Abstract
Diets high in saturated fatty acids (SFAs) are associated with the development of circadian dysregulation, obesity, and Type 2 diabetes mellitus. Conversely, polyunsaturated fatty acids (PUFAs) have recently been identified to improve insulin sensitivity, reduce weight gain, and relieve obesity-induced inflammation. While saturated fatty acids, such as the prevalent dietary fatty acid palmitate, have been implicated in circadian disruption, there is a paucity of studies regarding the effects of PUFAs on circadian parameters. Therefore, the immortalized murine neuronal model, mHypoE-37, was utilized to examine the effects of the SFA palmitate and omega-3 PUFA docosahexaenoic acid (DHA) on circadian rhythms. The mHypoE-37 neurons express the core clock genes, Bmal1, Per2, and Rev-erbα, in a circadian manner. 25 μM of palmitate significantly increased the transcriptional expression of Bmal1, without altering the expression of inflammatory markers TLR4, IκBα, and IL-6, nor the orexigenic neuropeptide AgRP, suggesting that the observed disruption of the molecular clock is the result of a mechanism distinct from that of hypothalamic cellular inflammation. Furthermore, treatment with the PUFA DHA resulted in alterations in the circadian expression profile of Bmal1, although differentially from the effects of palmitate. In the presence of DHA, the disruptive effects of palmitate on Bmal1 were less pronounced, suggesting a protective effect of DHA. These studies are the first to identify the potential for omega-3 PUFAs to protect against palmitate-mediated dysregulation of circadian parameters and will ultimately improve the understanding of circadian control mechanisms.
Keywords: circadian rhythms, cell biology, gene expression, saturated fatty acid, unsaturated fatty acid, hypothalamus
obesity has emerged as a major global health concern, affecting at least 10% of the world population (24). Many factors contribute to the development of obesity, such as genetics and dietary composition; however, the underlying cause leading to the obese state is excess energy intake. While obesity can modulate hypothalamic feeding and energy homeostasis, recent evidence suggests that obesity can also affect physiological processes other than energy homeostasis (27). It was recently demonstrated that mice fed a high-fat diet (HFD) developed atypical behavioral circadian rhythms (27). First discovered in the fruit fly, the molecular circadian clock acts through a negative feedback loop (28). In general, the cycle is divided into a positive and negative arm. The positive arm of the mammalian circadian cycle is composed of the transcription factors circadian locomotor output cycles kaput (Clock) and brain and muscle Arnt-like-1 (Bmal1) (5, 17). Following their transcription and translation, CLOCK and BMAL1 heterodimerize and enter the nucleus to initiate transcription of the Period (Per1–3) and Cryptochrome (Cry1, Cry2) genes (20, 29, 48, 54). Similarly, a PER-CRY complex is formed and subsequently enters the nucleus, inhibiting the activity of CLOCK-BMAL1, thus suppressing its own transcription, as well as other genes induced by CLOCK-BMAL1 (42, 54). The orphan nuclear receptor REV-ERBα forms an additional feedback loop, inhibiting the transcription of Bmal1 (41). Additionally, transcription of Rev-erbα is under the control of CLOCK-BMAL1. This self-sustained oscillator generates a nearly 24-h rhythm and is synchronized by external factors, including light, activity, and feeding (2, 33). Although the core molecular circadian clock only comprises a small number of genes, the clock proteins play a significant physiological role acting as transcription factors for an ∼10% of the entire transcriptome (12, 40). Circadian dysregulation can, therefore, lead to the onset of numerous pathologies, including obesity and metabolic disease (49).
Studies have demonstrated the detrimental effects of high-fat diets and, in particular, saturated fatty acids (SFAs), on energy homeostasis, obesity, and inflammation (26, 34). Mice subjected to an ad libitum high-fat diet show altered rhythms in core clock gene mRNA expression and in clock target genes involved in metabolic homeostasis (3, 19, 43). Conversely, disruptions in clock genes can cause alterations in normal energy homeostasis. For instance, Clock mutant mice, lacking one of the core elements of the molecular circadian clock, rapidly gained weight compared with wild-type mice and showed signs of metabolic disease (49), emphasizing the importance of the endogenous clock in energy homeostasis.
Interestingly, polyunsaturated fatty acids (PUFAs) have now been identified as putative agents to ameliorate the negative effects associated with obesity (8, 18). The effects exerted by both SFAs and PUFAs on obesity are accomplished, in part, through modulating hypothalamic cell biology. Although the bidirectional relationship between obesity and circadian rhythms has been recognized, few studies have examined the effects of SFA on the circadian molecular clock. Furthermore, the role of PUFAs on circadian rhythms remains to be elucidated. Importantly, little is known with respect to the role of fatty acids and circadian rhythms within the hypothalamus; thus, we have studied this issue using a clonal, hypothalamic cell line. The heterogeneity of the hypothalamus and previous lack of appropriate hypothalamic models has interfered with gaining insight into this relationship. We have addressed this issue by generating an array of immortalized, clonal hypothalamic neurons (4). Among these novel cell lines, the mHypoE-37 neuronal cells endogenously express key circadian molecular genes and will be used to test the general hypothesis that PUFAs can protect hypothalamic neurons against SFA-induced changes in circadian rhythms.
MATERIALS AND METHODS
Cell culture techniques.
mHypoE-37 neurons were grown in DMEM (Sigma-Aldrich, Oakville, Ontario, Canada) containing 5.5 mM glucose, supplemented with 5% FBS (Gibco, Burlington, Ontario, Canada), and 1% penicillin-streptomycin (Gibco), as previously described (4). Cultures were kept in standard cell culture conditions (37°C, 5% CO2, in humidified incubators). The cell line was previously characterized for a number of markers (4). The mHypoE-37 cell line was chosen due to its robust circadian clock gene rhythms, and its responsiveness to palmitate. This study used four distinct sets of experiments: 1) neurons that were untreated to examine basal expression patterns, 2) neurons treated with the saturated fatty acid palmitate, 3) neurons treated with the unsaturated fatty acid DHA, and 4) neurons pretreated with DHA followed by a combination of fatty acid treatments. In all experiments, 12 h prior to the serum shock, FBS-containing media were aspirated and replaced with media lacking FBS. Following serum starvation, cultures were shocked with 30% FBS (vol/vol) for 30 min; completion of the serum shock was considered time point 0. Serum starvation leads to quiescence of cells, and the subsequent serum shock synchronizes the cells (30). To determine basal expression, cells were placed into serum-containing media following the serum shock. RNA was then isolated every 3 h over a 36-h period. To determine the effects of fatty acids on inflammatory and orexigenic markers, cells were placed in culture media containing either 25 μM palmitate or water (Thermo Scientific, Nepean, Ontario, Canada), or 25 μM docosahexaenoic acid (DHA) or dimethylsulfoxide (DMSO, Sigma-Aldrich) immediately following serum shock. RNA was isolated every 3 h over a 36-h period. To determine the effects of DHA, after starvation, cells were pretreated with either 25 μM DHA (dissolved in <0.1% vol/vol DMSO) or DMSO vehicle (<0.1% vol/vol) for 1 h. The cells were then synchronized by a 30% FBS shock. Subsequently, the medium was replaced with treatment media containing either cotreatment of DHA and palmitate, or vehicle (DMSO and water, respectively). Thus, four groups of treatments were compared: vehicle control (DMSO-water), treatment with palmitate only (DMSO-palmitate), pretreatment with DHA only (DHA-water), and pretreatment with DHA followed by cotreatment of DHA and palmitate (DHA-palmitate). RNA was isolated every 3 h over a 24-h period. For a fifth experiment, cells were serum-starved for 12 h, followed by replacement with 5.5 mM glucose, 5% FBS media supplemented with either 25 μM palmitate, 25 μM DHA, water, or DMSO (<0.1% vol/vol). Protein was harvested before (time 0) and 30 min, 2 h, and 4 h after media change.
