protein kinase-like 1, isoform CRA_c | 7 | 0 |
| IPI00122069.1 | Prkcc | Protein kinase Cγ type | 23 | 8 |
| Ubiquitination and proteasome | ||||
| IPI00896727.1 | Cand1 | Cullin-associated NEDD8-dissociated protein 1 | 16 | 1 |
| IPI00758064.1 | Psmc3 | Proteasome 26 S subunit ATPase 3 | 4 | 0 |
| IPI00123313.1 | Uba1 | Ubiquitin-like modifier-activating enzyme 1 | 11 | 1 |
| Ion channel and associated | ||||
| IPI00845769.1 | Gpd1l | Glycerol-3-phosphate dehydrogenase 1-like protein isoform 2 | 19 | 3 |
| IPI00876341.1 | Vdac3 | Voltage-dependent anion-selective channel protein 3 | 39 | 15 |
| Extracellular matrix | ||||
| IPI00869394.1 | Bcan | Brevican | 4 | 0 |
| IPI00227126.1 | Tnr | Tenascin-R | 17 | 1 |
| Cytoskeletal and scaffolding | ||||
| IPI00921659.1 | Ank2 | Ankyrin-2 | 14 | 5 |
| IPI00111265.3 | Capza2 | F-actin-capping protein subunit alpha-2 | 9 | 0 |
| IPI00652858.1 | Emd | Emerin | 4 | 0 |
| IPI00229300.2 | Epb4.1l3 | Erythrocyte membrane protein band 4.1-like 3 | 20 | 4 |
| IPI00464296.5 | Epb4.1l3 | Erythrocyte membrane protein band 4.1-like 3 isoform 8 | 21 | 5 |
| IPI00816946.1 | Gphn | Gephyrin | 16 | 1 |
| IPI00882056.2 | Mapre2 | Microtubule-associated protein, RP/EB family, member 2 isoform 3 | 4 | 0 |
| IPI00656204.1 | Nckap1 | Nck-associated protein 1 isoform 2 | 5 | 0 |
| Protein translation | ||||
| IPI00831544.1 | Cyfip1 | Cytoplasmic FMR1-interacting protein 1 isoform 2 | 12 | 3 |
| IPI00405625.9 | Cyfip2 | Cytoplasmic FMR1-interacting protein 2 | 12 | 3 |
| IPI00318841.4 | Eef1g | Eukaryotic translation elongation factor 1 gamma | 8 | 0 |
| IPI00466069.3 | Eef2 | Eukaryotic translation elongation factor 2 | 5 | 1 |
| IPI00319956.1 | IMPACT | Imprinted and ancient gene protein homolog | 8 | 0 |
| IPI00880400.1 | Rps2 | Ribosomal protein S2 | 9 | 0 |
| IPI00880750.1 | Rps9 | Ribosomal protein S9 | 4 | 0 |
| IPI00130353.5 | Vars | Valyl-tRNA synthetase | 4 | 0 |
| Protein folding and glycosylation | ||||
| IPI00121575.1 | Alg2 | α1,3-Mannosyltransferase | 15 | 5 |
| IPI00317710.1 | Hspa4l | Heat shock 70-kDa protein 4L isoform 1 | 17 | 1 |
| IPI00319992.1 | Hspa5 | Heat shock 70-kDa protein 5 | 172 | 44 |
| IPI00224109.2 | Hsph1 | Heat shock protein 105-kDa isoform HSP 105-β | 9 | 0 |
| IPI00123342.4 | Hyou1 | Hypoxia up-regulated protein 1 | 18 | 3 |
| IPI00676130.1 | Tmx4 | Thioredoxin-related transmembrane protein 4 | 8 | 0 |
| Miscellaneous | ||||
| IPI00119114.2 | Acadl | Long-chain specific acyl-CoA dehydrogenase | 6 | 0 |
| IPI00119458.7 | Aldoc | Fructose-bisphosphate aldolase C | 63 | 11 |
| IPI00468999.4 | Cds2 | Phosphatidate cytidylyltransferase 2 | 7 | 0 |
| IPI00754739.1 | Chchd3 | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3 | 5 | 1 |
| IPI00126085.1 | Cyp46a1 | Cholesterol 24-hydroxylase | 3 | 0 |
| IPI00762654.1 | Erlin1 | ER lipid raft protein 1 | 7 | 1 |
| IPI00113223.2 | Fasn | Fatty acid synthase | 18 | 8 |
| IPI00118569.1 | Gna13 | Guanine nucleotide-binding protein subunit α-13 | 9 | 3 |
| IPI00229525.1 | Gnal | Guanine nucleotide-binding protein G(olf) subunit α | 7 | 3 |
| IPI00115607.3 | Hadhb | Trifunctional enzyme subunit β | 25 | 10 |
| IPI00114642.4 | Hist1h2bf | Histone H2B type 1-F/J/L | 6 | 2 |
| IPI00407339.7 | Hist1h4 | Histone H4 | 9 | 3 |
| IPI00953761.1 | Mgll | Monoglyceride lipase | 10 | 2 |
| IPI00608097.1 | Npepps | Aminopeptidase puromycin sensitive | 13 | 0 |
| IPI00121758.1 | Tardbp | TAR DNA-binding protein 43 | 9 | 0 |
| IPI00929787.1 | Tecr | Trans-2,3-enoyl-CoA reductase isoform 2 | 5 | 0 |
| Transgene tag | ||||
| CON_GFP | Green fluorescent protein | 90 | 1 | |
Over-representation Analysis
The possibility that certain classes of proteins were over-represented in the list of putative NL2 interactors versus all proteins identified was tested using the Over-Representation Analysis function in ErmineJ (32). We considered any term to be significantly over-represented if the adjusted p value (p values corrected for multiple hypothesis testing) was less than 0.05.
RESULTS
Generation of TAP-tagged NL2
We generated and pre-screened TAP-tagged versions of NL2 for proper localization in cultured rat hippocampal neurons prior to generating transgenic mice. Because YFP will be useful for imaging as well as biochemical purification, we began with YFP-tagged NL2. YFP-NL2 bears an extracellular YFP tag following a signal sequence at the mature N terminus of NL2 and localizes properly to inhibitory synapses in cultured neurons (12). We tried adding next to YFP the IgG binding unit of Protein A (ProtA) and calmodulin-binding peptide (CBP) tags separated by the tobacco etch virus (TEV) protease recognition site, a TAP-tagging strategy that worked well in yeast (21), plus the short FLAG tag. However, the resultant ProtA-TEV-CBP-FLAG-YFP-NL2 did not localize properly in cultured neurons, the YFP signal filled the processes and formed small non-synaptic puncta, presumably due to misfolding (not shown). Likewise, the smaller CBP-FLAG-YFP-NL2 also mislocalized in cultured neurons. It is possible that the CBP tag may not be suitable for the lumen of the secretory pathway and extracellular domain as required by the position of our tag. We next tested His6-FLAG-YFP-NL2 (HFY-NL2), which proved to be more suitable.
