Background: The number of GABA type-A receptors (GABAARs) affects the strength of GABAergic synapses.
Results: The E3 ubiquitin ligase RNF34 binds to and ubiquitinates the γ2 GABAAR subunit, tagging GABAARs for degradation.
Conclusion: RNF34 regulates the number of postsynaptic GABAARs.
Significance: This is the first identification of an E3 ubiquitin ligase involved in GABAAR trafficking.
Keywords: E3 Ubiquitin Ligase, Lysosome, Proteasome, Synapse, Yeast Two-hybrid, GABAA Receptors, RNF34, Interaction, Ubiquitination
Abstract
We have found that the large intracellular loop of the γ2 GABAA receptor (R) subunit (γ2IL) interacts with RNF34 (an E3 ubiquitin ligase), as shown by yeast two-hybrid and in vitro pulldown assays. In brain extracts, RNF34 co-immunoprecipitates with assembled GABAARs. In co-transfected HEK293 cells, RNF34 reduces the expression of the γ2 GABAAR subunit by increasing the ratio of ubiquitinated/nonubiquitinated γ2. Mutating several lysines of the γ2IL into arginines makes the γ2 subunit resistant to RNF34-induced degradation. RNF34 also reduces the expression of the γ2 subunit when α1 and β3 subunits are co-assembled with γ2. This effect is partially reversed by leupeptin or MG132, indicating that both the lysosomal and proteasomal degradation pathways are involved. Immunofluorescence of cultured hippocampal neurons shows that RNF34 forms clusters and that a subset of these clusters is associated with GABAergic synapses. This association is also observed in the intact rat brain by electron microscopy immunocytochemistry. RNF34 is not expressed until the 2nd postnatal week of rat brain development, being highly expressed in some interneurons. Overexpression of RNF34 in hippocampal neurons decreases the density of γ2 GABAAR clusters and the number of GABAergic contacts that these neurons receive. Knocking down endogenous RNF34 with shRNA leads to increased γ2 GABAAR cluster density and GABAergic innervation. The results indicate that RNF34 regulates postsynaptic γ2-GABAAR clustering and GABAergic synaptic innervation by interacting with and ubiquitinating the γ2-GABAAR subunit promoting GABAAR degradation.
Introduction
Ubiquitination is a post-translational protein modification, in which the 76-amino acid (aa)2 polypeptide ubiquitin or polymers of this molecule are conjugated to lysine residue(s) of substrate proteins. Ubiquitination relies on an enzymatic cascade consisting of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), ubiquitin ligases (E3s), and sometimes polyubiquitin ligases (E4s) (1, 2). The E3 ubiquitin ligases (E3 UBLs) are of particular interest because they confer substrate specificity. There are two main classes of E3 UBLs, the RING (really interesting new gene) and HECT (homologous to E6-associated protein C terminus) domain families (3–6). The RING family of E3 UBLs is the largest and contains ∼600 members (7). In this communication, we focus on RNF34, a RING E3 UBL (8, 9).
Protein ubiquitination plays a very important role in regulating synaptic transmission and plasticity, as well as neural development (7, 10–12). Pathological alterations of protein ubiquitination have been associated with some neurological diseases (7, 13). Several RING E3 UBLs regulate glutamatergic synapse assembly and stability by ubiquitinating scaffold proteins and neurotransmitter receptors (7, 11–13). Murine double minute 2 (MDM2) ubiquitinates the postsynaptic density protein 95 (PSD-95) in response to direct stimulation of NMDA receptor (14). Mindbomb2 (Mib2) interacts with and ubiquitinates the GluN2B subunit-containing NMDA receptors (R) in a phosphorylation-dependent manner (15). RNF167 selectively regulates AMPA receptor (R) surface expression and synaptic AMPAR currents (16). The seven in absentia homolog 1A (Siah1A) binds to the C-terminal domains of metabotropic glutamate receptors (mGluR1 and mGluR5) promoting their ubiquitination and proteasome degradation (17, 18).
Regarding GABAergic synapses, the epilepsy-related mutations A322D in the α1 and R177G in the γ2 GABAAR subunits cause misfolding of these subunits, retention in the endoplasmic reticulum (ER) and subsequent ER-associated degradation (ERAD) by the ubiquitin-proteasome system (19, 20). Ubiquitination of GABAARs is also involved in the regulation of the normal function of GABAergic synapses. Thus the β3 GABAAR subunit is ubiquitinated in an activity-dependent manner targeting GABAARs for proteasomal degradation (21). Also ubiquitination of the γ2L GABAAR subunit targets GABAARs for lysosomal degradation (22). Ubiquitination of the β3 and γ2 subunits occur in lysine-rich motifs within the large IL of these subunits (β3IL and γ2LIL), which is localized between TM3 and TM4. To the best of our knowledge, the E3 UBL(s) responsible for the ubiquitination of GABAARs remain to be identified.
In this communication, we show that the E3 UBL RNF34 regulates GABAergic synapse stability and postsynaptic GABAAR clustering by interacting with and ubiquitinating the γ2 GABAAR subunit.
EXPERIMENTAL PROCEDURES
Animals
All the animal handling procedures are in compliance with the regulations of the Institutional Animal Care and Use Committee of the University of Connecticut, and the guidelines from the National Institutes of Health were followed.
Antibodies
A rabbit (Rb) polyclonal antiserum to RNF34 (anti-RNF34) was generated after immunizing with a C-terminal peptide of the rat RNF34 protein (aa 368–381, CRQYVVRAVHVFKS). The antigenic peptide sequence is identical in human, rat, and mouse. The synthetic peptide was covalently coupled to keyhole limpet hemocyanin via an N-terminal cysteine and injected into a New Zealand rabbit in complete Freund's adjuvant for the initial immunization and incomplete Freund's adjuvant for subsequent immunizations. Four months after the first immunization, the antiserum was collected, and the anti-RNF34 antibody was affinity-purified on immobilized antigenic peptide. In all experiments, we used the affinity-purified anti-RNF34. The specificity of the anti-RNF34 antibody was demonstrated by the following. 1) Sequence specificity of the antigenic peptide was determined from Entrez and UniProt protein databases. 2) ELISA showed binding of anti-RNF34 to the antigenic peptide. 3) Immunoblots of homogenates and various subcellular fractions from rat forebrain showed a single 42-kDa protein band corresponding to the molecular weight of RNF34, and the immunoreactivity was displaced by antigenic peptide or purified His-RNF34 (aa 195–381) fusion protein. 4) The anti-RNF34 antibody reacted in immunoblots with purified His-RNF34 (aa 195–381) bacterial fusion protein (40-kDa protein band). 5) The antibody gave a strong RNF34 immunofluorescence signal in HEK293 cells transfected with a pCAGGS-RNF34 construct compared with the low immunofluorescence signal of neighboring nontransfected cells. 6) The anti-RNF34 immunofluorescence of cultured hippocampal neurons was displaceable by antigenic peptide or purified His-RNF34-C (aa 195–381) fusion protein.
The guinea pig (GP) anti-α1 (aa 1–15), Rb anti-α1 (aa 1–15), GP anti-γ2 (aa 1–15), Rb anti-γ2 (aa 1–15), and Rb anti-γ2(1–29) (aa 1–29) antibodies to rat GABAAR subunits were raised and affinity-purified in our laboratory. The mouse (Ms) mAbs to the N terminus of β2/3 GABAAR subunits (clone 62-3G1) and to γ2IL (clone KC4-8A7 that recognizes both γ2sIL and γ2LIL) were also made in our laboratory. The generation, affinity purification, and specificity of these GABAAR antibodies have been described elsewhere (23–43). The sheep anti-GAD was from Dr. Irwin J. Kopin (NINDS, National Institutes of Health, Bethesda, MD). The GP anti-GABA antiserum (catalogue no. AB175), Rb anti-GST (catalogue no. 06-332), and the Ms mAb to actin (clone C4, catalogue no. MAB1501) were from Millipore (Billerica, MA). The Ms mAb anti-HA tag (clone 16B12, catalogue no. MMS-101R) was from Covance (Princeton, NJ). The Ms mAb anti-His tag (clone N114/14, catalogue no. 75-169) and Ms mAb anti-α1 GABAAR subunit (clone N95/35, catalogue no. 75-136) were from NeuroMab (Davis, CA). The Ms mAb anti-ubiquitin (clone FK2, catalogue no. BML-PW8810) was from Enzo Life Sciences (Farmingdale, NY).
For the immunoblots in Fig. 6A only, the secondary goat anti-rabbit IgG antibody and rabbit peroxidase-antiperoxidase complexes were from MP Biomedicals (Aurora, OH). For the other immunoblots, IRDye 800CW-conjugated goat anti-rabbit or anti-mouse IgG and IRDye 680LT-conjugated goat anti-mouse IgG secondary antibodies were from Li-Cor Biosciences (Lincoln, NE).
FIGURE 6.
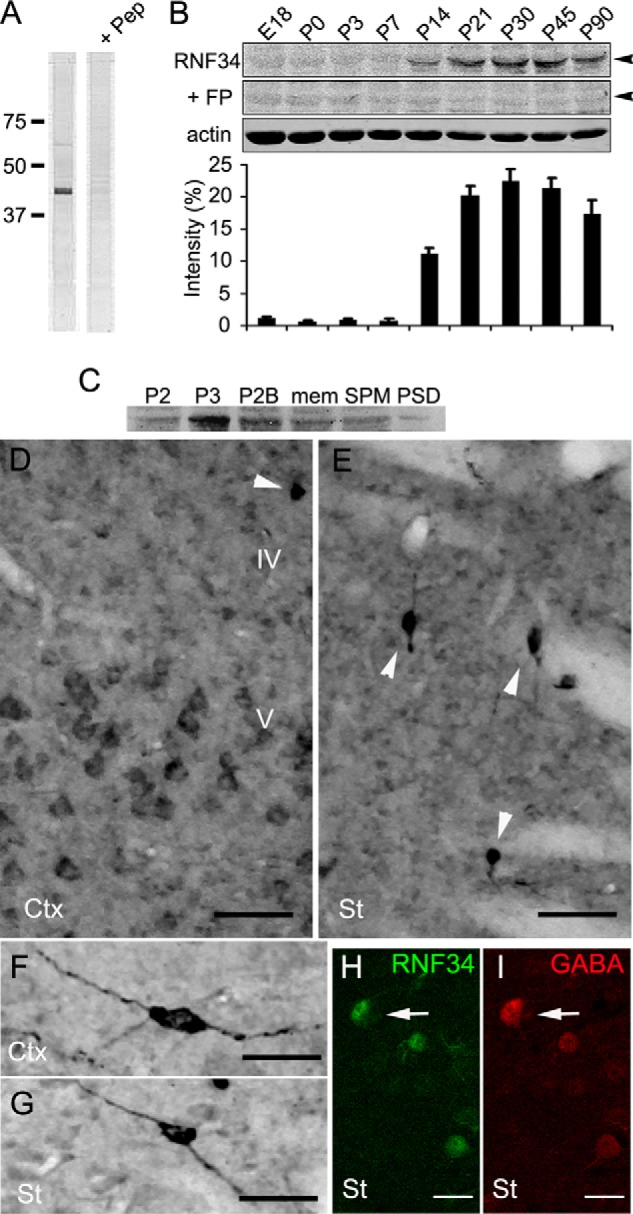
Expression and distribution of RNF34 in the rat brain. A, immunoblot with the Rb anti-RNF34 antibody of the rat forebrain crude synaptosomal (P2) fraction, shows a major protein band ∼42 kDa, which is displaced by 100 μg/ml of the antigenic peptide (+ Pep). 20 μg of the total proteins were loaded into each lane. B, immunoblots of rat forebrain homogenates of various ages from embryonic day 18 (E18) to postnatal day 90 (P90) with Rb anti-RNF34 and Ms anti-actin antibodies. 75 μg of total protein were loaded into each lane. The RNF34 antibody recognizes a 42-kDa protein band (arrowhead), whose immunoreactivity is displaced by incubating the antibody with 100 μg/ml of the purified bacterial His-RNF34 (aa 195–381) fusion protein (+ FP). The bar graph shows the quantification of the relative RNF34 protein expression levels by densitometry of the RNF34 protein band normalized to actin protein band density. C, immunoblot of various rat forebrain subcellular fractions with Rb anti-RNF34 antibody. The RNF34 protein band is present in all the fractions tested, including crude synaptosomal (P2), microsomal (P3), synaptosomal (P2B), total membrane (mem), SPM, and one Triton PSD fractions. 25 μg of the total proteins were loaded into each lane. D and E, RNF34 immunocytochemistry in layers IV and V of the cerebral cortex (Ctx) and corpus striatum (St). Arrowheads indicate neurons with high RNF34 immunoreactivity. F and G, individual neurons with high level of RNF34 immunoreactivity in the cerebral cortex and striatum, respectively. H and I, double label fluorescence confocal microscopy of corpus striatum from rat brain sections immunolabeled with anti-RNF34 (green in H) and anti-GABA (red in I). GABA+ interneurons show a high expression level of RNF34 protein (arrow). Scale bar, 50 μm (D and E), 25 μm (F and G), and 20 μm (H and I).
For immunofluorescence in cell cultures, Texas red-, Alexa Fluor 594- (Fig. 9, only), FITC-, and aminomethylcoumarin-conjugated species-specific anti-IgG secondary antibodies were raised in donkey (Jackson ImmunoResearch, West Grove, PA). For light microscopy immunocytochemistry, biotin-conjugated goat anti-Rb IgG secondary antibody and avidin-biotin-horseradish peroxidase complex were from Vector Laboratories (Burlingame, CA). For confocal microscopy of brain sections, Alexa Fluor 488- and 568 conjugated species-specific anti-IgG secondary antibodies were raised in goat (Invitrogen). For EM, the colloidal gold-labeled goat anti-mouse IgG (10 nm diameter) secondary antibody was from ICN (Irvine, CA), and the colloidal gold-labeled goat anti-Rb IgG (18 nm diameter) secondary antibody was from Jackson ImmunoResearch.
FIGURE 9.