Fatty acid preparation.
An initial 100-mM stock concentration of palmitate was made by heating 27.8 mg of sodium palmitate (Sigma-Aldrich) in 1 ml of molecular grade water (Thermo Scientific, Nepean, Ontario, Canada) at 60°C. DHA was diluted to an initial 100-mM stock solution using DMSO. Subsequently, a 25-mM working stock was made for both palmitate and DHA using water and DMSO, respectively. The final experimental concentration of 25 μM palmitate and DHA was made by diluting the working concentration in culture media with or without FBS. It should be noted that a dose curve from 10 to 100 μM of palmitate was performed prior to the circadian analysis to determine the sensitivity of the mHypoE-37 neurons to palmitate. It was found that at a concentration higher than 25 μM of palmitate, the cells underwent significant cell death, and therefore, this was used as the maximal concentration of palmitate in this study. This is in contrast to the mHypoE-44 cell line used previously in studies with palmitate that could withstand concentrations of palmitate from 200 to 1,000 μM (15, 34), indicating that specific neuronal cell types have distinct sensitivity to FFAs, likely depending upon the coexpressions of receptors and signal transduction machinery involved in FFA signaling.
Semiquantitative RT-PCR.
Total RNA was isolated as previously described (4). Reverse transcription and PCR amplification was performed using SuperScript III One-Step RT-PCR System (Invitrogen, Burlington, ON) with sequence-specific primers. PCR products were subjected to 2% agarose gel electrophoresis and were visualized under UV illumination using the Kodak Image Station 2000R.
Quantitative RT-PCR.
Total RNA extraction and reverse transcription were performed as previously described (4, 15, 16). In brief, RNA was isolated using a modified guanidinium thiocyonate method. Prior to the generation of cDNA, genomic DNA was eliminated using the Turbo DNase kit (Ambion, Streetsville, Ontario, Canada). Single-stranded cDNA required for real-time quantitative RT-PCR (qRT-PCR) was synthesized using the high-capacity cDNA reverse transcription kit (Applied Biosystems) as per the manufacturer's instructions. cDNA was amplified using a qRT-PCR master mix (Platinum SYBR Green qPCR SuperMix-UDG with ROX; Invitrogen) and gene-specific primers. Samples and reagents were loaded in triplicate into 384-well plates and placed in an Applied Biosystems (Applied Biosystems) Prism 7000 Sequence Detection System (SDS) machine for amplification and detection. Results were analyzed using the Applied Biosytems SDS version 2.4 software.
Western blot analysis.
Cells were treated as described above, and protein was harvested at 0, 30 min, 2 h, and 4 h after media change, using 1 × cell lysis buffer (Cell Signaling) supplemented with 1 mM PMSF, 1% phosphatase inhibitor cocktail 2 (Sigma-Aldrich), and 1% protease inhibitor. A total of 25–30 μg of protein was subjected to 8% SDS-PAGE, and transferred onto a 0.22-μm PVDF membrane (Bio-Rad, Mississauga, ON, Canada). Membranes were blocked for 1 h at room temperature in Tris-buffered saline with Tween-20 buffer (0.1% TBS-T) and 5% nonfat dry milk followed by an overnight incubation with phospho-specific or total antibodies (1:1,000; Cell Signaling Technology, Danvers, MA). Blots were then washed in 0.1% TBS-T and incubated with HRP-conjugated secondary antibodies (1:5,000; Cell Signaling Technology) for 1 h at room temperature. Bands were visualized by enhanced chemiluminescence using ECL Select Western blotting detection reagent (GE Healthcare Life Sciences, Pittsburgh, PA) and quantified using Kodak Image Station 2000R software. Obtained values were normalized to total protein or G-β accordingly.
Statistical analysis.
The relative mRNA expression of each replicate was calculated by normalizing and standardizing the gene of interest to the expression of Histone 3a (16). mRNA transcript levels were first tested for circadian (∼24 h) expression using CircWave v1.4 (39). CircWave uses a linear harmonic regression fit that describes the data by adding harmonics to the principal wave function; it assumes a period of 24 h. Then, the exact period (i.e., the time that is needed to fulfill a complete cycle), amplitude (i.e., the difference between the peak or trough and the mean value of a cosine curve), acrophase (i.e., the phase angle of the peak of a cosine curve), and mesor (i.e., the average value around which the variable oscillates) of each gene were determined using cosinor analysis with the statistical software OriginPro: Release 8.5 (OriginLab, Northampton, MA) using the following equation: y = mesor + amplitude·cos{[2·pi·(x-acrophase)]/period}. According to the nature of data, one-way ANOVA, followed by a post hoc test as indicated in the figure legend, or Student's t-test were conducted using GraphPad Prism 6 (GraphPad Software, San Diego, CA) to determine statistical differences between treatment groups. P values <0.05 were considered significant.
RESULTS
mHypoE-37 neurons exhibit robust circadian rhythms.
Evidence suggests that every tissue or cell possesses the ability to exhibit a circadian rhythm. Additionally, it is well known that transcript levels of core circadian genes oscillate in a ∼24-h rhythm. To test this theory and establish a functional cellular circadian model, the mHypoE-37 neurons were used, as this cell line expresses the core circadian genes Bmal1, Per2, Rev-erbα, Clock, and Cry1, in addition to genes involved in energy homeostasis and inflammation, such as AgRP, IL-6, and TLR4. Under confocal microscopy, mHypoE-37 cells exhibited classical neuronal morphology in culture.
We evaluated whether the mHypoE-37 neurons express the core circadian genes Bmal1, Per2, Rev-erbα, and Clock in a circadian manner by harvesting RNA from cultures over a 36-h time course. Bmal1, Per2, and Rev-erbα were all found to have period lengths of ∼24 h (23.73 ± 0.72 h, 22.88 ± 1.21 h, and 28.28 ± 1.80 h, respectively; n = 3) (Fig. 1A, i–iii, Table 1). All three genes showed consistent, robust rhythms in each replicate. The transcriptional profile of Clock did not oscillate in a circadian manner (period 35.25 ± 5.76 h; Fig. 1A, iv, Table 1). Further, Cry1 displayed a circadian rhythm (period 23.24 h ± 3.18 h; data not shown).
Fig. 1.
mHypoE-37 neurons exhibit robust circadian rhythms of core clock genes. The mHypoE-37 neurons were placed in serum-free media for 12 h prior to receiving a 30% serum shock. Following synchronization, total RNA was harvested every 3 h for 36 h. RNA was subsequently converted into cDNA and used for qRT-PCR with Bmal1 (i), Per2 (ii), Rev-erbα (iii), and Clock (iv)-specific primers. All mRNA values are normalized to Histone 3a expression. A: Bmal1, Per2, and Rev-erbα mRNA levels in the mHypoE-37 cell line are expressed in circadian manner with a period length of 23.73 ± 0.72 h, 22.88 ± 1.21 h, and 28.28 ± 1.80 h, respectively. Transcription of Clock is rhythmic, but not circadian. B: Superimposition of Bmal1-Per2 (left) and Bmal1-Rev-erbα (right) expression profiles reveals that the genes are cycling in opposing phases. Values are plotted as mean values ± SE of three individual experiments.
Table 1.