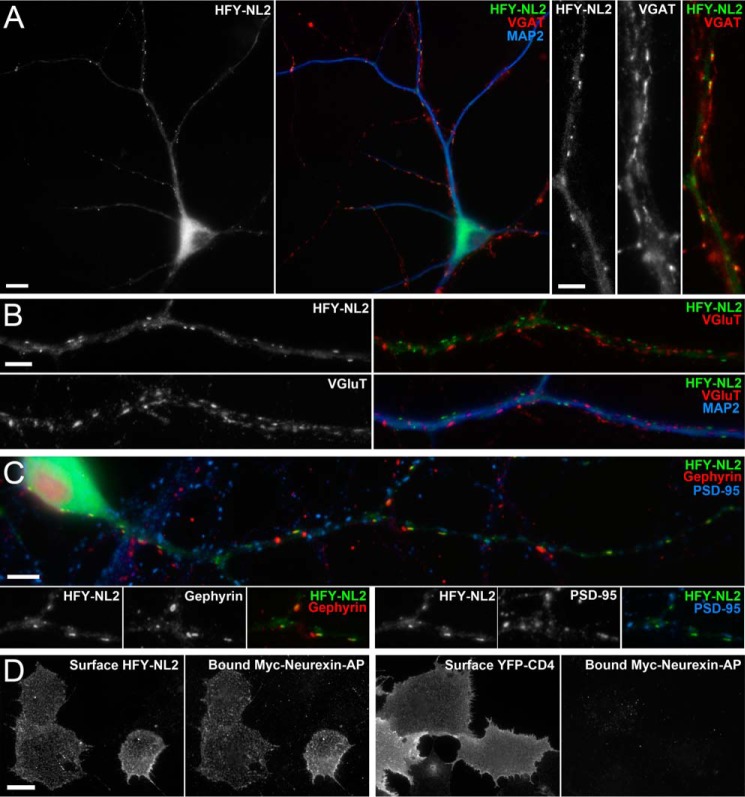
HFY-NL2 localized specifically to clusters at inhibitory postsynaptic sites on dendrites of transfected neurons (Fig. 1, A-C). HFY-NL2 clusters colocalized with gephyrin and were opposed to terminals labeled for the vesicular GABA transporter. HFY-NL2 clustered at distinct sites from excitatory synapses labeled for the excitatory postsynaptic scaffold PSD-95 family and vesicular glutamate transporter (Fig. 1, A–C). Thus HFY-NL2 exhibited appropriate targeting, similar to endogenous NL2, which clusters at inhibitory postsynaptic sites in cultured neurons and in vivo (10, 12).
FIGURE 1.
His6-FLAG-YFP-neuroligin-2 (HFY-NL2) localizes to inhibitory postsynaptic sites in cultured hippocampal neurons and binds neurexin. A–C, cultured neurons were transfected with HFY-NL2 expression vector at 9 DIV, then fixed and immunolabeled at 12 DIV. HFY-NL2 imaged by the unamplified YFP signal was restricted to the somatodendritic domain and formed clusters on dendrites labeled for microtubule-associated protein 2 (MAP2). HFY-NL2 clusters were opposed to inhibitory terminals labeled for the vesicular GABA transporter (VGAT) (A), but not to excitatory terminals labeled for the vesicular glutamate transporter (VGluT) (B). HFY-NL2 clusters colocalized with the inhibitory postsynaptic scaffold gephyrin but not with the excitatory postsynaptic scaffold PSD-95; some gephyrin puncta lacking HFY-NL2 are seen in crossing dendrites from untransfected neurons (C). D, COS7 cells expressing HFY-NL2 or YFP-CD4 as a negative control were incubated with the ectodomain of neurexin-1β fused to Myc-alkaline phosphatase, Myc-neurexin-AP. Whereas both constructs expressed well as detected by surface labeling with anti-GFP antibody, only HFY-NL2 mediated neurexin binding as detected with anti-Myc antibody. Scale bars: 10 μm in A (left panels); 5 μm in A (right panels), B and C; 20 μm in D.
The position of the tags at the mature N terminus of NL2 leaves the short intracellular domain unencumbered to interact with gephyrin and other partners. Interaction of the extracellular acetylcholinesterase homologous domain of NL2 with neurexin is critical for its function as a synapse organizing protein (33). Although we cannot rule out the possibility that the TAP tags may interfere with some extracellular interactions of NL2, we found that the key interaction with neurexin is preserved. In a cell-based binding assay, the purified recombinant extracellular domain of neurexin-1β fused to Myc-alkaline phosphatase (neurexin 1β-Myc-AP) bound strongly to COS cells expressing HFY-NL2 but not to COS cells expressing the unrelated protein YFP-CD4 as a negative control (Fig. 1D).
Low Level Expression of HFY-NL2 in Transgenic Mice
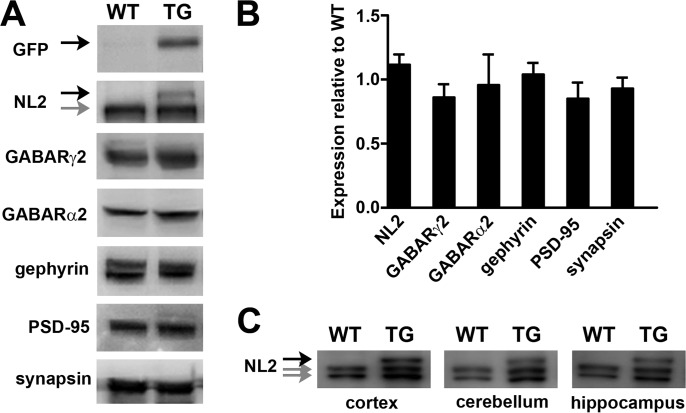
To generate transgenic mice with neuron-specific expression, HFY-NL2 was placed in a targeting vector under control of regulatory elements from the Thy1 gene previously used to drive neuron-specific expression of fluorescent proteins in numerous mouse lines (34). We focused further studies on one Thy1-HFY-NL2 transgenic line, which showed broad low level expression. A Western blot of whole brain homogenate for NL2 showed a higher molecular weight band also recognized by a GFP antibody in the transgenic sample in addition to the wild type band (Fig. 2A). Based on the relative intensities of the transgenic and wild type bands with the NL2 antibody, this Thy1-HFY-NL2 line expresses the transgene at mean 15.7 ± 1.6% (n = 3) of the endogenous level, i.e. the total level of transgenic plus wild type neuroligin-2 is increased by 15.7% in these mice. A number of other synaptic proteins showed no difference in levels in the Thy1-HFY-NL2 mice compared with wild type, including GABA receptor subunits γ2 and α2, gephyrin, excitatory postsynaptic scaffold PSD-95, and presynaptic protein synapsin (Fig. 2, A and B). The transgene was broadly expressed, including in the neocortex, hippocampus, and cerebellum (Fig. 2C). This Thy1-HFY-NL2 line has also been used for imaging inhibitory synapse development in retinal gangion cells (24). Furthermore, spontaneous inhibitory postsynaptic currents from whole cell recordings of these transgenic or wild type retinal ganglion cells showed no significant difference in frequency or amplitude (24). These Thy1-HFY-NL2 mice thrive and show no obvious deficits in survival, breeding, or home cage behavior. Thus, whereas NL2 overexpression in culture can alter synapse development (35) and a high level transgenic expression of HA-NL2 in the range of 66 to 440% of wild type level results in neurological deficits and lethality (36), no such phenotypes were observed in this low level Thy1-HFY-NL2 line. All these higher level Thy1-HA-NL2 transgenic lines exhibited a pronounced limb clasping phenotype (36), a phenotype never observed in our Thy1-HFY-NL2 line. Thus, in the Thy1-HFY-NL2 line, the TAP-tagged transgene is acting as a tracer without apparent alterations of synaptic protein levels, synaptic transmission, or overt behavior.