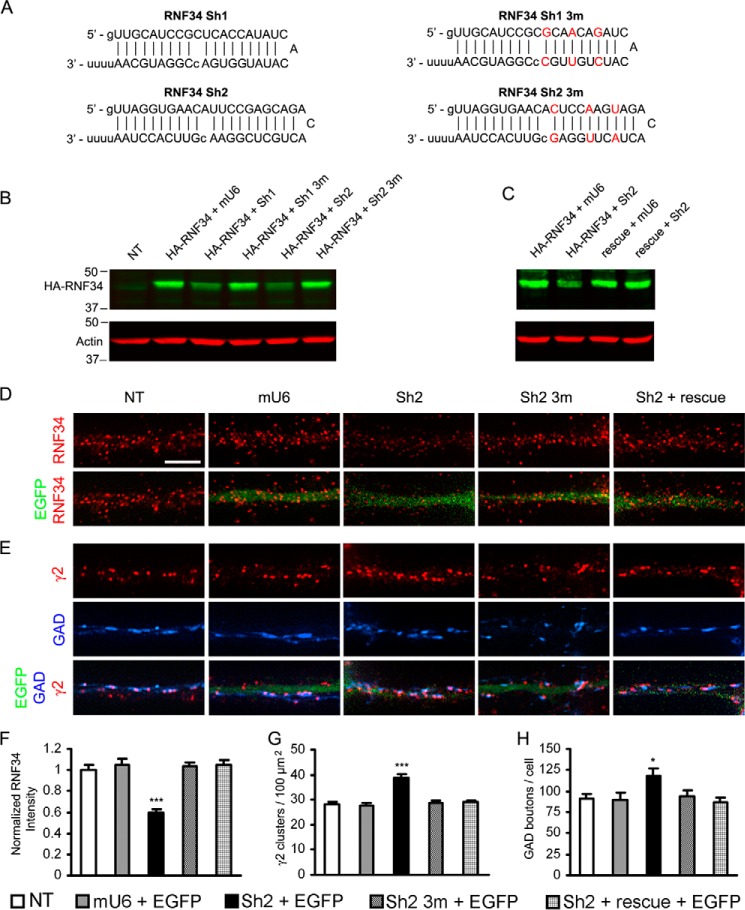
Knocking down endogenous RNF34 in cultured hippocampal neurons increases both the γ2-GABAAR cluster density and GABAergic innervation. A, RNF34 shRNAs (Sh1 and Sh2) used in this study. The three point mutations in Sh1 3m and Sh2 3m are shown in red. B, Sh1 and Sh2 significantly knocked down the protein expression of HA-RNF34 (in pRK5 plasmid), compared with HEK293 cells co-transfected with HA-RNF34 and various control plasmids (mU6, Sh1 3m, or Sh2 3m). Cell lysates were immunoblotted with Ms anti-HA and Ms anti-actin mAbs. C, Sh2 did not knock down the protein expression of the rescue mRNA, although Sh2 knocked down the protein expression of the HA-RNF34. D and E, representative immunofluorescence images of dendrites from hippocampal neurons that were nontransfected (NT) or co-transfected with EGFP and mU6 pro vector (mU6), or various Sh2 constructs (Sh2 or Sh2 3m), or Sh2 + rescue. Immunofluorescence with Rb anti-RNF34 (red in D), Rb anti-γ2 (red in E) or sheep anti-GAD (blue in E). Dendrites of transfected neurons were identified by EGFP fluorescence (green). Scale bar, 5 μm. F–H, quantification of the effect of knocking down RNF34 with Sh2 on the RNF34 fluorescence intensity (F), γ2-GABAAR cluster density (G), and GAD boutons contacting the transfected neurons (H). *, p < 0.05; ***, p < 0.001 in one-way ANOVA Tukey-Kramer multiple comparison test, n = 16 neurons for each group from four independent transfection experiments.
Yeast Two-hybrid (Y2H)
The detailed experimental procedure for Y2H has been described elsewhere (30, 31, 38). All vectors and yeast strains were from Dr. Roger Brent (University of California at San Francisco) or Origene Technologies (Rockville, MD). For the initial screening of a rat brain cDNA library, we used as bait the large IL between TM3 and TM4 of the γ2 short GABAAR subunit (γ2sIL, aa 318–404 of GenBankTM NP_899156). The γ2sIL with an added stop codon at the C terminus was inserted into pEG202 and expressed as a fusion protein with the DNA-binding protein LexA at its N terminus. The same procedure for bait construction was followed for α1IL, β1IL, β3IL, γ1IL, γ2LIL, γ3IL, and GluA3C, and various deletion constructs of the γ2sIL. The pSH17-4 plasmid, which contains the LexA DNA binding domain, was used as the positive control. The pRHFM1 plasmid, which contains the bicoid protein bait, or the empty bait plasmid pEG202 was used as negative controls. To map the RNF34-binding site for the γ2sIL, the full-length RNF34 and various truncated mutants with an added stop codon at the C terminus were subcloned into pJG4-5 vector. All the cloned DNAs were verified by DNA sequencing, and the protein expression for each of the constructs in yeast Saccharomyces cerevisiae EGY48 was confirmed by immunoblotting of the cell lysate with Ms anti-LexA or anti-HA mAbs.
Cloning of Full-length Rat RNF34 cDNA and Preparation of HA-tagged RNF34 Constructs
The Marathon-ready rat brain cDNA library (Clontech), containing full-length cDNA clones, was used as template in a PCR to clone the cDNA of rat RNF34 (GenBankTM NM_001004517). Two 26-base antisense primers recognizing two sequences in the 3′-UTR of RNF34 were used in two sequential PCRs: a 5′-rapid amplification of cDNA ends PCR using the 5′ library adaptor primer AP1 and a 3′-UTR RNF34 antisense primer (5′-GGCGAGTGGCCTCCACACCCACACTG-3′) followed by a nested rapid amplification of cDNA ends PCR using the 5′ library adaptor primer AP2 and a second 3′-UTR RNF34 antisense primer (5′-AGCCATGTGTCCACGGTTGACGGGCC-3′). A 1.5-kb DNA fragment containing the 5′-UTR, complete coding sequence, and partial 3′-UTR of RNF34 was generated, purified, cloned into pCR-XL-TOPO vector (Invitrogen), and sequenced. The RNF34 coding region (encoding aa 1–381) was amplified by PCR and subcloned into pCAGGS vector and pCAGGS-HA containing the HA tag at the N terminus. A similar procedure was followed for generating pCAGGS-HA-RNF34 ΔC. The pCAGGS-HA-RNF34 H351A (histidine to alanine) point mutant was generated by using pCAGGS-HA-RNF34 as template with the GeneTailor site-directed mutagenesis kit (Invitrogen), following the instructions of the manufacturer.
Generation of γ2 KR Mutants
The γ2s 7KR in pcDNA3.1 was derived from the GFP-γ2L K7R plasmid generously provided by Drs. Josef T. Kittler and I. Lorena Arancibia-Cárcamo from University College London, UK. The GFP-γ2L K7R plasmid had seven lysines (Lys-325, Lys-328, Lys-330, Lys-332, Lys-333, Lys-334, and Lys-335) of γ2L mutated into arginines (22). The DNA fragment that covers the seven mutations in the GFP-γ2L K7R plasmid was amplified by PCR using a pair of primers (sense primer, 5′-TCTGTTTGCTTCATCTTTGTGTTTTC-3′, and antisense primer, 5′-GTAGGGGCAGGGTTTCTCCTTCTTCT-3′). The γ2s 7KR was then generated using the two strands of the PCR-generated fragment as primers and γ2s WT-pcDNA3.1 as the template using the GeneTailor site-directed mutagenesis kit. Subsequently, γ2s 8KR (7KR + K373R), γ2s 9KR (8KR + K401R), and γ2s 10KR (9KR + K259R) mutants in pcDNA3.1 were generated with the GeneTailor site-directed mutagenesis kit.
shRNAs for RNF34 and Rescue mRNA
The procedure for making the shRNAs has been described elsewhere (34, 36, 38, 40). Two shRNA constructs targeting RNF34 were generated (Fig. 9A). The Sh1 shRNA targeted a sequence in the coding region (nucleotides 1049–1067, GenBankTM NM_001004075), whereas the Sh2 shRNA targeted the 3′-UTR (nucleotides 1623–1643) of rat RNF34. Each shRNA has an antisense strand that perfectly matches the target mRNA, a loop, and a sense strand that contains a mismatch in the middle to facilitate DNA sequencing (Fig. 9A). A pair of complementary DNA oligonucleotides encoding both arms of the shRNA were synthesized, annealed, and inserted into the mU6pro vector at BbsI and XbaI restriction sites. The corresponding DNA oligonucleotides that carry three point mutations were also made as control shRNAs (Fig. 9A).
To make the rescue mRNA for Sh2, the HA tag and the complete protein coding region of RNF34 corresponding to aa 1–381 were amplified by PCR using pCAGGS-HA-RNF34 as the template, and subcloned into pRK5 vector (rescue). This construct does not have the 3′-UTR, and therefore, the mRNA was not targeted by the Sh2 shRNA. We also subcloned it into pRK5, the HA-RNF34 that contains both the protein coding region and the 3′-UTR, with a similar procedure. The RNF34 constructs that were subcloned in the pRK5 vector and the shRNAs were used for the knockdown experiments in HEK293 cells or hippocampal neurons as shown in Fig. 9.
Fusion Protein Expression and Purification
The cDNAs corresponding to RNF34-C (aa 195–381) and RNF34 full-length (FL, aa 1–381) were subcloned into pET32a+ vector, which contains thioredoxin, His6, and S tags at the N terminus (∼18 kDa). RNF34-C* (aa 195–381, H351A) was generated from RNF34-C (aa 195–381) using the GeneTailor site-directed mutagenesis Kit. Escherichia coli BL 21 (DE3) cells were transformed, and the expression of His-tagged RNF34 fusion proteins was induced by isopropyl β-d-1-thiogalactopyranoside. Fusion proteins were purified from bacterial lysates with His60 Ni Superflow Resin, according to the manufacturer's instructions (Clontech). The cloning, expression, and purification of GST or GST-tagged γ2sIL (GST-γ2sIL) fusion protein have been described elsewhere (25).
In Vitro Pulldown Assay
Equal moles of purified His (8.6 μg), His-RNF34-C (aa 195–381, 19.3 μg), or His-RNF34-C* (aa 195–381, H351A, 19.3 μg) fusion proteins, were adsorbed to a 50-μl bed volume of His60Ni beads and incubated with purified GST (13 μg) or GST-γ2sIL (18 μg) in equilibration buffer (140 mm NaCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.5) with a protease inhibitor mixture (Roche Applied Science; catalogue no. 1697498) overnight at 4 °C. For the pulldown with His-RNF34 FL, due to the very low yield of the expression and purification of the bacterial His-RNF34 FL fusion protein, 2 ml of the His-RNF34 FL bacterial lysate were adsorbed to a 50-μl bed volume of His60Ni beads, followed by six washes and incubation with purified GST (13 μg) or GST-γ2sIL (18 μg). After six washes by centrifugation, the bound proteins were eluted from the beads at 4 °C with 250 mm imidazole in equilibration buffer. Eluates were analyzed by SDS-PAGE followed by immunoblotting with Rb anti-GST, Ms anti-γ2IL, and Ms anti-His antibodies.
In Vitro Ubiquitination Assay
The assay was carried out in a 50-μl reaction mixture containing 1× ubiquitination buffer and 1× Mg-ATP (catalogue no. SK-10), 315 ng of UBE1 (catalogue no. E-305), 650 ng of UbcH5a (catalogue no. E2–616), 10 μg of HA-ubiquitin (Ub, catalogue no. U-110) from Boston Biochem (Cambridge, MA), 2 mm DTT, 820 ng of purified GST-γ2IL, and 7.5 ng of purified His-RNF34 FL. In control reactions, purified GST-γ2IL or His-RNF34 FL was omitted, or 600 ng of GST was used in place of GST-γ2IL. The reaction mixture was incubated in a water bath at 30 °C for 2 h and then analyzed by SDS-PAGE followed by immunoblotting with Ms anti-Ub, Ms anti-γ2IL, or Rb anti-GST antibodies.
Preparation of Rat Forebrain Homogenates and Subcellular Fractionation
The preparation of rat forebrain homogenates and various subcellular fractions, including crude synaptosomal (P2), synaptosomal (P2B), microsomal (P3), synaptic plasma membrane (SPM), and “one Triton” postsynaptic density (PSD) fractions, has been described elsewhere (35, 37). For membrane preparation, the procedure has been described previously (37). Briefly, the forebrain (telencephalon) tissue of adult female Sprague-Dawley rats was homogenized with 10% sucrose in 50 mm Tris-HCl, pH 7.4 buffer, and centrifuged for 5 min at 1,000 × g. The supernatant was centrifuged at 100,000 × g for 1 h, and the pellet was suspended in 5 mm Tris-HCl, pH 7.4 buffer, and then subjected to homogenization in an all-glass Dounce homogenizer. The lysate was centrifuged at 12,000 × g for 30 min, and the pellet was suspended in 50 mm Tris-HCl, pH 7.4 buffer. All steps for preparation of brain homogenates and fractions were carried out at 4 °C. The samples were analyzed by SDS-PAGE, followed by immunoblotting with Rb anti-RNF34 antibody and corresponding secondary antibody.
Immunoblotting
For Fig. 6A only, immunoblots were done according to the method described elsewhere (44). For the other figures and panels, the immunoblot images were collected with a LI-COR Odyssey Infrared Imaging System (Li-Cor Biosciences), analyzed, and quantified with Odyssey software, version 3.0.
Co-immunoprecipitation of RNF34 and GABAARs from Brain Membrane Extracts
The experimental procedure has been described elsewhere (38). Briefly, the rat forebrain membrane fraction was incubated with RIPA buffer (10 mm Tris-HCl, 137 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.1% SDS, 1% Triton-X 100, 1% sodium deoxycholate, pH 7.4) containing 1 mm PMSF and a protease inhibitor mixture (10 μg/ml trypsin inhibitor, type I-S, 10 μg/ml trypsin inhibitor, type II-O, 1 mm benzamidine) for 1 h followed by centrifugation at 50,000 × g for 1 h. The supernatant was used for immunoprecipitations. 50 μl of protein A-Sepharose beads (GE Healthcare), suspended in 500 μl of 50 mm Tris-HCl, pH 7.4, were incubated with 50 μl of nonimmune GP serum, GP anti-γ2, GP anti-α1, preimmune (to α1) Rb serum, Rb anti-γ2, Rb anti-γ2(1–29), or Rb anti-α1 GABAAR subunit antiserum at 4 °C overnight, followed by cross-linking with disuccinimidyl suberate (Thermo Scientific, Rockford, IL). After washing with RIPA buffer, the beads were incubated with 1.l ml of the rat brain detergent extract (4 mg of protein) at 4 °C overnight and centrifuged, and the pellet was washed with RIPA buffer four times. The beads were incubated with 100 μl of 2× sample dissociation buffer (0.01 m Tris-HCl, pH 6.8, 20% glycerol, 10% β-mercaptoethanol, 2.3% SDS, 0.005% bromphenol blue) for 20 min at room temperature followed by centrifugation. The supernatant was collected and subjected to SDS-PAGE and immunoblotting with antibodies to RNF34, γ2, or α1 GABAAR subunits.
Cell Culture, Transfections, and Immunoprecipitation of Transfected Cell Extracts
Hippocampal (HP) neuronal cultures were prepared according to Higgins and Banker (45), as described elsewhere (26, 29, 46). Briefly, dissociated neurons from embryonic day 18 (E18) Sprague-Dawley rat hippocampi were plated at low density (3,000–8,000 cells per 18-mm diameter coverslip) and maintained in rat glial cell conditioned medium for up to 3 weeks in vitro, followed by immunofluorescence.