Summary of parameters derived from cosinor analysis
| Figure/Experiment | Gene | Curve fit R2 | Period ± SE | Difference with vehicle | P value | Amplitude ± SE | Difference with vehicle | P value | Acrophase ± SE | Difference with vehicle | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Figure 1: Basal expression | Bmal1 | 0.94 | 23.73 ± 0.72 | 0.41 ± 0.03 | 9.15 ± 0.11 | ||||||
| Per2 | 0.96 | 22.88 ± 1.21 | 0.51 ± 0.03 | 0.75 ± 1.29 | |||||||
| Rev-erbα | 0.91 | 28.28 ± 1.80 | 0.65 ± 0.06 | 16.85 ± 0.59 | |||||||
| Clock | 0.42 | 35.25 ± 5.76 | 0.30 ± 0.09 | 11.37 ± 1.60 | |||||||
| Figure 2: Palmitate treatment | |||||||||||
| water | Bmal1 | 0.87 | 29.02 ± 1.74 | 0.48 ± 0.05 | 7.77 ± 0.81 | ||||||
| palmitate | Bmal1 | 0.66 | 30.58 ± 2.15 | 1.56 ± 2.74 | 0.58 | 0.47 ± 0.09 | 0.01 ± 0.10 | 0.9 | 9.18 ± 1.22 | 1.35 ± 1.44 | 0.36 |
| water | Per2 | 0.88 | 24.90 ± 1.13 | 0.60 ± 0.07 | 0.99 ± 0.86 | ||||||
| palmitate | Per2 | 0.82 | 24.89 ± 1.07 | 0.01 ± 1.57 | 1 | 0.60 ± 0.09 | 0.0 ± 0.11 | 1 | 0.71 ± 0.85 | 0.27 ± 1.21 | 0.83 |
| water | Rev-erbα | 0.53 | 24.32 ± 1.35 | 0.49 ± 0.13 | 17.71 ± 0.77 | ||||||
| palmitate | Rev-erbα | 0.33 | 27.09 ± 1.43 | 2.77 ± 1.97 | 0.21 | 0.53 ± 0.20 | 0.04 ± 0.24 | 0.87 | 19.14 ± 0.77 | 1.43 ± 1.09 | 0.24 |
| Figure 5: DHA-palmitate | |||||||||||
| DMSO-water | Bmal1 | 0.95 | 26.29 ± 0.83 | 0.52 ± 0.04 | 7.49 ± 0.39 | ||||||
| DMSO-palmitate | Bmal1 | 0.94 | 25.67 ± 1.72 | 0.82 ± 0.08 | 8.32 ± 0.39 | ||||||
| DHA-water | Bmal1 | 0.98 | 32.84 ± 1.44 | 0.57 ± 0.03 | 5.16 ± 0.41 | ||||||
| DHA-palmitate | Bmal1 | 0.96 | 26.73 ± 1.75 | 0.67 ± 0.06 | 6.81 ± 0.60 | ||||||
| DHA-water versus DHA-palm | P = 0.013 | DHA-water versus DMSO-palm | P = 0.008 | DHA-water versus DHA palm | P = 0.02 | ||||||
| DHA-water versus DMSO-water | P = 0.009 | DMSO-water versus DMSO-palm | P = 0.003 | DHA-water versus DMSO-water | P = 0.003 | ||||||
| DHA-water versus DMSO-palm | P = 0.005 | DHA-water versus DMSO-palm | P < 0.001 | ||||||||
| DHA-palm versus DMSO-palm | P = 0.04 |
Values are expressed as means ± SE. mRNA expression levels were analyzed by quantitative RT-PCR and subsequently analyzed by cosinor analysis. This table gives a summary of the calculated period lengths, amplitudes and acrophases, per gene per experiment. Curve fit R2 indicates the goodness of the curve fit. P values indicate outcome of a Student's t-test between palmitate-treated cells and water controls. For the DHA-palmitate experiment, P values represent the significant outcomes of the post hoc Tukey's test after significance was reached in a two-way ANOVA for factors time and treatment (time: P < 0.001; treatment: P = 0.004; time × diet: P < 0.001).
Moreover, it is well established that the mRNA transcripts of the negative and positive arms of the circadian cycle oscillate in opposing phases (42, 54). Indeed, a superimposition of the transcriptional profiles of Bmal1-Per2 and Bmal1-Rev-erbα revealed that these genes cycle in opposing phases (Fig. 1B). Given that the mHypoE-37 cells possess the necessary molecular circadian machinery and cycle in a rhythmic manner reflective of the physiological arms of the circadian clock, we postulated that this cell line functions as a viable model for circadian molecular studies.
Saturated fatty acid palmitate significantly alters circadian rhythms.
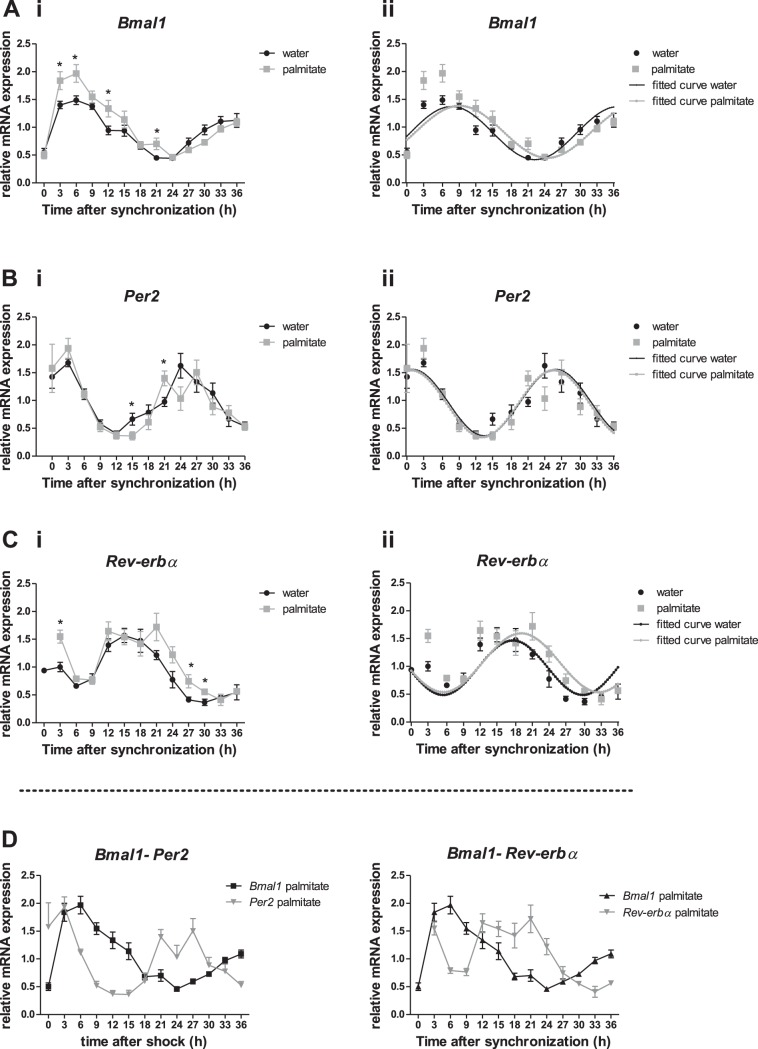
To explore the effect that SFAs have on the molecular circadian system, the mHypoE-37 neurons were treated with the 16-carbon, saturated fatty acid palmitate. Transcriptional analysis included an examination of the expression profile of each gene for changes in period length, amplitude, and acrophase. When analyzed using CircWave V1.4, Bmal1, Per2 and Rev-erbα mRNA expression significantly matched a 24-h cosine wave (P < 0.01, P < 0.05, and P < 0.05 respectively, Fig. 2, Table 1).
Fig. 2.