FIGURE 2.
Thy1-HFY-NL2 mice express low levels of the transgene and normal levels of other synaptic proteins. A, Western blot of brain lysate from Thy1-HFY-NL2 (TG) in comparison with wild type (WT) mice reveals expression of the HFY-NL2 transgene (black arrows) in addition to endogenous NL2 (gray arrow) and normal expression of other synaptic components. The transgene is expressed at mean 15.7 ± 1.6% (n = 3) of endogenous level. B, no differences were found in levels of the indicated proteins in TG compared with WT brain lysate (n = 3–6, p = 0.44 by two-way analysis of variance; NL2 indicates endogenous NL2). C, HFY-NL2 was expressed in cortex, cerebellum, and hippocampus (black arrow indicates HFY-NL2 transgene; gray arrows indicate endogenous NL2, which sometimes ran as a doublet).
Localization of HFY-NL2 to Inhibitory Synapses in Multiple Brain Regions
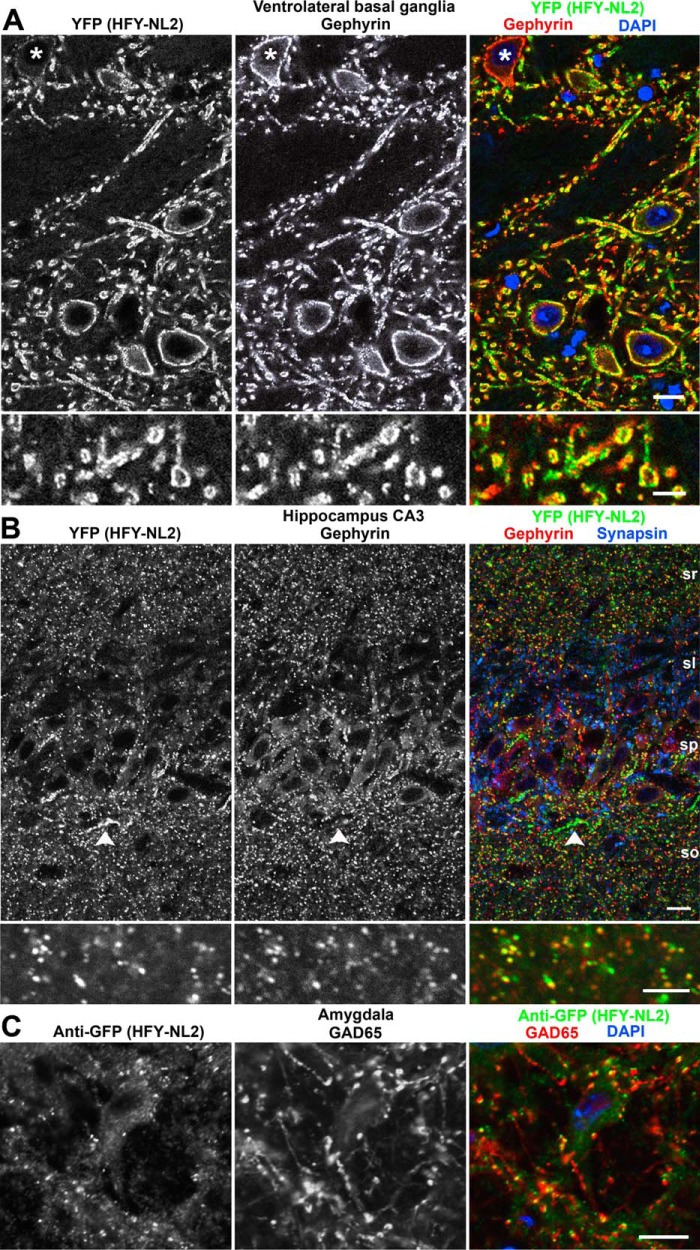
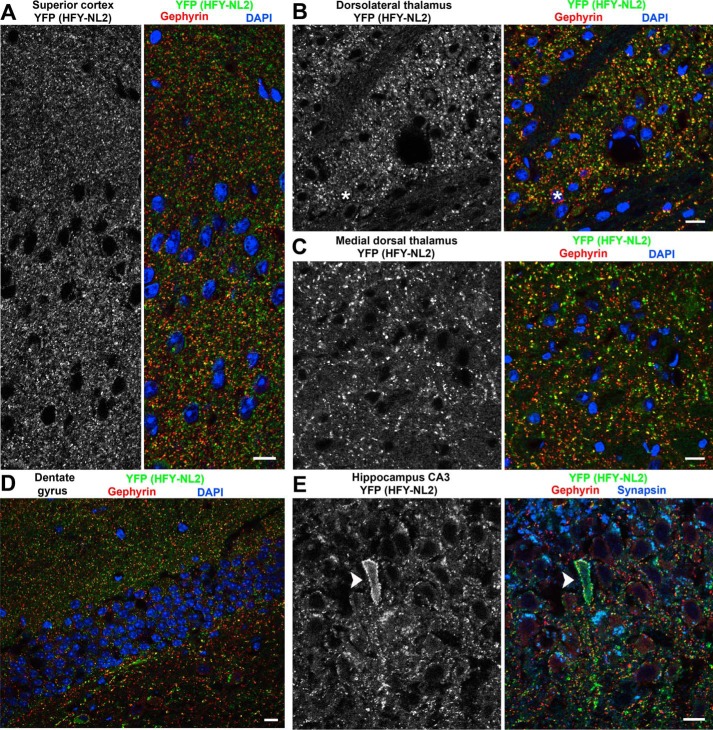
To interact with the appropriate molecular partners, and to be useful for imaging, the TAP-tagged NL2 must localize properly in brain in vivo. As previously reported for retina (24), indeed the HFY-NL2 localized specifically to inhibitory postsynaptic sites in multiple brain regions. The YFP channel was suitable for confocal imaging of HFY-NL2 without requiring amplification. HFY-NL2 showed strong colocalization with the inhibitory postsynaptic marker gephyrin in all brain regions (Figs. 3 and 4). The colocalization of YFP with gephyrin was particularly apparent at the large inhibitory postsynaptic sites formed in the ventral basal ganglia (Fig. 3A). Consistent with expression in multiple neuron classes from the Thy1 promoter (37), perisomatic HFY-NL2 was associated with presumptive interneurons outside the pyramidal cell layer as well as pyramidal cells of the hippocampus, The majority but not all inhibitory synapses were labeled with YFP, as a small subset of neurons did not express the transgene (e.g. Figs. 3A and 4B). Conversely, the YFP transgenic label was more pronounced than gephyrin at apparent axon initial segment synapses in hippocampal stratum pyramidale (Fig. 3B) and in rare individual neurons with high transgene expression (Fig. 4E). Clustering of HFY-NL2 opposite GABAergic terminals was shown by colabeling for the GABA synthetic enzyme and inhibitory presynaptic marker GAD65; because conditions for GAD65 labeling were not readily compatible with maintaining YFP fluorescence, anti-GFP antibody was used to detect HFY-NL2 (Fig. 3C). HFY-NL2 puncta showed significant overlap with GAD65 puncta, greater than the overlap observed after rotating one of the channels to simulate random overlap (p < 0.0001, t test, n = 44 fields). Importantly, HFY-NL2 was not detected at excitatory synapses. For example, in the hippocampal CA3 stratum lucidum region the large mossy fiber glutamatergic synapses immunopositive for synapsin lacked apposing HFY-NL2 or gephyrin (Fig. 3B).