For the overexpression of RNF34 constructs in hippocampal neurons, 11 days in vitro (11 DIV) HP neurons were transfected with pCAGGS-EGFP and pCAGGS-HA, or pCAGGS-HA-RNF34, or pCAGGS-HA-RNF34 ΔC, or pCAGGS-HA-RNF34 H351A constructs (2 μg total plasmids per coverslip). Two days after transfection, HP neurons were subjected to immunofluorescence. For the RNF34 knockdown experiment by shRNAs, 10 DIV HP neurons were transfected with pEGFP-N1 (0.6 μg) and mU6 (1.5 μg), or Sh2 (1.5 μg), or Sh2 3m (1.5 μg) per coverslip. For the rescue, an additional 0.65 μg of rescue HA-RNF34 in pRK5 was co-transfected per coverslip. Eleven days after transfection, DIV21 HP neurons were subjected to immunofluorescence.
HEK293 cells were cultured on either the 100 × 20-mm plastic dishes or poly-l-lysine-coated coverslips of 18 mm in diameter in DMEM (Invitrogen) with 10% FBS (Invitrogen). For immunoblotting analysis, the γ2s (WT or Lys-Arg mutants) alone (4 μg of plasmid) or a combination of α1β3γ2s (4 μg of each) GABAAR subunits together with 1 μg of Ub (47) and different RNF34 constructs (13–17 μg in a receptor subunit/RNF34 = 1:5 molar ratio) were transfected into HEK293 cells cultured on 100 × 20-mm plastic dishes. Three days after transfection, cells were harvested and extracted with RIPA containing protease inhibitors. After centrifugation, the supernatants were either subjected to SDS-PAGE and immunoblotting or used for immunoprecipitation experiments. For the latter, the RIPA extracts were precipitated with protein A-Sepharose beads that had been previously incubated with and cross-linked to the Rb anti-γ2(1–29) antiserum (or preimmune serum as control). After four washes with RIPA, proteins were eluted from the beads by incubation with 100 μl of 2× sample dissociation buffer for 20 min at room temperature, followed by centrifugation. The supernatants were then subjected to SDS-PAGE in sister gels and immunoblotting with Ms anti-Ub and Ms anti-γ2IL.
For immunofluorescence experiments in Fig. 4, D and E, HEK293 cells were co-transfected with 2 μg of total plasmids (pCAGGS-RNF34 + EGFP-Rab5 or EGFP-Rab7) per coverslip for protein expression. Twenty four hours after transfection, HEK293 cells were subjected to immunofluorescence procedures.
FIGURE 4.
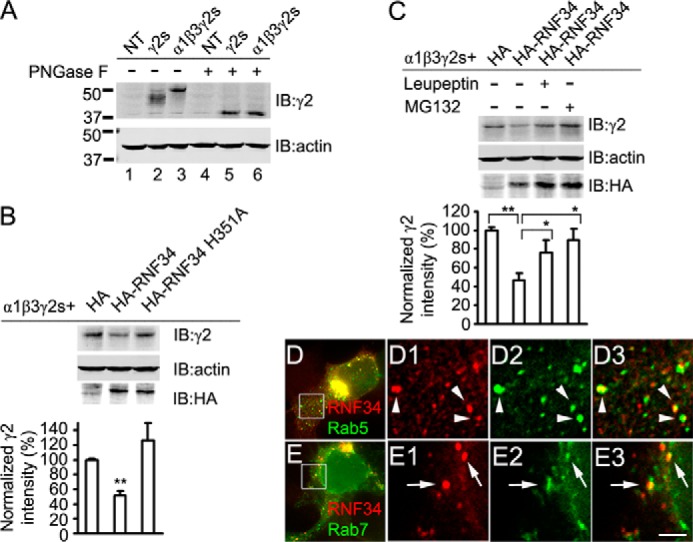
RNF34 induces the degradation of α1β3γ2s GABAARs via the lysosome and proteasome pathways. A, γ2s subunit, when in HEK293 cells, is co-expressed with α1 and β3 subunits (α1β3γ2s, lane 3), has slower mobility than when γ2s is expressed alone (γ2s, lane 2), as shown by immunoblotting with Rb anti-γ2 antibody. NT represents nontransfected HEK293 cells. After digestion with peptide-N-glycosidase F (PNGase F), γ2s showed faster mobility that was identical in cells transfected with γ2s alone or with α1β3γ2s (lanes 5 and 6, respectively). B, HA-RNF34, but not HA-RNF34 H351A, significantly reduces the expression levels of 50-kDa γ2s in the α1β3γ2s recombinant receptors. Data are presented as mean ± S.E., n = 8 independent experiments; **, p < 0.01 in one-way ANOVA Tukey-Kramer multiple comparison test. C, leupeptin or MG132 partially reverse the effect of HA-RNF34 on the expression of the recombinant γ2s. HEK293 cells were co-transfected with α1, β3, γ2s subunit, ubiquitin, and a control vector pCAGGS-HA (HA) or HA-RNF34. After 3 days, cells were lysed and immunoblotted with Rb anti-γ2, Ms anti-actin, and Ms anti-HA antibodies. To the indicated cultures (+), leupeptin or MG132 was added 12 h before harvesting. The bar graphs below B and C show the expression level of γ2 subunits (fluorescence intensity) normalized for that of actin and that of the HA control. Data are presented as mean ± S.E., n = 8 independent experiments; *, p < 0.05; **, p < 0.01 in one-way ANOVA Tukey-Kramer multiple comparison test. D, HEK293 cells were co-transfected with RNF34 and the early endosome marker EGFP-Rab5, followed by immunofluorescence with anti-RNF34 and EGFP fluorescence. D1–D3, high magnification images of the boxed area in D. RNF34 (D1, red) frequently co-localizes with EGFP-Rab5 (D2, green) as shown in the overlay (D3, arrowheads). E, HEK293 cells were co-transfected with RNF34 and the late endosome marker EGFP-Rab7. E1–E3, high magnification images of the boxed area in E. RNF34 (E1, red) frequently co-localizes with EGFP-Rab7 (E2, green) as shown in the overlay (E3, arrows). Scale bar, 10 μm (D and E), 2.5 μm (D1–D3 and E1–E3).
HP neurons and HEK293 cells (see below) were transfected with various plasmids using the CalPhos mammalian transfection kit (Clontech), following the instructions of the manufacturer.
Immunofluorescence of Cell Cultures
Immunofluorescence of HP neurons and transfected HEK293 cells was done as described elsewhere (26, 36–38, 43). Briefly, cells on glass coverslips were fixed in 4% paraformaldehyde, 4% sucrose in PBS for 15 min, followed by incubation with 50 mm NH4Cl in PBS for 10–15 min to quench the free aldehyde groups. Permeabilization was done by incubation with 0.25% Triton X-100 in PBS for 5 min, followed by incubation with 5% donkey normal serum in PBS for 30 min. The coverslips were then incubated overnight at 4 °C with a mixture of primary antibodies from different species in 0.25% Triton X-100/PBS, followed by incubation with a mixture of Texas red or Alexa Fluor 594 (in Fig. 9, only)-, FITC-, or aminomethylcoumarin fluorophore-conjugated species-specific anti-IgG secondary antibodies raised in donkey in 0.25% Triton X-100/PBS at room temperature for 1 h. The coverslips were then washed with PBS twice and mounted on glass slides with Prolong Gold antifade mounting solution (Invitrogen).
Light Microscopy Immunocytochemistry of Rat Brain Sections
This procedure has been described elsewhere (30–32, 36, 37, 39, 48). Briefly, a postnatal day 83 (P83) adult Sprague-Dawley rat was anesthetized with 60 mg/kg ketamine hydrochloride, 8 mg/kg xylazine, and 2 mg/kg acepromazine maleate. The anesthetized rats were perfused through the ascending aorta with 0.1 m phosphate buffer (PB), pH 7.4, and paraformaldehyde/lysine/periodate (PLP) fixative (4% w/v paraformaldehyde, 1.37% w/v lysine and 0.21% w/v sodium periodate in 0.1 m PB, pH 7.4). Brains were cryoprotected, frozen, and sectioned into 25-μm thick sagittal sections with a freezing microtome. After incubating with Rb anti-RNF34 antibody in 0.3% Triton X-100 in 0.1 m PB at 4 °C overnight, the free-floating brain sections were incubated with biotin-labeled anti-Rb IgG secondary antibodies followed by the avidin-biotin-horseradish peroxidase complex (ABC procedure, Vectastain Elite ABC kit, Vector Laboratories). The reaction product was visualized by incubating the sections with 3,3′-diaminobenzidine tetrahydrochloride in the presence of cobalt chloride, nickel ammonium sulfate, and H2O2. Sections were then washed and mounted onto gelatin-coated glass slides.
Immunofluorescence of Rat Brain Sections
The procedure has been described elsewhere (36, 37). Briefly, 25-μm-thick brain slices (prepared as described above) from a P90 Sprague-Dawley rat were subjected to an antigen retrieval procedure by incubation with pepsin (0.15 mg/ml in 0.2 n HCl) at 37 °C for 5 min to facilitate the exposure of the tissue antigens to the antibodies. The pepsin treatment did not affect the localization of RNF34 or the other protein markers tested in the brain when compared with nontreated sections. Brain sections were incubated with 5% normal goat serum, 0.3% Triton X-100 in 0.1 m PB for 1 h at room temperature, followed by incubation with a mixture of Rb anti-RNF34 and GP anti-GABA in 2% normal goat serum, 0.3% Triton X-100 in 0.1 m PB at 4 °C for 48 h. The brain sections were then incubated with a mixture of Alexa Fluor 488-conjugated anti-Rb and Alexa Fluor 568-conjugated anti-GP IgG secondary antibodies raised in goat dissolved in 2% normal goat serum, 0.3% Triton X-100 in 0.1 m PB at room temperature for 1 h. After washes with 0.1 m PB, the sections were mounted on gelatin-coated glass slides with Prolong Gold anti-fade mounting solution.
Postembedding EM Immunogold
The Lowicryl-embedded tissue blocks were kindly provided by Drs. Peter Somogyi and Zoltan Nusser, and the preparation of the blocks was described elsewhere (49). The tissue blocks were sectioned in our laboratory, and the ultrathin sections were subjected to the immunogold labeling procedure as described elsewhere (28, 33, 36, 37, 39). The tissue sections were incubated first with a mixture of affinity-purified Rb anti-RNF34 and Ms mAb to β2/3 GABAAR subunits, followed by incubation with goat anti-rabbit and goat anti-mouse IgG secondary antibodies labeled with colloidal gold particles of 18 and 10 nm in diameter, respectively. The tissue sections were counterstained with 2% uranyl acetate and then with 2% lead citrate. No immunolabeling was observed when the primary antibody was omitted. All sections were visualized with a Tecnai Biotwin 12-kV transmission electron microscope (FEI Corp., Eindhoven, Holland). EM images were stored in Adobe Photoshop 7.0, and contrast/brightness was adjusted.
Image Acquisition, Analysis, and Quantification
Fluorescence images of cultured HP neurons and HEK293 cells were collected using a Nikon Plan Apo 60×/1.40 objective on a Nikon Eclipse T300 microscope with a Photometrics CoolSnap HQ2 CCD camera, driven by IPLab 4.0 acquisition software (Scanalytics, Rockville, MD). For rat brain sections, fluorescence images were acquired on a Leica TCS SP2 laser confocal microscope using a HCX PL Apo 40×/1.25 oil CS objective lens and a pinhole set at 1 Airy unit. Images of single optical sections (0.5 μm thick) were collected.
For qualitative analysis, images were processed with Photoshop version 7.0 (Adobe, San Jose, CA) by adjusting brightness and contrast. For the quantification of the percentage of GABAergic synapses that have associated endogenous RNF34 clusters, 30 cultured HP pyramidal neurons from three independent experiments (15 synapses per neuron, 10 neurons per experiment) were randomly selected for analysis. Thus, a total of 450 GABAergic synapses defined as GAD+ and γ2+ were analyzed. To quantify the association of nonsynaptic γ2 clusters (GAD− and γ2+) with RNF34 clusters, a total of 635 nonsynaptic γ2 clusters were analyzed from 15 neurons (50 μm2 area per dendritic field, three dendritic fields per neuron, five neurons per experiment, and three independent experiments).
In the RNF34 overexpression or knockdown experiments, γ2 cluster density was analyzed from 16 randomly selected nontransfected or transfected neurons (50 μm2 area per dendritic field, six dendritic fields per neuron, four neurons per experiment, and four independent experiments). The number of GAD+ boutons per cell was counted in 16 randomly selected nontransfected or transfected neurons (four neurons per experiment and four independent experiments). The RNF34 fluorescence intensity in dendrites was analyzed from 16 neurons using the ImageJ software (National Institutes of Health, Bethesda, MD). For each neuron, the dendritic fluorescence intensity was calculated by averaging the fluorescence intensity of 5 dendritic fields (50 μm2 area per dendritic field). The background fluorescence (to set the zero immunofluorescence intensity value) was calculated by determining the average fluorescence intensity in five randomly chosen areas of the culture devoid of neurons and dendrites. The average RNF34 fluorescence intensity of neurons co-transfected with shRNA constructs (four neurons per experiment and four independent experiments) was normalized to that of nontransfected sister neurons from the same cultures (100% = 1.0). For RNF34 overexpression experiment, the number of counted γ2 clusters for each plasmid combination or nontransfected neurons was 312–580, and the number of counted GAD+ boutons was 616–987. In the RNF34 knockdown experiment, for each plasmid combination or nontransfected neurons, the total number of γ2 clusters was 1724–2189 and GAD+ boutons was 1393–1892. All values are given as means ± S.E. One-way ANOVA Tukey-Kramer multiple comparison test was used for statistical analysis.
RESULTS
Y2H and in Vitro Pulldown Show a Direct Interaction between RNF34 and the Large Intracellular Loop of the γ2s and γ2L GABAAR Subunits
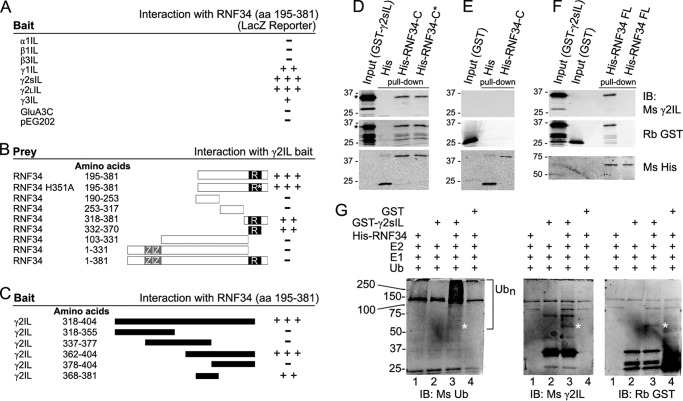
Clone GS29 was isolated from a rat brain cDNA library in a Y2H assay using the large IL of the γ2 GABAAR subunit short isoform (γ2sIL) as bait. Clone GS29 encoded approximately the C-terminal half of the E3 UBL RNF34, including the following: 1) a 561-bp open reading frame encoding the 187-aa C-terminal polypeptide of the RNF34 (aa 195–381); 2) the stop codon; 3) a 688-bp 3′-untranslated region (UTR); and 4) a poly(A) tail. Fig. 1A shows that in addition to γ2sIL, RNF34 (aa 195–381) also interacted with the ILs of the γ2L (γ2LIL), γ1 (γ1IL), and γ3 (γ3IL) GABAAR subunits. The amino acid sequences of the ILs of the γ subunits are highly conserved. However, RNF34 (aa 195–381) did not interact with the ILs of α1 (α1IL), β1 (β1IL), or β3 (β3IL) GABAAR subunits or the cytoplasmic 50-amino acid C terminus of the GluA3 AMPA receptor subunit (GluA3C) or the empty vector pEG202 (Fig. 1A).