Effects of saturated fatty acids on the expression profile of Bmal1, Per2, and Rev-erbα. The effects of fatty acids on the circadian system in the mHypoE-37 neurons were completed by exposure to a treatment of 25 μM palmitate following serum shock. Total RNA was harvested every 3 h for 36 h and subsequently used for quantitative RT-PCR (qRT-PCR). All values were normalized to Histone 3a. A, i, B, i, C, i: relative mRNA transcript levels of Bmal1, Per2, and Rev-erbα, respectively. Bmal1 transcript levels are upregulated in the palmitate-treated cells compared with controls. *P < 0.05 between palmitate and control group at the indicated time points, as determined by two-way ANOVA with post hoc Student's t-test. Values are plotted as means ± SE; n = 8–9 for Bmal1 and Per2, and n = 4 for Rev-erbα. A,ii, B,ii, C,ii: Transcript levels of the individual groups as shown in A,i, B,i, and C,i were subjected to cosinor analysis to determine period length, amplitude, and acrophase of each gene. Relative mRNA expression levels of Bmal1, Per2, and Rev-erbα, respectively (similar to A, i, B, i, and C, i), are plotted together with the obtained fitted curves from the cosinor analysis. Gray squares denote palmitate; black circles denote water. Solid gray line denotes fitted curve palmitate, while the solid black line denotes fitted-curve water. Analysis showed a phase delay of 1.4 h in Bmal1 and Rev-erbα rhythms in the palmitate treated group compared with controls, but this did not reach significance. D: superimposition of the relative mRNA expression levels of Bmal1-Per2 and Bmal1-Rev-erbα, after treatment with 25 μM of palmitate. Although Bmal1 continues to cycle in an opposing phase with Per2 and Rev-erbα, the expression profiles differ from the basal expression profiles, as shown in Fig. 1B.
Transcript levels of the examined genes throughout the 36-h time course were compared between the palmitate-treated and control group. A two-way ANOVA with post hoc t-test showed a significant upregulation of Bmal1 transcript levels at individual time points within the first 24 h after synchronization in the palmitate-treated group compared with the control group. This upregulation was significant at 3, 6, 12, and 21 h after synchronization (Fig. 2A, i). When comparing Per2 mRNA expression at individual time points, there was a significant downregulation at 15 h, and a significant upregulation at 21 h after synchronization in the palmitate-treated group compared with the control group (Fig. 2B, i). Rev-erbα transcript levels were significantly upregulated in the palmitate group compared with the control group at 3, 27, and 30 h after synchronization (Fig. 2C, i). Next, we used a cosinor analysis using OriginPro to describe and compare the circadian expression pattern between the groups. This cosinor analysis did not reveal significant differences in period or amplitude between control and palmitate-treated groups for Bmal1, Per2, and Rev-erbα (Fig. 2A, ii, B, ii, C, ii; Table 1) or Cry1 (data not shown). The circadian profiles were also examined for changes in acrophase, the time at which the cycle peaks. Palmitate exposure caused a phase delay of 1.4 h in both Bmal1 and Rev-erbα, but this did not reach significance (Fig. 2 and Table 1).
Taken together, these results suggest that palmitate can significantly alter the circadian clock at the molecular level, suggesting a dysregulation of basal circadian clock function.
Twenty-five micromoles of palmitate does not induce cellular inflammatory markers.
Fatty acids have recently been identified as a mediator of hypothalamic inflammation commonly associated with obesity (10). Therefore, the effect of palmitate on the transcript levels of key inflammatory genes was investigated. In the mHypoE-37 neurons, markers of cellular inflammation, TLR4, IκBα, and IL-6, were found to be expressed at levels detectable by qRT-PCR. Furthermore, qRT-PCR analysis revealed minor, insignificant increases in IL-6 and TLR4 transcript levels in the palmitate-treated cells compared with the control group, while nonappreciable changes were observed in IκBα (Fig. 3, A–C).
Fig. 3.
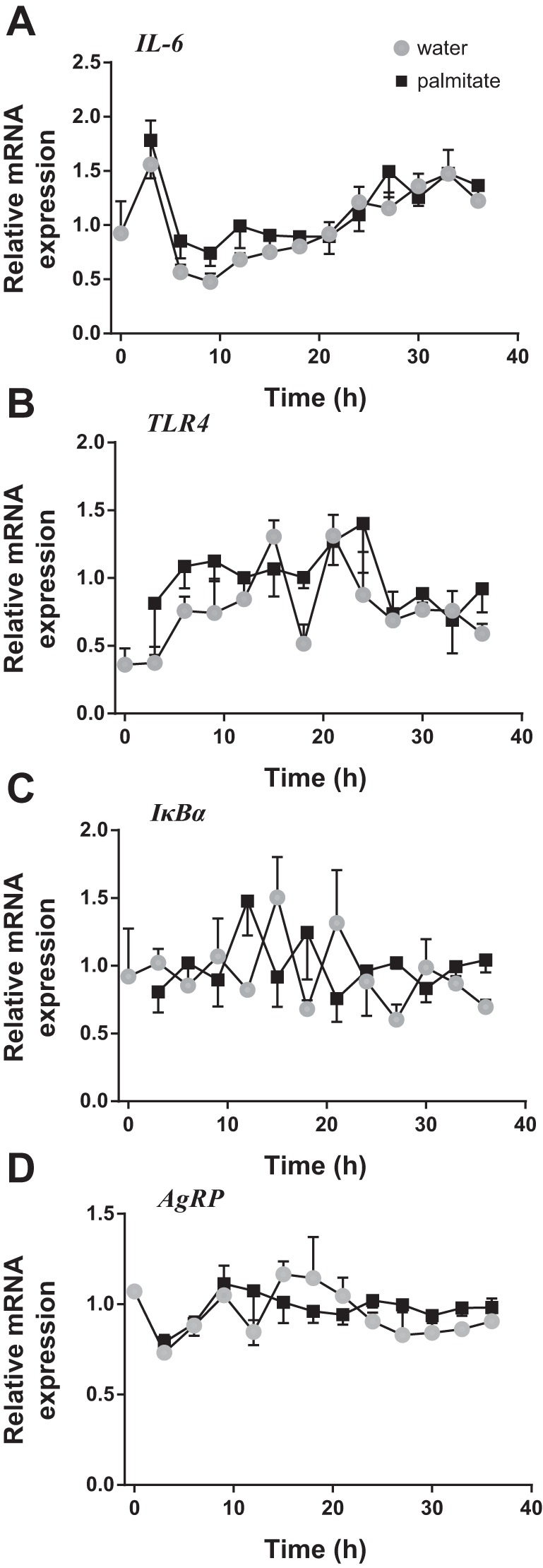
Effects of the saturated fatty acid palmitate on inflammatory and metabolic gene transcription. The mHypoE-37 cells were treated with 25 μM of palmitate following serum shock. Total RNA was harvested every 3 h for 36 h and subsequently used for qRT-PCR. All values were normalized to Histone 3a. A–D: exposure to palmitate did not alter the expression of any examined inflammatory genes, IL-6, TLR4, or IκBα, or the orexigenic neuropeptide AgRP. Gray circles denote water, while black squares denote palmitate. Values are plotted as means ± SE of four individual experiments.
Furthermore, fatty acids have been found to alter levels of specific neuropeptides involved in energy homeostasis (7, 36). As such, transcript levels of the orexigenic neuropeptide AgRP were examined following palmitate treatment. AgRP mRNA levels did not fluctuate, and expression levels were not significantly different between the palmitate-treated cells and the control group (Fig. 3D).
Twenty-five micromoles of DHA downregulates IL-6 mRNA expression.
As with the saturated fatty acid experiments, the effects of omega-3 fatty acids on the cellular inflammatory response were also studied. Transcript levels of the proinflammatory cytokine, IL-6, were found to be significantly downregulated between the hours of 21 and 33, inclusively (Fig. 4A). DHA neither altered the expression of IκBα nor TLR4 (Fig. 4, B and C). However, IκBα was slightly elevated during the second half of the time course, although this did not reach significance. Moreover, no changes were found in the transcription of AgRP throughout the duration of the time course (Fig. 4D).
Fig. 4.