FIGURE 3.
HFY-NL2 localizes to inhibitory synapses in transgenic mouse brain. Brain sections from perfused adult Thy1-HFY-NL2 mice were imaged for HFY-NL2 using the unamplified YFP signal and co-labeled for the nuclear marker DAPI and the inhibitory postsynaptic scaffold gephyrin. A, the large inhibitory synapses in the ventrolateral basal ganglia exhibit clear colocalization of HFY-NL2 with gephyrin. A minority of neurons (asterisk) do not express the transgene. B, HFY-NL2 colocalized well with gephyrin and overlapped with synapsin, a general presynaptic marker, in the hippocampal CA3 region stratum radiatum (sr), straum pyramidale (sp), and stratum oriens (so). Short strings of HFY-NL2 puncta at the border of stratum pyramidale and oriens likely correspond to axon initial segment synapses (arrowhead). In stratum lucidum (sl), the large mossy fiber glutamatergic synapses immunopositive for synapsin lacked HFY-NL2 or gephyrin. C, HFY-NL2 puncta detected with anti-GFP antibody overlapped with the GABAergic presynaptic marker GAD65, as shown here in the amygdala. Scale bars, 10 μm in the main panels, 5 μm in the enlarged regions.
FIGURE 4.
HFY-NL2 is broadly expressed at inhibitory synapses in multiple brain regions including cortex, thalamus, and hippocampus. Brain sections from perfused adult Thy1-HFY-NL2 mice were imaged for NFY-NL2 using the unamplified YFP signal and co-labeled for the nuclear marker DAPI and the inhibitory postsynaptic scaffold gephyrin. HFY-NL2 was detected at the majority of gephyrin-positive inhibitory synapses in superior cortex (A), dorsolateral thalamus (B), dentate gyrus (D) granule cell (gc), and molecular layers (ml), and hippocampal CA3 stratum pyramidale (E). Occasional neurons exhibited little or no transgene expression (asterisk in B) or high level expression (arrowhead in E). Overall expression of the transgene was more sparse in medial dorsal thalamus (C) and the hilus of the dentate gyrus (D) where a greater fraction of gephyrin puncta lacked HFY-NL2 YFP signal. Scale bars, 10 μm.
HFY-NL2 was expressed and appropriately co-clustered with gephyrin at the majority of inhibitory synapses in all regions of cortex, thalamus, and hippocampus, except for relatively low expression in the hilus of the dentate gyrus (Fig. 4). Although other brain regions were not examined in detail, expression was also observed more broadly. Especially considering the heterogeneity among GABAergic neurons and GABAergic synapses (38, 39), this widespread expression of HFY-NL2 may be important for detecting multiple potential interacting complexes, some of which may be region or cell-type specific.
Purification of HFY-NL2 Complexes
To optimize the purification, we first tested the solubility of HFY-NL2 from brain homogenate in multiple detergents at concentrations of 0.25–1%. HFY-NL2 solubility was in the 60–80% range efficient with DDM, Triton X-100, and Nonidet P-40, but <30% with digitonin, octyl β-d-glucopyranoside, polyoxyethylene (20), sorbitan monolaurate (Tween 20), or CHAPS. As a next test, we compared the yield of HFY-NL2 upon binding to a Ni-NTA column and elution with imidazole, the first step of purification based on the His6 tag. We obtained a better yield using DDM than either Triton X-100 or Nonidet P-40. Furthermore, DDM has been found superior to Triton X-100 and other detergents for maintaining interactions within membrane protein complexes such as active cytochrome oxidase and photosynthetic reaction centers (40). Thus we selected a buffer with 0.5% DDM for purification of HFY-NL2-associated complexes.
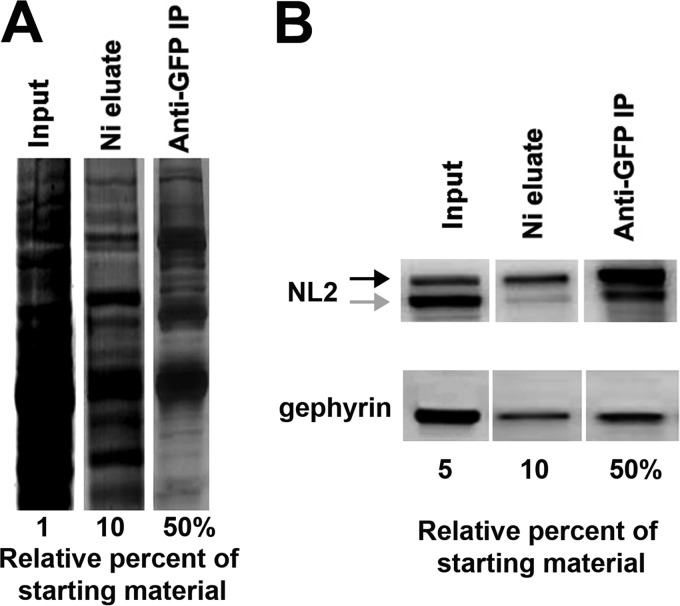
We used a two-step purification protocol, first binding to a Ni-NTA column via the His6 tag and then immunoprecipitation with biotinylated anti-GFP antibody and streptavidin-agarose via the YFP tag. Although HFY-NL2 was also designed with a FLAG tag between the His6 and YFP for a potential three-stage purification, HFY-NL2 did not immunoprecipitate well with M2 anti-FLAG antibody. The two-step purification resulted in an expected reduction in the overall protein amount and complexity assessed by SDS-PAGE and SYPRO Ruby staining (Fig. 5A). Furthermore, each step resulted in enrichment of the HFY-NL2 transgene assessed by Western blot (Fig. 5B). Some endogenous NL2 and gephyrin copurified with HFY-NL2, thus confirming the preservation of expected interactions based on the dimerization of NL2 (13, 33) and the association of NL2 with gephyrin (11).
FIGURE 5.
Isolation of HFY-NL2-associated proteins. Brain lysate from adult Thy1-HFY-NL2 mice was subjected to two rounds of affinity purification. Following the first step of binding to a Ni-NTA column via the His6 tag, the purified material (Ni eluate) was immunoprecipitated with biotinylated anti-GFP antibody and streptavidin-agarose via the YFP tag resulting in doubly purified material (Anti-GFP IP). A, SDS-PAGE and SYPRO Ruby stain shows a reduction in total amount and complexity of material during the two-step purification. B, Western blot shows increasing enrichment of the HFY-NL2 transgene (black arrow) as well as co-purification of some endogenous NL2 (gray arrow) and gephyrin.