FIGURE 1.
Characterization of the interaction between γ2IL and RNF34 and in vitro ubiquitination of γ2IL by RNF34. A–C, yeast two-hybrid. A, large intracellular loops (ILs) from various GABAA receptor subunits were used as bait to test for interaction with GS29, corresponding to RNF34 (aa 195–381). B, various fragments of RNF34 cDNA were tested as prey using γ2sIL as bait to determine the RNF34 domain that interacts with γ2sIL. ZZ and R represent the two zinc finger domains and the RING finger domain, respectively. The asterisk indicates the position of the H351A point mutation in the RING finger domain. C, various fragments of the γ2sIL were used as baits to map the domain that interacts with RNF34. D–F, in vitro pulldown of purified bacterial fusion proteins. D, GST-γ2sIL (35 kDa) was pulled down by immobilized His-RNF34-C (aa 195–381, 40 kDa) and His-RNF34-C* (aa 195–381, H351A, 40 kDa) but not by His (18 kDa). E, GST (27 kDa) was not pulled down by immobilized His-RNF34-C or His. F, GST-γ2sIL but not GST was pulled down by immobilized His-RNF34 FL (aa 1–381, 60 kDa). The GST-γ2sIL fusion protein reacted with both Ms anti-γ2IL and Rb anti-GST antibodies, whereas GST reacted with Rb anti-GST but not with Ms anti-γ2IL. His and His-tagged RNF34 fusion proteins were revealed with Ms anti-His. G, bacterial His-RNF34 fusion protein ubiquitinates GST-γ2sIL in an in vitro assay. Left immunoblot, smear corresponding to polyubiquitinated GST-γ2sIL (>55 kDa, Ubn) is observed when both His-RNF34 and GST-γ2sIL are present in the ubiquitination reaction (lane 3), as shown by immunoblotting (IB) with an anti-ubiquitin antibody (Ms Ub). No polyubiquitination smear is observed when GST is the substrate (lane 4). The smear is particularly strong at >100 kDa. Under the same reaction conditions, there is also an ∼55-kDa protein band (marked by *) corresponding to ubiquitinated GST-γ2sIL that reacted with Ms anti-Ub, Ms anti-γ2sIL, and Rb anti-GST in the three immunoblots (lane 3).
Further analysis showed that the RING domain of RNF34 (aa 332–370) was both necessary and largely sufficient for the interaction with γ2sIL (Fig. 1B), although the interaction was not as strong as that of RNF34 (aa 195–381). The H351A point mutation in the RING domain, which is known to inactivate the RNF34 ubiquitin ligase, did not affect the interaction of RNF34 (aa 195–381) with γ2sIL. However, the full-length RNF34 did not induce reporter expression. Presumably, the two zinc finger domains in the N terminus, which bind to membrane phospholipids, and the possible palmitoylation of the N terminus of RNF34 (8) keep the full-length RNF34 associated with membranes, preventing its entry to the nucleus and the activation of the reporter. This has also been the case for another γ2-subunit interacting protein, GABARAP, which has a tubulin binding domain in the N terminus, preventing the translocation of the full-length GABARAP into the nucleus and the interaction with γ2sIL in the Y2H assay (50).
We have also identified a domain in the γ2sIL (Fig. 1C) that strongly interacts with RNF34, corresponding to aa 362–404 of γ2s. A short 14-aa domain in the γ2sIL (ECLDGKDCASFFCC) was sufficient for interaction with RNF34, although the interaction intensity was lower than that of γ2sIL. This 14-aa domain is highly conserved among the ILs of the various GABAAR γ subunits.
We tested whether the interaction between RNF34 (aa 195–381) and γ2IL observed in the Y2H assay could also be observed in vitro in a bimolecular interaction assay. Immobilized bacterial His-RNF34-C (aa 195–381) and His-RNF34-C* (aa 195–381, H351A) fusion proteins (but not immobilized His) pulled down GST-γ2sIL (Fig. 1D), as revealed by immunoblotting with Ms anti-γ2IL and Rb anti-GST antibodies. Fig. 1E shows that the GST control was not pulled down by immobilized His-RNF34-C or His, indicating that the His or GST tags did not interfere with the interaction between γ2sIL and RNF34. The results showed that RNF34-C and RNF34-C* directly interact with γ2sIL.
We continued testing whether His-RNF34 FL interacts with GST-γ2sIL. For this purpose, His-RNF34 FL fusion protein was expressed in bacteria and purified. It is worth mentioning that the yield of purified His-RNF34 FL was much lower than that of His-RNF34-C. Nevertheless, the results (Fig. 1F) clearly showed that His-RNF34 FL pulled GST-γ2sIL down, but not GST, indicating that the full-length RNF34 interact with γ2sIL. These results and the co-precipitation of RNF34 with assembled GABAARs from brain extracts shown below, indicate that the full-length RNF34 interacts with assembled GABAARs.
RNF34 Ubiquitinates γ2sIL in Vitro
RING E3 UBLs recognize and bind to the substrates that they ubiquitinate. To test whether γ2sIL is a substrate for ubiquitination by RNF34, we performed an in vitro ubiquitination assay using the bacterial fusion proteins His-RNF34 FL (as E3 UBL) and GST-γ2sIL or GST (as substrates). The E3 UBL activity of the bacterial His-RNF34 FL fusion protein is very low. Nevertheless, a smear corresponding to polyubiquitinated GST-γ2sIL (Ubn) was observed only when RNF34 and all the other components of the ubiquitination machinery were present (Fig. 1G, left IB, lane 3). Under the same conditions, no polyubiquitination smear was observed when the protein substrate was GST instead of GST-γ2sIL (Fig. 1G, left IB, lane 4). Although the smear was observed above ∼55 kDa, it was particularly strong above ∼100 kDa.
Immunoblotting with anti-γ2IL or anti-GST antibodies revealed an ∼55-kDa protein band (marked with a *) only when all the components of the ubiquitination machinery and the GST-γ2IL were present (Fig. 1G, middle and right IB, lane 3). A faint protein band in the same position was also revealed with the anti-Ub Ab (Fig. 1G, left IB, lane 3). This ∼55-kDa protein band corresponds to ubiquitinated GST-γ2sIL (calculated having two Ub molecules, because the molecular mass of GST-γ2sIL is 35 kDa and that of HA-Ub monomer is 9.8 kDa).
These data indicate that the bacterial GST-γ2sIL fusion protein can indeed be ubiquitinated by the bacterial His-RNF34 fusion protein. Note that the polyubiquitination of GST-γ2sIL produces an amplification effect in the immunoblot signal with the anti-Ub Ab (Ubn). Such amplification of the signal does not occur when using immunoblots with the anti-γ2IL or anti-GST antibodies. These antibodies, however, show better single protein bands than when the protein signal is diluted in a smear. Also note that in the reported in vitro ubiquitination of other synaptic proteins with other bacterial E3 UBL fusion proteins, only a small fraction of the substrates becomes ubiquitinated (14, 51). Also note that a very small amount of His-RNF34 FL was used in the reaction mixture due to the very low purification yield. Because of the limitations of the in vitro ubiquitination assay, we assessed the ubiquitination of the γ2 GABAAR subunit by RNF34 in living cells.
RNF34 Ubiquitinates γ2s GABAAR Subunits in Co-transfected HEK293 Cells
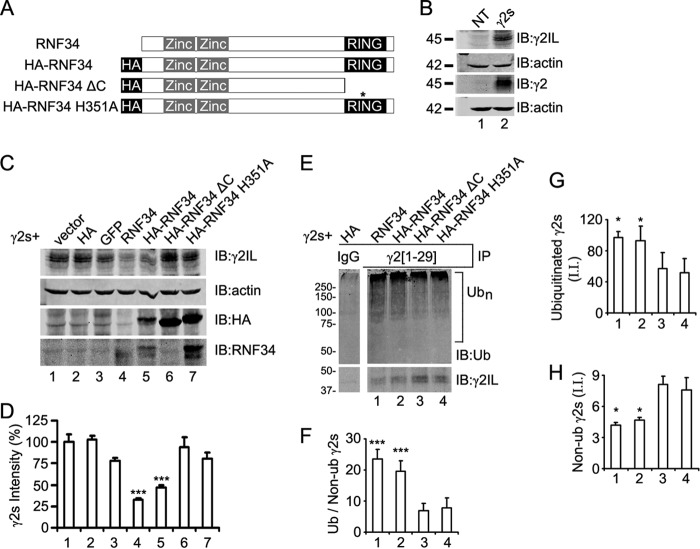
We have investigated whether RNF34 is involved in the ubiquitination of the γ2 GABAAR subunit in cultured cells. For this purpose, we made several HA-tagged RNF34 constructs (Fig. 2A). In the HA-RNF34 ΔC construct, the RING finger domain and the C terminus (aa 332–381) were deleted. In the HA-RNF34 H351A construct, histidine (His-351), which is critical for the E3 UBL activity of RNF34 and other RNF proteins, was mutated to alanine (9, 52, 53).
FIGURE 2.
RNF34 is an E3 ligase that ubiquitinates γ2 subunit in transfected HEK293 cells. A, schematic representation of the RNF34 constructs. The position of the HA tag, zinc finger domains, and RING domain is indicated. The asterisk indicates the position of the H351A point mutation. B, expression of the γ2s subunit in HEK293 cells co-transfected with γ2s and pCAGGS-HA (γ2s, lane 2) was verified by immunoblotting (IB) with Ms anti-γ2IL and Rb anti-γ2 antibodies. Both antibodies recognize the ∼45-kDa γ2s protein band. Nontransfected cells (NT, lane 1) showed no expression of γ2s. C, RNF34, both nontagged (lane 4) and HA-tagged (lane 5), significantly reduced the expression of the γ2s subunit. HEK293 cells were co-transfected with γ2s, ubiquitin, and pCAGGS (vector, lane 1), pCAGGS-HA (HA, lane 2), or pCAGGS-GFP (GFP, lane 3), or a RNF34 construct (RNF34, HA-RNF34, HA-RNF34 ΔC, or HA-RNF34 H351A, lanes 4–7, respectively). RIPA extracts were subjected to immunoblotting with Ms anti-γ2IL, Ms anti-actin, Ms anti-HA, or Rb anti-RNF34 antibodies. The expression of RNF34 constructs was verified by HA immunoreactivity (lanes 5–7) and Rb anti-RNF34 antibody to the C terminus (lanes 4–7). Note that Rb anti-RNF34 recognizes a C terminus epitope and does not recognize HA-RNF34 ΔC (lane 6). HA-RNF34 migrated slightly slower than RNF34 as shown by immunoblotting with Rb anti-RNF34. Actin was used as the loading control. The expected HA control protein band (∼1.5 kDa) corresponding to pCAGGS-HA (lane 2) ran out of the gel. D, quantification of the γ2s subunit protein band from the various lanes in C normalized for actin and vector transfection (100%). Data are presented as mean ± S.E., n = 4 independent experiments. ***, p < 0.001 in one-way ANOVA Tukey-Kramer multiple comparison test. E and F, RNF34 and HA-RNF34 significantly increase the ratio of ubiquitinated/nonubiquitinated γ2s subunits. RIPA extracts of HEK293 cells, which were co-transfected with γ2s, Ub, and RNF34 constructs, were immunoprecipitated with Rb anti-γ2(1–29) antibody or nonimmune serum (IgG), and the precipitates were subjected to SDS-PAGE and immunoblotting with Ms anti-ubiquitin (Ub) or Ms anti-γ2IL mAbs. The bar graph below F shows the quantification of the ratio of the ubiquitination smear fluorescence intensity (>55 kDa, Ubn) to the 45-kDa nonubiquitinated γ2s subunit fluorescence intensity. The lane corresponding to the nonimmune serum (IgG) is a nonadjacent lane from the same Western blot as the other lanes 1–4. G, quantification of the integrated intensity (I.I.) of the smear of ubiquitinated γ2s shown in E. H, quantification of the integrated intensity (I.I.) of the nonubiquitinated 45 kDa γ2s of E. Data in F–H are presented as mean ± S.E., n = 3 independent experiments, *, p < 0.05 and ***, p < 0.001 in one-way ANOVA Tukey-Kramer multiple comparison test.
We first verified that indeed we could detect the γ2s protein band in the immunoblots of HEK293 cells co-transfected with γ2s and pCAGGS-HA using two anti-γ2 Abs: Ms anti-γ2IL (γ2IL) and Rb anti-γ2 (γ2), as shown in Fig. 2B. Fig. 2, C and D, show that the expression of γ2s was significantly reduced in HEK293 cells co-transfected with γ2s and nontagged RNF34 (to 32.7 ± 2.3%, p < 0.001) or with γ2s and HA-RNF34 (to 47.4 ± 2.1%, p < 0.001), compared with that of HEK293 cells co-transfected with γ2s and the pCAGGS control (vector, 100 ± 9.3%) or other controls described below. No statistically significant difference in the expression of γ2s was observed in HEK293 cells co-transfected with γ2s and pCAGGS-HA (HA, 103.1 ± 4.1%), or pCAGGS-EGFP (EGFP, 77.9 ± 3.9%), or HA-RNF34 ΔC (94.1 ± 11.5%), or HA-RNF34 H351A (80.5 ± 7.0%), or the vector control (100 ± 9.3%). Note that the decrease in γ2s expression is strongest when active RNF34 or HA-RNF34 are co-expressed, even if their protein expression levels are lower than that of the enzymatically inactive forms HA-RNF34 ΔC and HA-RNF34 H351A, as revealed with anti-HA and anti-RNF34 Abs (Fig. 2C, lanes 4–7).