Effects of the unsaturated fatty acid DHA on inflammatory and metabolic gene transcription. The mHypoE-37 cell line was treated with 25 μM DHA following serum shock. Total RNA was harvested every 3 h for 36 h and subsequently used for qRT-PCR. All values were normalized to Histone 3a. A: exposure to 25 μM DHA significantly reduced the expression of IL-6 between the hours of 21 and 33, inclusively. However, DHA treatment did not alter the expression of TLR4, IκBα, or AgRP (B–D). Values are plotted as mean values ± SE of four individual experiments. *P < 0.05 between DHA-treated cells and DMSO controls.
Differential effects of DHA and palmitate on the expression levels of Bmal1.
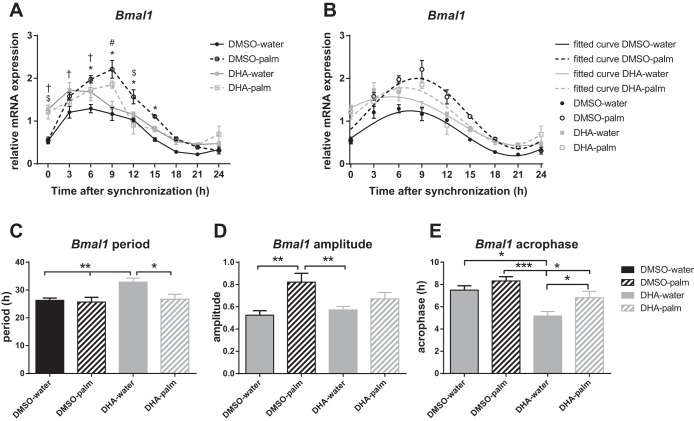
To evaluate the effects of the unsaturated fatty acid DHA and to assess whether DHA confers protective effects and prevents alterations in the circadian expression pattern of Bmal1 induced by palmitate, cells were pretreated with DHA (dissolved in DMSO) or DMSO for 1 h prior to synchronization. Following synchronization, cultures were treated with either a combination of DHA and palmitate, or palmitate alone. Given the significant upregulation of Bmal1 induced by palmitate (Fig. 2A, i), for the effects of DHA, the focus was on Bmal1.
The 1-h pretreatment with DHA increased levels of Bmal1, as the transcript level of Bmal1 at time point 0 h is significantly higher in the DHA-pretreated groups compared with the vehicle-pretreated groups (Fig. 5A). The upregulating effect of DHA on Bmal1 transcript levels was significant during the first 6 h after synchronization, compared with the vehicle (DHA-water vs. DMSO-water). Treatment with palmitate increased Bmal1 transcript levels compared with the vehicle-treated groups (DMSO-palm vs. DMSO-water) consistently throughout the entire 24 h, and this difference was statistically significant at the 6-, 9-, 12-, and 15-h time points. In contrast, when the cells were pretreated with DHA, treatment with palmitate only resulted in an upregulation of Bmal1 at the 9-h time point, whereas at the other time points, no differences were observed between the palmitate- and vehicle-treated groups (DHA-palm vs. DHA-water).
Fig. 5.
Differential effects of the unsaturated fatty acid DHA and palmitate on the expression levels and circadian expression profile of Bmal1. A: relative mRNA transcript levels of Bmal1 in the vehicle-treated group (DMSO-water, solid black circles), palmitate-treated group (DMSO-palmitate, open black circles), DHA-pretreated group (DHA-water, solid gray squares), and DHA-pretreated cells followed by DHA-palmitate cotreatment (DHA-palm; open gray squares). The mHypoE-37 neurons were pretreated for 1 h with either 25 μM DHA or DMSO vehicle, followed by synchronization. Next, medium was replaced into treatment media containing either cotreatment of 25 μM DHA and 25 μM palmitate, or vehicle (water). Total RNA was harvested every 3 h for 24 h and subsequently used for qRT-PCR. All values were normalized to Histone 3a. Values are plotted as means ± SE of four individual experiments. Symbols indicate significant differences with *P < 0.05, DMSO-palm vs. DMSO-water; #P < 0.05, DHA-palm vs. DHA-water; $P < 0.05, DMSO-palm vs. DHA-palm; †P < 0.05, DMSO-water vs DHA-water, as indicated by two-way ANOVA followed by post hoc Tukey's t-test. B: transcript levels of the individual groups as in 5A were subjected to cosinor analysis to determine period length, amplitude, and acrophase. Relative mRNA expression levels of Bmal1 (similar to 5A) are plotted together with the obtained fitted curves from the cosinor analysis. Statistical comparison between period length, amplitude, and acrophase is depicted in C, D, and E, respectively. C: Bmal1 period length for different treatment groups. DHA pretreatment lengthens the period compared with cells pretreated with vehicle, and compared with cells treated with palmitate after DHA-pretreatment. D: Bmal1 amplitude for different treatment groups. Palmitate significantly increases the amplitude compared with vehicle control. DHA pretreatment prevents the palmitate-induced increase in Bmal1 amplitude. E: Bmal1 acrophase for different treatment groups. DHA causes a phase advance compared with vehicle control. Palmitate treatment after DHA pretreatment results in a similar phase as the vehicle control. Values are plotted as mean values ± SE. *P < 0.05; **P < 0.01; ***P < 0.001, as indicated by one-way ANOVA followed by post hoc least significant difference test.
Within the palmitate-treated groups, pretreatment with DHA reduced Bmal1 expression 12 h after synchronization compared with the vehicle-treated group, but at the other time points no significant differences were observed, despite Bmal1 levels being consistently lower in the DHA-pretreated group (DHA-palm vs. DMSO-palm). However, the upregulation of Bmal1 by palmitate, as seen in the DMSO-palm group, was less pronounced when the cells were pretreated with DHA (DHA-palm), and the transcript levels rapidly decrease to levels similar to DHA-only treated cells (Fig. 5A). The peak expression of Bmal1 was delayed in the palmitate-treated groups as compared with the vehicle-treated groups (peak at 9 h in the DMSO-palm and DHA-palm group vs. 3 and 6 h in the DHA-water and DHA-palm group, respectively).
Differential effects of DHA and palmitate on the circadian expression pattern of Bmal1.
Fitted curves of the individual data points were calculated using a cosinor analysis (Fig. 5B and Table 1). Comparing the fitted curves, it was demonstrated that pretreatment with DHA significantly elongated the period of Bmal1 compared with the nonpretreated groups (DHA-water vs. DMSO-water: P = 0.009 and DMSO-palm: P = 0.005). This elongation was not observed in the DHA-pretreated group followed by palmitate treatment (Fig. 5C). The amplitude of Bmal1 expression in the palmitate-treated group was significantly higher than in the vehicle-treated group (DMSO-palm vs. DMSO-water: P = 0.003). In the DHA-pretreated groups, palmitate did not lead to an increased amplitude (DHA-palm vs. DHA-water: P = 0.226). DHA phase advanced the period of Bmal1 compared with the vehicle control, regardless of whether the pretreatment was followed by palmitate or vehicle (DMSO-water vs. DHA-water: P = 0.003; DMSO-palm vs. DHA-palm: P = 0.035) (Fig. 5D). Treatment with palmitate delayed the phase compared with vehicle control, but this was only statistically significant when palmitate was pretreated and cotreated with DHA (DMSO-water vs. DMSO-palm: P = 0.217; DHA-water vs. DHA-palm: P = 0.024). Compared with the vehicle-pretreated groups (DMSO-water and DMSO-palm), pretreatment with DHA caused a significant phase advance. However, DHA pretreatment followed by palmitate (DHA-palm) resulted in an acrophase similar to the vehicle control group (DHA-palm vs. DMSO-water: P = 0.301; Fig. 5E).