Characterization of HFY-NL2 Complexes
We performed the two-step purification side by side from the whole brain of adult age-matched and sex-matched Thy1-HFY-NL2 (TG) mice and wild type (WT) mice as a negative control for isolation of nonspecific components. Five independent isolations from TG and paired WT brains were performed and proteins were identified by LC-MS/MS. Putative specific interactors of NL2 (Table 1) were identified as described under “Experimental Procedures.” As an important validation of the set of specific interactors, this list contains well characterized components of GABAergic synapses including GABA receptor α1, β2, and γ2 subunits and gephyrin. Furthermore, a Gene Ontology analysis (32, 41) of the HFY-NL2-associated components in Table 1 as compared with the entire set of proteins isolated from TG or WT mice confirmed a significant enrichment in proteins involved in cell-cell signaling and synaptic transmission (Table 2).
TABLE 2.
Gene Ontology analysis of HFY-NL2 associated proteins
The set of HFY-NL2-associated proteins listed in Table 1 was compared with the entire set of proteins isolated from either transgenic or wild type mice in any of the five experiments to test for enrichment in any biological process Gene Ontology terms (41) using ermineJ (32). Adjustment was applied for multiple test correction. Terms with p value <0.001 are listed here.
| GO Term | Description | p value | Adjusted p value |
|---|---|---|---|
| GO:0007267 | Cell-cell signaling | 4.649E-05 | 0.043 |
| GO:0007268 | Synaptic transmission | 9.714E-05 | 0.045 |
| GO:0048666 | Neuron development | 3.921E-04 | 0.12 |
| GO:0031175 | Neuron projection development | 5.212E-04 | 0.12 |
| GO:0050808 | Synapse organization | 9.386E-04 | 0.17 |
Regulated Endocytosis of NL2
A striking aspect of the list of putative HFY-NL2-associated proteins is the number of components involved in membrane protein trafficking (Table 1). Among these, many have reported roles in the endocytic pathway, including AP-3, myosin VI, VPS35, Dnajc6, dynamin 1-like, dynamin 3, Rab21, SNAP91, and neurochondrin (42–46). As an initial functional test of whether NL2 undergoes dynamin-dependent regulated endocytosis, we expressed NL2 with an extracellular HA tag in cultured hippocampal neurons, labeled surface HA-NL2 with FITC-conjugated rat anti-HA Fab fragment, and followed the fate of this labeled NL2 upon application of soluble ligand. For these endocytosis assays, we switched from the HFY tag to an HA tag in the same position of NL2 to utilize the available monovalent Fab reagent, avoiding using divalent antibodies, which can alter protein trafficking when applied to live cells. After incubation with the FITC-conjugated rat anti-HA Fab fragment, neurons were then exposed to the NL2-binding extracellular domain of neurexin 1β fused to alkaline phosphatase (neurexin 1β-AP) or AP as a negative control, and any pre-labeled NL2 still present on the cell surface 1 h later was labeled with Alexa 568-conjugated anti-rat antibody. Neurexin 1β-AP as compared with the AP induced a loss of surface HA-NL2 and increased the endocytosed HA-NL2 (Fig. 6). Furthermore, treatment with dynasore, an inhibitor of dynamin GTPase activity and thus of dynamin-dependent endocytosis (47), blocked the endocytosis of HA-NL2 induced by neurexin 1β-AP. Thus, NL2 can undergo dynamin-dependent regulated endocytosis in neurons.
Colocalization of Endocytosed NL2 with VPS35 Retromer
An interesting protein found associated with HFY-NL2 (Table 1) is VPS35, a retromer component implicated in endosome to Golgi transport and endosome to plasma membrane recycling (45, 48). VPS35-containing retromer-associated endosomes were recently found to mediate frequent plasma membrane fusion events throughout neuronal dendrites (49). We wondered whether NL2 might traffic by this route and tested for colocalization of endocytosed HA-NL2 with VPS35. Indeed, punctate HA-NL2 that appeared in the soma and dendrites following surface labeling colocalized significantly with VPS35 (Fig. 7). Treatment with neurexin 1β-AP as compared with AP increased the area occupied by punctate HA-NL2 and HA-NL2 colocalized with VPS35, confirming the enhancement of endocytosis by soluble ligand. The fraction of punctate HA-NL2 present in VPS35-positive organelles was in the 49–59% range and was not altered by neurexin 1β-AP, indicating association of ligand-induced and basal endocytosed NL2 with VPS35-positive organelles.
DISCUSSION
Here we used tandem affinity purification and mass spectrometry from transgenic mice to identify proteins associated with the major inhibitory synaptic organizing protein NL2. Known components of inhibitory synapses were isolated, including GABA receptor subunits α1, β2, and γ2 and gephyrin, and multiple novel putative NL2-associated proteins. Gene Ontology analysis confirmed enrichment of proteins involved in cell-cell signaling and synaptic transmission. Isolation of a number of trafficking proteins suggested that NL2 may undergo regulated trafficking. In support, we show that recombinant NL2 in neurons undergoes dynamin-dependent internalization upon addition of a soluble ligand and that endocytosed NL2 colocalizes with VPS35 retromer. In addition, the YFP signal in Thy1-HFY-NL2 mice labels inhibitory postsynaptic sites in numerous brain regions and thus may prove useful for further imaging studies.
NL2 directly binds to gephyrin (11), whereas the association of most other proteins isolated here with NL2 may be indirect as part of a larger complex. Neuroligin-3 partially colocalizes with NL2 to inhibitory synapses and coimmunoprecipitates with NL2 (50), likely as part of a larger complex rather than a hetero-oligomer (51). The isolation of neuroligin-4 but not neuroligin-1 here is also consistent with the selectivity of neuroligin-4 for inhibitory synapses, albeit more for glycinergic than GABAergic (52), and of neuroligin-1 for excitatory synapses (53). In addition to binding NL2, gephyrin binds to multiple GABA receptor α subunits (54), thus constituting a link between NL2 and GABA receptors. The GABA receptor subunits isolated here, α1, β2, and γ2, are the most abundant and together comprise the major synaptic inhibitory receptor in the brain (55).
In addition to gephyrin, GABA receptors, and other neuroligins, several other proteins isolated here by association with HFY-NL2 have a previously demonstrated localization and/or function at inhibitory synapses. Neurofascin localizes specifically to inhibitory synapses on the axon initial segment (56), in contrast to NL2, which localizes more broadly to inhibitory synapses on the axon initial segment, perisomatic region, and dendrites. Functionally, in adult rat hippocampal neurons, altering levels of neurofascin altered the number and size of gephyrin and GABA receptor clusters and GABAergic innervation on the axon initial segment but not on dendrites (57). Ankyrin-2 borders the axon initial segment (58) and thus may also contribute to GABAergic synapse development in this specialized domain. Tenascin-R and brevican are major components of the perineuronal net that is prominent around the axon initial segment and around subsets of interneurons (59). Furthermore, tenascin-R−/− mice show reduced numbers and altered morphology of perisomatic asymmetric synapses and a corresponding reduction in inhibitory transmission (60, 61). Thus, prior evidence supports a role in GABA synapse development for multiple scaffolding, extracellular matrix, and cell adhesion proteins isolated here.