We examined whether the reduced protein expression level of γ2s induced by RNF34 was accompanied by increased ubiquitination of γ2s. Immunoprecipitation of HEK293 cell extracts with the Rb anti-γ2(1–29) antiserum (Fig. 2E) immunoprecipitated both nonubiquitinated 45-kDa and ubiquitinated forms of γ2s. The latter was revealed as a smear of ubiquitin immunoreactivity (>55 kDa) in the precipitates, corresponding to the conjugation of one or more ubiquitin molecules to γ2s. The smear was strongest at >75 kDa. No immunoprecipitation of ubiquitinated or nonubiquitinated γ2s was obtained with the preimmune serum (IgG, Fig. 2E). Increased intensity of the ubiquitinated γ2s and decreased intensity of the nonubiquitinated 45-kDa γ2s protein in the immunoprecipitates was observed when RNF34 or HA-RNF34 was co-transfected, compared with the enzymatically inactive HA-RNF34 ΔC or HA-RNF34 H351A forms. The ubiquitination activity of the two RNF34 constructs was determined by calculating the ratio of ubiquitinated γ2s to nonubiquitinated γ2s in the immunoprecipitates (22). Fig. 2F shows that RNF34 or HA-RNF34 (lanes 1 and 2) significantly increased the ratio (23.6 ± 3.1, p < 0.001 and 19.6 ± 3.4, p < 0.001, respectively) compared with either HA-RNF34 ΔC (6.8 ± 2.5, lane 3) or HA-RNF34 H351A (7.8 ± 3.2, lane 4).
The increase in the ratio of ubiquitinated to nonubiquitinated γ2s in the immunoprecipitates induced by RNF34 or HA-RNF34 compared with the E3 UBL inactive controls resulted from both an increase in γ2s ubiquitination (Fig. 2G, 97.1 ± 7.6 and 92.8 ± 18.8 in lanes 1 and 2 compared with 57.0 ± 20.9 and 51.9 ± 18.5 in lanes 3 and 4) and a decrease in nonubiquitinated γ2s in the immunoprecipitates (Fig. 2H, 4.2 ± 0.3 and 4.7 ± 0.2 in lanes 1 and 2 compared with 8.1 ± 0.8 and 7.6 ± 1.2 in lanes 3 and 4). The γ2s ubiquitination observed in the HA-RNF34 ΔC and HA-RNF34 H351A controls (Fig. 2E) corresponds to the ubiquitination of γ2s by endogenous E3 UBLs. Similar experiments using γ2L (data not shown) indicated that RNF34 is also able to ubiquitinate γ2L. These experiments show that RNF34 increases the pool of ubiquitinated γ2 (γ2s and γ2L) and that this effect requires the E3 UBL activity of RNF34, because deletion of the RING finger domain or the H351A mutation abolish the RNF34-induced increased ubiquitination of γ2.
Several Lysine Residues in the Large IL of the γ2 Subunit Are Targets of Ubiquitination by RNF34
It has been shown that seven lysines that are present in the IL of GFP-γ2L, when mutated into arginines (K7R), reduced the ubiquitination and lysosomal degradation of the γ2L subunit (22). We have investigated whether these and/or other lysines in γ2IL are involved in the ubiquitination of γ2 by RNF34. For this purpose, we have constructed several Lys → Arg mutants in the large IL of the γ2s, including γ2s 7KR (having seven mutated lysines, K325R, K328R, K330R, K332R, K333R, K334R, and K335R), γ2s 8KR (7KR + K373R), and γ2s 9KR (8KR + K401R). In the γ2s 9KR, all lysines of the large γ2sIL were mutated to arginines. In addition to all lysines in γ2s large IL, we mutated the single lysine (Lys-259) that is present in the γ2s small IL generating γ2s 10KR (9KR + K259R). We also made two single point Lys-Arg mutants in the 8th (K373R) or 9th (K401R) lysine residue of the large IL of γ2s subunit (Fig. 3A).
FIGURE 3.
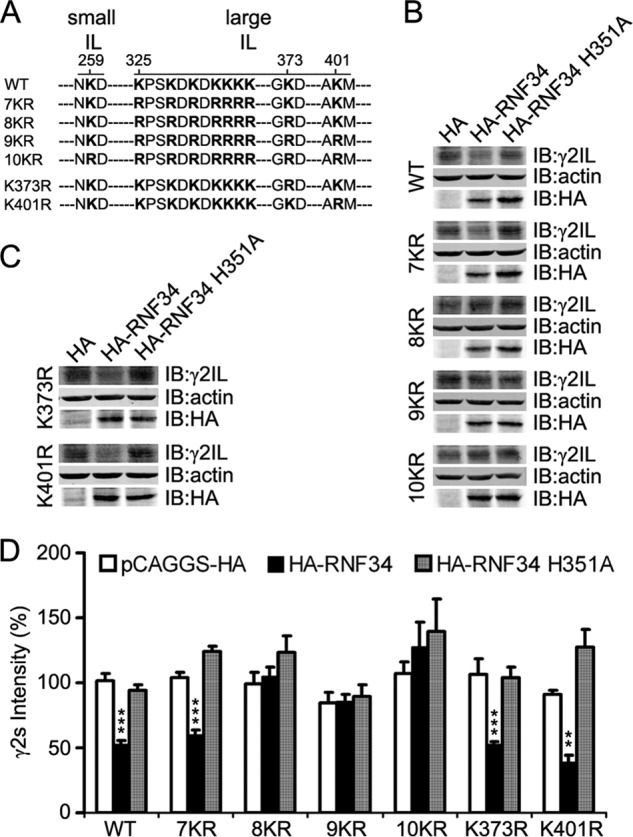
Combination of several lysine-to-arginine mutations in the IL makes the γ2s subunit resistant to the degradation by RNF34. A, illustration of the position of the lysine residue at the small intracellular loop (small IL, aa 259) and of the 9 lysine residues at the large intracellular loop (large IL, aa 325, 328, 330, 332, 333, 334, 335, 373, and 401) of the γ2s subunit that were mutated into arginines. The small IL (aa 258–260) is located between TM1 and TM2, and the large IL (aa 313–404) is between TM3 and TM4. B, HA-RNF34 significantly reduces the expression level of γ2s WT or 7KR, but it does not significantly affect the expression level of γ2s 8KR, 9KR, or 10KR, compared with the HA or HA-RNF34 H351A controls. HEK293 cells, co-transfected with a γ2s subunit (WT, 7KR, 8KR, 9KR, or 10KR), ubiquitin, and a RNF34 construct (HA-RNF34 or HA-RNF34 H351A) or pCAGGS-HA as control vector were lysed, subjected to SDS-PAGE, and immunoblotted (IB) with Ms anti-γ2IL, Ms anti-actin, or Ms anti-HA. C, expression level of γ2s K373R or K401R point mutants (the Lys-Arg point mutation of the 8th or 9th lysine residues in the large IL of γ2s subunit) was significantly reduced by HA-RNF34, but not by HA-RNF34 H351A. D, quantification of the γ2s subunit protein band intensity normalized for that of actin and pCAGGS-HA transfection control for WT. Data are presented as mean ± S.E., n = 4 independent experiments. **, p < 0.01; ***, p < 0.001, for each mutant, statistical significance was determined in one-way ANOVA Tukey-Kramer multiple comparison test.
Fig. 3, B–D, shows that HA-RNF34 significantly reduced the expression level not only of γ2s WT (52.7 ± 3.0%, p < 0.001) compared with HA control (100 ± 5.7%) but also significantly reduced the expression level of γ2s 7KR (59.9 ± 3.5%, p < 0.001), γ2s K373R (52.5 ± 2.5%, p < 0.001), and γ2s K401R (38.9 ± 5.7%, p < 0.01), compared with their corresponding HA controls. In contrast, HA-RNF34 did not significantly reduce the expression levels of γ2s 8KR (105.0 ± 7.4%), γ2s 9KR (85.5 ± 6.0%), or γ2s 10KR (127.8 ± 19.1%), compared with those of the corresponding HA controls (99.6 ± 8.8%, 84.7 ± 8.0%, or 107.3 ± 8.5%, p > 0.05). The HA-RNF34 H351A UBL-inactive mutant did not significantly reduce the expression level of either γ2s WT or any of the γ2s KR mutants when compared with their corresponding HA control (p > 0.05 for all). It is also worth noting that for all HA controls, the levels of γ2s WT, or γ2s 7KR, or γ2s 8KR, or γ2s 9KR, or γ2s 10KR, or γ2s K373R, or γ2s K401R were not significantly different from each other, indicating that the Lys → Arg mutations do not affect γ2s protein expression. Furthermore, the HA-RNF34 H351A mutation did not affect the expression level of this mutant when compared with that of HA-RNF34, as shown by immunoblotting with Ms anti-HA (Fig. 3, B and C).
Thus, neither the 7KR nor K373R mutations could prevent the RNF34-induced degradation of the mutated γ2s. However, the combination of both in the 8KR mutant prevented the degradation of this mutant. The combined results from Figs. 2 and 3 show that RNF34 ubiquitinates γ2s promoting its degradation and that the ubiquitination occurs in several lysines in the γ2sIL.
RNF34 Promotes the Degradation of Assembled GABAARs via the Lysosome- and Proteasome-dependent Pathways
When HEK293 cells are transfected only with the γ2s subunit (without α1 and β3) some of the expressed γ2s goes to the cell surface, but much of the γ2s remain in the ER (54, 55). When γ2s is co-transfected with α1 and β3, much of the γ2s assembles with α1 and β3 forming functional heteromeric α1β3γ2s receptors that exit the ER and are translocated to the cell surface (38, 54–57). When HEK293 cells were transfected with γ2s only, the γ2s protein migrate significantly faster (∼45 kDa) than when cells were co-transfected with γ2s, α1, and β3 (∼50 kDa, Fig. 4A, lanes 2 and 3, respectively). After treating the cell extracts with the glycosidase peptide-N-glycosidase F, which cleaves N-linked oligosaccharides from glycoproteins, the γ2s subunits showed the same mobility (∼38 kDa) in the presence or absence of α1 and β3 (Fig. 4A, lanes 5 and 6). These results indicate that γ2s is differentially N-glycosylated when it is assembled with α1 and β3 than when α1 and β3 are absent. This is likely due to the differential glycosylation of γ2s during translocation from the ER to the cell surface, which is favored by the assembly into α1β3γ2s heteropentamers.
We also tested whether RNF34 reduces the level of assembled GABAARs by monitoring the mature glycosylated γ2s in α1β3γ2s recombinant receptors. Fig. 4B shows that HA-RNF34 significantly reduces the expression levels of the ∼50 kDa γ2s subunit in the α1β3γ2s recombinant receptors (51.3 ± 6.1%, p < 0.01), compared with cells co-transfected with HA (100 ± 2.4%) or the UBL inactive mutant HA-RNF34 H351A (126.9 ± 23.2%). Upon treatment of the transfected HEK293 cells for 12 h with leupeptin or MG132 before cell harvesting, the reduction of the expression level of γ2s subunit induced by HA-RNF34 (46.1 ± 8.2%, p < 0.01) was partially reversed (76 ± 13.6%, p < 0.05 for leupeptin treatment; 89.2 ± 12.3%, p < 0.05 for MG132 treatment; Fig. 4C). Note that HEK293 cells were treated only for 12 h with leupeptin or MG132 to prevent the toxicity of these drugs. The results indicate that RNF34 induced the degradation of assembled γ2s-containing GABAARs via both the lysosome and proteasome pathways.
The effect of leupeptin suggests that part of the assembled GABAARs present at the surface, after endocytosis, are degraded via the endosome-lysosome pathway. Moreover, endosomal GABAARs can also go into the proteasome degradation pathway. We investigated whether RNF34 is indeed localized in endosomes by co-transfecting HEK293 cells with RNF34 and EGFP-Rab5 or EGFP-Rab7. It has been shown that EGFP-Rab5 and EGFP-Rab7 target to early and late endosomes, respectively (58). RNF34 forms clusters (Fig. 4, D and E), which are frequently co-localizing with EGFP-Rab5 (Fig. 4, D1–D3, arrowheads) or EGFP-Rab7 (Fig. 4, E1–E3, arrows) clusters. The results show that RNF34 is in part localized at endosomes (early and late endosomes), which is consistent with RNF34 being involved in ubiquitination and degradation of membrane GABAARs.
RNF34 and GABAARs Co-immunoprecipitate from Brain Membrane Extracts
We studied whether the interaction between RNF34 and GABAARs that we have observed in Y2H and in vitro also occurs in the brain. We found that GABAARs and RNF34 co-immunoprecipitated from rat forebrain membrane extracts. The antisera to the γ2 GABAAR subunit, such as the two anti-γ2 Abs raised in rabbit (Rb γ2 and Rb γ2(1–29), Fig. 5A) or an anti-γ2 Ab raised in guinea pig (GP γ2, Fig. 5B) co-precipitated the 42-kDa RNF34 protein together with the GABAARs γ2 (45 kDa) and α1 (51 kDa) subunits. The co-precipitation of α1 with γ2 indicates that heteromeric assemblies of GABAAR subunits were co-precipitated with RNF34. The corresponding control with nonimmune rabbit or guinea pig serum (IgG) did not precipitate RNF34 or GABAARs.
FIGURE 5.
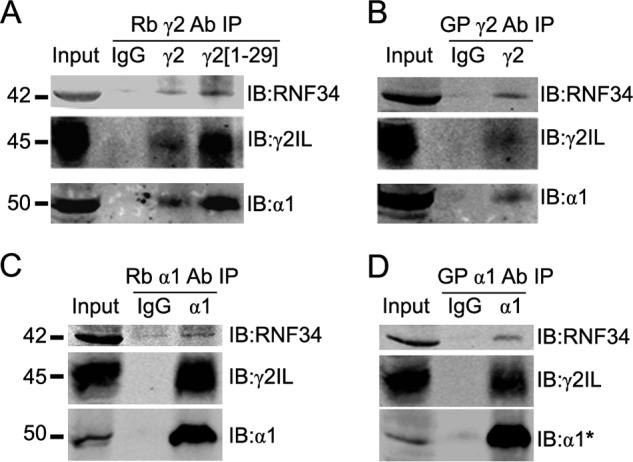
RNF34 co-immunoprecipitates with brain GABAARs. A and B, various antisera to the γ2 GABAAR subunit, two raised in rabbit (Rb γ2 and Rb γ2(1–29), in A) and one in guinea pig (GP γ2, in B) co-precipitate RNF34 (42 kDa) with GABAARs from rat forebrain membrane extracts. GABAARs in the precipitate were revealed by the presence of γ2 (45 kDa) and α1 (50 kDa) GABAAR subunits. C and D, antisera to the α1 GABAAR subunit raised in rabbit (Rb α1, in C) or guinea pig (GP α1, in D) co-precipitate RNF34 and GABAARs, as shown by the presence of RNF34, γ2, and α1 subunits in the precipitate. In all panels, the presence of RNF34 and GABAARs in the precipitate was detected by immunoblotting with Rb anti-RNF34, Ms anti-γ2IL, Ms anti-α1, with the exception of α1 in D where Rb anti-α1* was used.
Moreover, the antisera to the α1 GABAAR subunits that were raised in rabbit (Rb α1, Fig. 5C) or guinea pig (GP α1, Fig. 5D) co-precipitated RNF34 together with GABAARs γ2 and α1 subunits, confirming that RNF34 co-precipitates with assembled GABAARs. No precipitation was obtained with nonimmune rabbit or guinea pig serum (IgG). The data indicate that in the brain, RNF34 is associated with GABAARs.