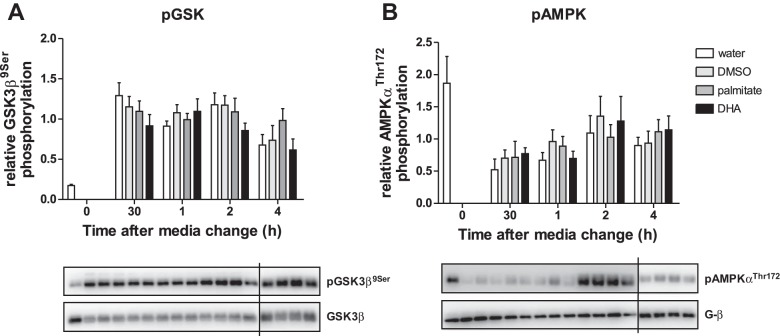
Phosphorylation of GSK and AMPK is not affected by palmitate or DHA.
To study putative mechanisms by which palmitate exerts its effects on the clock genes, we focused on glycogen synthase kinase 3β (GSK3β) and AMPK. AMPK, a heterotrimeric protein kinase that functions as a central regulator of metabolic processes, has been shown to mediate circadian regulation (reviewed in Ref. 21). GSK3β is activated by SFAs and can ultimately affect the circadian transcription of Bmal1 (13, 53). After 12 h of serum starvation, replenishing the cells with fresh media increased levels of phosphorylated GSK3β, hence inactivating GSK3β, and reduced levels of phosphorylated AMPK, hence activating AMPK. At 2 h after media change, there was a slight downregulation of pGSK3β by DHA, indicating activation of GSK3β, but this effect was not statistically significant (Fig. 6). Neither palmitate nor DHA had a significant effect on the temporal phosphorylation levels of AMPK after 4 h (data not shown).
Fig. 6.
Treatment with 25 μM of palmitate or 25 μM of DHA does not cause alterations in phosphorylation levels of GSK3β and AMPK. The mHypoE-37 cells were serum-starved for 12 h prior to media change with 25 μM palmitate, 25 μM DHA, water, or DMSO. Cell lysates were collected at 30 min, 1 h, 2 h, and 4 h after the medium was changed. For Western blot analysis, protein expression was normalized to total protein (for pGSK3β) or G-β (for pAMPK) levels. Graphs display means ± SE of four or five individual experiments. No differences were detected in phosphorylation levels of GSK3β (A) or AMPK (B) with palmitate or DHA treatment. Representative Western blots are presented for each histogram. The representative Western blots are loaded in the same order as the histograms above, whereas the vertical line represents data taken from a second gel/Western blot split due to the number of experimental parameters analyzed.
DISCUSSION
The use of defined cell lines to study the regulation of circadian rhythms allows for a thorough examination of the direct effects of nutrients on specific hypothalamic neuronal populations expressing unique complements of neuropeptides, receptor, and signaling components. While basic levels of clock gene expression can be studied in vivo after HFD exposure, it is difficult to delineate specific mechanisms involved in altered rhythms due to substantial extraneous physiological inputs. The mHypoE-37 cell line strongly expressed Bmal1, Per2, Rev-erbα, and Cry1 mRNA transcripts, which were found to cycle with a ∼24-h period, and was, therefore, found to be a valid model for circadian studies. The transcriptional profile of Clock was also analyzed, but was not found to possess circadian properties. Nonrhythmic expression of Clock has been reported in many other cell types, including the GT1–7 neurons, primary retinal tissue, and even primary SCN tissue (17, 22). The proper time-dependent expression was confirmed by superimposing the transcriptional profiles of Bmal1-Per2 and Bmal1-Rev-erbα, whereby the transcripts cycle in opposing phases (Fig. 1). Overall, given that the mHypoE-37 cell line possesses the necessary cellular circadian machinery and that the core circadian genes are expressed in the correct temporal-rhythmic manner, these data validate this cell line as a functional model for circadian molecular studies.
Studies have been shown that both palmitate and DHA can cross the blood-brain barrier and can reach the hypothalamus (23, 46). Thus, treating the mHypo-37 neuronal cells with palmitate and/or DHA is a valid model to test the effects of these fatty acids on hypothalamic clock gene expression. In 2007, Kohsaka et al. (27) demonstrated in mice that a HFD disrupted behavioral and molecular circadian rhythms, indicating that a HFD can affect the central clock, as well as peripheral clocks. In a similar fashion, the mHypoE-37 neurons, indeed, displayed altered circadian rhythms following exposure with the SFA palmitate. Of particular importance, palmitate induced an increase in the expression levels of Bmal1 during the first 24 h, with significant increases at 3, 6, 12, and 21 h after synchronization compared with the control group. Furthermore, palmitate caused a phase delay of 1.4 h in Bmal1 and Rev-erbα transcript levels (Fig. 2). This is in line with a study in which a HFD caused alterations in the expression levels of Rev-erbα in adipose tissue. Although this difference was not statistically significant, a shift of this magnitude may be physiologically relevant. REV-ERBα protein, which modulates Bmal1 transcription, participates in the regulation of many metabolic processes, including gluconeogenesis and cholesterol homeostasis. Furthermore, daily expression of Rev-erbα controls the circadian rhythm in hepatic lipid metabolism via recruitment of histone deacetylase 3 (14). Although we present a model here for hypothalamic, peripheral clocks, the alterations in Rev-erbα caused by palmitate, could be a putative mechanism through which fatty acids can modulate alterations in genes involved in metabolism, through disruptions in the molecular clock.
Increased levels of Bmal1 mRNA are expected to result in higher levels of BMAL1 protein levels and would lead to an increase in Per mRNA transcription. Interestingly, transcript levels of Per2 were not affected by palmitate. Resilience of Per transcription has been described before. Lee et al. (31) reported only modest increases in Per mRNA levels after overexpression of CLOCK-BMAL in fibroblasts, and compared with other direct target genes, including Rev-erbα, Per mRNA levels are less easily disturbed (11). Superimposition of Bmal1 and Per2 expression profiles after palmitate treatment indicated that the two genes continuously cycle in opposing phases, but the ratio between the two genes at each time point changed (Fig. 2D). The ratio between PER-CRY and CLOCK-BMAL1 is important for robustness of circadian rhythms (31). Although palmitate impacted molecular circadian rhythms, palmitate did not alter the expression of either the examined inflammatory markers (IL-6, TLR4, and IκBα) or the orexigenic neuropeptide AgRP mRNA [in accordance with our previous study in the mHypoE-44 cell line (15)], suggesting that the hypothalamic molecular clock is susceptible to palmitate-induced changes, even before the induction of the inflammatory response by high levels of palmitate. Furthermore, it suggests that disruption of the molecular clock is the result of a mechanism distinct from that of hypothalamic cellular inflammation or ER stress-induced cell death (15, 34). SFAs can exert their cellular effects through numerous mechanisms, including activation of specific signaling molecules (34, 37). With respect to signaling molecules, GSK3β is activated by SFAs and known to modulate acetylation of proteins (13, 53). Activation of GSK3β can also alter the stability of REV-ERBα, thereby reducing the suppression of Bmal1 (53). In the present study, we did not find significant effects of palmitate or DHA on the phosphorylation of GSK, indicating that the alterations in Bmal1 and Rev-erbα were independent of GSK activation.
An additional link between metabolism and circadian rhythms is formed by the peroxisome proliferator-activated receptors (PPARs) (51, 52). PPARs are transcription factors activated by various nutrients, including fatty acids and their derivatives. Although PPARs are expressed mostly in tissues with a high rate of fatty oxidation, they have also been found in the brain (35). The transcription of Ppar is driven by BMAL1, and PPARs in their turn activate the transcription of Bmal (6, 32). As PPARs regulate Bmal1 transcription, it can be speculated that nutrients, and, in particular, fatty acids, can influence the molecular clock through PPAR. Conversely to SFA, the omega-3 PUFAs have recently gained much attention for their potential to combat obesity and Type 2 diabetes mellitus (9, 25, 44). However, there is a paucity of studies examining their effects on circadian rhythms. This study is the first to examine the effects of omega-3 PUFAs on the molecular circadian clock. To assess the effects of DHA on the molecular clock, and to test the potential protective ability of DHA, the mHypoE-37 neurons were pretreated with DHA to determine potential rescue from palmitate-mediated disruption of basal circadian patterns. Given that palmitate most significantly altered the expression profile of Bmal1, for DHA, we focused our attention on Bmal1.