A number of components associated with membrane protein trafficking were isolated with HFY-NL2. Many of these have reported roles in the endocytic pathway, including AP-3, myosin VI, VPS35, Dnajc6, dynamin 1-like, dynamin 3, Rab21, SNAP91, and neurochondrin (42–46). We further show that HA-NL2 can undergo dynamin-dependent endocytosis in neurons, triggered by incubation with the soluble ectodomain of neurexin 1β, and that endocytosed HA-NL2 is present in VPS35 retromer-containing organelles. Although neurexins are expressed mainly as transmembrane proteins, secreted forms of neurexin 3 can be generated by alternative splicing (62). Furthermore, multiple subtypes of neurexins are subject to proteolytic processing by metalloproteases and presenilins resulting in ectodomain shedding (63, 64). Thus the soluble neurexin 1β ectodomain used here to trigger endocytosis may mimic the shed ectodomain. An important future direction will be to characterize more thoroughly the regulation of NL2 trafficking. Neuroligin-1 undergoes rapid internalization upon chemical induction of long term depression (65), and interestingly both neuroligin-1 and NL2 are also subject to ectodomain shedding (66, 67). The presence of endocytosed NL2 in VPS35 retromer-containing organelles, a major route of recycling in dendrites (49), suggests that sorting and trafficking of endocytosed NL2 may be regulated by the retromer.
A limitation of this study is that only a subset of inhibitory synaptic components were isolated. The lack of enrichment of Slitrk3 (68) and the dystrophin glycoprotein complex (69, 70) might reflect the presence of distinct complexes mediating inhibitory synaptic adhesion. However, we cannot rule out a more technical explanation, especially because neurexins and multiple GABA receptor subunits were not enriched either. Their absence may be due to the labile nature of inhibitory synaptic complexes, even in the presence of the relatively mild detergent DDM. Rather than standard methods, chemical cross-linking was used to first achieve coimmunoprecipitation of gephyrin with NL2 from brain (11). Similarly, although gephyrin is now generally accepted to interact directly with GABA receptors, albeit with lower affinity than glycine receptors, classical methods were not successful to demonstrate such an interaction (54). While this study was underway, two studies taking a similar approach as ours to identify synaptic components were reported. Proteomic analysis of excitatory postsynaptic sites using a knock-in mouse expressing TAP-tagged PSD-95 identified 118 associated proteins (23). In contrast, proteomic analysis of inhibitory postsynaptic sites using a transgenic mouse expressing the singly tagged Venus-GABA receptor α1 identified 18 associated proteins (71). Thus, unlike excitatory postsynaptic complexes, many of which survive isolation with 2% SDS, inhibitory synaptic complexes appear to be particularly labile making it difficult to generate a complete catalogue of components.
Conversely, some components listed in Table 1 are likely contaminants and not bona fide components of NL2 complexes. However, putative NL2-associated proteins cannot be dismissed outright because they have housekeeping functions. As a prime example, gephyrin at first may have seemed an unlikely candidate as a major scaffolding protein of inhibitory synapses because of its dual essential function in molybdenum cofactor biosynthesis (54).
In addition to its utility for TAP-MS proteomics, the HFY-NL2 transgenic line may also be useful for further imaging studies. Whereas spiny excitatory synapses can be imaged in many brain regions using a fluorescent cell fill for spines, inhibitory postsynaptic specializations are not apparent just with a cell fill but require a molecular tag. Thy1-HFY-NL2 mice express the tagged NL2 at a low level, essentially as a tracer, in multiple brain regions and the YFP signal is suitable for imaging without amplification. This line may be useful to bypass the necessity for in utero electroporation of tagged constructs for in vivo imaging of inhibitory synapses (72) and for large scale mapping of inhibitory synapses using recently developed tissue clearing techniques (73).
Acknowledgments
We thank Xiling Zhou and Nazarine Fernandes for excellent technical assistance and Dr. Austen Milnerwood for advice on VPS35.
This work was supported, in whole or in part, by Canadian Institutes of Health Research Operating Grants 69096 (to A. M. C.) and 77688 (to L. J. F.), National Institutes of Health Grants EY10699 (to R. O. W.) and EY02422 (to R. S. M.), a Bluma Tischler Postdoctoral Fellowship (to Y. K.), and Canada Research Chair Awards (to A. M. C., R. S. M., and L. J. F.).
- NL2
- neuroligin-2
- TAP
- tandem affinity purification
- DIV
- days in vitro
- AP
- alkaline phosphatase
- DDM
- n-dodecyl-β-d-maltoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- LTQ
- linear-trapping quadrupole
- TG
- transgenic
- ProtA
- IgG binding unit of Protein A
- CBP
- calmodulin-binding peptide
- TEV
- tobacco etch virus
- HFY-NL2
- His6-FLAG-YFP-neuroligin-2
- DMSO
- dimethyl sulfoxide
- PSM
- peptide spectrum match.