For revealing RNF34 in the immunoprecipitation experiments described above and for the immunocytochemistry experiments described below, we raised a novel rabbit polyclonal antibody (Rb anti-RNF34) to the C terminus of rat RNF34 (aa 368–381) and prepared affinity-purified antibody on immobilized antigen peptide. Immunoblots of a rat forebrain crude synaptosomal (P2) fraction showed that the affinity-purified Rb anti-RNF34 antibody specifically recognized a 42-kDa protein (Fig. 6A), which corresponds to the molecular weight of the rat RNF34 protein calculated by its amino acid sequence (42,680 Da). The immunoreactivity was blocked by preincubation of Rb anti-RNF34 antibody with the antigenic peptide (Fig. 6A).
Developmental Expression and Cellular and Subcellular Localization of RNF34 in the Rat Brain
Some GABAergic interneurons show high expression levels of RNF34. We have used the affinity-purified Rb anti-RNF34 antibody to study the expression of RNF34 during brain development. Immunoblots of homogenates of the forebrains from rats of various ages show that the RNF34 protein is not expressed until the 2nd postnatal week coinciding with the peak of synaptogenesis, reaching the maximum expression at P30 (Fig. 6B). The immunoreactivity of Rb anti-RNF34 antibody was displaced by the purified fusion protein His-RNF34 (aa 195–381, Fig. 6B) or antigenic peptide (data not shown).
RNF34 is highly enriched in the microsomal (P3) fraction, although it is also present in other fractions, including crude synaptosomal (P2), synaptosomal (P2B), total membrane, SPM, and postsynaptic density (PSD) fractions (Fig. 6C). RNF34 carries the N-terminal putative signal sites for palmitoylation (8), which is consistent with its presence in membrane fractions (membrane and SPM). Moreover, RNF34 contains a FYVE domain that can serve as a membrane-targeting or endosome-localizing signal (9, 59), which is consistent with the enrichment of RNF34 in the microsomal fraction.
Light microscopy immunocytochemistry of adult rat brain sagittal sections showed that RNF34 is ubiquitously expressed in most regions of the brain, including cerebral cortex (Fig. 6D) and corpus striatum (Fig. 6E). At the cellular level, the immunoreactivity concentrated on the perikaryon of neurons such as the large pyramidal neurons of layer V of the cerebral cortex (Fig. 6D). Some interneurons showed very strong immunoreactivity in cerebral cortex (Fig. 6, D, arrowheads, and F) and corpus striatum (Fig. 6, E, arrowheads, and G). Double labeling of rat brain sections with Rb anti-RNF34 and GP anti-GABA antibodies confirmed the high expression of RNF34 in some GABAergic interneurons (GABA+) in corpus striatum (Fig. 6, H and I, arrows) and cerebral cortex (data not shown). The RNF34 immunoreactivity was blocked by purified fusion protein His-RNF34 (aa 195–381, data not shown).
GABAergic Synapses Often Have RNF34 Associated with Them
We have studied the expression and localization of RNF34 in 21 DIV cultured HP neurons by immunofluorescence. At this culture age, most pyramidal neurons and interneurons show RNF34 clusters both in dendrites (Fig. 7A) and in soma (data not shown). The RNF34 cluster immunofluorescence was blocked by the purified fusion protein His-RNF34 (aa 195–381) or antigenic peptide (data not shown). Although the majority of the RNF34 clusters were not associated with synapses, a significant proportion of GABAergic synapses had associated RNF34 clusters. Triple-label immunofluorescence with anti-RNF34, anti-GAD, and anti-γ2 GABAAR subunit antibodies showed that 75 ± 2% of the GABAergic synapses (GAD+ and γ2+) had associated RNF34 clusters (Fig. 7, A, and A1–A4, arrowheads). Many nonsynaptic (GAD−) γ2 clusters also showed colocalizing RNF34 clusters, although the proportion of nonsynaptic γ2 clusters that colocalized with RNF34 was significantly smaller (51 ± 2%, p < 0.001 by Student's t test) than when presynaptic GAD+ terminals were juxtaposed to γ2 clusters. This result shows that although the presence of a presynaptic GAD terminal is not necessary for the colocalization of RNF34 with γ2, the colocalization of RNF34 with γ2 clusters significantly increased at GABAergic synapses.
FIGURE 7.
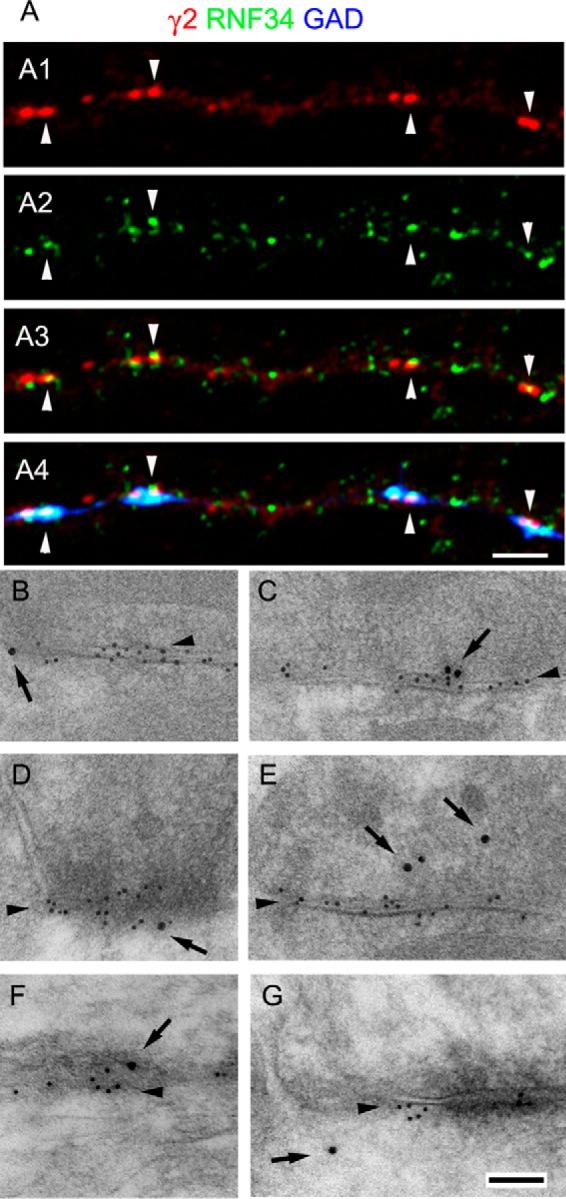
GABAergic synapses frequently have RNF34 associated with them. A, triple-labeled immunofluorescence of 21 DIV cultured hippocampal neurons with GP anti-γ2 GABAAR subunit (red, A1), Rb anti-RNF34 (green, A2), and sheep anti-GAD (blue) antibodies. A3 shows the overlay of γ2 and RNF34 fluorescence, whereas A4 shows the overlay of the three fluorescence channels. Arrowheads indicate GABAergic synapses (γ2+ and GAD+) that have co-localizing RNF34 clusters. Scale bar, 2.5 μm (A1–A4). B–G, postembedding EM immunogold double-labeling of rat brain cerebellum (B, C, and E) and cerebral cortex (D, F, and G) with Rb anti-RNF34 and mouse anti-β2/3 GABAAR subunits. The β2/3 immunolabeling (smaller gold particles) is indicated by arrowheads. GABAergic synapses are identified by their morphology and the concentration of β2/3 GABAAR subunits. The RNF34 immunolabeling (larger gold particles, arrows) is localized at or near GABAergic synapses (B–G), GABAergic presynaptic terminals (C, E, and F), and GABAergic postsynapses (B, D, and G). The goat anti-rabbit IgG and goat anti-mouse IgG secondary antibodies were conjugated to 18 and 10 nm diameter colloidal gold particles, respectively. Scale bar, 100 nm (B–G).
Gold particles corresponding to anti-RNF34 (Fig. 7, B–G, large gold particles, arrows) were also associated with GABAergic synapses, as shown in double-label immunogold experiments with GABAAR β2/3 subunits (Fig. 7, B–G, small gold particles, arrowheads). RNF34 immunogold particles were found at GABAergic synapses (Fig. 7, B–F, arrows) or near GABAergic synapses (Fig. 7G, arrow), either presynaptically (Fig. 7, C and E, arrows) or postsynaptically (Fig. 7, B and D, arrows). In the latter case, the RNF34 immunogold particles were associated with the postsynaptic membrane (Fig. 7B, arrow), the postsynaptic density (Fig. 7D, arrow), or the postsynaptic cytoplasm (Fig. 7G, arrow).
The EM immunogold data are consistent with the notion that RNF34 is localized at synapses (pre- and post-synaptically), nonsynaptic membranes, and cytoplasm, probably at cytoplasmic organelles such as endosomes. Thus, the immunofluorescence and immunogold data show that although RNF34 is not exclusively localized at GABAergic synapses, the latter frequently show the presence of RNF34. GABAAR clusters frequently have RNF34 associated with them. Moreover, the high expression of RNF34 in some GABAergic interneurons is consistent with the presynaptic localization of some RNF34 (in the axon of these GABAergic interneurons).
Hippocampal Neurons Overexpressing RNF34 Show Reduced Endogenous γ2-GABAAR Cluster Density and GABAergic Innervation
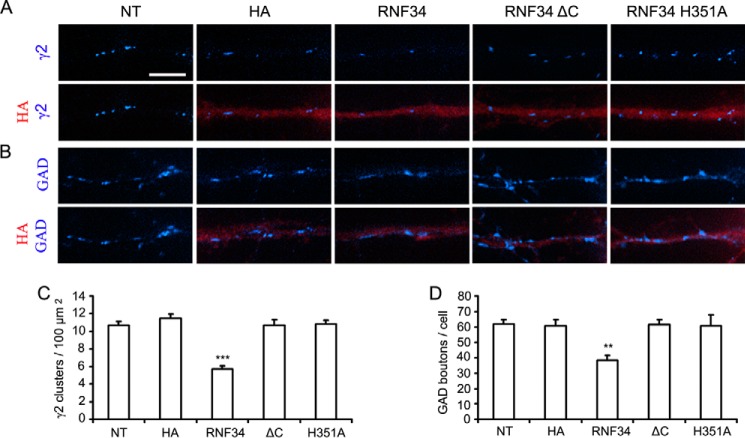
Cultured HP neurons transfected with HA-RNF34 exhibited a significant decrease in the density of endogenous γ2-GABAAR clusters (5.7 ± 0.4 clusters/100 μm2), compared with that of nontransfected neurons (10.7 ± 0.5 clusters/100 μm2, p < 0.001) or neurons transfected with HA (11.5 ± 0.5 clusters/100 μm2, p < 0.001, Fig. 8, A and C). However, neurons transfected with the inactive E3 UBL HA-RNF34 ΔC (10.7 ± 0.7 clusters/100 μm2) or HA-RNF34 H351A (10.8 ± 0.4 clusters/100 μm2) showed no significant difference in the density of endogenous γ2-GABAAR clusters compared with nontransfected controls or neurons transfected with HA.
FIGURE 8.
Cultured hippocampal neurons overexpressing RNF34 show reduced density of endogenous γ2-GABAAR clusters and reduced GABAergic innervations. A and B, representative immunofluorescence images of dendrites from hippocampal neurons that were nontransfected (NT) or transfected with EGFP and vector control (HA) or EGFP and various RNF34 constructs (HA-RNF34, HA-RNF34 ΔC, or HA-RNF34 H351A, all in pCAGGS plasmid). Primary antibodies were Ms anti-HA (red) in A and B, Rb anti-γ2 (blue in A), or sheep anti-GAD (blue in B). Neurons transfected with the RNF34 constructs or HA were identified by anti-HA fluorescence (red), which coincided with the fluorescence of the co-transfected EGFP (not shown for economy of space). The HA-RNF34 constructs were made in pCAGGS plasmid, and overexpression results were determined 2 days after transfection (13 DIV). Scale bar, 5 μm. C and D, quantification of the effect of RNF34 constructs on the density of γ2 clusters and the number of GAD boutons innervating the transfected neurons. Data are presented as mean ± S.E., **, p < 0.01; ***, p < 0.001 in one-way ANOVA Tukey-Kramer multiple comparison test, n = 16 neurons for each group from four independent transfection experiments.
Neurons transfected with HA-RNF34 also exhibited decreased number of endogenous GAD boutons contacting the transfected cells (38.5 ± 3.1 GAD boutons/cell, Fig. 8, B and D) compared with that of nontransfected neurons (61.7 ± 3.1 GAD boutons/cell, p < 0.01), or neurons transfected with HA (60.8 ± 3.9 GAD boutons/cell, p < 0.01), or HA-RNF34 ΔC (61.4 ± 3.3 GAD boutons/cell, p < 0.01), or HA-RNF34 H351A (60.7 ± 7.0 GAD boutons/cell, p < 0.01).
These results show that overexpression of RNF34 significantly decreases the number of γ2-GABAAR clusters in the transfected neurons and the number of GABAergic contacts that the transfected neurons receive, and these effects depend on the E3 UBL activity of RNF34.
Knocking Down Endogenous RNF34 in Hippocampal Neurons Increases Both γ2-GABAAR Cluster Density and GABAergic Innervation
We made short hairpin RNAs (Sh1 and Sh2) that specifically target RNF34 mRNA in the coding region and the 3′-UTR, respectively. The corresponding shRNAs with three point mutations (Sh1 3m and Sh2 3m) were used as controls (Fig. 9A).
The knockdown effect of Sh1 and Sh2 on RNF34 expression was determined in HEK293 cells co-transfected with HA-RNF34 and shRNAs (Fig. 9, B and C). The HA-RNF34 protein expression was significantly knocked down by Sh1 (51.3 ± 4.3%, p < 0.05) and even more by Sh2 (29.2 ± 3.9%, p < 0.001) but not by Sh1 3m (104.7 ± 13%, p > 0.05) or Sh2 3m (88.5 ± 8.7%, p > 0.05) controls, compared with HEK293 cells co-transfected with HA-RNF34 and mU6 vector (100 ± 4.8%). Because of the stronger knockdown effect of Sh2, we decided to use Sh2 in subsequent experiments and designed a rescue mRNA for Sh2. Because Sh2 targets the 3′-UTR of RNF34 mRNA, we used for rescue an mRNA that contains the coding region of RNF34 but not the 3′-UTR (rescue). In another experiment, we tested the effect of Sh2 on the rescue construct. Sh2 did not affect rescue expression (108 ± 17.2%, p > 0.05), compared with HEK293 cells co-transfected with rescue and mU6 vector (107.1 ± 15%) as shown in Fig. 9C. The internal positive control showed that Sh2 significantly knocked down HA-RNF34 (30.1 ± 2.7%, p < 0.001) compared with HEK293 cells co-transfected with HA-RNF34 and mU6 vector (100 ± 8.6%). Actin was used as the loading control, and no effect on actin expression was observed by Sh1 or Sh2.