In Fig. 5, we show that palmitate increased the amplitude of Bmal1, whereas no effects were observed on period length or on acrophase. The increased amplitude in the DMSO-palmitate group is comparable to the observed upregulation of Bmal1, as presented in Fig. 2. The difference between these two groups is, that in the first experiment, palmitate was dissolved in water, whereas in this experiment, it was dissolved in DMSO. In contrast to palmitate, DHA alone had no effect on the amplitude of Bmal1, whereas it increased period length and phase advanced Bmal1 expression compared with the vehicle control (DHA-water vs. DMSO-water). These results show that both palmitate and DHA can affect the circadian expression pattern of Bmal1, although the effects on amplitude, period, and acrophase are different between the two fatty acids.
Furthermore, a putative protective effect of DHA was studied by pretreating the cells with DHA followed by cotreatment with DHA and palmitate. Compared with cells that only received DHA (pre)-treatment, DHA followed by DHA-palmitate cotreatment (DHA-palm) caused a shortened period and a phase delay, whereas no difference in amplitude was observed. This suggests that palmitate and DHA have differential effects on the expression profile of Bmal1 in this hypothalamic cell model. Although both palmitate and DHA increased Bmal1 transcript levels, the effect of palmitate was later, and lasted longer than the effect of DHA (Fig. 5A). Interestingly, the significant upregulation of Bmal1 after palmitate treatment was reduced after pretreatment with DHA. This suggests that DHA may protect the cells from palmitate-induced increases in Bmal1 expression. Furthermore, palmitate after DHA pretreatment resulted in a phase advance compared with palmitate without DHA pretreatment. In contrast to DHA alone, palmitate alone did not alter the acrophase of Bmal1, suggesting that DHA has an overriding effect on the phase of Bmal1, when given together with palmitate. In summary, both palmitate and DHA were able to significantly, but differentially, alter the circadian expression profile of Bmal1 in this neuronal cell model. When given together, the putative detrimental effects on circadian parameters were reduced.
The mechanisms through which DHA and palmitate affect Bmal1 are still elusive. As described, PPARs are activated by fatty acids, including omega-3 PUFAs, and can stimulate the expression of Bmal1. How this may differentially alter the circadian expression pattern of Bmal1 requires further analysis. Activation of PPARγ by DHA may explain the robust downregulation in IL-6 mRNA expression following DHA treatment, as omega-3 PUFAs have been shown to significantly reduce inflammation, including the transcription and secretion of IL-6 (1, 38). Omega-3 PUFAs are thought to suppress inflammation through PPARγ by both its activity as a transcription factor and as an inhibitor of NF-κB (47, 50).
We hypothesized that AMPK could be involved in the protective effect of DHA on the circadian clock for several reasons. DHA can induce activation of AMPK, leading to a decreased cellular concentration of palmitate, thus removing the action of palmitate on the circadian clock. Furthermore, increased AMPK activity can inhibit the transcriptional activation of both PPARα and PPARγ, thereby reducing the induction of Bmal1 expression (45). In this study, phosphorylation levels of AMPK were not affected by palmitate or DHA, indicating that the disruptive effects of palmitate within this hypothalamic cell model are independent of AMPK activation. The rapid decrease in AMPK phosphorylation levels after media change indicates that the cells are able to detect changes in nutrient levels and that AMPK activity changes accordingly. It has been shown in another hypothalamic cell line, mHypoE-44, that a substantially higher concentration of palmitate (300 μM) increased the phosphorylation levels of AMPK at 24 h (15). Although this could be true for the mHypoE-37 cells, this would not explain the early increase in Bmal1 transcript levels and suggests that palmitate affects the molecular circadian clock independently of AMPK.
This study presents a novel hypothalamic cell model for the study of the transcriptional and molecular events underlying cellular circadian rhythms, and the potential alterations caused by exposure to dietary components, specifically fatty acids. Herein, the mHypoE-37 cell line has been shown to possess robust rhythmic expression of the circadian clock genes, Bmal1, Per2, and Rev-erbα, representative of those found in vivo. Additionally, this study is the first to demonstrate that both the saturated fatty acid, palmitate, and the omega-3 polyunsaturated fatty acid, DHA, are able to affect clock gene expression in hypothalamic cells. Furthermore, this study suggests that DHA can ameliorate the disruptive effects of palmitate on the core clock gene Bmal1.
Perspectives and Significance
Disruptions of normal rhythmicity in clock genes can lead to many metabolic disruptions, including altered gluconeogenesis, lipogenesis, and metabolic syndrome. In this study, we demonstrated that palmitate, a saturated fatty acid that is highly abundant in the human diet, is able to alter gene expression of core clock genes in a cellular model of hypothalamic neurons. Although DHA also altered the rhythmic expression pattern of Bmal1, the effect was different from palmitate, and the combination of DHA and palmitate resulted in less pronounced alterations in Bmal1. Although the mechanism via which this occurs is still elusive, we speculate that alterations in the core clock mechanism could lead to alterations in the expression of other factors, including the nuclear hormone receptor PPARγ, which is known to be involved in lipid oxidation. The exact role of PPARγ in the hypothalamus is not yet clear, and studies need to be conducted to assess the involvement of hypothalamic PPARγ in energy homeostasis. Overall, these results contribute to the growing understanding of the direct effects that fatty acids may exert on the cellular circadian system, specifically in hypothalamic neurons linked to energy homeostasis.
GRANTS
We acknowledge funding from the Natural Sciences and Engineering Research Council (NSERC), Canadian Institutes for Health Research (CIHR), and Canada Foundation for Innovation and Canada Research Chairs Program (to D. D. Belsham). J. A. Greco was supported by a CIHR Sleep and Biological Rhythms Training Grant Studentship. J. E. Oosteman. was supported by an Academic Medical Center Ph.D. scholarship.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: J.A.G., J.E.O., and D.D.B. conception and design of research; J.A.G. and J.E.O. performed experiments; J.A.G., J.E.O., and D.D.B. analyzed data; J.A.G., J.E.O., and D.D.B. interpreted results of experiments; J.A.G., J.E.O., and D.D.B. prepared figures; J.A.G., J.E.O., and D.D.B. drafted manuscript; J.A.G., J.E.O., and D.D.B. edited and revised manuscript; J.A.G., J.E.O., and D.D.B. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Jennifer Chalmers, Alice Treen, and Dean Tran for their generous support with the experiments. Many thanks to Dr. Leigh Wellhauser for invaluable insight and technical assistance.