REFERENCES
- 1. Kennedy M. B. (2000) Signal-processing machines at the postsynaptic density. Science 290, 750–754 [DOI] [PubMed] [Google Scholar]
- 2. Sheng M., Hoogenraad C. C. (2007) The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu. Rev. Biochem. 76, 823–847 [DOI] [PubMed] [Google Scholar]
- 3. Husi H., Ward M. A., Choudhary J. S., Blackstock W. P., Grant S. G. (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 3, 661–669 [DOI] [PubMed] [Google Scholar]
- 4. Li K. W., Hornshaw M. P., Van Der Schors R. C., Watson R., Tate S., Casetta B., Jimenez C. R., Gouwenberg Y., Gundelfinger E. D., Smalla K. H., Smit A. B. (2004) Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 279, 987–1002 [DOI] [PubMed] [Google Scholar]
- 5. Collins M. O., Husi H., Yu L., Brandon J. M., Anderson C. N., Blackstock W. P., Choudhary J. S., Grant S. G. (2006) Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, 16–23 [DOI] [PubMed] [Google Scholar]
- 6. Bayés A., Grant S. G. (2009) Neuroproteomics: understanding the molecular organization and complexity of the brain. Nat. Rev. Neurosci. 10, 635–646 [DOI] [PubMed] [Google Scholar]
- 7. Luscher B., Fuchs T., Kilpatrick C. L. (2011) GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 70, 385–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vithlani M., Terunuma M., Moss S. J. (2011) The dynamic modulation of GABA(A) receptor trafficking and its role in regulating the plasticity of inhibitory synapses. Physiol. Rev. 91, 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sassoè-Pognetto M., Panzanelli P., Sieghart W., Fritschy J. M. (2000) Colocalization of multiple GABA(A) receptor subtypes with gephyrin at postsynaptic sites. J. Comp. Neurol. 420, 481–498 [DOI] [PubMed] [Google Scholar]
- 10. Varoqueaux F., Jamain S., Brose N. (2004) Neuroligin 2 is exclusively localized to inhibitory synapses. Eur. J. Cell Biol. 83, 449–456 [DOI] [PubMed] [Google Scholar]
- 11. Poulopoulos A., Aramuni G., Meyer G., Soykan T., Hoon M., Papadopoulos T., Zhang M., Paarmann I., Fuchs C., Harvey K., Jedlicka P., Schwarzacher S. W., Betz H., Harvey R. J., Brose N., Zhang W., Varoqueaux F. (2009) Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron 63, 628–642 [DOI] [PubMed] [Google Scholar]
- 12. Graf E. R., Zhang X., Jin S. X., Linhoff M. W., Craig A. M. (2004) Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dean C., Scholl F. G., Choih J., DeMaria S., Berger J., Isacoff E., Scheiffele P. (2003) Neurexin mediates the assembly of presynaptic terminals. Nat. Neurosci. 6, 708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Sullivan G. A., Hofer W., Betz H. (2009) Inhibitory postsynaptic membrane specializations are formed in gephyrin-deficient mice. Neurosci. Lett. 458, 106–110 [DOI] [PubMed] [Google Scholar]
- 15. Patrizi A., Scelfo B., Viltono L., Briatore F., Fukaya M., Watanabe M., Strata P., Varoqueaux F., Brose N., Fritschy J. M., Sassoè-Pognetto M. (2008) Synapse formation and clustering of neuroligin-2 in the absence of GABAA receptors. Proc. Natl. Acad. Sci. U.S.A. 105, 13151–13156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blundell J., Tabuchi K., Bolliger M. F., Blaiss C. A., Brose N., Liu X., Südhof T. C., Powell C. M. (2009) Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 8, 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wöhr M., Silverman J. L., Scattoni M. L., Turner S. M., Harris M. J., Saxena R., Crawley J. N. (2013) Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav. Brain Res. 251, 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woo J., Kwon S. K., Nam J., Choi S., Takahashi H., Krueger D., Park J., Lee Y., Bae J. Y., Lee D., Ko J., Kim H., Kim M. H., Bae Y. C., Chang S., Craig A. M., Kim E. (2013) The adhesion protein IgSF9b is coupled to neuroligin 2 via S-SCAM to promote inhibitory synapse development. J. Cell Biol. 201, 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pettem K. L., Yokomaku D., Takahashi H., Ge Y., Craig A. M. (2013) Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development. J. Cell Biol. 200, 321–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Völkel P., Le Faou P., Angrand P. O. (2010) Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry. Biochem. Soc. Trans. 38, 883–887 [DOI] [PubMed] [Google Scholar]
- 21. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 22. Angrand P. O., Segura I., Völkel P., Ghidelli S., Terry R., Brajenovic M., Vintersten K., Klein R., Superti-Furga G., Drewes G., Kuster B., Bouwmeester T., Acker-Palmer A. (2006) Transgenic mouse proteomics identifies new 14-3-3-associated proteins involved in cytoskeletal rearrangements and cell signaling. Mol. Cell Proteomics 5, 2211–2227 [DOI] [PubMed] [Google Scholar]
- 23. Fernández E., Collins M. O., Uren R. T., Kopanitsa M. V., Komiyama N. H., Croning M. D., Zografos L., Armstrong J. D., Choudhary J. S., Grant S. G. (2009) Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol. Syst. Biol. 5, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soto F., Bleckert A., Lewis R., Kang Y., Kerschensteiner D., Craig A. M., Wong R. O. (2011) Coordinated increase in inhibitory and excitatory synapses onto retinal ganglion cells during development. Neural. Dev. 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaech S., Banker G. (2006) Culturing hippocampal neurons. Nat. Protoc. 1, 2406–2415 [DOI] [PubMed] [Google Scholar]
- 26. Scheiffele P., Fan J., Choih J., Fetter R., Serafini T. (2000) Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669 [DOI] [PubMed] [Google Scholar]
- 27. Siddiqui T. J., Pancaroglu R., Kang Y., Rooyakkers A., Craig A. M. (2010) LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J. Neurosci. 30, 7495–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Graf E. R., Kang Y., Hauner A. M., Craig A. M. (2006) Structure function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J. Neurosci. 26, 4256–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 30. Chan Q. W., Howes C. G., Foster L. J. (2006) Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell Proteomics 5, 2252–2262 [DOI] [PubMed] [Google Scholar]
- 31. Ishihama Y., Rappsilber J., Andersen J. S., Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 32. Lee H. K., Braynen W., Keshav K., Pavlidis P. (2005) ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics 6, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krueger D. D., Tuffy L. P., Papadopoulos T., Brose N. (2012) The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr. Opin. Neurobiol. 22, 412–422 [DOI] [PubMed] [Google Scholar]
- 34. Feng G., Mellor R. H., Bernstein M., Keller-Peck C., Nguyen Q. T., Wallace M., Nerbonne J. M., Lichtman J. W., Sanes J. R. (2000) Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 [DOI] [PubMed] [Google Scholar]
- 35. Craig A. M., Kang Y. (2007) Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 17, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hines R. M., Wu L., Hines D. J., Steenland H., Mansour S., Dahlhaus R., Singaraja R. R., Cao X., Sammler E., Hormuzdi S. G., Zhuo M., El-Husseini A. (2008) Synaptic imbalance, stereotypies, and impaired social interactions in mice with altered neuroligin 2 expression. J. Neurosci. 28, 6055–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee W. C., Huang H., Feng G., Sanes J. R., Brown E. N., So P. T., Nedivi E. (2006) Dynamic remodeling of dendritic arbors in GABAergic interneurons of adult visual cortex. PLoS Biol. 4, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klausberger T., Somogyi P. (2008) Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321, 53–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fritschy J. M., Panzanelli P., Tyagarajan S. K. (2012) Molecular and functional heterogeneity of GABAergic synapses. Cell Mol. Life Sci. 69, 2485–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. VanAken T., Foxall-VanAken S., Castleman S., Ferguson-Miller S. (1986) Alkyl glycoside detergents: synthesis and applications to the study of membrane proteins. Methods Enzymol. 125, 27–35 [DOI] [PubMed] [Google Scholar]