Endogenous RNF34 is fully expressed in 21 DIV HP neurons (Fig. 7A). In the rat brain, RNF34 was expressed postnatally by the 2nd postnatal week (Fig. 6A). In HP cultures, RNF34 immunofluorescence signal was detected after 17 DIV. Therefore, for the following knockdown experiments, neuronal cultures were transfected at 10 DIV, and the effects were studied at 21 DIV, when nontransfected cells show strong endogenous RNF34 expression.
Fig. 9, D and F, shows that neurons co-transfected with Sh2 exhibited a significant reduction in the immunofluorescence intensity of endogenous RNF34 (59.2 ± 3.6%, p < 0.001), compared with nontransfected neurons (100 ± 4.4%) and other controls shown below. Neurons transfected with control plasmids, mU6 vector (105.3 ± 5.4%) or Sh2 3m (103.4 ± 3.9%) showed no reduction in RNF34 immunofluorescence intensity, compared with nontransfected neurons (p < 0.05). The knockdown effect of Sh2 on endogenous RNF34 expression was prevented by the rescue mRNA (104.9 ± 4.1%, p < 0.001 compared with Sh2). Note that the rescue RNF34 was constructed in pRK5 plasmid to achieve a moderate level of expression in neurons. Under the transfection conditions, this construct acted as rescue restoring control levels of RNF34 expression. In all neuronal transfections, EGFP was co-transfected as indicator of the transfected neurons.
Knocking down endogenous RNF34 expression in cultured hippocampal neurons with Sh2 significantly increased the density of γ2-GABAAR clusters (38.9 ± 1.6 clusters/100 μm2, p < 0.001, Fig. 9, E and G), compared with nontransfected neurons (28.1 ± 1.2 clusters/100 μm2) or neurons transfected with mU6 vector (27.8 ± 1.1 clusters/100 μm2) or Sh2 3m (28.8 ± 1.0 clusters/100 μm2). The up-regulation of γ2 cluster density induced by Sh2 was prevented by the rescue mRNA (28.9 ± 0.9 clusters/100 μm2, p > 0.05 compared with the other controls and p < 0.01 compared with Sh2). Similarly, knocking down endogenous RNF34 expression with Sh2 significantly increased the number of presynaptic GAD+ boutons contacting these neurons (118.3 ± 9.0 GAD boutons/cell, p < 0.05, Fig. 9, E and H), compared with nontransfected neurons (90.5 ± 5.9 GAD boutons/cell) or neurons transfected with the control plasmids mU6 vector (90.1 ± 7.6 GAD boutons/cell) or Sh2 3m (93.6 ± 7.3 GAD boutons/cell). The increase in the number of presynaptic GAD+ boutons by Sh2 was blocked by the rescue mRNA (87.1 ± 5.5 GAD boutons/cell, p > 0.05 compared with the other controls and p < 0.05 compared with Sh2). These results show that knocking down endogenous RNF34 significantly increases the number of γ2-GABAAR clusters and GABAergic contacts in the transfected neurons.
DISCUSSION
The interaction between the E3 UBL RNF34 and the γ2 GABAAR subunit was initially revealed by Y2H screening of a rat brain cDNA library. We found that a C-terminal half (aa 195–381) of RNF34 strongly interacts with the large IL of γ2 GABAARs. That the two proteins directly interacted with each other was confirmed by an in vitro pulldown assay with bacterially expressed fusion proteins. The notion that the two proteins interact in the rat brain was supported by the co-precipitation of RNF34 and assembled GABAARs from rat brain membrane extracts.
There are several lines of evidence supporting the notion that RNF34 ubiquitinates the γ2 subunit of GABAARs. In HEK293 cells, RNF34 significantly reduced the expression level of the γ2s GABAAR subunit. This effect was accompanied by an increased ratio of ubiquitinated to nonubiquitinated γ2s in the immunoprecipitates and was prevented either by deleting the C terminus containing the RING domain (RNF34 ΔC) or by the point-mutation H351A in the RING domain. Deletion of the RING domain both inactivates the E3 UBL and prevents the binding of RNF34 ΔC to the γ2IL; however, the H351A mutation inactivates the E3 UBL (RNF34 H351A (9)) without affecting the binding of RNF34 to γ2IL (Fig. 1). Additional support for the notion that RNF34 ubiquitinates the γ2 subunit was derived from mutating several lysines of the γ2sIL into arginines (γ2s 8KR, 9KR, or 10KR) which prevented RNF34-induced degradation of γ2. Also, bacterially expressed His-RNF34 was able to in vitro ubiquitinate GST-γ2sIL but not GST. The E3 UBL activity of the bacterial fusion protein was very low but measurable. Other reports using bacterially expressed E3 UBL for in vitro ubiquitinating synaptic proteins also show low activity, and only a very small fraction of the synaptic substrates become ubiquitinated (14, 51).
Previous studies by us and others have shown that α, β, and γ2 subunits could assemble into functional GABAARs and target to the cell membrane when they were co-expressed in HEK293 cells (38, 54–57). Our data indicate that RNF34 reduces the protein expression level of the γ2 subunit, regardless of whether γ2 is expressed alone or expressed together with α1 and β3 subunits forming assembled α1β3γ2 pentameric GABAARs at the cell surface. The treatment of HEK293 cells co-expressing α1β3γ2 subunits with leupeptin or MG132 partially reversed the effect on GABAAR expression caused by RNF34, which strongly suggests the involvement of both lysosomal and proteasomal degradation of the γ2 subunits that had been ubiquitinated by RNF34.
Our results also showed that RNF34, although not exclusively, is associated with GABAergic synapses. Immunofluorescence of cultured hippocampal neurons showed that RNF34 is associated with 75% of GABAergic synapses (γ2+ and GAD+) and with 51% nonsynaptic γ2 clusters. Postembedding EM immunogold also revealed the presence of RNF34 in GABAergic synapses, both pre- and postsynaptically. Consistent with a postsynaptic localization, RNF34 is present in the SPM fraction and the classical one-Triton PSD fraction, which contains postsynaptic densities from both GABAergic and glutamatergic synapses (35). The presence of RNF34 in GABAergic presynaptic terminals observed by EM is consistent with the high expression of RNF34 in GABAergic interneurons (Fig. 7). A synaptic role of RNF34 is also supported by our observation that during development the RNF34 expression appears and increases during the 2nd and 3rd postnatal week coinciding with the time course of synaptogenesis (60). The presence of RNF34 in membrane and synaptic fractions as well as in endosomes (discussed below) suggests that RNF34 plays an important role in the ubiquitination of synaptic proteins regulating their endocytosis, recycling, and degradation.
Additional evidence consistent with the notion that RNF34 plays a functional role in regulating γ2 GABAAR expression and GABAergic synaptic function and stability was derived from overexpression and knockdown experiments in neurons. It has been shown that the γ2 GABAAR subunit is essential for the postsynaptic localization of GABAARs (34, 61, 62). Therefore, the ubiquitination and degradation of the γ2 GABAARs play an important role in controlling the number of postsynaptic GABAARs. Thus, overexpression of RNF34 significantly reduced the postsynaptic clustering of γ2 GABAARs in cultured hippocampal neurons. Moreover, it also reduced the presynaptic GABAergic innervation that these neurons received. We and others have previously shown that reduced postsynaptic clustering of γ2 GABAARs in neurons is accompanied by reduced presynaptic innervation that these neurons receive (34, 63). However, knocking down the endogenous RNF34 expression by shRNAs increased the density of postsynaptic γ2-GABAAR clusters as well as the number of presynaptic GAD+ boutons. In our experiments, the transfected neurons are postsynaptic, and the observed effects resulted from altering RNF34 expression levels in the postsynaptic neurons. Taken together, our data indicate that in hippocampal neurons RNF34 regulates the expression and clustering of GABAARs and also the stability of GABAergic synapses. This is not to imply that RNF34 is the only E3 UBL involved in the ubiquitination and degradation of GABAARs. It has been shown that GABAAR subunits other than γ2 are subjected to ubiquitination (21) by still unidentified E3 UBL(s). Moreover, we cannot exclude the involvement of other E3 UBL in the ubiquitination of γ2.
The ubiquitination of γ2 by RNF34 and subsequent degradation of γ2 could explain by themselves the effects observed after overexpressing and knocking down RNF34 in neurons. Nevertheless, E3 UBLs could ubiquitinate multiple protein targets. It is likely that, in addition to γ2, RNF34 ubiquitinates other synaptic and nonsynaptic proteins. As indicated above, much of the RNF34 is not associated with GABAergic synapses.
RNF34 (also named hRFI, momo, and CARP1) was initially isolated from human esophageal cancer (64). RNF34 has E3 UBL activity and RNF34 mRNA is highly expressed in brain and testis compared with other tissues (8, 9). In studies done on non-neural tissues, RNF34 had anti-apoptotic activity, against both death receptor- and mitochondrion-mediated apoptosis, by ubiquitinating protein substrates such as caspases 8/10 and p53, targeting them for proteasome-mediated degradation (9, 52, 53, 59, 65–68). To the best of our knowledge, this is the first report of a synaptic role for RNF34.
We have found that RNF34 is enriched in the microsomal fraction, which contains endosomes and ER among other micro-organelles. Support for a role of RNF34 in endosomes is derived from the presence in RNF34 of an N-terminal FYVE (Fab1, YOTB, Vac1, and EEA1) domain (9, 59, 69). The FYVE domain of RNF34 preferentially binds in vitro to phosphatidylinositol 3-phosphate and phosphatidylinositol 5-phosphate and with lower binding affinity to phosphatidylinositol 4-phosphate (9). The binding of RNF34 to phosphatidylinositol 3-phosphate, which is mainly localized in endosomes (70), is consistent with an endosomal function of RNF34. Moreover, our immunofluorescence data show an association of some RNF34 with the early and late endosome markers Rab5 and Rab7, respectively. Others have previously shown that a lysine-rich motif (Lys-325, Lys-328, Lys-330, Lys-332, Lys-333, Lys-334, and Lys-335) within the γ2IL is involved in the ubiquitination and targeting of γ2 GABAARs for lysosomal degradation (22). The presence of RNF34 in endosomes and the ubiquitination of these and other lysines of γ2IL by RNF34 is consistent with this E3 UBL involvement in the reported ubiquitination and lysosomal degradation of γ2 GABAARs. It is also consistent with our observed partial reversal of the RNF34 effect on γ2 expression by leupeptin. We have also observed a partial reversal by MG132, indicating that ubiquitination of γ2 by RNF34 can also be derived in proteasomal degradation.
Further mapping data by Y2H shows that RNF34 specifically binds to the C-terminal region (aa 362–404) of γ2sIL. This protein region contains two motifs involved in the interaction of the γ2 subunit with other proteins as follows: the GODZ-binding motif (aa 368–381 (71)) and the GABARAP binding motif (aa 386–403 (50)). Further analysis of the RNF34-binding motif reveals that there is no overlap between the GABARAP and RNF34-binding motifs in γ2sIL. However, the 14-aa GODZ-binding motif of γ2sIL (aa 368–381) is also involved in the interaction with RNF34. This raises the possibility that these two types of post-translational modifications of the γ2 subunit, involving palmitoylation by GODZ, which promotes the translocation of GABAARs to the cell surface and clustering (63, 71), and ubiquitination by RNF34, which promotes GABAAR degradation, might work in concert regulating the stability and clustering of GABAARs.
In summary, our results, consistent with the notion that RNF34, a RING domain E3 ubiquitin ligase, are as follows: 1) interact with and ubiquitinate the γ2 GABAAR subunit; 2) are present, although not exclusively, in GABAergic synapses; 3) regulate the expression and clustering of GABAARs; and 4) regulate GABAergic synapse stability.
Acknowledgments
We thank Drs. Josef T. Kittler and I. Lorena Arancibia-Cárcamo for providing the EGFP-γ2L K7R plasmid. We thank Dr. Cecilia Bucci at Universita del Salento, Italy, for EGFP-Rab5 and EGFP-Rab7 plasmids. We thank Drs. Peter Somogyi (MRC Anatomical Neuropharmacology Unit, Oxford, UK) and Zoltan Nusser (Institute of Experimental Medicine, Budapest, Hungary) for providing the Lowicryl-embedded block of brain tissue used in Fig. 7.
This work was supported, in whole or in part, by National Institutes of Health Grant R01NS038752 from NINDS (to A. L. D.).