REFERENCES
- 1.Babcock TA, Novak T, Ong E, Jho DH, Helton WS, Espat NJ. Modulation of lipopolysaccharide-stimulated macrophage tumor necrosis factor-alpha production by omega-3 fatty acid is associated with differential cyclooxygenase-2 protein expression and is independent of interleukin-10. J Surg Res 107: 135–139, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre A. Resetting of Circadian Time in Peripheral Tissues by Glucocorticoid Signaling. Science 289: 2344–2347, 2000 [DOI] [PubMed] [Google Scholar]
- 3.Barnea M, Madar Z, Froy O. High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology 150: 161–168, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Belsham DD, Cai F, Cui H, Smukler SR, Salapatek AM, Shkreta L. Generation of a phenotypic array of hypothalamic neuronal cell models to study complex neuroendocrine disorders. Endocrinology 145: 393–400, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009–1017, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol 20: 1715–1727, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Morris MJ. Differential responses of orexigenic neuropeptides to fasting in offspring of obese mothers. Obesity (Silver Spring) 17: 1356–1362, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Clarke SD. Polyunsaturated fatty acid regulation of gene transcription: a mechanism to improve energy balance and insulin resistance. Br J Nutr 83 Suppl 1: S59–66, 2000 [DOI] [PubMed] [Google Scholar]
- 9.Cunnane SC, McAdoo KR, Horrobin DF. n-3 Essential fatty acids decrease weight gain in genetically obese mice. Br J Nutr 56: 87–95, 1986 [DOI] [PubMed] [Google Scholar]
- 10.De Souza CT, Araujo EP, Bordin S, Ashimine R, Zollner RL, Boschero AC, Saad MJ, Velloso LA. Consumption of a fat-rich diet activates a proinflammatory response and induces insulin resistance in the hypothalamus. Endocrinology 146: 4192–4199, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50: 465–477, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Duffield GE. DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocrinol 15: 991–1002, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Eom TY, Jope RS. GSK3β N-terminus binding to p53 promotes its acetylation. Mol Cancer 8: 14, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fick LJ, Fick GH, Belsham DD. Palmitate alters the rhythmic expression of molecular clock genes and orexigenic neuropeptide Y mRNA levels within immortalized, hypothalamic neurons. Biochem Biophys Res Commun 413: 414–419, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Fick LJ, Fick GH, Belsham DD. Rhythmic clock and neuropeptide gene expression in hypothalamic mHypoE-44 neurons. Mol Cell Endocrinol 323: 298–306, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280: 1564–1569, 1998 [DOI] [PubMed] [Google Scholar]
- 18.Hainault I, Carolotti M, Hajduch E, Guichard C, Lavau M. Fish oil in a high lard diet prevents obesity, hyperlipemia, and adipocyte insulin resistance in rats. Ann NY Acad Sci 683: 98–101, 1993 [DOI] [PubMed] [Google Scholar]
- 19.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15: 848–860, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishida N, Noji S, Ono K, Koyama E, Nohno T, Taniguchi S, Tokunaga A, Fujii T, Mitsui Y. Diurnal regulation of per repeat mRNA in the suprachiasmatic nucleus in rat brain. Neurosci Lett 122: 113–116, 1991 [DOI] [PubMed] [Google Scholar]
- 21.Jordan SD, Lamia KA. AMPK at the crossroads of circadian clocks and metabolism. Mol Cell Endocrinol 366: 163–169, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamphuis W, Cailotto C, Dijk F, Bergen A, Buijs RM. Circadian expression of clock genes and clock-controlled genes in the rat retina. Biochem Biophys Res Commun 330: 18–26, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Kasser TR, Deutch A, Martin RJ. Uptake and utilization of metabolites in specific brain sites relative to feeding status. Physiol Behav 36: 1161–1165, 1986 [DOI] [PubMed] [Google Scholar]
- 24.Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int J Obes (Lond) 32: 1431–1437, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Khalfoun B, Thibault F, Watier H, Bardos P, Lebranchu Y. Docosahexaenoic and eicosapentaenoic acids inhibit in vitro human endothelial cell production of interleukin-6. Adv Exp Med Biol 400B: 589–597, 1997 [PubMed] [Google Scholar]
- 26.Kim F, Pham M, Luttrell I, Bannerman DD, Tupper J, Thaler J, Hawn TR, Raines EW, Schwartz MW. Toll-like receptor-4 mediates vascular inflammation and insulin resistance in diet-induced obesity. Circ Res 100: 1589–1596, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6: 414–421, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A 68: 2112–2116, 1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205, 1999 [DOI] [PubMed] [Google Scholar]
- 30.Larsson O, Zetterberg A, Engström W. Cell-cycle-specific induction of quiescence achieved by limited inhibition of protein synthesis: counteractive effect of addition of purified growth factors. J Cell Sci 73: 375–387, 1985 [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. The Journal of biological chemistry 286: 7033–7042, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest 116: 571–580, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 5: 407–441, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer CM, Belsham DD. Palmitate attenuates insulin signaling and induces endoplasmic reticulum stress and apoptosis in hypothalamic neurons: rescue of resistance and apoptosis through adenosine 5′ monophosphate-activated protein kinase activation. Endocrinology 151: 576–585, 2010 [DOI] [PubMed] [Google Scholar]
- 35.Moreno S, Farioli-Vecchioli S, Ceru MP. Immunolocalization of peroxisome proliferator-activated receptors and retinoid X receptors in the adult rat CNS. Neuroscience 123: 131–145, 2004 [DOI] [PubMed] [Google Scholar]
- 36.Morris MJ, Chen H, Watts R, Shulkes A, Cameron-Smith D. Brain neuropeptide Y and CCK and peripheral adipokine receptors: temporal response in obesity induced by palatable diet. Int J Obes (Lond) 32: 249–258, 2008 [DOI] [PubMed] [Google Scholar]
- 37.Nakamura S, Takamura T, Matsuzawa-Nagata N, Takayama H, Misu H, Noda H, Nabemoto S, Kurita S, Ota T, Ando H, Miyamoto K, Kaneko S. Palmitate induces insulin resistance in H4IIEC3 hepatocytes through reactive oxygen species produced by mitochondria. The Journal of biological chemistry 284: 14809–14818, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, Li P, Lu WJ, Watkins SM, Olefsky JM. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142: 687–698, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms 21: 350–361, 2006 [DOI] [PubMed] [Google Scholar]
- 40.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320, 2002 [DOI] [PubMed] [Google Scholar]
- 41.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43: 527–537, 2004 [DOI] [PubMed] [Google Scholar]
- 42.Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet 38: 312–319, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman H, Genzer Y, Cohen R, Chapnik N, Madar Z, Froy O. Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB J 26: 3493–3502, 2012 [DOI] [PubMed] [Google Scholar]
- 44.Shimura T, Miura T, Usami M, Ishihara E, Tanigawa K, Ishida H, Seino Y. Docosahexanoic acid (DHA) improved glucose and lipid metabolism in KK-Ay mice with genetic non-insulin-dependent diabetes mellitus (NIDDM). Biol Pharm Bull 20: 507–510, 1997 [DOI] [PubMed] [Google Scholar]
- 45.Sozio MS, Lu C, Zeng Y, Liangpunsakul S, Crabb DW. Activated AMPK inhibits PPAR-α and PPAR-γ transcriptional activity in hepatoma cells. Am J Physiol Gastrointest Liver Physiol 301: G739–G747, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spector AA. Plasma free fatty acid and lipoproteins as sources of polyunsaturated fatty acid for the brain. J Mol Neurosci 16: 159–165, discussion 215–121, 2001 [DOI] [PubMed] [Google Scholar]
- 47.Szanto A, Nagy L. The many faces of PPARgamma: anti-inflammatory by any means? Immunobiology 213: 789–803, 2008 [DOI] [PubMed] [Google Scholar]
- 48.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282: 1490–1494, 1998 [DOI] [PubMed] [Google Scholar]
- 49.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043–1045, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanden Berghe W, Vermeulen L, Delerive P, De Bosscher K, Staels B, Haegeman G. A paradigm for gene regulation: inflammation, NF-kappaB and PPAR. Adv Exp Med Biol 544: 181–196, 2003 [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Lazar MA. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol 28: 2213–2220, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, Symons JD, Schnermann JB, Gonzalez FJ, Litwin SE, Yang T. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab 8: 482–491, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science 311: 1002–1005, 2006 [DOI] [PubMed] [Google Scholar]
- 54.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105: 683–694, 2001 [DOI] [PubMed] [Google Scholar]