- 41. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McMahon H. T., Boucrot E. (2011) Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 [DOI] [PubMed] [Google Scholar]
- 43. Simpson J. C., Griffiths G., Wessling-Resnick M., Fransen J. A., Bennett H., Jones A. T. (2004) A role for the small GTPase Rab21 in the early endocytic pathway. J. Cell Sci. 117, 6297–6311 [DOI] [PubMed] [Google Scholar]
- 44. Hasson T. (2003) Myosin VI: two distinct roles in endocytosis. J. Cell Sci. 116, 3453–3461 [DOI] [PubMed] [Google Scholar]
- 45. Seaman M. N. (2012) The retromer complex: endosomal protein recycling and beyond. J. Cell Sci. 125, 4693–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oku S., Takahashi N., Fukata Y., Fukata M. (2013) In silico screening for palmitoyl substrates reveals a role for DHHC1/3/10 (zDHHC1/3/11)-mediated neurochondrin palmitoylation in its targeting to Rab5-positive endosomes. J. Biol. Chem. 288, 19816–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. (2006) Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 10, 839–850 [DOI] [PubMed] [Google Scholar]
- 48. van Weering J. R., Cullen P. J. (2014) Membrane-associated cargo recycling by tubule-based endosomal sorting. Semin. Cell Dev. Biol. 31, 40–47 [DOI] [PubMed] [Google Scholar]
- 49. Choy R. W., Park M., Temkin P., Herring B. E., Marley A., Nicoll R. A., von Zastrow M. (2014) Retromer mediates a discrete route of local membrane delivery to dendrites. Neuron 82, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Budreck E. C., Scheiffele P. (2007) Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur. J. Neurosci. 26, 1738–1748 [DOI] [PubMed] [Google Scholar]
- 51. Poulopoulos A., Soykan T., Tuffy L. P., Hammer M., Varoqueaux F., Brose N. (2012) Homodimerization and isoform-specific heterodimerization of neuroligins. Biochem. J. 446, 321–330 [DOI] [PubMed] [Google Scholar]
- 52. Hoon M., Soykan T., Falkenburger B., Hammer M., Patrizi A., Schmidt K. F., Sassoè-Pognetto M., Löwel S., Moser T., Taschenberger H., Brose N., Varoqueaux F. (2011) Neuroligin-4 is localized to glycinergic postsynapses and regulates inhibition in the retina. Proc. Natl. Acad. Sci. U.S.A. 108, 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Song J. Y., Ichtchenko K., Südhof T. C., Brose N. (1999) Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc. Natl. Acad. Sci. U.S.A. 96, 1100–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tretter V., Mukherjee J., Maric H. M., Schindelin H., Sieghart W., Moss S. J. (2012) Gephyrin, the enigmatic organizer at GABAergic synapses. Front. Cell Neurosci. 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Whiting P. J. (2003) GABA-A receptor subtypes in the brain: a paradigm for CNS drug discovery? Drug Discov. Today 8, 445–450 [DOI] [PubMed] [Google Scholar]
- 56. Hedstrom K. L., Xu X., Ogawa Y., Frischknecht R., Seidenbecher C. I., Shrager P., Rasband M. N. (2007) Neurofascin assembles a specialized extracellular matrix at the axon initial segment. J. Cell Biol. 178, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kriebel M., Metzger J., Trinks S., Chugh D., Harvey R. J., Harvey K., Volkmer H. (2011) The cell adhesion molecule neurofascin stabilizes axo-axonic GABAergic terminals at the axon initial segment. J. Biol. Chem. 286, 24385–24393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Galiano M. R., Jha S., Ho T. S., Zhang C., Ogawa Y., Chang K. J., Stankewich M. C., Mohler P. J., Rasband M. N. (2012) A distal axonal cytoskeleton forms an intra-axonal boundary that controls axon initial segment assembly. Cell 149, 1125–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McRae P. A., Porter B. E. (2012) The perineuronal net component of the extracellular matrix in plasticity and epilepsy. Neurochem. Int. 61, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Saghatelyan A. K., Dityatev A., Schmidt S., Schuster T., Bartsch U., Schachner M. (2001) Reduced perisomatic inhibition, increased excitatory transmission, and impaired long-term potentiation in mice deficient for the extracellular matrix glycoprotein tenascin-R. Mol. Cell Neurosci. 17, 226–240 [DOI] [PubMed] [Google Scholar]
- 61. Nikonenko A., Schmidt S., Skibo G., Brückner G., Schachner M. (2003) Tenascin-R-deficient mice show structural alterations of symmetric perisomatic synapses in the CA1 region of the hippocampus. J. Comp. Neurol. 456, 338–349 [DOI] [PubMed] [Google Scholar]
- 62. Ushkaryov Y. A., Südhof T. C. (1993) Neurexin III α: extensive alternative splicing generates membrane-bound and soluble forms. Proc. Natl. Acad. Sci. U.S.A. 90, 6410–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bot N., Schweizer C., Ben Halima S., Fraering P. C. (2011) Processing of the synaptic cell adhesion molecule neurexin-3β by Alzheimer disease α- and γ-secretases. J. Biol. Chem. 286, 2762–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saura C. A., Servián-Morilla E., Scholl F. G. (2011) Presenilin/γ-secretase regulates neurexin processing at synapses. PLoS One 6, e19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schapitz I. U., Behrend B., Pechmann Y., Lappe-Siefke C., Kneussel S. J., Wallace K. E., Stempel A. V., Buck F., Grant S. G., Schweizer M., Schmitz D., Schwarz J. R., Holzbaur E. L., Kneussel M. (2010) Neuroligin 1 is dynamically exchanged at postsynaptic sites. J. Neurosci. 30, 12733–12744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Peixoto R. T., Kunz P. A., Kwon H., Mabb A. M., Sabatini B. L., Philpot B. D., Ehlers M. D. (2012) Transsynaptic signaling by activity-dependent cleavage of neuroligin-1. Neuron 76, 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Suzuki K., Hayashi Y., Nakahara S., Kumazaki H., Prox J., Horiuchi K., Zeng M., Tanimura S., Nishiyama Y., Osawa S., Sehara-Fujisawa A., Saftig P., Yokoshima S., Fukuyama T., Matsuki N., Koyama R., Tomita T., Iwatsubo T. (2012) Activity-dependent proteolytic cleavage of neuroligin-1. Neuron 76, 410–422 [DOI] [PubMed] [Google Scholar]
- 68. Takahashi H., Katayama K., Sohya K., Miyamoto H., Prasad T., Matsumoto Y., Ota M., Yasuda H., Tsumoto T., Aruga J., Craig A. M. (2012) Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction. Nat. Neurosci. 15, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Knuesel I., Mastrocola M., Zuellig R. A., Bornhauser B., Schaub M. C., Fritschy J. M. (1999) Short communication: altered synaptic clustering of GABAA receptors in mice lacking dystrophin (mdx mice). Eur. J. Neurosci. 11, 4457–4462 [DOI] [PubMed] [Google Scholar]
- 70. Pribiag H., Peng H., Shah W. A., Stellwagen D., Carbonetto S. (2014) Dystroglycan mediates homeostatic synaptic plasticity at GABAergic synapses. Proc. Natl. Acad. Sci. U.S.A. 111, 6810–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Heller E. A., Zhang W., Selimi F., Earnheart J. C., Ślimak M. A., Santos-Torres J., Ibañez-Tallon I., Aoki C., Chait B. T., Heintz N. (2012) The biochemical anatomy of cortical inhibitory synapses. PLoS One 7, e39572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen J. L., Villa K. L., Cha J. W., So P. T., Kubota Y., Nedivi E. (2012) Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron 74, 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kim S. Y., Chung K., Deisseroth K. (2013) Light microscopy mapping of connections in the intact brain. Trends Cogn. Sci. 17, 596–599 [DOI] [PubMed] [Google Scholar]
- 74. Takahashi H., Arstikaitis P., Prasad T., Bartlett T. E., Wang Y. T., Murphy T. H., Craig A. M. (2011) Postsynaptic TrkC and presynaptic PTPsigma function as a bidirectional excitatory synaptic organizing complex. Neuron 69, 287–303 [DOI] [PMC free article] [PubMed] [Google Scholar]