- aa
- amino acid
- EGFP
- enhanced green fluorescent protein
- GAD
- glutamic acid decarboxylase
- γ2IL
- γ2 subunit IL
- γ2L
- γ2 subunit long isoform
- γ2s
- γ2 subunit short isoform
- GP
- guinea pig
- HP
- hippocampal
- IL
- large intracellular loop of a GABAAR subunit
- Ms
- mouse
- NT
- nontransfected
- PB
- phosphate buffer
- Rb
- rabbit
- RING
- really interesting new gene
- TM
- transmembrane domain
- Ub
- ubiquitin
- UBL
- ubiquitin ligase
- Y2H
- yeast two-hybrid
- ANOVA
- analysis of variance
- Ab
- antibody
- R
- receptor
- RIPA
- radioimmune precipitation assay buffer
- FL
- full-length
- PSD
- postsynaptic density
- DIV
- days in vitro
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Hoppe T. (2005) Multiubiquitylation by E4 enzymes: “one size” doesn't fit all. Trends Biochem. Sci. 30, 183–187 [DOI] [PubMed] [Google Scholar]
- 3. Lovering R., Hanson I. M., Borden K. L., Martin S., O'Reilly N. J., Evan G. I., Rahman D., Pappin D. J., Trowsdale J., Freemont P. S. (1993) Identification and preliminary characterization of a protein motif related to the zinc fingeret. Proc. Natl. Acad. Sci. U.S.A. 90, 2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 5. Deshaies R. J., Joazeiro C. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 6. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 7. Mabb A. M., Ehlers M. D. (2010) Ubiquitination in postsynaptic function and plasticity. Annu. Rev. Cell Dev. Biol. 26, 179–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Araki K., Kawamura M., Suzuki T., Matsuda N., Kanbe D., Ishii K., Ichikawa T., Kumanishi T., Chiba T., Tanaka K., Nawa H. (2003) A palmitoylated RING finger ubiquitin ligase and its homologue in the brain membranes. J. Neurochem. 86, 749–762 [DOI] [PubMed] [Google Scholar]
- 9. McDonald E. R., 3rd, El-Deiry W. S. (2004) Suppression of caspase-8- and -10-associated RING proteins results in sensitization to death ligands and inhibition of tumor cell growth. Proc. Natl. Acad. Sci. U.S.A. 101, 6170–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ehlers M. D. (2003) Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 6, 231–242 [DOI] [PubMed] [Google Scholar]
- 11. Lin A. W., Man H. Y. (2013) Ubiquitination of neurotransmitter receptors and postsynaptic scaffolding proteins. Neural Plast. 2013, 432057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamada T., Yang Y., Bonni A. (2013) Spatial organization of ubiquitin ligase pathways orchestrates neuronal connectivity. Trends Neurosci. 36, 218–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tai H. C., Schuman E. M. (2008) Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9, 826–838 [DOI] [PubMed] [Google Scholar]
- 14. Colledge M., Snyder E. M., Crozier R. A., Soderling J. A., Jin Y., Langeberg L. K., Lu H., Bear M. F., Scott J. D. (2003) Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40, 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jurd R., Thornton C., Wang J., Luong K., Phamluong K., Kharazia V., Gibb S. L., Ron D. (2008) Mind bomb-2 is an E3 ligase that ubiquitinates the N-methyl-d-aspartate receptor NR2B subunit in a phosphorylation-dependent manner. J. Biol. Chem. 283, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lussier M. P., Herring B. E., Nasu-Nishimura Y., Neutzner A., Karbowski M., Youle R. J., Nicoll R. A., Roche K. W. (2012) Ubiquitin ligase RNF167 regulates AMPA receptor-mediated synaptic transmission. Proc. Natl. Acad. Sci. U.S.A. 109, 19426–19431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishikawa K., Nash S. R., Nishimune A., Neki A., Kaneko S., Nakanishi S. (1999) Competitive interaction of seven in absentia homolog-1A and Ca2+/calmodulin with the cytoplasmic tail of group 1 metabotropic glutamate receptors. Genes Cells 4, 381–390 [DOI] [PubMed] [Google Scholar]
- 18. Moriyoshi K., Iijima K., Fujii H., Ito H., Cho Y., Nakanishi S. (2004) Seven in absentia homolog 1A mediates ubiquitination and degradation of group 1 metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 101, 8614–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gallagher M. J., Ding L., Maheshwari A., Macdonald R. L. (2007) The GABAA receptor α1 subunit epilepsy mutation A322D inhibits transmembrane helix formation and causes proteasomal degradation. Proc. Natl. Acad. Sci. U.S.A. 104, 12999–13004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Todd E., Gurba K. N., Botzolakis E. J., Stanic A. K., Macdonald R. L. (2014) GABAA receptor biogenesis is impaired by the γ2 subunit febrile seizure-associated mutation, GABRG2(R177G). Neurobiol. Dis. 69, 215–224 [DOI] [PubMed] [Google Scholar]
- 21. Saliba R. S., Michels G., Jacob T. C., Pangalos M. N., Moss S. J. (2007) Activity-dependent ubiquitination of GABA(A) receptors regulates their accumulation at synaptic sites. J. Neurosci. 27, 13341–13351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arancibia-Cárcamo I. L., Yuen E. Y., Muir J., Lumb M. J., Michels G., Saliba R. S., Smart T. G., Yan Z., Kittler J. T., Moss S. J. (2009) Ubiquitin-dependent lysosomal targeting of GABA(A) receptors regulates neuronal inhibition. Proc. Natl. Acad. Sci. U.S.A. 106, 17552–17557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Blas A. L., Vitorica J., Friedrich P. (1988) Localization of the GABAA receptor in the rat brain with a monoclonal antibody to the 57,000 Mr peptide of the GABAA receptor/benzodiazepine receptor/Cl-channel complex. J. Neurosci. 8, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vitorica J., Park D., Chin G., de Blas A. L. (1988) Monoclonal antibodies and conventional antisera to the GABAA receptor/benzodiazepine receptor/Cl− channel complex. J. Neurosci. 8, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fernando L. P., Khan Z. U., McKernan R. M., De Blas A. L. (1995) Monoclonal antibodies to the human γ2 subunit of the GABAA/benzodiazepine receptors. J. Neurochem. 64, 1305–1311 [DOI] [PubMed] [Google Scholar]
- 26. Christie S. B., Miralles C. P., De Blas A. L. (2002) GABAergic innervation organizes synaptic and extrasynaptic GABAA receptor clustering in cultured hippocampal neurons. J. Neurosci. 22, 684–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christie S. B., Li R. W., Miralles C. P., Yang B-, De Blas A. L. (2006) Clustered and nonclustered GABAA receptors in cultured hippocampal neurons. Mol. Cell. Neurosci. 31, 1–14 [DOI] [PubMed] [Google Scholar]
- 28. Riquelme R., Miralles C. P., De Blas A. L. (2002) Bergmann glia GABA(A) receptors concentrate on the glial processes that wrap inhibitory synapses. J. Neurosci. 22, 10720–10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Christie S. B., De Blas A. L. (2003) GABAergic and glutamatergic axons innervate the axon initial segment and organize GABAA receptor clusters of cultured hippocampal pyramidal cells. J. Comp. Neurol. 456, 361–374 [DOI] [PubMed] [Google Scholar]
- 30. Charych E. I., Yu W., Li R., Serwanski D. R., Miralles C. P., Li X., Yang B. Y., Pinal N., Walikonis R., De Blas A. L. (2004) A four PDZ domain-containing splice variant form of GRIP1 is localized in GABAergic and glutamatergic synapses in the brain. J. Biol. Chem. 279, 38978–38990 [DOI] [PubMed] [Google Scholar]
- 31. Charych E. I., Yu W., Miralles C. P., Serwanski D. R., Li X., Rubio M., De Blas A. L. (2004) The brefeldin A-inhibited GDP/GTP exchange factor 2, a protein involved in vesicular trafficking, interacts with the β subunits of the GABA receptors. J. Neurochem. 90, 173–189 [DOI] [PubMed] [Google Scholar]
- 32. Charych E. I., Li R., Serwanski D. R., Li X., Miralles C. P., Pinal N., De Blas A. L. (2006) Identification and characterization of two novel splice forms of GRIP1 in the rat brain. J. Neurochem. 97, 884–898 [DOI] [PubMed] [Google Scholar]
- 33. Li R. W., Serwanski D. R., Miralles C. P., Li X., Charych E., Riquelme R., Huganir R. L., de Blas A. L. (2005) GRIP1 in GABAergic synapses. J. Comp. Neurol. 488, 11–27 [DOI] [PubMed] [Google Scholar]
- 34. Li R. W., Yu W., Christie S., Miralles C. P., Bai J., Loturco J. J., De Blas A. L. (2005) Disruption of postsynaptic GABA receptor clusters leads to decreased GABAergic innervation of pyramidal neurons. J. Neurochem. 95, 756–770 [DOI] [PubMed] [Google Scholar]
- 35. Li X., Serwanski D. R., Miralles C. P., Bahr B. A., De Blas A. L. (2007) Two pools of Triton X-100-insoluble GABA(A) receptors are present in the brain, one associated to lipid rafts and another one to the post-synaptic GABAergic complex. J. Neurochem. 102, 1329–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X., Serwanski D. R., Miralles C. P., Nagata K., De Blas A. L. (2009) Septin 11 is present in GABAergic synapses and plays a functional role in the cytoarchitecture of neurons and GABAergic synaptic connectivity. J. Biol. Chem. 284, 17253–17265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Serwanski D. R., Miralles C. P., Fiondella C. G., Loturco J. J., Rubio M. E., De Blas A. L. (2010) Synaptic and non-synaptic localization of protocadherin-γC5 in the rat brain. J. Comp. Neurol. 518, 3439–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Y., Xiao H., Chiou T. T., Jin H., Bonhomme B., Miralles C. P., Pinal N., Ali R., Chen W. V., Maniatis T., De Blas A. L. (2012) Molecular and functional interaction between protocadherin-γC5 and GABAA receptors. J. Neurosci. 32, 11780–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Serwanski D. R., Miralles C. P., Christie S. B., Mehta A. K., Li X., De Blas A. L. (2006) Synaptic and nonsynaptic localization of GABAA receptors containing the α5 subunit in the rat brain. J. Comp. Neurol. 499, 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu W., Jiang M., Miralles C. P., Li R. W., Chen G., de Blas A. L. (2007) Gephyrin clustering is required for the stability of GABAergic synapses. Mol. Cell. Neurosci. 36, 484–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu W., Charych E. I., Serwanski D. R., Li R. W., Ali R., Bahr B. A., De Blas A. L. (2008) Gephyrin interacts with the glutamate receptor interacting protein 1 isoforms at GABAergic synapses. J. Neurochem. 105, 2300–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu W., De Blas A. L. (2008) Gephyrin expression and clustering affects the size of glutamatergic synaptic contacts. J. Neurochem. 104, 830–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chiou T. T., Bonhomme B., Jin H., Miralles C. P., Xiao H., Fu Z., Harvey R. J., Harvey K., Vicini S., De Blas A. L. (2011) Differential regulation of the postsynaptic clustering of γ-aminobutyric acid type A (GABAA) receptors by collybistin isoforms. J. Biol. Chem. 286, 22456–22468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Blas A. L., Cherwinski H. M. (1983) Detection of antigens on nitrocellulose paper immunoblots with monoclonal antibodies. Anal. Biochem. 133, 214–219 [DOI] [PubMed] [Google Scholar]
- 45. Higgins D., Banker G. (1998) in Culturing Nerve Cell (Banker G., Goslin K., eds) 2nd Ed., pp. 37–78, MIT Press, Cambridge, MA [Google Scholar]
- 46. Christie S. B., de Blas A. L. (2002) α5 Subunit-containing GABA(A) receptors form clusters at GABAergic synapses in hippocampal cultures. Neuroreport 13, 2355–2358 [DOI] [PubMed] [Google Scholar]
- 47. Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., Dawson T. M. (2005) Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 25, 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Blas A. L. (1984) Monoclonal antibodies to specific astroglial and neuronal antigens reveal the cytoarchitecture of the Bergmann glia fibers in the cerebellum. J. Neurosci. 4, 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nusser Z., Sieghart W., Somogyi P. (1998) Segregation of different GABAA receptors to synaptic and extrasynaptic membranes of cerebellar granule cells. J. Neurosci. 18, 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang H., Bedford F. K., Brandon N.J., Moss S. J., Olsen R. W. (1999) GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature 397, 69–72 [DOI] [PubMed] [Google Scholar]
- 51. Pavlopoulos E., Trifilieff P., Chevaleyre V., Fioriti L., Zairis S., Pagano A., Malleret G., Kandel E. R. (2011) Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell 147, 1369–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang W., Rozan L. M., McDonald E. R., 3rd, Navaraj A., Liu J. J., Matthew E. M., Wang W., Dicker D. T., El-Deiry W. S. (2007) CARPs are ubiquitin ligases that promote MDM2-independent p53 and phospho-p53Ser20 degradation. J. Biol. Chem. 282, 3273–3281 [DOI] [PubMed] [Google Scholar]
- 53. Yang W., Dicker D. T., Chen J., El-Deiry W. S. (2008) CARPs enhance p53 turnover by degrading 14–3-3σ and stabilizing MDM2. Cell Cycle 7, 670–682 [DOI] [PubMed] [Google Scholar]
- 54. Connolly C. N., Krishek B. J., McDonald B. J., Smart T. G., Moss S. J. (1996) Assembly and cell surface expression of heteromeric and homomeric γ-aminobutyric acid type A receptors. J. Biol. Chem. 271, 89–96 [DOI] [PubMed] [Google Scholar]
- 55. Connolly C. N., Uren J. M., Thomas P., Gorrie G. H., Gibson A., Smart T. G., Moss S. J. (1999) Subcellular localization and endocytosis of homomeric γ2 subunit splice variants of gamma-aminobutyric acid type A receptors. Mol. Cell. Neurosci. 13, 259–271 [DOI] [PubMed] [Google Scholar]
- 56. Tretter V., Ehya N., Fuchs K., Sieghart W. (1997) Stoichiometry and assembly of a recombinant GABAA receptor subtype. J. Neurosci. 17, 2728–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kittler J. T., Wang J., Connolly C. N., Vicini S., Smart T. G., Moss S. J. (2000) Analysis of GABAA receptor assembly in mammalian cell lines and hippocampal neurons using γ2 subunit green fluorescent protein chimeras. Mol. Cell. Neurosci. 16, 440–452 [DOI] [PubMed] [Google Scholar]
- 58. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 59. Yang W., El-Deiry W. S. (2007) CARPs are E3 ligases that target apical caspases and p53. Cancer Biol. Ther. 6, 1676–1683 [DOI] [PubMed] [Google Scholar]
- 60. Craig A. M., Graf E. R., Linhoff M. W. (2006) How to build a central synapse: clues from cell culture. Trends Neurosci. 29, 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Essrich C., Lorez M., Benson J. A., Fritschy J. M., Lüscher B. (1998) Postsynaptic clustering of major GABAA receptor subtypes requires the γ2 subunit and gephyrin. Nat. Neurosci. 1, 563–571 [DOI] [PubMed] [Google Scholar]
- 62. Schweizer C., Balsiger S., Bluethmann H., Mansuy I. M., Fritschy J. M., Mohler H., Lüscher B. (2003) The γ2 subunit of GABA(A) receptors is required for maintenance of receptors at mature synapses. Mol. Cell. Neurosci. 24, 442–450 [DOI] [PubMed] [Google Scholar]
- 63. Fang C., Deng L., Keller C. A., Fukata M., Fukata Y., Chen G., Lüscher B. (2006) GODZ-mediated palmitoylation of GABA(A) receptors is required for normal assembly and function of GABAergic inhibitory synapses. J. Neurosci. 26, 12758–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sasaki S., Nakamura T., Arakawa H., Mori M., Watanabe T., Nagawa H., Croce C. M. (2002) Isolation and characterization of a novel gene, hRFI, preferentially expressed in esophageal cancer. Oncogene 21, 5024–5030 [DOI] [PubMed] [Google Scholar]
- 65. Konishi T., Sasaki S., Watanabe T., Kitayama J., Nagawa H. (2005) Exogenous expression of hRFI induces multidrug resistance through escape from apoptosis in colorectal cancer cells. Anticancer Res. 25, 2737–2741 [PubMed] [Google Scholar]
- 66. Konishi T., Sasaki S., Watanabe T., Kitayama J., Nagawa H. (2005) Overexpression of hRFI (human ring finger homologous to inhibitor of apoptosis protein type) inhibits death receptor-mediated apoptosis in colorectal cancer cells. Mol. Cancer Ther. 4, 743–750 [DOI] [PubMed] [Google Scholar]
- 67. Konishi T., Sasaki S., Watanabe T., Kitayama J., Nagawa H. (2006) Overexpression of hRFI inhibits 5-fluorouracil-induced apoptosis in colorectal cancer cells via activation of NF-κB and upregulation of BCL-2 and BCL-XL. Oncogene 25, 3160–3169 [DOI] [PubMed] [Google Scholar]
- 68. Sasaki S., Watanabe T., Kobunai T., Konishi T., Nagase H., Sugimoto Y., Oka T., Nagawa H. (2006) hRFI overexpressed in HCT116 cells modulates Bcl-2 family proteins when treated with 5-fluorouracil. Oncol. Rep. 15, 1293–1298 [DOI] [PubMed] [Google Scholar]
- 69. Stenmark H., Aasland R., Toh B. H., D'Arrigo A. (1996) Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 271, 24048–24054 [DOI] [PubMed] [Google Scholar]
- 70. Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Keller C. A., Yuan X., Panzanelli P., Martin M. L., Alldred M., Sassoè-Pognetto M., Lüscher B. (2004) The γ2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J. Neurosci. 24, 5881–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]